Abstract
Potassium (K+) is an essential nutrient required by plants in large quantities, but changes in soil concentrations may limit K+ acquisition by roots. It is not known how plant root cells sense or signal the changes that occur after the onset of K+ deficiency. Changes in the kinetics of Rb+ uptake in Arabidopsis roots occur within 6 h after K+ deprivation. Reactive oxygen species (ROS) and ethylene increased when the plants were deprived of K+. ROS accumulated in a discrete region of roots that has been shown to be active in K+ uptake and translocation. Suppression of an NADPH oxidase in Arabidopsis (rhd2), which is involved in ROS production, prevented the up-regulation of genes that are normally induced by K+ deficiency, but the induction of high-affinity K+ transport activity was unchanged. Application of H2O2 restored the expression of genes induced by K+ deficiency in rhd2 and was also sufficient to induce high-affinity K+ transport activity in roots grown under K+-sufficient conditions. ROS production is an early root response to K+ deficiency that modulates gene expression and physiological changes in the kinetics of K+ uptake.
Potassium (K+) is essential to and required in large quantities by plants. To ensure the adequate supply, plants have a number of redundant mechanisms for K+ acquisition and translocation (1–3). Whereas much is known about K+ transport mechanisms, little is known about how plant cells sense and respond to changes in the K+ concentrations encountered in their environment. At the whole-plant level, K+ deficiency is known to enhance root respiration (4). This response suggests a shift in plant metabolism required for mineral acquisition or compensation for increased stress. External K+ concentrations alter the kinetics of uptake, which may be due to allosteric regulation by internal changes in K+ concentrations (5). Specific proteins in the plasma membrane and tonoplast were shown to be produced in response to K+ deficiency, indicating the synthesis of new transport proteins (6–8). Whereas changes in respiration and membrane transport occur in response to K+ deficiency, the cell biological events that trigger these changes have not been elucidated.
In Escherichia coli, some of the molecules involved in the sensing and signaling of K+ deficiency have been elucidated (9). KdpD is a sensor kinase that undergoes autophosphorylation and transfers a phosphoryl group to a response regulator KdpE (10). The response regulator controls the expression of an operon coding for the high-affinity K+ uptake system in E. coli. The sensor kinase is thought to transduce changes in turgor caused by low K+ (11). It is not clear whether similar mechanisms are involved in transducing K+ deprivation in plants.
Roots are the primary organs involved in mineral acquisition for plants and function at the interface with the rhizosphere. Although genes whose expression is related to external changes in nutrient composition have been identified, the cascade of cellular responses involved in sensing and signaling nutrient deficiency has not been elucidated (12–18). To identify the initial cellular responses to K+ deprivation, we performed preliminary microarray experiments on Arabidopsis roots and developed the working hypothesis that reactive oxygen species (ROS) production and ethylene may be involved in root response to K+ deprivation. In this study, we used uptake kinetics, biochemical analyses, an NADPH oxidase mutant, and a set of K+-responsive genes to identify ROS as one component of the signal transduction pathway in roots deprived of K+. To our knowledge, no comparable study on nutrient deprivation has implicated a link between ROS and nutrient deficiency.
Materials and Methods
Plant Material and Growth Conditions. Plants were grown in nutrient solutions by using a rock wool system (19) for 6–8 weeks at 22°C with 8 h daylight at 200 μmol·m–2·s–1. Maize seeds (Zea mays L. cv. FR27 × FRM017) were also planted in rock wool plugs in the same nutrient solution as Arabidopsis and grown at 22°C, humidity 50%, 16 h light/8 h dark, and 200 μmol·m–2·s–1. Two-week-old maize plants were used for the H2O2 measurement.
Root Growth Assay and Rb+ Uptake Assay. For the root growth assay, Arabidopsis was planted on nutrient containing (19) 1.2% Seakem LE Agarose (BMA, Rockland, ME) and 2% sucrose. Ten seedlings were transferred to full nutrient or K+-deprived plates, and their growth was measured every day for 10 days. The result of the root growth assays is the average of three experiments.
On the day of each Rb+ uptake experiment, control solutions were changed in the morning to solutions with and without K+ as described (20). After 6 h, plants were moved to beakers to equilibrate with the Rb+ at the different concentrations used for 5 min. Trace amounts of 86Rb+ were added to start a 10-min uptake period. After the uptake period, roots were desorbed for 10 min in 0.5 mM CaSO4 at 22°C. Roots were then blotted, weighed, and placed in scintillation vials containing scintillant. Radioactivity in the roots was counted by using a Beckman LS6500 scintillation counter. The experiments were repeated on two different days with plants from four different hydroponic tanks (n = 4). From each tank subsamples of two to three plants were used at each concentration at which uptake was measured. Data were fitted with a one-site binding model (hyperbola) by using the equation Y = (Vmax· X)/(Km + X); where X was the Rb+ concentration and Y was the uptake rate. The nonlinear regression was performed by using prism software (GraphPad, San Diego), which finds the values of Vmax and Km at which the sum of squares are the smallest and calculates the SE for those kinetic parameters.
RT-PCR Analysis and H2O2 Treatment. RNA was extracted from the roots of plants that were deprived of K+ or supplied with sufficient K+ for 6 and 30 h by using RNAwiz. cDNAs were synthesized by using the SuperScript first-strand synthesis system (Invitrogen) and an oligo(dT)18 primer. For the H2O2 treatments, 1 mM (Sigma) was added to the nutrient medium for 6 or 30 h. For the diphenylene iodonium (DPI) experiment (Sigma), 100 μM DPI was supplied in the nutrient medium for 2 h before K+ deprivation. Plants were then transferred to K+-deficient or -sufficient nutrient solution for 6 h before harvest. Each PCR/experiment was performed three times with different cDNA sets from independent biological replicates. All PCR products were sequenced to ensure that the correct transcript was detected. photoshop 7.0 (Adobe Systems, San Jose, CA) was used to determine the mean relative intensity of each amplification product.
Ethylene Measurement. Six-week-old hydroponic cultured Arabidopsis was transferred to 40-ml vials containing K+ -sufficient or -deficient nutrient medium for 6 and 30 h. One milliliter of the headspace was taken from the vials, and ethylene concentration was measured with a Shimadzu (Kyoto) GC-8A gas chromatograph. The data were analyzed by ANOVA, and means were compared by using Student's t test. Experiments were repeated three times.
H2O2 Measurement and Localization. Six-week-old plants were used to measure H2O2 production in controls and after K+ deficiency by using an Amplex red hydrogen peroxide/peroxidase assay kit (Molecular Probes). The data were analyzed by ANOVA, and means were compared by using Student's t test. Experiments were performed three times for Arabidopsis and maize.
Five-day-old plants were used to localize the generation of ROS in the root. For experiments with 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester; (Molecular Probes), plants were incubated on K+-deficient or -sufficient plates for 30 h (21). All fluorescence images were obtained with a Nikon SMZ1500 microscope and a QImaging (Burnaby, BC, Canada) Retiga cooled mono 12-bit camera.
Results
K+ Deficiency Affects Lateral Root Growth and Alters the Kinetics of Uptake. Arabidopsis seedlings were transferred to plates with and without K+ 3 days after germination. Final primary root length was the same 10 days after transfer to plates containing low K+ and sufficient K+ (Table 1). However, lateral root length and the numbers of lateral roots decreased in plants on low K+ medium (Table 1). These data show that in the long term, root growth is affected by the lack of K+.
Table 1. Root growth under K+-deficient and -sufficient conditions.
| Condition | Final primary root length, cm | Lateral root length, cm per cm of primary root | Total no. lateral roots |
|---|---|---|---|
| +K+ | 3.23 ± 0.06 | 1.04 ± 0.02 | 15 ± 0.4 |
| -K+ | 3.21 ± 0.09 | 0.46 ± 0.03 | 7 ± 0.5 |
| NS | P < 0.001 | P < 0.001 |
Means ± SE are shown. Results are the average of three experiments (n = 4 plates, 5 plants per plate). Comparison between roots grown with and without K+ were performed by one-way ANOVA. NS, no significant differences. P values shown indicate highly significant differences between treatments in lateral roots.
To demonstrate that after 6 h, K+ deprivation alters root function, we measured the kinetics of K+ uptake by using Rb+ as an analog for K+ (22). Classical studies (2) have shown that large differences in kinetic parameters are induced in many plant species by changes in the composition of the growth medium. In Arabidopsis, kinetic uptake studies of K+-deprived plants have not been published. Our studies using Rb+ uptake show that a component of high-affinity uptake is induced after 6 h of K+ deprivation. Fig. 1A shows that the kinetics of uptake after deprivation were saturable at low concentrations of Rb+ with a Km of 25 ± 9 μM. In contrast, the plants grown with sufficient amounts of K+ showed linear rates of K+ uptake (Fig. 1B) over the same concentration range.
Fig. 1.
Kinetics of Rb+ uptake in Arabidopsis roots. (A) Rb+ uptake rates in Col-0 roots starved of K+ for 6 h showed a saturable kinetic component; Km = 25 ± 9 μM. (B) Rb+ uptake rates in Col-0 roots grown under K+-sufficient conditions do not show saturable kinetics over the concentration range tested. Mean uptake rates at each concentration are shown ± SE; n = 4 for each concentration.
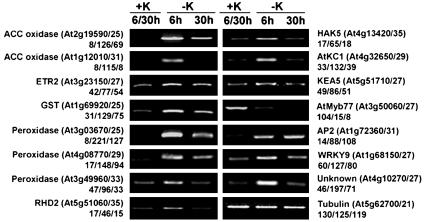
Identification of Genes Whose Expression Changes upon K+ Deprivation. Preliminary microarray experiments of changes in Arabidopsis root gene expression identified genes that were related to ROS production, ethylene synthesis, K+ transport, and transcription. From these experiments, we identified up-regulated genes related to ROS metabolism and ethylene synthesis (Fig. 2). The expression of a gene encoding the K+ transporter HAK5 increased, which confirms previous results (20). Two other genes related to K+ transport KEA5 and AtKC1 were also up-regulated (Fig. 2). An ethylene-responsive AP2 transcription factor, a WRKY-type transcription factor (WRKY9), and a Myb-type transcription factor (AtMyb77), were up- or down-regulated (Fig. 2). One unknown gene At4g10270 was strongly up-regulated by K+ deficiency and is homologous to a tomato wound-inducible gene (Fig. 2). We also measured the changes in gene expression at early time points for the genes in Fig. 2. We found no change in gene expression in this group of genes (Fig. 2) after 30 min. After 2 h, the expression of many of these genes was up-regulated, including At2g19590, At3g03670, At3g23150, At1g72360, At1g68150, At4g32650, and a single gene, At3g50060, was down-regulated. Plants used in these experiments were analyzed by using inductively coupled plasma-MS, which showed that the mineral composition (except for K+) was not altered by the short-term treatments.
Fig. 2.
Changes in gene expression upon K+ deprivation monitored by RT-PCR. RNA was extracted from Arabidopsis roots grown under K+-sufficient conditions (+K), 6 h after K+ deprivation (6 h), and 30 h after K+ deprivation (30 h). β-Tubulin was used for cDNA normalization. The gene number follows a description of the gene function, and the number of PCR cycles used for amplification follows the gene number. Mean relative intensity for each RT-PCR amplification product is shown below the gene number or description.
K+ Deficiency Alters in Vivo Ethylene Level. Because several genes encoding enzymes that are involved in ethylene production were up-regulated by K+ deprivation (Fig. 2), we determined that ethylene production increased after K+ starvation by measuring ethylene production in plants 6 and 30 h after K+ deprivation. The result of three experiments showed that ethylene production increased by a factor of 1.5–2 after K+ starvation (Fig. 3).
Fig. 3.
Ethylene production in Arabidopsis plants grown under hydroponic conditions with K+ or without K+ for 6 and 30 h. Experiments were performed three times, and the mean (n = 6 plants ± SE) is shown from one experiment. Mean comparison showed that ethylene levels were significantly higher than control levels at 6 (P < 0.01) and 30 h (P < 0.001) after deprivation.
H2O2 Production Increases After K+ Deficiency. Several genes related to ROS metabolism were significantly up-regulated by K+ starvation (Fig. 2), and therefore H2O2 levels were measured in roots after K+ starvation by using multiple plants in three sets of independent biological replicates. Leaves and roots of Arabidopsis and maize that were deprived of K+ were analyzed with Amplex red reagent (10-acetyl-3,7-dihydrophenoxazine). Maize was used to determine whether a monocot with large roots and seed stores responds in a way similar to Arabidopsis. The production of H2O2 in roots was higher at 6 h than at 30 h. The level of H2O2 in Arabidopsis leaves increased only slightly at 6 h, but increased by a factor of two 30 h after deprivation (Fig. 4). The trends in H2O2 production in Arabidopsis and maize were similar, but Arabidopsis had a higher amount of H2O2 in leaves than maize, and Arabidopsis H2O2 levels in the roots were lower (Fig. 4). The increase in H2O2 after K+ deprivation suggested that it might play a role in cellular signaling of K+ deprivation.
Fig. 4.
H2O2 production after K+ deprivation in Arabidopsis and maize. Experiments were performed three times for Arabidopsis and maize, and one representative set of results are shown. +K, Plants grown with 1.75 mM KCl; –K, plants that were deprived of K+. The data were analyzed by ANOVA. *, P < 0.05 and **, P < 0.01 between treatments.
To investigate whether ROS are localized to a specific region of the root, an ROS-sensitive dye was loaded into root cells. Roots were deprived of K+ for 30 h. ROS increased after K+ deprivation in a discrete region of the root just behind the elongation zone (Fig. 5 C and D). In contrast, only small amounts of ROS accumulated in K+-sufficient roots (Fig. 5 A and B) in the same region as in K+-deficient plants, just behind the zone of elongation. There was a significant increase in ROS accumulation in plants grown under K+-deficient conditions as compared with K+-sufficient conditions.
Fig. 5.
Localization of ROS in Arabidopsis roots during K+ deprivation. Pseudocolor images of bright field (white) and fluorescence (red) are shown for K+-sufficient (A and B) roots and for K+-deficient roots with 50 μM 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (C and D) and without it (E and F). Fluorescence indicates the presence of ROS. (Scale bar, 5 mm.) Ten control and 10 K+-deprived roots showed similar results.
H2O2 Modulates Gene Expression After K+ Deprivation. Biochemical assays confirmed that ROS levels increased in response to K+ deprivation in a specific location in roots, just behind the zone of elongation. To test whether H2O2 might regulate gene expression in response to low K+, we deprived plants of K+ and then studied the effects on the expression of a selected number of K+-responsive genes in wild-type Arabidopsis Col-0 with an NADPH oxidase inhibitor and in the Arabidopsis mutant rhd2, in which a single NADPH oxidase (At5g51060) is not functional (21). The expression of this NADPH oxidase increases upon K+ deprivation (Fig. 2).
Treatment with DPI caused a higher basal level of expression of most genes tested as compared with the non-DPI-treated controls, but abolished or greatly reduced the up-regulation of six of the nine genes tested (Fig. 6). In contrast to the wild type, in rhd2 plants we found that two peroxidases, the WRKY9 transcription factor, the K+ transporters KEA5 and Hak5, and an unknown gene, were not up-regulated in response to K+ deprivation. This result suggests that H2O2 produced by the RHD2 NADPH oxidase regulates the expression of these genes in response to K+ deficiency. We also found that the regulation of certain genes including a GST, an AP2 transcription factor, and an 1-aminocyclopropane-a-carboxylic acid (ACC) oxidase were the same in rhd2 mutant as compared to the wild type (Fig. 6), and therefore may not be directly modulated by H2O2.
Fig. 6.
Differential gene expression by K+ deprivation and the NADPH oxidase inhibitor DPI and the NADPH oxidase mutant rhd2. RT-PCR analysis of selected K+-responsive genes in rhd2 and Col-0 plants 6 and 30 h after K+ deprivation with and without H2O2 or added DPI. The roots of hydroponic cultured plants were treated with 1 mM H2O2 for 6 or 30 h, and 100 μM DPI for 2 h before K+ starvation. β-tubulin was used for cDNA normalization. The gene number follows a description of the gene function and the number of PCR cycles used for amplification. +K, Plants grown with sufficient K+ (1.75 mM); –K, plants deprived of K+. Mean relative intensity for each RT-PCR product is shown in Table 2, which is published as supporting information on the PNAS web site.
To further test the effects of H2O2 on the expression of selected genes, H2O2 was applied to roots for 6 and 30 h in K+-sufficient and -deficient conditions. In some cases, application of H2O2 induced the expression of these K+-responsive genes in the rhd2 mutant even under K+-sufficient conditions (Fig. 6). In other cases, H2O2 was required, but was not sufficient, for the induction of a peroxidase, Hak5, and an unknown gene at 6 and 30 h after deprivation. For these three genes, K+ deficiency and H2O2 are necessary for increased gene expression. These results show that H2O2 and K+ deprivation play an important role in the regulation of gene expression in response to K+ deficiency.
Relationship Between the Kinetics of Rb+ Uptake and H2O2. In response to K+ deprivation, the induction of some genes was altered in the rhd2 mutant. To test whether these changes in gene expression led to changes in the induction of high-affinity K+ uptake, we measured the kinetics of Rb+ uptake, which is commonly used as an analog for K+. After 6 h of K+ deprivation, we found that a component of high-affinity uptake was induced in rhd2 with Km = 27 ± 8 μM (Fig. 7A), which was similar to the Km for Rb+ uptake in Col-0 after 6 h of K+ deprivation (Fig. 1 A).
Fig. 7.
Kinetics of Rb+ uptake in Arabidopsis roots. (A) Rb+ uptake rates in rhd2 roots starved of K+ for 6 h showed a saturable kinetic component; Km = 27 ± 8 μM. (B) Rb+ uptake rates in Col-0 roots grown under K+-sufficient conditions and treated with 1 mM H2O2 for 30 h showed a saturable kinetic component; Km = 31 ± 20 μM. Mean uptake rates at each concentration are shown ± SE; n = 4 for each concentration.
To determine whether the application of H2O2 altered the kinetics of K+ uptake, we added 1 mM H2O2 to the nutrient solutions containing sufficient (1.75 mM K+) K+ in which Col-0 plants were growing. After 30 h of exposure to H2O2, the kinetics of Rb+ uptake by the roots of Col-0 plants were measured. We observed that the addition of H2O2 induced a high-affinity component of Rb+ uptake (Km = 31 ± 20 μM) even though the plants were growing in K+-sufficient conditions (Fig. 7B). The Km for Rb+ uptake over the concentration range tested was similar in the H2O2-treated plants (Fig. 1C) and in plants that were deprived of K+ (Fig. 1 A). These results suggest that H2O2 is sufficient for the induction of a component of high-affinity Rb+ uptake. Variation in Vmax of uptake between experiments could be attributed to the differences in the age of plants used in the different experiments (6–8 weeks old).
Discussion
Overall, the short-term effects of K+ deprivation are a subtle stress, because plant cells may have mechanisms in place to ensure that the cytoplasmic K+ concentration is maintained homeostatically (23, 24). The removal of K+ from the growth medium does not impose a large osmotic stress, but it leads to changes in the membrane polarization (25). To confirm that cellular changes occur after a brief period of K+ deprivation, we conducted classical kinetic uptake studies 6 h after deprivation. These studies on K+-deprived plants have been performed on other plant species, but there are no available results for such experiments on Arabidopsis. After 6 h of K+ deprivation, a high-affinity uptake component was induced in Arabidopsis roots; this finding confirmed that cellular changes occurred after a brief period of deprivation.
To understand how K+ deficiency alters root development over a longer period of deprivation, we measured root growth and lateral development. Our data showed that lateral root growth and number were reduced (Table 1). In contrast, low phosphate availability increased lateral root growth more than primary root growth (26, 27). Lateral root initiation was repressed after transfer to low-nitrogen plates by a high sucrose-to-nitrogen ratio in areas of new root growth (28, 29). This response was not due to nitrogen starvation alone, because lowering the sucrose concentration restored lateral root initiation even under low-nitrogen conditions. However, lateral root number was reduced in older regions of the root under nitrogen starvation (28).
Root development is affected by plant hormones as well as nutrients. Several studies have reported that increased ethylene levels inhibit root growth (30–33). Ethylene and auxin are essential for the development of root hairs in response to iron deficiency but are apparently not required for root hair development induced by phosphate deficiency (34). ACC is a precursor of ethylene that has been shown to inhibit lateral root formation (27). This precursor increased elongation in high phosphorus, but decreased elongation in low phosphorus (35). Nothing is known about the role that hormones play in root growth responses to K+ deficiency. Our results showed that lateral root growth was inhibited after K+ deprivation, and in the short term the expression of genes related to ethylene production and ethylene production increased. Because the ethylene precursor ACC inhibits lateral root number (27), it is also possible that the higher levels of ethylene under K+ deprivation led to reduced lateral root initiation in the response to K+ deprivation.
Changes in the expression of genes related to K+ transport were evident after K+ deprivation. The up-regulation of Hak5, which encodes a K+ ion transporter upon K+ deprivation was confirmed (20). The homologue of Hak5 in tomato was also regulated by nitrate, phosphate, and iron deprivation (12). The function of this transporter is unknown, but has been shown to transport K+ with high affinity (36). In addition, AtKC1, which is a K+ channel (37, 38) that is expressed in roots, and KEA5, which is suggested to be a K+/H+ antiporter (1), were also up-regulated, highlighting the changes in K+ transport across various membranes that occurs when plants are deprived of K+. Previous reports (38) have shown that AtKC1 is regulated by salt stress, but not strongly by K+ deprivation.
H2O2 has been shown to play a role in many different signaling pathways, including: ABA-mediated stomatal closure (39), auxin-regulated root gravitropism (40), responses to wounding, systemin, and methyl jasmonate (41, 42), and programmed cell death (43). Because recent studies confirm that ROS, notably H2O2, is a signaling molecule in plants, we tested whether ROS plays a role in signaling after K+ deprivation. Our studies show that H2O2 production increases after K+ deprivation and that the increase in ROS production is localized to a specific region of the root. The region of ROS accumulation in K+-starved roots was similar to the region where antioxidants such as ascorbate and dehydroascorbate accumulated in onion roots under control conditions (44). Increased ascorbate free radicals in this specific region were demonstrated to increase the uptake of sucrose, glucose, and nitrate in onion roots (45). In another study, (46) oxygen-free radicals in Arabidopsis roots were shown to cause a transient increase in K+ efflux. Although ROS may play different roles in ion transport, it is interesting to note that the region just behind the elongation zone (where we observed high concentrations of ROS) may be the most active in uptake and translocation to the shoots (47). The addition of 1 mM H2O2 to plants growing in K+-sufficient medium led to the induction of a component of high-affinity K+ uptake, which supports the conclusion that H2O2 is involved in K+ signaling after deprivation and leads to changes in transport processes.
H2O2 may play an important role in modulating the induction of some of the genes in Arabidopsis in response to K+ deficiency. The NADPH oxidase inhibitor DPI abolished or greatly reduced the increased expression of K+ ion transporter genes (Hak5 and KEA5), WRKY9, two peroxidases, and an unknown protein that were normally up-regulated after K+ deprivation. Recently Foreman et al. (21) showed that the basis for a root-hair-deficient mutant in Arabidopsis was the inactivation of NADPH oxidase (At5g51060, rhd2). In rhd2, the increased expression of several genes after K+ deprivation was abolished 6 and 30 h after deprivation. Those genes include: two K+ transporters, the WRKY9 transcription factor, two peroxidases, and an unknown protein. The expression of these genes in rhd2 could be restored by H2O2 or H2O2 in combination with K+ deprivation. The results with DPI and the rhd2 mutant were similar, and both results indicate that H2O2 plays a role in controlling the expression of certain genes in response to K+ deprivation. We also found that a component of high-affinity K+ uptake was induced in the rhd2 mutant by K+ deficiency. Therefore, the subset of genes that are controlled by the H2O2 may not be required for the induction of high-affinity K+ uptake. In the rhd2 mutant, many genes are still induced by K+ deficiency, and therefore the genes that are still induced by K+ deprivation may be involved in high-affinity K+ uptake. Alternatively, the component of high-affinity K+ uptake induced by K+ deprivation in the rhd2 mutant after 6 h of K+ deprivation may be due to the posttranslational modification of an existing transporter.
The cellular events that occur when plant root cells are deprived of K+ are poorly understood. We showed that preliminary gene expression data could lead to testable biochemical and physiological hypotheses related to cellular signaling in response to K+ deprivation. From these biochemical and physiological studies, we found that certain cellular responses to K+ deficiency in Arabidopsis roots are activated by ROS and perhaps ethylene. At this time, the specificity of the ROS-induced responses are not known, in part because little is known about nutrient signaling cascades in plants. We propose that these signals regulate changes in root function and growth. In the future, it will be important to understand how protein function is modulated by factors other than gene expression when plants are deprived of K+.
Supplementary Material
Acknowledgments
We thank Janet Oriatti for assistance in editing the manuscript; U. Hammes, E. Nielsen, and L. Xiong for critical comments; Teresa Thiel (University of Missouri, St. Louis) for the use of GC; Brett Lahner, David Salt, Lauren McIntyre, and Lisa Bono at Purdue University (West Lafayette, IN); and Howard Berg for microscopy assistance. R.S. was supported in part by a fellowship from the Korean Science and Engineering Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ROS, reactive oxygen species; DPI, diphenylene iodonium; ACC, 1-aminocyclopropane-a-carboxylic acid.
References
- 1.Maser, P., Thomine, S., Schroeder, J. I., Ward, J. M., Hirschi, K., Sze, H., Talke, I. N., Amtmann, A., Maathuis, F. J. M., Sanders, D., et al. (2001) Plant Physiol. 126, 1646–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kochian, L. V. & Lucas, W. J. (1988) in Advances in Botanical Research, ed. Callow, J. A. (Academic, London), Vol. 15, pp. 93–178. [Google Scholar]
- 3.Very, A. A. & Sentenac, H. (2003) Annu. Rev. Plant Biol. 54, 575–603. [DOI] [PubMed] [Google Scholar]
- 4.Singh, P. & Blanke, M. M. (2000) Plant Growth Regul. 32, 77–81. [Google Scholar]
- 5.Glass, A. D. M. (1976) Plant Physiol. 58, 33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glass, A. D. M. (1983) Annu. Rev. Plant Physiol. 34, 311–326. [Google Scholar]
- 7.Fernando, M., Mehroke, J. & Glass, A. D. M. (1992) Physiol. Plant. 100, 1269–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernando, M., Kulpa, J., Siddiqi, M. Y. & Glass, A. D. M. (1990) Plant Physiol. 92, 1128–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poolman, B., Blount, P., Folgering, J. H. A., Friesen, R. H. E., Moe, P. C. & van der Heide, T. (2002) Mol. Microbiol. 44, 889–902. [DOI] [PubMed] [Google Scholar]
- 10.Laimins, L. A., Rhoads, D. B., Altendorf, K. & Epstein, W. (1978) Proc. Natl. Acad. Sci. USA 75, 3216–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung, K., Veen, M. & Altendorf, K. (2000) J. Biol. Chem. 275, 40142–40147. [DOI] [PubMed] [Google Scholar]
- 12.Wang, Y. H., Garvin, D. F. & Kochian, L. V. (2002) Plant Physiol. 130, 1361–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond, J. P., Bennett, M. J., Bowen, H. C., Broadley, M. R., Eastwood, D. C., May, S. T., Rahn, C., Swarup, R., Woolaway, K. E. & White, P. J. (2003) Plant Physiol. 132, 578–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, R., Guegler, K., LaBrie, S. T. & Crawford, N. M. (2000) Plant Cell 12, 1491–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, Y. H., Garvin, D. F. & Kochian, L. V. (2001) Plant Physiol. 127, 345–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, R., Okamoto, M., Xing, X. & Crawford, N. M. (2003) Plant Physiol. 132, 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thimm, O., Essigmann, B., Kloska, S., Altmann, T. & Buckhout, T. J. (2001) Plant Physiol. 127, 1030–1043. [PMC free article] [PubMed] [Google Scholar]
- 18.Maruyama-Nakashita, A., Inoue, E., Watanabe-Takahashi, A., Yamaya, T. & Takahashi, H. (2003) Plant Physiol. 132, 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibeaut, D. M., Hulett, J., Cramer, G. R. & Seemann, J. R. (1997) Plant Physiol. 115, 317–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahn, S. J., Shin, R. & Schachtman, D. P. (2004) Plant Physiol. 134, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foreman, J., Demidchik, V., Bothwell, J. H. F., Mylona, P., Miedema, H., Torres, M. A., Linstead, P., Costa, S., Brownlee, C., Jones, J. D. G., et al. (2003) Nature 422, 442–446. [DOI] [PubMed] [Google Scholar]
- 22.Epstein, E., Rains, D. W. & Elzam, O. E. (1963) Proc. Natl. Acad. Sci. USA 49, 684–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker, D. J., Leigh, R. A. & Miller, A. J. (1996) Proc. Natl. Acad. Sci. USA 93, 10510–10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leigh, R. A. & Wyn Jones, R. G. (1984) New Phytol. 97, 1–13. [Google Scholar]
- 25.Spalding, E. P., Hirsch, R. E., Lewis, D. R., Qi, Z., Sussman, M. R. & Lewis, B. D. (1999) J. Gen. Physiol. 113, 909–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williamson, L. C., Ribrioux, P. C. P., Fitter, A. H. & Leyser, H. M. O. (2001) Plant Physiol. 126, 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez-Bucio, J., Hernandez-Abreu, E., Sanchez-Calderon, L., Nieto-Jacobo, M. F., Simpson, J. & Herrera-Estrella, L. (2002) Plant Physiol. 129, 244–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malamy, J. E. & Ryan, K. S. (2001) Plant Physiol. 127, 899–909. [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, H. & Forde, B. G. (2000) J. Exp. Bot. 51, 51–59. [PubMed] [Google Scholar]
- 30.Beaudoin, N., Serizet, C., Gosti, F. & Giraudat, J. (2000) Plant Cell 12, 1103–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen, H. & Grossmann, K. (2000) Plant Physiol. 124, 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Locke, J. M., Bryce, J. H. & Morris, P. C. (2000) J. Exp. Bot. 51, 1843–1849. [DOI] [PubMed] [Google Scholar]
- 33.Spollen, W. G., LeNoble, M. E., Samuels, T. D., Bernstein, N. & Sharp, R. E. (2000) Plant Physiol. 122, 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schimdt, W. & Schikora, A. (2001) Plant Physiol. 125, 2078–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma, Z., Baskin, T. I., Brown, K. M. & Lynch, J. P. (2003) Plant Physiol. 131, 1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubio, F., Santa-Maria, G. E. & Rodriguez-Navarro, A. (2000) Physiol. Plant. 109, 34–43. [Google Scholar]
- 37.Reintanz, B., Szyroki, A., Ivashikina, N., Ache, P., Godde, M., Becker, D., Palme, K. & Hedrich, R. (2002) Proc. Natl. Acad. Sci US. 99, 4079–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pilot, G., Gaymard, F., Mouline, K., Cherel, I. & Sentenac, H. (2003) Plant Mol. Biol. 51, 773–787. [DOI] [PubMed] [Google Scholar]
- 39.Pei, Z. M., Murata, Y., Benning, G., Thomine, S., Klusener, B., Allen, G. J., Grill, E. & Schroeder, J. L. (2000) Nature 406, 731–734. [DOI] [PubMed] [Google Scholar]
- 40.Joo, J. H., Bae, Y. S. & Lee, J. S. (2001) Plant Physiol. 126, 1055–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orozco-Cardenas, M. L., Narvaez-Vasquez, J. & Ryan, C. A. (2001) Plant Cell 13, 179–191. [PMC free article] [PubMed] [Google Scholar]
- 42.Orozco-Cardenas, M. & Ryan, C. A. (1999) Proc. Natl. Acad. Sci. USA 96, 6553–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Pinto, M. C., Tommasi, F. & De Gara, L. (2002) Plant Physiol. 130, 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Del Carmen Cordoba-Pedregosa, M., Cordoba, F., Villalba, J. M. & Gonzalez-Reyes, J. A. (2003) Plant Physiol. 131, 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gonzalez-Reyes, J. A., Hidalgo, A., Caler, J. A., Palos, R. & Navas, P. (1994) Plant Physiol. 104, 271–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Demidchik, V., Shabala, S. N., Coutts, K. B., Tester, M. A. & Davies, J. M. (2003) J. Cell Sci. 116, 81–88. [DOI] [PubMed] [Google Scholar]
- 47.Moritsugu, M., Shibasaka, M. & Kawasaki, T. (1993) Soil Sci. Plant Nutr. 39, 299–307. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.