Abstract
Common fragile sites (CFSs) are regions of the genome with a predisposition to DNA double-strand breaks in response to intrinsic (oncogenic) or extrinsic replication stress. CFS breakage is a common feature in carcinogenesis from its earliest stages. Given that a number of oncogenes and tumor suppressors are located within CFSs, a question that emerges is whether fragility in these regions is only a structural “passive” incident or an event with a profound biological effect. Furthermore, there is sparse evidence that other elements, like non-coding RNAs, are positioned with them. By analyzing data from various libraries, like miRbase and ENCODE, we show a prevalence of various cancer-related genes, miRNAs, and regulatory binding sites, such as CTCF within CFSs. We propose that CFSs are not only susceptible structural domains, but highly organized “functional” entities that when targeted, severe repercussion for cell homeostasis occurs.
Keywords: Fragile sites, miRNAs, Genomic instability, DNA elements, DNA repair, Carcinogenesis
Introduction
Activated oncogenes are a key feature of cancer development from its earliest stages [1]. One of their major effects is the induction of DNA damage via replication stress (RS) [2]. Specifically, oncogene-induced DNA replication stress (OIRS) leads to the formation of DNA double-strand breaks (DSBs), due to replication forks (RFs) collapse, fueling genomic instability (GI) [2, 3]. In early precancerous lesions, the collapse of DNA RFs occurs preferentially at specific loci termed fragile sites (FSs) [4]. As a result, FSs exhibit breaks, gaps, and rearrangements, collectively termed FSs expression. This is due to the activation of pathways responsible for fork collapse resolution and completion of DNA replication, which involve recombinogenic processes and DNA DSBs production [2].
An important issue is whether the instability manifested at these sites has any wider biological impact on cancer development. As further discussed in the manuscript, although FSs are heterogeneous in their expression patterns, they possess unique features that make them vulnerable to structural destabilization under RS conditions. Are these regions simply prone to DNA damage due to their intrinsic characteristics, conferring only to GI? Sparse evidence indicates that FSs enclose genes and non-coding RNAs, like microRNAs (miRs), while their expression could be epigenetically modulated by histones, implying that they are regions of the genome of a higher organization level (Fig. 1) [5–9]. Important bioinformatic resources are currently available and can be exploited to define potential topological associations between CFSs and these elements. Notably, the miRbase is constantly expanding while the ENCODE project [10] has deposited information on a vast range of binding elements and genomic modifications, including histone marks (like H3K79me2, H3K9ac, H3K4me3, and H3K27ac) that have a prominent influence on the expression process of the genome. As the pattern of instability at FSs in human tumors is variable, suggesting that it also depends on the cell type, this further complicates the role of FSs in malignancy. Last but not least, if such sites contribute to cancer development, why have they not been evolutionary selected for elimination? Could there be a higher reason that makes them at the same time vulnerable “units” of the genome with potentially meaningful function? Attempting to address these questions, in the current work we conducted an extensive review on the nature of their heterogeneity that accounts for their preferential instability. Next, by applying bioinformatic tools on data from the latest miRbase and the ENCODE project, we reveal that these sites are enriched in various (coding and non-coding) elements, such as cancer-related genes, miRs, and binding elements, as well as specific variations in histone modifications (Fig. 1). Based on these findings, we propose that these sites may represent unique “functional” units of the genome that may have a complex role upon OIRS with implications both in normal cell survival and cancer progression.
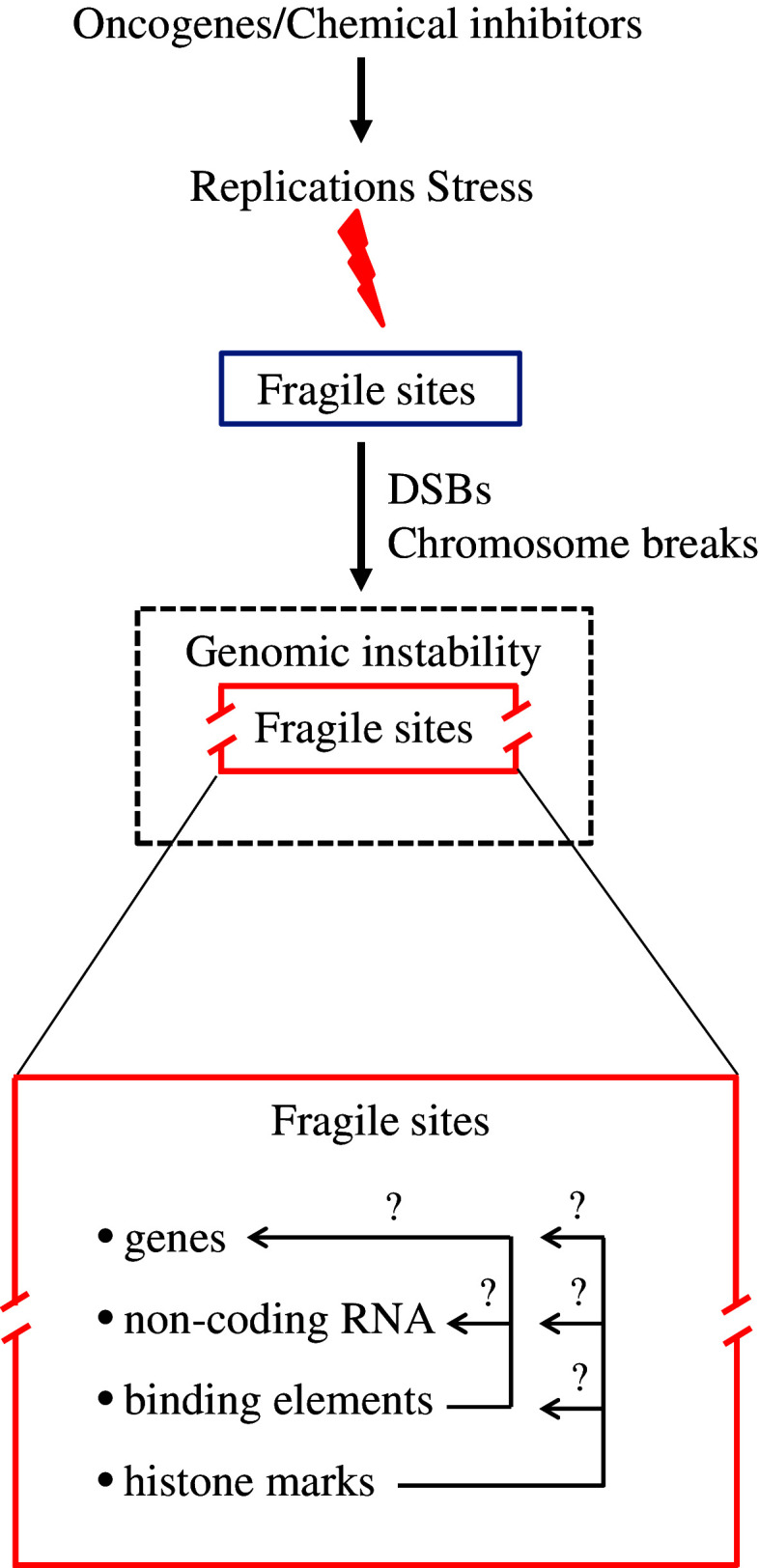
Fig. 1.
CFSs are not only vulnerable structural domains but may also be functional units of the genome that are sensitive to replication stress. CFS’s stability is affected by replications stress (RS). During cancer development, they are affected from the earliest precancerous lesions due to oncogene-induced replication stress (OIRS). Breakage at CFSs (broken red rectangle) may not only confer to genomic instability (GI) (dashed black rectangle), but could also have wider biological implications by affecting elements located within them (question mark). DSBs DNA double-strand breaks
Heterogeneity of fragile sites
FSs have been assigned in two classes, defined as rare fragile sites (RFSs) and common fragile sites (CFSs). Rare FSs are mainly induced by folate deficiency, correspond to dinucleotide or trinucleotide repeats, usually CGGn, and are found in less than 5 % of the human population and in specific families [11]. Their fragility is due to expansions of the micro- or mini-satellites sequences that they contain and in some cases are responsible for inherited diseases [11]. Therefore, RFSs will not be further discussed in this work.
Historically, CFSs were recognized as recurrent hotspots of double-stranded DNA breaks in cultured lymphocytes from healthy individuals [12]. They are present in all individuals, are part of the normal chromosomes, but exhibit different frequencies of expression in a population (reviewed in [13, 14]). CFSs are typically vulnerable to extrinsic replication stress, most notably to aphidicolin (APH), an inhibitor of DNA polymerase α, δ and ε, but are otherwise quiescent under normal conditions. This observation has been gradually broadened to include breakage patterns resulting from various replication inhibitors, such as nucleotide analogs (5-azacytidine, bromodeoxyuridine) or antitumor antibiotics (distamycin) and RS resulting from folate deficiency. Dietary and environmental factors like caffeine, cigarette smoke, and hypoxia may also enhance FS expression [15]. Recently, the induction of stress during the early S phase in B lymphocytes by hydroxyurea has been found to provoke DNA damage in a distinct pattern, corresponding to a new class of “early replicating fragile sites” (ERFSs) [16]. They occur primarily in early replicating DNA, close to replication origins, and are mainly situated in actively transcribed gene clusters (coding regions) [17]. This contrasts with CFSs, like FRA3B, which are most sensitive during their replication in late S phase [18]. Nevertheless, ERFSs seem also to arise from RF collapse and are similarly sensitive to ATR inhibition and oncogene-induced stress (see A. Nussenzweig chapter in this issue). OIRS is expected to induce instability at both ERFS and CFS, as suggested in two independent studies [16, 19]. Recent reports using the phosphorylated form of histone H2AX, the γ-H2AX, as a marker of DSB induction showed that ERFS were enriched for H2AX and γ-H2AX, while CFSs and heterochromatin lacked both, also suggesting differential DNA damage response at these sites [20]. Notably, both ERFSs and CFSs are rich in CpG-rich regions [17, 19], implying that these classes of FSs may either share structural similarities or the exact classification of their members as early and late replicating ones may need further re-assessment. Surprisingly, telomeric regions also appear to exhibit fragility in a similar manner as CFSs upon replication stress, including APH treatment [21].
It has been shown that CFS expression patterns depend not only on culture conditions but also on cell type [22]. Although traditionally studied almost exclusively in lymphocytes, different CFSs have been observed in fibroblasts [22, 23], breast, and colon epithelial cell lines [24, 25] and erythroid cell lines [25]. Considerable overlap exists between experiments, but the relative frequency of CFS breaks varies significantly. For example, FRA3B is the most frequent fragile locus in lymphocytes, but does not seem to be fragile in epithelial cell lines [24, 26], possibly due to the plasticity of replication programs in different cell lineages or because of a putative “housekeeping” role of FHIT. The second most fragile site, FRA16D, is very frequently affected in epithelial breast cancer cell lines (20–25 %), but only occasionally in colon epithelial cells (~5 %). Clearly, a complete characterization of CFS breakage probabilities will require a panel of different cell types.
CFSs are found in different individuals and are conserved across different species, including the mouse, the rat, and many mammals [27, 28], and across kingdoms, such as in the yeast S. cerevisiae [29]. Evolutionary conservation could argue in favor of a meaningful function if CFSs are considered outliers compared with the overall fragility of the genome in general. Nevertheless, variation between individuals can be significant. In a study of 20 normal adults [30], only FRA3B and FRA16D were found to be fragile in all individuals, and only 42 % of CFSs (19 of 45 identified) were present in the majority of individuals. In the earliest studies, less than 20 CFSs would explain more than 80 % of gaps and breaks [12]. A similar distribution was found in a population study of Deer mice, where high-frequency CFSs constituted approximately 26 % of the population total breaks and 38 % of CFSs were only found in single individuals.
Fragility of CFS
The issue of fragility at CFSs is a matter of intensive investigation. CFSs replicate either late in S-phase or initiate replication in mid-S phase, but exhibit a significant delay in completing it. Under conditions of RS, they may remain unreplicated even during G2-phase and up to mitosis leading eventually to their instability (as discussed in next section and reviewed in [13, 14]). Several features responsible for their replication sensitivity have not been revealed until now. These include intrinsic structural characteristics, the presence and overlap with large genes, differences in replication features, and epigenetic modulation [13, 14].
At the structural level, CFSs have the propensity to form secondary non-B structures that interfere with the movement of the replication fork thus leading to its collapse and associated DNA breaks [31]. Specifically, at sequence level, CFS are enriched in long stretches of AT dinucleotide-rich repeats that may form stable secondary cruciform DNA structures inducing fork stalling during DNA replication and in general incomplete or delayed DNA replication [31, 32]. In an earlier study, we performed a whole-genome analysis of CFS sequences and observed that they are on average rich in GC and Alu sequences [19]. The Alu family is a family of short interspersed repetitive elements (SINE) of about 300 bp containing mid and terminal poly A-stretches [33]. Interestingly, these elements are the most abundant mobile elements, and thus potentially recombinogenic in the human genome, and are implicated in various inherited human diseases and in cancer [34].
CFSs have been associated with genes extending over long genomic regions (“large genes”) [5]. The FHIT gene in FRA3B and WWOX in FRA16D are striking examples, measuring approximately 1.5 and 1.1 Mb respectively, compared with a mean of 10–15 kb for protein coding genes. The PARK2 gene, at approximately 1.4 Mb is also associated with FRA6E and may be down-regulated in ovarian tumors [30]. Intriguingly, genes over 800 kb may be prone to form RNA:DNA hybrid loops (termed R-loops) at sites of replication–transcription collision [35]. R-loops are structures formed by the association of the nascent transcript with the DNA template strand leaving unpaired the complementary non-coding DNA strand. Replication of large genes is time consuming and exposes the replication machinery to a risk of collision with the transcriptional machinery. In such an occurrence, the elongating RNA polymerase is blocked, leading to increased R-loop formation at Pol II pause sites. As a result, collision events may induce CFS breakage and a consequent enhancement of genomic instability [35].
Recent data have shed new light on the dynamics of the replication process at CFSs, providing several new mechanistic aspects explaining their instability (reviewed in [13, 14]). In the first one, it was shown that stability of FRA16C is perturbed under RS, as RFs progress more slowly and stall upon accounting AT-rich regions within this site. While in the bulk genome dormant origins are activated to complete replication, these are not available within FRA16C leading to delayed replication and instability [14] (also see B. Kerem chapter in this issue). A second report studying the mechanistic fragility of FRA3B, showed that fork speed slowing and stalling is similar with respect to the bulk genome, even under RS [26] (also see M. Debatisse chapter in this issue). In this case, the inability to complete replication was attributed to a large 700-kb core within this site that was found to be poor in origins. To accomplish replication of this site, origins from a long distance, located in the flanking regions, are required to come in and cover its length. The density and timing of origin firing events in the flanking regions seem to dictate the timing of FRA3B replication completion, thus influencing its stability. In a third mechanistic model regarding the FRA6E site, both replication arrest and paucity of origin activation lead to RS sensitivity, providing a functional combination of the two previous models [36]. Interestingly, some CFSs like the FRA3B do not exhibit stably these replication features in each cell type, suggesting that CFSs may demonstrate different patterns of instability, which are tissue specific. This may also explain why in various malignancies distinct profiles of GI are observed at CFSs that characterize each type of cancer. It would be interesting in the future to define the replication behavior of each CFS according to the specific cell type. That would be helpful in defining and possibly predicting the precise patterns of GI that take place during cancer development.
The replication density and timing of the genome has been proposed to be highly flexible and epigenetically controlled rather than directed by specific sequence motifs [13]. Therefore, an epigenetic control of CFSs replication stability may also apply. In support of this is the observed H3K9/14 hypoacetylation pattern displayed by the six most expressed CFSs in lymphoblastoid cells [9]. This histone modification has been reported to be associated with chromatin compactness and increased breakage. Also, regions with evenly spaced nucleosomes, an unusual chromatin structure preferentially formed at promoters and regulatory binding sites, have also been observed in FRA3B [37].
Overall, it seems that there is no single mechanism that can explain the fragility of CFSs but rather a multitude. They depend on several characteristics, including structural properties of the FSs as well as dynamic features governing their replication that apply in a given cell type. Interestingly, they are not necessarily mutually exclusive and often can function in complementary ways [13, 14]. The only common shared aspect by all these mechanisms is that they can eventually lead to a mechanical breakage of CFSs.
Maintenance of fragile site integrity
Instability at FSs is a recognized signature of DNA damage induced by replication stress [2] and it is detected from the earliest premalignant stages [3, 4, 19]. Replication checkpoints are activated in response to the stress induced at CFSs [38]. Central to these checkpoints are the DNA damage response kinases, ATM and ATR, which respectively sense DNA double-strand breaks and RF integrity [38]. Specific targeting of these kinases in cellular models revealed that ATR disruption or hypomorphic mutations lead to chromosomal instability within CFS even under normal replication, a phenomenon that is aggravated after low doses of APH [39]. In addition, dual inhibition of ATM and ATR using caffeine has been found to significantly increase CFS breakage compared with ATR deficiency alone, denoting also a role for ATM and a possible interplay with ATR in CFS protection [40, 41]. Inactivation of several down-stream components of the ATR network like Chk1, HUS1, Claspin, and SMC1 revealed similar effects, although not as efficient as ATR loss (Table 1) [31, 42–44].
Table 1.
Factors involved in control of CFSs stability and genome integrity
| Factor | Function | Reference |
|---|---|---|
| ATR | Main kinase-activating replication checkpoint | [39, 75] |
| ATM | Complementary acting kinase | [76] |
| Chk1 | Main ATR-downstream kinase involved in activation of the replication checkpoint | [31] |
| Chk2 | ATM-downstream kinase | [76] |
| HUS1 | Participates in the Rad9-Rad1-Hus1 (9-1-1) complex and is homologous to the PCNA clamp. The complex phosphorylates ATR substrates upon loading to sites of DNA damage | [42] |
| Claspin | Encodes for an adaptor protein which binds to BRCA1 and Chk1 and facilitates the ATR-dependent phosphorylation of both proteins during DNA replication stress in human cells | [43] |
| SMC1 | Member of the family of “structural maintenance of chromosomes” proteins participating in chromosome condensation, sister chromatid cohesion, and DNA repair. Prevents the collapse of stalled replication fork in an ATR-dependent manner and is required for S-phase checkpoint activation | [77] |
| BRCA1 | ATR substrate, implicated in the activation of the G2/M checkpoint, homologous recombination, and DSB repair | [78] |
| FANCD2 | Component of the Fanconi anemia (FA) pathway, phosphorylated by ATR. Plays a role not only in HR-dependent replication recovery, but also in regulating CFSs stability | [79] |
| Polymerase η | Involved in DNA synthesis of complex sequences, like repetitive and secondary structure that impede replication performing, the so-called ‘by-pass’ function | [80] |
| Rev3 | Catalytic subunit of Polζ that is required for maintaining fragile site stability in human cells | [81] |
| Polymerase κ | Involved in DNA synthesis of complex sequences, like repetitive and secondary structure that impede replication performing | [82] |
| WRN | RecQ helicase regulated in an ATR and ATM-dependent manner, which prevents DSBs formation at perturbed forks after replication stress. Promotes stability of arrested RFs and their efficient restart | [83] |
| BLM | RecQ helicase contributing to restarting stalled forks through unwinding DNA structures and/or homologous recombination, to maintenance of pyrimidine pools balance, regulation of fork speed and decatenation of UFBs at CFSs | [49, 50] |
| RECQ1 | Member of the RecQ family of DNA helicases. Promotes fork recovery and repair | [84] |
| Topoisomerase I and II | Alleviate DNA secondary structures during replication by cleavage and re-ligation. Can facilitate oncogenic rearrangements at induced CFSs | [85] |
| MUS81-EME1 | Endonuclease involved in resolving HJ-dependent replication intermediates. Required for fork repair and resolving UFBs | [41, 50] |
| PICH | PICH (Plk1 interacting checkpoint) is a helicase/translocase involved in resolving UFBs in mitosis | [49] |
| SNM1B/APOLLO | A nuclease component of the FA pathway involved in homology-directed repair. Also facilitates DNA localization of FANCD2 and BRCA1 | [86] |
| Rad51 | Component of the HR pathway involved in DSB repair and HJ mediated RF restart | [47] |
| DNA-PKcs | NHEJ pathway component | [47] |
| Ligase IV | NHEJ pathway component | [47] |
These checkpoints are vital as they ensure that DNA is replicated and chromosomes are prepared for mitosis [38]. Nevertheless, CFSs exhibit an increased vulnerability to RS leading to the activation of repair mechanisms [31]. The frequently observed presence of sister chromatid exchanges (SCEs) at the majority of CFSs breaks after APH treatment suggests that homologous recombination (HR) plays a major role in response to DSBs induced under conditions of RS [45]. The FANCD2 component of the Fanconi anemia (FA) pathway has been shown to play a role not only in HR-dependent replication recovery, but also in regulating CFSs stability (Table 1) (also reviewed in [31]). Similarly, activation of BRCA1 and other DSB repair proteins like RAD51 have also been found to be vital for maintaining CFSs stability (Table 1) [46]. Apart from HR the non-homologous end join pathway is also essential for chromosomal stability at these sites [47]. Specifically, by knocking down Rad51, DNA-PKcs, or Ligase IV, a significantly increased expression of CFSs under RS has been demonstrated. Notably, MDC1 and γ-H2AX foci were formed and co-localized with those of Rad51 and DNA-PKcs, while γ-H2AX and phospho-DNA-PKcs foci localized at expressed FSs on metaphase chromosomes.
Other components implicated in resolving replication over specific CFSs region include specialized polymerases (Table 1) [48]. These polymerases, like DNA polymerase eta (Pol η), mainly deal with DNA synthesis of complex sequences, like repetitive and secondary structures that impede replication, by performing the so-called ‘by-pass’ function. Depletion of these specialized polymerases has been shown to lead to persistence of unreplicated CFSs in mitosis. Various helicases/translocases have been proposed to promote fork restart at CFSs in non-redundant ways, like the BLM, WRN, and RECQ1 through Holliday junction-mediated fork remodeling that is independent of DSB formation (Table 1) [41]. Their main purpose seems to be resetting of structural intermediates arising from HR as well as unwinding of DNA secondary structures, in order to facilitate replication fork restart. Alternatively, nucleases, such as the structural endonuclease MUS81-EME1 and the FA pathway nuclease SNM1B/APOLLO, are responsible for DSB-mediated fork restart and/or elimination of permanently collapsed forks by cleavage of replication intermediates and consequent DNA synthesis (Table 1) [41].
Many of the above-described factors have been shown to stabilize CFSs during S-phase replication. Nevertheless, recent observations have shown that under RS, non-fully replicated or interlinked DNA at CFSs may escape S and G2–M checkpoints and enter mitosis (reviewed in [13, 49, 50]). Attempts to segregate these intermediates lead to sister chromatid entanglement followed by non-disjunction, ultimately leading to formation of ultra-fine bridges (UFBs) in anaphase cells. UFBs are defined by FANCD2/FANCI FA proteins binding to their edges, while BLM and PICH (Plk1-interacting checkpoint helicase) attach along the bridge. These persistent replication intermediates have been shown to be processed by the MUS81-EME1 nuclease in early M-phase, possibly with the help of ERCC1, to provide a controlled production of DNA breaks, aiming to allow undisturbed disjunction of sister chromatids. In case of failure, UFBs are formed as mentioned during anaphase. At this stage, BLM helicase and PICH translocase assisted by topoisomerase IIIa (TOPIIIa) and the BLM-associated proteins RMI1/2 function as a second line of defense by decatenating these structures and permitting chromatid segregation [50]. If unresolved UFBs still persist, they will eventually lead to chromosome miss-segregation by uneven distribution of DNA between the daughter cells and micronuclei formation. The transmitted errors at CFSs will be shielded in 53BP1 nuclear bodies in the emerging G1-daughter cells and possibly replicated in the S-phase by high-fidelity polymerases. These results pose a new light on CFSs cleavage, and led to the proposal that apart from being detrimental in initiating genomic instability, it can also serve as a mechanism for controlled production of DNA breaks that rather maintain than compromise genome integrity.
Several questions though emerge from this model that has been established with extrinsic factors (chemical inhibitors) that induce RS. How does this model apply and/or differentiate pre-malignant cells that are known to undergo OIRS? Which of the two types of damage, MUS81-EME1 cleavage or the decatenation inability, confers to the cancer-associated genome instability? A tempting but speculative model is that during premalignant stages, MUS81-EME1 cleavage activity is probably aberrantly increased, leading to a high frequency of DSBs, particularly at CFSs. This could be mediated by the active oncogenes that are present at such stages [2] and which in turn increase the activity of CDKs that regulate MUS81-EME1 expression [50]. As long as the checkpoints and their p53 effector are intact, the damage is repaired or the antitumor barriers of apoptosis and senescence eliminate such cells [2]. A similar scenario regarding gross genome damage due to decatenation inability may apply. Notably, it has been recently shown that under moderate RS, CFSs breaks can escape from efficient ATR checkpoint surveillance, leading to mitotic tolerance of such aberrations [44]. Such a pool of cells could undergo selection for loss of checkpoint function(s) and eventually accumulate DNA damage, probably through both MUS81-EME1 processing and chromatid non-disjunctions, provided that they are compatible with survival. Eventually, progression to full malignancy will ensue. This scenario fully concurs with our previous findings showing a prevalence of CFSs breakage along with the presence of UFBs and micronuclei in U2OS cells experiencing OIRS due to sustained expression of the replication licensing factor (RLF) Cdt1 [51]. Importantly, clones of these cells that “escaped” from the antitumor barriers, after prolonged Cdt1 expression, acquired a highly invasive potential. In a paradoxical way, this recently described model of deliberate CFSs controlled breakage to protect genome integrity of cells, may apply to malignant cells in the sense that it allows them to survive at the expense of genome integrity. Further expanding on this model, an emerging question concerns the effect exerted from the CFSs’ instability on the various elements like genes and non-coding RNAs that are located within them.
Functional elements in fragile sites
Several major publications arising from the ENCODE project [10] have underlined the importance of non-coding DNA. Non-coding regions of the genome have been found to participate in biochemical reactions with regulatory potential, such as transcription factor binding, epigenetic modifications, or long-distance interactions. Numerous functions have been attributed to non-coding RNA, including the expanding and best understood family of microRNAs (miRs) that are involved in post-transcriptional regulation of messenger RNA [52]. As a result of this progress, CFSs content may be now understood at a finer scale and the implications of CFS breakage will have to be reexamined carefully.
Fragile sites and cancer-associated genes
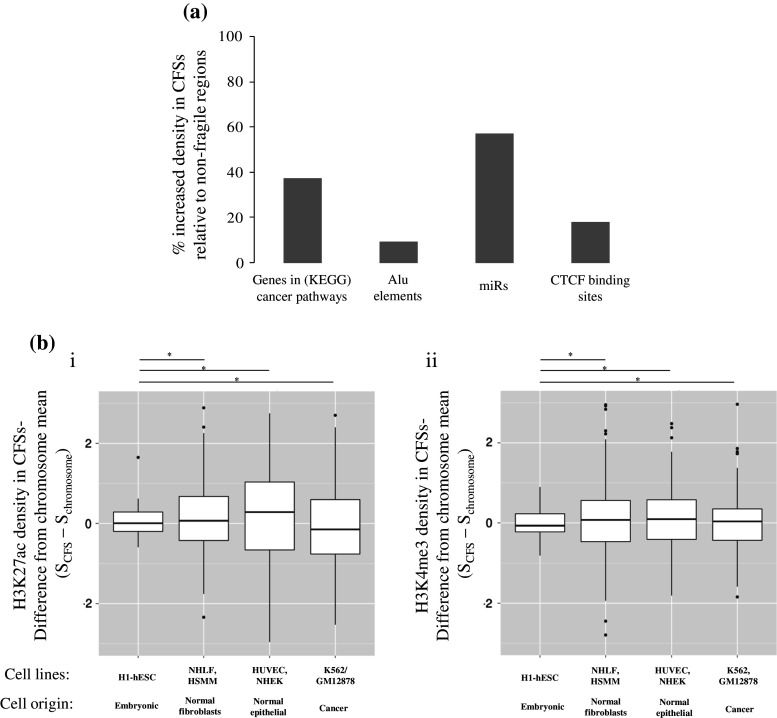
While available data point to an overlap between genes and CFSs [5], there is only one report showing that CFSs are denser in protein coding genes, with their distribution among fragile versus non-fragile regions varying among the chromosomes [7]. At the same time, a systematic review for the density of genes present within these genome areas is not available. To address this question, we retrieved a list of 327 genes participating in pathways in cancer from the Kyoto Encyclopedia of Genes and Genomes (KEGG)1 and investigated their association with both cytogenetically and molecularly mapped CFSs. We found that 110 cancer-related genes (33.6 % of all cancer-related genes) are located within CFSs (Table 2). Based on this, the density of cancer-related genes in a cytogenetically defined CFS compared to the rest of the genome is 37.2 % higher (Fig. 2a).
Table 2.
Cytogenetic defined common fragile sites (CFSs) association with cancer genes and miRs
| FRA | Chr | Start | End | Band | Type | Frequency | Cancer genes | miRNAs | Reference |
|---|---|---|---|---|---|---|---|---|---|
| FRA1A | 1 | 1 | 2,8000,000 | 1p36 | Aphidicolin | Common | DVL1, PIK3CD, MTOR, CASP9, CDC42, WNT4, E2F2 | hsa-mir-6859-1, hsa-mir-1302-2, hsa-mir-6723, hsa-mir-200b, hsa-mir-200a, hsa-mir-429, hsa-mir-6726, hsa-mir-6727, hsa-mir-6808, hsa-mir-4251, hsa-mir-551a, hsa-mir-4417, hsa-mir-4689, hsa-mir-4252, hsa-mir-6728, hsa-mir-34a, hsa-mir-5697, hsa-mir-1273d, hsa-mir-6729, hsa-mir-7846, hsa-mir-4632, hsa-mir-6730, hsa-mir-3675, hsa-mir-3972, hsa-mir-4695, hsa-mir-1290, hsa-mir-6084, hsa-mir-1256, hsa-mir-4418, hsa-mir-6127, hsa-mir-4684, hsa-mir-4253, hsa-mir-3115, hsa-mir-4419a, hsa-mir-378f, hsa-mir-6731, hsa-mir-4425, hsa-mir-3917, hsa-mir-1976 | [90] |
| FRA1B | 1 | 5,0700,001 | 6,1300,000 | 1p32 | Aphidicolin | Common | JUN | hsa-mir-4421, hsa-mir-6500, hsa-mir-761, hsa-mir-1273f, hsa-mir-5095, hsa-mir-1273g, hsa-mir-4781, hsa-mir-4422, hsa-mir-4711 | [8] |
| FRA1C | 1 | 68,900,001 | 69,700,000 | 1p31.2 | Aphidicolin | Common | |||
| FRA1D | 1 | 84,900,001 | 94,700,000 | 1p22 | Aphidicolin | Common | hsa-mir-4423, hsa-mir-7856, hsa-mir-760 | [91] | |
| FRA1E | 1 | 99,700,001 | 102,200,000 | 1p21.2 | Aphidicolin | Common | hsa-mir-553 | [92] | |
| FRA1F | 1 | 142,600,001 | 155,000,000 | 1q21 | Aphidicolin | Common | ARNT, TPM3, CKS1B | hsa-mir-3118-1, hsa-mir-3118-2, hsa-mir-3118-3, hsa-mir-6077-1, hsa-mir-6736, hsa-mir-5087, hsa-mir-6077-2, hsa-mir-6878, hsa-mir-4257, hsa-mir-554, hsa-mir-8083, hsa-mir-6737, hsa-mir-5698, hsa-mir-190b, hsa-mir-4258 | [91] |
| FRA1G | 1 | 172,900,001 | 176,000,000 | 1q25.1 | Aphidicolin | Common | |||
| FRA1H | 1 | 224,100,001 | 236,600,000 | 1q42 | 5-Azacytidine | Common | WNT9A, WNT3A, EGLN1 | hsa-mir-320b-2, hsa-mir-4742, hsa-mir-6741, hsa-mir-5008, hsa-mir-3620, hsa-mir-6742, hsa-mir-4666a, hsa-mir-1182, hsa-mir-4427, hsa-mir-4671, hsa-mir-4753, hsa-mir-1537 | [93] |
| FRA1I | 1 | 243,700,001 | 249,250,621 | 1q44 | Aphidicolin | Common | hsa-mir-3916, hsa-mir-3124 | ||
| FRA1J | 1 | 128,900,001 | 142,600,000 | 1q12 | 5-Azacytidine | Common | [94] | ||
| FRA1K | 1 | 185,800,001 | 198,700,000 | 1q31 | Aphidicolin | Common | TPR, PTGS2 | hsa-mir-4426, hsa-mir-1278, hsa-mir-4735 | [95] |
| FRA1L | 1 | 61,300,001 | 84,900,000 | 1p31 | Aphidicolin | Common | JAK1 | hsa-mir-3116-1, hsa-mir-3116-2, hsa-mir-6068, hsa-mir-4794, hsa-mir-3671, hsa-mir-101-1, hsa-mir-3117, hsa-mir-1262, hsa-mir-186, hsa-mir-7156 | |
| FRA1M | 1 | 94,700,001 | 99,700,000 | 1p21.3 | Folic acid | Rare | hsa-mir-378g, hsa-mir-2682, hsa-mir-137 | ||
| FRA2A | 2 | 96,800,001 | 102,700,000 | 2q11.2 | Folic acid | Rare | hsa-mir-3127, hsa-mir-5696 | ||
| FRA2B | 2 | 110,200,001 | 114,400,000 | 2q13 | Folic acid | Rare | PAX8 | hsa-mir-4267, hsa-mir-4436b-1, hsa-mir-4436b-2, hsa-mir-4435-2, hsa-mir-4771-2, hsa-mir-1302-3 | [96] |
| FRA2C | 2 | 16,700,001 | 19,200,000 | 2p24.2 | Aphidicolin | Common | [97] | ||
| FRA2D | 2 | 52,900,001 | 55,000,000 | 2p16.2 | Aphidicolin | Common | hsa-mir-4431, hsa-mir-3682 | [27] | |
| FRA2E | 2 | 68,600,001 | 75,000,000 | 2p13 | Aphidicolin | Common | TGFA | hsa-mir-3126, hsa-mir-1285-2 | [98] |
| FRA2F | 2 | 135,100,001 | 136,800,000 | 2q21.3 | Aphidicolin | Common | hsa-mir-5590, hsa-mir-128-1 | [8] | |
| FRA2G | 2 | 169,700,001 | 18,3000,000 | 2q31 | Aphidicolin | Common | ITGA6 | hsa-mir-933, hsa-mir-10b, hsa-mir-7704, hsa-mir-1246, hsa-mir-4444-1, hsa-mir-3128, hsa-mir-6512, hsa-mir-1258, hsa-mir-4437 | [99, 100] |
| FRA2H | 2 | 183,000,001 | 189,400,000 | 2q32.1 | Aphidicolin | Common | ITGAV | hsa-mir-548ae-1, hsa-mir-561 | [100] |
| FRA2I | 2 | 197,400,001 | 209,000,000 | 2q33 | Aphidicolin | Common | CASP8, FZD7, FZD5 | hsa-mir-3130-2, hsa-mir-3130-1, hsa-mir-2355, hsa-mir-7845, hsa-mir-1302-4, hsa-mir-4775 | [101] |
| FRA2J | 2 | 237,300,001 | 243,199,373 | 2q37.3 | Aphidicolin | Common | hsa-mir-6811, hsa-mir-4440, hsa-mir-4441, hsa-mir-4269, hsa-mir-2467, hsa-mir-4786, hsa-mir-149, hsa-mir-3133 | [102] | |
| FRA2K | 2 | 144,100,001 | 148,700,000 | 2q22.3 | Folic acid | Rare | [103] | ||
| FRA2L | 2 | 83,300,001 | 90,500,000 | 2p11.2 | Folic acid | TCF7L1 | hsa-mir-6071, hsa-mir-4779, hsa-mir-4771-1, hsa-mir-4435-1, hsa-mir-4780, hsa-mir-4436a | [104] | |
| FRA2S | 2 | 144,100,001 | 154,900,000 | 2q22.3-2q23.3 | Aphidicolin | Common | hsa-mir-4773-2, hsa-mir-4773-1 | [105] | |
| FRA3A | 3 | 23,900,001 | 26,400,000 | 3p24.2 | Aphidicolin | Common | RARB | hsa-mir-4792, hsa-mir-4442 | |
| FRA3B | 3 | 58,600,001 | 63,700,000 | 3p14.2 | Aphidicolin | Common | [106] | ||
| FRA3C | 3 | 182,700,001 | 187,900,000 | 3q27 | Aphidicolin | Common | DVL3 | hsa-mir-4448, hsa-mir-1224, hsa-mir-5588, hsa-mir-548aq, hsa-mir-1248 | [107] |
| FRA3D | 3 | 148,900,001 | 160,700,000 | 3q25 | Aphidicolin | Common | hsa-mir-5186, hsa-mir-3919, hsa-mir-15b, hsa-mir-16-2 | [107] | |
| FRA4A | 4 | 6,000,001 | 11,300,000 | 4p16.1 | Aphidicolin | Common | hsa-mir-4798, hsa-mir-4274, hsa-mir-95, hsa-mir-548i-2, hsa-mir-3138 | ||
| FRA4B | 4 | 527,00,001 | 59,500,000 | 4q12 | BrdU | Common | PDGFRA, KIT | hsa-mir-4449 | |
| FRA4C | 4 | 139,500,001 | 141,500,000 | 4q31.1 | Aphidicolin | Common | [108] | ||
| FRA4D | 4 | 11,300,001 | 35,800,000 | 4p15 | Aphidicolin | Common | hsa-mir-572, hsa-mir-5091, hsa-mir-218-1, hsa-mir-7978, hsa-mir-573, hsa-mir-4275 | ||
| FRA4F | 4 | 88,000,001 | 98,800,000 | 4q22 | Aphidicolin | Common | hsa-mir-5705 | [109] | |
| FRA5A | 5 | 28,900,001 | 42,500,000 | 5p13 | BrdU | Common | SKP2 | hsa-mir-4279, hsa-mir-579, hsa-mir-580, hsa-mir-3650 | |
| FRA5B | 5 | 92,300,001 | 98,200,000 | 5q15 | BrdU | Common | hsa-mir-2277, hsa-mir-583 | ||
| FRA5C | 5 | 130,600,001 | 136,200,000 | 5q31.1 | Aphidicolin | Common | TCF7 | hsa-mir-6830, hsa-mir-3936, hsa-mir-1289-2, hsa-mir-3661, hsa-mir-4461, hsa-mir-5692c-1 | [110] |
| FRA5D | 5 | 92,300,001 | 98,200,000 | 5q15 | Aphidicolin | Common | hsa-mir-2277, hsa-mir-583 | ||
| FRA5E | 5 | 18,400,001 | 28,900,000 | 5p14 | Aphidicolin | Common | [95] | ||
| FRA5F | 5 | 98,200,001 | 109,600,000 | 5q21 | Aphidicolin | Common | hsa-mir-548p | ||
| FRA5G | 5 | 168,500,001 | 180,915,260 | 5q35 | Folic acid | Rare | FGF18, MAPK9 | hsa-mir-585, hsa-mir-378e, hsa-mir-3912, hsa-mir-5003, hsa-mir-8056, hsa-mir-4634, hsa-mir-1271, hsa-mir-4281, hsa-mir-1229, hsa-mir-340, hsa-mir-8089, hsa-mir-4638 | |
| FRA6A | 6 | 13,400,001 | 15,200,000 | 6p23 | Folic acid | Rare | [111] | ||
| FRA6B | 6 | 4,200,001 | 71,00000 | 6p25.1 | Aphidicolin | Common | hsa-mir-3691, hsa-mir-7853, hsa-mir-5683 | ||
| FRA6C | 6 | 25,200,001 | 27,000,000 | 6p22.2 | Aphidicolin | Common | [112] | ||
| FRA6D | 6 | 70,000,001 | 75,900,000 | 6q13 | BrdU | Common | hsa-mir-30c-2, hsa-mir-30a, hsa-mir-4282 | ||
| FRA6E | 6 | 16,1000,001 | 164,500,000 | 6q26 | Aphidicolin | Common | [113] | ||
| FRA6F | 6 | 105,500,001 | 114,600,000 | 6q21 | Aphidicolin | Common | LAMA4, HDAC2 | hsa-mir-587 | [114] |
| FRA6G | 6 | 88,000,001 | 93,100,000 | 6q15 | Aphidicolin | Common | hsa-mir-4464, hsa-mir-4643 | ||
| FRA6H | 6 | 30,400,001 | 46,200,000 | 6p21 | Aphidicolin | Common | RXRB, PPARD, CDKN1A, VEGFA, HSP90AB1 | hsa-mir-877, hsa-mir-4640, hsa-mir-6891, hsa-mir-6832, hsa-mir-4646, hsa-mir-1236, hsa-mir-6721, hsa-mir-6833, hsa-mir-3135b, hsa-mir-219a-1, hsa-mir-6873, hsa-mir-6834, hsa-mir-5004, hsa-mir-3934, hsa-mir-7159, hsa-mir-1275, hsa-mir-6835, hsa-mir-7111, hsa-mir-5690, hsa-mir-3925, hsa-mir-4462, hsa-mir-4641, hsa-mir-6780b, hsa-mir-4647, hsa-mir-4642, hsa-mir-586 | [115] |
| FRA7A | 7 | 54,000,001 | 58,000,000 | 7p11.2 | Folic acid | Rare | EGFR | hsa-mir-4283-1, hsa-mir-3147 | |
| FRA7B | 7 | 1 | 7,300,000 | 7p22 | Aphidicolin | Common | PDGFA, RAC1 | hsa-mir-339, hsa-mir-4655, hsa-mir-6836, hsa-mir-4648, hsa-mir-4656, hsa-mir-589, hsa-mir-6874, hsa-mir-3683 | [116] |
| FRA7C | 7 | 35,000,001 | 37,200,000 | 7p14.2 | Aphidicolin | Common | hsa-mir-1200 | ||
| FRA7D | 7 | 43,300,001 | 45,400,000 | 7p13 | Aphidicolin | Common | hsa-mir-6837, hsa-mir-6838, hsa-mir-4649, hsa-mir-4657 | ||
| FRA7E | 7 | 91,100,001 | 92,800,000 | 7q21.2 | Aphidicolin | Common | CDK6 | hsa-mir-1285-1 | [117] |
| FRA7F | 7 | 98,000,001 | 107,400,000 | 7q22 | Aphidicolin | Common | PIK3CG | hsa-mir-3609, hsa-mir-25, hsa-mir-93, hsa-mir-106b, hsa-mir-4658, hsa-mir-6840, hsa-mir-6875, hsa-mir-4653, hsa-mir-4285, hsa-mir-548o, hsa-mir-5090, hsa-mir-4467 | |
| FRA7G | 7 | 114,600,001 | 117,400,000 | 7q31.2 | Aphidicolin | Common | MET, WNT2 | hsa-mir-6132 | [118] |
| FRA7H | 7 | 130,400,001 | 132,600,000 | 7q32.3 | Aphidicolin | Common | hsa-mir-29a, hsa-mir-29b-1 | [119] | |
| FRA7I | 7 | 147,900,001 | 159,138,663 | 7q36 | Aphidicolin | Common | SHH | hsa-mir-671, hsa-mir-3907, hsa-mir-153-2, hsa-mir-595, hsa-mir-5707 | [120] |
| FRA7J | 7 | 59,900,001 | 77,500,000 | 7q11 | Aphidicolin | Common | FZD9 | hsa-mir-4283-2, hsa-mir-6839, hsa-mir-4650-1, hsa-mir-3914-1, hsa-mir-3914-2, hsa-mir-4650-2, hsa-mir-4284, hsa-mir-590, hsa-mir-4651 | |
| FRA7K | 7 | 107,400,001 | 114,600,000 | 7q31.1 | Aphidicolin | Common | LAMB1, LAMB4 | hsa-mir-3666 | [121] |
| FRA8A | 8 | 101,600,001 | 106,200,000 | 8q22.3 | Folic acid | Rare | FZD6 | hsa-mir-7705, hsa-mir-5680, hsa-mir-3151, hsa-mir-548a-3 | |
| FRA8B | 8 | 93,300,001 | 99,000,000 | 8q22.1 | Aphidicolin | Common | CCNE2 | hsa-mir-8084, hsa-mir-378d-2, hsa-mir-3150b, hsa-mir-3150a | |
| FRA8C | 8 | 117,700,001 | 127,300,000 | 8q24.1 | Aphidicolin | Common | hsa-mir-3610, hsa-mir-548az, hsa-mir-4663, hsa-mir-548aa-1, hsa-mir-548d-1, hsa-mir-6844, hsa-mir-4662b, hsa-mir-4662a | [91] | |
| FRA8D | 8 | 139,900,001 | 146364,022 | 8q24.3 | Aphidicolin | Common | PTK2 | hsa-mir-151a, hsa-mir-1302-7, hsa-mir-4472-1, hsa-mir-4664, hsa-mir-937, hsa-mir-6845, hsa-mir-661, hsa-mir-6846, hsa-mir-6847, hsa-mir-7112-1, hsa-mir-7112-2, hsa-mir-6848, hsa-mir-939, hsa-mir-1234, hsa-mir-6849, hsa-mir-6893, hsa-mir-6850 | [66] |
| FRA8E | 8 | 117,700,001 | 127,300,000 | 8q24.1 | Distamycin A | Rare | hsa-mir-3610, hsa-mir-548az, hsa-mir-4663, hsa-mir-548aa-1, hsa-mir-548d-1, hsa-mir-6844, hsa-mir-4662b, hsa-mir-4662a | [122] | |
| FRA9A | 9 | 19,900,001 | 33,200,000 | 9p21 | Folic acid | Rare | CDKN2A, CDKN2B | hsa-mir-4473, hsa-mir-4474, hsa-mir-491, hsa-mir-31, hsa-mir-876, hsa-mir-873 | [123] |
| FRA9B | 9 | 114,900,001 | 117,700,000 | 9q32 | Folic acid | Rare | hsa-mir-455 | ||
| FRA9C | 9 | 19,900,001 | 33,200,000 | 9p21 | BrdU | Common | CDKN2A, CDKN2B | hsa-mir-4473, hsa-mir-4474, hsa-mir-491, hsa-mir-31, hsa-mir-876, hsa-mir-873 | |
| FRA9D | 9 | 90,400,001 | 91,800,000 | 9q22.1 | Aphidicolin | Common | hsa-mir-4289 | [91] | |
| FRA9E | 9 | 114,900,001 | 117,700,000 | 9q32 | Aphidicolin | Common | hsa-mir-455 | [124] | |
| FRA9F | 9 | 50,700,001 | 65,900,000 | 9q12 | 5-Azacytidine | Common | |||
| FRA9G | 9 | 16,600,001 | 18,500,000 | 9p22.2 | Aphidicolin | Common | [125] | ||
| FRA10A | 10 | 89,500,001 | 97,000,000 | 10q23.3 | Folic acid | Rare | PTEN, FAS | hsa-mir-4679-2, hsa-mir-4679-1, hsa-mir-107 | [126] |
| 10 | 99,300,001 | 101,900,000 | 10q24.2 | hsa-mir-1287, hsa-mir-4685, hsa-mir-6507 | |||||
| FRA10AC1 | 10 | 89,500,001 | 97,000,000 | 10q23.3 | Folic acid | Rare | PTEN, FAS | hsa-mir-4679-2, hsa-mir-4679-1, hsa-mir-107 | [127] |
| 10 | 99,300,001 | 101,900,000 | 10q24.2 | hsa-mir-1287, hsa-mir-4685, hsa-mir-6507 | |||||
| FRA10B | 10 | 111,900,001 | 114,900,000 | 10q25.2 | BrdU | Rare | hsa-mir-4680, hsa-mir-548e, hsa-mir-6715b, hsa-mir-6715a, hsa-mir-4295 | [128] | |
| FRA10C | 10 | 52,900,001 | 70,600,000 | 10q21 | BrdU | Common | CCDC6, CTNNA3 | hsa-mir-605, hsa-mir-548f-1, hsa-mir-3924, hsa-mir-1296, hsa-mir-7151, hsa-mir-1254-1 | [129] |
| FRA10D | 10 | 70,600,001 | 74,900,000 | 10q22.1 | Aphidicolin | Common | hsa-mir-7152, hsa-mir-4676 | [126] | |
| FRA10E | 10 | 111,900,001 | 114,900,000 | 10q25.2 | Aphidicolin | Common | hsa-mir-4680, hsa-mir-548e, hsa-mir-6715b, hsa-mir-6715a, hsa-mir-4295 | [126] | |
| FRA10F | 10 | 119,100,001 | 127,500,000 | 10q26.1 | Aphidicolin | Common | FGFR2, CTBP2 | hsa-mir-4681, hsa-mir-4682, hsa-mir-3941, hsa-mir-4296 | [130] |
| FRA10G | 10 | 42,300,001 | 52,900,000 | 10q11.2 | Aphidicolin | Common | RET, MAPK8, NCOA4 | hsa-mir-5100, hsa-mir-3156-1, hsa-mir-4294 | [129] |
| FRA11A | 11 | 68,400,001 | 70,400,000 | 11q13.3 | Folic acid | Rare | CCND1, FGF19, FGF4, FGF3, FADD | hsa-mir-3164, hsa-mir-548k | [131] |
| FRA11B | 11 | 114,500,001 | 121,200,000 | 11q23.3 | Folic acid | Rare | CBL | hsa-mir-6716, hsa-mir-4492, hsa-mir-3656, hsa-mir-6756 | [132, 133] |
| FRA11C | 11 | 16,200,001 | 21,700,000 | 11p15.1 | Aphidicolin | Common | hsa-mir-3159, hsa-mir-4486, hsa-mir-4694 | [91] | |
| FRA11D | 11 | 26,100,001 | 27,200,000 | 11p14.2 | Aphidicolin | Common | |||
| FRA11E | 11 | 31,000,001 | 36,400,000 | 11p13 | Aphidicolin | Common | hsa-mir-1343, hsa-mir-3973 | [134] | |
| FRA11F | 11 | 85,600,001 | 88,300,000 | 11q14.2 | Aphidicolin | Common | FZD4 | hsa-mir-6755, hsa-mir-3166 | [135] |
| FRA11G | 11 | 114,500,001 | 121,200,000 | 11q23.3 | Aphidicolin | Common | CBL | hsa-mir-6716, hsa-mir-4492, hsa-mir-3656, hsa-mir-6756 | [133] |
| FRA11H | 11 | 63,400,001 | 77,100,000 | 11q13 | Aphidicolin | Common | VEGFB, BAD, RELA, GSTP1, CCND1, FGF19, FGF4, FGF3, FADD, WNT11 | hsa-mir-7155, hsa-mir-1237, hsa-mir-192, hsa-mir-194-2, hsa-mir-6750, hsa-mir-6749, hsa-mir-6879, hsa-mir-6751, hsa-mir-612, hsa-mir-4690, hsa-mir-4489, hsa-mir-3163, hsa-mir-6860, hsa-mir-6752, hsa-mir-7113, hsa-mir-4691, hsa-mir-6753, hsa-mir-3164, hsa-mir-548k, hsa-mir-3664, hsa-mir-6754, hsa-mir-3165, hsa-mir-139, hsa-mir-4692, hsa-mir-548al, hsa-mir-4696, hsa-mir-326 | [136] |
| FRA11I | 11 | 16,200,001 | 21,700,000 | 11p15.1 | Distamycin A | Rare | hsa-mir-3159, hsa-mir-4486, hsa-mir-4694 | ||
| FRA12A | 12 | 46,400,001 | 54,900,000 | 12q13.1 | Folic acid | Rare | WNT10B, WNT1 | hsa-mir-4698, hsa-mir-4494, hsa-mir-6505, hsa-mir-1291, hsa-mir-4701, hsa-mir-1293, hsa-mir-6757, hsa-mir-196a-2, hsa-mir-615, hsa-mir-3198-2, hsa-mir-148b | [137] |
| FRA12B | 12 | 80,300,001 | 92,600,000 | 12q21.3 | Aphidicolin | Common | KITLG | hsa-mir-617, hsa-mir-618, hsa-mir-4699 | |
| FRA12C | 12 | 114,300,001 | 120,700,000 | 12q24.2 | BrdU | Rare | hsa-mir-620, hsa-mir-4472-2, hsa-mir-1178, hsa-mir-4498 | [138] | |
| FRA12D | 12 | 112,300,001 | 114,300,000 | 12q24.13 | Folic acid | Rare | hsa-mir-3657, hsa-mir-6861, hsa-mir-1302-1, hsa-mir-7106, hsa-mir-6762 | ||
| FRA12E | 12 | 109,000,001 | 133,851,895 | 12q24 | Aphidicolin | Common | FZD10 | hsa-mir-4496, hsa-mir-619, hsa-mir-4497, hsa-mir-6760, hsa-mir-6761, hsa-mir-3657, hsa-mir-6861, hsa-mir-1302-1, hsa-mir-7106, hsa-mir-6762, hsa-mir-620, hsa-mir-4472-2, hsa-mir-1178, hsa-mir-4498, hsa-mir-4700, hsa-mir-7107, hsa-mir-4304, hsa-mir-8072, hsa-mir-3908, hsa-mir-6880, hsa-mir-5188, hsa-mir-4419b, hsa-mir-3612, hsa-mir-6763 | |
| FRA13A | 13 | 34,000,001 | 35,500,000 | 13q13.2 | Aphidicolin | Common | [139] | ||
| FRA13B | 13 | 55,300,001 | 73,300,000 | 13q21 | BrdU | Common | hsa-mir-5007, hsa-mir-3169, hsa-mir-548x-2, hsa-mir-4704 | ||
| FRA13C | 13 | 59,600,001 | 62,300,000 | 13q21.2 | Aphidicolin | Common | hsa-mir-3169 | [140] | |
| FRA13D | 13 | 95,000,001 | 101,700,000 | 13q32 | Aphidicolin | Common | hsa-mir-4501, hsa-mir-3170, hsa-mir-623, hsa-mir-4306 | ||
| FRA13E | 13 | 73,300,001 | 79,000,000 | 13q22 | Aphidicolin | Common | hsa-mir-3665 | [115] | |
| FRA14B | 14 | 58,100,001 | 67,900,000 | 14q23 | Aphidicolin | Common | HIF1A, MAX | hsa-mir-5586, hsa-mir-548h-1, hsa-mir-7855, hsa-mir-4706, hsa-mir-4708, hsa-mir-625 | |
| FRA14C | 14 | 67,900,001 | 70,200,000 | 14q24.1 | Aphidicolin | Common | hsa-mir-5694 | ||
| FRA15A | 15 | 59,100,001 | 67,500,000 | 15q22 | Aphidicolin | Common | DAPK2, MAP2K1, SMAD3 | hsa-mir-2116, hsa-mir-8067, hsa-mir-6085, hsa-mir-190a, hsa-mir-422a, hsa-mir-1272, hsa-mir-4511, hsa-mir-4311, hsa-mir-4512 | [141] |
| FRA16A | 16 | 14,800,001 | 16,800,000 | 16p13.11 | Folic acid | Rare | hsa-mir-3179-1, hsa-mir-3670-1, hsa-mir-3180-1, hsa-mir-6511a-1, hsa-mir-6770-1, hsa-mir-1972-1, hsa-mir-6511b-2, hsa-mir-3180-4, hsa-mir-6506, hsa-mir-484, hsa-mir-3179-2, hsa-mir-3670-2, hsa-mir-3180-2, hsa-mir-6511a-2, hsa-mir-6770-2, hsa-mir-6511a-3 | [142] | |
| FRA16B | 16 | 66,700,001 | 70,800,000 | 16q22.1 | Distamycin A | Rare | CDH1 | hsa-mir-328, hsa-mir-6773, hsa-mir-1538, hsa-mir-140, hsa-mir-1972-2 | [143] |
| FRA16C | 16 | 66,700,001 | 70,800,000 | 16q22.1 | Aphidicolin | Common | CDH1 | hsa-mir-328, hsa-mir-6773, hsa-mir-1538, hsa-mir-140, hsa-mir-1972-2 | [144] |
| FRA16D | 16 | 79,200,001 | 81,700,000 | 16q23.2 | Aphidicolin | Common | hsa-mir-4720, hsa-mir-7854, hsa-mir-6504 | [143] | |
| FRA16E | 16 | 24,200,001 | 28,100,000 | 16p12.1 | Distamycin A | Rare | hsa-mir-1273h, hsa-mir-548w | [145] | |
| FRA17A | 17 | 10,700,001 | 16,000,000 | 17p12 | Distamycin A | Rare | hsa-mir-744, hsa-mir-1269b, hsa-mir-548h-3, hsa-mir-4731 | [123] | |
| FRA17B | 17 | 57,600,001 | 58,300,000 | 17q23.1 | Aphidicolin | Common | hsa-mir-21, hsa-mir-4737 | [112] | |
| FRA18A | 18 | 32,700,001 | 37,200,000 | 18q12.2 | Aphidicolin | Common | hsa-mir-3975, hsa-mir-187, hsa-mir-3929, hsa-mir-4318 | [146] | |
| FRA18B | 18 | 53,800,001 | 61,600,000 | 18q21.3 | Aphidicolin | Common | BCL2 | hsa-mir-122, hsa-mir-3591 | [147] |
| FRA18C | 18 | 66,800,001 | 68,700,000 | 18q22.2 | Aphidicolin | Common | [148] | ||
| FRA19A | 19 | 32,400,001 | 59,128,983 | 19q13 | 5-Azacytidine | Common | CEBPA, AKT2, EGLN2, TGFB1, CBLC, FGF21, BAX, FLT3LG, KLK3, BIRC8, PRKCG | hsa-mir-6887, hsa-mir-5196, hsa-mir-4530, hsa-mir-6719, hsa-mir-641, hsa-mir-6796, hsa-mir-6797, hsa-mir-4323, hsa-mir-8077, hsa-mir-4531, hsa-mir-8085, hsa-mir-6088, hsa-mir-330, hsa-mir-642a, hsa-mir-642b, hsa-mir-769, hsa-mir-320e, hsa-mir-3190, hsa-mir-3191, hsa-mir-6798, hsa-mir-4324, hsa-mir-150, hsa-mir-5088, hsa-mir-6799, hsa-mir-6800, hsa-mir-4749, hsa-mir-4750, hsa-mir-4751, hsa-mir-8074, hsa-mir-99b, hsa-let-7e, hsa-mir-125a, hsa-mir-6801, hsa-mir-643, hsa-mir-512-1, hsa-mir-512-2, hsa-mir-1323, hsa-mir-498, hsa-mir-520e, hsa-mir-515-1, hsa-mir-519e, hsa-mir-520f, hsa-mir-515-2, hsa-mir-519c, hsa-mir-1283-1, hsa-mir-520a, hsa-mir-526b, hsa-mir-519b, hsa-mir-525, hsa-mir-523, hsa-mir-518f, hsa-mir-520b, hsa-mir-518b, hsa-mir-526a-1, hsa-mir-520c, hsa-mir-518c, hsa-mir-524, hsa-mir-517a, hsa-mir-519d, hsa-mir-521-2, hsa-mir-520d, hsa-mir-517b, hsa-mir-520g, hsa-mir-516b-2, hsa-mir-526a-2, hsa-mir-518e, hsa-mir-518a-1, hsa-mir-518d, hsa-mir-516b-1, hsa-mir-518a-2, hsa-mir-517c, hsa-mir-520h, hsa-mir-521-1, hsa-mir-522, hsa-mir-519a-1, hsa-mir-527, hsa-mir-516a-1, hsa-mir-1283-2, hsa-mir-516a-2, hsa-mir-519a-2, hsa-mir-371a, hsa-mir-371b, hsa-mir-372, hsa-mir-373, hsa-mir-935, hsa-mir-4752, hsa-mir-8061, hsa-mir-7975, hsa-mir-6804, hsa-mir-6802, hsa-mir-6803, hsa-mir-6805, hsa-mir-6806, hsa-mir-4754, hsa-mir-6807 | |
| FRA19B | 19 | 1 | 20,000,000 | 19p13 | Folic acid | Rare | FGF22, APC2, DAPK3, MAP2K2, PIK3R2 | hsa-mir-1302-10, hsa-mir-4745, hsa-mir-3187, hsa-mir-1909, hsa-mir-1227, hsa-mir-6789, hsa-mir-4321, hsa-mir-7108, hsa-mir-7850, hsa-mir-637, hsa-mir-4746, hsa-mir-7-3, hsa-mir-4747, hsa-mir-6885, hsa-mir-6790, hsa-mir-3940, hsa-mir-6791, hsa-mir-6792, hsa-mir-4999, hsa-mir-5589, hsa-mir-4322, hsa-mir-1181, hsa-mir-1238, hsa-mir-638, hsa-mir-4748, hsa-mir-199a-1, hsa-mir-6793, hsa-mir-6886, hsa-mir-7974, hsa-mir-5684, hsa-mir-6794, hsa-mir-5695, hsa-mir-6515, hsa-mir-24-2, hsa-mir-27a, hsa-mir-23a, hsa-mir-181c, hsa-mir-181d, hsa-mir-1199, hsa-mir-639, hsa-mir-6795, hsa-mir-1470, hsa-mir-3188, hsa-mir-3189, hsa-mir-640 | |
| FRA20A | 20 | 17,900,001 | 21,300,000 | 20p11.23 | Folic acid | Rare | hsa-mir-3192 | ||
| FRA20B | 20 | 920,0001 | 12,100,000 | 20p12.2 | Aphidicolin | Common | hsa-mir-6870 | ||
| FRA22A | 22 | 37,600,001 | 51,304,566 | 22q13 | Folic acid | Rare | RAC2, PDGFB, RBX1, EP300, WNT7B | hsa-mir-658, hsa-mir-659, hsa-mir-6820, hsa-mir-4534, hsa-mir-4766, hsa-mir-1281, hsa-mir-6889, hsa-mir-33a, hsa-mir-378i, hsa-mir-1249, hsa-mir-4762, hsa-mir-3619, hsa-let-7a-3, hsa-mir-4763, hsa-let-7b, hsa-mir-3201, hsa-mir-4535, hsa-mir-3667, hsa-mir-6821 | [149] |
| FRA22B | 22 | 29,600,001 | 32,200,000 | 22q12.2 | Aphidicolin | Common | hsa-mir-3653, hsa-mir-6818, hsa-mir-3200, hsa-mir-3928, hsa-mir-7109 | ||
| FRAXA | X | 142,100,001 | 147,100,000 | Xq27.3 | Folic acid | Rare | hsa-mir-892c, hsa-mir-890, hsa-mir-888, hsa-mir-892a, hsa-mir-892b, hsa-mir-891b, hsa-mir-891a, hsa-mir-513c, hsa-mir-513b, hsa-mir-513a-1, hsa-mir-513a-2, hsa-mir-506, hsa-mir-507, hsa-mir-508, hsa-mir-514b, hsa-mir-509-2, hsa-mir-509-3, hsa-mir-509-1, hsa-mir-510, hsa-mir-514a-1, hsa-mir-514a-2, hsa-mir-514a-3 | [150] | |
| FRAXB | X | 6,000,001 | 9,500,000 | Xp22.31 | Aphidicolin | Common | hsa-mir-4770, hsa-mir-4767, hsa-mir-651 | [119] | |
| FRAXC | X | 98,300,001 | 102,600,000 | Xq22.1 | Aphidicolin | Common | [119] | ||
| FRAXD | X | 140,300,001 | 142,100,000 | Xq27.2 | Aphidicolin | Common | [151] | ||
| FRAXE | X | 147,100,001 | 155,270,560 | Xq28 | Folic acid | Rare | IKBKG | hsa-mir-2114, hsa-mir-4330, hsa-mir-224, hsa-mir-452, hsa-mir-105-1, hsa-mir-767, hsa-mir-105-2, hsa-mir-3202-1, hsa-mir-3202-2, hsa-mir-718, hsa-mir-6858, hsa-mir-664b, hsa-mir-1184-1, hsa-mir-1184-2, hsa-mir-1184-3 | [152] |
| FRAXF | X | 147,100,001 | 155,270,560 | Xq28 | Folic acid | Rare | IKBKG | hsa-mir-2114, hsa-mir-4330, hsa-mir-224, hsa-mir-452, hsa-mir-105-1, hsa-mir-767, hsa-mir-105-2, hsa-mir-3202-1, hsa-mir-3202-2, hsa-mir-718, hsa-mir-6858, hsa-mir-664b, hsa-mir-1184-1, hsa-mir-1184-2, hsa-mir-1184-3 | [153] |
A total of 125 CFSs were collected from GeneCards V3 [87] and from manual bibliographic search. Data for 1,871 human miRs were downloaded from miRBase 20 [53]. Genomic data of exon genes were downloaded from Ensembl Biomart [88]. A list of 327 genes that participate in Pathways in cancer (KEGG PATHWAY: hsa05200) was retrieved from KEGG [89]. PHP scripts parsed the data to populate a MySQL (http://www.mysql.com) relational database. Other scripts queried the database to produce a list of CFSs with their associated cancer-related genes and microRs. The main results found here are: (i) the total length of the cytogenetic bands of the 125 CFSs is 835.22 Mbps (27 % of the entire haploid genome), while 110 cancer-related genes (33.6 % of all cancer-related genes) and 686 miRs (36.7 % of all miRs) are located within these CFSs, (ii) the total length of the 26 CFSs with precise genomic boundaries is 79.3 Mbps (2.6 % of the entire haploid genome), and (iii) nine cancer-related genes (2.8 % of all cancer-related genes) and 44 miRs (2.4 % of all miRNAs) are located within these molecularly defined CFSs (Table 2). Based on the above, the density of cancer-related genes and miRs in cytogenetically defined CFS compared to the rest of the genome is 37.2 and 56.7 % higher, respectively
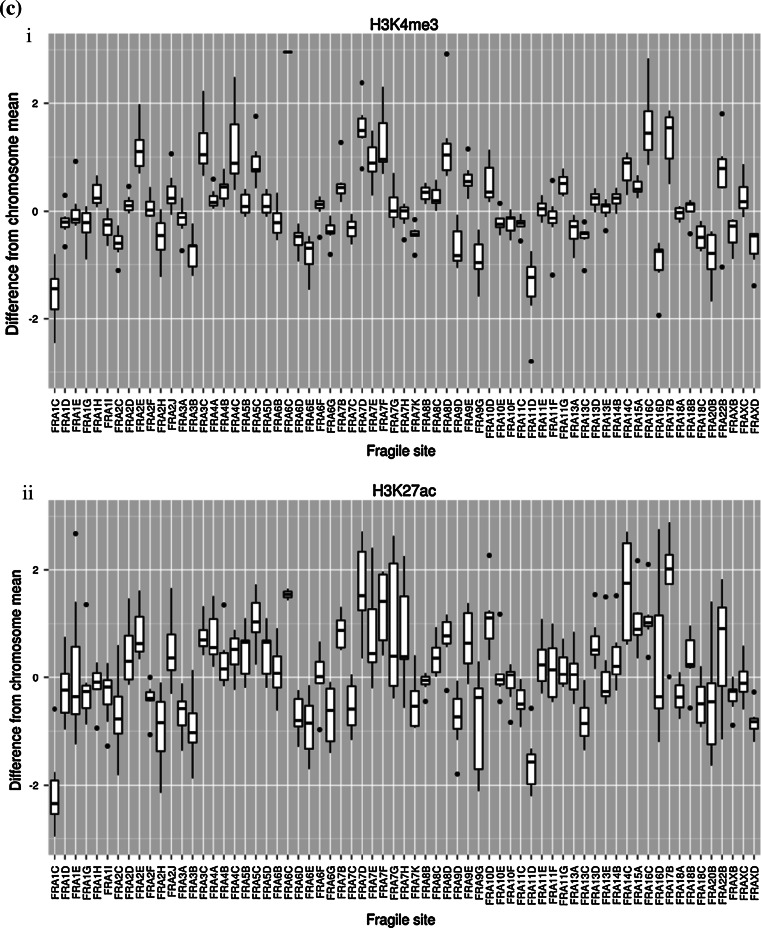
Fig. 2.
Frequency of cancer-related genes, repetitive elements, miRs, binding elements, and histone marks in CFSs. a CFSs exhibit a higher density of cancer-related genes (obtained from Kyoto Encyclopedia of Genes and Genomes), Alu repetitive elements [19], miRs, and the CTCF binding element relative to non-fragile regions. b CFSs exhibit a differential density of the histone marks (i) Histone 3 lysine 27 acetylation (H3K27ac) and (ii) Histone 3 lysine 4 trimethylation (H3K4me3), relative to non-fragile regions that is cell type origin-dependent (data concerning histone modifications derived from ChIP-seq experiments belonging to the ENCODE project were downloaded from the UCSC server (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeRegMarkH3k4me3/, http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeRegMarkH3k27ac/). Specifically, we obtained bigWig files for H3k4me3 and H3k27ac modifications in the GM12878, H1-hesc, HSMM, HUVEC, K562, NHEK, and NHLF cell types. Information concerning regions of interest was extracted with the bigWigSummary utility, also available from the UCSC server. Specifically, the average signal was calculated for every chromosome in every cell line by repeatedly invoking: bigWigSummary-type = mean “bigwigfile” chrN start end. Similarly, the average signal was calculated for cytogenetically and molecularly mapped fragile sites. For every defined fragile site, the mean histone modification signal of the corresponding chromosome was subtracted from the mean signal of the fragile region. Signal difference from mean for site i = mean(FSi) − mean(chromosomeFSi). The chromosome means varied within each cell type (data not shown), as did the histone modifications between cell types) (S histone signal). c Frequency of histone marks per CFSs. Each CFS exhibits a differential density of the histone marks. (i) Histone 3 lysine 4 trimethylation (H3K4me3) and (ii) Histone 3 lysine 27 acetylation (H3K27ac), relative to non-fragile regions averaged over all cell types presented in Fig. 2 (using the data generated for Fig. 2b, we also plotted a boxplot for individual cytogenetically defined CFSs in all 7 cell lines mentioned above with respect to H3K4me3 and H3K27ac. Significant heterogeneity between CFSs can be observed)
Fragile sites and microRNA genes
According to an early study, miRs are particularly frequent in CFSs [6]. Out of the 186 miRs known at the time, 35 were found in, or very close (<3 Mb) to, CFSs, occurring at a density (number of miRs per length) that was estimated to be approximately 9 % higher than in non-FSs. A newer analysis has found approximately 33.8 % of 715 miRs within CFSs, corresponding to a relative 50 % (26–85 %) higher density in regard to non-FSs, but the relation seems to vary between chromosomes [7] with some, like chromosome 16 and 19, having many more fragile than non-fragile miRAs, and others, like chromosome 14, having a lower incidence of miRNAs in fragile regions. Given that the number of known miRs has more than doubled since then, we have repeated this analysis with the recent version of miRBase (v20, [53]) and have found 686 miRs out of 1,871 (36.7 %) within cytogenetically defined fragile sites (Table 2) (corresponding incidence in molecularly mapped CFSs is shown in Table 3). Thus, the relative density of miR in cytogenetically defined CFSs is 57 % higher than in the rest of the genome (Fig. 2a; Table 2). Specific pertinent examples include tumor suppressors like hsa-mir-34a in FRA1A and oncomirs like hsa-mir-21 in FRA17B. In addition, more than 50 % of microRNAs seem to be clustered in relatively short regions, up to 50 kb, often containing multiple miR isoforms belonging to the same family [54]. When mapped, approximately 28 % of miR clusters overlap with known FSs. Rearrangements within these regions can disrupt multiple miRs in a single hit and produce complex phenotypic changes.
Table 3.
A list of molecularly delimited common fragile sites (CFSs) obtained from a manual search of the literature
| FRA | Chr | Start | End | BAC/STS from | BAC/STS to | Type | Frequency | Cancer genes | miRNAs | References |
|---|---|---|---|---|---|---|---|---|---|---|
| FRA1E | 1 | 97,749,961 | 98,119,925 | RP4-538N4 | RP11-128G13 | Aphidicolin | Common | [92] | ||
| FRA1H | 1 | 216,110,179 | 226,742,379 | RP11-22M7 | RP11-118H4 | 5AZA | Common | TGFB2 | hsa-mir-215, hsa-mir-194-1, hsa-mir-664a, hsa-mir-320b-2, hsa-mir-4742, hsa-mir-6741 | [154] |
| FRA2Ccen | 2 | 18,524,760 | 19,272,430 | RP11-720L11 | RP11-78J22 | Aphidicolin | Common | [97] | ||
| FRA2Ctel | 2 | 14,934,889 | 15,681,539 | RP11-526G2 | RP11-32P22 | Aphidicolin | Common | [97] | ||
| FRA2G | 2 | 169,498,510 | 170,313,244 | RP11-285F23 | RP11-724O16 | Aphidicolin | Common | [155] | ||
| FRA2H | 2 | 186,716,111 | 187,251,132 | RP11-561J1 | RP11-639N24 | Aphidicolin | Common | [156] | ||
| FRA3B | 3 | 59,623,632 | 63,846,635 | RP11-70P20 | RP11-50F24 | Aphidicolin | Common | [157] | ||
| FRA4F | 4 | 90,208,191 | 97,312,924 | RP11-549C16 | RP11-145G20 | Aphidicolin | Common | [109] | ||
| FRA6E | 6 | 160,275,245 | 166,079,958 | D6S1581 | D6S1719 | Aphidicolin | Common | [158] | ||
| FRA6F | 6 | 111,577,970 | 112,568,919 | RP5-1112D6 | RP1-142L7 | Aphidicolin | Common | [159] | ||
| FRA6H | 6 | 27,804,038 | 37,156,663 | RP1-193B12 | RP11-588I14 | Aphidicolin | Common | RXRB, PPARD, CDKN1A | hsa-mir-877, hsa-mir-4640, hsa-mir-6891, hsa-mir-6832, hsa-mir-4646, hsa-mir-1236, hsa-mir-6721, hsa-mir-6833, hsa-mir-3135b, hsa-mir-219a-1, hsa-mir-6873, hsa-mir-6834, hsa-mir-5004, hsa-mir-3934, hsa-mir-7159, hsa-mir-1275, hsa-mir-6835, hsa-mir-7111, hsa-mir-5690, hsa-mir-3925 | [115] |
| FRA7B | 7 | 1 | 12,392,296 | Telomere | RP11-507C1 | Aphidicolin | Common | PDGFA, RAC1 | hsa-mir-339, hsa-mir-4655, hsa-mir-6836, hsa-mir-4648, hsa-mir-4656, hsa-mir-589, hsa-mir-6874, hsa-mir-3683 | [116] |
| FRA7E | 7 | 80,508,751 | 84,935,939 | D7S1934 | SHGC-104456 | Aphidicolin | Common | HGF | [117] | |
| FRA7G | 7 | 115,894,865 | 116,072,877 | D7S486 | D7S522 | Aphidicolin | Common | [160] | ||
| FRA7H | 7 | 130,413,791 | 130,857,950 | D7S786 | D7S649 | Aphidicolin | Common | hsa-mir-29a, hsa-mir-29b-1 | [161] | |
| FRA7I | 7 | 144,671,106 | 146,121,417 | SHGC-153624 | sWSS2627 | Aphidicolin | Common | [120] | ||
| FRA7K | 7 | 110,657,856 | 111,031,412 | WI-5281 | SHGC-78648 | Aphidicolin | Common | [121] | ||
| FRA8C | 8 | 124,285,269 | 128,421,245 | RP11-468O2 | RP11-382A18 | Aphidicolin | Common | hsa-mir-548aa-1, hsa-mir-548d-1, hsa-mir-6844, hsa-mir-4662b, hsa-mir-4662a | [162] | |
| FRA9G | 9 | 17,135,038 | 17,503,917 | Within c9orf39/CNTLN | Aphidicolin | Common | [125] | |||
| FRA10F | 10 | 125,391,503 | 128,256,865 | RP11-391M7 | RP11-179O22 | Aphidicolin | Common | CTBP2 | hsa-mir-4296, hsa-mir-4484 | [130] |
| FRA11E | 11 | 32,086,458 | 34,028,916 | RP1-17K7 | RP13-786C16 | Aphidicolin | Common | [134] | ||
| FRA11G | 11 | 113,688,799 | 118,157,704 | RP11-667M19 | RP11-832A4 | Aphidicolin | Common | ZBTB16 | [133] | |
| FRA13A | 13 | 35,546,088 | 36,184,897 | RP11-307O13 | RP11-270C18 | Aphidicolin | Common | [139] | ||
| FRA13E | 13 | 73,285,774 | 76,386,498 | RP11-342J4 | RP11-29G8 | Aphidicolin | Common | [115] | ||
| FRA16D | 16 | 78,420,359 | 78,731,553 | AH009490.2 | FRA16D seq | Aphidicolin | Common | [163] | ||
| FRAXB | X | 6,595,111 | 7,548,235 | DXS1130 | DXS237 | Aphidicolin | Common | hsa-mir-4767 | [164] |
The list of molecularly mapped CFSs was compiled by performing a systematic search of the literature for each one of the known cytogenetically mapped fragile sites (n = 125). Whenever the placement of STS markers or BACs on GRC37/hg19 was unknown, it was verified by megaBLAST (default parameters) of the complete sequence against the human chromosome sequences. Only matches with greater than 90 % coverage were considered for CFS placement. Twenty-six (26) CFSs with a precise mapping were identified (see Table 4), whose coordinates were then mapped to the reference genome version GRC37. Whenever the precise location of BACs was unknown, it was verified by alignment with NCBI Megablast (default settings). The annotation of the reference human genome was obtained from UCSC (URL: hgdownload.cse.ucsc.edu/goldenPath/hg19/) in December 2013. The utility “bigWigSummary” was used to extract mean scores for molecularly mapped fragile regions and randomly selected non-fragile control regions. The BAC/STS limits have been extracted from the cited publication and coordinates mapped on GRC37. Note that FRA2C has been divided into two separate, molecularly defined hotspots
Fragile sites and regions with regulatory potential
Other DNA elements, such as regions with regulatory potential, may also be contained or overlap with CFSs. As an example, CTCF binding sites from ENCODE ChIP-seq are distributed throughout the genome. CTCF is a critical “weaver” of chromatin structure and function, and can provide an anchor point for nucleosome positioning [55]. Indirectly, CTCF can influence the accessibility of chromatin and plays diverse roles in chromatin insulation, gene regulation, imprinting, intra/interchromosomal interactions, nuclear compartmentalization, and alternative splicing [56, 57]. In some cases, distal fragments bound by CTCF have been found to mediate long-range interactions by loop formation and could modulate transcription at distant sites [58]. We examined the percentage of all potential CTCF binding sites within molecularly mapped CFSs and found that this ranges between 2.76 and 3.20 % in different cell lines (Table 4). Relative to the total length of fragile segments, this corresponds to an 18 % (10–25 %)-fold increase in the number of potential CTCF binding sites (Fig. 2a). Although it is impossible to know whether CTCF binding at these sites exerts a meaningful effect, its presence seems in accordance with the observation that many CFSs are gene-rich. Even more, CFSs rearrangements could influence gene expression further away on the same chromosome.
Table 4.
| Name | CTCF bs (count) | CTCF bs (%) | CTCF bs/kbp | H3K27ac | 99-way cons. score |
|---|---|---|---|---|---|
| FRA1E | 97 | 0.002 | 0.261 | 1.32 | 0.133 |
| FRA1H | 17,338 | 0.372 | 1.631 | 3.07 | – |
| FRA2Ctel | 700 | 0.015 | 0.938 | 2.53 | 0.071 |
| FRA2Ccen | 1,108 | 0.024 | 1.486 | 1.56 | 0.111 |
| FRA2G | 1,501 | 0.032 | 1.698 | 4.22 | 0.114 |
| FRA2H | 132 | 0.003 | 0.255 | 1.44 | 0.103 |
| FRA3B | 4,156 | 0.089 | 0.982 | 1.57 | 0.105 |
| FRA4F | 4,765 | 0.102 | 0.671 | 1.63 | 0.085 |
| FRA6H | 33,735 | 0.724 | 3.607 | 8.14 | 0.12 |
| FRA6F | 2,104 | 0.045 | 2.123 | 2.93 | 0.126 |
| FRA6E | 6,274 | 0.135 | 1.081 | 2.47 | 0.062 |
| FRA7B | 23,331 | 0.5 | 1.907 | 3.13 | – |
| FRA7E | 4,253 | 0.091 | 0.961 | 1.6 | 0.082 |
| FRA7K | 265 | 0.006 | 0.709 | 2.56 | 0.097 |
| FRA7G | 265 | 0.006 | 1.489 | 2.46 | 0.098 |
| FRA7H | 1,458 | 0.031 | 3.283 | 4.38 | 0.089 |
| FRA7I | 435 | 0.009 | 0.3 | 1.98 | 0.065 |
| FRA8C | 8,599 | 0.184 | 2.079 | 3.26 | 0.082 |
| FRA9G | 168 | 0.004 | 0.455 | 1.2 | 0.142 |
| FRA10F | 5,878 | 0.126 | 2.051 | 2.57 | 0.071 |
| FRA11E | 4,494 | 0.096 | 2.314 | 4.05 | 0.123 |
| FRA11G | 9,545 | 0.205 | 2.136 | 2.57 | 0.126 |
| FRA13A | 193 | 0.004 | 0.302 | 1.23 | 0.108 |
| FRA13E | 2,950 | 0.063 | 0.951 | 1.78 | 0.104 |
| FRA16D | 378 | 0.008 | 1.215 | 1.87 | 0.118 |
| FRAXB | 718 | 0.015 | 0.753 | 1.54 | 0.056 |
CTCF binding sites, studied in 89 cell lines, are shown respectively as absolute counts, with respect to the total number of CTCF binding sites in the whole genome and as a frequency per kb. Average H3k27 acetylation scores from ChIP-seq analysis of the K562 cell line have been calculated for each fragile site. Average conservation score between human and 99 vertebrates was obtained from the UCSC browser (http://genome.ucsc.edu/cgi-bin/hgTrackUi?db=hg19&g=cons100way). We compiled a list of molecularly mapped CFSs by performing a systematic search of the literature for each one of the known cytogenetically mapped CFSs (n = 125). Twenty-six CFSs with a precise mapping were identified (see Table 3), and their coordinates were then mapped to the reference genome version GRC37. Whenever the precise location of BACs was unknown, it was verified by alignment with NCBI Megablast (default settings). The annotation of the reference human genome was obtained from UCSC (URL: hgdownload.cse.ucsc.edu/goldenPath/hg19/) in December 2013. The utility “bigWigSummary” was used to extract mean scores for molecularly mapped fragile regions and randomly selected non-fragile control regions
Fragile sites and histone modifications
Epigenetic modifications, such as histone methylation and acetylation, can also take place within FS. It appears that H3K9/14 hypoacetylation is a global feature of CFSs in a lymphoblastoid cell line [9] and could impede replication progression. Several enzymes modify key histone residues with relatively high specificity and regulate, indirectly, transcription, repair, and replication. Indeed, histone modifications vary significantly between cell types and are well correlated with transcription levels in the ENCODE data (Figure 2 in [10]), especially for H3K79me2, H3K9ac, H3K4me3, and H3K27ac. Histone 3 lysine 27 acetylation (H3K27ac) co-localizes with active enhancers [59] and regions with open chromatin structure and could play a role in protecting against replication–transcription collisions and R-loop formation. Intriguingly, H3K27ac varies between cell types and CFSs (Fig. 2b, c). The average ChIP-seq acetylation signal and the number of signal peaks (data not shown) within cytogenetic CFSs are slightly lower than the corresponding chromosome mean in K562 cells, but higher in HUVEC cells and equivocal for other cell types. Such variability may explain the plasticity of CFSs and their differential expression between individuals, cell types, and culture conditions.
Histone 3 lysine 4 trimethylation is also associated with active promoters [60] and correlates well with transcription in the ENCODE data [10]. It appears that H3K4me3 may also contribute to the DNA damage response and repair of DSBs in yeast cells [61], mediating cellular responses to genotoxic stresses, and interacting with the human tumor suppressor ING1, which is required for DNA repair and apoptotic activities [62]. A recent study of ERFSs [9] has shown that replication stall, as identified by anti-replication protein A ChIP, preferentially co-localizes with H3K4me3 (see supplemental figure 1 in [16]). Although cytogenetically defined CFSs as a whole do not show a large deviation from the mean, some sites in particular, like FRA3B and FRA16D, seem to be on average poor in H3K4me3 while others, like FRA2E, FRA3C, and FRA7D seem to be on average rich in H3K4me3 (Fig. 2c). When the cell lines employed were grouped according to their origin (cancerous versus embryonic versus normal epithelial versus normal mesenchymal), a pattern regarding the density of the H3K27ac and H3K4me3 within CFSs relative to non-fragile sites could be discerned (Fig. 2b). Cancer and embryonic cells (K562, GM12878, H1-hESC) displayed lower signals of H3K27ac and H3K4me3 relative to the non-fragile regions, whereas a significantly different distribution was noticed in the other cell line groups (p < 0.001, ANOVA). Despite the small number of cell lines examined, a possible functional link between histone modifications and the other elements (genes, non-coding RNAs, regulatory sequences) positioned within the CFS cannot be excluded (Fig. 1). This potential interplay may be even more complex during carcinogenesis. Oncogenes may distress this functional cross-talk by altering the epigenome of a particular region. As an example, oncogenic Cdc6 was shown to act as “molecular switch” at certain tumor-suppressor loci by regulating CTCF binding. The latter led to suppression of the genes encoded and simultaneous firing of adjacent dormant origins. If such a scenario takes place within CFS that are rich in CTCF sites (Fig. 2a), and depending on the cellular context, the density and timing of firing origins can be altered affecting replication dynamics [56, 57].
Our current understanding of CFSs is traditionally based on a static mapping, often cytogenetic and imprecise, which cannot fully capture the interaction of non-coding DNA, regulatory elements, and histone modifications with vulnerability to RS. Even though the current CFS mapping successfully predicts response to extrinsic stress and OIRS, a more accurate model of fragility will eventually have to integrate experimental data at the nucleotide resolution with other non-coding elements. This is an intriguing area for further study.
Fragile sites in carcinogenesis
Extending the concept of CFSs in carcinogenesis has been a subject of active research since the discovery of an association between cancer breakpoints and FSs [63]. Overall, it appears that CFSs are generally sensitive to innate RS occurring naturally in various tumors and cell lines [64]. Multiple clusters of homozygous deletions, usually small, have been detected over known CFSs in an exhaustive survey of cancer genomes but their expression profile is variable [65]. For example, FRA2F, FRA3B, FRA4F, FRA5H, and FRA16D were most affected while others, like FRA2B and FRA4B, were least affected. Recurrent alterations have been identified in FRA3B and FRA16D in several cancer types, leading to further investigation of the FHIT and WWOX genes, respectively, in mouse models (see K. Huebner and R. Aqeilan chapters in this issue). Fragile sites FRA10C and FRA10G may be involved in the formation of the oncogenic RET/PTC rearrangement in papillary thyroid carcinoma [15]. Specifically, in RET/PTC1, the FRA10G-localized RET is rearranged with the FRA10C-localized tumor suppressor gene CCDC6, while in RET/PTC3 it is rearranged with NCO4 that is located in FRA10G. In a similar manner, the MYC oncogene is flanked by CFSs FRA8C and FRA8D, that may facilitate adjacent integration of HPV18 [66] or MYC amplification [67]. Viral DNA integration in the genome of a host cell can lead to cancer development and CFSs provide preferential hotspots for this [68]. Particularly, HPV16 E6 and E7 oncogenic products have been shown to induce replication stress and DSBs in the host cell. This occurs preferentially at CFSs allowing viral genome integration at these sites [68].
Despite the abundance of CFS breaks in cancer, it would be inappropriate to assume that alterations of genes residing within CFSs always confer a clonal advantage in cancer development [65] without evidence of selection or at least convincing causative models. Clearly, breakage probability (passenger alterations) as a consequence of fragility and clonal selection (driver alterations) in cancer development are two separate phenomena that should not be confused.
Nevertheless, the impact of CFS instability in cancer should not be easily dismissed or oversimplified. CFSs breakpoints have been detected in preneoplastic lesions in human and mouse models [4, 19] well before the emergence of the malignant phenotype. Briefly, exposure of xenografted normal human skin to growth factors preferentially induces CFS instability. Similarly, hyperplastic mouse urothelium from HRAS transgenic mice showed numerous copy number alterations in fragile areas. CFS instability is an early manifestation and can be attributed to experimentally controlled, oncogene-induced stress in these studies, in a way that more closely resembles carcinogenesis than APH-induced stress. Therefore, it could be argued that CFS alterations are frequent in cancer, as described above [65], not just because of a higher breakage probability but also because of an earlier involvement, even before the complete deregulation of the cellular machinery.
Furthermore, any double-strand break can have dire consequences, such as the initiation of a breakage-fusion-bridge cycle, especially when subtelomeric and peri-centromeric CFSs are disrupted simultaneously [69]. Through this mechanism, CFS breaks can amplify oncogenes, delete tumor suppressors or, most importantly, initiate persistent chromosomal instability. Massive accumulation of localized chromosomal rearrangements in a single time-point, termed chromothripsis (literally: shattering of the chromosome) and chromoanasynthesis, has recently been identified in several cancer types [70]. Indirect evidence suggests that CFSs may have an important role in this process [71] by stalling the RF, favoring RF collapse and, in extreme cases, chromosome pulverization leading to clustering of chromosomal breaks [13]. Indeed, chromosome fragmentation distal to the CFS has been observed under the microscope in some cases [71] and could be a triggering factor for chromothripsis. On the other hand, multi-step, recurrent CFS alterations could be difficult to discriminate from single-step rearrangements, rendering the identification of chromothripsis even more difficult. In that scenario, CFS stability and localization is an important parameter in the bioinformatic algorithms that are applied to define and model such cancer rearrangements. In addition, CFSs can contribute to the clustered shuttering of the chromosomes also during the process of premature chromatin condensation (PCC) [72, 73] in which interphase chromosomes or late replicating chromosome zones like CFS or extranuclear bodies micronuclei are ‘induced’ to condensation by various mitotic factors [74]. This reveals more possibilities for CFSs to act as contributors to GI through chromosome breakage. An interesting scenario suggested that the G2–M mammalian checkpoint can fail to delay mitotic onset as it may not be sensitive enough to detect a few remaining long-replicating forks, thus allowing chromatin condensation of late replicating CFS regions, resulting in multiple DNA breaks [26, 44].
CFSs as “functional” units: a new perception
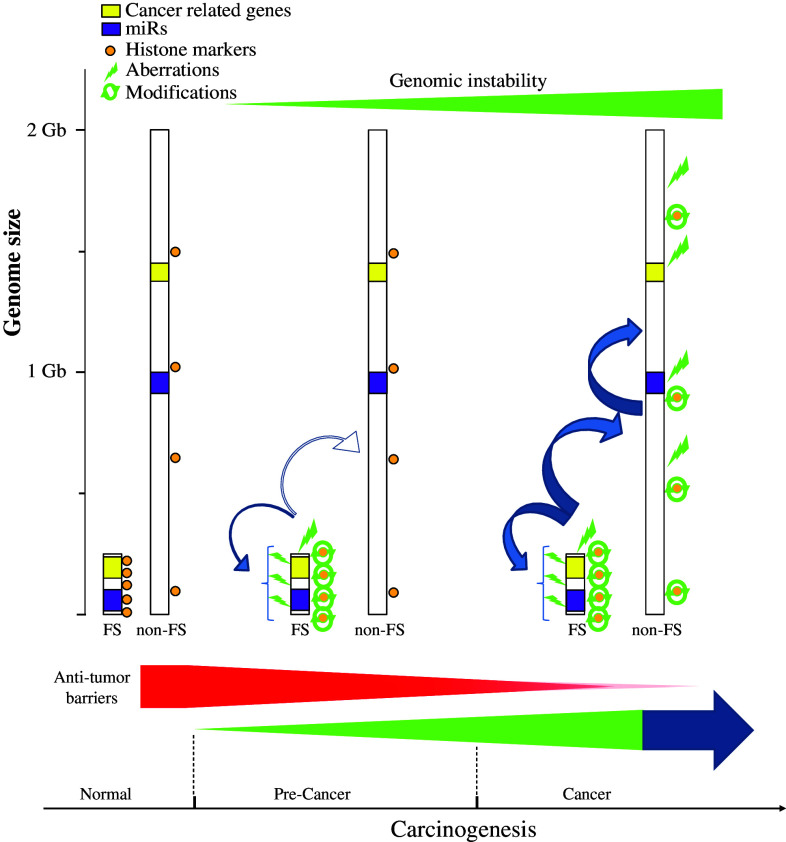
Common fragile sites have long been considered vulnerable breakage sites in the genome in response to RS from extrinsic factors. Their fragility has also been associated with GI in cancer development. As we have previously shown, CFSs are preferentially affected from the earliest precancerous lesions, in response to OIRS [2, 4]. In the current work, we first performed a review on the heterogeneity and fragility mechanisms affecting these sites. Next, by applying bioinformatic tools and exploiting available information in various databases, like the KEGG, miRbase, and ENCODE, we show a prevalence of various cancer-related genes, miRs, binding elements, and histone modifications in CFSs (Figs. 1, 3). The presence of such a wide spectrum of coding and non-coding elements changes the view on CFSs content and their nature itself. Given that CFSs are altered from the earliest stages in cancer, their impact on cancer development may be more profound than simply participating in the emergence of GI. On one hand, cancer-related genes and miRs may be affected from such early precancerous stages, therefore possibly exerting a strong pressure for malignant progression (Fig. 3). On the other hand, this pressure is also reinforced by alterations and imbalances in the binding elements and histone patterns, respectively, in the CFSs. Furthermore, collectively, all these alterations may further affect in an “avalanche” mode not only the stability of the CFSs, but overall of the genome (Figs. 1, 3). Therefore, as the anti-tumor barriers are gradually overwhelmed, this avalanche effect may function in a positive feedback mode to promote cancer. An important question that emerges is why CFSs are not selected for elimination from the genome, but are rather conserved features in mammals? A tempting but speculative answer is that by locating a set of important coding and non-coding elements in regions that replicate late and/or with delay and thus are prone to instability, they may function as alarm sensors scattered throughout the genome in various chromosomes, to signify detrimental effects from the RS on the cell. As long as the mammalian checkpoints and repair mechanisms are not compromised, cells can monitor and protect their genome and functional integrity through such a dynamic interaction. Nevertheless, this imposes the risk that if the checkpoints and the anti-tumor barriers gradually fail, tumor promotion ensues (Fig. 3). As we were able to examine only a small subset of binding elements and histone modifications from the ENCODE and the miRbase is constantly expanding, in the future more in-depth studies are required to obtain a comprehensive picture of CFSs and on their role in cancer. Overall, CFSs may not be merely structural domains vulnerable only to breakage but highly organized “functional” units that may have deeper biological consequences for the cell when affected.
Fig. 3.
Model proposing that CFS apart from contributing to GI exert wider biological effects during cancer development. CFSs are preferentially affected from the earliest precancerous lesions, in response to OIRS, conferring to GI. A wide spectrum of coding and non-coding elements are present within CFSs. Cancer-related genes and miRs may be affected from such early precancerous stages, therefore possibly exerting a strong pressure for malignant progression. This pressure is also reinforced by alterations and imbalances in the binding elements and histone patterns, respectively, in the CFSs. Furthermore, collectively, all of these alterations may further affect in an “avalanche” mode not only the stability of the CFSs, but overall of the genome. As the anti-tumor barriers are gradually overwhelmed, this avalanche effect may function in a positive feedback mode to promote cancer
Acknowledgments
Financial support was provided from the European Union’s Seventh Framework Programme for research, technological development and demonstration under grant agreement No 284460. Dr. Georgakilas was supported by an EU Marie Curie Reintegration Grant MC-CIG-303514, Greek National funds through the Operational Program ‘Educational and Lifelong Learning of the National Strategic Reference Framework (NSRF)-Research Funding Program: THALES (Grant number MIS 379346) and COST Action CM1201 ‘Biomimetic Radical Chemistry’. Dr A. Kotsinas is a recipient of an Empeirikeion Foundation fellowship.
Conflict of interest
All authors have no conflict of interest to declare.
Footnotes
A. G. Georgakilas, P. Tsantoulis, and A. Kotsinas contributed equally to this manuscript.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Sci Mar. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 3.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability—an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 4.Gorgoulis VG, Vassiliou L-VF, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, DiTullio RA, Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035):907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 5.Dereli-Oz A, Versini G, Halazonetis TD. Studies of genomic copy number changes in human cancers reveal signatures of DNA replication stress. Mol Oncol. 2011;5(4):308–314. doi: 10.1016/j.molonc.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagana A, Russo F, Sismeiro C, Giugno R, Pulvirenti A, Ferro A. Variability in the incidence of miRNAs and genes in fragile sites and the role of repeats and CpG islands in the distribution of genetic material. PLoS ONE. 2010;5(6):e11166. doi: 10.1371/journal.pone.0011166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith DI, Zhu Y, McAvoy S, Kuhn R. Common fragile sites, extremely large genes, neural development and cancer. Cancer Lett. 2006;232(1):48–57. doi: 10.1016/j.canlet.2005.06.049. [DOI] [PubMed] [Google Scholar]
- 9.Jiang Y, Lucas I, Young DJ, Davis EM, Karrison T, Rest JS, Le Beau MM. Common fragile sites are characterized by histone hypoacetylation. Hum Mol Genet. 2009;18(23):4501–4512. doi: 10.1093/hmg/ddp410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Encode Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sutherland GR, Jacky PB, Baker EG. Heritable fragile sites on human chromosomes. XI. Factors affecting expression of fragile sites at 10q25, 16q22, and 17p12. Am J Hum Genet. 1984;36(1):110–122. [PMC free article] [PubMed] [Google Scholar]
- 12.Glover T, Berger C, Coyle J, Echo B. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum Genet. 1984;67(2):136–142. doi: 10.1007/BF00272988. [DOI] [PubMed] [Google Scholar]
- 13.Debatisse M, Le Tallec B, Letessier A, Dutrillaux B, Brison O. Common fragile sites: mechanisms of instability revisited. Trends Genet. 2012;28(1):22–32. doi: 10.1016/j.tig.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 14.Ozeri-Galai E, Bester AC, Kerem B. The complex basis underlying common fragile site instability in cancer. Trends Genet. 2012;28(6):295–302. doi: 10.1016/j.tig.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Dillon LW, Burrow AA, Wang YH. DNA instability at chromosomal fragile sites in cancer. Curr Genomics. 2010;11(5):326–337. doi: 10.2174/138920210791616699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barlow JH, Faryabi RB, Callén E, Wong N, Malhowski A, Chen HuaΒ T, Gutierrez-Cruz G, Sun H-W, McKinnon P, Wright G, Casellas R, Robbiani DavideΒ F, Staudt L, Fernandez-Capetillo O, Nussenzweig A. Identification of early replicating fragile sites that contribute to genome instability. Cell. 2013;152(3):620–632. doi: 10.1016/j.cell.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mortusewicz O, Herr P, Helleday T. Early replication fragile sites: where replication–transcription collisions cause genetic instability. EMBO J. 2013;32(4):493–495. doi: 10.1038/emboj.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Beau MM, Rassool FV, Neilly ME, Espinosa R, 3rd, Glover TW, Smith DI, McKeithan TW. Replication of a common fragile site, FRA3B, occurs late in S phase and is delayed further upon induction: implications for the mechanism of fragile site induction. Hum Mol Genet. 1998;7(4):755–761. doi: 10.1093/hmg/7.4.755. [DOI] [PubMed] [Google Scholar]
- 19.Tsantoulis PK, Kotsinas A, Sfikakis PP, Evangelou K, Sideridou M, Levy B, Mo L, Kittas C, Wu XR, Papavassiliou AG, Gorgoulis VG. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene. 2007;27(23):3256–3264. doi: 10.1038/sj.onc.1210989. [DOI] [PubMed] [Google Scholar]
- 20.Seo J, Kim K, Chang DY, Kang HB, Shin EC, Kwon J, Choi JK. Genome-wide reorganization of histone H2AX toward particular fragile sites on cell activation. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138(1):90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murano I, Kuwano A, Kajii T. Fibroblast-specific common fragile sites induced by aphidicolin. Hum Genet. 1989;83(1):45–48. doi: 10.1007/BF00274145. [DOI] [PubMed] [Google Scholar]
- 23.Le Tallec B, Dutrillaux B, Lachages AM, Millot GA, Brison O, Debatisse M. Molecular profiling of common fragile sites in human fibroblasts. Nat Struct Mol Biol. 2011;18:1421–1423. doi: 10.1038/nsmb.2155. [DOI] [PubMed] [Google Scholar]
- 24.Hosseini SA, Horton S, Saldivar JC, Miuma S, Stampfer MR, Heerema NA, Huebner K. Common chromosome fragile sites in human and murine epithelial cells and FHIT/FRA3B loss-induced global genome instability. Genes Chromosomes Cancer. 2013;52(11):1017–1029. doi: 10.1002/gcc.22097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Tallec B, Millot GA, Blin ME, Brison O, Dutrillaux B, Debatisse M. Common fragile site profiling in epithelial and erythroid cells reveals that most recurrent cancer deletions lie in fragile sites hosting large genes. Cell Rep. 2013;4(3):420–428. doi: 10.1016/j.celrep.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470(7332):120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- 27.Helmrich A, Stout-Weider K, Hermann K, Schrock E, Heiden T. Common fragile sites are conserved features of human and mouse chromosomes and relate to large active genes. Genome Res. 2006;16(10):1222–1230. doi: 10.1101/gr.5335506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elder FF, Robinson TJ. Rodent common fragile sites: are they conserved? Evidence from mouse and rat. Chromosoma. 1989;97(6):459–464. doi: 10.1007/BF00295030. [DOI] [PubMed] [Google Scholar]
- 29.Raveendranathan M, Chattopadhyay S, Bolon YT, Haworth J, Clarke DJ, Bielinsky AK. Genome-wide replication profiles of S-phase checkpoint mutants reveal fragile sites in yeast. EMBO J. 2006;25(15):3627–3639. doi: 10.1038/sj.emboj.7601251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denison SR, Simper RK, Greenbaum IF. How common are common fragile sites in humans: interindividual variation in the distribution of aphidicolin-induced fragile sites. Cytogenet Genome Res. 2003;101(1):8–16. doi: 10.1159/000073411. [DOI] [PubMed] [Google Scholar]
- 31.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 32.Franchitto A. Genome instability at common fragile sites: searching for the cause of their instability. Biomed Res Int. 2013;2013:730714. doi: 10.1155/2013/730714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Häsler J, Strub K. Alu elements as regulators of gene expression. Nucleic Acids Res. 2006;34(19):5491–5497. doi: 10.1093/nar/gkl706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 35.Helmrich A, Ballarino M, Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell. 2011;44(6):966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 36.Palumbo E, Matricardi L, Tosoni E, Bensimon A, Russo A. Replication dynamics at common fragile site FRA6E. Chromosoma. 2010;119(6):575–587. doi: 10.1007/s00412-010-0279-4. [DOI] [PubMed] [Google Scholar]
- 37.Mulvihill DJ, Wang YH. Two breakpoint clusters at fragile site FRA3B form phased nucleosomes. Genome Res. 2004;14(7):1350–1357. doi: 10.1101/gr.2304404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432(7015):316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 39.Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111(6):779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 40.Ozeri-Galai E, Lebofsky R, Rahat A, Bester AC, Bensimon A, Kerem B. Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Mol Cell. 2011;43(1):122–131. doi: 10.1016/j.molcel.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 41.Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol. 2010;11(10):683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- 42.Zhu M, Weiss RS. Increased common fragile site expression, cell proliferation defects, and apoptosis following conditional inactivation of mouse Hus1 in primary cultured cells. Mol Biol Cell. 2007;18(3):1044–1055. doi: 10.1091/mbc.E06-10-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Focarelli ML, Soza S, Mannini L, Paulis M, Montecucco A, Musio A. Claspin inhibition leads to fragile site expression. Genes Chromosomes Cancer. 2009;48(12):1083–1090. doi: 10.1002/gcc.20710. [DOI] [PubMed] [Google Scholar]
- 44.Koundrioukoff S, Carignon S, Techer H, Letessier A, Brison O, Debatisse M. Stepwise activation of the ATR signaling pathway upon increasing replication stress impacts fragile site integrity. PLoS Genet. 2013;9(7):e1003643. doi: 10.1371/journal.pgen.1003643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Glover TW, Stein CK. Induction of sister chromatid exchanges at common fragile sites. Am J Hum Genet. 1987;41(5):882–890. [PMC free article] [PubMed] [Google Scholar]
- 46.Arlt MF, Durkin SG, Ragland RL, Glover TW. Common fragile sites as targets for chromosome rearrangements. DNA Repair (Amst) 2006;5(9–10):1126–1135. doi: 10.1016/j.dnarep.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 47.Schwartz M, Zlotorynski E, Goldberg M, Ozeri E, Rahat A, le Sage C, Chen BP, Chen DJ, Agami R, Kerem B. Homologous recombination and nonhomologous end-joining repair pathways regulate fragile site stability. Genes Dev. 2005;19(22):2715–2726. doi: 10.1101/gad.340905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bergoglio V, Boyer AS, Walsh E, Naim V, Legube G, Lee MY, Rey L, Rosselli F, Cazaux C, Eckert KA, Hoffmann JS. DNA synthesis by Pol eta promotes fragile site stability by preventing under-replicated DNA in mitosis. J Cell Biol. 2013;201(3):395–408. doi: 10.1083/jcb.201207066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pellicioli A, Muzi-Falconi M. A blooming resolvase at chromosomal fragile sites. Nat Cell Biol. 2013;15(8):883–885. doi: 10.1038/ncb2812. [DOI] [PubMed] [Google Scholar]
- 50.Minocherhomji S, Hickson ID. Structure-specific endonucleases: guardians of fragile site stability. Trends Cell Biol. 2013 doi: 10.1016/j.tcb.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 51.Liontos M, Koutsami M, Sideridou M, Evangelou K, Kletsas D, Levy B, Kotsinas A, Nahum O, Zoumpourlis V, Kouloukoussa M, Lygerou Z, Taraviras S, Kittas C, Bartkova J, Papavassiliou AG, Bartek J, Halazonetis TD, Gorgoulis VG. Deregulated overexpression of hCdt1 and hCdc6 promotes malignant behavior. Cancer Res. 2007;67(22):10899–10909. doi: 10.1158/0008-5472.CAN-07-2837. [DOI] [PubMed] [Google Scholar]
- 52.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8(2):93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 53.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database issue):D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mathelier A, Carbone A. Large scale chromosomal mapping of human microRNA structural clusters. Nucleic Acids Res. 2013;41(8):4392–4408. doi: 10.1093/nar/gkt112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4(7):e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holwerda SJ, de Laat W. CTCF: the protein, the binding partners, the binding sites and their chromatin loops. Philos Trans R Soc Lond B Biol Sci. 2013;368(1620):20120369. doi: 10.1098/rstb.2012.0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sideridou M, Zakopoulou R, Evangelou K, Liontos M, Kotsinas A, Rampakakis E, Gagos S, Kahata K, Grabusic K, Gkouskou K, Trougakos IP, Kolettas E, Georgakilas AG, Volarevic S, Eliopoulos AG, Zannis-Hadjopoulos M, Moustakas A, Gorgoulis VG. Cdc6 expression represses E-cadherin transcription and activates adjacent replication origins. J Cell Biol. 2011;195(7):1123–1140. doi: 10.1083/jcb.201108121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489(7414):109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130(1):77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Faucher D, Wellinger RJ. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 2010;6(8):e1001082. doi: 10.1371/journal.pgen.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peña PV, Hom RA, Hung T, Lin H, Kuo AJ, Wong RPC, Subach OM, Champagne KS, Zhao R, Verkhusha VV, Li G, Gozani O, Kutateladze TG. Histone H3K4me3 binding is required for the DNA repair and apoptotic activities of ING1 tumor suppressor. J Mol Biol. 2008;380(2):303–312. doi: 10.1016/j.jmb.2008.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hecht F, Glover TW. Cancer chromosome breakpoints and common fragile sites induced by aphidicolin. Cancer Genet Cytogenet. 1984;13(2):185–188. doi: 10.1016/0165-4608(84)90060-8. [DOI] [PubMed] [Google Scholar]
- 64.DiTullio RA, Jr, Mochan TA, Venere M, Bartkova J, Sehested M, Bartek J, Halazonetis TD. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat Cell Biol. 2002;4(12):998–1002. doi: 10.1038/ncb892. [DOI] [PubMed] [Google Scholar]
- 65.Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, Buck G, Chen L, Beare D, Latimer C, Widaa S, Hinton J, Fahey C, Fu B, Swamy S, Dalgliesh GL, Teh BT, Deloukas P, Yang F, Campbell PJ, Futreal PA, Stratton MR. Signatures of mutation and selection in the cancer genome. Nature. 2010;463(7283):893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferber MJ, Eilers P, Schuuring E, Fenton JA, Fleuren GJ, Kenter G, Szuhai K, Smith DI, Raap AK, Brink AA. Positioning of cervical carcinoma and Burkitt lymphoma translocation breakpoints with respect to the human papillomavirus integration cluster in FRA8C at 8q24.13. Cancer Genet Cytogenet. 2004;154(1):1–9. doi: 10.1016/j.cancergencyto.2004.01.028. [DOI] [PubMed] [Google Scholar]
- 67.Gibaud A, Vogt N, Brison O, Debatisse M, Malfoy B. Characterization at nucleotide resolution of the homogeneously staining region sites of insertion in two cancer cell lines. Nucleic Acids Res. 2013;41(17):8210–8219. doi: 10.1093/nar/gkt566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thorland EC, Myers SL, Gostout BS, Smith DI. Common fragile sites are preferential targets for HPV16 integrations in cervical tumors. Oncogene. 2003;22(8):1225–1237. doi: 10.1038/sj.onc.1206170. [DOI] [PubMed] [Google Scholar]
- 69.Coquelle A, Pipiras E, Toledo F, Buttin G, Debatisse M. Expression of fragile sites triggers intrachromosomal mammalian gene amplification and sets boundaries to early amplicons. Cell. 1997;89(2):215–225. doi: 10.1016/s0092-8674(00)80201-9. [DOI] [PubMed] [Google Scholar]
- 70.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal S, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Quail MA, Burton J, Swerdlow H, Carter NP, Morsberger LA, Iacobuzio-Donahue C, Follows GA, Green AR, Flanagan AM, Stratton MR, Futreal PA, Campbell PJ. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144(1):27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mackinnon RN, Campbell LJ. Chromothripsis under the microscope: a cytogenetic perspective of two cases of AML with catastrophic chromosome rearrangement. Cancer Genet. 2013;206(6):238–251. doi: 10.1016/j.cancergen.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 72.El Achkar E, Gerbault-Seureau M, Muleris M, Dutrillaux B, Debatisse M. Premature condensation induces breaks at the interface of early and late replicating chromosome bands bearing common fragile sites. Proc Natl Acad Sci USA. 2005;102(50):18069–18074. doi: 10.1073/pnas.0506497102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hashash N, Johnson AL, Cha RS. Topoisomerase II- and condensin-dependent breakage of MEC1ATR-sensitive fragile sites occurs independently of spindle tension, anaphase, or cytokinesis. PLoS Genet. 2012;8(10):e1002978. doi: 10.1371/journal.pgen.1002978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ravi M, Nivedita K, Pai GM. Chromatin condensation dynamics and implications of induced premature chromosome condensation. Biochimie. 2013;95(2):124–133. doi: 10.1016/j.biochi.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 75.Casper AM, Durkin SG, Arlt MF, Glover TW. Chromosomal instability at common fragile sites in Seckel syndrome. Am J Hum Genet. 2004;75(4):654–660. doi: 10.1086/422701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ozeri-Galai E, Schwartz M, Rahat A, Kerem B. Interplay between ATM and ATR in the regulation of common fragile site stability. Oncogene. 2008;27(15):2109–2117. doi: 10.1038/sj.onc.1210849. [DOI] [PubMed] [Google Scholar]
- 77.Musio A, Montagna C, Mariani T, Tilenni M, Focarelli ML, Brait L, Indino E, Benedetti PA, Chessa L, Albertini A, Ried T, Vezzoni P. SMC1 involvement in fragile site expression. Hum Mol Genet. 2005;14(4):525–533. doi: 10.1093/hmg/ddi049. [DOI] [PubMed] [Google Scholar]
- 78.Arlt MF, Xu B, Durkin SG, Casper AM, Kastan MB, Glover TW. BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function. Mol Cell Biol. 2004;24(15):6701–6709. doi: 10.1128/MCB.24.15.6701-6709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Howlett NG, Taniguchi T, Durkin SG, D’Andrea AD, Glover TW. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet. 2005;14(5):693–701. doi: 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- 80.Boyer AS, Grgurevic S, Cazaux C, Hoffmann JS. The human specialized DNA polymerases and non-B DNA: vital relationships to preserve genome integrity. J Mol Biol. 2013;425(23):4767–4781. doi: 10.1016/j.jmb.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 81.Bhat A, Andersen PL, Qin Z, Xiao W. Rev3, the catalytic subunit of Polzeta, is required for maintaining fragile site stability in human cells. Nucleic Acids Res. 2013;41(4):2328–2339. doi: 10.1093/nar/gks1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Walsh E, Wang X, Lee MY, Eckert KA. Mechanism of replicative DNA polymerase delta pausing and a potential role for DNA polymerase kappa in common fragile site replication. J Mol Biol. 2013;425(2):232–243. doi: 10.1016/j.jmb.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Franchitto A, Pichierri P. Understanding the molecular basis of common fragile sites instability: role of the proteins involved in the recovery of stalled replication forks. Cell Cycle. 2011;10(23):4039–4046. doi: 10.4161/cc.10.23.18409. [DOI] [PubMed] [Google Scholar]
- 84.Lu X, Parvathaneni S, Hara T, Lal A, Sharma S. Replication stress induces specific enrichment of RECQ1 at common fragile sites FRA3B and FRA16D. Mol Cancer. 2013;12(1):29. doi: 10.1186/1476-4598-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dillon LW, Pierce LC, Lehman CE, Nikiforov YE, Wang YH. DNA topoisomerases participate in fragility of the oncogene RET. PLoS ONE. 2013;8(9):e75741. doi: 10.1371/journal.pone.0075741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mason JM, Das I, Arlt M, Patel N, Kraftson S, Glover TW, Sekiguchi JM. The SNM1B/APOLLO DNA nuclease functions in resolution of replication stress and maintenance of common fragile site stability. Hum Mol Genet. 2013;22(24):4901–4913. doi: 10.1093/hmg/ddt340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, Sirota-Madi A, Olender T, Golan Y, Stelzer G, Harel A, Lancet D (2010) GeneCards Version 3: the human gene integrator. Database (Oxford) 2010:baq020. doi:10.1093/database/baq020 [DOI] [PMC free article] [PubMed]
- 88.Kinsella RJ, Kahari A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, Kersey P, Flicek P (2011) Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011:bar030. doi:10.1093/database/bar030 [DOI] [PMC free article] [PubMed]
- 89.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Iakovleva TK, Monakhov AS, Moiseenko VM. Chromosomal restructurings and the distribution of chromosome fragile sites in the peripheral blood lymphocytes in a breast cancer patient after cytostatic therapy. Tsitologiia. 1992;34(2):77–83. [PubMed] [Google Scholar]
- 91.Moriarty HT, Webster LR. Fragile sites and bladder cancer. Cancer Genet Cytogenet. 2003;140(2):89–98. doi: 10.1016/s0165-4608(02)00650-7. [DOI] [PubMed] [Google Scholar]
- 92.Hormozian F, Schmitt JG, Sagulenko E, Schwab M, Savelyeva L. FRA1E common fragile site breaks map within a 370kilobase pair region and disrupt the dihydropyrimidine dehydrogenase gene (DPYD) Cancer Lett. 2007;246(1–2):82–91. doi: 10.1016/j.canlet.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 93.Pelliccia F, Limongi MZ, Gaddini L, Rocchi A. Assignment of FRA1H common fragile site to human chromosome band 1q42.1 proximal to the nuclear NAD+ ADP-ribosyltransferase gene (ADPRT) and to the main 5S rRNA gene locus. Cytogenet Cell Genet. 1998;82(1–2):121–122. doi: 10.1159/000015084. [DOI] [PubMed] [Google Scholar]
- 94.Lamparska K, Clark J, Babilonia G, Bedell V, Yip W, Smith SS. 2′-Deoxyriboguanylurea, the primary breakdown product of 5-aza-2′-deoxyribocytidine, is a mutagen, an epimutagen, an inhibitor of DNA methyltransferases and an inducer of 5-azacytidine-type fragile sites. Nucleic Acids Res. 2012;40(19):9788–9801. doi: 10.1093/nar/gks706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lai LA, Kostadinov R, Barrett MT, Peiffer DA, Pokholok D, Odze R, Sanchez CA, Maley CC, Reid BJ, Gunderson KL, Rabinovitch PS. Deletion at fragile sites is a common and early event in Barrett’s esophagus. Mol Cancer Res. 2010;8(8):1084–1094. doi: 10.1158/1541-7786.MCR-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.IJdo JW, Baldini A, Wells RA, Ward DC, Reeders ST. FRA2B is distinct from inverted telomere repeat arrays at 2q13. Genomics. 1992;12(4):833–835. doi: 10.1016/0888-7543(92)90319-n. [DOI] [PubMed] [Google Scholar]
- 97.Blumrich A, Zapatka M, Brueckner LM, Zheglo D, Schwab M, Savelyeva L. The FRA2C common fragile site maps to the borders of MYCN amplicons in neuroblastoma and is associated with gross chromosomal rearrangements in different cancers. Hum Mol Genet. 2011;20(8):1488–1501. doi: 10.1093/hmg/ddr027. [DOI] [PubMed] [Google Scholar]
- 98.Luo WJ, Takakuwa T, Ham MF, Wada N, Liu A, Fujita S, Sakane-Ishikawa E, Aozasa K. Epstein-Barr virus is integrated between REL and BCL-11A in American Burkitt lymphoma cell line (NAB-2) Lab Invest. 2004;84(9):1193–1199. doi: 10.1038/labinvest.3700152. [DOI] [PubMed] [Google Scholar]
- 99.Limongi MZ, Pelliccia F, Rocchi A. Assignment of the human nebulin gene (NEB) to chromosome band 2q24.2 and the alpha 1 (III) collagen gene (COL3A1) to chromosome band 2q32.2 by in situ hybridization; the FRA2G common fragile site lies between the two genes in the 2q31 band. Cytogenet Cell Genet. 1997;77(3–4):259–260. doi: 10.1159/000134590. [DOI] [PubMed] [Google Scholar]
- 100.Limongi MZ, Pelliccia F, Gaddini L, Rocchi A. Clustering of two fragile sites and seven homeobox genes in human chromosome region 2q31–>q32.1. Cytogenet Cell Genet. 2000;90(1–2):151–153. doi: 10.1159/000015651. [DOI] [PubMed] [Google Scholar]
- 101.Saitoh T, Hirai M, Katoh M. Molecular cloning and characterization of human Frizzled-5 gene on chromosome 2q33.3-q34 region. Int J Oncol. 2001;19(1):105–110. doi: 10.3892/ijo.19.1.105. [DOI] [PubMed] [Google Scholar]
- 102.Parmentier M, Passage E, Mattei MG, Vassart G. A new human hypervariable locus (K29) maps to the q37.3 region of chromosome 2 and reveals a fingerprint. Genomics. 1991;11(3):760–762. doi: 10.1016/0888-7543(91)90086-t. [DOI] [PubMed] [Google Scholar]
- 103.Mulatinho MV, de Carvalho Serao CL, Scalco F, Hardekopf D, Pekova S, Mrasek K, Liehr T, Weise A, Rao N, Llerena JC., Jr Severe intellectual disability, omphalocele, hypospadia and high blood pressure associated to a deletion at 2q22.1q22.3: case report. Mol Cytogenet. 2012;5(1):30. doi: 10.1186/1755-8166-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schuffenhauer S, Lederer G, Murken J. A heritable folate-sensitive fragile site on chromosome 2p 11.2 (FRA2L) Chromosome Res. 1996;4(3):252–254. doi: 10.1007/BF02254969. [DOI] [PubMed] [Google Scholar]
- 105.Pelliccia F, Bosco N, Rocchi A. Breakages at common fragile sites set boundaries of amplified regions in two leukemia cell lines K562—molecular characterization of FRA2H and localization of a new CFS FRA2S. Cancer Lett. 2010;299(1):37–44. doi: 10.1016/j.canlet.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 106.Li XZ, Yan ZA, Zhou XT. The effect of 1-beta-d-arabinofuranosyl-cytosine on the expression of the common fragile site at 3p14. Hum Genet. 1986;74(4):444–446. doi: 10.1007/BF00280503. [DOI] [PubMed] [Google Scholar]
- 107.Darai-Ramqvist E, Sandlund A, Muller S, Klein G, Imreh S, Kost-Alimova M. Segmental duplications and evolutionary plasticity at tumor chromosome break-prone regions. Genome Res. 2008;18(3):370–379. doi: 10.1101/gr.7010208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bolivar J, Garcia-Cozar FJ, Astola A, Iglesias C, Pendon C, Valdivia MM. Genomic structure and chromosome location of the human gene encoding the zinc finger autoantigen ZNF330. Cytogenet Cell Genet. 2001;93(3–4):234–238. doi: 10.1159/000056989. [DOI] [PubMed] [Google Scholar]
- 109.Rozier L, El-Achkar E, Apiou F, Debatisse M. Characterization of a conserved aphidicolin-sensitive common fragile site at human 4q22 and mouse 6C1: possible association with an inherited disease and cancer. Oncogene. 2004;23(41):6872–6880. doi: 10.1038/sj.onc.1207809. [DOI] [PubMed] [Google Scholar]
- 110.Zimonjic DB, Durkin ME, Keck-Waggoner CL, Park SW, Thorgeirsson SS, Popescu NC. SMAD5 gene expression, rearrangements, copy number, and amplification at fragile site FRA5C in human hepatocellular carcinoma. Neoplasia. 2003;5(5):390–396. doi: 10.1016/s1476-5586(03)80041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Olavesen MG, Davies AF, Broxholme SJ, Wixon JL, Senger G, Nizetic D, Campbell RD, Ragoussis J. An integrated map of human chromosome 6p23. Genome Res. 1995;5(4):342–358. doi: 10.1101/gr.5.4.342. [DOI] [PubMed] [Google Scholar]
- 112.Thorland EC, Myers SL, Persing DH, Sarkar G, McGovern RM, Gostout BS, Smith DI. Human papillomavirus type 16 integrations in cervical tumors frequently occur in common fragile sites. Cancer Res. 2000;60(21):5916–5921. [PubMed] [Google Scholar]
- 113.Kahkonen M. Population cytogenetics of folate-sensitive fragile sites. I. Common fragile sites. Hum Genet. 1988;80(4):344–348. doi: 10.1007/BF00273649. [DOI] [PubMed] [Google Scholar]
- 114.Morelli C, Sherratt T, Trabanelli C, Rimessi P, Gualandi F, Greaves MJ, Negrini M, Boyle JM, Barbanti-Brodano G. Characterization of a 4-Mb region at chromosome 6q21 harboring a replicative senescence gene. Cancer Res. 1997;57(19):4153–4157. [PubMed] [Google Scholar]
- 115.Fechter A, Buettel I, Kuehnel E, Schwab M, Savelyeva L. Cloning of genetically tagged chromosome break sequences reveals new fragile sites at 6p21 and 13q22. Int J Cancer. 2007;120(11):2359–2367. doi: 10.1002/ijc.22564. [DOI] [PubMed] [Google Scholar]
- 116.Bosco N, Pelliccia F, Rocchi A. Characterization of FRA7B, a human common fragile site mapped at the 7p chromosome terminal region. Cancer Genet Cytogenet. 2010;202(1):47–52. doi: 10.1016/j.cancergencyto.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 117.Zlotorynski E, Rahat A, Skaug J, Ben-Porat N, Ozeri E, Hershberg R, Levi A, Scherer SW, Margalit H, Kerem B. Molecular basis for expression of common and rare fragile sites. Mol Cell Biol. 2003;23(20):7143–7151. doi: 10.1128/MCB.23.20.7143-7151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shridhar V, Sun QC, Miller OJ, Kalemkerian GP, Petros J, Smith DI. Loss of heterozygosity on the long arm of human chromosome 7 in sporadic renal cell carcinomas. Oncogene. 1997;15(22):2727–2733. doi: 10.1038/sj.onc.1201448. [DOI] [PubMed] [Google Scholar]
- 119.Smith DI, Huang H, Wang L. Common fragile sites and cancer (review) Int J Oncol. 1998;12(1):187–196. [PubMed] [Google Scholar]
- 120.Ciullo M, Debily MA, Rozier L, Autiero M, Billault A, Mayau V, El Marhomy S, Guardiola J, Bernheim A, Coullin P, Piatier-Tonneau D, Debatisse M. Initiation of the breakage-fusion-bridge mechanism through common fragile site activation in human breast cancer cells: the model of PIP gene duplication from a break at FRA7I. Hum Mol Genet. 2002;11(23):2887–2894. doi: 10.1093/hmg/11.23.2887. [DOI] [PubMed] [Google Scholar]
- 121.Helmrich A, Stout-Weider K, Matthaei A, Hermann K, Heiden T, Schrock E. Identification of the human/mouse syntenic common fragile site FRA7K/Fra12C1–relation of FRA7K and other human common fragile sites on chromosome 7 to evolutionary breakpoints. Int J Cancer. 2007;120(1):48–54. doi: 10.1002/ijc.22049. [DOI] [PubMed] [Google Scholar]
- 122.Takahashi E, Hori T, O’Connell P, Leppert M, White R. Mapping of the MYC gene to band 8q24.12—q24.13 by R-banding and distal to fra(8)(q24.11), FRA8E, by fluorescence in situ hybridization. Cytogenet Cell Genet. 1991;57(2–3):109–111. doi: 10.1159/000133124. [DOI] [PubMed] [Google Scholar]
- 123.Kahkonen M, Tengstrom C, Alitalo T, Matilainen R, Kaski M, Airaksinen E. Population cytogenetics of folate-sensitive fragile sites. II. Autosomal rare fragile sites. Hum Genet. 1989;82(1):3–8. doi: 10.1007/BF00288261. [DOI] [PubMed] [Google Scholar]
- 124.Callahan G, Denison SR, Phillips LA, Shridhar V, Smith DI. Characterization of the common fragile site FRA9E and its potential role in ovarian cancer. Oncogene. 2003;22(4):590–601. doi: 10.1038/sj.onc.1206171. [DOI] [PubMed] [Google Scholar]
- 125.Sawinska M, Schmitt JG, Sagulenko E, Westermann F, Schwab M, Savelyeva L. Novel aphidicolin-inducible common fragile site FRA9G maps to 9p22.2, within the C9orf39 gene. Genes Chromosomes Cancer. 2007;46(11):991–999. doi: 10.1002/gcc.20484. [DOI] [PubMed] [Google Scholar]
- 126.Cannizzaro LA, Aronson MM, Thiesen HJ. Human zinc finger gene ZNF23 (Kox16) maps to a zinc finger gene cluster on chromosome 16q22, and ZNF32 (Kox30) to chromosome region 10q23-q24. Hum Genet. 1993;91(4):383–385. doi: 10.1007/BF00217362. [DOI] [PubMed] [Google Scholar]
- 127.Sarafidou T, Kahl C, Martinez-Garay I, Mangelsdorf M, Gesk S, Baker E, Kokkinaki M, Talley P, Maltby EL, French L, Harder L, Hinzmann B, Nobile C, Richkind K, Finnis M, Deloukas P, Sutherland GR, Kutsche K, Moschonas NK, Siebert R, Gecz J, European Collaborative Consortium for the Study of ADLTE Folate-sensitive fragile site FRA10A is due to an expansion of a CGG repeat in a novel gene, FRA10AC1, encoding a nuclear protein. Genomics. 2004;84(1):69–81. doi: 10.1016/j.ygeno.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 128.Voiculescu I, Back E, Schempp W. Homozygous condition for a BrdU-requiring fragile site on chromosome 12. Hum Genet. 1991;86(4):416–417. doi: 10.1007/BF00201849. [DOI] [PubMed] [Google Scholar]
- 129.Gandhi M, Dillon LW, Pramanik S, Nikiforov YE, Wang YH. DNA breaks at fragile sites generate oncogenic RET/PTC rearrangements in human thyroid cells. Oncogene. 2010;29(15):2272–2280. doi: 10.1038/onc.2009.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ma K, Qiu L, Mrasek K, Zhang J, Liehr T, Quintana LG, Li Z. Common fragile sites: genomic hotspots of DNA damage and carcinogenesis. Int J Mol Sci. 2012;13(9):11974–11999. doi: 10.3390/ijms130911974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hagemeijer A, Lafage M, Mattei MG, Simonetti J, Smit E, de Lapeyriere O, Birnbaum D. Localization of the HST/FGFK gene with regard to 11q13 chromosomal breakpoint and fragile site. Genes Chromosomes Cancer. 1991;3(3):210–214. doi: 10.1002/gcc.2870030307. [DOI] [PubMed] [Google Scholar]
- 132.Simmers RN, Sutherland GR. Further localization of ETS1 indicates that the chromosomal rearrangement in Ewing sarcoma does not occur at fra(11)(q23) Hum Genet. 1988;78(2):144–147. doi: 10.1007/BF00278185. [DOI] [PubMed] [Google Scholar]
- 133.Fechter A, Buettel I, Kuehnel E, Savelyeva L, Schwab M. Common fragile site FRA11G and rare fragile site FRA11B at 11q23.3 encompass distinct genomic regions. Genes Chromosomes Cancer. 2007;46(1):98–106. doi: 10.1002/gcc.20389. [DOI] [PubMed] [Google Scholar]
- 134.Bester AC, Schwartz M, Schmidt M, Garrigue A, Hacein-Bey-Abina S, Cavazzana-Calvo M, Ben-Porat N, Von Kalle C, Fischer A, Kerem B. Fragile sites are preferential targets for integrations of MLV vectors in gene therapy. Gene Ther. 2006;13(13):1057–1059. doi: 10.1038/sj.gt.3302752. [DOI] [PubMed] [Google Scholar]
- 135.Reshmi SC, Huang X, Schoppy DW, Black RC, Saunders WS, Smith DI, Gollin SM. Relationship between FRA11F and 11q13 gene amplification in oral cancer. Genes Chromosomes Cancer. 2007;46(2):143–154. doi: 10.1002/gcc.20394. [DOI] [PubMed] [Google Scholar]
- 136.Bester AC, Kafri M, Maoz K, Kerem B. Infection with retroviral vectors leads to perturbed DNA replication increasing vector integrations into fragile sites. Sci Rep. 2013;3:2189. doi: 10.1038/srep02189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tommerup N, Leffers H. Assignment of human KH-box-containing genes by in situ hybridization: hNRNPK maps to 9q21.32-q21.33, PCBP1 to 2p12-p13, and PCBP2 to 12q13.12-q13.13, distal to FRA12A. Genomics. 1996;32(2):297–298. doi: 10.1006/geno.1996.0121. [DOI] [PubMed] [Google Scholar]
- 138.Amarose AP, Huttenlocher PR, Sprudzs RM, Laitsch TJ, Pettenati MJ. A heritable fragile 12q24.13 segregating in a family with the fragile X chromosome. Hum Genet. 1987;75(1):4–6. doi: 10.1007/BF00273829. [DOI] [PubMed] [Google Scholar]
- 139.Savelyeva L, Sagulenko E, Schmitt JG, Schwab M. The neurobeachin gene spans the common fragile site FRA13A. Hum Genet. 2006;118(5):551–558. doi: 10.1007/s00439-005-0083-z. [DOI] [PubMed] [Google Scholar]
- 140.Vincent JB, Neves-Pereira ML, Paterson AD, Yamamoto E, Parikh SV, Macciardi F, Gurling HM, Potkin SG, Pato CN, Macedo A, Kovacs M, Davies M, Lieberman JA, Meltzer HY, Petronis A, Kennedy JL. An unstable trinucleotide-repeat region on chromosome 13 implicated in spinocerebellar ataxia: a common expansion locus. Am J Hum Genet. 2000;66(3):819–829. doi: 10.1086/302803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhu Y, McAvoy S, Kuhn R, Smith DI. RORA, a large common fragile site gene, is involved in cellular stress response. Oncogene. 2006;25(20):2901–2908. doi: 10.1038/sj.onc.1209314. [DOI] [PubMed] [Google Scholar]
- 142.Simmers RN, Mulley JC, Hyland VJ, Callen DF, Sutherland GR. Mapping the human alpha globin gene complex to 16p13.2—pter. J Med Genet. 1987;24(12):761–766. doi: 10.1136/jmg.24.12.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fratini A, Simmers RN, Callen DF, Hyland VJ, Tischfield JA, Stambrook PJ, Sutherland GR. A new location for the human adenine phosphoribosyltransferase gene (APRT) distal to the haptoglobin (HP) and fra(16)(q23)(FRA16D) loci. Cytogenet Cell Genet. 1986;43(1–2):10–13. doi: 10.1159/000132291. [DOI] [PubMed] [Google Scholar]
- 144.Simmers RN, Sutherland GR, West A, Richards RI. Fragile sites at 16q22 are not at the breakpoint of the chromosomal rearrangement in AMMoL. Science. 1987;236(4797):92–94. doi: 10.1126/science.3470945. [DOI] [PubMed] [Google Scholar]
- 145.Dooley TP, Mitchison HM, Munroe PB, Probst P, Neal M, Siciliano MJ, Deng Z, Doggett NA, Callen DF, Gardiner RM, et al. Mapping of two phenol sulphotransferase genes, STP and STM, to 16p: candidate genes for Batten disease. Biochem Biophys Res Commun. 1994;205(1):482–489. doi: 10.1006/bbrc.1994.2691. [DOI] [PubMed] [Google Scholar]
- 146.Karkera JD, Balan KV, Yoshikawa T, Lipman TO, Korman L, Sharma A, Patterson RH, Sani N, Detera-Wadleigh SD, Wadleigh RG. Systematic screening of chromosome 18 for loss of heterozygosity in esophageal squamous cell carcinoma. Cancer Genet Cytogenet. 1999;111(1):81–86. doi: 10.1016/s0165-4608(98)00225-8. [DOI] [PubMed] [Google Scholar]
- 147.Capello D, Scandurra M, Poretti G, Rancoita PM, Mian M, Gloghini A, Deambrogi C, Martini M, Rossi D, Greiner TC, Chan WC, Ponzoni M, Moreno SM, Piris MA, Canzonieri V, Spina M, Tirelli U, Inghirami G, Rinaldi A, Zucca E, Favera RD, Cavalli F, Larocca LM, Kwee I, Carbone A, Gaidano G, Bertoni F. Genome wide DNA-profiling of HIV-related B-cell lymphomas. Br J Haematol. 2010;148(2):245–255. doi: 10.1111/j.1365-2141.2009.07943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Debacker K, Winnepenninckx B, Ben-Porat N, FitzPatrick D, Van Luijk R, Scheers S, Kerem B, Frank Kooy R. FRA18C: a new aphidicolin-inducible fragile site on chromosome 18q22, possibly associated with in vivo chromosome breakage. J Med Genet. 2007;44(5):347–352. doi: 10.1136/jmg.2006.044628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shen Y, Baker E, Callen DF, Sutherland GR, Willson TA, Rakar S, Gough NM. Localization of the human GM-CSF receptor beta chain gene (CSF2RB) to chromosome 22q12.2->q13.1. Cytogenet Cell Genet. 1992;61(3):175–177. doi: 10.1159/000133401. [DOI] [PubMed] [Google Scholar]
- 150.Culjkovic B, Stojkovic O, Savic D, Zamurovic N, Nesic M, Major T, Keckarevi D, Romac S, Zamurovi B, Vukosavic S. Comparison of the number of triplets in SCA1, MJD/SCA3, HD, SBMA, DRPLA, MD, FRAXA and FRDA genes in schizophrenic patients and a healthy population. Am J Med Genet. 2000;96(6):884–887. doi: 10.1002/1096-8628(20001204)96:6<884::aid-ajmg41>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 151.Ledbetter SA, Ledbetter DH. A common fragile site at Xq27: theoretical and practical implications. Am J Hum Genet. 1988;42(5):694–702. [PMC free article] [PubMed] [Google Scholar]
- 152.Sharma D, Gupta M, Thelma BK. Expansion mutation frequency and CGG/GCC repeat polymorphism in FMR1 and FMR2 genes in an Indian population. Genet Epidemiol. 2001;20(1):129–144. doi: 10.1002/1098-2272(200101)20:1<129::AID-GEPI11>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 153.Shaw MA, Chiurazzi P, Romain DR, Neri G, Gecz J. A novel gene, FAM11A, associated with the FRAXF CpG island is transcriptionally silent in FRAXF full mutation. Eur J Hum Genet. 2002;10(11):767–772. doi: 10.1038/sj.ejhg.5200881. [DOI] [PubMed] [Google Scholar]
- 154.Curatolo A, Limongi ZM, Pelliccia F, Rocchi A. Molecular characterization of the human common fragile site FRA1H. Genes Chromosomes Cancer. 2007;46(5):487–493. doi: 10.1002/gcc.20432. [DOI] [PubMed] [Google Scholar]
- 155.Limongi MZ, Pelliccia F, Rocchi A. Characterization of the human common fragile site FRA2G. Genomics. 2003;81(2):93–97. doi: 10.1016/s0888-7543(03)00007-7. [DOI] [PubMed] [Google Scholar]
- 156.Brueckner LM, Sagulenko E, Hess EM, Zheglo D, Blumrich A, Schwab M, Savelyeva L. Genomic rearrangements at the FRA2H common fragile site frequently involve non-homologous recombination events across LTR and L1(LINE) repeats. Hum Genet. 2012;131(8):1345–1359. doi: 10.1007/s00439-012-1165-3. [DOI] [PubMed] [Google Scholar]
- 157.Becker NA, Thorland EC, Denison SR, Phillips LA, Smith DI. Evidence that instability within the FRA3B region extends four megabases. Oncogene. 2002;21(57):8713–8722. doi: 10.1038/sj.onc.1205950. [DOI] [PubMed] [Google Scholar]
- 158.Denison SR, Callahan G, Becker NA, Phillips LA, Smith DI. Characterization of FRA6E and its potential role in autosomal recessive juvenile parkinsonism and ovarian cancer. Genes Chromosomes Cancer. 2003;38(1):40–52. doi: 10.1002/gcc.10236. [DOI] [PubMed] [Google Scholar]
- 159.Morelli C, Karayianni E, Magnanini C, Mungall AJ, Thorland E, Negrini M, Smith DI, Barbanti-Brodano G. Cloning and characterization of the common fragile site FRA6F harboring a replicative senescence gene and frequently deleted in human tumors. Oncogene. 2002;21(47):7266–7276. doi: 10.1038/sj.onc.1205573. [DOI] [PubMed] [Google Scholar]
- 160.Huang H, Qian J, Proffit J, Wilber K, Jenkins R, Smith DI. FRA7G extends over a broad region: coincidence of human endogenous retroviral sequences (HERV-H) and small polydispersed circular DNAs (spcDNA) and fragile sites. Oncogene. 1998;16(18):2311–2319. doi: 10.1038/sj.onc.1200202. [DOI] [PubMed] [Google Scholar]
- 161.Mishmar D, Rahat A, Scherer SW, Nyakatura G, Hinzmann B, Kohwi Y, Mandel-Gutfroind Y, Lee JR, Drescher B, Sas DE, Margalit H, Platzer M, Weiss A, Tsui LC, Rosenthal A, Kerem B. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc Natl Acad Sci U S A. 1998;95(14):8141–8146. doi: 10.1073/pnas.95.14.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ferber MJ, Thorland EC, Brink AA, Rapp AK, Phillips LA, McGovern R, Gostout BS, Cheung TH, Chung TK, Fu WY, Smith DI. Preferential integration of human papillomavirus type 18 near the c-myc locus in cervical carcinoma. Oncogene. 2003;22(46):7233–7242. doi: 10.1038/sj.onc.1207006. [DOI] [PubMed] [Google Scholar]
- 163.Ried K, Finnis M, Hobson L, Mangelsdorf M, Dayan S, Nancarrow JK, Woollatt E, Kremmidiotis G, Gardner A, Venter D, Baker E, Richards RI. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum Mol Genet. 2000;9(11):1651–1663. doi: 10.1093/hmg/9.11.1651. [DOI] [PubMed] [Google Scholar]
- 164.Arlt MF, Miller DE, Beer DG, Glover TW. Molecular characterization of FRAXB and comparative common fragile site instability in cancer cells. Genes Chromosomes Cancer. 2002;33(1):82–92. doi: 10.1002/gcc.10000. [DOI] [PubMed] [Google Scholar]