Abstract
Neurotrophins are recognized widely as essential factors in the developing nervous system. Previously, we demonstrated that neurotrophin 3 activation of TrkC inhibits Schwann cell myelination and enhances the migration of primary Schwann cells through the signaling pathway regulated by the Rho GTPases Rac1 and Cdc42. Here, we show that neurotrophins activate divergent signaling pathways to promote or inhibit Schwann cell migration. Endogenous brain-derived neurotrophic factor acting through p75NTR inhibits Schwann cell migration dramatically by Src kinase-dependent activation of the guanine-nucleotide exchange factor Vav2 and RhoA. Together, these results suggest that neurotrophins and their receptors differentially regulate Schwann cell migration through the signaling pathways that depend on Rho GTPases.
The formation of peripheral nervous system myelin is a complex, dynamic process involving two different cell types, the Schwann cell and the neuron, which involve a series of glial–neuronal interactions controlling the various stages leading to the formation of myelin (1, 2). Schwann cells proliferate, migrate along axons, ensheath individual axons, and eventually form the myelin sheath. We recently reported (3, 4) that brain-derived neurotrophic factor (BDNF) enhances peripheral nervous system myelination and that it does so by interacting with one of the neurotrophin receptors, p75NTR, on Schwann cells. p75NTR is a member of the tumor necrosis factor/nerve growth factor receptor superfamily (5, 6). In contrast, neurotrophin 3 (NT3) inhibits peripheral nervous system myelination by its interaction with one of the neurotrophin tyrosine kinase receptors, TrkC, which is also present on the Schwann cell (3, 4). These results provide increasing evidence that the involvement of neurotrophins in glial–neuronal interactions extend further than just their role in supporting the survival and differentiation of neurons.
Interestingly, although NT3 inhibits myelination, its activation of TrkC stimulates Schwann cell migration (7). This signaling pathway involves the Rho GTPases, Rac1 and Cdc42, as well as the activation of c-Jun N-terminal kinase (7). The most well characterized Rho GTPases include RhoA, Rac1, and Cdc42, and it is well established that the reorganization of the cytoskeleton to govern morphological changes, cellular focal adhesion, and migration is regulated by these Rho GTPases (8, 9). GTPases cycle between active (GTP-bound) and inactive (GDP-bound) conformations; however, the interconversions proceed slowly and are rate limiting. Therefore, GTPases are regulated positively by guanine nucleotide exchange factors (GEFs), which catalyze the replacement of GDP with GTP, and they are attenuated by GTPase-activating proteins (GAPs), which enhance the endogenous GTPase activity. To date, >60 GEFs have been identified in mammalian cells. Certain GEFs are specific for each Rho GTPase, whereas others show broader specificity. For example, Lbc and Lfc are specific for RhoA, whereas ephexin acts as the GEF for RhoA, Rac1, and Cdc42. When active, Rho GTPases control the cytoskeleton by the activation of their downstream effectors, such as Rho-kinase.
Because Schwann cell migration precedes the myelination process and, as noted above, NT3 enhances Schwann cell migration (7) and inhibits myelination (3, 4), is it possible that the reverse holds true and that inhibition of cell migration contributes to the premature differentiation of Schwann cells to form myelin? Here, we show that the interaction of BDNF with p75NTR, which enhances the myelination process (3, 4), does indeed inhibit Schwann cell migration strikingly. In this instance, the interaction of BDNF with p75NTR activates a different signaling pathway, which starts with the Src family tyrosine kinase but also involves the Rho GTPase, RhoA. Although RhoA, Rac1, and Cdc42 possess functions that are seemingly redundant (altering actin dynamics and focal adhesion), it is well established that they may function in a mutually antagonistic manner (8, 9) because they appear to do so in controlling Schwann cell migration. These results also provide an example of a functional role for the Src kinase/RhoA-GEF Vav2/RhoA signaling cascade in the nervous system.
Methods
Materials and Plasmids. For details on the materials and plasmids used, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Cell Culture. Primary Schwann cells were prepared from sciatic nerves of Sprague–Dawley rats or 129S3/SvImJ mice (wild type or p75NTR–/–; The Jackson Laboratory) at postnatal day 2, as described (7). Dorsal root ganglion (DRG) neurons that were dissociated from rat embryos at gestational day 15 were purified and cultured on collagen-coated dishes for 2–3 weeks, as described (3, 4). DRG neurons were used for collecting conditioned media, purifying axon membranes, and assaying for Schwann cell migration. Cos-7 cells were cultured and experiments were performed, as described (7). Unless indicated otherwise, cells were pretreated with or without C3 exoenzyme (10 μg/ml, 18 h), Y-27632 (10 μM, 45 min), PP1 (0.5 μM, 45 min), or PP3 (0.5 μM, 45 min) before stimulation with BDNF (100 ng/ml, 20 min). To confirm cell viability under these experimental conditions, cells were stained with 0.4% trypan blue. Trypan blue-incorporating cells were <1% in each experiment.
Small Interfering RNA (siRNA) Preparation and Transfection. The siRNAs were transfected into Schwann cells by using the Oligofectamine reagent (Invitrogen), according to the manufacturer's protocol. The medium was replaced 24 h after transfection, and cells were cultured in Sato medium containing 1 mg/ml BSA for 24 h (7). Transfection efficiencies for siRNA were ≈94 ± 2.3% for p75NTR and 78 ± 5.5% for Vav2. See Supporting Materials and Methods for details.
Plasmid Transfection. Cos-7 cells were transfected by the method of calcium phosphate precipitation. By using the pEGFP-C1 plasmid as the control, we found that transfection efficiency typically exceeded 98%. The final amount of the transfected DNA for a 6-cm dish was adjusted to 15 μg by addition of empty vector, pCMV. pCMV-RhoA (3 μg), pUSE-c-Src (1 μg), or pCMV-Vav2 (0.25 μg) were cotransfected with pCMV-p75NTR (0.5 μg) or p3XFLAG-CMV-Vav2ΔCat (0.5 μg) into Cos-7 cells. The medium was replaced 24 h after transfection, and cells were cultured in DMEM containing 1% FBS and 1 mg/ml BSA for 24 h (7).
Cell Migration Assays. Cell migration was routinely measured by using 24-well Boyden chambers (BD Biosciences) as described (7). Briefly, polyethylene terephthalate (8-μm pore size) filters were coated with axonal membranes from DRG neurons (10), collagen, fibronectin, or laminin. Schwann cells (2.5 × 105) or Cos-7 cells (7.5 × 105) in 750 μl of medium were loaded into the upper chambers, which were inserted into the tissue-culture wells. The tissue-culture wells contained 500 μl of conditioned media from DRG neurons with anti-nerve growth factor antibody (1 μg/ml; Chemicon) and Fc chimeras (2.5 μg/ml), or media with BDNF (250 ng/ml) or NT3 (25 ng/ml). After incubation at 37°C for 6–7 h, the filters were stained with Giemsa solution. Representative experiments are shown in GFigs. 1–5. The migrating cells at the bottom of the filters were counted at four fields per filter in two to four independent experiments.
Schwann cell migration was assayed also by using fasciculated DRG axons and reaggregated Schwann cells to mimic physiological conditions. DRG neurons were plated onto one side of a collagen-coated dish with parallel lanes etched throughout, and they were allowed to extend axons in the respective lanes. The axons became fasciculated after 2–3 weeks in culture. Schwann cell reaggregates were formed by plating Schwann cells on a nonpermissive substrate (bacterial dishes) overnight with gentle agitation every 2–3 h. The reaggregated Schwann cells were then plated onto the fasciculated axons, and individual Schwann cells were allowed to migrate out of the reaggregates along the axons in the presence of BDNF (100 ng/ml) or NT3 (10 ng/ml) for 6 and 12 h. Cells were immunostained, and images were collected with a Eclipse E800 fluorescence microscope (Nikon) and RT Slider SPOT CCD camera (Diagnostic Instruments, Sterling Heights, MI). The distance of migration was calculated by measuring the size of the reaggregates over time, subtracting the average initial size of the reaggregates, and dividing the remaining distance in half. Experiments were performed in quadruplicate, and the values shown represent the mean ± SEM from separate experiments. Student's t test was carried out for inter-group comparisons. The following primary antibodies were used: anti-S100β (DAKO) and antineurofilament (Sigma).
Immunoblotting. Cells were lysed in 200 μl (35 mm) or 600 μl (60 mm) of lysis buffer, as described (7). The proteins in the cell lysates were denatured and then separated on SDS/PAGE gels. The electrophoretically separated proteins were transferred to nitrocellulose membranes, blocked, and immunoblotted. The bound antibodies were detected by using the enhanced chemiluminescence (ECL) system (Amersham Biosciences).
Assay for Src Kinase. Activation of Src kinase in the cell lysates was detected by immunoblotting with an antibody against the antiphosphorylated form (pTyr416). To compare the total amount of Src kinase, immunoblotting was also performed by using anti-Src kinase antibody. Three to four separate experiments were performed, and a representative experiment is shown in Figs. 1, 2, 3, 4, and 5. The band intensity in the immunoblot was semiquantified. Levels of the phosphorylated forms were normalized to the total amount of kinase.
Fig. 1.
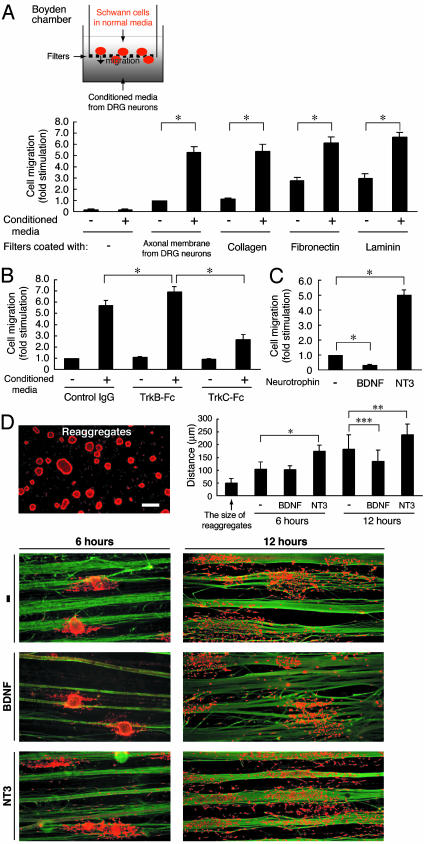
Schwann cell migration is modulated by neurotrophins. The migration of primary Schwann cells was measured by using Boyden chambers (A–C) and on fasciculated DRG axons (D). Filters were coated with DRG axonal membranes (A–C) and various extracellular substrates (A). After incubation with conditioned media from DRG neurons (A), conditioned media containing TrkB-Fc or TrkC-Fc (B), or media containing BDNF or NT3 (C), migration was assayed. (D) Schwann cell reaggregates on fasciculated DRG axons were incubated with BDNF or NT3, and the distance of migration was measured. Axons were stained for neurofilament (green) and the Schwann cells were stained with S100β (red). Scale bar indicates 50 μm. Data were evaluated by using Student's t test. *, P < 0.002; **, P < 0.01; ***, P < 0.02.
Fig. 2.
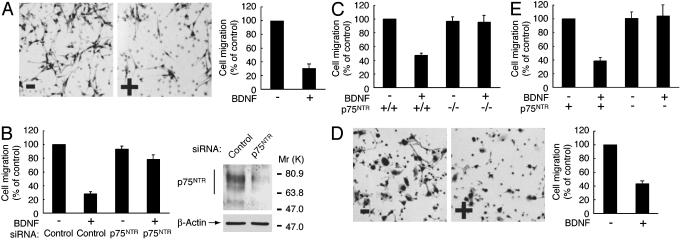
BDNF inhibits migration through p75NTR. After incubation with (+) or without (–) BDNF, migration of primary Schwann cells isolated from sciatic nerves of rats (A and B), p75NTR+/+ or p75NTR–/– mice (C), and Cos-7 cells transfected with (D and E) or without (E) p75NTR was assayed by using collagen-coated Boyden chambers. (B) Schwann cells were transfected with control or p75NTR siRNA. To confirm the effects of siRNAs, lysates from transfected cells were immunoblotted with anti-p75NTR or β-actin antibody.
Fig. 3.
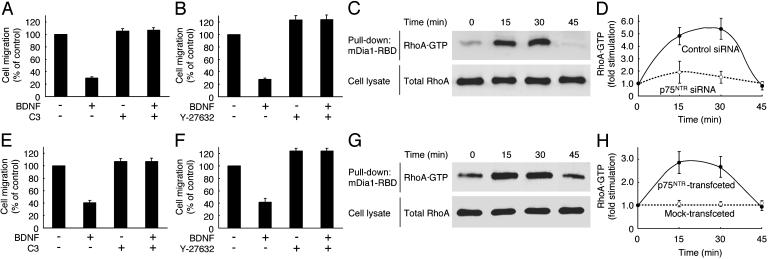
Involvement of the RhoA signaling pathway in the BDNF inhibition of migration. Schwann cells (A and B), and Cos-7 cells (E and F) transfected with p75NTR, were pretreated with C3 exoenzyme (A and E) or Y-27632 (B and F). After incubation with BDNF, migration was assayed by using collagen-coated Boyden chambers (A, B, E, and F). After stimulation with BDNF (0–45 min), RhoA activity was measured by pull-down assay using GST-mDia1-Rho-binding domain in the lysates from Schwann cells (C) and Cos-7 cells (G) transfected with RhoA and p75NTR. The total RhoA in the cell lysates is also shown (C and G). RhoA activity was measured in Schwann cells (D) transfected with control or p75NTR siRNA, and Cos-7 cells (H) were transfected with RhoA ± p75NTR.
Fig. 4.
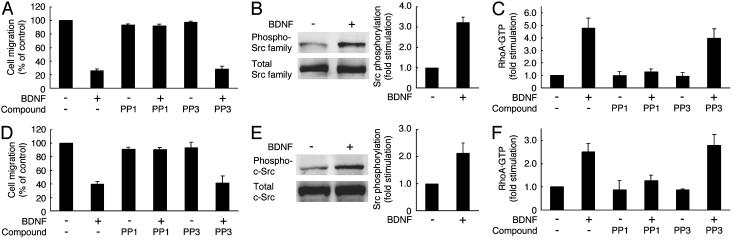
BDNF inhibits migration through Src kinase. Schwann cells (A) and Cos-7 cells (D) transfected with p75NTR were pretreated with PP1 or PP3. After incubation with BDNF, migration was assayed by using collagen-coated Boyden chambers (A and D). Src activity was determined by immunoblotting with an antiphosphorylated Src antibody in the cell lysates of Schwann cells (B), and Cos-7 cells (E) were transfected with c-Src and p75NTR. The total Src in the cell lysates is shown also (B and E). Schwann cells (C) and Cos-7 cells (F) transfected with RhoA and p75NTR were pretreated with PP1 or PP3, and RhoA activity was then measured.
Fig. 5.
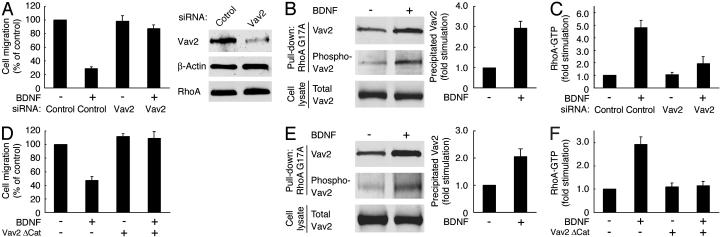
Migration is regulated by Src kinase-dependent Vav2 activation. (A) Schwann cells were transfected with control or Vav2 siRNA. (D) Cos-7 cells were transfected with or without Vav2ΔCat and p75NTR. After incubation with BDNF, migration was assayed by using collagen-coated Boyden chambers (A and D). To confirm the effects of siRNAs, lysates from transfected cells were immunoblotted with anti-Vav2, RhoA, or β-actin antibody (A). Active Vav2 was detected by the pull-down assay using GST-RhoAG15A from the lysates of Schwann cells (B), and Cos-7 cells (E) were transfected with Vav2 and p75NTR. The total Vav2 in the cell lysates is shown also (B and E). Schwann cells were transfected with a control or the Vav2 siRNA (C); Cos-7 cells were transfected with or without Vav2ΔCat, RhoA, and p75NTR (F); and RhoA activity was then measured.
Pull-Down Assay for Vav2. Vav2 that was coprecipitated with GST-RhoAG17A was immunoblotted with an antibody against Vav2 or the phosphorylated form. To compare the total amount of Vav2, immunoblotting was also performed by using an anti-Vav2 antibody. Three or four separate experiments were performed, and a representative experiment is shown in Figs. 1, 2, 3, 4 and 5. The band intensity in the immunoblots was semiquantified, and levels of the affinity-precipitated Vav2 were normalized to the amount of total Vav2.
Pull-Down Assay for RhoA. To detect GTP-bound RhoA, we performed the pull-down assay by using GST-mDia1-Rho-binding domain (7). Three to four separate experiments were performed, and a representative experiment is shown in Figs. 1, 2, 3, 4 and 5. The band intensity in the immunoblot was semiquantified, and levels of RhoA·GTP were normalized to the amount of total GTPase.
Statistical Analysis. Values shown represent the mean ± SD from separate experiments. Student's t test was carried out for inter-group comparisons.
Results
Endogenous Neurotrophins Are Regulators of Schwann Cell Migration. Previously, we demonstrated that DRG neurons are capable of secreting the neurotrophins BDNF and NT3 (3). NT3 inhibits Schwann cell myelination (3) and stimulates migration through the TrkC/Rac1/Cdc42/c-Jun N-terminal kinase pathway (7). In an attempt to study these potential factors involved in the regulation of Schwann cell migration, conditioned media from DRG neurons were applied to primary Schwann cells. By using Boyden chambers coated with axonal membranes from DRG neurons, we plated primary Schwann cells onto the filters and allowed them to migrate into the lower chamber along a concentration gradient of the conditioned media for 6–7 h. Fig. 1A shows that conditioned media from DRG neurons contain potential factor(s) that regulate Schwann cell migration powerfully.
To examine whether neurotrophins in the conditioned media from DRG neurons are potential factors that regulate Schwann cell migration, we added the BDNF scavenger TrkB-Fc or the NT3 scavenger TrkC-Fc to the conditioned media. Removal of BDNF with TrkB-Fc enhanced Schwann cell migration above control levels, whereas removal of NT3 with TrkC-Fc inhibited the migration effect by 2-fold (Fig. 1B), indicating that BDNF and NT3 are major endogenous factors that oppositely regulate Schwann cell migration. Also, exogenous BDNF reduced Schwann cell migration dramatically, whereas addition of exogenous NT3 enhanced migration by 5-fold (Fig. 1C). The effects of BDNF and NT3 on migration were much greater without the addition of conditioned media. This result is due to numerous factors, besides neurotrophins, that may influence cell migration, such as the neuregulins, which are factors that enhance Schwann cell migration and also inhibit the myelination process (11, 12).
To further investigate the effects of these neurotrophins on Schwann cell migration, we developed a migration assay by using reaggregated Schwann cells on fasciculated DRG axons to mimic more physiological conditions. Consistent with our previous findings, BDNF significantly inhibited Schwann cell migration from the reaggregates after stimulation with BDNF for 12 h. In contrast, NT3 stimulated migration at both 6 and 12 h (Fig. 1D). These results suggest that the endogenous neurotrophins BDNF and NT3 are key regulators of Schwann cell migration because they inhibit and promote migration, respectively.
BDNF Inhibits Schwann Cell Migration Through p75NTR. To examine the effects of extracellular matrix proteins on Schwann cell migration, Boyden chambers were coated with collagen, fibronectin, or laminin (Fig. 1 A). Fibronectin and laminin significantly enhanced Schwann cell migration by 3-fold in the absence of the conditioned media from DRG neurons (13, 14), whereas the collagen-coated filters were similar to the filters coated with DRG axonal membranes (Fig. 1 A). Therefore, because collagen modestly stimulated migration (Figs. 1 A and 2A), as compared with fibronectin and laminin, collagen-coated filters were used in the following experiments. Although NT3 stimulates Schwann cell migration through TrkC (7), we set out to study the signaling pathway coupling BDNF to the inhibition of migration. Primary Schwann cells express high levels of the neurotrophin receptors, p75NTR and the full-length TrkC (4). To test whether p75NTR is involved in the BDNF-induced inhibition of Schwann cell migration, we used RNA interference. Expression of p75NTR in Schwann cells was markedly down-regulated by transfection with p75NTR siRNA, whereas expression of β-actin was unaffected, as revealed by immunoblotting (Fig. 2B). The knockdown of p75NTR abolished the BDNF-induced inhibition of Schwann cell migration (Fig. 2B). To confirm these findings, we used Schwann cells from p75NTR–/– mice. Migration of wild-type Schwann cells was inhibited by BDNF, whereas the migration of p75NTR–/– Schwann cells was unaffected (Fig. 2C). Additionally, in Cos-7 cells, a transient transfection system with p75NTR was used to confirm that p75NTR mediates the BDNF-induced inhibition of migration. Although Cos-7 cells do not express any neurotrophin receptors (7), BDNF inhibited migration of Cos-7 cells expressing p75NTR (Fig. 2 D and E). Together, these results indicate that p75NTR mediates the BDNF-induced inhibition of migration.
BDNF Inhibition of Schwann Cell Migration Depends on RhoA Signaling. Because it has been reported that the small GTPase RhoA acts downstream of certain repulsive receptors (8, 9, 15), we examined whether BDNF inhibits migration through RhoA. Pretreatment with C3 exoenzyme (16), which ADP-ribosylates and inhibits RhoA, reversed the BDNF-induced inhibition of migration of Schwann cells (Fig. 3A) and Cos-7 cells transfected with p75NTR (Fig. 3E). Additionally, Y-27632 (17), a specific inhibitor of RhoA effector Rho-kinase, suppressed the effects of BDNF (Fig. 3 B and F), suggesting that BDNF inhibits migration through the RhoA signaling pathway.
To determine whether BDNF directly activates endogenous RhoA in Schwann cells, we performed a pull-down assay to detect RhoA·GTP by using Rho-binding domain. Endogenous RhoA was activated after stimulation with BDNF (Fig. 3 C and D), whereas knockdown of p75NTR inhibited this activation (Fig. 3D). Similarly, RhoA was activated by treatment with BDNF in Cos-7 cells transfected with p75NTR and RhoA (Fig. 3 G and H). These results indicate that BDNF/p75NTR inhibits migration through the RhoA signaling pathway.
Src Kinase Acts Upstream of RhoA Signaling. Because Src kinase often acts upstream of RhoA (18, 19), we investigated the involvement of Src kinase in the BDNF inhibition of migration. Pretreatment with PP1 (20), a specific inhibitor of Src kinase, abolished the BDNF inhibition of migration of Schwann cells (Fig. 4A) and Cos-7 cells transfected with p75NTR (Fig. 4D). On the other hand, PP3 (20), which is a PP1 analogue incapable of inhibiting Src kinase, had no effect on BDNF inhibition of migration (Fig. 4 A and D). These results suggest that Src kinase is involved also in the BDNF inhibition of migration.
To clarify whether BDNF simulates the intrinsic activity of Src kinase, we used an antibody specific for the autophosphorylated Src kinase. We detected the activation of Src kinase, as demonstrated by autophosphorylation, in response to BDNF treatment in Schwann cells (Fig. 4B) and Cos-7 cells transfected with p75NTR and c-Src (Fig. 4E). However, it remains unclear which Src kinase is responsible for BDNF stimulation because c-Src, Fyn, and c-Yes of the Src family are all expressed in Schwann cells (data not shown). Because Fyn shows comparatively higher expression in Schwann cells, Fyn is a likely candidate in the BDNF inhibition of Schwann cell migration.
Next, we confirmed whether BDNF activates RhoA through Src kinase. PP1, but not PP3, inhibited BDNF-induced activation of RhoA in Schwann cells (Fig. 4C) and Cos-7 cells transfected with p75NTR and RhoA (Fig. 4F). These results indicate that BDNF inhibits migration through the signaling pathway that depends on Src kinase, which is upstream of RhoA activation.
Src Kinase-Dependent Activation of Vav2 Mediates BDNF Inhibition of Migration. GEFs, such as Vav, Vav2, and Vav3, have been shown to be directly tyrosine-phosphorylated and activated by Src kinase (8, 9, 21). Vav shows limited expression in lymphocytes, whereas Vav2 and Vav3 are widely expressed in various tissues (9, 21). Because Vav2 is the predominant Vav isoform in Schwann cells (data not shown) and has the potential to activate RhoA, we tested whether Vav2 is involved in the BDNF inhibition of Schwann cell migration. Transfection of Vav2 siRNA into Schwann cells abolished this effect, whereas control siRNA did not (Fig. 5A), suggesting that Vav2 is essential in the BDNF inhibition of Schwann cell migration. Expression of Vav2 in Schwann cells was markedly inhibited by Vav2 siRNA, whereas that of β-actin and RhoA were unaffected, as revealed by immunoblotting (Fig. 5A). In Cos-7 cells, catalytic Dbl-homology domain-deficient Vav2 (Vav2ΔCat) blocked BDNF inhibition of migration (Fig. 5D). The GEF mutant lacking the catalytic domain is known to display a dominant-inhibitory effect (22).
To clarify whether Vav2 is activated by BDNF stimulation, we carried out a pull-down assay to detect active Vav2 by using RhoAG17A, a guanine-nucleotide-free form of RhoA (23). Active GEFs preferentially interact with guanine-nucleotide-free forms of the small GTPases (8, 9, 23). A glycine to alanine point mutation of residue 17 in RhoA decreases its nucleotide binding (22). After stimulation with BDNF, endogenous Vav2 precipitated with RhoAG17A increased in Schwann cells, as seen similarly in Cos-7 cells transfected with p75NTR and Vav2 (Fig. 5 B and E). Furthermore, affinity-precipitated Vav2 was tyrosine-phosphorylated at Tyr-172, the residue that is phosphorylated by Src kinase and critical for this activation (20). Together, these results suggest that BDNF leads to Src kinase-dependent activation of Vav2.
Next, we confirmed whether BDNF activates RhoA through Vav2. BDNF activation of RhoA was inhibited by transfection of Vav2 siRNA in Schwann cells (Fig. 5C). Similarly, Vav2ΔCat inhibited the BDNF-induced RhoA activation in Cos-7 cells transfected with p75NTR and RhoA (Fig. 5F). These results indicate that BDNF inhibits migration through Src kinase/Vav2/RhoA.
Discussion
Myelination by Schwann cells consists of three major phases, including proliferation, premyelination, and myelination (1, 2). The proliferative stage is characterized by cell division and migration of Schwann cells, whereas the premyelinating and myelinating stages exhibit the characteristic dynamic morphological changes as the Schwann cells form myelin. We have demonstrated (3, 4) that BDNF enhances myelination through p75NTR in Schwann cells. BDNF levels remain constant throughout the proliferative, premyelinating, and early myelinating stages (3). Similarly, the expression of p75NTR is present at high levels at the onset of myelination in Schwann cell/DRG neuronal cocultures as well as sciatic nerves during postnatal development (4), suggesting that BDNF and p75NTR play multiple roles in the myelination process of Schwann cells. Here, we show that endogenous BDNF inhibits migration of primary Schwann cells through the functional p75NTR.
Studies addressing the role of p75NTR in Schwann cell migration (24, 25) have reported modest effects in the enhancement of cell migration. Although these results seem to conflict with our findings, there are factors that may contribute to our different conclusions. Migration assays vary tremendously depending on the substrate used, the concentration of the factors, as well as the purity and density of cells. In our study, we demonstrate that neurotrophins directly affect the Schwann cell to either promote or inhibit migration. The Boyden chambers are reliable and highly reproducible assays that allow us to examine the effects of neurotrophins on Schwann cell migration by using various substrates. Additionally, the Schwann cell/DRG neuronal migration assay allows us to confirm these results on axons. These assays, combined with technical advances in molecular biology (RNA interference) and protein biochemistry (neurotrophin scavengers, inhibitors, pull-down assays, etc.), allow us to examine individual steps in the complex signaling cascades and illustrate direct neurotrophin effects on Schwann cell migration clearly.
Our study reveals a p75NTR signaling cascade that is mediated by Src kinase-dependent activation of Vav2 and, in turn, the activation of the RhoA signaling pathway in primary Schwann cells and Cos-7 cells expressing p75NTR. It has been reported (26) that neurotrophin activation of p75NTR leads to inactivation of basal RhoA activity in chick ciliary neurons. In this scenario, the inactivation of RhoA results in promoting neurite outgrowth, presumably by activating Rac1 and Cdc42, which are Rho GTPases that are regulated oppositely from RhoA. The intracellular domain of p75NTR is composed of a long juxtamembrane region and a type II death domain sequence, each of which associates with multiple proteins (5, 6). The difference in the pathway linking p75NTR to RhoA may be due to the recruitment of various factors to different intracellular regions of p75NTR depending on the cellular context. In the case of Schwann cells, the juxtamembrane region of p75NTR is known to interact with tumor necrosis factor receptor-associated factor (TRAF) 6 in a neurotrophin-dependent manner (27). Additionally, TRAF6 forms a stable complex with Src kinase (28). It is possible that p75NTR activates Vav2/RhoA through the TRAF6–Src kinase complex in Schwann cells.
In summary, endogenous neurotrophins and their receptors differentially regulate Schwann cell migration before initiation of myelination in the peripheral nervous system, by using apparently antagonistic Rho GTPases (8, 9). These effects depend on the concentration of available NT3 and BDNF. Simultaneous activation of p75NTR and TrkC with high concentration of NT3 (100 ng/ml) results in an inhibition of migration (data not shown), illustrating predominant p75NTR signaling over TrkC. On the basis of the findings described above, we summarize the proposed signaling pathways in Fig. 6, which is published as supporting information on the PNAS web site. NT3/TrkC stimulates Schwann cell migration through Rho GTPases Rac1 and Cdc42 and the downstream c-Jun N-terminal kinase cascade, thereby inhibiting myelination. In contrast, BDNF/p75NTR inhibits migration through Src kinase/Vav2/RhoA/Rho-kinase and ultimately promotes the initiation of myelination. Further study is necessary to clarify the detailed molecular network surrounding Rho GTPases in each stage of myelination. It is possible that the mechanisms responsible not only for Schwann cell migration but also the ensheathment of axons and the initiation of myelin formation may all require signals from Rho GTPases. Such studies could possibly help to elucidate how neurotrophins regulate different aspects of the myelination program through Rho GTPases, during development and also remyelination. Defining these fundamental signaling pathways necessary for establishing the appropriate environment for myelination will relate specifically to cell transplantation studies and remyelination paradigms.
Supplementary Material
Acknowledgments
We thank Drs. L. Chen, H. Maruta, Y. Miyamoto, and K. Nishimura for helpful discussions. This work was supported by National Institute of Neurological Disorders and Stroke Grant NS 04270; grants from the Muscular Dystrophy Association and the McGowan Charitable Trust (to E.M.S); and a National Research Service Award (to J.R.C.).
Abbreviations: BDNF, brain-derived neurotrophic factor; NT3, neurotrophin 3; GEF, guanine nucleotide exchange factor; DRG, dorsal root ganglion; siRNA, small interfering RNA.
References
- 1.Bunge, M. B., Williams, A. K. & Wood, P. M. (1982) Dev. Biol. 92, 449–460. [DOI] [PubMed] [Google Scholar]
- 2.Bunge, R. P. (1993) Curr. Opin. Neurobiol. 3, 805–809. [DOI] [PubMed] [Google Scholar]
- 3.Chan, J. R., Cosgaya, J. M., Wu, Y. J. & Shooter, E. M. (2001) Proc. Natl. Acad. Sci. USA 95, 10459–10464. [Google Scholar]
- 4.Cosgaya, J. M., Chan, J. R. & Shooter, E. M. (2002) Science 298, 1245–1248. [DOI] [PubMed] [Google Scholar]
- 5.Huang, E. J. & Reichardt, L. F. (2001) Annu. Rev. Neurosci. 24, 677–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gentry J. J., Barker P. A. & Carter B. D. (2004) Prog. Brain Res. 146, 25–39. [DOI] [PubMed] [Google Scholar]
- 7.Yamauchi, J., Chan, J. R. & Shooter, E. M. (2003) Proc. Natl. Acad. Sci. USA 100, 14421–14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaibuchi, K., Kuroda, S. & Amano, M. (1999) Annu. Rev. Biochem. 68, 459–486. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt, A. & Hall, A. (2002) Genes Dev. 16, 1587–1609. [DOI] [PubMed] [Google Scholar]
- 10.Grimes, M. L., Zhou, J., Beattie, E. C., Yuen, E. C., Hall, D. E., Valletta, J. S., Topp, K. S., LaVail, J. H., Bunnett, N. W. & Mobley, W. C. (1996) J. Neurosci. 16, 7950–7964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahanthappa, N. K., Anton, E. S. & Matthew, W. D. (1996) J. Neurosci. 16, 4673–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zanazzi, G., Einheber, S., Westreich, R., Hannocks, M. J., Bedell-Hogan, D., Marchionni, M. A. & Salzer, J. L. (2001) J. Cell Biol. 152, 1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baron-Van Evercooren, A., Kleinman, H. K., Seppa, H. E., Rentier, B. & Dubois-Dalcq, M. (1982) J. Cell Biol. 93, 211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarthy, J. B., Palm, S. L. & Furcht, L. T. (1983) J. Cell Biol. 97, 772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ridley, A. J., Schwartz, M. A., Burridge, K., Firtel, R. A., Ginsberg, M. H., Borisy, G., Parsons, J. T. & Horwitz, A. R. (2003) Science 302, 1704–1709. [DOI] [PubMed] [Google Scholar]
- 16.Sah, V. P., Seasholtz, T. M., Sagi, S. A. & Brown, J. H. (2000) Annu. Rev. Pharmacol. Toxicol. 40, 459–489. [DOI] [PubMed] [Google Scholar]
- 17.Uehata, M., Ishizaki, T., Satoh, H., Ono, T., Kawahara, T., Morishita, T., Tamakawa, H., Yamagami, K., Inui, J., Maekawa, M. & Narumiya, S. (1997) Nature 389, 990–994. [DOI] [PubMed] [Google Scholar]
- 18.DeMali, K. A., Wennerberg, K. & Burridge, K. (2003) Curr. Opin. Cell Biol. 15, 572–582. [DOI] [PubMed] [Google Scholar]
- 19.Koshimizu, T., Tanoue, A., Hirasawa, A., Yamauchi, J. & Tsujimoto, G. (2003) Pharmacol. Ther. 98, 235–244. [DOI] [PubMed] [Google Scholar]
- 20.Susva, M., Missbach, M. & Green, J. (2000) Trends Pharmacol. Sci. 21, 489–495. [DOI] [PubMed] [Google Scholar]
- 21.Schuebel, K. E., Movilla, N., Rosa, J. L. & Bustelo, X. R. (1998) EMBO J. 17, 6608–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyamoto, Y., Yamauchi, J. & Itoh, H. (2003) J. Biol. Chem. 278, 29890–29900. [DOI] [PubMed] [Google Scholar]
- 23.Arthur, W. T., Ellerbroek, S. M., Der, C. J., Burridge, K. & Wennerberg, K. (2002) J. Biol. Chem. 277, 42964–42972. [DOI] [PubMed] [Google Scholar]
- 24.Bentley, C. A. & Lee, K. F. (2000) J. Neurosci. 20, 7706–7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anton, E. S., Weskamp, G., Reichardt, L. F. & Matthew, W. D. (1994) Proc. Natl. Acad. Sci. USA 91, 2795–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamashita, T., Tucker, K. L. & Barde, Y. A. (1999) Neuron 24, 585–593. [DOI] [PubMed] [Google Scholar]
- 27.Khursigara, G., Orlinick, J. R. & Chao, M. V. (1999) J. Biol. Chem. 274, 2597–2600. [DOI] [PubMed] [Google Scholar]
- 28.Wong, B. R., Besser, D., Kim, N., Arron, J. R., Vologodskaia, M., Hanafusa, H. & Choi, Y. (1999) Mol. Cell 4, 1041–1049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.