Abstract
IL-1 and IL-18 are members of the IL-1 family of ligands, and their receptors are members of the IL-1 receptor family. Although several biological properties overlap for these cytokines, differences exist. IL-18 uniquely induces IFN-γ from T lymphocytes and natural killer cells but does not cause fever, whereas fever is a prominent characteristic of IL-1 in humans and animals. In the present study, human epithelial cells were stably transfected with the IL-18 receptor β chain and responded to IL-18 with increased production of IL-1α, IL-6, and IL-8. Five minutes after exposure to either cytokine, phosphorylation of mitogen activated protein kinase (MAPK) p38 was present; specific inhibition of p38 MAPK reduced IL-18 activity to background levels. Whereas IL-1β induced the expression of the NF-κB-reporter gene and was suppressed by competitive inhibition of NF-κB binding, IL-18 responses were weak or absent. In contrast to IL-1β, IL-18 also did not activate degradation of the NF-κB inhibitor. After 4 h, both cytokines induced comparable levels of mRNA for the chemokine IL-8 but, in the same cells, steady-state levels of cyclooxygenase (COX)-2 mRNA were high after IL-1β but low or absent after IL-18. After 30 h, IL-18-induced COX-2 appeared in part to be IL-1 dependent. Similarly, low levels of prostaglandin E2 were measured in IL-18-stimulated A549 cells and freshly obtained primary human monocytes and mouse macrophages. We conclude that in epithelial cells, IL-18 signal transduction is primarily via the MAPK p38 pathway rather than NF-κB, which may explain the absence of COX-2 and the failure of IL-18 to cause fever.
Prostaglandins (PG) are involved in several inflammatory processes and are produced via cyclooxygenase (COX) and PG synthases. Three isoforms of COX have been identified: COX-1, -2, and -3 (1). COX-1 displays the characteristics of a housekeeping gene, constitutively expressed in many tissues, and maintains basal levels of PGs for normal cell function. COX-3 is expressed primarily in the brain as a constitutive enzyme; acetomenophen/paracetamol inhibits this isoform of COX and may account for its antipyretic property. COX-2, however, is inducible by proinflammatory stimuli, particularly the cytokine IL-1 (2). COX-2-deficient mice, for example, do not develop fever to endotoxin (3), IL-1β (4), or IL-6 (5), whereas COX-1-deficient mice do exhibit fever after systemic or intracerbroventricular injections of these pyrogens (3–5). In addition to fever, the importance of PG E2 (PGE2) is best appreciated in malignant transformation in patients at high risk for developing malignant adenomas of the intestinal tract.
IL-18 is a proinflammatory cytokine that belongs to the IL-1 family of ligands (6, 7). The IL-18 receptors, although distinct from IL-1 receptors, also belong to the IL-1 receptor family. The IL-18 receptor (IL-18R) complex consists of two receptor chains: a ligand-binding chain termed the IL-18Rα chain and a coreceptor termed IL-18Rβ chain; both chains are required for signaling. Using specific neutralization of IL-18 activity, there is a role for IL-18 in rheumatoid arthritis (8, 9), ischemic renal (10), and heart disease (11), as well as atherosclerosis (12).
IL-1 consistently induces COX-2 gene expression and PGE2 synthesis in several cell lines (13) and in primary human blood monocytes (14, 15). Although IL-18 has been reported to induce COX-2 gene expression in the articular chondrocytes (16), IL-18 does not cause fever (5, 17, 18), which depends on COX-2 (3, 4). Humans, who are highly responsive to the rapid pyrogenic response of i.v. injected IL-1β at 10 ng/kg, manifests weak and late fevers to i.v. IL-18 at 10 μg/kg. To study the differences between IL-1 and IL-18, we generated a stable transfectant of the IL-18Rβ chain in A549 cells (A549-Rβ) and compared the responses between IL-1β and IL-18.
Materials and Methods
Cell Culture and Cytokines. A549 and A549-Rβ cells were cultured in F12-K culture medium (Cellgro, Waukesha, WI) supplemented with 10% FBS. Recombinant human and mouse IL-18 and mouse IL-12 were obtained from R & D Systems. Recombinant human IL-1β was supplied by Sclavo (Siena, Italy). The IL-1 receptor antagonist (IL-1Ra) was supplied by C. K. Edwards (Amgen Biologicals). The NF-κB inhibitor (SN-50) and the p38 inhibitor (SB203580) were purchased from Calbiochem. Freshly obtained human peripheral blood mononuclear cells (PBMC) and freshly isolated resident mouse peritoneal macrophages, respectively, were prepared and cultured as described (15, 19).
Generation of IL-18Rβ Stable Transfectants. IL-18Rβ cDNA ORF was obtained by PCR by using a human T-B lymphoblast cDNA library (Stratagene). The IL-18Rβ cDNA was inserted into the mammalian expression vector pTARGET (Promega) by using TA cloning (pTARGET). A549 cells were transfected in the same medium containing 8 μg of plasmid (pTARGET-IL-18Rβ) and 20 μg of Lipofectamine 2000 (Invitrogen) for 6 h. Subsequently, cells were cultured in fresh medium for 24 h before adding 0.5 mg/ml G-418 (Sigma). After 2-wk culture in the presence of G-418, cells were cloned by using limiting dilutions.
RT-PCR. Total RNA was isolated from A549 cells by using by Tri-Reagent (Sigma). The sense primer for IL-8 was 5′-GTCAGTGCATAAAGACATACTC-3′ and the reverse primer was 5′-AGGAATCTTGTATTGCATCTGG-3′. The primer for COX-2 and GAPDH as well as the methods for the PCR were reported (15).
Analysis of Cytokines. The liquid-phase electrochemiluminescence method (Origen Analyzer, Igen, Gaithersburg, MD) was used to measure IL-8 in cell culture media as described (20). Enhanced chemiluminescence for human IL-1α was developed by using the combination of ruthenylated mouse anti-human IL-1α (R & D Systems) and biotinylated polyclonal affinity purified goat anti-human IL-1α (R & D Systems). After incubation, A549-Rβ cell supernatants were removed. To prepare lysates, the cells were washed with ice-cold PBS, lysed in 1% Triton, frozen, thawed, and centrifuged for 10 min. Nitric oxide was measured as described (19).
Analysis of PGE2. Enhanced chemiluminescence with acetylcholinesterase-conjugated tracer was used for quantification of PGE2 levels (21, 22). The PGE2 EIA kit was purchased from Cayman Chemical (Ann Arbor, MI). The sensitivity of the assay was 25 pg/ml.
Western Blot Analysis. Extracts of cell pellets were prepared by either direct lysis in a buffer (0.5% Triton/50 mM β-glycerophosphate, pH 7.2/0.1 mM sodium vanadate/2 mM MgCl2/1 mM EGTA/1 mM DTT/0.1 mM phenylmethylsulfonyl urea/2 μg/ml leupeptin/4 μg/ml aprotinin) or trypsinization in EDTA followed by treatment with lysis buffer. The lysates were resolved by 10% SDS/PAGE and transferred to nitrocellulose membrane. The membranes were blocked in Tris-buffered saline (10 mM Tris·Cl, pH 7.4) containing 0.5% Tween 20 and 5% nonfat dry milk (or 1% BSA for anti-human phospho-p38 Ab) and then incubated with the first specific Ab in blocking solution for 18 h, washed, and incubated for 1 h with the developing second Ab. Polyclonal anti-human COX-2, anti-human actin, anti-human NF-κB inhibitor (IκB), and anti-human phospho-p38 were purchased from Signaling Technology (Beverly, MA). Donkey anti-rabbit and donkey anti-goat peroxidase-conjugated Abs were purchased from Jackson ImmunoReasearch.
Dual-Luciferase Assay. A549-Rβ cells were transiently cotransfected with either NF-κB or activator protein-1 (AP-1)-reporter vectors (Stratagene) and the pRL-TK Renilla vector (Promega) at a ratio of 50:1, respectively, by using Lipofectamine 2000 (Invitrogen). After 24 h, the cells were stimulated with IL-1β or IL-18, and after 24 h, supernatants were collected for IL-8 assay. Cell lysis (20 μl) was measured for firefly luciferase activity (Luciferase Assay reagent, Promega) and for Renilla luciferase activity (Stop & Glo reagent, Promega) (23).
Statistical Analysis. Data are expressed as mean ± SEM. Differences between group means were determined by Student's t test by using statview (Brain Power, Calabasas, CA).
Results
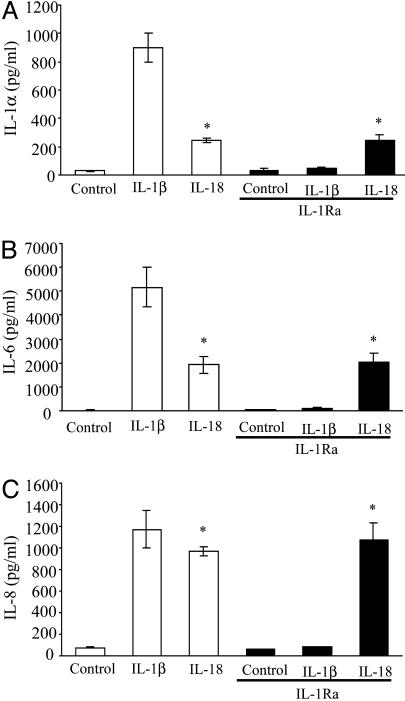
IL-18 Induction of IL-1α, IL-6, and IL-8 and Phosphorylation of p38-MAPK. A549-Rβ cells were stimulated with either IL-1β or IL-18, and the levels of IL-1α, IL-6, and IL-8 were measured. As shown in Fig. 1, both IL-1β and IL-18 induced intracellular IL-1α and secreted IL-6 and IL-8. We also observed IL-1α in the supernatants of these cultures but at one-tenth the intracellular concentration. To establish that IL-18 was stimulating cytokine production independent of IL-1, saturating concentrations of IL-1Ra were added. Whereas IL-1Ra suppressed IL-1β induction of IL-1α, there was no effect on IL-18-induced IL-6 and IL-8 production.
Fig. 1.
IL-18-induced cytokines is independent of IL-1. A549-Rβ cells were stimulated with either IL-1β (10 ng/ml) or IL-18 (50 ng/ml) in the absence or presence of IL-1Ra (10 μg/ml), and after 24 h, the supernatants were assayed for IL-6 (B) and IL-8 (C). The cells were lysed and assayed for intracellular IL-1α (A). The data are the mean ± SEM of nine experiments. *, P < 0.05 compared to control.
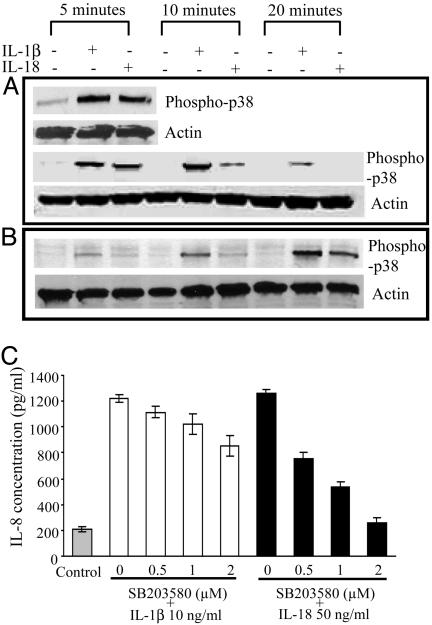
Stimulation of A549-Rβ cells with either IL-1β or IL-18 revealed phosphorylated-p38 by immunoblotting. As shown in Fig. 2A, treatment with IL-1β or IL-18 induced p38 phosphorylation, respectively, each lasting for 10 min, which progressively decreased. In Fig. 2B, the cells were lysed with Triton X-100 without previous trypsin treatment, and phosphorylation was observed after 20 min for both cytokines. We next studied the inhibitory effect of the p38-MAPK inhibitor SB203580 on IL-18- and IL-1β-induced IL-8 production. As shown in Fig. 2C, IL-8 production in response to IL-18 was significantly inhibited in a dose-dependent manner (50% inhibition at 1 μM), whereas IL-1β-induced IL-8 production was inhibited by only 20% at 2 μM.
Fig. 2.
Phosphorylation and inhibition of p38 MAPK. (A and B) Western blot analysis by antiphospho-p38 Ab. Cell extracts were prepared after stimulation with IL-1β (10 ng/ml) and IL-18 (50 ng/ml) for indicated times by trypsinization (A). Direct lysis in Triton buffer was used to prepare cell lysates (B). (C) A549-Rβ cells were preincubated for 1 h with increasing concentrations (0.5–2 μM) of the p38 inhibitor, SB203580, and stimulated with IL-1β or IL-18 at the same concentration in A and B. After 24 h, the supernatants were removed and assayed for IL-8 concentration by enhanced chemiluminescence. Data represent the mean ± SEM (n = 3).
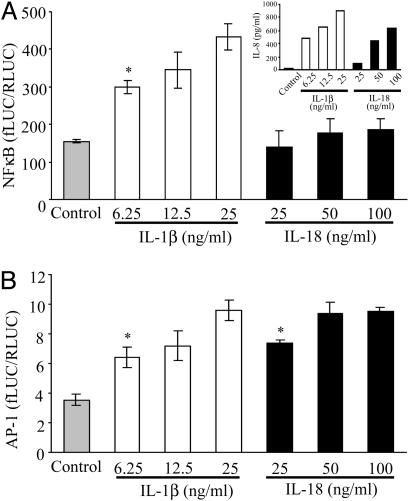
IL-18 Cell Signal Pathway in A549-Rβ Cells. We compared cell-signaling pathways by IL-1β and IL-18 for activation of NF-κB and AP-1. A549-Rβ cells were transiently cotransfected with either a NF-κB or AP-1 firefly-luciferase-reporter plasmid and a Renilla-luciferase vector. After recovery for 18 h, the transfectants were stimulated with IL-1β or IL-18 for 24 h, and the supernatants were removed for the IL-8 assay. The cells were harvested for luciferase assay. As shown in Fig. 3A Inset, both cytokines induced IL-8, but IL-18 did not induce NF-κB-reporter activity. IL-1β-induced NF-κB-reporter activity was 3-fold more than that of unstimulated cells. At the lowest concentration of IL-18 tested (25 ng/ml), there was a doubling of AP-1-reporter activity (P < 0.05) comparable to the lowest concentration (6.25 ng/ml) of IL-1β.
Fig. 3.
Effect of IL-1β or IL-18 on NF-κB and AP-1-signaling pathways. A549-Rβ cells were transiently cotransfected with NF-κB(A) or AP-1 (B) luciferase reporter plasmids or control pRL-TK Renilla vector at a ratio of 50:1 and cultured for 24 h. After stimulation with the indicated concentrations of IL-1β or IL-18 for 24 h, supernatants were collected for IL-8 assay (A Inset), and cells were harvested for luciferase assay. The data obtained were evaluated as firefly luciferase activity/Renilla luciferase activity. The data represent the mean ± SEM, n = 6. *, P < 0.05 compared to control.
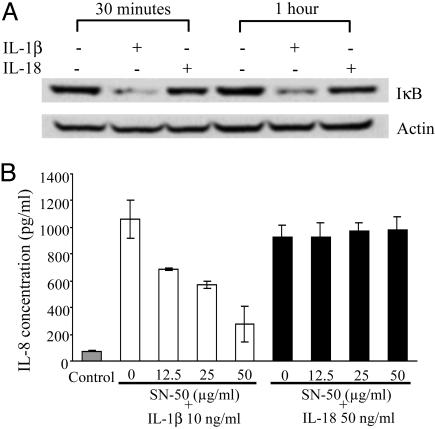
Consistent with the weak response of IL-18-induced NF-κB-reporter activity, there was little or no degradation in IκB degradation as assessed by immunoblotting (Fig. 4A). In contrast, IL-1β rapidly induced complete degradation of IκB and was sustained for 1 h. The specific competitive peptide inhibitor of NF-κB, SN-50, was used to test the effect of IL-1β and IL-18 on IL-8 production in A549-Rβ cells. This inhibitor dose-dependently reduced IL-1β-induced IL-8 production (50% reduction between 25 and 50 μg/ml) whereas there was no effect of the inhibitor on IL-18-induced IL-8 at each concentration studied (Fig. 4B).
Fig. 4.
IκB degradation after IL-1β or IL-18 stimulation. (A) Western blot analysis of IκB was performed with cell extracts from A549-Rβ, immediately following stimulation with IL-1β (10 ng/ml) or IL-18 (50 ng/ml) for indicated times. The amount of protein loading was determined by probing the same filter with antiactin Ab. (B) A549-Rβ cells were preincubated for 1 h with increasing concentrations of SN-50 and then stimulated with either IL-1β (10 ng/ml) or IL-18 (50 ng/ml). After 24 h, the supernatants were collected for IL-8 assay. The data represent the mean ± SEM, n = 3.
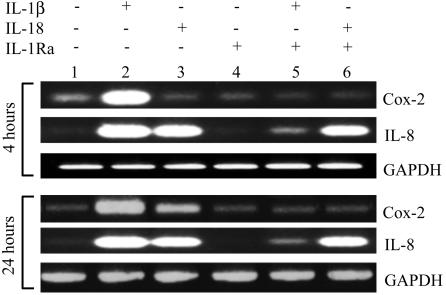
A Comparison of IL-1β and IL-18 on Steady-State COX-2 mRNA Levels. Because NF-κB activation is linked to COX-2 induction, A549-Rβ cells were stimulated with IL-18 or IL-1β for 4 or 24 h and harvested for COX-2 RT-PCR. As shown in Fig. 5 Upper, IL-1β increased COX-2 expression above basal levels after 4 h of exposure, whereas IL-18 had no effect. Nevertheless, these same cells at the same time point revealed an increase in the level of IL-8 mRNA that was nearly the same as induced by IL-1β (lane 3). IL-1β-induced IL-8 mRNA was markedly reduced by the presence of IL-1Ra, whereas IL-18-induced IL-8 gene expression was unaffected by blocking IL-1 receptors (lane 6). After 24 h of exposure (Fig. 5 Lower), IL-18-induced COX-2 had clearly taken place (lane 3) but appeared to be IL-1 dependent, because IL-1Ra reduced the level to that unstimulated cells (lane 6). However, similar to IL-18-induced IL-8 at 4 h, this activity on A549-Rβ cells (lane 6) was IL-1 independent at 24 h. Not shown is the finding that IL-18 did not induce IL-8 gene expression or protein in A549 cells lacking the IL-18 receptor β chain.
Fig. 5.
Steady-state COX-2 and IL-8 mRNA levels. A549-Rβ were stimulated with IL-1β or IL-18 in the absence or presence of IL-1Ra (10 μg/ml) added 1 h before cytokine stimulation. Total RNA was extracted and analyzed by RT-PCR for COX-2 and IL-8 after 4 h (Upper) or 24 h (Lower). The data are from a single experiment representative of four similar studies.
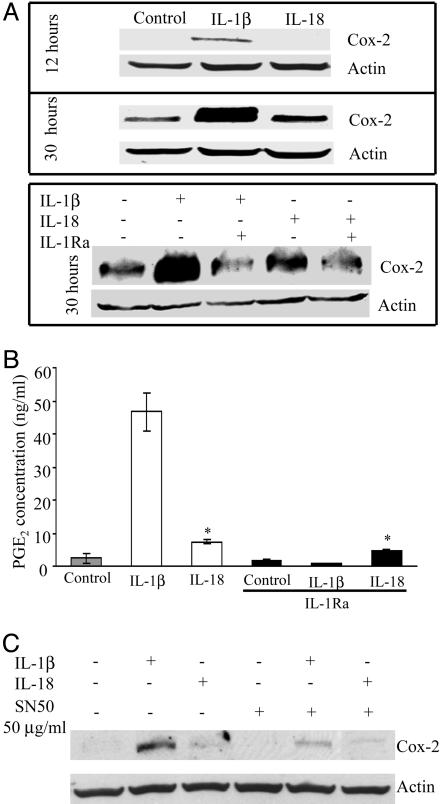
COX-2 Protein and PGE2 Levels After IL-18 Stimulation. A549-Rβ cells were treated with IL-18 for increasing time intervals, and COX-2 production was monitored by Western blot. As shown in Fig. 6A Upper, COX-2 protein, although considerably less than COX-2 protein induced by IL-1β, was elevated by IL-18 after 30 h of stimulation, but not present at 12 h. Consistent with the results of RT-PCR, COX-2 protein induced by IL-18 was reduced by pretreatment of the cells with IL-1Ra (Fig. 6A Lower).
Fig. 6.
COX-2 and PGE2 production in IL-1β- or IL-18-stimulated cells. (A) A549-Rβ cells were stimulated with IL-1β (10 ng/ml) or IL-18 (50 ng/ml), and Western blot analysis was performed after 12 (A) or 30 (B) h. (B) PGE2 levels in supernatants of cells stimulated with the same concentrations of cytokines for 48 h in the absence or presence of IL-1Ra (10 μg/ml). The data represent the mean ± SEM, n = 3. *, P < 0.05 compared to control. (C) Immunoblot of COX-2 at 24 h in A549-Rβ cells pretreated for 1 h with SN-50 and then stimulated with IL-1β or IL-18.
Cells were treated with IL-18 or IL-1β, in the presence or absence of IL-1Ra, and after 48 h supernatants were assayed for PGE2 levels. As shown in Fig. 6B, IL-1β induced a 25-fold increase in PGE2. IL-18-induced PGE2 production was 6-fold lower than PGE2 induced by IL-1β. The level of IL-1β-induced PGE2 was completely reduced by IL-1Ra, whereas IL-18-induced PGE2 was reduced by 40% by IL-1Ra. After 48 h, the concentration of PGE2 in the supernatants of unstimulated cells was 2.1 ng/ml compared to 7.33 ng/ml in cells stimulated with IL-18. In the presence of saturating concentrations of IL-1Ra, the level was reduced to 4.73 ng/ml, suggesting that not all of the PGE2 induced by IL-18 after 48 h is IL-1 dependent. Nevertheless, IL-18-induced COX-2 and PGE2 appear, for the most part, to be a weak response and partially IL-1 dependent.
To further confirm the importance of NF-κB activation in IL-1β COX-2 production, we tested the effect of the specific competitive peptide inhibitor of NF-κB, SN50, on IL-1β- or IL-18-induced COX-2 protein. As shown in Fig. 6C, the response to IL-1β was significantly inhibited by SN-50. The low level of induction by IL-18 was also affected by SN50.
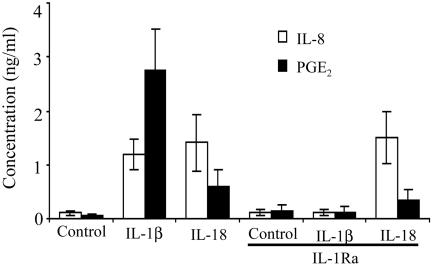
Studies in Primary Cells. Freshly obtained human PBMC were stimulated with IL-1β or IL-18 in the absence or presence of saturating concentrations of IL-1Ra and after 24 h, the level of IL-8 was determined. As shown in Fig. 7, there were comparable levels of IL-8 induced by either cytokine, but only IL-1β stimulation was reduced by IL-1Ra. The induction of PGE2 revealed a 51-fold elevation over unstimulated cells (0.054 vs. 2.76 ng/ml) for IL-1β but a 10-fold stimulation for IL-18 (0.054 vs. 0.59 ng/ml). Of this weak response to IL-18, 45% was due to coinduction of IL-1.
Fig. 7.
Effect of IL-18 on IL-8 and PGE2 production in primary human cells. Human PBMC from three donors were incubated with IL-1β (10 ng/ml) or IL-18 (50 ng/ml) in the absence or presence of IL-1Ra (10 μg/ml). After 24 h, IL-8 was measured in the supernatants, and PGE2 was measured after 48 h. Mean levels (± SEM) are shown.
In freshly obtained mouse resident peritoneal macrophages, 50 ng/ml of mouse IL-18 induced PGE2: unstimulated controls, 392; IL-18, 475; IL-18 plus IL-1Ra, 395; IL-18 plus IL-12, 888; and IL-18 plus IL-12 plus IL-1Ra, 1,008 (pg/ml, mean ± SEM). There was a 2-fold increase in nitric oxide induced by 50 ng/ml of mouse IL-18 in these cells (2.1 vs. 4.3 mM).
Discussion
IL-1β, IL-18, COX-2, and Fever. The present study was performed to examine the differences between IL-1β and IL-18 for the induction of COX-2, because injection of IL-18 does not cause fever in animals (5, 17, 18). These data are in sharp contrast to the ability of IL-1 to produce fever in animals and humans at exceedingly low doses of a few nanograms per kilogram (13). In humans, a late pyrogenic response is seen only at 10 μg/kg and likely represents a response to IL-18–IL-1 or TNFα. In subjects with genetic defects in caspase-1 activity resulting in spontaneous secretion of IL-1β, there is a prompt reduction in fever after a single injection of IL-1Ra (24, 25). IL-1-induced fever depends on COX-2 and not COX-1-induced PGE2 (3–5). In human synovial cells, half-maximal stimulation of PGE2 by IL-1β is achieved at 1.3 ± 0.24 pM (18–27 pg/ml) (26). Therefore, the failure of IL-18 to induce fever is linked to COX-2. Although one could argue that the lack of IL-18 receptors on the specialized endothelial cells of the hypothalamic thermoregulatory center explains the lack of fever, direct injection of IL-18 into the brain also does not evoke a febrile response (5). Moreover, there is not a dearth of evidence that the brain is highly responsive to endogenous as well as exogenous IL-18 (18, 27), and that the brain expresses IL-18 receptors (28–30).
There are conflicting data regarding the effects of IL-18 on COX-2 or PGE2 production (15, 16). Olee et al. (16) reported that IL-18 induced gene expression for COX-2 in human chondrocytes, whereas Reznikov et al. (15) was unable to demonstrate COX-2 expression in primary human PBMC or macrophage cultures. In PBMC, the failure to detect IL-18-induced COX-2 is compromised by the ability of IL-18 to induce IFN-γ in those cultures. IFN-γ inhibits IL-1β-induced PGE2 (31). In the study by Olee et al. (16), it remains unclear whether IL-18-induced COX-2 was IL-1 dependent. Therefore, to compare IL-1β and IL-18 effects of COX-2, we constructed an IL-18-responding cell line (A549-Rβ) by stable transfection of IL-18Rβ. The importance of the IL-18Rβ chain for responsiveness to IL-18 is well established.
IL-18 did not induce COX-2 gene expression in A549-Rβ cells after 4h of exposure, whereas we observed a 10-fold increase by IL-1β. However, after 24 h, COX-2 was observed in IL-18-stimulated cells, but this was nearly entirely IL-1 dependent. After 30 h, IL-18-induced COX-2 protein was present, but this activity was also IL-1 dependent. Because we demonstrated that IL-18 induced IL-1α in A549-Rβ cells (Fig. 1), it is likely that the increase in COX-2 after IL-18 reflects endogenous IL-1α activity from A549-Rβ cells via activation of NF-κB, extracellular signal-regulated protein kinase, MAPK p38, and protein kinase C, each pathway contributing to maximal COX-2 induction (32).
IL-18 and NF-κB. It appears that in A549-Rβ cells, IL-18 by itself does not directly induce COX-2 or weakly so because of an apparent inability of NF-κB activation. NF-κB is required for regulation of many genes, particularly COX-2 (33) as well as TNF-α, IL-1, and IL-6 (34). In A549-Rβ, IL-18 did not induce luciferase activity driven by the NF-κB-reporter vector. There was no IκB degradation, and it was not affected by NF-κB-specific inhibitor SN-50. In contrast, IL-1β was active in each of these parameters. In fact, 90% of IL-1β-induced IL-8 was inhibited by SN-50. SN-50 is a cell-permeable peptide containing the nuclear localization sequence of the p50 subunit that competes with the binding of the NF-κB complex in the nucleus (35). SN-50 attenuated both IL-1β-induced IL-8 levels and IL-1β-COX-2 protein, whereas the inhibitor had no effect on IL-18-induced IL-8. In human synovial fibroblasts, IL-18 induced the doubling of IL-8 production, which was inhibited by only 44% by using antisense p65 (36). However, in human T cells, IL-18 activity was reduced by inhibitors of NF-κB but not MAPK p38 (37). These data are consistent with an 8-fold increase in COX-2 transcription by IL-1β and with promoter-based studies implicating NF-κB in the activation of the COX-2 promoter in other cell lines (38–41).
The failure of IL-18 to induce COX-2 in cells of epithelial origin should be contrasted with reports of IL-18 activation of NF-κB in nonepithelial cells such as chondrocytes, T helper cells, and EL-4 cells (16, 42, 43). Whereas IL-1β activates COX-2 via NF-κB in parent A549 cells (33), COX-2 expression induced by phorbol esters was independent of NF-κB in the same cells (39). Consistent with the present study, the activation of NF-κB differs between IL-18 and IL-1β in synovial fibroblasts (36).
IL-18 May Preferentially Activate p38-MAPK and AP-1. In human myelomonocytic cells, IL-18 activates AP-1 (44, 45). Superoxide production from freshly obtained human neutrophils is increased by IL-18 and this depends on active MAPK p38 (46). Previous reports also indicate that p38-MAPK/AP-1 is essential for IL-1β-induced IL-8 gene expression in vascular smooth muscle cells (47). In rheumatoid arthritis-derived synovial fibroblasts, three distinct signaling pathways have been described for IL-18, and two are p38-MAPK and AP-1 (48).
High Concentrations of IL-18 Are Needed vs. Low Concentrations of IL-1β. Most studies on the effects of human IL-18 in immunecompetent cells require a costimulant. The costimulants are IL-2, IL-12, or IL-15, but most commonly, IL-12 is required for IL-18-induced IFN-γ (49). In general, IL-1 activates a variety of cells and does not require costimulants, and IL-1 is active in the low picomol range (26). In contrast, the direct effects of IL-18 in nonimmunocompetent cells use nanomolar concentrations of IL-18. For example, IL-18-induced phosphatidylinositol 3-kinase activation and MAPK p38 phosphorylation in synovial fibroblasts required 10 nM (180 ng/ml) (48), and production of superoxide in human neutrophils required 100 ng/ml (46). Also, human neutrophil production of IL-1α did not reach statistical significance even at 100 ng/ml, whereas IL-8 production required 10–100 ng/ml (50). A doubling of IL-8 required the same concentration (36), and in human vascular smooth muscle cells, 100 ng/ml was used (47). IL-18-induced migration of HL-60 cells doubled only by using 100 ng/ml of IL-18 (51), and the same concentration was used to stimulate IFN-γ in K562 transfected with both chains of the IL-18-receptor complex (52). Gene expression for serum amyloid A was induced by IL-18 at 100 ng/ml (53), and 100 ng/ml was required to stimulate TNFα from human blood T cells (37). In mouse microglial cells, 500 ng/ml of mouse IL-18 stimulated IL-1β, IL-1α, and IL-6 release (54). The sources of recombinant human or mouse IL-18 differ (1, 20, 30, 37, 46, 48, 50, 53), and yet 100-fold less IL-18 from the same sources are effective in stimulating IFN-γ in natural killer cells, primary T lymphocytes and T cell lines by using IL-12 or IL-15 as the costimulants (6, 49, 55).
Costimulants increase the expression of the IL-18 receptors on immunocompetent cells (56, 57). Although the A549-Rβ cell line used in the present study was a stable transfectant, we cannot exclude that in the absence of a costimulant, IL-18 signaling does not activate NF-κB sufficiently to trigger COX-2. Nevertheless, IL-18 remains an inflammatory cytokine, because induction of IL-8 and IL-6 is a direct result of IL-18 signaling, is via the activation of the p38-MAPK/AP-1 signal pathway, and hence asserts the important role of IL-18 in the inflammatory responses.
IL-18 Responses in Primary Cells. The data between A549-Rβ cells and freshly obtained human PBMC remain consistent. In PBMC, the level of PGE2 production in response to IL-18 is low (10-fold increase) compared to that of IL-1β (51-fold), and 45% of the total PGE2 in IL-18-stimulated PBMC is actually secondary to IL-1. In primary mouse macrophages, mouse IL-18 stimulated low levels of PGE2 (392 vs. 475 pg/ml). However, in the presence of mouse IL-12, this level increased to 888 pg/ml. Similar to most studies, IL-18 requires a costimulant, which functions to increase receptor expression. These results are consistent with the concept that IL-18 is a weak activator of intracellular signaling resulting in PGE2 in primary as well as A549-Rβ cells. It appears that the default mode for IL-18 responsiveness is low expression of IL-18 receptors as a possible evolutionary advantage for preventing a dominant T helper-1 immune reaction during infection. The constitutive and high level of secretion of the IL-18-binding protein (58) may also reflect evolutionary pressure to suppress the T helper-1 response and reduce the chance of autoimmune disease.
Acknowledgments
We thank Ariel Werman for helpful suggestions. National Institutes of Health Grants AI-15614, HL-68743, and CA-046934 supported these studies.
Abbreviations: COX, cyclooxygenase; PG, prostaglandin; PGE2, PGE2; IκB, NF-κB inhibitor; IL-18R, IL-18 receptor; IL-1Ra, IL-1R antagonist; PBMC, peripheral blood mononuclear cells; AP-1, activator protein-1; MAPK, mitogen-activated protein kinase.
References
- 1.Chandrasekharan, N. V., Dai, H., Roos, K. L., Evanson, N. K., Tomsik, J., Elton, T. S. & Simmons, D. L. (2002) Proc. Natl. Acad. Sci. USA 99, 13926–13931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maier, J. A., Hla, T. & Maciag, T. (1990) J. Biol. Chem. 265, 10805–10808. [PubMed] [Google Scholar]
- 3.Li, S., Wang, Y., Matsumura, K., Ballou, L. R., Morham, S. G. & Blatteis, C. M. (1999) Brain Res. 825, 86–94. [DOI] [PubMed] [Google Scholar]
- 4.Li, S., Ballou, L. R., Morham, S. G. & Blatteis, C. M. (2001) Brain Res. 910, 163–173. [DOI] [PubMed] [Google Scholar]
- 5.Li, S., Goorha, S., Ballou, L. R. & Blatteis, C. M. (2003) Brain Res. 992, 76–84. [DOI] [PubMed] [Google Scholar]
- 6.Okamura, H., Tsutsi, H., Komatsu, T., Yutsudo, M., Hakura, A., Tanimoto, T., Torigoe, K., Okura, T., Nukada, Y., Hattori, K., et al. (1995) Nature 378, 88–91. [DOI] [PubMed] [Google Scholar]
- 7.Ushio, S., Namba, M., Okura, T., Hattori, K., Nukada, Y., Akita, K., Tanabe, F., Konishi, K., Micallef, M., Fujii, M., et al. (1996) J. Immunol. 156, 4274–4279. [PubMed] [Google Scholar]
- 8.Banda, N. K., Vondracek, A., Kraus, D., Dinarello, C. A., Kim, S. H., Bendele, A., Senaldi, G. & Arend, W. P. (2003) J. Immunol. 170, 2100–2105. [DOI] [PubMed] [Google Scholar]
- 9.Plater-Zyberk, C., Joosten, L. A., Helsen, M. M., Sattonnet-Roche, P., Siegfried, C., Alouani, S., van De Loo, F. A., Graber, P., Aloni, S., Cirillo, R., et al. (2001) J. Clin. Invest. 108, 1825–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melnikov, V. Y., Ecder, T., Fantuzzi, G., Siegmund, B., Lucia, M. S., Dinarello, C. A., Schrier, R. W. & Edelstein, C. L. (2001) J. Clin. Invest. 107, 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pomerantz, B. J., Reznikov, L. L., Harken, A. H. & Dinarello, C. A. (2001) Proc. Natl. Acad. Sci. USA 98, 2871–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mallat, Z., Corbaz, A., Scoazec, A., Graber, P., Alouani, S., Esposito, B., Humbert, Y., Chvatchko, Y. & Tedgui, A. (2001) Circ. Res. 89, E41–E45. [DOI] [PubMed] [Google Scholar]
- 13.Dinarello, C. A. (1996) Blood 87, 2095–2147. [PubMed] [Google Scholar]
- 14.Dinarello, C. A. & Thompson, R. C. (1991) Immunol. Today 12, 404–410. [DOI] [PubMed] [Google Scholar]
- 15.Reznikov, L. L., Kim, S. H., Westcott, J. Y., Frishman, J., Fantuzzi, G., Novick, D., Rubinstein, M. & Dinarello, C. A. (2000) Proc. Natl. Acad. Sci. USA 97, 2174–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olee, T., Hashimoto, S., Quach, J. & Lotz, M. (1999) J. Immunol. 162, 1096–1100. [PubMed] [Google Scholar]
- 17.Gatti, S., Beck, J., Fantuzzi, G., Bartfai, T. & Dinarello, C. A. (2002) Am. J. Physiol. 282, R702–R709. [DOI] [PubMed] [Google Scholar]
- 18.Kubota, T., Fang, J., Brown, R. A. & Krueger, J. M. (2001) Am. J. Physiol. 281, R828–R838. [DOI] [PubMed] [Google Scholar]
- 19.Leoni, F., Zaliani, A., Bertolini, G., Porro, G., Pagani, P., Pozzi, P., Dona, G., Fossati, G., Sozzani, S., Azam, T., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 2995–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puren, A. J., Fantuzzi, G., Gu, Y., Su, M. S.-S. & Dinarello, C. A. (1998) J. Clin. Invest. 101, 711–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pradelles, P., Grassi, J. & Maclouf, J. (1990) Methods Enzymol. 187, 24–34. [DOI] [PubMed] [Google Scholar]
- 22.Fadok, V. A., Bratton, D. L., Konowal, A., Freed, P. W., Westcott, J. Y. & Henson, P. M. (1998) J. Clin. Invest. 101, 890–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grentzmann, G., Ingram, J. A., Kelly, P. J., Gesteland, R. F. & Atkins, J. F. (1998) RNA 4, 479–486. [PMC free article] [PubMed] [Google Scholar]
- 24.Hawkins, P. N., Lachmann, H. J. & McDermott, M. F. (2003) N. Engl. J. Med. 348, 2583–2584. [DOI] [PubMed] [Google Scholar]
- 25.Hawkins, P. N., Lachmann, H. J., Aganna, E. & McDermott, M. F. (2004) Arthritis Rheum. 50, 607–612. [DOI] [PubMed] [Google Scholar]
- 26.Chin, J., Rupp, E., Cameron, P. M., MacNaul, K. L., Lotke, P. A., Tocci, M. J., Schmidt, J. A. & Bayne, E. K. (1988) J. Clin. Invest. 82, 420–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yatsiv, I., Morganti-Kossmann, M. C., Perez, D., Dinarello, C. A., Novick, D., Rubinstein, M., Otto, V. I., Rancan, M., Kossmann, T., Redaelli, C. A., et al. (2002) J. Cereb. Blood Flow Metab. 22, 971–978. [DOI] [PubMed] [Google Scholar]
- 28.Prinz, M. & Hanisch, U. K. (1999) J. Neurochem. 72, 2215–2218. [DOI] [PubMed] [Google Scholar]
- 29.Hedtjarn, M., Leverin, A. L., Eriksson, K., Blomgren, K., Mallard, C. & Hagberg, H. (2002) J. Neurosci. 22, 5910–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wheeler, R. D., Culhane, A. C., Hall, M. D., Pickering-Brown, S., Rothwell, N. J. & Luheshi, G. N. (2000) Brain Res. Mol. Brain Res. 77, 290–293. [DOI] [PubMed] [Google Scholar]
- 31.Browning, J. L. & Ribolini, A. (1987) J. Immunol. 138, 2857–2863. [PubMed] [Google Scholar]
- 32.Mifflin, R. C., Saada, J. I., Di Mari, J. F., Adegboyega, P. A., Valentich, J. D. & Powell, D. W. (2002) Am. J. Physiol. 282, C824–C834. [DOI] [PubMed] [Google Scholar]
- 33.Jung, Y. J., Isaacs, J. S., Lee, S., Trepel, J. & Neckers, L. (2003) FASEB J. 17, 2115–2117. [DOI] [PubMed] [Google Scholar]
- 34.Xiao, C. W., Ash, K. & Tsang, B. K. (2001) Endocrinology 142, 557–563. [DOI] [PubMed] [Google Scholar]
- 35.Lin, Y. Z., Yao, S. Y., Veach, R. A., Torgerson, T. R. & Hawiger, J. (1995) J. Biol. Chem. 270, 14255–14258. [DOI] [PubMed] [Google Scholar]
- 36.Morel, J. C., Park, C. C., Kumar, P. & Koch, A. E. (2001) Lab. Invest. 81, 1371–1383. [DOI] [PubMed] [Google Scholar]
- 37.Dai, S. M., Matsuno, H., Nakamura, H., Nishioka, K. & Yudoh, K. (2004) Arthritis Rheum. 50, 432–443. [DOI] [PubMed] [Google Scholar]
- 38.Newton, R., Kuitert, L. M., Bergmann, M., Adcock, I. M. & Barnes, P. J. (1997) Biochem. Biophys. Res. Commun. 237, 28–32. [DOI] [PubMed] [Google Scholar]
- 39.Catley, M. C., Chivers, J. E., Cambridge, L. M., Holden, N., Slater, D. M., Staples, K. J., Bergmann, M. W., Loser, P., Barnes, P. J. & Newton, R. (2003) FEBS Lett. 547, 75–79. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto, K., Arakawa, T., Ueda, N. & Yamamoto, S. (1995) J. Biol. Chem. 270, 31315–31320. [DOI] [PubMed] [Google Scholar]
- 41.Schmedtje, J. F., Jr., Ji, Y. S., Liu, W. L., DuBois, R. N. & Runge, M. S. (1997) J. Biol. Chem. 272, 601–608. [DOI] [PubMed] [Google Scholar]
- 42.Matsumoto, S., Tsuji-Takayama, K., Aizawa, Y., Koide, K., Takeuchi, M., Ohta, T. & Kurimoto, M. (1997) Biochem. Biophys. Res. Commun. 234, 454–457. [DOI] [PubMed] [Google Scholar]
- 43.Thomassen, E., Bird, T. A., Renshaw, B. R., Kennedy, M. K. & Sims, J. E. (1998) J. Interferon Cytokine Res. 18, 1077–1088. [DOI] [PubMed] [Google Scholar]
- 44.Baggiolini, M., Dewald, B. & Moser, B. (1997) Annu. Rev. Immunol. 15, 675–705. [DOI] [PubMed] [Google Scholar]
- 45.Kojima, H., Aizawa, Y., Yanai, Y., Nagaoka, K., Takeuchi, M., Ohta, T., Ikegami, H., Ikeda, M. & Kurimoto, M. (1999) J. Immunol. 162, 5063–5069. [PubMed] [Google Scholar]
- 46.Wyman, T. H., Dinarello, C. A., Banerjee, A., Gamboni-Robertson, F., Hiester, A. A., England, K. M., Kelher, M. & Silliman, C. C. (2002) J. Leukocyte Biol. 72, 401–409. [PubMed] [Google Scholar]
- 47.Jung, Y. D., Fan, F., McConkey, D. J., Jean, M. E., Liu, W., Reinmuth, N., Stoeltzing, O., Ahmad, S. A., Parikh, A. A., Mukaida, N. & Ellis, L. M. (2002) Cytokine 18, 206–213. [DOI] [PubMed] [Google Scholar]
- 48.Morel, J. C., Park, C. C., Zhu, K., Kumar, P., Ruth, J. H. & Koch, A. E. (2002) J. Biol. Chem. 277, 34679–34691. [DOI] [PubMed] [Google Scholar]
- 49.Nakanishi, K., Yoshimoto, T., Tsutsui, H. & Okamura, H. (2001) Annu. Rev. Immunol. 19, 423–474. [DOI] [PubMed] [Google Scholar]
- 50.Leung, B. P., Culshaw, S., Gracie, J. A., Hunter, D., Canetti, C. A., Campbell, C., Cunha, F., Liew, F. Y. & McInnes, I. B. (2001) J. Immunol. 167, 2879–2886. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, B., Wu, K. F., Cao, Z. Y., Rao, Q., Ma, X. T., Zheng, G. G. & Li, G. (2004) Leuk. Res. 28, 91–95. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, B., Cao, Z. Y., Wu, K. F. & Lin, Y. M. (2003) Leuk. Res. 27, 971–972. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka, F., Migita, K., Kawabe, Y., Aoyagi, T., Ida, H., Kawakami, A. & Eguchi, K. (2004) Life Sci. 74, 1671–1679. [DOI] [PubMed] [Google Scholar]
- 54.Wheeler, R. D., Brough, D., Le Feuvre, R. A., Takeda, K., Iwakura, Y., Luheshi, G. N. & Rothwell, N. J. (2003) J. Neurochem. 85, 1412–1420. [DOI] [PubMed] [Google Scholar]
- 55.Dinarello, C. A. (1999) J. Allergy Clin. Immunol. 103, 11–24. [DOI] [PubMed] [Google Scholar]
- 56.Kim, S. H., Reznikov, L. L., Stuyt, R. J., Selzman, C. H., Fantuzzi, G., Hoshino, T., Young, H. A. & Dinarello, C. A. (2001) J. Immunol. 166, 148–154. [DOI] [PubMed] [Google Scholar]
- 57.Yoshimoto, T., Takeda, K., Tanaka, T., Ohkusu, K., Kashiwamura, S., Okamura, H., Akira, S. & Nakanishi, K. (1998) J. Immunol. 161, 3400–3407. [PubMed] [Google Scholar]
- 58.Novick, D., Kim, S.-H., Fantuzzi, G., Reznikov, L., Dinarello, C. A. & Rubinstein, M. (1999) Immunity 10, 127–136. [DOI] [PubMed] [Google Scholar]