Abstract
In mice, ≈1,000 odorant receptor (OR) genes are expressed in olfactory sensory neurons (OSNs). Homeodomain sites can be recognized in the promoter and upstream regions of several OR genes. Here, using the yeast one-hybrid system and electrophoretic mobility shift assay, we report that Lhx2, a LIM-homeodomain protein, binds to the homeodomain site in the mouse M71 OR promoter region. In Lhx2-deficient mice, the morphology of the olfactory epithelium is grossly normal. However, expression of OMP is abolished and that of GAP43 is severely reduced, indicating that no mature and few immature OSNs are produced. M71 and other OR genes also are not expressed. OSN development appears to be arrested between the terminal differentiation into neurons and the transition to immature neurons. Thus, Lhx2 is required for complete development of OSNs in mice.
In the mammalian olfactory epithelium (OE), olfactory sensory neurons (OSNs) detect chemical stimuli in the external environment by expressing odorant receptor (OR) genes (1, 2). These genes encode proteins with a putative seven-transmembrane domain structure, a defining characteristic of G protein-coupled receptors. It is believed that an individual OSN expresses a single OR gene (3) from the ≈1,000 OR genes in the mouse genome (4), but the strength of the evidence for this one-neuron–one-receptor hypothesis has been questioned (5). An OR gene is expressed from a single allele in a given cell (6). The OE is thus a complex mosaic of ≈2,000 OSN populations, each defined as expressing one allele from a choice of 2 ×≈1,000 genes. OSNs expressing the same OR gene typically reside within one of at least four zones in the OE (7, 8), where they are interspersed with OSNs expressing other OR genes. By targeted mutagenesis, all OSNs expressing the same OR were shown to project their axons to the same glomeruli in a remarkably precise manner, and these OR-specific projections to the olfactory bulb are influenced by the specificity of the expressed OR (9, 10). The onset of OR expression occurs before the first contact between OSN axons and the forebrain in embryos, and OSNs express OR genes in the absence of the axonal target, the olfactory bulb (11).
The mechanisms underlying OR gene choice and expression are not understood, but irreversible DNA rearrangements have been excluded (12, 13). Sequence analyses of human, mouse, and rat OR genes have revealed a variety of motifs upstream of the transcription start site, including O/E-like, E-box, Ikaros, homeodomain, and methylation-sensitive vMYB motifs (14). However, an involvement in OR gene regulation has not been shown for any of these motifs. Short (9-kb) transgenes of two OR genes, MOR23 and M71, replicate most OR expression features (15). A region of 405 bp upstream of the MOR23 transcription start site is required for transgene expression. In this and other upstream OR regions, common sequence motifs of homeodomain and O/E-like sites have been recognized (15).
Here, we have conducted yeast one-hybrid screening (16) to identify transcription factors that bind to the homeodomain site in the M71 promoter region. We identify Lhx2, a LIM-homeodomain protein (17), as a protein binding to the M71-homeodomain site. A knockout of the Lhx2 gene (18) results in failure of OR expression and incomplete development of OSNs.
Materials and Methods
Yeast One-Hybrid Assay. The MATCHMAKER one-hybrid system (Clontech) was used. Five tandem repeats of ACTAATTGTT, the putative homeodomain site in the mouse M71 promoter region, were ligated into the EcoRI–XbaI sites of pHISi and the EcoRI–SalI sites of pLacZi to generate pHISi-HD and pLacZi-HD, respectively. These two bait constructs were linearized and sequentially integrated into the genome of YM4271 yeast to obtain a strain carrying both pHISi-HD and pLacZi-HD. A library was prepared by subcloning oligo-dT-primed cDNA, prepared from the OE of 5-week-old female C57BL/6 mice, into the EcoRI–XhoI sites of the pAD-GAL4–2.1 phagemid vector (Stratagene). Approximately 1.4 × 107 clones were screened, and double-positive clones for His+ and LacZ+ were selected both by growth on minimal synthetic dropout plates (–Leu, –His) supplemented with 30 mM 3-amino-1,2,4-triazole, and by a β-galactosidase colony-lift filter assay.
Expression of Recombinant GST-Lhx2. Full-length Lhx2 cDNA was cloned into the BamHI and SalI sites of pGEX3T-1 (Amersham Biosciences), in which a GST-tag was introduced into the N terminus of recombinant Lhx2. GST-Lhx2 was expressed in Escherichia coli BL21 (DE3) and purified by using glutathione Sepharose 4B (Amersham Biosciences).
Electrophoretic Mobility-Shift Assays (EMSA). The following sets of complemented synthetic oligonucleotides were used for making DNA probes. M71 homeodomain: 5′-GAGAAACATGAACTAATTGTTACTTTAGAGA-3′ and 5′-GTCTCTAAAGTAACAATTAGTTCATGTTTCT-3′ (the homeodomain is underlined); mutant M71 homeodomain: 5′-GAGAAACATGAACTGGTTGTTACTTTAGAGA-3′ and 5′-GTCTCTAAAGTAACAACCAGTTCATGTTTCT-3′ (mutated nucleotides are in bold). A 50-μM concentration of each set of oligonucleotides was mixed and heated at 95°C for 2 min, and at 65°C for 30 min, and then cooled slowly to room temperature to make double-stranded DNA probes. Five picomoles of DNA probes was labeled by filling the 3′ end with [γ-32P]dCTP by using the (exo-) Klenow fragment of DNA polymerase I (NEB). Labeled probes were purified by using ProbeQuant G-50 Micro Columns (Amersham Biosciences) and ethanol precipitation to remove excess unincorporated [γ-32P]dCTP.
Lhx2 (100 ng) and 0.2 pmol of labeled probe were mixed in EMSA buffer (20 mM Hepes-KOH, pH 7.9/20% glycerol/100 mM KCl/20 mM MgCl2/0.2 mM EDTA/0.5 mM PMSF/1 mM 2-mercaptoethanol) supplemented with 100 ng of sonicated salmon sperm DNA, and 10 μg of BSA, in the presence or absence of nonlabeled probe as specific competitor. Mixtures were incubated at room temperature for 20 min and loaded on an 8% nondenaturing polyacrylamide gel to separate bound and unbound probe. For EMSA with inhibitory anti-Lhx2 antibody (Abcam, Cambridge, U.K.), 6% gels were used.
Mutant Mice. Lhx2+/– mice were a kind gift from H. Westphal (18). The mouse strains OMP-taulacZ, P2-IRES-taulacZ, M71-IRES-taulacZ, and M72-IRES-taulacZ have been described (9, 15, 19). For embryo staging, midday of the vaginal plug is embryonic day (E) 0.5.
In Situ Hybridization. Probes were prepared from plasmids kindly provided by F. Guillemot (National Institute for Medical Research, London): Mash1, NeuroD, and SCG10. Other probes were prepared from cDNA for Lhx2 (nucleotides 453–2150 from GenBank accession no. NM_010710), Neurogenin1 (Ngn1) (nucleotides 111–811 from U63841), GAP43 (nucleotides 147–860 from NM_008083), and OMP (nucleotides 820–2891 from U01213).
Detection of Mitotic Progenitors and Apoptotic Cells. To detect progenitors in mitosis and apoptotic cells, immunostaining with mouse monoclonal anti-BrdUrd clone BU33 (Sigma) and rabbit monoclonal anti-active caspase-3 clone C92–605 (BD Biosciences Pharmingen) were used, respectively. For BrdUrd pulse–chase labeling, pregnant Lhx2 heterozygous mutant mice carrying E16.5 embryos were injected i.p. with 5-bromo-2-deoxyuridine (Sigma) (50 mg/kg of body weight) for 1 hr, after which the heads of the embryos were collected and fixed in 4% paraformaldehyde/PBS for 30 min on ice. After the cryoprotection with 30% sucrose/PBS, heads were embedded in OCT compound (Sakura Finetek, Torrance, CA). Cryosections (12 μm) were collected on SuperFrost/plus microscope slides (Fisher), dried for 1 hr at room temperature, postfixed in 4% paraformaldehyde for 15 min, and washed three times with PBS for 5 min each. For immunostaining with anti-BrdUrd antibody, slides were incubated in 2 M HCl/PBS for 30 min and washed three times with PBS for 5 min each. Slides were blocked with the Vector Avidin/Biotin Blocking Kit (Vector Laboratories). Immunostaining with anti-BrdUrd antibody was performed with the Vector M.O.M. Immunodetection Kit (Vector Laboratories). For immunostaining with anti-active caspase-3, slides were treated with the Vector Avidin/Biotin Blocking Kit (Vector Laboratories), and immunostaining was performed with the Vectastain detection kit (Vector Laboratories). Immunodetected cells were counted on 10 coronal sections (12 μm thick) that were collected every 10 sections in wild-type embryos and every 8 sections in Lhx2-deficient embryos, to cover sections from the beginning of OE to the section in which the olfactory bulb appears.
Results
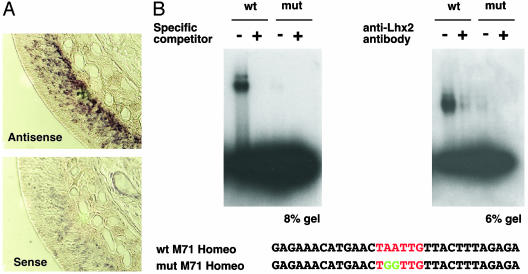
Lhx2 Is an OR-Homeodomain Site-Binding Protein. To identify proteins binding to the homeodomain site in the M71 promoter region, a mouse OE cDNA library was screened by the yeast one-hybrid system. From 1.4 × 107 clones, we identified 286 candidate-positive clones. Of these, 272 clones represent nine homeodomain proteins: Lhx2 (169 clones), Cart1 (34 clones), Dlx5 (20 clones), Prrx1 (18 clones), Alx3 (14 clones), Prrx2 (7 clones), Emx2 (6 clones), Barx1 (3 clones), and Dlx3 (1 clone). We examined expression of these genes in the OE by in situ hybridization. Emx2 and Lhx2 are well expressed, whereas expression of the other seven genes was not detected with digoxygenin-labeled probes. Expression of Emx2 is observed in both apical and basal portions of the OE (data not shown). Lhx2 is expressed strongly in basal cells (progenitor or neuronal precursor cells) and gradually decreases in apical cells (immature and mature neurons) (Fig. 1A). OR genes are expressed both in immature and mature neurons (20), suggesting that OR gene choice occurs during terminal differentiation to neurons from neuronal precursors or during the immature-neuron stage. LIM-homeodomain genes have distinct roles in neuronal development, particularly in the specification of neuronal identity (21). Because OR gene choice is a critical event in determining the identity of an OSN, Lhx2 is a promising candidate among the nine homeodomain proteins discovered in the screening. We therefore focused on Lhx2 for further studies.
Fig. 1.
Lhx2 gene expression in the olfactory epithelium and EMSA analyses. (A) In situ hybridization with RNA probes for Lhx2 on coronal sections of the OE from 25-day-old mice. Lhx2 is expressed in a graded fashion: strong expression in the basal portion to weak expression in mature OSNs. Lhx2 is not expressed in sustentacular cells. (B) Binding of Lhx2 protein to the homeodomain in the M71 promoter region was biochemically examined by using EMSA. Recombinant GST-Lhx2 binds to the M71 homeodomain site (wt), but not to the mutated homeodomain site (mut), on the left side of B (8% gel). Addition of anti-Lhx2 antibody, which is directed against the DNA binding domain of Lhx2, inhibits binding of Lhx2 to the homeodomain site, on the right side of B (6% gel). Sequences of probes for EMSA, wt and mut, are shown at the bottom. The homeodomain site is shown in red, and mutated sites are in green.
By EMSA, recombinant GST-Lhx2 protein binds to the M71-homeodomain site specifically (Fig. 1B). This binding is inhibited by an antibody against Lhx2, which recognizes its DNA-binding domain, probably by steric hindrance of the antibody. These results suggest that Lhx2 may be involved in regulating M71 gene choice and/or expression.
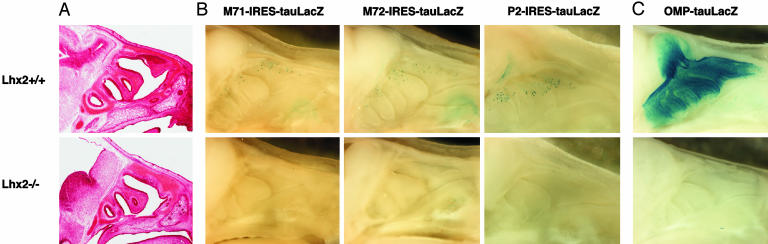
No OR Gene Expression in Lhx2-Deficient Embryos. Next, we analyzed Lhx2-knockout mice. These mutants have defects in the development of the eyes, forebrain, and erythrocytes and die a few days before birth because of severe anemia (18). We analyzed mutant embryos at E16.5, when OSNs already express OR genes (11). The olfactory turbinates of Lhx2–/– embryos appear to develop grossly normally, except for a slightly smaller size in comparison with wild-type embryos (Fig. 2A). No olfactory bulb structure is observed in Lhx2–/– embryos (Fig. 2 A).
Fig. 2.
Lack of OR gene and OMP expression in Lhx2–/– embryos. (A) Sagittal sections (12 μm) of heads show that the structure of the OE appears normal except for a slightly smaller OE in Lhx2–/– embryos (E16.5). (B) M71, M72, and P2 OR gene expression was examined by using M71-IRES-tauLacZ, M72-IRES-tauLacZ, and P2-IRES-tauLacZ heterozygous embryos at E16.5. Whole-mount preparations of sagittally transected mouse heads show that tauLacZ (M71, M72, and P2) expression is not detected in Lhx2–/– embryos. (C) Mature OSNs were visualized by using OMP-tauLacZ heterozygous embryos at E17.5. No mature neurons are present in Lhx2–/– embryos.
Expression of OR genes was examined by introducing several OR-IRES-tauLacZ mutations into the Lhx2–/– background; OSNs expressing a particular OR gene can thus be visualized by X-Gal staining. No β-galactosidase activity is observed for the tagged OR genes M71, M72, and P2 in Lhx2–/– embryos, whereas Lhx2+/+ embryos express these genes at E16.5 (Fig. 2B). OR gene expression was also examined by in situ hybridization; the OR genes tested, M71, M72, P2, MOR23, M12, M50, and MOR251–4, are not expressed (data not shown).
The absence of OR gene expression may indicate a defect in OSN development. We crossed Lhx mutant mice with OMP-tauLacZ mice, in which mature OSNs express tauLacZ. Fig. 2C shows that tauLacZ expression is absent in OMP-tauLacZ+/–, Lhx2–/– embryos. The lack of mature OMP-expressing cells reveals a profound defect in a late stage of OSN development.
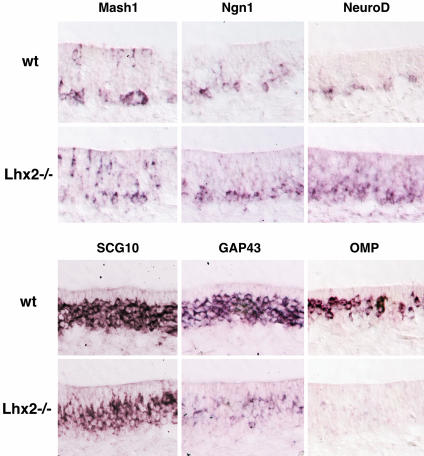
Incomplete Development of OSNs in Lhx2-Deficient Embryos. To define the step in OSN development that is arrested in Lhx2 mutant embryos, we performed in situ hybridization with RNA probes for Mash1 (as a marker for neuronal progenitors), Ngn1 (neuronal precursors), NeuroD (differentiating cells/postmitotic neurons), SCG10 (pan-neuronal), GAP43 (immature neurons), and OMP (mature neurons) (Fig. 3). Expression of Mash1 and Ngn1 is similar in Lhx2–/– and wild-type embryos, indicating that the early stage of neurogenesis, fate determination to neurons, proceeds normally. NeuroD-positive cells are increased in number and also occupy an intermediate region of the mutant OE, in addition to the basal region in which NeuroD is normally expressed. Although cells expressing SCG10 are decreased, this pan-neuronal marker is expressed in mutants, indicating that terminal differentiation into neurons is not blocked. GAP43-positive cells are severely reduced in number, and no OMP expression occurs in mutants, as was observed in OMP-tauLacZ+/–, Lhx2–/– embryos (Fig. 2C). These results indicate that the defect resulting from the Lhx2 mutation occurs between the stage of terminal differentiation to neurons and the stage of GAP43-positive, immature OSNs.
Fig. 3.
Incomplete development of OSNs in Lhx2–/– embryos. The developmental defect in the OE of Lhx2–/– embryos (E16.5) was characterized by in situ hybridization with RNA probes for Mash1 (neuronal progenitors), Ngn1 (neuronal precursors), NeuroD (differentiating neurons/postmitotic neurons), SCG10 (for pan-neurons), GAP43 (immature neurons), and OMP (mature neurons). Mash1 is expressed in cells located in apical, intermediate, and basal positions, in both wild-type and Lhx2–/– embryos. Ngn1 is expressed in basal cells, in both wild-type and mutant embryos. NeuroD is expressed in cells at intermediate positions, as well as in basal cells, in Lhx2–/– mice. SCG10 is expressed in an intermediate portion in both wild type and Lhx2–/–. GAP43 expression is severely reduced in Lhx2–/–, and OMP expression is abolished.
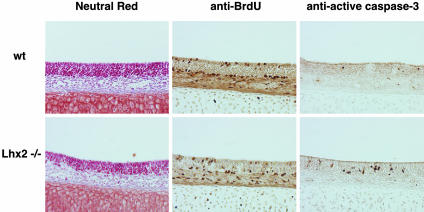
Increased Number of Apoptotic Cells in Lhx2–/– Mice. The mutant OE is slightly smaller and thinner than that of the wild type (Figs. 2 A and 4). This phenotype could be due to failure of maturation of OSNs resulting in increased apoptosis rather than to defects in the production of progenitor cells, because the early stages of neurogenesis (neuronal progenitors and precursors) appear normal (Fig. 3). Therefore, we counted dividing cells and apoptotic cells (Fig. 4). Dividing progenitor cells, marked by BrdUrd incorporation, are slightly reduced: a 37% decrease in apical cells (216 ± 21 cells in mutant and 338 ± 46.5 in wild type, n = 3; see Materials and Methods for details) and a 29% decrease in basal cells (286 ± 10.0 in mutant and 401 ± 21.7 in wild type, n = 3). By contrast, apoptotic cells, stained by anti-active caspase-3, are increased by 280% (778 ± 27.8 cells in mutant and 203 ± 32.3 cells in wild type, n = 3). Thus, the production of neuronal progenitors and precursors is normal as observed by in situ hybridization with markers (Fig. 3), and defective transition into immature neurons leads to increased apoptosis, which results in a thinner OE.
Fig. 4.
Decreased proliferative cells and increased apoptotic cells in Lhx2–/– embryos. The OE of Lhx2–/– embryos is thinner than that of wild type (neutral red staining). Immunohistochemistry for BrdUrd after a pulse labeling reveals progenitor cells in S phase. Proliferative progenitors are present in apical and basal positions in both wild-type and mutant embryos. The number of BrdUrd-positive cells in mutant embryos is slightly decreased. Immunohistochemistry for active caspase-3 shows an increased number of apoptotic cells in Lhx2–/– embryos.
Discussion
Lhx2 Binds to a Homeodomain Site in the M71 Promoter Region. Transgenic analyses of M71 and MOR23 promoter regions have revealed conserved sequences: homeodomain sites and O/E-like sites (15). In this study, we screened for proteins binding to the homeodomain in the M71 promoter region with the one-hybrid method, thus identifying nine homeodomain proteins. Among these, Emx2 and Lhx2 are expressed well in the OE. Expression of Emx2 occurs in patches both in the apical and basal portions of the OE. By contrast, Lhx2 is expressed strongly in the basal cell layer, from which OSNs are generated. Lhx2 is of particular interest for three reasons: it is the most frequent clone (60% of total positive clones), its expression pattern in the OE is consistent with a role in OR gene choice, and the function of LIM-homeodomain proteins in neuronal development is to specify neuronal identity (21). OR gene choice also represents a determination of neuronal identity: an individual OSN is thought to express a single OR gene, which determines both odorant specificity and glomerular inner vation. Binding of Lhx2 to the M71 OR-homeodomain site gives rise to the hypothesis that Lhx2 regulates M71 gene choice directly and may thus be required for an OSN to assume an identity. Although we demonstrate an interaction between Lhx2 and the M71 OR-homeodomain site in vitro, a direct linkage in vivo remains unclear. Lhx2 mutant mice do not express any of the tested OR genes, suggesting that Lhx2 is a general determinant, directly or indirectly, of OR gene expression. Because OR genes are expressed in the absence of the olfactory bulb (11), the lack of OR gene expression in Lhx2–/– embryos is unlikely to be the consequence of defective bulb development.
Hoppe et al. (22) performed one-hybrid screening with the mOR262–6 upstream region, which contains homeodomain, O/E-like, and E-box motifs, and thus identified Ptx-1, BEN, O/E-2, Alx3, Lhx2, and AP-2β. Functional involvement of this upstream region in mOR262–6 expression has not been shown, and binding of Lhx2 to the putative mOR262–6 promoter region has not been demonstrated. Yet these findings suggest that Lhx2 may bind to multiple putative OR promoter regions.
OR Gene Expression Controlled by Proximal and Long-Distance Regions. In Lhx2 mutant mice, none of the tested OR genes is expressed. Do all OR genes have a homeodomain site in their control region? A region of 405 bp upstream of the MOR23 transcription start site is required for transgene expression, and this region, as well as the M71 promoter region, contains a combination of two conserved motifs, a homeodomain site and an O/E-like site (15). This combination is also found upstream of the transcription start site of other OR genes, such as the M72 and mOR262 family (15, 22), although they have not been experimentally proven to be part of the control region. By contrast, Serizawa et al. (23) identified by yeast artificial chromosome transgenic technology a cis-acting DNA region, reportedly 75 kb upstream of the MOR28 gene, that is required for transgene expression. This 2-kb region, designated the H region, is proposed to activate one gene from the downstream OR gene cluster (the MOR28 gene and neighboring OR genes). Deletion of the H region abolishes expression of all OR genes from the cluster in the transgenes. Transgenic mice with 3 kb upstream of exon 1 of MOR28 did not show expression, but inclusion of the H region affords expression.
What is the common feature between the proximal regions in MOR23 and M71 (hundreds of base pairs upstream) and the distal region in MOR28 (many kilobases upstream)? We searched the H region in the mouse genome sequence (UCSC Genome Browser at http://genome.ucsc.edu), using the reported definition of human/mouse homology and 2.1-kb ScaI fragment size (23). We found at least one set of homeodomain and O/E-like sites in the H region, which, however, is 55–60 kb upstream of MOR28 in the database (Fig. 5A). Lhx2 binds in vitro to the homeodomain site in the M71-proximal control region, as well as to such a site in the MOR28 H region (Fig. 5B). O/E1 binds to O/E-like sites in both regions (data not shown). There could be additional homeodomain and O/E-like sites in the H region, and the in vivo role of any of these sites in OR expression remains to be shown. Nonetheless, the common occurrence of these sites in proximal and distal control regions is consistent with a broader role in OR gene regulation.
Fig. 5.
Homeodomain and O/E-like sites in OR control regions. (A) Sequence analysis of the M71 proximal control region and MOR28 H region shows a combination of conserved motifs: homeodomain and O/E-like sites. Lower sequences show oligonucleotides used for EMSA. The homeodomain site is in red, and mutated nucleotides are in green. (B) EMSA shows that GST-Lhx2 binds to the M71 and H region homeodomain sites.
Development of Olfactory Sensory Neurons. The molecular mechanisms that control the orderly series of developmental steps leading to mature OSNs remain poorly defined. To test the role of a gene in OSN development, the consequences of a gene knockout can be characterized in mice. In the olfactory system, there is such evidence for only a few transcription factors, Mash1 (24), Ngn1 (25, 26), and Hes1 and Hes5 (27). Mash1-knockout mice exhibit an early arrest of neurogenesis, and few OSNs mature (24, 26, 28). Ngn1-knockout mice have a severe but transient neurogenesis defect at an early stage of OE development (around E12.5), but normal numbers of OSNs are observed after E15.5 (26). By contrast, Hes1 and Hes5 regulate olfactory neurogenesis negatively (27).
Starting from the finding that Lhx2 binds to the homeodomain site in the M71 promoter region, we have now identified Lhx2 as a third transcription factor with a positive regulatory role in OSN development. A deficiency in Lhx2 results in the absence of mature OSNs, and OR gene expression cannot be detected. The defect of OSN development occurs between terminally differentiating cells and immature neurons. Thus, Lhx2 is a determinant of a late stage of OSN development. As Lhx2 binds to the M71 homeodomain site and deficiency of Lhx2 abolishes the expression of M71 and other OR genes, Lhx2 may specify OSN identity by enabling it to choose an OR gene. It is not clear whether Lhx2 is also required for continuous OR expression, once the gene has been chosen. Failure to complete OSN development would then be a direct consequence from a disruption of the mechanisms regulating OR gene choice and/or expression. It is also possible that Lhx2 is required for OMP and GAP43 gene expression. Lhx2 may regulate genes that are essential to complete neuronal development, such that neurons do not reach the stage in which OR gene choice occurs. The two views are not mutually exclusive: Lhx2 may control both OR gene choice and OSN development via distinct mechanisms.
Acknowledgments
We thank H. Westphal for his generous gift of Lhx2 heterozygous mice; F. Guillemot for in situ hybridization probes; T. McClintock for critical reading of the manuscript; K. Ishiguro for advice on the gel-shift assays; and A. Vassalli, P. Feinstein, A. Rothman, T. Ishii, and I. Rodriguez for useful comments. J.H. was the recipient of a long-term fellowship from the Human Frontier Science Program Organization. P.M. acknowledges the generous grant support from the National Institutes of Health and the National Institute on Deafness and Other Communication Disorders.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: EMSA, electrophoretic mobility-shift assay; En, embryonic day n; OE, olfactory epithelium; OMP, olfactory marker protein; OR, odorant receptor; OSN, olfactory sensory neuron.
References
- 1.Buck, L. & Axel, R. (1991) Cell 65, 175–187. [DOI] [PubMed] [Google Scholar]
- 2.Mombaerts, P. (2004) Nat. Rev. Neurosci. 5, 263–278. [DOI] [PubMed] [Google Scholar]
- 3.Malnic, B., Hirono, J., Sato, T. & Buck, L. B. (1999) Cell 96, 713–723. [DOI] [PubMed] [Google Scholar]
- 4.Zhang, X., Rodriguez, I., Mombaerts, P. & Firestein, S. (2004) Genomics 5, 802–811. [DOI] [PubMed] [Google Scholar]
- 5.Mombaerts, P. (2004) Curr. Opin. Neurobiol. 14, 31–36. [DOI] [PubMed] [Google Scholar]
- 6.Chess, A., Simon, I., Cedar, H. & Axel, R. (1994) Cell 78, 823–834. [DOI] [PubMed] [Google Scholar]
- 7.Ressler, K. J., Sullivan, S. L. & Buck, L. B. (1993) Cell 73, 597–609. [DOI] [PubMed] [Google Scholar]
- 8.Vassar, R., Ngai, J. & Axel, R. (1993) Cell 74, 309–318. [DOI] [PubMed] [Google Scholar]
- 9.Mombaerts, P., Wang, F., Dulac, C., Chao, S. K., Nemes, A., Mendelsohn, M., Edmondson, J. & Axel, R. (1996) Cell 87, 675–686. [DOI] [PubMed] [Google Scholar]
- 10.Wang, F., Nemes, A., Mendelsohn, M. & Axel, R. (1998) Cell 93, 47–60. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan, S. L., Bohm, S., Ressler, K. J., Horowitz, L. F. & Buck, L. B. (1995) Neuron 15, 779–789. [DOI] [PubMed] [Google Scholar]
- 12.Eggan, K., Baldwin, K., Tackett, M., Osborne, J., Gogos, J., Chess, A., Axel, R. & Jaenisch, R. (2004) Nature 428, 44–49. [DOI] [PubMed] [Google Scholar]
- 13.Li, J., Ishii, T., Feinstein, P. & Mombaerts, P. (2004) Nature 428, 393–399. [DOI] [PubMed] [Google Scholar]
- 14.Lane, R. P., Cutforth, T., Young, J., Athanasiou, M., Friedman, C., Rowen, L., Evans, G., Axel, R., Hood, L. & Trask, B. J. (2001) Proc. Natl. Acad. Sci. USA 98, 7390–7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vassalli, A., Rothman, A., Feinstein, P., Zapotocky, M. & Mombaerts, P. (2002) Neuron 35, 681–696. [DOI] [PubMed] [Google Scholar]
- 16.Wang, M. M. & Reed, R. R. (1993) Nature 364, 121–126. [DOI] [PubMed] [Google Scholar]
- 17.Xu, Y., Baldassare, M., Fisher, P., Rathbun, G., Oltz, E. M., Yancopoulos, G. D., Jessell, T. M. & Alt, F. W. (1993) Proc. Natl. Acad. Sci. USA 90, 227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porter, F. D., Drago, J., Xu, Y., Cheema, S. S., Wassif, C., Huang, S. P., Lee, E., Grinberg, A., Massalas, J. S., Bodine, D. et al. (1997) Development (Cambridge, U.K.) 124, 2935–2944. [DOI] [PubMed] [Google Scholar]
- 19.Zheng, C., Feinstein, P., Bozza, T., Rodriguez, I. & Mombaerts, P. (2000) Neuron 26, 81–91. [DOI] [PubMed] [Google Scholar]
- 20.Iwema, C. L. & Schwob, J. E. (2003) J. Comp. Neurol. 459, 209–222. [DOI] [PubMed] [Google Scholar]
- 21.Hobert, O. & Westphal, H. (2000) Trends Genet. 16, 75–83. [DOI] [PubMed] [Google Scholar]
- 22.Hoppe, R., Frank, H., Breer, H. & Strotmann, J. (2003) Genome Res. 13, 2674–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serizawa, S., Miyamichi, K., Nakatani, H., Suzuki, M., Saito, M., Yoshihara, Y. & Sakano, H. (2003) Science 302, 2088–2094. [DOI] [PubMed] [Google Scholar]
- 24.Guillemot, F., Lo, L. C., Johnson, J. E., Auerbach, A., Anderson, D. J. & Joyner, A. L. (1993) Cell 75, 463–476. [DOI] [PubMed] [Google Scholar]
- 25.Ma, Q., Kintner, C. & Anderson, D. J. (1996) Cell 87, 43–52. [DOI] [PubMed] [Google Scholar]
- 26.Cau, E., Casarosa, S. & Guillemot, F. (2002) Development (Cambridge, U.K.) 129, 1871–1880. [DOI] [PubMed] [Google Scholar]
- 27.Cau, E., Gradwohl, G., Casarosa, S., Kageyama, R. & Guillemot, F. (2000) Development (Cambridge, U.K.) 127, 2323–2332. [DOI] [PubMed] [Google Scholar]
- 28.Murray, R. C., Navi, D., Fesenko, J., Lander, A. D. & Calof, A. L. (2003) J. Neurosci. 23, 1769–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]