Abstract
Since their discovery as cell-division factors in plant tissue culture about five decades ago, cytokinins have been hypothesized to play a central role in the regulation of cell division and differentiation in plants. To test this hypothesis in planta, we isolated Arabidopsis plants lacking one, two, or three of the genes encoding a subfamily of histidine kinases (CRE1, AHK2, and AHK3) that function as cytokinin receptors. Seeds were obtained for homozygous plants containing mutations in all seven genotypes, namely single, double, and triple mutants, and the responses of germinated seedlings in various cytokinin assays were compared. Both redundant and specific functions for the three different cytokinin receptors were observed. Plants carrying mutations in all three genes did not show cytokinin responses, including inhibition of root elongation, inhibition of root formation, cell proliferation in and greening of calli, and induction of cytokinin primary-response genes. The triple mutants were small and infertile, with a reduction in meristem size and activity, yet they possessed basic organs: roots, stems, and leaves. These results confirm that cytokinins are a pivotal class of plant growth regulators but provide no evidence that cytokinins are required for the processes of gametogenesis and embryogenesis.
Since the discovery of kinetin in 1956 as a degradation product of DNA that promotes cell division in plants (1), a considerable amount of biochemical, physiological, and, most recently, genetic research has focused on elucidating the diverse roles that cytokinins play in plant growth and development. Perturbations of cytokinin levels in plants via over-expression of bacterial cytokinin synthesis genes (2–4), recovery of mutant plants with a higher-than-normal cytokinin content (5), and characterization of loss-of-function mutants of the cytokinin receptor CYTOKININ RESPONSE 1 (CRE1) (6–9) have implicated cytokinins in a wide variety of processes, including cell division, organ formation and regeneration, senescence, apical dominance, vascular development, response to pathogens, and nutrient mobility. These numerous roles for cytokinins, coupled with the failure of mutant screens to yield plants with nondetectable cytokinin levels, led to the longstanding belief that cytokinins are essential for plant growth and development.
Plants respond to cytokinin through a multistep phosphorelay system, consisting of sensor histidine kinase (HK) proteins, histidine phosphotransfer (HPt) proteins, and effector response regulator (RR) proteins. Over-expression and loss-of-function analyses of particular HK, HPt, and RR proteins in Arabidopsis (8–13), combined with transient expression assays in protoplasts (14), have led to a model for cytokinin signaling (for a review, see refs. 15 and 16), beginning with perception of cytokinins by HK proteins.
The Arabidopsis genome encodes six nonethylene receptor HKs: CRE1/WOL/AHK4, AHK2, AHK3, AtHK1, CKI1, and CKI2/AHK5. Among them, CRE1, Arabidopsis HK2 (AHK2), and Arabidopsis HK3 (AHK3) (hereafter called the CRE family) are highly homologous at the amino acid level, especially within the putative cytokinin-binding extracellular domain (≈60% identity). CRE1 was the first cytokinin-signaling component identified. A substantial body of evidence supports a role for CRE1 as a cytokinin receptor: plants carrying loss-of-function mutations in the CRE1 gene have a reduced sensitivity to cytokinin (8, 9), CRE1 initiates a phosphorelay in response to cytokinins when expressed in heterologous systems (8, 9, 17), and fission yeast expressing CRE1 bind active cytokinins in a specific and saturable manner (18). Similar to CRE1, AHK2 (M.H. and T.K., unpublished data) and AHK3 (18) are also activated by cytokinins when expressed in yeast and bacteria, respectively.
CKI1 was first identified as a gene that induces constitutive cytokinin responses when overexpressed in callus tissue (19). However, all subsequent efforts to detect cytokinin receptor activity of CKI1 have yielded negative results (refs. 14 and 18 and T.K., unpublished work). CKI2 is the only HK lacking a putative extracellular domain, and loss-of-function mutants of CKI2 have no noticeable phenotype (Y.H. and T.K., unpublished work). AtHK1 has been implicated in osmosensing (20), and the HK activity of AtHK1 in yeast is unchanged by cytokinins. These results indicate that CRE-family members are cytokinin receptors, but that the other nonethylene receptor HKs are less likely to perform a cytokinin-sensing role.
To elucidate the in planta roles of the three CRE-family members, as well as the role of cytokinin signaling in plant development, we identified loss-of-function alleles for CRE1, AHK2, and AHK3; created plants containing all possible mutant allele combinations; and characterized their responses in a series of cytokinin assays. These observations are discussed in relation to the relative contributions of each family member to cytokinin action, as well as the overall role of cytokinins in plant growth and development.
Materials and Methods
Plant Materials and Growth Conditions. Unless otherwise indicated, sterilized seeds of Arabidopsis thaliana, ecotype Columbia (Col), were incubated at 4°C for 2 days on germination medium (GM) containing full-strength Murashige and Skoog salts (MS) (21); 0.05% (wt/vol) Mes-KOH (pH 5.7); 1% (wt/vol) sucrose; 100 mg/ml inositol; 10 mg/ml thiamine HCl; 1 mg/ml nicotinic acid; 1 mg/ml pyridoxine HCl; and 0.3% (wt/vol) Phytagel (Sigma– Aldrich); and grown at 22°C under constant illumination (100 μmol/m2·s). For plastochron measurements, the number of leaves >0.5 mm in width from 11-day-old seedlings was counted under a dissecting microscope. For adventitious root formation assays as well as callus and shoot induction assays, 11-day-old plants grown aseptically under dim light (2.5 μmol/m2·s) were used. For visualization of root nuclei, roots from 21-day-old plants were fixed with 3.7% (vol/vol) formaldehyde, stained with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI), and photographed on an Olympus BX50 (Melville, NY) microscope with a Roper Scientific Coolsnap HQ/OL digital camera. For analysis of shoot apical meristem (SAM) morphology, seeds were germinated on full-strength MS, 4.5% (wt/vol) sucrose, 0.05% (wt/vol) Mes, and 0.8% (wt/vol) agar (MS + 4.5 suc), and grown for 7 days. Seedlings were fixed, embedded, and sectioned according to Mähönen et al. (6). Three-micrometer sections were stained with 0.05% (wt/vol) toluidine blue in water and photographed on an Olympus Provis microscope with an Olympus DP70 digital camera.
Cytokinin Response Assays. For the root elongation assay, seedlings grown under constant light for 8 days on GM supplemented with 1/5,000 volume of appropriate concentrations of benzyl adenine (BA) dissolved in DMSO were removed from plates, and root lengths were measured. Plants that had not germinated within 2 days of culture were excluded from the analysis. For the adventitious root formation assay, plants were separated into upper and lower portions by bisecting the hypocotyl with fine scissors. The upper portions were inserted into GM supplemented with 1/5,000 volume of appropriate concentrations of trans (t)-zeatin dissolved in DMSO. After 11 days, the presence or absence of adventitious roots near the cut site was observed under a dissecting microscope. For the callus induction assay, hypocotyls were excised with fine scissors and cultured for 24 days on GM supplemented with 30 ng/ml 2,4-dichloriphenoxy-aceticacid (2,4-D) and varying concentrations of kinetin. All plates contained 0.02% (vol/vol) DMSO. For the shoot formation assay, excised hypocotyls were cultured for 7 days on GM supplemented with 500 ng/ml 2,4-D and 50 ng/ml kinetin to induce callus formation. Calli were then moved onto GM supplemented with 0.3 μg/ml indolebutyric acid and varying concentrations of t-zeatin and cultured for 14 days.
Expression Patterns of Cytokinin Receptor Genes. For RNA gel blot analysis, 5 μg of total RNA was used per lane. DNA fragments corresponding to a region of CRE1, AHK2, or AHK3 were amplified by PCR. Primer sequences are available in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. After the T7 promoter was ligated to the 3′ end of each amplified fragment, radiolabeled ribo-probes were synthesized by using the MAXIscript T7 Kit (Ambion, Austin, TX). Hybridizations were conducted in PerfectHyb (Toyobo, Dojima, Osaka) according to the manufacturer's instructions. For expression patterns of reporter genes, the promoter sequence for each of the CRE-family genes was cloned in-frame upstream of the β-glucuronidase (GUS) gene. Details of the reporter gene cloning are available in the Supporting Materials and Methods. CRE1::GUS and AHK3::GUS were introduced into Arabidopsis, ecotype Wassilewskija (Ws), by the floral dip method (22). AHK2::GUS was similarly introduced into Arabidopsis (Col). Expression patterns for each gene were determined according to Miyawaki et al. (23).
Screening for T-DNA Insertion Mutants. Multiple alleles of T-DNA insertion mutants were identified. One set of mutants (cre1–10, ahk2–1, ahk3–1) is in the ecotype Ws, whereas another set of mutants (cre1–12, ahk2–2, and ahk3–3) is in the ecotype Col. Double and triple mutants were generated in the same backgrounds. Additional mutant alleles identified in the ecotype Ws include ahk3–2 and cre1–11. Details of the screening process are provided in Supporting Materials and Methods.
Expression of Cytokinin Receptor Genes in Mutant Backgrounds. Details of the RT-PCR analysis of CRE-family genes in cre1–12, ahk2–2, and ahk3-3 mutant plants are available in Supporting Materials and Methods.
Expression of Cytokinin Primary-Response Genes. For real-time quantitative PCR (qRT-PCR) analysis of cytokinin-inducible gene expression, seeds were germinated on MS + 4.5suc and grown for 6 days. Cytokinin treatment was carried out by incubating seedlings in an MS + 4.5suc solution without agar and supplemented with 10 μM BA for 30 min. Before RNA preparation, three WT Col and five cre1–12 ahk2–2 ahk3–3 (Col) seedlings were pooled and stored in RNAlater solution (Qiagen, Valencia, CA). Total RNA was extracted by using the RNeasy Plant Mini Kit (Qiagen). TaqMan RT-PCR reagents (Applied Biosystems) were used to synthesize double-stranded cDNA. Unlabeled gene-specific primers and 6-carboxy-fluoroscein-labeled gene-specific TaqMan Minor Groove Binder probes were used for qRT-PCR with ABI prism 7700 (Applied Biosystems). The number of ARR5 and ARR15 (Arabidopsis Response Regulator) transcripts present in two biological replicates each of WT, and cre1–12 ahk2–2 ahk3–3 seedlings, with or without BA, was determined three separate times. Fold induction of the ARR5 and ARR15 cytokinin primary-response gene transcripts was calculated relative to the SHORT ROOT (SHR) transcript (24), according to the manufacturer's instructions (ABI Prism 7700 Sequence Detection System, User Bulletin #2). Primer and probe sequences are available in Supporting Materials and Methods.
Flow Cytometry. Experiments were performed according to Shpak et al. (25), with some modifications. Plants were grown vertically on GM plates, with 1.8% (wt/vol) purified agar. Whole roots of 33-day-old plants were finely chopped in 0.5 ml of ice-cold extraction buffer [15 mM Hepes/1 mM EDTA/80 mM KCl/20 mM NaCl/300 mM sucrose/0.5% (vol/vol) Triton X-100/0.5 mM spermine/0.1% (vol/vol) 2-mercaptoethanol), passed through 33-μm nylon mesh, and centrifuged at 3,000 × g for 1 min. The pellet was resuspended in 100 μl of staining buffer (1/10,000 dilution of SYBR green I (Molecular Probes)/50 μg/ml RNase A/3.7% (vol/vol) formaldehyde in the extraction buffer) and subjected to FACScan (Becton Dickinson) by using the FL2 channel with a photomultiplier voltage of 300 V.
Results
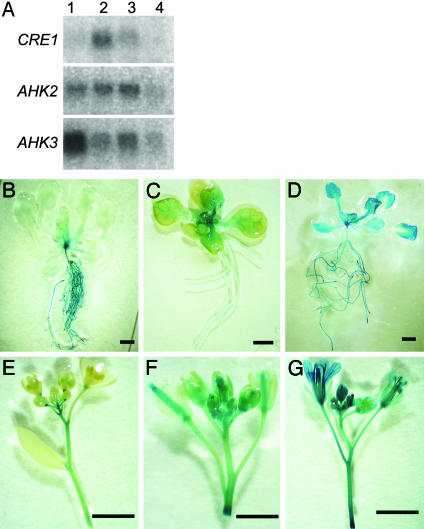
Expression Patterns of Cytokinin Receptor Genes. RNA gel blot hybridization experiments confirmed that the three CRE-family genes have distinct expression patterns (Fig. 1A). CRE1 expression was highest in the root and low in rosette leaves. AHK2 was expressed to about the same degree in rosette leaves and the root. AHK3 expression was highest in rosette leaves, moderate in the root, and low in the silique. Expression of all three cytokinin receptor genes was detected in flowers. Differential expression of the cytokinin receptor genes in root and shoot tissue was confirmed by expression of the GUS reporter gene directed by regulatory sequences from CRE1, AHK2, or AHK3. CRE1::GUS activity was high in the root, moderate in the inflorescence stems and pedicels, and low in the leaves (Fig. 1 B and E). AHK2::GUS activity was high in leaf veins, petioles, inflorescence stems, flowers, and siliques, and moderate in the roots (Fig. 1 C and F). AHK3::GUS was expressed ubiquitously in root and shoot tissues including leaves, inflorescence stems, and flowers (Fig. 1 D and G).
Fig. 1.
Expression patterns of the CRE-family genes. (A) RNA gel blot hybridizations (5 μg of RNA) from different tissues of WT plants, probed with gene-specific probes for CRE1, AHK2, or AHK3. 1, rosette leaves; 2, roots; 3, floral bunches; 4, siliques. (B) Expression of the GUS reporter gene under control of regulatory sequences from cytokinin-receptor genes. (B and E) CRE1::GUS activity. (C and F) AHK2::GUS activity. (D and G) AHK3::GUS activity. (Bars = 2 mm.)
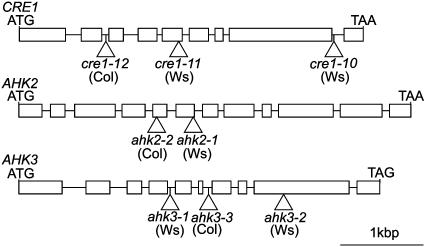
Expression of Cytokinin Receptor Genes in the Mutant Background. Multiple independent T-DNA insertion alleles for all members of the Arabidopsis CRE cytokinin receptor family (AHK2, AHK3, and CRE1) were identified (Fig. 2). To determine whether full-length transcripts of CRE1, AHK2, and AHK3 were present in the T-DNA insertion mutants, RT-PCR analysis was performed on RNA prepared from plants carrying a single mutation in the CRE-family genes by using gene-specific primers flanking the T-DNA insertion site (see Fig. 10, which is published as supporting information on the PNAS web site). Even with saturating numbers of PCR cycles, the full-length transcripts of CRE1 and AHK2 were not detected in cre1–12 and ahk2–2 mutants, respectively (Fig. 10A). The full-length transcript of AHK3 was not detected in the ahk3–3 mutant with the same number of PCR cycles that yielded near-saturating amplification of AHK3 in WT plants (Fig. 10A). However, the transcript was detected with a greater number of PCR cycles (data not shown). The full-length transcripts of CRE1, AHK2, or AHK3 were not detected in cre1–10 or cre1–11, ahk2–1, ahk3–1, or ahk3–2 mutants, respectively (data not shown). Recovery of Arabidopsis plants containing multiple independent T-DNA insertion alleles for each of the CRE-family genes, lacking expression of a full-length transcript, indicates that these T-DNA insertion mutants are null alleles. The greatly reduced level of AHK3 expression detected in the ahk3–3 mutant is unlikely to contribute to cytokinin signaling, because the cytokinin-response phenotypes of the ahk3–3 mutant are the same as those observed for the ahk3–1 and ahk3–2 null mutants. Likewise, the cytokinin-response phenotypes of the cre1–12 ahk3–3 and ahk2–2 ahk3–3 double mutants are the same as those observed for the cre1–10 ahk3–1 and ahk2–1 ahk3–1 double mutants, respectively.
Fig. 2.
Description of the CRE1, AHK2, and AHK3 T-DNA insertion alleles. Boxes represent exons; horizontal bars, introns; and triangles, T-DNA integration sites.
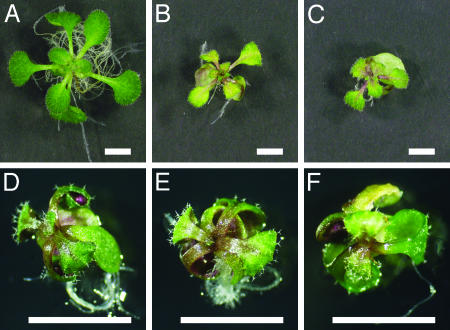
Overall Appearance of Cytokinin Receptor Mutants. When grown on soil, plants with a single mutation in AHK2, AHK3, or CRE1 grew normally (see Fig. 11, which is published as supporting information on the PNAS web site). When grown on soil, plants carrying the cre1–12 ahk2–2, cre1–12 ahk3–3 (Fig. 11), cre1–10 ahk2–1, and cre1–10 ahk3–1 (not shown) mutations exhibited no noticeable phenotype. The ahk2–2 ahk3–3 (Fig. 11) and ahk2–1 ahk3–1 (not shown) double mutants had smaller leaves and shorter stems than did the WT plants. This result indicates that AHK2 and AHK3 functions dominate in the shoot. Roots of double mutants for any mutant combination were normal. Surprisingly, triple mutants were recovered in both the Ws background (cre1–10 ahk2–1 ahk3–1, and cre1–11 ahk2–1 ahk3–1) (not shown) and the Col background (cre1–12 ahk2–2 ahk3–3) (Figs. 3D and 11), indicating that seeds can germinate and seedlings can grow for a limited period without any of the CRE-family genes being expressed. The shoot and root growth of triple mutants was very slow, and leaf numbers were decreased (see below). The triple mutants occasionally produced an inflorescence stem with abnormal and nonfunctional flowers, but did not produce seeds. Supplementing media with 1 μg/ml t-zeatin severely inhibited the growth of WT seedlings but did not affect the growth of triple mutants (Fig. 3), suggesting that the triple mutants lack a mechanism for cytokinin perception. The insensitivity of the triple mutants to cytokinins was verified with several cytokinin-response assays, as shown below.
Fig. 3.
Phenotype of the triple mutant grown with or without t-zeatin. (A–C) Thirteen-day-old WT seedlings from the ecotype Col. (D–F) Thirteen-day-old cre1–12 ahk3–3 ahk2–2 (Col) triple mutants. Plants were grown on plates with 0ng/ml (A and D), 100 ng/ml (B and E), 1,000 ng/ml t-zeatin (C and F). (Bars = 5 mm.)
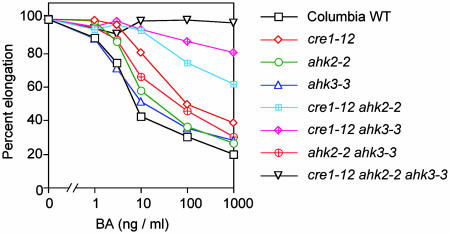
Cytokinin Sensitivity in the Root Growth Assay. Exogenous cytokinins normally inhibit root elongation. As previously reported (8), mutations in CRE1 caused a reduced sensitivity to cytokinin (cre1–12, Fig. 4; cre1–10 and cre1–11, not shown). The ahk2–2 or ahk3–3 mutants exhibited normal or slightly reduced sensitivity (Fig. 4). Additive effects were seen in the double mutants; that is, cre1–12 ahk2–2, cre1–12 ahk3–3, and ahk2–2 ahk3–3 double mutants were less sensitive to cytokinin than was either single mutant (Fig. 4). Similarly, cre1–10 ahk2–1 and cre1–10 ahk3–1 double mutants were less sensitive than was either single mutant (not shown). The triple mutant had a shorter root with respect to WT plants, and the root length was not affected by BA up to 1 μg/ml. These data indicate that CRE1, AHK2, and AHK3 have redundant functions in cytokinin signaling in roots.
Fig. 4.
Elongation of roots of cytokinin-receptor mutants in the presence of increasing concentrations of BA. Root length of each genotype without cytokinin was set at 100%. Lengths of roots in the absence of BA were: WT, 2.58 ± 0.29 (mean ± SD); cre1–12, 2.65 ± 0.32; ahk2–2, 2.18 ± 0.28; ahk3–3, 2.81 ± 0.38; cre1–12 ahk2–2, 2.57 ± 0.38; cre1–12 ahk3–3, 3.01 ± 0.35; ahk2–2 ahk3–3, 2.69 ± 0.35; and cre1–12 ahk2–2 ahk3–3, 0.65 ± 0.09.
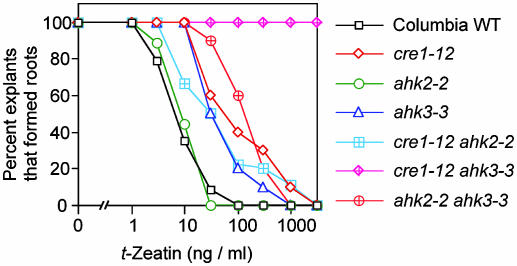
Cytokinin Sensitivity in the Adventitious Root Formation Assay. Cytokinins normally inhibit adventitious root formation near the cut end of hypocotyls (26). The cre1–12 and ahk3–3 single mutants were less sensitive to cytokinins in the adventitious root formation assay, whereas the ahk2–2 mutant exhibited a normal sensitivity (Fig. 5). The effect of mutations in both CRE1 and AHK3 was synergistic; that is, cre1–12 ahk3–3 roots were completely resistant to all cytokinin concentrations tested (up to a nonphysiological concentration of 3 μg/ml t-zeatin) (Fig. 5). These data indicate that CRE1 and AHK3 are key regulators of cytokinin-induced inhibition of adventitious root formation in Arabidopsis.
Fig. 5.
Cytokinin inhibition of adventitious root formation in cytokinin-receptor mutants. The percent of explants per genotype producing adventitious roots on increasing concentrations of t-zeatin is presented. At least eight plants were used for each data point.
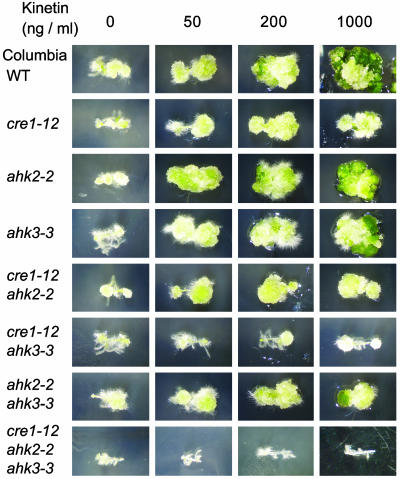
Cytokinin Sensitivity in the Callus Induction Assay. Cytokinins normally stimulate cell division and greening of calli (1). Similar to previously published work (8), cytokinin-induced cell division and greening of hypocotyl-derived calli were partially inhibited in cre1–12 mutants (Fig. 6), as well as in cre1–10 and cre1–11 mutants (data not shown). The ahk2–2 and ahk3–3 mutants responded normally to cytokinin in this assay (Fig. 6). Similar results were seen with ahk2–1, ahk3–1, and ahk3–2 mutants (data not shown). Mutations in AHK2 and AHK3, in combination with a mutation in CRE1, enhanced the effect of the cre1–12 mutation (Fig. 6). Finally, the triple cytokinin receptor mutant showed no significant response in this assay (Fig. 6). These results indicate that CRE1, AHK2, and AHK3 have redundant function in callus induction.
Fig. 6.
Induction of callus formation on hypocotyl segments, from cytokinin-receptor mutants, on different concentrations of kinetin.
Cytokinin Sensitivity in the Shoot Formation Assay. Cytokinins induce shoot formation and inhibit root formation on calli (27). The cre1–12, ahk2–2, and ahk3–3 single mutants exhibited normal or slightly reduced sensitivity to cytokinins in a shoot induction assay (see Fig. 12, which is published as supporting information on the PNAS web site). Similar results were seen for the cre1–10, cre1–11, ahk2–1, ahk3–1, and ahk3–2 single mutants (data not shown). Additive effects were observed for all CRE-family mutant combinations (Fig. 12). This result indicates that CRE1, AHK2, and AHK3 have redundant functions in cytokinin-induced shoot formation.
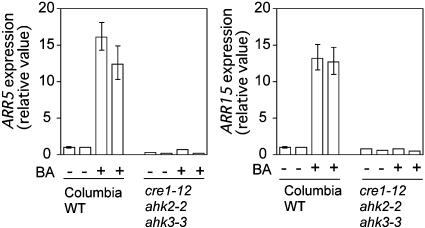
Cytokinin Induction of Primary-Response Genes Is Absent in the Triple Mutant. Cytokinins normally induce the transcription of type A response regulator genes in Arabidopsis (28, 29). To determine whether induction of these cytokinin primary-response genes was compromised in the triple mutant, reverse transcription, and real-time quantitative PCR analysis was performed on RNA prepared from WT and cre1–12 ahk2–2 ahk3–3 triple mutants, before and after a 30-min cytokinin treatment. Cytokinin treatment of WT seedlings induced transcription of the ARR5 and ARR15 transcripts by ≈14- and 13-fold, respectively (Fig. 7). Cytokinin treatment of the triple mutant produced no change in ARR5 or ARR15 transcript levels (Fig. 7).
Fig. 7.
Induction of cytokinin primary-response genes in the triple mutant. Fold induction of the ARR5 and ARR15 (Arabidopsis Response Regulator) transcripts in WT (Col) and the cre1–12 ahk3–3 ahk2–2 (Col) triple mutant, in response to BA, is presented. Results are based on real-time quantitative PCR analysis, and normalized to the SHR (SHORT ROOT) transcript level.
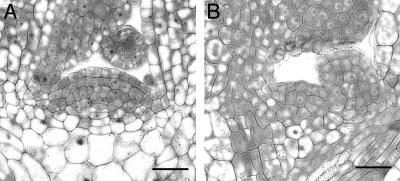
The Size and Activity of SAMs Are Decreased in the Triple Mutant. The diameter of the SAM was almost three times smaller in the triple mutant (29 ± 7 μm; n = 5) when compared to WT (82 ± 7 μm; n = 9) (Fig. 8). Fewer cell layers were seen in the SAM of triple mutants, as well as fewer cells per layer. One function of the SAM is to produce leaf primordia. Typically, plants with an enlarged SAM have a shorter interval of leaf production, or more rapid plastochron, than plants with a smaller SAM (5, 30, 31). As expected from the reduced SAM, the triple mutants had a prolonged plastochron with respect to WT plants. The leaf number of the cre1–12 ahk2–2 ahk3–3 (Col) triple mutants was 4.0 ± 0.0 (mean ± standard deviation, n = 7) after 11 days of culture, whereas that of WT (Col) was 9.13 ± 0.83 (n = 7). The leaf numbers of single and double mutants were not significantly different from those of WT.
Fig. 8.
Microscopic analysis of median longitudinal sections from the SAM of WT (Col) (A) and the cre1–12 ahk2–2 ahk3–3 (Col) mutant (B). (Bar = 25 μm.)
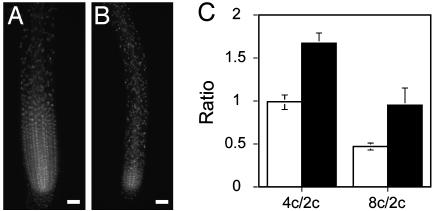
The size and activity of the root apical meristem of the triple mutant were also decreased with respect to WT plants (Fig. 9). The reduction in activity was seen in fluorescence-activated cell sorting experiments of root cells whose DNA had incorporated SYBR green I dye (see Fig. 13, which is published as supporting information on the PNAS web site). As shown in Fig. 9C, the triple mutant had a reduced diploid (2C) content of DNA with respect to WT plants, indicating that root cells of the triple mutant are delayed in the transition from G2→M phase of the cell cycle.
Fig. 9.
Root meristem activity of the triple mutant. (A and B) One microgram per milliliter 4′,6-diamidino-2-phenylindole staining of root cells of WT (Col) (A) and cre1–12 ahk2–2 ahk3–3 (Col) triple mutant (B). (Bars = 50 μm.) (C) tetraploid (4C/2C) and octaploid (8C/2C) ratios ± standard deviation (n = 4). Open column, WT; filled column, cre1–12 ahk2–2 ahk3–3 mutant.
Discussion
The main purpose of this study was to determine whether CRE-family members are the only cytokinin receptors in Arabidopsis by identifying a plant lacking expression of all three genes, and noting any developmental consequences, should this plant be viable. Indeed, plants homozygous for a T-DNA insertion in CRE1, AHK2, and AHK3 were recovered. The severe developmental abnormalities of these plants confirm that cytokinins are key regulators of plant growth and development. However, the phenotype of the triple mutant calls into question the longstanding belief that cytokinins are essential plant hormones, because an embryo is formed and a seedling produced without expression of the three known cytokinin receptors. It should be noted that our results do not eliminate the possibility that additional yet-unknown cytokinin receptors function in gametogenesis and embryogenesis.
Functions of CRE-Family Genes in the Root. The resistance of cre1 mutants to cytokinin-induced inhibition of root growth and adventitious root formation indicates that CRE1 functions dominate in the root. Mutations in either AHK2 or AHK3 had only minor effects on cytokinin-induced inhibition of root growth. However, either of these mutations in combination with the cre1 mutation enhanced the effect of cre1 mutation, suggesting that AHK2, AHK3, and CRE1 have redundant functions in the root. The important roles cytokinins play in root development can best be seen in the triple mutant, which has shorter, narrower roots than do WT plants (data not shown).
The size and activity of the root apical meristem were markedly reduced in the triple mutant. Cytokinins were first discovered for their role in promoting cell division (1) and have since been implicated in stimulating both the G1→S phase transition (32) as well as the G2→M phase transition of the cell cycle (33). The reduced meristem activity seen in the triple mutant roots seems to be a consequence of a delay in the G2→M phase, as demonstrated by FACS experiments. Interestingly, a sharp increase in the levels of zeatin, zeatin riboside, and zeatin riboside-5′-monophosphate was reported in tobacco cell cultures at the G2→M phase transition (34).
Our data are consistent with a role for cytokinins as positive regulators of root apical meristem activity. This is inconsistent with the findings of Werner et al. (35, 36), who demonstrated that over-expression of several members of the Arabidopsis cytokinin oxidase family led to an increase in root meristem size and activity. A possible explanation for this difference is to assume that cytokinins have two opposing effects on root growth, one inhibitory and one stimulatory. An inhibitory effect on root elongation could be mediated by cytokinin-induced ethylene production, because it is known that cytokinins induce ethylene production, ethylene inhibits root growth, and roots of ethylene-resistant mutants are also resistant to cytokinins (37). Another role for cytokinins in roots could be as a stimulator of cell division. Most likely, cell-division defects in the root occur only when cytokinin signaling is severely inhibited, as seen in the triple mutant.
CRE1, AHK2, and AHK3 Functions in the Shoot. The observation that ahk2 ahk3 double mutants have shorter inflorescence stems and smaller leaves than do WT plants, yet normal root growth, indicates that AHK2 and AHK3 functions dominate in the shoot. The severely stunted growth of the aerial portions of triple cytokinin receptor mutants indicates a redundant function for CRE1 in the shoot as well. Histological analyses showed that the SAM of the triple mutant was 3-fold smaller than normal. The organization of the SAM also appeared to be disrupted, because only the outermost layer of the tunica (L1) was recognizable. The interval of leaf formation was prolonged in the triple mutant, providing evidence of reduced SAM activity. Thus, our results support the finding of Werner et al. (35, 36) that cytokinins play a positive role in the SAM.
CRE1, AHK2, and AHK3 Function in de Novo Organ Formation. Since the late 1950s, scientists have recognized the ability of relative concentrations of the hormones cytokinin and auxin to induce plant cells to form particular tissues: undifferentiated callus, shoot structures, root structures, or a whole plant (21, 27). The ability to respond appropriately to cytokinin in an organ induction assay was retained in the cre1, ahk2, and ahk3 single mutants, suggesting redundant functions for the CRE-family members in organ formation. Among the single mutants, callus formation was most compromised in the cre1 mutant, indicating that CRE1 may play an important role in the process of dedifferentiation. The ability to appropriately respond to cytokinin in callus and shoot induction assays was lost in all three double mutant combinations, indicating that no single cytokinin receptor is sufficient for organ formation. Strikingly, the triple mutant did not respond to cytokinins at all, indicating that there may be no other cytokinin receptors that function in cell division and differentiation under tissue culture conditions.
What Is the Role of Cytokinins in the Formation of a Basic Vegetative Body Plan? The complete lack of cytokinin responses in the triple mutant, including the absence of cytokinin primary-response gene induction, could suggest that no other mechanism for cytokinin sensing exists in these plants. The retarded growth and sterility of the triple mutants indicate that cytokinins are very important growth regulators. In light of the inability of the triple mutant to form organs in tissue culture, that these plants can germinate and produce the basic plant organs induced in vitro by cytokinin and auxin is surprising. One possible explanation for this result is that, despite the well documented requirement for cytokinin in organogenesis during tissue culture, cytokinin-mediated regulation may not be required in planta for the formation of a basic vegetative body plan. Less radical interpretations include that there is another cytokinin receptor important for the earliest stages of plant development, or that the cytokinin production and responses of maternal tissues are sufficient for gametogenesis, embryogenesis, and germination of diploid offspring.
Supplementary Material
Acknowledgments
We thank Ryoko Kajita for RNA purification, Kirsi Törmäkangas for plant materials, and Dana B. Steien and Mirkka Kivimaki for excellent technical assistance. This work was supported by Grants-in-Aid for Scientific Research 12142207 and 15107001 (to T. Kakimoto); a Japan Society for the Promotion of Science Research Fellowship for Young Scientists (to M.H.); National Research Service Award T32 GM072125 from the National Institute of General Medical Sciences (to M.S.P.); research funds provided to M.R.S. by the University of Wisconsin–Madison Graduate School, the National Science Foundation, and the U.S. Department of Energy; and research funds provided to Y. Helariutta from the Academy of Finland.
Abbreviations: HK, histidine kinase; AHK2, Arabidopsis HK2; AHK3, Arabidopsis HK3; BA, benzyl adenine; Col, Columbia; CRE1, cytokinin response 1; SAM, shoot apical meristem; GM, germination medium; MS, Murashige and Skoog salts; GUS, β-glucuronidase; t-zeatin, trans-zeatin.
References
- 1.Miller, C. O., Skoog, F., Okumura, F. S., Saltza, M. H. V. & Strong, F. M. (1956) J. Am. Chem. Soc. 78, 1375–1380. [Google Scholar]
- 2.Rupp, H.-M., Frank, M., Werner, T., Strnad, M. & Schmülling, T. (1999) Plant J. 18, 557–563. [DOI] [PubMed] [Google Scholar]
- 3.Smigocki, A., J. W. Neal, J., McCanna, I. & Douglass, L. (1993) Plant Mol. Biol. 23, 325–335. [DOI] [PubMed] [Google Scholar]
- 4.Li, Y., Hagen, G. & Guilfoyle, T. J. (1992) Dev. Biol. 153, 386–395. [DOI] [PubMed] [Google Scholar]
- 5.Chaudhury, A. M., Letham, S., Craig, S. & Dennis, E. S. (1993) Plant J. 4, 907–916. [Google Scholar]
- 6.Mähönen, A. P., Bonke, M., Kauppinen, L., Riikonen, M., Benfey, P. N. & Helariutta, Y. (2000) Genes Dev. 14, 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franco-Zorrilla, J. M., Martin, A. C., Solano, R., Rubio, V., Leyva, A. & Paz-Ares, J. (2002) Plant J. 32, 353–360. [DOI] [PubMed] [Google Scholar]
- 8.Inoue, T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., Tabata, S., Shinozaki, K. & Kakimoto, T. (2001) Nature 409, 10601–10603. [DOI] [PubMed] [Google Scholar]
- 9.Ueguchi, C., Sato, S., Kato, T. & Tabata, S. (2001) Plant Cell Physiol. 42, 751–755. [DOI] [PubMed] [Google Scholar]
- 10.To, J. P. C., Haberer, G., Ferreira, F. J., Deruère, J., Mason, M. G., Schaller, G. E., Alonso, J. M., Ecker, J. R. & Kieber, J. J. (2004) Plant Cell 16, 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakai, H., Honma, T., Aoyama, T., Sato, S., Kato, T., Tabata, S. & Oka, A. (2001) Science 294, 1519–1521. [DOI] [PubMed] [Google Scholar]
- 12.Osakabe, Y., Miyata, S., Urao, T., Seki, M., Shinozaki, K. & Yamaguchi-Shinozaki, K. (2002) Biochem. Biophys. Res. Commun. 293, 806–815. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki, T., Ishikawa, K., Yamashino, T. & Mizuno, T. (2002) Plant Cell Physiol. 43, 123–129. [DOI] [PubMed] [Google Scholar]
- 14.Hwang, I. & Sheen, J. (2001) Nature 413, 383–389. [DOI] [PubMed] [Google Scholar]
- 15.Kakimoto, T. (2003) Annu. Rev. Plant Biol. 54, 605–627. [DOI] [PubMed] [Google Scholar]
- 16.Hutchison, C. E. & Kieber, J. J. (2002) Plant Cell 14, S47–S59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki, T., Miwa, K., Ishikawa, K., Yamada, H., Aiba, H. & Mizuno, T. (2001) Plant Cell Physiol. 42, 107–113. [DOI] [PubMed] [Google Scholar]
- 18.Yamada, H., Suzuki, T., Terada, K., Takei, K., Ishikawa, K., Miwa, K., Yamashino, T. & Mizuno, T. (2001) Plant Cell Physiol. 42, 1017–1023. [DOI] [PubMed] [Google Scholar]
- 19.Kakimoto, T. (1996) Science 274, 982–985. [DOI] [PubMed] [Google Scholar]
- 20.Urao, T., Yakubov, B., Satoh, R., Yamaguchi-Shinozaki, K., Seki, M., Hirayama, T. & Shinozaki, K. (1999) Plant Cell 11, 1743–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murashige, T. & Skoog, F. (1962) Physiol. Plant. 15, 473–497. [Google Scholar]
- 22.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- 23.Miyawaki, K., Matsumoto-Kitano, M. & Kakimoto, T. (2004) Plant J. 37, 128–138. [DOI] [PubMed] [Google Scholar]
- 24.Helariutta, Y., Fukaki, H., Wysocka-Diller, J., Nakajima, K., Jung, J., Sena, G., Hauser, M. T. & Benfey, P. N. (2000) Cell 101, 555–567. [DOI] [PubMed] [Google Scholar]
- 25.Shpak, E. D., Lakeman, M. B. & Torii, K. U. (2003) Plant Cell 15, 1095–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuroha, T., Kato, H., Asami, T., Yoshida, S., Kamada, H. & Satoh, S. (2002) J. Exp. Bot. 53, 2193–2200. [DOI] [PubMed] [Google Scholar]
- 27.Skoog, F. & Miller, C. O. (1957) Symp. Soc. Exp. Biol. 11, 118–131. [PubMed] [Google Scholar]
- 28.D'Agostino, I. B., Deruère, J. & Kieber, J. J. (2000) Plant Physiol. 124, 1706–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiba, T., Yamada, H. & Mizuno, T. (2002) Plant Cell Physiol. 43, 1059–1066. [DOI] [PubMed] [Google Scholar]
- 30.Itoh, J.-I., Kitano, H., Matsuoka, M. & Nagato, Y. (2000) Plant Cell 12, 2161–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conway, L. J. & Poethig, R. S. (1997) Proc. Natl. Acad. Sci. USA 94, 10209–10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riou-Khamlichi, C., Huntley, R., Jacqmard, A. & Murray, J. A. H. (1999) Science 283, 1541–1544. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, K., Letham, D. S. & John, P. C. L. (1996) Planta 200, 2–12. [DOI] [PubMed] [Google Scholar]
- 34.Redig, P., Shaul, O., Inze, D., Montagu, M. V. & Onckelen, H. V. (1996) FEBS Lett. 391, 175–180. [DOI] [PubMed] [Google Scholar]
- 35.Werner, T., Motyka, V., Strnad, M. & Schmülling, T. (2001) Proc. Natl. Acad. Sci. USA 98, 10487–10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Werner, T., Motyka, V., Laucou, V., Smets, R., Onckelen, H. V. & Schmülling, T. (2003) Plant Cell 15, 2532–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cary, A. J., Liu, W. & Howell, S. H. (1995) Plant Physiol. 107, 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.