Abstract
During catalytic activation of the spliceosome, snRNP remodeling events occur, leading to the formation of a 35S U5 snRNP that contains a large group of proteins, including Prp19 and CDC5, not found in 20S U5 snRNPs. To investigate the function of 35S U5 proteins, we immunoaffinity purified human spliceosomes that had not yet undergone catalytic activation (designated BΔU1), which contained U2, U4, U5, and U6, but lacked U1 snRNA. Comparison of the protein compositions of BΔU1 and activated B* spliceosomes revealed that, whereas U4/U6 snRNP proteins are stably associated with BΔU1 spliceosomes, 35S U5-associated proteins (which are present in B*) are largely absent, suggesting that they are dispensable for complex B formation. Indeed, immunodepletion/complementation experiments demonstrated that a subset of 35S U5 proteins including Prp19, which form a stable heteromeric complex, are required prior to catalytic step 1 of splicing, but not for stable integration of U4/U6.U5 tri-snRNPs. Thus, comparison of the proteomes of spliceosomal complexes at defined stages can provide information as to which proteins function as a group at a particular step of splicing.
Keywords: mass spectrometry, proteomics, Prp19/CDC5, 35S U5 snRNP, spliceosome
Introduction
Pre-mRNA splicing is catalyzed by the spliceosome, a highly dynamic RNP machine. The main components of the major spliceosome are the U1, U2, U4, U5, and U6 snRNPs (Burge et al, 1999). These RNA–protein complexes together with a large number of non-snRNP splicing factors totaling well over 120 distinct proteins (Hartmuth et al, 2002; Jurica et al, 2002; Makarov et al, 2002; Rappsilber et al, 2002; Zhou et al, 2002) associate with the pre-mRNA in an ordered manner (Burge et al, 1999). First, the U1 snRNP interacts with the 5′ splice site, followed by the stable association of U2 snRNP with the branch site to form spliceosomal complex A. The pre-assembled 25S U4/U6.U5 tri-snRNP is then recruited to form complex B. The latter is structurally rearranged to form the catalytically activated spliceosome, which subsequently catalyzes the first transesterification reaction, generating complex C. After the second step of splicing, the mRNA is released, the post-spliceosomal complex disassembles, and the snRNPs are recycled for new rounds of splicing. Thus, spliceosome formation goes through many intermediate stages, the most stable of which (e.g., A, B, and C complexes) can be detected biochemically, for example, by native gel electrophoresis. Many additional stable intermediates, such as the recently purified activated spliceosome B*, also appear to exist (Makarov et al, 2002; reviewed by Brow, 2002).
The most decisive step during the spliceosome maturation process is the conversion of complex B, which does not contain an active site, into the catalytically activated spliceosome B*, which is poised to catalyze both steps of splicing. Activation of the spliceosome entails major structural changes, including the displacement of U1 from the 5′ splice site, unwinding of the U4 and U6 base-pairing interaction, and the subsequent base pairing of U6 with the 5′ splice site and the U2 snRNA (Nilsen, 1994; Staley and Guthrie, 1998). These rearrangements result in the dissociation of the U1 and U4 snRNPs during the activation step. The resulting RNA network forms the core of the newly formed catalytic center of the spliceosome.
While spliceosome activation is well characterized at the RNA level, the role of proteins at this stage, as well as RNP remodeling events accompanying activation, is only poorly understood. Proteins of the DEAD/H-box family of RNA/RNP unwindases appear to be one of the driving forces that mediate RNA rearrangements at this step. For example, Prp28/U5-100K has been implicated in displacing U1 from the 5′ splice site (Staley and Guthrie, 1999; Chen et al, 2001), whereas Brr2/U5-200K appears to facilitate U4/U6 unwinding (Laggerbauer et al, 1998; Raghunathan and Guthrie, 1998). In yeast, evidence has been provided that Prp19p plays an essential role in spliceosome activation (Tarn et al, 1993a, 1993b). Prp19p is present in a heteromeric protein complex that consists of at least eight proteins (the NTC, nineteen complex) (Tarn et al, 1994; Tsai et al, 1999; Chen et al, 2002), and more recent data suggest that it is part of a very large RNP complex containing U2, U5, and U6 snRNA, as well as many additional proteins (Ohi et al, 2002). The NTC complex acts subsequent to U4 dissociation, stabilizing the association of U5 and U6 with the activated spliceosome (Chan et al, 2003). In humans, hPrp19p co-purifies with CDC5 in a larger complex containing approximately 30 proteins, and this CDC5/Prp19 complex was reported to play a role in the second step of splicing (Ajuh et al, 2000).
We recently immunoaffinity purified the activated B* spliceosome using antipeptide antibodies against the splicing factor SKIP (Makarov et al, 2002). We intentionally purified spliceosomal complexes under stringent conditions (i.e., in the presence of heparin) so that only stably bound proteins would be present. Consistent with the absence of U4, the purified activated spliceosome lacked all U4/U6-associated proteins. In addition, a group of 16 proteins (see Table I), including CDC5, Prp19 and additional proteins that were previously shown to be physically associated with Prp19 (Tarn et al, 1994; Tsai et al, 1999; Ben-Yehuda et al, 2000; Chen et al, 2002; Ohi and Gould, 2002), were also stably associated with the activated spliceosome, as well as a novel 35S form of the U5 snRNP (Makarov et al, 2002). These results indicated that the U5 snRNP is remodeled during activation, and that the 16 proteins that are stably associated with it within the activated spliceosome may play a role in activation or potentially at an earlier stage, for example during tri-snRNP integration into the spliceosome.
Table 1.
Comparison of the proteome of the purified BΔU1 spliceosome with that of the activated spliceosome (complex B*), the 35S U5 snRNP, and the 25S U4/U6.U5 tri-snRNP particle
 |
| Proteins in the dark blue box, namely NPW38BP, NPW38, NCoR-1, hPrp4 kinase, FBP-21, TBLR-1, SMC1, and p220/NPAT, are not found in the activated spliceosome. Most of the 35S U5 snRNP proteins highlighted red are either absent or significantly under-represented (indicated by italics) in the BΔU1 complex. The protein compositions of complex B*, the 35S U5 snRNP, and the tri-snRNP, as well as the color code for the various groups of proteins, was taken from Makarov et al (2002). In this manuscript, the group of proteins highlighted red are referred to as 35S U5 snRNP proteins. For accession numbers of proteins from complexes BΔU1 and B*, the 35S U5 snRNP, and tri-snRNP, see Table S2 in Supplementary data. |
To learn more about the driving forces behind spliceosome activation, and in particular the contribution of 35S U5-associated proteins to this process, we set out to isolate spliceosomal complexes prior to activation but after stable recruitment of the U4/U6.U5 tri-snRNP. Indeed, using antibodies against the U4/U6-specific 61K protein, we affinity purified spliceosomal B complexes that contain stably integrated, intact U4/U6.U5 tri-snRNPs but lack U1 snRNPs (henceforth denoted BΔU1). The protein composition of these complexes differed dramatically from that of the activated spliceosome; all U4/U6-specific proteins were present in the BΔU1 complexes, whereas essentially all 35S U5-specific proteins were largely absent. We addressed a possible role of 35S U5-specific proteins in spliceosome activation by performing immunodepletion/complementation assays in splicing active Hela nuclear extract, and demonstrate that a subset of them, including CDC5 and Prp19, function after tri-snRNP addition but prior to/during the first catalytic step of splicing.
Results
Purification of BΔU1 spliceosomes
To isolate spliceosomes just prior to their activation, but after stable integration of the U4/U6.U5 tri-snRNP, we used antipeptide antibodies against the U4/U6 protein 61K (hPrp31), which had previously been shown to be required for formation of stable tri-snRNPs (Makarova et al, 2002). Splicing complexes were allowed to form after incubation of pre-mRNA (MINX) with splicing active HeLa nuclear extract. After addition of heparin, immunoprecipitation was performed with anti-61K antibodies at different time points. Unspliced pre-mRNA was efficiently precipitated after 10 min (when B complex is efficiently formed) and also at later time points, whereas no precipitation of splicing intermediates or products was observed even at time points when efficient splicing had occurred (not shown). Thus, there is a window during spliceosome assembly (i.e., after tri-snRNP addition but prior to activation) when the 61K protein, and thus the U4/U6.U5 tri-snRNP as well (see below), is stably associated with the spliceosome, such that it remains bound even in the presence of heparin. Notably, under the same stringent conditions, 25S U4/U6.U5 tri-snRNPs present in the extract completely dissociate (not shown).
For biochemical analyses, we immunoaffinity purified preparative amounts of spliceosomes formed after 10 min of splicing. Bound spliceosomal complexes were then eluted with antigenic 61K peptide and further purified by glycerol gradient centrifugation. Purified spliceosomes contained stoichiometric amounts of uncleaved pre-mRNA, U2, U4, U5, and U6 snRNA but lacked U1 snRNA (Figure 1A, lane 1). Thus, in contrast to the activated spliceosome, which was isolated under identical conditions but with anti-SKIP antibodies (Figure 1A, lane 2), spliceosomes purified with anti-61K antibodies contain U4 snRNA. They, therefore, resemble complex B, but differ from it in that they lack the U1 snRNP. We therefore refer to these complexes as BΔU1 spliceosomes.
Figure 1.
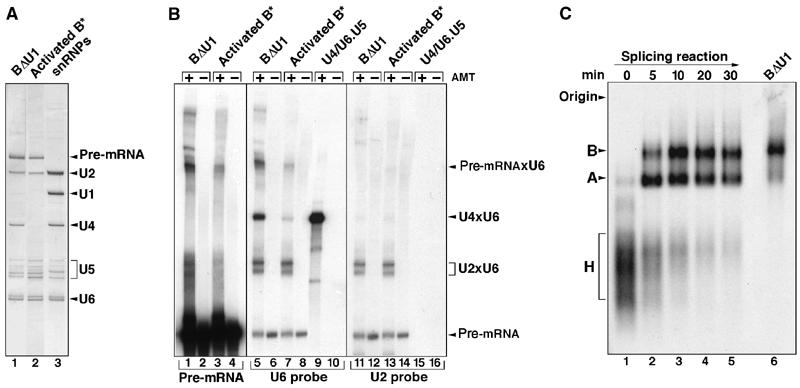
Characterization of immunoaffinity-purified BΔU1 spliceosomes. (A) RNA composition of BΔU1 versus activated B* spliceosomes. RNAs extracted from gradient-purified BΔU1 (lane 1), B* (lane 2) or total snRNPs (lane 3) were analyzed by denaturing PAGE and visualized by silver staining. (B) Identification of RNA base-pairing interactions in BΔU1. Gradient-fractionated BΔU1 (lanes 1–2, 5–6, and 11–12), B* (lanes 3–4, 7–8, and 13–14), or U4/U6.U5 tri-snRNPs (lanes 9–10 and 15–16) were incubated in the absence (even lanes) or presence (odd lanes) of psoralen (AMT) prior to UV irradiation. RNA was analyzed by denaturing PAGE, followed by either autoradiography to directly detect the 32P-labeled pre-mRNA present in the BΔU1 and B* spliceosomes (lanes 1–4), or by Northern blotting with 32P-labeled probes specific for U6 (lanes 5–10) or U2 (lanes 11–16). (C) Migration behavior of purified BΔU1 spliceosomes on native agarose gels. 32P-labeled MINX pre-mRNA was incubated under splicing conditions with HeLa nuclear extract for 0–30 min (lanes 1–5), and spliceosome assembly was analyzed on a 2% agarose gel, together with immunoaffinity-purified BΔU1 (lane 6). The positions of the H, A, and B complexes are indicated on the left.
To determine whether U4 is base paired with U6 in BΔU1 spliceosomes, psoralen crosslinking studies were performed with affinity-purified complexes that had been fractionated on a glycerol gradient. Gradient fractions containing BΔU1 or B* complexes, or purified tri-snRNPs (which served as a positive control for a U4/U6 crosslink), were UV irradiated in the presence or absence of psoralen (AMT), and RNA–RNA crosslinks were analyzed by Northern blotting, probing sequentially for U6 and U2 snRNA (Figure 1B). Multiple U6 snRNA-containing crosslinks were observed, one of which (designated U4 × U6) was clearly present in BΔU1 complexes but only weakly detected in the activated spliceosome (Figure 1B, compare lanes 5 and 7). This band was the major crosslink obtained with purified U4/U6.U5 tri-snRNPs (lane 9), demonstrating that it is a U4/U6 crosslink. Thus, U4 and U6 are still base paired in BΔU1 spliceosomes, confirming that they have not undergone catalytic activation.
Two of the U6-containing bands were also detected with a probe specific for the U2 snRNA (lanes 11 and 13), indicating that they represent U2/U6 crosslinks. Based on their migration behavior, the major U2/U6 crosslinks observed in both BΔU1 and B* appear to correspond to the well-characterized U2/U6 interaction between complementary regions of the 5′ end of U2 and the 3′ end of U6 snRNA (Hausner et al, 1990). This so-called U2/U6 helix II base-pairing interaction does not require U4/U6 unwinding. As previous crosslinking studies analyzing spliceosomes were performed in the presence of nuclear extract and large amounts of a U2/U6 helix II crosslink are observed in extracts even in the absence of splicing (Hausner et al, 1990), it was previously not possible to determine precisely at what stage of spliceosome assembly this base-pairing interaction first occurs. Thus, our psoralen crosslinking studies performed with purified spliceosomal complexes suggest that U2/U6 helix II is formed at the time of B complex formation.
A crosslink containing U6 that precisely co-migrated with a pre-mRNA-containing crosslink (designated Pre-mRNAxU6) was also observed (Figure 1B, compare lanes 1 and 5), indicating that U6 contacts the pre-mRNA in BΔU1 spliceosomes. The fact that this band was not observed after probing for U2 (lane 11), and that the signals of the non-crosslinked 32P-labeled pre-mRNA are of equal intensity in both the U2 and U6 panels, confirms that the band designated Pre-mRNAxU6 in lane 5 does not contain solely 32P-labeled pre-mRNA. Although the crosslinking site has not been precisely mapped, the presence of this crosslink in activated B* spliceosomes (lanes 3 and 7) raises the possibility that it may represent a base-pairing interaction between the region of U6 snRNA containing the ACAGA box and the 5′ end of the intron (for a summary of known RNA interactions in spliceosome, see Nilsen, 1994; Staley and Guthrie, 1998). We note that a U6/5′ splice site interaction can potentially occur concomitantly with U4/U6 base pairing.
Consistent with their designation as B complexes, which have been reported to sediment approximately as a 50S particle (Frendewey and Keller, 1985), purified BΔU1 spliceosomes sedimented as a 45S complex on glycerol gradients (not shown). They also exhibited a migration behavior similar to spliceosomal B complexes formed in vitro in Hela nuclear extract, when analyzed by native gel electrophoresis (Figure 1C, cf lane 6 with lanes 1–5). Note that, under the conditions used for native gel electrophoresis (i.e., in the presence of heparin), the U1 snRNP has been reported to dissociate from the spliceosomal B complex (Konarska and Sharp, 1986). By eluting this complex from the gel and analyzing its snRNA composition, we have confirmed that the spliceosomal complex designated B that we observe after native gel electrophoresis of splicing reactions in fact represents a BΔU1 spliceosome; it contains U2, U4, U5, and U6 snRNA, but U1 is clearly absent (not shown).
In the presence of nuclear extract in which endogenous snRNPs had been destroyed by microccocal nuclease treatment, the purified BΔU1 complex catalyzed both steps of splicing, whereas naked pre-mRNA did not (Figure 2). These experiments demonstrate that the purified BΔU1 spliceosome does not require complementation with U4/U6.U5 tri-snRNPs or U1 snRNPs for its activity, and that it is thus functionally committed for subsequent activation and the catalytic steps of splicing.
Figure 2.
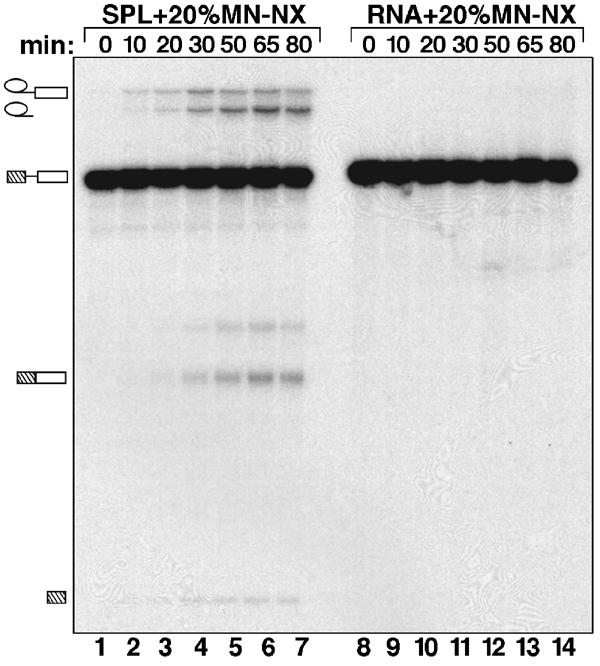
BΔU1 spliceosomes catalyze splicing in the absence of exogenously added snRNPs. Immunoaffinity-purified BΔU1 spliceosomes (SPL) (lanes 1–7) or 32P-labeled MINX pre-mRNA (RNA) (lanes 8–14) were incubated for 0–80 min (as indicated) under splicing conditions in the presence of 20% micrococcal nuclease-treated nuclear extract. RNAs were analyzed by denaturing PAGE and visualized by autoradiography. The positions of the pre-mRNA, and the splicing intermediates and products, are indicated on the left.
Protein composition of BΔU1 versus activated spliceosomes
To determine its protein composition, the purified BΔU1 complex was fractionated by glycerol gradient centrifugation and proteins isolated from the 45S peak fractions were separated by SDS–PAGE and characterized by MALDI mass spectrometry (MS). More than 70 proteins were identified and are summarized in Table I. Thus, compared to the activated B* spliceosome (Table I; Makarov et al, 2002), BΔU1 contains significantly fewer proteins. Consistent with the absence of U1 snRNA, no U1 snRNP-specific proteins (i.e., 70K, A and C) were identified. Most U2 snRNP proteins, such as A′, B″, SF3a and SF3b subunits, SPF30 and hPrp43 that were previously found in isolated 17S U2 snRNPs and the purified complex A (Hartmuth et al, 2002; Will et al, 2002), were present in the BΔU1 complex.
The protein compositions of the BΔU1 and activated B* spliceosome differ significantly with respect to the presence/absence of two groups of proteins (Table I). First, BΔU1 contains the U4/U6-specific proteins 90K, 61K, 60K, 15.5K, and 20K (CypH), while these are essentially absent in the activated B* complex (Table I). This finding, together with the fact that U4 and U6 are still base-paired, as evidenced by psoralen crosslinking (see above), strongly supports the idea that an intact U4/U6 snRNP is present in the BΔU1 spliceosome. Moreover, the U4/U6.U5 tri-snRNP also appears to be essentially intact in BΔU1. That is, the only U5 protein missing is 100K (hPrp28). Furthermore, both the U4/U6-61K and U5-102K proteins, which interact with each other and mediate stable tri-snRNP formation (Makarova et al, 2002), are present, suggesting that the two particles are still bridged in BΔU1.
The second major difference in protein composition between BΔU1 and B* is that all of the 35S U5-specific proteins, including Prp19 and CDC5 (highlighted red in Table I) that are found in the activated spliceosome, are absent or under-represented in BΔU1 (i.e., the sequence coverage is two- to three-fold lower compared to that of U2 or U5 proteins). To perform a more accurate comparison, activated and BΔU1 spliceosomes were immunoaffinity purified in parallel under the same stringent conditions, and the amount of material loaded onto the gel was normalized. LC-MSMS, focusing on the U4/U6 snRNP proteins and 35S U5 proteins, was subsequently performed and confirmed that the 35S U5 proteins are largely absent in the BΔU1 complex; only hPrp19 and SKIP were identified by two peptides each and CDC5 and hSyf3 by one peptide (Supplementary Table S1). In contrast, nearly all of the 35S U5 proteins, with the exception of hIsy1, were clearly more abundant in the purified B* complex. For example, eight peptides were obtained for hPrp19 and CDC5, and 11 for SKIP. The difference in the abundance of the U4/U6 snRNP proteins between BΔU1 and B* was similarly pronounced but in the opposite way; that is, two to 11 peptides were obtained for the U4/U6 proteins in the BΔU1 spliceosome, while none were obtained from B* (Supplementary Table S1). For comparison, the sequence coverage of U5 snRNP proteins, such as 220K, 200K, 116K, and 102K, was very similar in both complexes, confirming that equal molar amounts of both spliceosomes had been analyzed (Supplementary Table S1).
To provide more quantitative information about the absence or presence of the 35S U5-associated proteins, as well as U4/U6-specific proteins, we performed immunoblotting experiments with selected antibodies (Figure 3). We normalized the amount of each complex applied to the blot according to the amount of 32P-labeled pre-mRNA. Indeed, antibodies against the stably associated U5-116K and U5-40K proteins confirmed that equal molar amounts of each spliceosome were present (Figure 3). Subsequent immunoblotting experiments demonstrated that the U4/U6-specific 90K and 60K proteins, as well as the U2-associated protein SPF30, are present in BΔU1 but not in the activated spliceosome. Conversely, the 35S U5 proteins Prp19, CDC5L, Syf1, Syf3, and SKIP were essentially absent from BΔU1 but present in the activated spliceosome. Thus, whereas the U4/U6 proteins are stably associated with the spliceosome prior to activation, they dissociate or are destabilized at the time of activation. In contrast, the 35S U5-associated group of proteins become stably integrated into the spliceosome during transition from the B complex to the activated spliceosome. This observation suggests that the latter group of proteins may play a major role in the activation of the spliceosome rather than in the stable integration of the tri-snRNP. However, it is also conceivable that, despite the fact that they may only be loosely associated with the BΔU1 spliceosome (and thus lost due to heparin treatment), one or more of the 35S U5 snRNP proteins may play a role in tri-snRNP integration and thus B complex formation.
Figure 3.
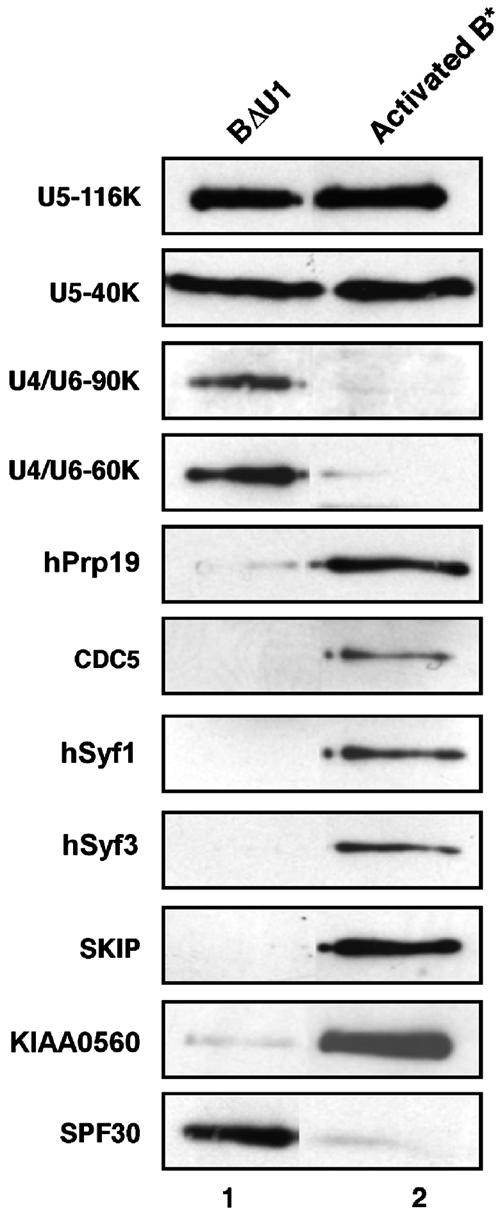
Protein composition of BΔU1 versus activated B* spliceosomes. BΔU1 and B* spliceosomes were immunoaffinity purified under identical conditions and subjected to glycerol gradient centrifugation. Proteins from BΔU1 or B* were separated by SDS–PAGE, blotted onto a membrane and immunostained with affinity-purified antibodies against selected proteins, as indicated on the right.
Spliceosomal complex B forms in the absence of a subset of 35S U5 proteins
To determine whether one or more of the 35S U5-specific proteins play a role in tri-snRNP integration, we carried out immunodepletion/complementation studies with HeLa nuclear extract. For this purpose, we raised antibodies against the 35S U5 protein CDC5. To define more precisely which proteins are stably associated with CDC5 and thus potentially co-immunodepleted, we first performed immunoaffinity chromatography with nuclear extract and antipeptide antibodies raised against CDC5. The bound complexes were eluted with an excess of CDC5 peptide and the eluate fractionated on a 5–20% glycerol gradient. Proteins were isolated from fractions across the gradient and analyzed by SDS–PAGE (Figure 4). A group of seven proteins co-migrated in fractions 13–15, indicating that they form a complex that sediments with a Svedberg value of 14S. Proteins present in the 14S peak were analyzed by MALDI-MS and identified as CDC5, Hsp73, β-catenin-like 1, PRL1, Prp19, AD002, and SPF27. MS analyses further revealed that the proteins Npw38 and Npw38BP co-migrate at the top of the gradient, suggesting that they were co-isolated with the CDC5/Prp19 complex but then dissociated from it during centrifugation.
Figure 4.
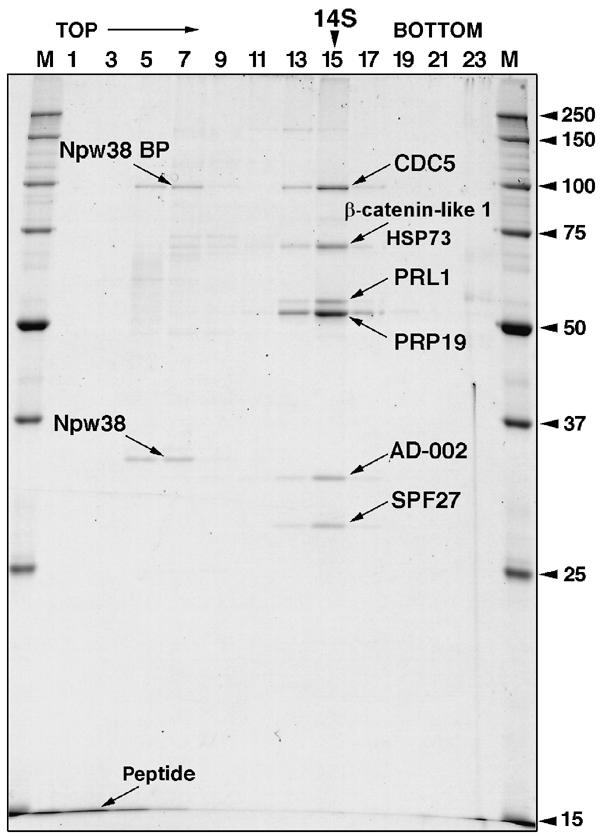
Sedimentation behavior and protein composition of CDC5/Prp19-containing complexes. Complexes were immunoaffinity purified from HeLa nuclear extract with anti-CDC5 antibodies and subjected to 5–20% glycerol gradient centrifugation. Proteins from uneven fractions were separated by SDS–PAGE and stained with Coomassie. The following proteins (indicated with arrows) were identified by MS: CDC5 (NP_001244), HSP73 (NP_006588), β-catenin-like 1 (NP_110517), PRL1 (NP_002660), Prp19 (NP_055317), AD002 (AAF14858), SPF27 (NP_005863), Npw38 (NP_005701), and Npw38BP (NP_057396).
With the exception of the β-catenin-like 1 protein, 14S CDC5/Prp19 complex consists primarily of a subset of 35S U5 proteins (i.e., CDC5, Hsp73, PRL1, Prp19, AD002, and SPF27) (Makarov et al, 2002). To determine whether this subset of 35S U5-associated proteins play a role in splicing, we immunodepleted nuclear extract of the CDC5/Prp19 complex using anti-CDC5 and anti-AD002 antibodies. The level of each protein was significantly reduced in their respective immunodepleted extract when compared with the mock-depleted extract, as evidenced by immunoblotting (Figure 5A). Consistent with the fact that they are present together in a stable complex, depletion of CDC5 also led to a reduction in AD002 and vice versa (Figure 5A, lanes 2 and 3). In vitro splicing was subsequently performed with mock-depleted extract and extract depleted with both anti-CDC5 and anti-AD002 antibodies. As shown in Figure 5B, splicing activity was significantly reduced in the CDC5/AD002-depleted as compared to mock-depleted extract (cf lanes 1–4 and 5–8). Significantly, addition of immunoaffinity-purified 14S CDC5/Prp19 complex (i.e., gradient fraction 14 from Figure 4) partially restored splicing activity to the depleted extract (lanes 8–12).
Figure 5.
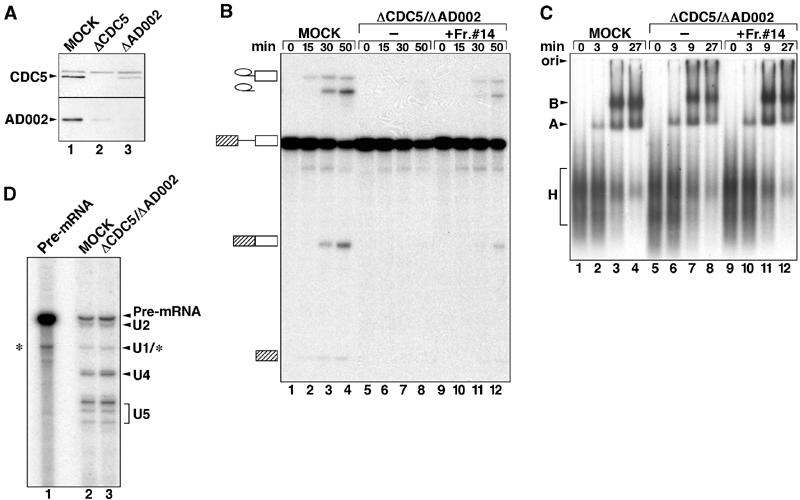
Spliceosomal B complex forms in the absence of the CDC5/PRP19 complex. (A) Nuclear extracts are efficiently immunodepleted of CDC5 and AD002. Western blot of mock-depleted (lane 1), CDC5-depleted (lane 2) or AD002-depleted (lane 3) extract probed with anti-CDC5 (upper panel) or anti-AD002 (lower panel) antibodies. (B) Time course of splicing of [32P] MINX pre-mRNA in mock-depleted extract (MOCK, lanes 1–4) or extract depleted with CDC5 and AD002 (ΔCDC5/ΔAD002, lanes 5–8), or CDC5/AD002-depleted extract complemented with purified CDC5/PRP19 complex (fraction no. 14, from the gradient in Figure 4) (lanes 9–12). RNA was analyzed by denaturing PAGE and visualized by autoradiography. The pre-mRNA, and splicing intermediates and products are indicated on the left. (C) The CDC5/PRP19 complex is not required for B-complex formation. Spliceosome assembly in mock-depleted extract, CDC5/AD002-depleted extract, or CDC5/AD002-depleted extract complemented with the CDC5/PRP19 complex (lanes marked as in (B)) was analyzed at the indicated times by native gel electrophoresis and visualized by autoradiography. The position of the H, A, and B complexes is indicated on the left. (D) Spliceosomes formed in CDC5/AD002-depleted extract contain U4 snRNA. Spliceosomes were immunoprecipitated from mock- (lane 2) or CDC5/AD002-depleted extract (lane 3) with anti-61K antibodies. RNA was isolated, end labeled with 32P-pCp, fractionated by denaturing PAGE, and visualized by autoradiography. The identities of the RNAs are indicated on the right. Internally radiolabeled MINX pre-mRNA (lane 1) was analyzed in parallel. Note that the band migrating at the position of U1 may also contain a pre-mRNA degradation product (*).
To determine at what step of the splicing reaction the 14S CDC5/Prp19 complex first acts, we analyzed splicing complex formation in mock- and CDC5/AD002-depleted extract by agarose gel electrophoresis. Relative to the mock-depleted extract (Figure 5C, lanes 1–4), no significant decrease in the formation of A or B complex was observed with the CDC5/AD002-depleted extract (Figure 5C, lanes 5–8). Due to the fact that the catalytic steps of splicing occur very rapidly with the MINX pre-mRNA, C complex is difficult to detect (Das and Reed, 1999).
To determine whether or not the complex designated B that we observe in the absence of the 14S CDC5/Prp19 complex has undergone activation (i.e., release of U4), we performed immunoprecipitation experiments under the same conditions used to isolate BΔU1 spliceosomes with anti-61K antibodies. After allowing for splicing complex formation, spliceosomes were immunoprecipitated from mock- or CDC5/AD002-depleted extract and their RNA composition was analyzed by 3′-end labeling with 32P-pCp, followed by denaturing PAGE. The RNA compositions of spliceosomes immunoprecipitated from mock- and CDC5/AD002-depleted extract were essentially identical (Figure 5D); when antibodies were preblocked with cognate peptide, essentially no RNA was precipitated (not shown). Importantly, in addition to U2 and U5 (due to inefficient end labeling, the U6 snRNA is difficult to detect), a similar level of U4 snRNA was detected in both cases. These results demonstrate that the majority of spliceosomes present have not undergone activation and, thus, spliceosomal complex B can form even in the absence of the CDC5/Prp19 complex, but splicing is blocked prior to step 1 of splicing.
Discussion
To gain insight into the multiple RNP rearrangements and changes in protein composition of the spliceosome during its maturation, catalytic activity, and subsequent disassembly, we have set out to isolate various spliceosomal complexes under identical stringent conditions at defined assembly/functional stages. Using antipeptide antibodies directed against spliceosomal proteins that are transiently stably associated with the spliceosome, we have now been able to isolate two spliceosomal complexes (BΔU1 and the activated spliceosome) under the same conditions, allowing a meaningful comparison of their protein compositions via MS. These results demonstrate that, in addition to the highly stable spliceosomal complexes detected by standard biochemical methods, there exist additional spliceosomal intermediates that transiently form configurations of high stability, and that such intermediates can be isolated by targeting the suitable epitope for immunoaffinity selection. This method can theoretically be extended to isolate additional spliceosomal complexes and thus generate additional ‘snapshots' of the spliceosome's changing protein composition and RNP structure.
Composition of the BΔU1 spliceosome
The protein composition of the BΔU1 spliceosome differed markedly from that of the activated B* spliceosome in that U4/U6-associated proteins (which were not found in B*) were present, but a group of 16 35S U5-associated proteins that are stably associated with B* were largely absent. Only minimal amounts of a subset of these proteins were detected by MS or immunoblotting. That some proteins appear to be present at a very low level in BΔU1 could reflect the existence of a subpopulation of spliceosomes in which the 61K protein is still stably associated and several 35S U5 snRNP proteins have also been stably integrated. BΔU1 contained a group of eight proteins, including Npw38, Npw38BP, N-CoR1, hPrp4 kinase, FBP-21, TBLR-1, SMC1, and p220(NPAT), not detected in the pre-spliceosome or activated spliceosome (Table I) (Hartmuth et al, 2002; Makarov et al, 2002). Thus, these proteins could potentially function specifically at this stage of spliceosome formation. Consistent with these proteins being present in the BΔU1 spliceosome, hPrp4 kinase was previously shown to interact with N-CoR (Dellaire et al, 2002). Interestingly, the Saccharomyces pombe ortholog of hPrp4 kinase has been shown to be essential for splicing and to phosphorylate the S. pombe prp1 protein (human U5-102K protein) (Schwelnus et al, 2001). Thus, hPrp4 kinase, via phosphorylation of the U5-102K protein, could play a role in tri-snRNP rearrangement during spliceosome activation.
Interestingly, anti-61K affinity-purified spliceosomes lacked the U1 snRNP. The absence of U1 may simply be due to the presence of heparin during the isolation procedure, and thus anti-61K affinity-purified spliceosomes likely represent spliceosomal complex B that has been depleted of U1. Alternatively, the BΔU1 spliceosome could represent a bona fide spliceosome intermediate in which the U1 snRNP/5′ splice site interaction has been actively disrupted, but the U4/U6 base-pairing interaction remains intact. The detection of a pre-mRNA/U6 snRNA crosslink (Figure 1B) is a first indication that this may indeed be the case. However, more detailed analyses are required to map the precise U6 crosslinking site on the pre-mRNA. In this respect, it is interesting to note that, in the minor U12-type spliceosome, interaction of U6atac with the 5′ splice site was observed prior to complete unwinding of the U4atac/U6atac base-pairing interaction (Frilander and Steitz, 2001). Complementation experiments with micrococcal nuclease-treated nuclear extract demonstrated that BΔU1 can be chased into a catalytically active complex in the absence of snRNPs. Thus, the presence of U1 is not required for the subsequent activation of the spliceosome. Displacement of U1 from the 5′ splice site is a prerequisite for the subsequent base-pairing interaction of the U6 snRNA, and appears to be facilitated by the DEAD-box protein hPrp28/U5-100K (Staley and Guthrie, 1999; Chen et al, 2001). Intriguingly, hPrp28p/U5-100K is also no longer stably associated with the BΔU1 spliceosome, and thus may dissociate together with U1 after catalyzing its displacement from the 5′ splice site.
Isolation of a well-defined, human 14S CDC5/Prp19 heteromeric complex
Using antibodies against CDC5, we isolated a heteromeric, 14S protein complex consisting of CDC5, Prp19, Hsp73, β-catenin-like 1, PRL1, AD002, and SPF27. This CDC5/Prp19 complex contains only a subset of the 35S U5 proteins that are stably integrated first in the activated spliceosome. Whether additional 35S U5 proteins associate and/or are stably integrated into the spliceosome as a larger group, together with the CDC5/Prp19 heptameric complex, is presently not clear. Previous studies characterizing the human CDC5/Prp19 complex did not allow precise conclusions regarding its protein composition (Ajuh et al, 2000). In the latter studies, several CDC5-containing complexes were isolated from HeLa nuclear extract either by immunoaffinity chromatography or by size exclusion, followed by anion exchange chromatography; in each case approximately 30 proteins appeared to co-purify with CDC5 (Table II). Only six of these proteins, that is, CDC5, Prp19, PRL1, Hsp73 (CCAP1), SPF27, and CCAP6, were found in all preparations, and thus were denoted core components of the CDC5-containing complex (Ajuh et al, 2000; Table II). While five of these are also found in our 14S CDC5/Prp19 complex, CCAP6 is not. Furthermore, AD002, which is present in our complex, was not previously reported to be a core component of CDC5 complex, although it co-purifies with CDC5 (AD002 is denoted CCAP2 by Ajuh et al, 2000).
Table 2.
Comparison of Prp19/CDC5 complexes
| 14S CDC5/hPrp19 complex (this study)a | Human CDC5 complex (Ajuh et al, 2000)b | Prp19 complex (Chen et al, 2002)c | Prp19 complex (Ohi et al, 2002)d |
|---|---|---|---|
| hPrp19 | hPrp19/CCAP3e | Prp19 | Prp19 |
| CDC5 | CDC5Le | Ntc85/Cef1/Cdc5 | Cef1/Cdc5 |
| PRL1 | PRL1/hPrp46e | Cwc1/Prp46 | |
| AD-002 | AD-002/CCAP2 | ||
| HSP73 | HSP73/CCAP1e | ||
| SPF27 | SPF27e | Ntc25/Snt309f | Snt309f |
| β-catenin like | |||
| CCAP4 | |||
| CCAP5 | |||
| CCAP6e | |||
| CCAP7 | |||
| CCAP8 | |||
| ASF/SF2 | |||
| PSF | |||
| SRm160 | |||
| SC35 | |||
| NF45 | |||
| TOPIIa | |||
| U1A | |||
| hnRNPG | |||
| PP2c (WIP1) | |||
| DDP kinase | |||
| PP1-α | |||
| Sm proteins | Sm proteins | ||
| U2-specific proteins | U2-specific proteins | ||
| U5-specific proteins | |||
| Ntc77/Clf1/Syf3 | Clf1/Syf3 | ||
| Ntc90/Syf1 | Syf1 | ||
| Ntc30/Isy1 | Isy1 | ||
| Ntc31/Syf2 | Syf2 | ||
| Ntc20 | Ntc20 | ||
| Slu7 | |||
| Ecm2 | |||
| Spp2 | |||
| Cwc2, Cwc14, Cwc15 | |||
| Cwc16 Cwc21, Cwc22 | |||
| Cwc23, Cwc24, Cwc25 Cwc26, Cwc27 | |||
| |
|
|
Prp17, Prp22, Prp45 |
| a Complexes were immunoaffinity purified from HeLa nuclear extract with anti-CDC5 antibodies at 200 mM salt and further purified by glycerol gradient centrifugation. | |||
| b Complexes were purified from HeLa nuclear extracts either by immunoaffinity chromatography with anti-CDC5 antibodies at 150 mM salt or by tandem chromatographic steps involving gel filtration and ion exchange chromatography (Ajuh et al, 2000). | |||
| c Complexes were isolated from S. cerevisiae total cell extracts at 60 mM salt by tagging the Prp19 protein with an HA-tag and precipitating with anti-HA antibodies (Tarn et al, 1994). | |||
| d Complexes were isolated from S. cerevisiae total cell extracts using the TAP method (150 mM salt) after tagging the Prp19 protein (Ohi et al, 2002). | |||
| e Stably associated core protein in the CDC5 protein complex (Ajuh et al, 2000). | |||
| f Snt309 is a functional counterpart of S. pombe Cwf7 that is homologous to SPF27 (Ohi and Gould, 2002). | |||
The organization of the human 14S CDC5/Prp19 complex differs considerably from the NTC complex in S. cerevisiae (Table II). The latter complex has not been characterized in its entirety; it consists of at least eight proteins with several additional proteins also thought to be present (Chan et al, 2003). Of the known eight NTC components, only two, Prp19 and CDC5/Cef1, are found in our 14S CDC5/Prp19 complex. A third candidate is Snt309, which may be a functional counterpart of SPF27 (Ohi and Gould, 2002). That the NTC complex likely consists of a much larger group of proteins is further suggested by more recent studies (Ohi et al, 2002). Using the TAP method, Prp19 in S. cerevisiae was shown to be part of a large RNP complex containing numerous proteins (Table II) as well as U2, U5, and U6 snRNAs. A comparison of this complex with our 14S CDC5/Prp19 complex reveals that their protein compositions are significantly different.
Interestingly, Npw38 and Npw38BP, which interact with one another (Komuro et al, 1999), were reproducibly co-isolated with the CDC5/Prp19 complex, but appeared to dissociate during glycerol gradient centrifugation. Thus, these proteins, which have no apparent orthologs in S. cerevisiae (Supplementary Table S1), likely exhibit affinity for one or more components of the CDC5/Prp19 complex, and may play a role in recruiting this complex to the spliceosome, at least in higher eucaryotes. Indeed, Npw38 and Npw38BP are stably associated with the BΔU1 spliceosome and could thus act as a docking site for the CDC5/Prp19 complex. Npw38 has been shown to interact with the U5-15K (Dib1) protein (Zhang et al, 2000), and both of these proteins together with Npw38BP are no longer stably associated with the activated spliceosome (Table I), suggesting that they are destabilized together during activation.
Function of the human CDC5/Prp19 complex
Previous functional analyses of the CDC5/Prp19 complex in both yeast and humans were performed with a poorly defined, large group of proteins (see above) (Ajuh et al, 2000; Chan et al, 2003). Thus, these studies do not allow clear-cut conclusions as to precisely which components of the CDC5/Prp19 complex are required for its function. We have performed depletion/complementation studies with a well-defined complex containing CDC5, Prp19, Hsp73, β-catenin-like 1, PRL1, AD002, and SPF27, and thus can narrow down the number of essential Prp19/CDC5 proteins to a very small subset. The CDC5/Prp19 complex as defined here does not play a role in tethering the tri-snRNP within the spliceosome, as evidenced by the fact that this group of proteins is not found in the BΔU1 complex, where the tri-snRNP is stably associated. Moreover, by physically depleting this complex, we have further demonstrated that these proteins are not required for the initial integration of the tri-snRNP. The 17S U2-associated protein SPF30, which is present in the BΔU1 spliceosome, likely plays an important role in recruiting/tethering the U4/U6.U5 tri-snRNP complex; indeed SPF30 has been shown to interact with the U4/U6-90K protein (Meister et al, 2001; Rappsilber et al, 2001). Interestingly, SPF30 is no longer stably associated with the activated spliceosome, consistent with the fact that the U4 snRNA and the U4/U6-associated proteins (15.5K, 20K, 60K, 61K, and 90K) dissociate or are destabilized at this time. This finding underscores how comparison of the stable proteomes of spliceosomal complexes at defined stages may help identify those proteins that are interaction partners and/or function as a group at a particular step of the splicing process.
Depletion/complementation studies provided evidence that the CDC5/Prp19 heptameric complex is required for the first step of splicing. Thus, one or more members of this complex initially function in a time window encompassing the spliceosome activation step (i.e., after B-complex formation but prior to step one of splicing). These results differ from previous reports where immunodepletion of human CDC5 and its associated proteins led predominantly to a block in the second step of splicing (Ajuh et al, 2000). One possible explanation for this difference is that a more efficient depletion of the CDC5/Prp19 complex was achieved in our hands due to the double depletion with both anti-CDC5 and anti-Ad002 antibodies. Although spliceosomal B complex was observed in the absence of the CDC5/Prp19 complex, it is presently not clear whether it functions prior to, during, or after U4/U6 unwinding. A role in activation and/or the first step of splicing, but not in tri-snRNP integration, likely also holds for the nine other 35S U5 proteins that are present in the activated spliceosome but were not present in the immunoaffinity-purified CDC5/Prp19 complex. Although direct evidence that they function at this stage is lacking, the fact that they are stably integrated into the spliceosome at the same time as proteins of the CDC5/Prp19 complex is a first indication that they may also function at a similar step. Additional studies are clearly required to address this question in more detail.
In yeast, the NTC complex acts subsequent to U4 dissociation, stabilizing the association of U5 and U6 with the activated spliceosome (Chan et al, 2003). Recent studies in yeast have also indicated that the U6-associated Lsm proteins (Lsm2–8) are destabilized during activation of the spliceosome and that the NTC complex is required for their destabilization (Chan et al, 2003). Our MS, as well as initial immunoblotting data, suggests that Lsm proteins are not destabilized during spliceosome activation in humans. That is, several Lsm proteins were detected by MS both in BΔU1 and activated B* spliceosomes, despite the fact that they are generally difficult to detect by MS (Table I). In addition, in an initial immunoblotting experiment with antibodies against LSm4, we did not observe a decrease in signal between the BΔU1 and B*-activated spliceosome; in both complexes, significant amounts of Lsm4 were detected (not shown). However, additional experiments are required to clarify whether the situation in humans indeed differs from that in yeast.
The ability to isolate both BΔU1 and activated spliceosomes under identical conditions allows us to address other interesting questions. For example, what triggers U4 release and other RNP rearrangements during spliceosome activation? Aside from RNA helicase activity, post-translational modifications might also be a driving force that alters the stability of protein–protein interactions within the spliceosome during its activation. To address this question, we have begun to compare the modification states of proteins in the BΔU1 versus activated spliceosome.
Materials and methods
In vitro splicing
HeLa nuclear extract was prepared according to Dignam et al (1983). In vitro splicing, analytical immunoprecipitation of spliceosomes, and micrococcal nuclease (MN) digestions were performed as described (Makarova et al, 2001, 2002). For analysis of splicing complexes, heparin (5 mg/ml) was added to 5 μl of a splicing reaction or 10 μl of immunoaffinity-purified BΔU1 spliceosomes to a final concentration of 1 mg/ml, and samples were placed on ice. Complexes were analyzed on 2% agarose gels (Das and Reed, 1999).
Preparative immunoaffinity purification of spliceosomes and MS
Antibodies raised against a peptide (aa 484–497) of U4/U6-61K or the splicing factor SKIP (Makarov et al, 2002) were affinity purified using a SulfoLink column (Pierce) containing the cognate peptide. A 4.0-ml splicing reaction containing 40% HeLa nuclear extract and 10–20 nM 32P-labeled MINX pre-mRNA (2000–4000 cpm/pmol) was incubated at 30°C for 10 min. Heparin was added to a final concentration of 0.5 mg/ml and incubation at 30°C continued for 5 min. Subsequent procedures were performed at 4°C. The splicing reaction was diluted 10-fold with IP buffer (20 mM HEPES, pH 7.9, 150 mM NaCl, 1.5 mM MgCl2, 0.5 mM DTT) containing 0.05% NP-40 and incubated for 1 h with 1 ml of protein A–sepharose (PAS) preblocked with 0.5 mg/ml of BSA and 50 μg/ml of yeast tRNA. The reaction was then incubated for 2 h with 1 ml of PAS charged with affinity-purified anti-61K or anti-SKIP antibodies. Beads were washed 6 × with IP buffer containing 0.05% NP-40 and the bound material was eluted by incubating for 1 h with 2.5 ml IP buffer containing 5% glycerol, 0.05% NP-40, and 0.6 mg/ml of cognate peptide. The eluate was loaded onto a 10–30% glycerol gradient containing IP buffer and centrifuged in a TST41.14 rotor (Sorvall) for 14 h at 31 000 rpm. Gradients were fractionated into 24 0.5-ml aliquots. Five fractions containing the peak of the BΔU1 or B* spliceosome were combined and complexes were pelletted by centrifugation in a TH660 rotor (Sorvall) for 4 h at 60 000 rpm. Pellets were resuspended in SDS–PAGE loading buffer, subjected to 10/13% SDS–PAGE, and stained with Coomassie. Individual bands were excised from the gel and analyzed by both MALDI-MS and LC-MSMS (Hartmuth et al, 2002). MS analyses of BΔU1 were performed twice: once alone and then subsequently in direct comparison with an equal molar amount of B*.
Isolation of the CDC5/Prp19 complex
PAS beads (0.4 ml) were charged with affinity-purified antibodies against a peptide of CDC5 (aa 106–124), preblocked with 0.5 mg/ml of BSA and 50 μl/ml of yeast tRNA, and incubated with 1 ml of HeLa nuclear extract in C buffer (Dignam et al, 1983) for 2 h at 4°C. Beads were washed 6 × with G200 buffer (20 mM HEPES, pH 7.9, 200 mM NaCl, 1.5 mM MgCl2, 0.05% NP-40). The bound material was eluted with 400 μl of IP buffer containing 200 mM NaCl, 0.5 mg/ml antigenic peptide, and 0.5 mM DTT, and loaded on a 5–20% glycerol gradient containing IP buffer. Gradients were centrifuged for 14.5 h at 35 000 rpm in a TH-660 rotor. 12S U1 and 20S U5 snRNPs served as S-value markers.
Psoralen crosslinking
A 1.25 ml splicing reaction containing 32P-labeled MINX pre-mRNA was incubated for 10 min at 30°C, heparin was added to a final concentration of 0.5 mg/ml, and the reaction incubated for an additional 5 min. The splicing reaction was diluted eight-fold with IP buffer, precleaned by incubating with 300 μl PAS preblocked with BSA and tRNA, and then incubated with 300 μl PAS charged with anti-61K antibodies (to isolate BΔU1) or anti-SKIP antibodies (to isolate activated B* spliceosomes). Complexes were eluted with cognate peptide as described above and further purified by centrifugation on a 10–30% glycerol gradient prepared with IP buffer. Peak fractions from each gradient were combined and split into two aliquots. Psoralen (AMT) was added to one aliquot to a final concentration of 25 μg/ml and incubated on ice for 10 min. Both the psoralen-treated and nontreated aliquots were subsequently irradiated for 15 min at 365 nm on ice. After proteinase K digestion, RNA was recovered by phenol–chloroform extraction and ethanol precipitation, and separated on a 5% polyacrylamide, 7 M urea gel (Hausner et al, 1990). The 32P-labeled pre-mRNA was detected directly by autoradiography, whereas the U2 and U6 snRNAs were visualized by Northern blotting (Hartmuth et al, 2002). Psoralen crosslinking was performed in parallel with anti-m3G affinity-purified, gradient-fractionated U4/U6.U5 tri-snRNPs (Laggerbauer et al, 1998).
Immunoblotting
Proteins from BΔU1 or B* spliceosomes were separated by SDS–PAGE, transferred to Hybond P membrane and immunostained with affinity-purified antibodies, using an ECL detection kit (Amersham). Antibodies were raised in rabbits against the following antigens: U5-116K (aa 1–117), U5-40K (full length); U4/U6-90K (aa 287–302); U4/U6-60K (aa 58–94), Prp19 (aa 176–191), CDC5 (aa 106–124), hSyf1 (aa 6–21), hSyf3 (aa 47–60), SKIP (aa 516–531), and KIAA0560 (aa 1470–1485).
Immunodepletion/complementation assays
HeLa nuclear extract (0.5 ml) in C buffer was incubated 2 × with 75 μl of PAS beads charged with affinity-purified antibodies against CDC5 or a peptide of AD002 (aa 71–86) for 2 h at 4°C with HOT rotation. For double depletion, incubation with anti-CDC5 beads was followed by anti-AD002 beads. The extract was then dialyzed for 4 h against D buffer (Dignam et al, 1983) in a Slide-A-Lyzer 3.5K (Pierce). Mock-depleted extracts were treated in an identical manner, except that antibody was omitted. For complementation, prior to dialysis, 100 μl of depleted extract was mixed with 56 μl of gradient fraction no. 14 (Figure 4) containing the CDC5/Prp19 complex at a concentration of ∼0.1 mg/ml, and then dialyzed as above. Splicing reactions (10–20 μl) contained 35% mock-, depleted- or CDC/Prp19-complemented extract and 10 nM MINX 32P-labeled pre-mRNA (10 000–50 000 cpm/pmol).
Analysis of spliceosomal complexes formed in CDC5/AD002-depleted extract
A 150 μl splicing reaction containing mock- or CDC5/AD002-depleted extract and 10 nM MINX pre-mRNA was incubated at 30°C for 10 min and subsequently treated with heparin as described above. The reaction was diluted eight-fold with IP buffer containing 0.05% NP-40, incubated for 2 h at 4°C with 30 μl PAS charged with anti-61K antibodies, and washed 6 × with IP buffer containing 0.05% NP-40. RNA was isolated from the immunoprecipitated spliceosomal complexes, 3′-endlabeled with [32P]pCp, analyzed on a 14% polyacrylamide/7 M urea gel, and visualized by autoradiography.
Supplementary Material
Supplementary Material
Acknowledgments
We thank Monika Raabe, Gabi Heyne, Irene Öchsner, and Peter Kempkes for excellent technical assistance, and Klaus Hartmuth for helpful advice and discussions regarding psoralen crosslinking. We are grateful to Utz Fischer for kindly providing antibodies against SPF30. This work was supported by grants from the Deutsche Forschungsgemeinschaft (Lu294/12-1), BMBF (031U215B), Ernst Jung Stiftung, and the Fonds der Chemischen Industrie to RL, and a grant from the BMBF (031U215A) to MW.
References
- Ajuh P, Kuster B, Panov K, Zomerdijk JC, Mann M, Lamond AI (2000) Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. EMBO J 19: 6569–6581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S, Dix I, Russell CS, McGarvey M, Beggs JD, Kupiec M (2000) Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae. Genetics 156: 1503–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow DA (2002) Allosteric cascade of spliceosome activation. Annu Rev Genet 36: 333–360 [DOI] [PubMed] [Google Scholar]
- Burge CB, Tuschl T, Sharp PA (1999) Splicing of precursors to mRNAs by the spliceosomes. In The RNA World, Gesteland C, Atkins (eds), 2nd edn, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press pp 525–560 [Google Scholar]
- Chan SP, Kao DI, Tsai WY, Cheng SC (2003) The Prp19p-associated complex in spliceosome activation. Science 302: 279–282 [DOI] [PubMed] [Google Scholar]
- Chen CH, Yu WC, Tsao TY, Wang LY, Chen HR, Lin JY, Tsai WY, Cheng SC (2002) Functional and physical interactions between components of the Prp19p-associated complex. Nucleic Acids Res 30: 1029–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Stands L, Staley JP, Jackups RR, Latus LJ, Chang TH (2001) Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol Cell 7: 227–232 [DOI] [PubMed] [Google Scholar]
- Das R, Reed R (1999) Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA 5: 1504–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaire G, Makarov EM, Cowger JJ, Longman D, Sutherland HG, Lührmann R, Torchia J, Bickmore WA (2002) Mammalian PRP4 kinase copurifies and interacts with components of both the U5 snRNP and the N-CoR deacetylase complexes. Mol Cell Biol 22: 5141–5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendewey D, Keller W (1985) Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell 42: 355–367 [DOI] [PubMed] [Google Scholar]
- Frilander MJ, Steitz JA (2001) Dynamic exchanges of RNA interactions leading to catalytic core formation in the U12-dependent spliceosome. Mol Cell 7: 217–226 [DOI] [PubMed] [Google Scholar]
- Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, Lührmann R (2002) Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc Natl Acad Sci USA 99: 16719–16724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausner TP, Giglio LM, Weiner AM (1990) Evidence for base-pairing between mammalian U2 and U6 small nuclear ribonucleoprotein particles. Genes Dev 4: 2146–2156 [DOI] [PubMed] [Google Scholar]
- Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ (2002) Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8: 426–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro A, Saeki M, Kato S (1999) Association of two nuclear proteins, Npw38 and NpwBP, via the interaction between the WW domain and a novel proline-rich motif containing glycine and arginine. J Biol Chem 274: 36513–36519 [DOI] [PubMed] [Google Scholar]
- Konarska MM, Sharp PA (1986) Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell 46: 845–855 [DOI] [PubMed] [Google Scholar]
- Laggerbauer B, Achsel T, Lührmann R (1998) The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc Natl Acad Sci USA 95: 4188–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Lührmann R (2002) Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298: 2205–2208 [DOI] [PubMed] [Google Scholar]
- Makarova OV, Makarov EM, Liu S, Vornlocher HP, Lührmann R (2002) Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing. EMBO J 21: 1148–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova OV, Makarov EM, Lührmann R (2001) The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP are essential for the assembly of mature spliceosomes. EMBO J 20: 2553–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Hannus S, Plottner O, Baars T, Hartmann E, Fakan S, Laggerbauer B, Fischer U (2001) SMNrp is an essential pre-mRNA splicing factor required for the formation of the mature spliceosome. EMBO J 20: 2304–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW (1994) RNA–RNA interactions in the spliceosome: unraveling the ties that bind. Cell 78: 1–4 [DOI] [PubMed] [Google Scholar]
- Ohi MD, Gould KL (2002) Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA 8: 798–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Link AJ, Ren L, Jennings JL, McDonald WH, Gould KL (2002) Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol Cell Biol 22: 2011–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan PL, Guthrie C (1998) RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol 8: 847–855 [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Ajuh P, Lamond AI, Mann M (2001) SPF30 is an essential human splicing factor required for assembly of the U4/U5/U6 tri-small nuclear ribonucleoprotein into the spliceosome. J Biol Chem 276: 31142–31150 [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M (2002) Large-scale proteomic analysis of the human spliceosome. Genome Res 12: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwelnus W, Richert K, Opitz F, Gross T, Habara Y, Tani T, Kaufer NF (2001) Fission yeast Prp4p kinase regulates pre-mRNA splicing by phosphorylating a non-SR-splicing factor. EMBO Rep 2: 35–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C (1998) Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C (1999) An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell 3: 55–64 [DOI] [PubMed] [Google Scholar]
- Tarn WY, Hsu CH, Huang KT, Chen HR, Kao HY, Lee KR, Cheng SC (1994) Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J 13: 2421–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn WY, Lee KR, Cheng SC (1993a) Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc Natl Acad Sci USA 90: 10821–10825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn WY, Lee KR, Cheng SC (1993b) The yeast PRP19 protein is not tightly associated with small nuclear RNAs, but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol Cell Biol 13: 1883–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WY, Chow YT, Chen HR, Huang KT, Hong RI, Jan SP, Kuo NY, Tsao TY, Chen CH, Cheng SC (1999) Cef1p is a component of the Prp19p-associated complex and essential for pre-mRNA splicing. J Biol Chem 274: 9455–9462 [DOI] [PubMed] [Google Scholar]
- Will CL, Urlaub H, Achsel T, Gentzel M, Wilm M, Lührmann R (2002) Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J 21: 4978–4988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lindblom T, Chang A, Sudol M, Sluder AE, Golemis EA (2000) Evidence that dim1 associates with proteins involved in pre-mRNA splicing, and delineation of residues essential for dim1 interactions with hnRNP F and Npw38/PQBP-1. Gene 257: 33–43 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, Reed R (2002) Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


