Abstract
A porcine enteric calicivirus (PEC), strain Cowden in the family Caliciviridae (genus Sapovirus), can be propagated in a continuous cell line, LLC-PK cells, but only in the presence of an intestinal content fluid filtrate from gnotobiotic pigs. This cell culture system is presently the only in vitro model among caliciviruses that cause gastrointestinal disease, including members of the genera Sapovirus and Norovirus. We report here the identification of bile acids as active factors in intestinal content fluid essential for PEC growth. Bile acids that allowed PEC growth induced an increase in cAMP concentration in LLC-PK cells that was associated with down-regulation of IFN-mediated signal transducer and activator of transcription 1 phosphorylation, a key element in innate immunity. In addition, cAMP/protein kinase A pathway inhibitors, suramin, MDL12330A, or H89 suppressed bile acid-mediated PEC replication. We propose a mechanism for enteric calicivirus growth dependent on bile acids, ubiquitous molecules present in the intestine at the site of the virus replication that involves the protein kinase A cell-signaling pathway and a possible down-regulation of innate immunity.
Bile acids play an essential role as carriers of dietary lipids for absorption, and recent studies indicate that they function also as hormone-like regulatory molecules with specific cellular receptors (1). Bile acids regulate expression of various transport proteins and enzymes involved in their synthesis through binding and activating nuclear receptors, such as farnesoid X receptor (2, 3), pregnane X receptor (4, 5) and vitamin D receptor (6). Recently, a G protein-coupled receptor for bile acids was identified (7, 8). Bile acids induced an increase in intracellular cAMP concentrations in the presence of the G protein-coupled receptor, and it was suggested that this effect might be involved in the immunosuppressive activities associated with bile acids (7).
Caliciviruses (Family Caliciviridae) are small, nonenveloped viruses of 27–35 nm in diameter (9). They possess a single-strand, plus-sense genomic RNA of 7–8 kb. Four genera have been established in the family: Vesivirus, Lagovirus, Norovirus, and Sapovirus (10). Caliciviruses in the genera Norovirus and Sapovirus are enteric pathogens in humans and animals. Recent studies (9, 11–13) showed that noroviruses are responsible for >90% of nonbacterial gastroenteritis outbreaks and are associated with an estimated 23 million cases of gastroenteritis in the U.S. each year. It has been difficult to study pathogenesis and immunity to the noroviruses and sapoviruses, because with the exception of porcine enteric calicivirus (PEC), they cannot be propagated in cell culture. Recently, it was reported that a murine norovirus showed significantly higher virulence in signal transducer and activator of transcription (STAT)1–/– knockout mice, which indicated a major role for innate immunity in mediating resistance to calicivirus pathogenicity (14).
PEC strain Cowden, a member of the genus Sapovirus, was first detected in 1980 (15). The Cowden strain of PEC was adapted to growth in cell culture by serial passage in a continuous cell line (LLC-PK), but growth depended on the presence of an intestinal content (IC) fluid filtrate, obtained from uninfected gnotobiotic pigs, in the cell culture medium (16, 17). Recently (18), we showed that the IC effects on the growth of Cowden PEC in cell culture were associated with the induction of a protein kinase A (PKA) signaling pathway by IC, which suggested a novel mechanism for this virus-host relationship dependent on a specific cellular environment.
Here, we report the identification of bile acids as active factors in IC essential for growth of PEC in cultured cells. We also show that bile acids induce the PKA signaling pathway and down-regulate IFN-mediated STAT1 activation. Because STAT1 is an essential element for antiviral innate immunity by IFNs, we propose that this down-regulation by bile acids might allow PEC replication in vitro and in vivo.
Materials and Methods
Cells, Viruses, and Antisera. The cell-culture-adapted Cowden PEC strain (16, 17) was passaged in LLC-PK cells in the presence of 1% IC. Guinea pig hyperimmune antiserum against recombinant PEC virus-like particles (VLPs) (19) and pig antiserum against Cowden PEC were described (20). PEC virus infection was studied on confluent monolayers of LLC-PK cells grown in 6- or 12-well plates or in 150-cm2 flasks. Viruses were inoculated at 0.5 or 0.05 multiplicity of infection (moi) based on tissue culture 50% infective dose titer, and incubated at 37°C for 1 h. Virus-inoculated cells were then washed twice and incubated in the presence or absence of IC or other reagents at 37°C for the desired time. Virus detection methods included immunofluorescence assay (18), ELISA (18), radioimmunoprecipitation assay (RIPA) (21), and tissue culture 50% infective dose assay (22). Viral replication complexes (RCs) were isolated from PEC-infected cells similar to the protocol used for feline calicivirus RCs (23), and the RCs were analyzed in an RNA replication assay as described (23).
The Effects of Various Bile Acids on PEC Replication in LLC-PK Cells. The bile acids (Sigma) used in this study included cholic acid (CA), glycocholic acid (GCA), chenodeoxycholic acid (CDCA), glycochenodeoxycholic acid (GCDCA), taurochenodeoxycholic acid (TCDCA), deoxycholic acid (DCA), glycodeoxycholic acid (GDCA), lithocolic acid (LCA), taurolithocolic acid (TLCA), ursodeoxycholic acid (UDCA), and tauroursodeoxycholic acid (TUDCA). The conjugated bile acids were prepared in sterile water, and unconjugated bile acids were prepared in ethanol, both as 50-mM stock solutions. The cytotoxicity of each bile acid at various concentrations in LLC-PK cells was measured by microscopic observation and Trypan blue staining of cells. Each bile acid was incubated with virus-inoculated cells at final concentrations ranging from 1 μM to 1 mM, and virus growth was detected by immunofluorescence staining, RIPA, and ELISA. For PEC growth kinetics, the virus-inoculated cells were collected at 24, 48, 72, 96, and 120 h after inoculation. To control bile acid-mediated cytotoxic effects on LLC-PK cells, bilirubin (0.5 μM) or α-tocopherol (100 μM; ref. 24) was added to the medium. The concentration of bile acids in IC was measured with a colorimetric total bile acids assay kit (Bio-Quant, San Diego). To deplete bile acids in IC, cholestryamine resin (Sigma) was incubated in a 2% (wt/vol) suspension with IC at room temperature for 5 min. After the resin was removed by centrifugation (20,000 × g for 5 min), the concentration of bile acids in the supernatant was measured. The bile acid-depleted IC was examined for its ability to support PEC replication in LLC-PK cells at concentrations up to 5%.
Transfection of Viral RNA into LLC-PK Cells. Cell culture-propagated Cowden PEC virions were pelleted by ultracentrifugation (100,000 × g for 2 h), and viral RNA was extracted with the RNeasy kit (Qiagen, Valencia, CA). The viral RNA (0.1 μg) was transfected into 2-day-old LLC-PK cells in six-well plates by using Lipofectamine 2000 (Invitrogen), and the transfected cells were incubated at 37°C with IC (1%), GCDCA (100 μM), or TCDCA (100 μM) for up to 120 h. Controls included cells incubated with Lipofectamine 2000 alone and RNA-transfected cells maintained without IC or bile acids. Virus growth was monitored by ELISA.
Isolation and Characterization of a Field Strain (LL14) of PEC in LLC-PK Cells. A more recently circulating PEC stain was identified, isolated in the presence of IC, and designated as the LL14 strain. The LL14 strain was purified through three rounds of plaque isolation (in the presence of IC), and the genome sequence (except for the 5′ and 3′ nontranslated regions) was determined from RT-PCR products and cDNA clones.
Quantitation of cAMP. The confluent LLC-PK cells grown in six-well plates were treated with mock fluid, IC, or each individual bile acid for 20 min in the presence of 0.2 mM 3-isobutyl-1-methylxanthine (Sigma). The amount of intracellular cAMP was determined with the direct cAMP enzyme immunoassay kit (Sigma) according to the manufacturer's direction.
Modulators for Cell Signal Transduction. The PKA pathway inhibitors, which included G protein inhibitor, suramin, adenylate cyclase inhibitor MDL12,330A [cis-N-(2-Phenylcyclopentyl)-azacyclotridec-1-en-2-amine monohydrochloride], and PKA inhibitor H89 {N-(2-[Bromocinnamulamino]ethyl)-5-isoquinolinesulfonamide} were examined for their effect on bile acid-mediated growth of PEC. After inoculation of LLC-PK cells with PEC (at an moi of 0.5 or 0.05) for 1 h, infected cells were incubated with medium containing various concentrations of each inhibitor with IC (1%), GCDC (100 μM), or TCDC (100 μM). The cytotoxicity of the inhibitors in LLC-PK cells was monitored at various time points after treatment of confluent cells with each inhibitor by using a cytotoxicity detection kit (LDH; Roche Applied Science, Indianapolis). Also, cell growth in the presence of each inhibitor was monitored by counting viable trypsinized cells after treatment for 24, 48, 72, and 96 h.
Analysis of STAT1 Phosphorylation. The phosphorylation status of STAT1 by IFN-γ or IFN-α and bile acids was assessed by immunoblot analysis using PhosPhoPlus STAT1 (Tyr-701) antibody kit (Cell Signaling Technology, Beverly, MA). The LLC-PK cells (5 × 106 cells) in six-well plates were preincubated with either medium, individual bile acid (100 μM), IC (1%), IC (1%) plus H89 (20 μM), or GCDCA (100 μM) plus H89 (20 μM) for 10 min, then either medium, recombinant porcine IFN-γ or recombinant IFN type 1 (human IFN-αA plus IFN-αD fusion protein; 50 units/ml each, Serotec, Raleigh, NC) was added for 20 min. The treated cells were lysed in 0.2 ml of SDS sample buffer (with 1% 2-mercaptoethanol) for SDS/PAGE. Immunoblot analysis by using anti-STAT1 or antiphosphorylated STAT1 (Tyr-701) antibodies was performed according to the manufacturer's procedure.
pGAS-TA-Luc Plasmids and Transfection. One- or 2-day-old LLC-PK cells in six-well plates were transfected with 1 μg of pGAS-TA-Luc (Clontech) by using Lipofectamine 2000 (Invitrogen). After 18–20 h, the cells were incubated with either medium, individual bile acids (100 or 200 μM), or IC (1%) for 10 min, followed by incubation with medium or INF-γ (50 units/ml) for 20 min. Fresh medium was then added and the cells were incubated at 37°C for 6 h. Cells were lysed with 0.2 ml of lysis buffer and luciferase levels were assayed by using a luciferase assay kit (Promega).
Results
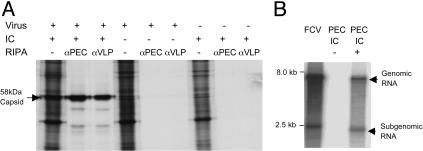
Requirement of IC for PEC Protein and RNA Synthesis. De novo synthesis of viral proteins was monitored by radiolabeling of infected cells in the presence or absence of IC followed by a RIPA with antisera against PEC (Fig. 1A). Moreover, RCs isolated from infected cells grown in the presence or absence of IC were tested for their ability to synthesize RNA (Fig. 1B). Neither detectable viral protein synthesis nor RNA synthesis occurred in the absence of IC, confirming an absolute requirement for IC in the cell culture medium for PEC growth.
Fig. 1.
Requirement of IC for PEC protein and RNA synthesis. (A) 35S-radiolabeled proteins from mock- or virus-infected LLC-PK cells maintained in the presence (1%) or absence of IC precipitated with PEC-specific antibodies. At 24 h after virus or mock infection, the cells were radiolabeled with [35S]-Met and [35S]-Cys for 4 h, and collected in RIPA buffer. RIPA was performed with either pig hyperimmune antiserum against native virions (αPEC) or guinea pig hyperimmune antiserum raised against recombinant PEC VLPs (αVLP). (B) RCs were isolated from virus-infected cells maintained in the presence (1%) or absence of IC for 24 h. An in vitro replication assay was performed in the presence of [α-32P]CTP and the synthesized RNA was purified and analyzed under denaturing conditions. Feline calicivirus RCs isolated from FCV-infected CFRK cells (23) were included as a control.
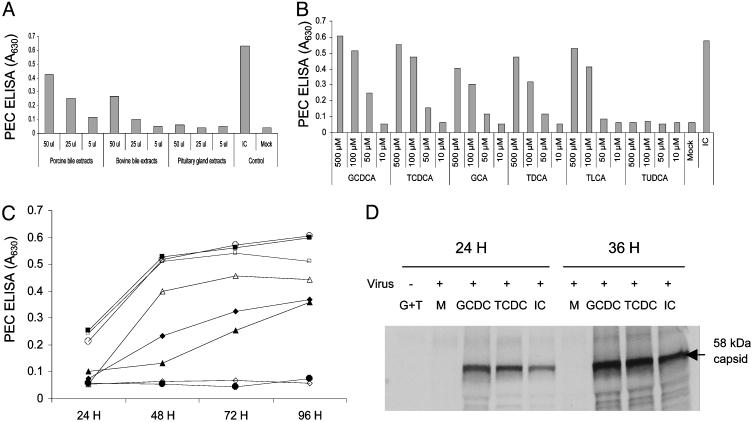
The Effects of Various Bile Acids on PEC Replication in LLC-PK Cells. Our initial studies on the identification of the active factor(s) in IC required for PEC growth indicated that the active factor was a small molecule (<1 kDa) that was resistant to proteinase and nuclease treatment (see Supporting Text and Fig. 6, which are published as supporting information on the PNAS web site). Moreover, the presence of porcine or bovine bile extract (from Sigma) in the growth medium allowed limited virus replication (Fig. 2A), which suggested the active factor may be related to bile acids. Commercially available bile acids, including unconjugated bile acids (CA, CDCA, DCA, LCA, and UDCA) and conjugated bile acids (GCA, GCDCA, TCDCA, GDCA, TLCA, and TUDCA) were each examined for its ability to support PEC replication. All bile acids but UDCA and TUDCA enabled PEC replication in LLC-PK cells, but with various efficiencies, depending on the concentration, ranging from 50 to 500 μM (Fig. 2 B–D, and Fig. 7, which is published as supporting information on the PNAS web site). None of the bile acids induced PEC replication in LLC-PK cells at 10 μM, which is the physiological blood concentration of bile acids outside the enterohepatic circulation (Fig. 2B). The UDCA (data not shown) and TUDCA did not support virus replication, even at concentrations as high as 500 μM (Fig. 2B).
Fig. 2.
An active factor is present in bile extract, and the effects of various bile acids on PEC growth. (A) Ability of various extracts to support PEC growth in cell culture. Porcine bile extract and bovine bile extract were solubilized in sterile water (2%) and bovine pituitary extract was solubilized in sterile water at 25 mg/ml. After virus inoculation, cells were incubated in the presence of various concentrations (5–50 μl) of each extract, and PEC antigen was measured by ELISA after 120 h. (B) PEC growth at various concentrations (500–10 μM) of indicated conjugated bile acids, and IC (1%). PEC antigen was measured 120 h after virus inoculation by ELISA. (C) PEC growth kinetics in the presence of mock fluid (•), IC (○), and various conjugated bile acids [GCDCA (▪), TCDCA (□), GCA (▴), TLCA (▵), TDCA (♦), and TUDCA (⋄)]. After PEC infection, cells were incubated with 100 μM of various conjugated bile acids or IC (1%), and PEC antigen was measured by ELISA. (D) RIPA with hyperimmunized pig serum raised against PEC VLPs was used for the detection of de novo synthesis of 35S-labeled PEC proteins during viral infection in the presence of mock fluid (M), GCDCA (100 μM), TCDCA (100 μM), or IC (1%) for period of 24 or 36 h. The first lane contains LLC-PK cells incubated with a mixture of 100 μM each of GCDCA and TCDCA (G plus T) without virus inoculation.
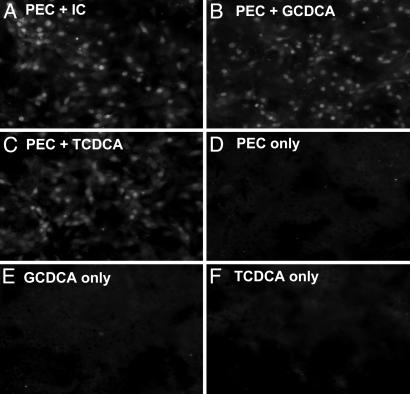
All bile acids examined caused cytotoxic effects at concentrations of 25 μM or higher in LLC-PK cells (data not shown). These cytotoxic effects were prevented by adding an antioxidant (bilirubin or α-tocopherol), which was consistent with a previous report of the inhibitory effects of antioxidants on bile acid-induced apoptosis (24). Without the antioxidant, each conjugated bile acid (100 μM) could still support PEC replication, but the cytotoxic effect of the bile acid resulted in lower virus growth (data not shown). Because GCDCA and TCDCA were most effective in supporting PEC replication (Fig. 2 B and C), we used them in subsequent experiments. As noted above, bilirubin (0.5 μM) or α-tocopherol (100 μM) was used in all experiments. A RIPA with the porcine antiserum against Cowden PEC showed de novo synthesis of viral capsid protein at 24 and 36 h after virus inoculation in the presence of GCDCA, TCDCA, or IC. In the absence of IC or these bile acids (Fig. 2D), there was no evidence of viral protein synthesis in LLC-PK cells. Immunofluorescence staining of PEC-infected LLC-PK cells incubated with IC, GCDCA (100 μM), or TCDCA (100 μM), confirmed the presence of viral antigen in cells and the distribution of antigen-positive cells was similar in wells with IC, GCDCA, or TCDCA (Fig. 3). The concentration of total bile acids in various IC preparations ranged from 30 to 50 μM in the working dilution (1%) used for growth of PEC. The treatment of IC with cholestyramine resin (bile acid-binding resin) depleted the bile acids as measured by the colorimetric assay, and the bile acid-depleted IC did not support PEC replication in LLC-PK cells at concentration as high as 5% (data not shown).
Fig. 3.
Immunofluorescence assay of PEC-infected cells. Mock- or PEC-infected LLC-PK cells were incubated with medium only, or medium containing IC (1%), GCDCA (100 μM), or TCDCA (100 μM) for 24 h as indicated. Cells were fixed with 100% cold methanol and were incubated with guinea pig αPEC VLP serum. Bound antibodies were detected with FITC-conjugated goat anti-guinea pig IgG.
Bile Acids Support Recovery of Virus from LLC-PK Cells Transfected with Viral RNA and Allow Growth of PEC Field Strain (LL14). We examined whether bile acids could support the replication of Cowden PEC initiated from infectious RNA (18) to rule out possible carryover of IC in the virus stock. When purified viral RNA (0.1 μg) was transfected into LLC-PK cells, virus growth was observed only when IC, GCDCA, or TCDCA was present in the medium as determined by cytopathic effect, immunofluorescence staining, and ELISA (Fig. 8A, which is published as supporting information on the PNAS web site). To prove further that bile acids are an active factor in IC that support PEC replication, we isolated and characterized an additional PEC strain (LL14) in LLC-PK cells. The LL14 genome showed an overall identity of 95% and 98% to the Cowden strain in nucleotides and amino acids, respectively, in the coding region (Fig. 9, which is published as supporting information on the PNAS web site; GenBank accession no. AY425671 for the LL14 strain). The LL14 strain required the presence of IC or bile acids in the cell culture medium for growth, similar to the Cowden strain, although the kinetics of virus growth was slower than that of the Cowden strain (Fig. 8B).
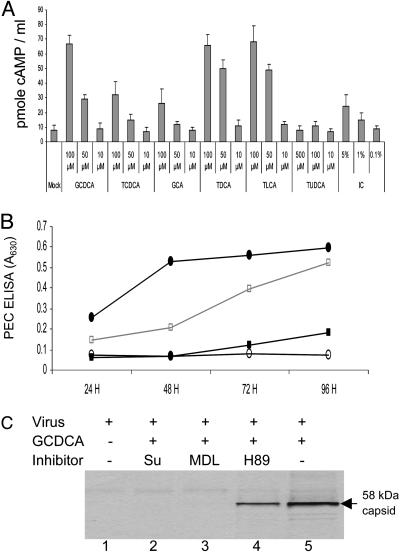
Induction of Intracellular cAMP by Bile Acids and the Effects of Modulators for Cell Signal Transduction on Bile Acid-Associated Virus Growth. In an extension of our previous study (18) that linked a PKA pathway to PEC growth in the presence of IC, we examined whether bile acids could induce an increase in cAMP levels in LLC-PK cells. All bile acids that allowed PEC growth increased levels of intracellular cAMP up to 6- to 7-fold in LLC-PK cells in the presence of 3-isobutyl-1-methylxanthine, an inhibitor of phosphodiesterase (Fig. 4A). The minimum concentration required for the induction of cAMP varied among the bile acids, but all bile acids except TUDCA induced cAMP at 100 μM (Fig. 4A). At a 10-μM concentration, none of the bile acids we tested induced intracellular cAMP or supported PEC growth (Fig. 2B). Of interest, UDCA (data not shown) and TUDCA, which did not support PEC growth, did not increase cAMP levels when used at concentrations as high as 500 μM (Figs. 2B and 4A). In comparison with IC, the individual bile acids induced cAMP to higher levels (Fig. 4A).
Fig. 4.
Effects of bile acids or IC on induction of cAMP in LLC-PK cells and of various PKA signaling pathway inhibitors on bile acid-mediated PEC growth. (A) cAMP induction by various concentrations of conjugated bile acids in LLC-PK cells. Confluent 3- to 4-day-old LLC-PK cell monolayers were incubated with various concentrations of GCDCA, TCDCA, GCA, TDCA, TLCA, TUDCA, or IC as indicated for 20 min in the presence of 3-isobutyl-1-methylxanthine, an inhibitor of phosphodiesterase. Cells were lysed, and the concentration of cAMP was measured. (B) The effects of G protein (suramin), adenylate cyclase (MDL12,330A), and PKA (H89) inhibitors on PEC growth in the presence of GCDCA was examined. After virus inoculation of 3- to 4-day-old LLC-PK cells, cells were incubated with 100 μM of GCDCA alone (•) or 100 μM of GCDCA in the presence of 200 μM of suramin (○), 20 μM of MDL12,330A (▪), or 20 μM of H89 (□). PEC antigen was assayed by ELISA. (C) PEC-infected cells were incubated with medium only (lane 1), 100 μM of GCDCA alone (lane 5), or 100 μM GCDCA with suramin (200 μM, lane 2), MDL12,330A (20 μM, lane 3), or H89 (20 μM, lane 4). PEC-infected cells were 35S-Met- and 35S-Cys-labeled at 48 h after virus infection for 4 h, and cell lysates were analyzed by RIPA with αPEC VLP serum.
The effect of inhibitors of the PKA signaling pathway on bile acid-mediated PEC growth was examined. Cowden PEC was inoculated onto LLC-PK cells at an moi of 0.5 or 0.05 with the bile acid GCDCA (100 μM) in the presence or absence of various concentrations of the PKA pathway inhibitors, suramin, MDL12,330A, or H89 (Fig. 4 B and C). We examined PKA pathway inhibitors because we found previously that the inhibitors of pathways other than PKA did not affect PEC growth in the presence of IC (18). Suramin, which uncouples G proteins from G protein-coupled receptors, completely blocked virus replication when added at concentrations of 200 μM daily during the experiment (Fig. 4 B and C). Adenylate cyclase inhibitor MDL12,330A also blocked virus replication up to 48 h when 20 μM was added daily (Fig. 4 B and C). The PKA inhibitor H89 showed a partial inhibitory effect on virus growth at 20 μM (Fig. 4 B and C). When PEC was inoculated at a low moi (0.05 moi), the inhibitory activity was more evident even with lower concentrations of suramin, MDL12,330A, and H89 (data not shown). The highest concentration of each inhibitor used in this study did not show evidence for nonspecific cytotoxic effects on LLC-PK cells, and did not affect cell growth.
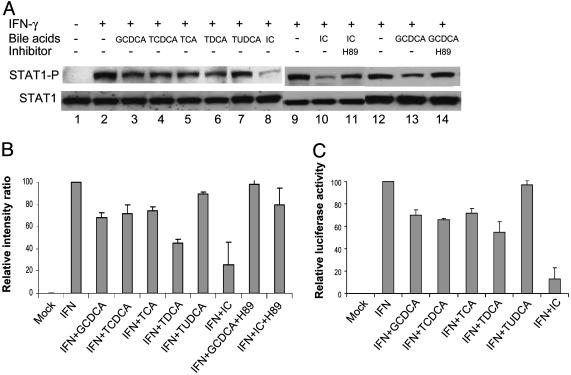
Effects of Bile Acids on IFN Induction of STAT1. Several reports have shown that activation of the PKA/cAMP pathway could cause down-regulation of STAT1 activation (25–29). We next examined whether bile acids could affect IFN-mediated STAT1 activation in LLC-PK cells. Individual bile acids at a concentration of 100 or 200 μM (with the exception of TUDCA) could inhibit IFN-γ or IFN-α (data not shown) mediated STAT1 phosphorylation by 30–50% (Fig. 5 A and B). IC fluid (1%) showed higher levels of inhibition of STAT1 activation than individual bile acids, with up to 75% inhibition (Fig. 5 A and B). However, TUDCA showed little effect on STAT1 activation (Fig. 5 A and B). Addition of adenylate cyclase inhibitor, MDL12,330, or PKA inhibitor H89 to cells treated with IC or GCDCA restored STAT1 activation by IFN-γ, to ≈80% (from ≈25% with IC without inhibitors) or 96% (from ≈68% with GCDCA alone), respectively (Fig. 5 A and B). The negative effects of bile acids on STAT1 activation by IFN-γ were also confirmed in the pGAS-TA-luc reporter system (ref. 30 and Fig. 5C). Each bile acid (except TUDCA) or IC inhibited IFN-γ induction of luciferase expression at levels ranging from 30% to 80% (Fig. 5C).
Fig. 5.
Inhibition of IFN-γ induced STAT1 activation by bile acids. (A) Immunoblot analysis of STAT1 activation (phosphorylation status of STAT1) by medium only (lane 1), IFN-γ (lanes 2, 9, and 12), IFN-γ with individual bile acids (GCDCA, TCDCA, TCA, TDCA, and TUDCA, 100 μM each, lanes 3–7), or IC (1%, lanes 8 and 10), IFN-γ with IC (1%) plus H89 (20 μM, lane 11), or IFN-γ with GCDCA (100 μM) plus H89 (20 μM, lane 14). (B) Relative value (as presented by the value of IFN-γ treatment as 100%) of immunoblot analysis of STAT1 activation with various treatments. The intensity of each band in A was measured by scanning and represented as the ratio of STAT1 to phosphorylated STAT1. (C) Relative values (as represented by the value of IFN-γ treatment as 100%) of luciferase activity after pGAS-TA-luc transfection and treatments of medium only, IFN-γ, IFN-γ with individual bile acids (GCDCA, TCDCA, TCA, TDCA, or TUDCA, 100 μM each), or IC (1%). Bars represent SE of at least three independent experiments.
Discussion
The original rationale for including IC in cell culture medium to promote PEC replication was based on an effort to mimic in vivo conditions in the intestinal tract, where the virus was known to replicate (20, 31). For rotaviruses, proteases in IC can activate virus by cleaving the outer capsid protein, VP4, and this procedure allows virus penetration and entry into the cell (32). However, IC-mediated PEC growth in cell culture was different from the protease-mediated rotavirus growth: instead, it was associated with the induction of a PKA pathway by IC (18). In this report, we identified bile acids as active factors in IC that allow PEC replication in LLC-PK cells, and like proteases (trypsin) for rotaviruses, bile acids could replace IC in the PEC cell culture system.
Innate immunity is an important host mechanism to control virus infection before the development of adaptive immune responses. Since their discovery in 1956, IFNs have been recognized as key components in antiviral innate immunity (33). STAT1 is an essential element for both type I (IFNα/β) and II (IFNγ) IFN responses because its activation by phosphorylation when IFNs bind to their cognate receptors is involved in a cascade of antiviral activities (33). Many viruses have developed defensive mechanisms against IFN signaling to avoid innate immunity, including a mechanism involved in the degradation of STAT1 by viral proteins (33, 34). Recent reports (25–29) have shown evidence for cross-talk between PKA and STAT1 pathways: induction of PKA in several experimental systems was shown to down-regulate STAT1 activation. In this study, we observed that induction of PKA by bile acids was associated with down-regulation of STAT1 in LLC-PK cells. This observation suggests that bile acids may allow PEC to replicate in cultured cells by a mechanism that involves inhibition of innate immunity. Our data are consistent with studies by Podevin et al. (35), in which bile acids inhibited the antiviral activities of IFN-α in hepatic cells, and those by Karst et al. (14), in which murine norovirus showed increased pathogenicity in STAT1–/– mice.
Because bile acids (≈300 mM) stored in the gall bladder are released into the duodenum and are actively reabsorbed in the ileum, the concentration of bile acids is much higher in the proximal intestinal tract (duodenum and jejunum; ref. 36). This higher concentration of bile acids might explain the greater susceptibility of the proximal intestinal tract to PEC pathogenesis when compared with the distal intestinal tract (ileum) (20, 31). In the presence of bile acids, susceptible cells (which may have receptors for both bile acids and PEC) are infected with viruses and die, resulting in villus atrophy. These morphological changes disrupt the absorptive capacity of the intestinal tissue, resulting in diarrhea. It is interesting to speculate on the evolution of viruses that have adapted to an environment containing high concentrations of bile acids. If environmental factors (bile acids) provide negative regulation of innate immunity in susceptible target cells, enteric viruses such as PEC may exploit this mechanism to overcome innate immunity and establish a productive infection in the gut. The strong growth restriction of PEC in cultured cells in the absence of bile acids suggests that the virus itself cannot overcome innate immunity, but instead depends on exogenous factors to maintain cells in a susceptible state. Interestingly, even in the presence of viral receptors on cells, certain viruses are severely limited for growth in cultured cells if they lack a mechanism for counteraction of cellular innate immunity (34, 37, 38). However, we cannot rule out a role for other potential mechanisms associated with the dependence of PEC replication in LLC-PK cells on the presence of IC or bile acids. Although IC inhibited the activation of STAT1 by IFNs with the highest efficiency among the reagents we examined, it induced only a moderate increase in the detectable level of cAMP compared with the bile acids that supported virus replication. Because IC is a complex mixture of molecules, some components in IC might inhibit cAMP induction in the direct assay we used, which required the presence of 3-isobutyl-1-methylxanthine and an induction period of ≈20 min. Alternatively, IC may contain molecules other than bile acids that inhibit STAT1 activation by means of other mechanisms. Although the identification of at least one active component, bile acids, in the IC can now allow further studies of the mechanism of its action with defined reagents, it will be of interest to further elucidate other components of IC that are present at the site of viral replication in the gut and examine their influence on virus growth. In summary, we propose a mechanism for enteric calicivirus growth mediated by bile acids, ubiquitous molecules present in the intestine at the site of the virus replication, that is associated with inhibition of innate immunity. Our findings could be applicable to the development of cell culture systems for noncultivable viruses, including other enteric caliciviruses.
Supplementary Material
Acknowledgments
We thank Albert Z. Kapikian for continuing support of our work and Tanaji Mitra for technical assistance and helpful discussions. This work was supported in part by National Institutes of Health, National Institute of Allergy and Infectious Diseases Grant RO1AI 49716 (to L.J.S.).
Abbreviations: IC, intestinal content; STAT, signal transducer and activator of transcription; PEC, porcine enteric calicivirus; VLP, virus-like particle; moi, multiplicity of infection; RIPA, radioimmunoprecipitation assay; RC, replication complex; CA, cholic acid; GCA, glycocholic acid; CDCA, chenodeoxycholic acid; GCDCA, glycochenodeoxycholic acid; TCDCA, taurochenodeoxycholic acid; DCA, deoxycholic acid; GDCA, glycodeoxycholic acid; LCA, lithocolic acid; TLCA, taurolithocolic acid; UDCA, ursodeoxycholic acid; TUDCA, tauroursodeoxycholic acid.
Data deposition: The sequence for the LL14 strain reported in this paper has been deposited in the GenBank database (accession no. AY425671).
References
- 1.Johnson, L. R. (1998) in Essential Medical Physiology, ed. Johnson, L. R. (Lippincott-Raven, New York), pp. 445–472.
- 2.Parks, D. J., Blanchard, S. G., Bledsoe, R. K., Chandra, G., Consler, T. G., Kliewer, S. A., Stimmel, J. B., Willson, T. M., Zavacki, A. M., Moore, D. D. & Lehmann, J. M. (1999) Science 284, 1365–1368. [DOI] [PubMed] [Google Scholar]
- 3.Makishima, M., Okamoto, A. Y., Repa, J. J., Tu, H., Learned, R. M., Luk, A., Hull, M. V., Lustig, K. D., Mangelsdorf, D. J. & Shan, B. (1999) Science 284, 1362–1365. [DOI] [PubMed] [Google Scholar]
- 4.Jones, S. A., Moore, L. B., Shenk, J. L., Wisely, G. B., Hamilton, G. A., McKee, D. D., Tomkinson, N. C., LeCluyse, E. L., Lambert, M. H., Willson, T. M., et al. (2000) Mol. Endocrinol. 14, 27–39. [DOI] [PubMed] [Google Scholar]
- 5.Staudinger, J. L., Goodwin, B., Jones, S. A., Hawkins-Brown, D., MacKenzie, K. I., LaTour, A., Liu, Y., Klaassen, C. D., Brown, K. K., Reinhard, J., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 3369–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makishima, M., Lu, T. T., Xie, W., Whitfield, G. K., Domoto, H., Evans, R. M., Haussler, M. R. & Mangelsdorf, D. J. (2002) Science 296, 1313–1316. [DOI] [PubMed] [Google Scholar]
- 7.Kawamata, Y., Fujii, R., Hosoya, M., Harada, M., Yoshida, H., Miwa, M., Fukusumi, S., Habata, Y., Itoh, T., Shintani, Y., et al. (2003) J. Biol. Chem. 278, 9435–9440. [DOI] [PubMed] [Google Scholar]
- 8.Maruyama, T., Miyamoto, Y., Nakamura, T., Tamai, Y., Okada, H., Sugiyama, E., Itadani, H. & Tanaka, K. (2002) Biochem. Biophys. Res. Commun. 298, 714–719. [DOI] [PubMed] [Google Scholar]
- 9.Green, K. Y., Chanock, R. M. & Kapikian, A. Z. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Williams & Wilkins, Philadelphia), Vol. 1, pp. 841–874. [Google Scholar]
- 10.Green, K. Y., Ando, T., Balayan, M. S., Berke, T., Clarke, I. N., Estes, M. K., Matson, D. O., Nakata, S., Neill, J. D., Studdert, M. J. & Thiel, H. J. (2000) J. Infect. Dis. 181, Suppl. 2, S322–S330. [DOI] [PubMed] [Google Scholar]
- 11.Fankhauser, R. L., Noel, J. S., Monroe, S. S., Ando, T. & Glass, R. I. (1998) J. Infect. Dis. 178, 1571–1578. [DOI] [PubMed] [Google Scholar]
- 12.Green, K. Y. (1997) Arch. Virol. Suppl. 13, 153–165. [DOI] [PubMed] [Google Scholar]
- 13.Mead, P. S., Slutsker, L., Dietz, V., McCaig, L. F., Bresee, J. S., Shapiro, C., Griffin, P. M. & Tauxe, R. V. (1999) Emerg. Infect. Dis. 5, 607–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karst, S. M., Wobus, C. E., Lay, M., Davidson, J. & Virgin, H. W., IV (2003) Science 299, 1575–1578. [DOI] [PubMed] [Google Scholar]
- 15.Saif, L. J., Bohl, E. H., Theil, K. W., Cross, R. F. & House, J. A. (1980) J. Clin. Microbiol. 12, 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flynn, W. T. & Saif, L. J. (1988) J. Clin. Microbiol. 26, 206–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parwani, A. V., Flynn, W. T., Gadfield, K. L. & Saif, L. J. (1991) Arch. Virol. 120, 115–122. [DOI] [PubMed] [Google Scholar]
- 18.Chang, K. O., Kim, Y., Green, K. Y. & Saif, L. J. (2002) Virology 304, 302–310. [DOI] [PubMed] [Google Scholar]
- 19.Guo, M., Qian, Y., Chang, K. O. & Saif, L. J. (2001) J. Clin. Microbiol. 39, 1487–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flynn, W. T., Saif, L. J. & Moorhead, P. D. (1988) Am. J. Vet. Res. 49, 819–825. [PubMed] [Google Scholar]
- 21.Sosnovtsev, S. V., Sosnovtseva, S. A. & Green, K. Y. (1998) J. Virol. 72, 3051–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burleson, F. G., Chambers, T. M. & Wiedbrauk, D. L. (1992) Virology, A Laboratory Manual (Academic, New York).
- 23.Green, K. Y., Mory, A., Fogg, M. H., Weisberg, A., Belliot, G., Wagner, M., Mitra, T., Ehrenfeld, E., Cameron, C. E. & Sosnovtsev, S. V. (2002) J. Virol. 76, 8582–8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yerushalmi, B., Dahl, R., Devereaux, M. W., Gumpricht, E. & Sokol, R. J. (2001) Hepatology 33, 616–626. [DOI] [PubMed] [Google Scholar]
- 25.Delgado, M. (2003) J. Biol. Chem. 278, 27620–27629. [DOI] [PubMed] [Google Scholar]
- 26.Kanda, N. & Watanabe, S. (2003) J. Invest. Dermatol. 120, 411–419. [DOI] [PubMed] [Google Scholar]
- 27.Lee, E. H. & Rikihisa, Y. (1998) Infect. Immun. 66, 2514–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sengupta, T. K., Schmitt, E. M. & Ivashkiv, L. B. (1996) Proc. Natl. Acad. Sci. USA 93, 9499–9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.David, M., Petricoin, E., III, & Larner, A. C. (1996) J. Biol. Chem. 271, 4585–4588. [DOI] [PubMed] [Google Scholar]
- 30.Shuai, K., Schindler, C., Prezioso, V. R. & Darnell, J. E., Jr. (1992) Science 258, 1808–1812. [DOI] [PubMed] [Google Scholar]
- 31.Guo, M., Hayes, J., Cho, K. O., Parwani, A. V., Lucas, L. M. & Saif, L. J. (2001) J. Virol. 75, 9239–9251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Estes, M. K. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Williams & Wilkins, Philadelphia), Vol. 2, pp. 1747–1786. [Google Scholar]
- 33.Samuel, C. E. (2001) Clin. Microbiol. Rev. 14, 778–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young, D. F., Andrejeva, L., Livingstone, A., Goodbourn, S., Lamb, R. A., Collins, P. L., Elliott, R. M. & Randall, R. E. (2003) J. Virol. 77, 2174–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Podevin, P., Rosmorduc, O., Conti, F., Calmus, Y., Meier, P. J. & Poupon, R. (1999) Hepatology 29, 1840–1847. [DOI] [PubMed] [Google Scholar]
- 36.McLeod, G. M. & Wiggins, H. S. (1968) Lancet 1, 873–876. [DOI] [PubMed] [Google Scholar]
- 37.Strong, J. E., Coffey, M. C., Tang, D., Sabinin, P. & Lee, P. W. (1998) EMBO J. 17, 3351–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mangeat, B., Turelli, P., Caron, G., Friedli, M., Perrin, L. & Trono, D. (2003) Nature 424, 99–103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.