Summary
ATOH8 has previously been shown to be an iron-regulated transcription factor, however its role in iron metabolism is not known. ATOH8 expression in HEK293 cells resulted in increased endogenous HAMP mRNA levels as well as HAMP promoter activity. Mutation of the E-box or SMAD response elements within the HAMP promoter significantly reduced the effects of ATOH8, indicating that ATOH8 activates HAMP transcription directly as well as through bone morphogenic protein (BMP) signalling. In support of the former, Chromatin immunoprecipitation assays provided evidence that ATOH8 binds to E-box regions within the HAMP promoter while the latter was supported by the finding that ATOH8 expression in HEK293 cells led to increased phosphorylated SMAD1,5,8 levels. Liver Atoh8 levels were reduced in mice under conditions associated with increased erythropoietic activity such as hypoxia, haemolytic anaemia, hypotransferrinaemia and erythropoietin treatment and increased by inhibitors of erythropoiesis. Hepatic Atoh8mRNA levels increased in mice treated with holo transferrin, suggesting that Atoh8 responds to changes in plasma iron. ATOH8 is therefore a novel transcriptional regulator of HAMP, which is responsive to changes in plasma iron and erythroid activity and could explain how changes in erythroid activity lead to regulation of HAMP.
Keywords: ATOH8; HAMP; iron; erythropoiesis; pSMAD1,5,8
Erythropoiesis is essential for life and is by far the body's largest user of iron, consuming almost two-thirds of the body's total iron. Increased erythropoietic activity has a rapid and dramatic effect on iron metabolism, which has been well documented (Finch, 1994). Increased erythropoietic activity generally results in an increase in the reticulocyte fraction in the blood with a concomitant fall in plasma iron as iron is used up by the developing erythrocytes. Increased erythropoietic activity is a powerful suppressor of the iron hormone hepcidin (HAMP) levels thereby allowing for more iron to be made available for erythropoiesis through increased intestinal iron absorption and iron release from macrophages via regulation of the hepcidin–ferroportin axis (Nicolas et al, 2002a; Weinstein et al, 2002). The suppression of HAMP by increased erythropoietic drive is not well understood and occurs even in conditions where liver iron levels are high and which would normally lead to increased HAMP levels, such as in β-thalassaemia (Nemeth & Ganz, 2006) and hypotransferrinaemia (Bartnikas et al, 2010). Thus the erythropoietic regulator appears to be capable of overiding the iron stores regulator of hepcidin.
Hepatic HAMP mRNA levels are regulated by three major stimuli: (i) tissue and serum iron concentration; (ii) inflammatory signals and (iii) erythropoieitic activity. Regulation of HAMP appears to occur mainly at the transcriptional level via various response elements within the HAMP promoter, such as the bone morphogenic proteins response elements (BMP-REs), signal transducer and activation of transcription 3 (STAT-3), cAMP response element binding protein (CREB), hepatocyte nuclear factor 4 (HNF4) and enhancer boxes (E-boxes) binding elements (Courselaud et al, 2002; Bayele et al, 2006; Wrighting & Andrews, 2006). The bone morphogenic protein (BMP) pathway is involved in regulating the responses of HAMP to changes in tissue iron via changes in hepatic BMP6 levels (Meynard et al, 2009; Ramos et al, 2011) and inflammatory signals act via STAT-3 resulting in activation of HAMP (Wrighting & Andrews, 2006).
Less is known about how changes in erythropoietic activity lead to altered hepatic HAMP levels, for example, what signals are sensed by the liver as a result of changes in erythropoiesis as well as the nature of the signal transduction mechanism are unclear. Both changes in serum iron and/or release of soluble factors from developing erythrocytes have been evoked as potential indicators of altered erythropoietic activity. The level of plasma holo-transferrin (transferrin saturation) changes rapidly as a result of altered erythropoietic activity and is a key modulator of liver HAMP levels (Bartnikas et al, 2010; Li et al, 2010) and thus could be one signal, however the signal transduction pathway leading to HAMP regulation has not been defined. On the other hand, soluble factors, such as growth differentiation factor 15 (GDF15) or twisted gastrulation factor 1 (TWSG1), produced by erythroid precursors, and bone morphogenic protein binding endothelial cell precursor-derived regulator (BMPER), produced by endothelial cells, have all been postulated to play a role in the suppression of HAMP (Tanno et al, 2007, 2009; Patel et al, 2012) by, in most cases, inhibiting BMP signalling. However, the roles of these molecules in the regulation of HAMP in other forms of anaemia and conditions with altered erythropoiesis have not been shown. Thus the molecular basis of erythropoietic regulation of HAMP remains unclear.
Atoh8 (or Math6) was originally identified as a distant mammalian homologue of the drosophila neural gene Atonal (Inoue et al, 2001). The Atoh8 mRNA encodes a basic helix loop helix (bHLH) transcription factor and is ubiquitiously expressed in mouse at least in embryonic tissues (Lynn et al, 2008). Atoh8 has been implicated in development of various tissues although its physiological function remains unknown (Lynn et al, 2008; Yao et al, 2010). Early studies had suggested that knock-out of Atoh8 was embryonic lethal in mice (Lynn et al, 2008) however, using an alternative targeting strategy, recent work has shown that mice survive Atoh8 ablation with no obvious phenotype (Rawnsley et al, 2013). bHLH or E-box proteins, such as ATOH8, bind to a palindromic (canonical) core consensus DNA sequence 5′-CANNTG-3′ known as an E-box element, where NN is usually CG or TG (Blackwell et al, 1993). Two canonical E-boxes have been described within the core human HAMP promoter with the sequence 5′-CACGTG-3′ and have been shown to bind other bHLH factors, USF1 and 2, as well as MYC and MAX (Bayele et al, 2006). Atoh8 was first linked with iron metabolism by Kautz et al (2008) who found that hepatic Atoh8 mRNA levels were up-regulated in mice chronically fed a high iron diet and down-regulated in those fed an iron-deficient diet. Thus Atoh8 appears to be the only known iron-regulated bHLH transcription factor. In addition we noted that Atoh8 mRNA was strongly down-regulated in liver expression microarrays of Tfrhpx/hpx mice (Patel et al, 2012), a mutant with a very high degree of liver iron overload, chronic anaemia and very low Hamp1 levels. Given that the regulation of Atoh8 was similar to Hamp1 we hypothesized that ATOH8 may be a transcriptional regulator of HAMP.
Here we report that ATOH8 can activate HAMP transcription and regulate cellular levels of pSMAD1,5,8. Moreover, Atoh8 mRNA and protein levels were regulated in mouse liver under various conditions with altered erythropoietic activity, providing a mechanistic link between erythropoiesis and HAMP transcription.
Materials and methods
Animals
Hypotransferrinaemic mice (HPX or Trfhpx/hpx) were bred and maintained as previously described (Simpson et al, 1991). Normal littermates (mixture of Trf+/+ and Trfhpx/+) were used as controls. Hypoxia was induced by placing 7-week-old male CD1 mice in a hypobaric chamber for 24–72 h, as previously described (Raja et al, 1989); controls of the same gender and age were maintained under normoxic conditions. Hamp1−/− mice and wild type (WT) littermates (all female C57BL/6/129 mixed background, aged 5–7 weeks old) were injected intraperitoneally with 60 mg/kg body weight of neutralized phenylhydrazine (PHZ) or saline solution twice on consecutive days as previously described (Masaratana et al, 2011) and sacrificed 3 d after the last injection. For erythropoietin (EPO), Carboplatin, Apo and Holo transferrin treatments, male 6-week-old C57BL/6 mice were switched to a diet containing <4 ppm iron (TD.80396; Harlan Teklad, Madison, WI, USA) for 10 d to reduce the effect of the high iron chow diet on HAMP expression as previously described (Pak et al, 2006). Mice received intraperitoneal injection of either 200 units of EPO (Jansen Cilag Ltd, High Wycombe, UK), 2·5 mg of carboplatin or 200 units of EPO with 2·5 mg of carboplatin (Sigma-Aldrich, Gillingham, UK) dissolved in 100 μl saline on three consecutive days. Control mice received 100 μl of saline. Mice were sacrificed 24 h after the last injection. Apo and Holo transferrin (10 mg) dissolved in 100 μl of saline was injected i.p (control mice received saline alone). Mice were sacrificed 6 h later. Serum iron was measured with a liquid ferrozine-based Fe reagent (Thermo Electron, Melbourne, Vic., Australia). Tissue non-haem iron was determined as previously described (Masaratana et al, 2012). All animal experiments were performed under the authority of a UK Home Office license.
Cell culture and HAMP promoter assays
HEK-293 cells were obtained from the American type culture collection (ATCC, Teddington, UK) and cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich, Gillingham, UK) and 10% heat-inactivated fetal bovine serum (FBS; Sigma-Aldrich), penicillin-streptomycin and glutamine (Sigma-Aldrich). Cell cultures were maintained at 37°C under 95% air/5% CO2. Promoter assays employed approximately 0·9 kb of the human HAMP promoter (WT) cloned in the pGL3-basic luciferase reporter vector (Promega, Southampton, UK). E-box and BMP-RE mutated versions of this vector were kindly provided by Dr Pavle Matak (Department of Pharmacology and Cancer Biology, Duke University Medical Center, Durham NC, details can be found in Table SI). Reporter constructs were co-transfected into cells with a TK-renilla (3:1 ratio) using Fugene-6 (Roche Diagnostics, Burgess Hill, UK). Human ATOH8 –DDK (Flag) tagged plasmid (Origene technologies, Rockville, MD, USA) was co-transfected along with the reporter plasmids. Luminescence was detected using Dual-Luciferase Reporter Assay system and measured by luminometer (Promega).
Western blotting and immunohistochemistry
Whole cell lysates were extracted from mouse liver or cultured HEK 293 cells by homogenization in 500 μl of radioimmunoprecipitation assay buffer (10 mmol/l Tris, 150 mmol/l NaCl, 1 mmol/l EDTA, 1% Nonidet P-40, 0·1% sodium dodecyl sulphate [SDS]) and protease inhibitor cocktail (1:200 dilution; Sigma Aldrich). The homogenates were centrifuged at 1000 × g at 4°C for 5 min. Nuclear protein from cells and tissues was extracted using the NE-PER nuclear and cytoplasmic extraction kit (Thermo Fisher Scientific, Loughborough, UK) according to the manufacturer's instructions. Protein was quantified using a protein assay (BioRad, Hemel Hempstead, UK) and resolved using pre-cast 10-12% reducing SDS polyacrylamide gel electrophoresis (SDS-PAGE; BioRad) before transfer to polyvinylidene difluoride (PVDF) membrane using a Trans blot Turbo (BioRad). Anti-ATOH8 and anti-DDK (FLAG) (Origene technologies) and pSMAD 1,5,8 (Cell Signaling Technology, Danvers, MA, USA) were used to detect the respective proteins. SMAD1 (Santa Cruz Biotechnology, Heidelberg, Germany) or beta actin (Sigma-Aldrich) were used as controls for protein loading. Blots were visualized by chemiluminescence (Thermo Fisher Scientific, Loughborough, UK). Immunohistochemistry was performed on cryostat sections of mouse liver as previously described (Patel et al, 2012) using anti-ATOH8 and fluorescein isothicyanate- conjugated secondary (Dako, Ely, UK). Sections were counterstained with propidium iodide (Vector Laboratories, Peterborough, UK) and images captured using Leica LS-2 confocal microscope (Leica Microsytems, Milton Keynes, UK).
Quantitative polymerase chain reaction (qPCR)
One microgram of total liver RNA was reverse transcribed using a Transcriptor High Fidelity cDNA kit (Roche Diagnostics). All primers were designed using Universal Probe Library system (Roche Diagnostics) and qPCR was performed using an ABI PRISIM 7900 HT PCR machine (Applied Biosystems, Paisley, UK). Results were normalized to the housekeeping RNA Rpl19. Fold change was calculated using the method of Livak and Schmittgen (2001). In the case of HAMP, qPCR (Fig1B) results were normalized to the housekeeping RNA RPL19 expressed as the negative of ΔcT. Details of primer sequences used are presented in Table SI.
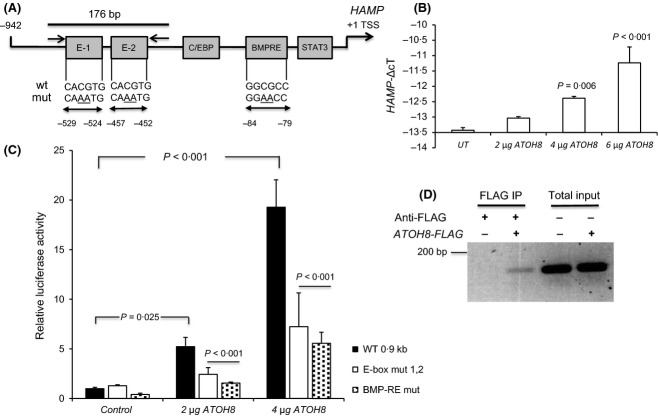
Figure 1.
ATOH8 regulates HAMP transcription. (A) Schematic showing HAMP promoter and locations of bone morphogenic proteins response element (BMP-RE) and E-box region and mutations introduced. (B) Quantitative polymerase chain reaction (qPCR) assay of endogenous HAMP levels (normalized to RPL19 plotted as –ΔcT values) in HEK 293 cells after transfection with 2, 4 or 6 μg ATOH8-FLAG plasmid. (C) HAMP promoter luciferase reporter assays in HEK 293 cells transfected with wild type (WT) HAMP, E-box mutant (E-box mut 1,2) or BMP-RE mutant (BMP-RE mut) after co-transfection with 2 or 4 μg of ATOH8-FLAG. Promoter activity was expressed relative to WT promoter activity without ATOH8 co-transfection. Luciferase assays shown are means ± SD derived from a single experiment with three biological replicates and experiment shown is representative of at least three similar experiments. (D) CHIP assay: chromatin DNA was immunoprecipitated from untransfected HEK 293 cells or cells transfected with ATOH8-FLAG. Immunoprecipitation (IP) was performed using Anti-FLAG antibody. PCR (40 cycles) was performed on the IP material and 10% of the total input using primers flanking the E-box region (Fig 1A) and products run on a 1·5% agarose gel stained with ethidium bromide. Statistical analysis was performed using 1 or 2-way anova with Tukey's post hoc test.
Chromatin immunoprecipitation (CHIP) assays
Chromatin immunoprecipitation assays were performed using a commercially available kit (Thermo Fisher Scientific). Chromatin DNA was prepared from untransfected HEK 293 cells and cells transfected with ATOH8-FLAG following the manufacturer's protocol and immunoprecipitated using Anti-FLAG antibody (Origene technologies, Rockville, MD, USA). PCR (40 cycles) was performed using the primers (5′ CCAGTTACCAGAGCCACATC 3′ and 5′ CAGGAGTGTCTGCATGTTG 3′), generating a 176 bp fragment encompassing the E-box 1 and 2 region (Fig1A). Control PCRs were performed using 10% of the input DNA.
Statistical analysis
Data are presented as means ± SD. Statistical differences were determined using spss (IBM, Portsmouth, UK) where appropriate using either 1-way analysis of variance (anova) followed by Tukey's post hoc test or two-tailed Students t-test. 2-way analysis of variance (2-way anova) was used to test for significance between two or more groups and Bonferroni post-hoc test for interactions. A P value of <0·05 was considered as significant.
Results
ATOH8 regulates HAMP transcription and pSMAD1,5,8 levels in vitro
To test whether ATOH8 could play a role in regulating HAMP transcription, HEK-293 cells were transfected with increasing amounts of an ATOH8-FLAG tagged expression plasmid. Endogenous HAMP mRNA levels were increased by up to fourfold (P < 0·006) following transfection with increasing amounts of ATOH8-FLAG (Fig1B). In accord with this, HAMP promoter activity was increased by around 20- fold (P < 0·001) in cells transfected with ATOH8 (Fig1C) and 0·9-kb of the human HAMP promoter fused to the luciferase gene (Fig1A). In both cases the effect of ATOH8 was dose-dependent.
We next investigated the effect of mutation of the two E-box elements within the HAMP promoter previously shown to bind other bHLH proteins (Bayele et al, 2006). Mutation of the internal dinucleotide within the E-box elements from 5′-CACGTG-3′ to 5′-CAAATG-3′ (Fig1A) abolishes nuclear factor binding(Chen et al, 2012). Mutation of both E-boxes attenuated ATOH8-dependent HAMP promoter activity by more than 50% (P < 0·001) when compared to the WT promoter treated with ATOH8 (Fig1C). In addition, mutation of the BMP response element (BMP-RE) resulted in a 50% (P < 0·001) reduction in ATOH8 dependent HAMP promoter activation compared to WT treated with ATOH8 (Fig1C). To provide evidence for promoter occupancy by ATOH8 we performed CHIP assays using HEK 293 cells transfected with ATOH8-FLAG. Using the Anti-FLAG antibody as the immunoprecipitation (IP) antibody we were able to amplify a 176 bp band encompassing the E-box 1 and 2 regions of the HAMP promoter from cells transfected with ATOH8-FLAG but not untransfected cells (Fig1D).
We next investigated whether ATOH8 transfection affected pSMAD 1,5,8 levels in HEK 293 cells. ATOH8 transfection significantly increased pSMAD 1,5,8 levels (P = 0·001, Fig2C) in a dose-dependent fashion (Fig2A,B). Hence, ATOH8 appears to regulate HAMP in two ways, firstly by acting directly on the HAMP promoter via E-boxes and indirectly through increased pSMAD1,5,8 levels. This may explain why mutation of E-boxes does not fully repress HAMP promoter activity.
Figure 2.
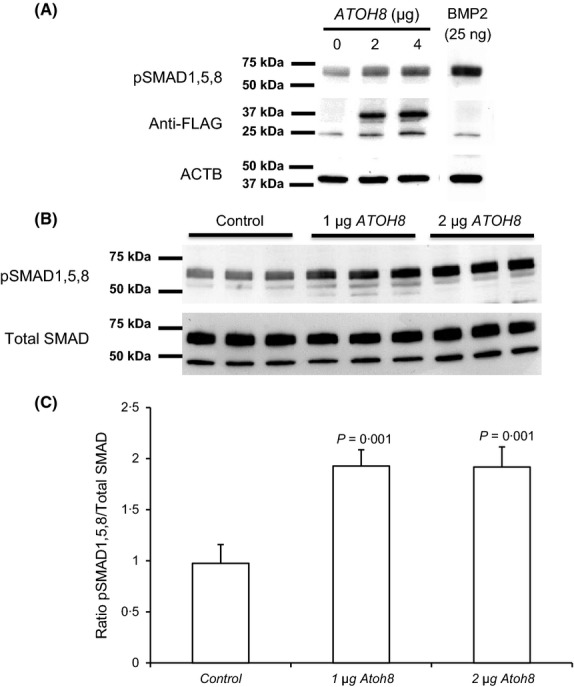
ATOH8 transfection increases pSMAD1,5,8 levels. (A) Western blot showing pSMAD 1,5,8 levels in HEK 293 cells after transfection with increasing amounts of ATOH8-FLAG (0, 2 and 4 μg plasmid DNA) compared with 25 ng of BMP2 as a positive control; lower panels show same blot re-probed with Anti-FLAG and ACTB (β-actin) antibodies. All lanes in A were run on the same gel, blotted and processed together and are from the same exposure. (B) Western blot of pSMAD 1,5,8 levels in HEK cells after transfection with ATOH8 in comparison with total SMAD. (C) Densitometry of Western blots in B showing ratio of pSMAD1,5,8 to total SMAD. Statistical analysis was performed using 1-way anova with Tukey's post hoc test.
Regulation of hepatic ATOH8 levels in mouse models with altered erythropoietic activity
We confirmed the significant down regulation of Atoh8 in liver of Tfrhpx/hpx mice by qPCR (P = 0·001, Fig3A). In addition, reduced ATOH8 protein was evident by both Western blotting and immunohistochemistry (Fig3B,C) in Tfrhpx/hpx mice compared to controls. Thus it appears that ATOH8 upregulation by iron was overridden in Tfrhpx/hpx mice in a similar fashion to regulation of liver Hamp1 levels.
Figure 3.
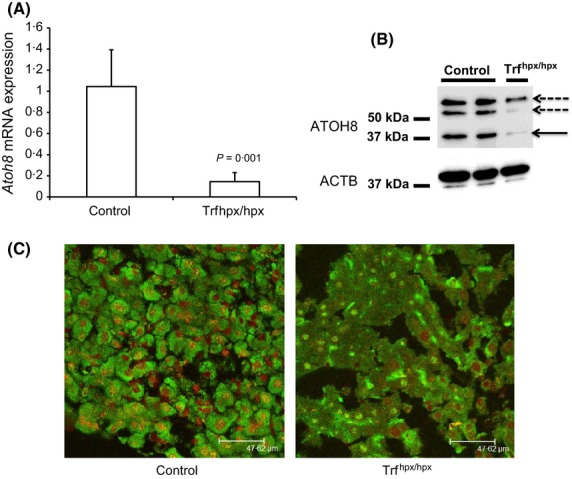
Expression of Atoh8 in HPX mouse liver. (A) Q-PCR shows relative Atoh8mRNA levels as (normalized to Rpl19, plotted as fold change relative to control) in 10- to 11-week-old male Trfhpx/hpx mice compared to control (Trfhpx/+) mice (P = 0·001 Student's ‘t’ test, n = 3 for each group). (B) Western blots for ATOH8 protein in liver extracts from two control (Trfhpx/+) and one Trfhpx/hpx mouse. Solid arrow indicates predicted molecular weight of ATOH8 (∽37 kDa), dashed arrows indicate possible homo- or heterodimers. (C) ATOH8 immunostaining (visualized in green) in liver sections from male 7-week-old Trfhpx/hpx compared to an age- and sex-matched control (Trfhpx/+); counterstain is propidium iodide (red).
We reasoned that the reduction in ATOH8 in Tfrhpx/hpx mouse liver may be driven by increased erythroid activity. Treatment of rats or mice with PHZ leads to increased erythropoietic rate and suppression of Hamp1 usually after a lag period of 3 d (Frazer et al, 2004; Latunde-Dada et al, 2006; Masaratana et al, 2012). In mice injected with PHZ there was an approximate 27-fold increase in the percentage of blood reticulocytes (Raja et al, 1989), reduced or absent serum iron and a 2–3 fold increase in liver non-haem iron. In normal mice (C57BL/6/129 mixed background) sacrificed 3 d after PHZ treatment, ATOH8 protein levels decreased by around eightfold (P = 0·001, Fig4A,B) while Atoh8 mRNA levels were reduced by around twofold (P > 0·018) (Fig4C). We considered the possibility that ATOH8 may be regulated downstream of Hamp1, however similar reductions in ATOH8 protein and mRNA levels were observed in Hamp1−/− mice treated with PHZ (Fig4A,B,C), suggesting that regulation of ATOH8 is upstream of Hamp1. The response of liver ATOH8 protein and mRNA levels to PHZ was also similar in CD1 and C57BL/6 mice and in both male and female mice (data not shown). Smad7 and Id1 levels were suppressed by PHZ treatment in control and Hamp1−/− mice, however the decrease was only statistically significant in the case of Smad7 (Fig S1). Thus in another model with increased erythropoiesis and liver iron loading, hepatic ATOH8 levels followed the same downward direction as Hamp1.
Figure 4.
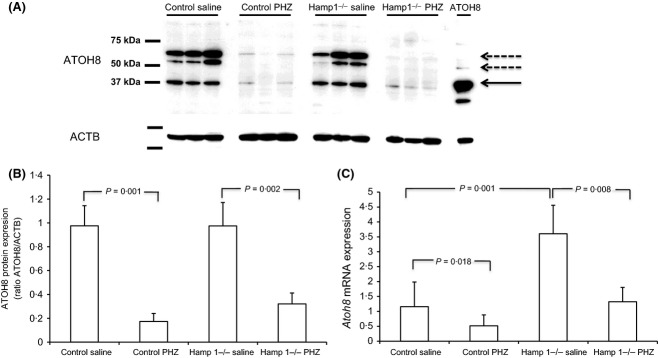
Liver ATOH8 levels in control and Hamp1−/− mice with induced haemolytic anaemia. (A) Western blot showing ATOH8 protein levels in liver extracts from individual mice (three per group). Last lane shows extract from HEK293 cells transfected with ATOH8 plasmid as a positive control. Solid arrow indicates predicted molecular weight of ATOH8 (∽37 kDa), dashed arrows indicate possible homo- or heterodimers, lower panel shows same blot re-probed for ACTB (β-actin). (B) Densitometry of Western blots performed in panel A showing ratio of ATOH8 to ACTB. (C) qPCR shows Atoh8mRNA levels in saline-injected control mice (n = 7) versus control mice injected with PHZ (n = 8) and saline-injected Hamp1−/− mice (n = 6) versus Hamp1−/− injected with PHZ (n = 7). Control mice were wild type littermates. (Atoh8 levels were normalized to Rpl19 and plotted as fold change relative to control). Values are means ± SD. Statistical comparisons were made using 1-way anova with Tukey's post hoc test.
Given that PHZ injection results in other effects, such as release of haem and increased oxidative stress, we investigated the effects of other more physiological modulators of erythropoiesis, such as exposure of mice to hypoxia and EPO injection on liver Atoh8 levels. In mice, EPO injection and exposure to hypoxia lead to increased erythropoiesis and suppression of liver Hamp1 mRNA (Nicolas et al, 2002a; Pak et al, 2006). On the other hand, injection of the cytotoxic agent carboplatin, which inhibits erythropoiesis, results in increased liver Hamp1 (Pak et al, 2006; Bartnikas et al, 2010). As previously shown, we found that liver Hamp1 mRNA was suppressed by EPO treatment and increased by carboplatin treatment (Fig S2A). EPO treatment significantly reduced liver Atoh8 mRNA levels by around twofold compared to control mice (P = 0·01), whereas mice treated with carboplatin alone or EPO and carboplatin had significantly higher Atoh8 mRNA levels (P < 0·01) compared to control or EPO-treated mice (Fig5A). Mice treated with carboplatin alone showed the highest induction of Atoh8 mRNA levels (approximately fourfold induction, P < 0·001).
Figure 5.
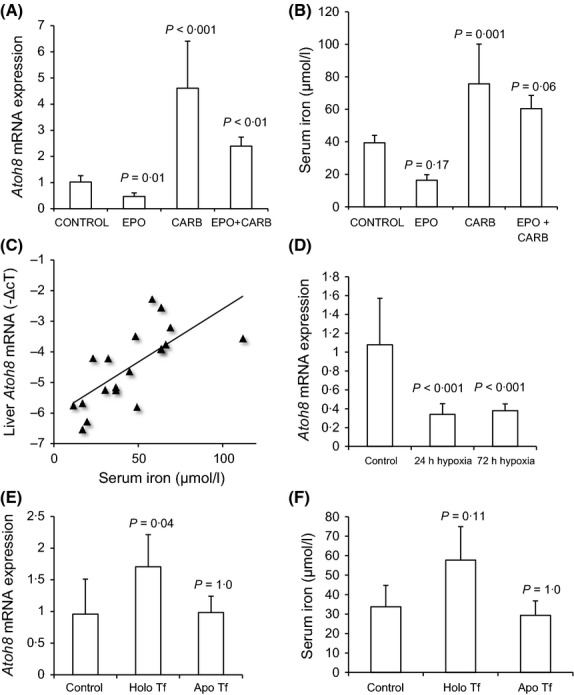
Effect of Hypoxia, EPO, Carboplatin and Transferrin on liver Atoh8 mRNA levels. (A) qPCR shows relative Atoh8 levels (normalized to Rpl19; plotted fold-change relative to control). Control (saline-injected; n = 7), EPO (n = 4), carboplatin (CARB; n = 4) and EPO and Carboplatin injected mice (EPO + CARB; n = 4). (B) serum iron levels in treated mice. (C) Correlation between serum iron and Atoh8mRNA in all mice (Pearson correlation coefficient, R2 = 0·70, P = 0·013, n = 19). (D) qPCR of liver Atoh8mRNA levels in control (normoxia) and in mice exposed to hypoxia for 24 and 72 h (Atoh8 levels normalized to Rpl19, plotted as fold-change relative to control) (E) Atoh8 levels in control (saline injected) (n = 7), Holo-transferrin (n = 4) or Apo-transferrin (n = 4) (Atoh8 levels normalized to Rpl19; plotted as fold-change relative to control) (F) serum iron levels in treated mice. Data are presented as mean ± SD. Statistical comparisons were made using 1-way anova with Tukey's post hoc test.
Serum iron levels generally tracked liver Atoh8 levels (Fig5B) although some of the treatments did not reach statistical significance (EPO treatment and EPO with carboplatin). There was a significant correlation between serum iron and Atoh8 mRNA levels when all experimental groups were taken together (R2 = 0·70, P = 0·013, n = 19, Fig5C). Liver non-haem iron concentration was not significantly affected by EPO or carboplatin treatments alone but increased significantly with carboplatin and EPO treatment (P < 0·001, Fig S3) and there was no correlation between liver iron concentration and Atoh8 mRNA levels (R2 = 0·41, P = 0·07, n = 15). Liver Id1 and Smad7 mRNA levels were not affected by EPO treatment although levels of both were increased by carboplatin treatment (Fig S4). In contrast to Atoh8 and Hamp1,Id1 and Smad7 levels were increased further by carboplatin plus EPO treatments (Fig S4).
Hypoxia is a well-known physiological stimulator of erythropoiesis that is known to suppress liver HAMP levels in both mice and man (Nicolas et al, 2002a; Talbot et al, 2012). Exposure of mice to 24 or 72 h hypoxia reduced liver Atoh8 mRNA levels by around threefold (P < 0·001, Fig5D). Thus, Atoh8 was regulated by altered erythropoietic activity in the same direction as Hamp1 and hepatic Atoh8 mRNA levels correlated with serum iron.
Holo-transferrin has a direct effect on liver Atoh8 mRNA levels
Holo-transferrin is thought to be a key regulator of liver Hamp1, therefore we investigated whether diferric transferrin had any direct effect on liver Atoh8 mRNA levels. C57BL/6 mice were treated with 10 mg of holo-transferrin via i.p injection and sacrificed 6 h later, a treatment previously shown to increase liver Hamp1 levels without changing liver iron (Ramos et al, 2011). As previously shown (Ramos et al, 2011) liver Hamp1 mRNA levels were significantly induced by holo-transferrin but not apo-transferrin (Fig S2B). Treatment of mice with holo-transferrin resulted in an approximate twofold increase in Atoh8 mRNA levels (P = 0·037) whereas apo-transferrin had no effect (Fig5E). Serum iron increased in holo-transferrin injected mice although this did not reach statistical significance (P = 0·11, Fig5F). There was no significant effect of holo- or apo-transferrin on liver non-haem iron levels (Fig S3) or on hepatic Smad7 and Id1 mRNA levels (Fig S4A,B). Thus serum levels of diferric transferrin can directly regulate liver Atoh8 mRNA levels.
Discussion
This study establishes ATOH8 as a novel candidate transcriptional regulator of hepatic HAMP levels and cellular pSMAD1,5,8 levels. ATOH8 stimulated HAMP transcription while mutation of E-boxes within the HAMP promoter attenuated ATOH8-dependent HAMP transcriptional responses and CHIP assays provided additional evidence that ATOH8 binds to these E-boxes both in vivo and in vitro. Although MYC, MAX, USF1 and 2 can bind the same E-boxes in vitro (Bayele et al, 2006), the physiological role of these proteins in iron metabolism in vivo remains unclear. In the case of USF1 and 2 with the well-known exception of a Usf2 knockout mouse in which the Hamp1 locus was also disrupted (Nicolas et al, 2001), selective knock out of either Usf1 (Nicolas et al, 2001) or Usf2 (Nicolas et al, 2002b) in mice has no effect on liver Hamp1 levels or iron metabolism. Furthermore, analysis of the published array data supplied by Kautz et al (2008) for iron loaded and iron deficient mouse liver (Data available at the National Center for Biotechnology Information [NCBI] Geo database (Edgar et al, 2002), accession GSE10421) in C57 and DBA strains shows that liver Usf1 mRNA was not iron-regulated in either strain whereas Usf2 mRNA was decreased by iron loading in both strains of mice but also decreased in iron deficiency. In the HPX mouse liver, we found no change in Usf1 and an increase in Usf2 mRNA levels (data not shown). In contrast, hepatic Atoh8 and Hamp1 mRNA levels correlated in vivo in mice over a wide range of conditions of altered iron metabolism (iron overload, iron deficiency, hypotransferrinaemia, hypoxia, PHZ, EPO and carboplatin treatment). Moreover, downregulation of ATOH8 by increased erythropoietic drive occurred in Hamp1 null mice, a scenario consistent with ATOH8 being an upstream regulator of Hamp1. In vivo, there is likely to be competition between the various E-box proteins for binding to the HAMP promoter and which protein binds would depend on hepatic expression levels, DNA binding affinity as well as other tissue and gene-specific factors. At present it is unclear what the nature of the higher molecular weight bands found on ATOH8 liver Western blots (Figs3B and 4A) are, however they appear to be regulated in the same manner as the 37 kDa ATOH8 band. bHLH proteins, such as MYC and MAX homo or heterodimerize with each other in order to bind DNA (Blackwell et al, 1993). It is possible that these higher molecular weight bands are SDS-resistant dimers with other bHLH proteins although further work will be required to identify these.
ATOH8 also regulated pSMAD1,5,8 levels, providing an additional mechanism by which ATOH8 could influence HAMP levels. This was supported by the finding that mutation of the BMP-RE in the HAMP promoter also attenuated ATOH8-dependent HAMP transcription. It has been suggested that increased erythropoietic activity in mice after PHZ treatment can attenuate BMP6 signalling and decrease liver Hamp1 levels without any change in pSMAD1,5,8 levels (Frazer et al, 2012). We speculate that the reduction in liver ATOH8 as observed following acute PHZ treatment could negate the effect of increased BMP6 levels on pSMAD1,5,8, levels and reduce E-box-dependent transcriptional activation of HAMP.
Previous work has established that hypoxia and EPO suppresses HAMP indirectly through stimulation of erythropoiesis while inhibition of erythropoiesis with carboplatin leads to increases in HAMP (Pak et al, 2006; Talbot et al, 2012). Liver Atoh8 levels responded to these stimuli in a similar manner and direction to HAMP, suggesting Atoh8 responds to the same systemic cues as HAMP. What these cues are remains to be fully elucidated.
Transferrin saturation correlates directly with erythropoietic activity (Frazer et al, 2004) while numerous studies in vivo show that serum diferric transferrin levels correlate with liver Hamp1 levels in mice (Wilkins et al, 2006; Bartnikas et al, 2010; Li et al, 2010; Ramos et al, 2011). It is thought that increased diferric transferrin levels leads to stabilization of TfR2, possibly due to binding of Hfe (Robb & Wessling-Resnick, 2004; Schmidt et al, 2008), generating an as yet unidentified signal leading to increased HAMP levels. Our data, showing that that holo-transferrin also directly regulates liver Atoh8 levels, suggest that this signalling pathway may involve ATOH8 (Fig6). However ATOH8 levels were also suppressed in Hamp1−/− mice after PHZ treatment where plasma iron remains high in the former (Masaratana et al, 2012) Thus it is possible that other as yet unidentified erythroid factor (s) released from rapidly developing erythrocytes or the bone marrow also regulate Atoh8 levels (Fig6). Interestingly, hepatic Atoh8 levels were not increased in Hfe knockout mice, in contrast to other iron loaded models (Kautz et al, 2008, 2009). This indicates that HFE may also be required for regulation of ATOH8. Further work is required to uncover the link between iron sensing molecules and ATOH8.
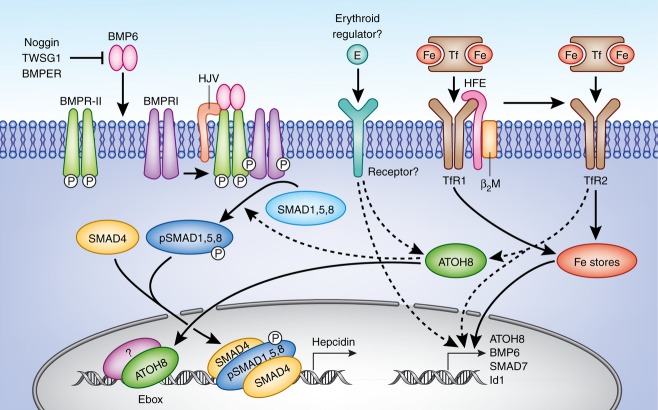
Figure 6.
Working model Hepatic BMP6, ATOH8, SMAD7 and ID1 levels are increased by increased tissue iron stores, however only ATOH8 levels are decreased by increased erythropoietic activity. ATOH8 regulates HAMP levels via direct transcriptional activation of the HAMP gene via binding to E-boxes (possibly as a heterodimer with other as yet unidentified bHLH or bHLH-ZIP proteins) and by modulation of cellular pSMAD1,5,8 levels. Reductions in hepatic ATOH8 levels under increased erythroid activity lead to reduced pSMAD1,5,8 levels and E-box dependent HAMP transcription. Dashed arrows indicate hypothetical connections between ATOH8 and other proteins. ATOH8 may be a component of signal transduction pathways linking HAMP transcription with levels of diferric transferrin (via iron-sensing molecules such as HFE and TfR2) and with erythroid activity (by as yet unidentified erythroid regulators).
To date, none of the other factors known to be involved in BMP signalling, including BMPs2, 4 and 9, Alk2 (ACVR1), Alk3 (BMPR1A), Hjv (HFE2) and TMPRSS6, have been shown to be iron-regulated or regulated by changes in erythropoietic activity in vivo. SMAD7 is a known inhibitor of the BMP signalling pathway while ID1 is an HLH transcription factor of unknown function, which acts as a dominant negative inhibitor of other bHLH proteins (Pesce & Benezra, 1993) because it lacks a basic DNA binding domain but can still form heterodimers (Langlands et al, 1997; Bounpheng et al, 1999). SMAD7 and ID1 are iron regulated (Kautz et al, 2008) and therefore could influence HAMP transcription under increased erythropoietic activity. However, given that levels of both Smad7 and Id1 mRNA decreased in livers of mice treated with PHZ, and were unaffected by holo-transferrin or EPO treatments, its seems unlikely that either are of major importance in HAMP suppression under enhanced erythropoietic drive.
BMP6 has been dubbed the iron stores regulator as several studies have suggested that tissue iron rather than serum iron is the dominant regulatory factor for BMP6 (Ramos et al, 2011; Frazer et al, 2012). Recently, it has been revealed that liver Hamp1 levels increased markedly in Bmp6 knockout mice following chronic iron loading, indicating other pathways in addition to BMP6 are involved in the regulation of Hamp1 by iron (Ramos et al, 2011). A plausible explanation, based on our data, is that the effects of iron on Hamp1 expression in the absence of BMP6 are mediated by stimulation of the Atoh8 pathway. Thus BMP6 may modulate Hamp1 in response to changes tissue iron whereas Atoh8 may regulate responses to serum iron and/or changes in erythropoietic activity that dominate under certain circumstances. Given that ATOH8 affects at least two pathways which regulate HAMP transcription (BMP signalling and E-Box dependent transcription) ATOH8 could have a strong influence hepatic Hamp1. This would allow Hamp1 responses to various stimuli, explaining how suppression occurs by increased erythropoietic activity even in the face of liver iron loading and increased BMP6 levels.
In summary, we identify ATOH8 as a novel transcriptional regulator of HAMP via two independent mechanisms: E-box dependent transcriptional activation; and regulation of cellular pSMAD1,5,8 levels. The regulation of liver ATOH8 levels observed in mice with altered erythropoiesis suggests ATOH8 as a novel physiological regulator of HAMP. ATOH8 may link erythropoietic activity and iron-sensing molecules to HAMP transcription and will open up new avenues of research leading to improved therapies and management of iron overload disorders, such as haemochromatosis and β-thalassaemia.
Acknowledgments
This work was funded by a Kings College Graduate School Bursary supported by the BBSRC (NP), the Anandamahidol Foundation (PM) and an Early Career Fellowship awarded by the Wellcome Trust-DBT India Alliance to JV. We acknowledge the help of Yen Fei Wong and thank Pavle Matak for providing the HAMP promoter constructs and Sophie Vaulont for the Hamp1−/− mice.
Authorship
N. Patel, P. Masaratana, O. Latunde-Dada, and J. Varghese performed research and analysed data. R.J. Simpson and Molly Jacob analysed data and wrote the paper, A.T. McKie performed research, analysed data and wrote the paper.
Conflict of interest disclosure
The authors have no conflicting interests.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig S1. Hepatic Id1 and Smad7 mRNA levels in control and Hamp1−/− mice with induced haemolytic anemia.
Fig S2. Effect of modulators of erythropoiesis and changes in transferrin levels on hepatic Hamp1 mRNA expression.
Fig S3. Effect of modulators of erythropoiesis and changes in transferrin levels on liver non-heme iron levels.
Fig S4. Effect of modulators of erythropoiesis and changes in transferrin levels on hepatic Id1 and Smad7 expression.
Table SI. Primer sequences.
References
- Bartnikas TB, Andrews NC, Fleming MD. Transferrin is a major determinant of hepcidin expression in hypotransferrinemic mice. Blood. 2010;117:630–637. doi: 10.1182/blood-2010-05-287359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayele HK, McArdle H, Srai SKS. Cis and trans regulation of HAMP expression by upstream stimulatory factor. Blood. 2006;108:4237–4245. doi: 10.1182/blood-2005-07-027037. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Huang J, Ma A, Kretzner L, Alt FW, Eisenman RN, Weintraub H. Binding of myc proteins to canonical and noncanonical DNA sequences. Molecular and Cellular Biology. 1993;13:5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounpheng MA, Melnikova IN, Dimas JJ, Christy BA. Identification of a novel transcriptional activity of mammalian Id proteins. Nucleic Acids Research. 1999;27:1740–1746. doi: 10.1093/nar/27.7.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Hsu R, Li Z, Kogut PC, Du Q, Rouser K, Camoretti-Mercado B, Solway J. Upstream stimulatory factor 1 activates GATA5 expression through an E-box motif. Biochemical Journal. 2012;446:89–98. doi: 10.1042/BJ20111942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courselaud B, Pigeon C, Inoue Y, Inoue J, Gonzalez FJ, Leroyer P, Gilot D, Boudjema K, Guguen-Guillouzo C, Brissot P, Loréal O, Ilyin G. C/EBPα regulates hepatic transcription of HAMP, an antimicrobial peptide and regulator of iron metabolism. Journal of Biological Chemistry. 2002;277:41163–41170. doi: 10.1074/jbc.M202653200. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch C. Regulators of iron balance in humans. Blood. 1994;84:1697–1702. [PubMed] [Google Scholar]
- Frazer DM, Inglis HR, Wilkins SJ, Millard KN, Steele TM, McLaren GD, McKie AT, Vulpe CD, Anderson GJ. Delayed HAMP response explains the lag period in iron absorption following a stimulus to increase erythropoiesis. Gut. 2004;53:1509–1515. doi: 10.1136/gut.2003.037416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer DM, Wilkins SJ, Darshan D, Badrick AC, McLaren GD, Anderson GJ. Stimulated erythropoiesis with secondary iron loading leads to a decrease in HAMP despite an increase in bone morphogenetic protein 6 expression. British Journal of Haematology. 2012;157:615–626. doi: 10.1111/j.1365-2141.2012.09104.x. [DOI] [PubMed] [Google Scholar]
- Inoue C, Bae S-K, Takatsuka K, Inoue T, Bessho Y, Kageyama R. Math6, a bHLH gene expressed in the developing nervous system, regulates neuronal versus glial differentiation. Genes to Cells. 2001;6:977–986. doi: 10.1046/j.1365-2443.2001.00476.x. [DOI] [PubMed] [Google Scholar]
- Kautz L, Meynard D, Monnier A, Darnaud V, Bouvet R, Wang R-H, Deng C, Vaulont S, Mosser J, Coppin H, Roth M-P. Iron regulates phosphorylation of Smad1/5/8 and gene expression of Bmp6, Smad7, Id1, and Atoh8 in the mouse liver. Blood. 2008;112:1503–1509. doi: 10.1182/blood-2008-03-143354. [DOI] [PubMed] [Google Scholar]
- Kautz L, Meynard D, Besson-Fournier C, Darnaud V, Al Saati T, Coppin H, Roth MP. BMP/Smad signaling is not enhanced in Hfe-deficient mice despite increased Bmp6 expression. Blood. 2009;114:2515–2520. doi: 10.1182/blood-2009-02-206771. [DOI] [PubMed] [Google Scholar]
- Langlands K, Yin X, Anand G, Prochownik EV. Differential interactions of Id proteins with basic-helix-loop-helix transcription factors. Journal of Biological Chemistry. 1997;272:19785–19793. doi: 10.1074/jbc.272.32.19785. [DOI] [PubMed] [Google Scholar]
- Latunde-Dada GO, McKie AT, Simpson RJ. Animal models with enhanced erythropoiesis and iron absorption. Biochimica et Biophysica Acta (BBA) – Molecular Basis of Disease. 2006;1762:414–423. doi: 10.1016/j.bbadis.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Li H, Rybicki AC, Suzuka SM, von Bonsdorff L, Breuer W, Hall CB, Cabantchik ZI, Bouhassira EE, Fabry ME, Ginzburg YZ. Transferrin therapy ameliorates disease in [beta]-thalassemic mice. Nature Medicine. 2010;16:177–182. doi: 10.1038/nm.2073. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lynn FC, Sanchez L, Gomis R, German MS, Gasa R. Identification of the bHLH factor Math6 as a novel component of the embryonic pancreas transcriptional network. PLoS ONE. 2008;3:e2430. doi: 10.1371/journal.pone.0002430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaratana P, Laftah AH, Latunde-Dada GO, Vaulont S, Simpson RJ, McKie AT. Iron absorption in HAMP1 knockout mice. British Journal of Nutrition. 2011;105:1583–1591. doi: 10.1017/S0007114510005507. [DOI] [PubMed] [Google Scholar]
- Masaratana P, Latunde-Dada GO, Patel N, Simpson RJ, Vaulont S, McKie AT. Iron metabolism in HAMP1 knockout mice in response to phenylhydrazine-induced hemolysis. Blood Cells, Molecules, & Diseases. 2012;49:85–91. doi: 10.1016/j.bcmd.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Meynard D, Kautz L, Darnaud V, Canonne-Hergaux F, Coppin H, Roth MP. Lack of the bone morphogenetic protein BMP6 induces massive iron overload. Nature Genetics. 2009;41:478–481. doi: 10.1038/ng.320. [DOI] [PubMed] [Google Scholar]
- Nemeth E, Ganz T. HAMP and iron-loading anemias. Haematologica. 2006;91:727–732. [PubMed] [Google Scholar]
- Nicolas G, Bennoun M, Devaux I, Beaumont C, Grandchamp B, Kahn A, Vaulont S. Lack of HAMP gene expression and severe tissue iron overload in upstream stimulatory factor 2 (USF2) knockout mice. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8780–8785. doi: 10.1073/pnas.151179498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas G, Chauvet C, Viatte L, Danan JL, Bigard X, Devaux I, Beaumont C, Kahn A, Vaulont S. The gene encoding the iron regulatory peptide HAMP is regulated by anemia, hypoxia, and inflammation. Journal of Clinical Investigation. 2002a;110:1037–1044. doi: 10.1172/JCI15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas G, Bennoun M, Porteu A, Mativet S, Beaumont C, Grandchamp B, Sirito M, Sawadogo M, Kahn A, Vaulont S. Severe iron deficiency anemia in transgenic mice expressing liver HAMP. Proceedings of the National Academy of Sciences of the United States of America. 2002b;99:4596–4601. doi: 10.1073/pnas.072632499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak M, Lopez MA, Gabayan V, Ganz T, Rivera S. Suppression of HAMP during anemia requires erythropoietic activity. Blood. 2006;108:3730–3735. doi: 10.1182/blood-2006-06-028787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N, Masaratana P, Diaz-Castro J, Latunde-Dada GO, Qureshi A, Lockyer P, Jacob M, Arno M, Matak P, Mitry RR, Hughes RD, Dhawan A, Patterson C, Simpson RJ, McKie AT. BMPER protein is a negative regulator of HAMP and is up-regulated in hypotransferrinemic mice. Journal of Biological Chemistry. 2012;287:4099–4106. doi: 10.1074/jbc.M111.310789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesce S, Benezra R. The loop region of the helix-loop-helix protein Id1 is critical for its dominant negative activity. Molecular and Cellular Biology. 1993;13:7874–7880. doi: 10.1128/mcb.13.12.7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja KB, Simpson RJ, Pippard MJ, Peters TJ. In vivo studies on the relationship between intestinal iron (Fe3+) absorption, hypoxia and erythropoiesis in the mouse. British Journal of Haematology. 1988;68:373–378. doi: 10.1111/j.1365-2141.1988.tb04217.x. [DOI] [PubMed] [Google Scholar]
- Raja KB, Simpson RJ, Peters TJ. Effect of exchange transfusion of reticulocytes on in vitro and in vivo intestinal iron (Fe3+) absorption in mice. British Journal of Haematology. 1989;73:254–259. doi: 10.1111/j.1365-2141.1989.tb00261.x. [DOI] [PubMed] [Google Scholar]
- Ramos E, Kautz L, Rodriguez R, Hansen M, Gabayan V, Ginzburg Y, Roth M-P, Nemeth E, Ganz T. Evidence for distinct pathways of HAMP regulation by acute and chronic iron loading in mice. Hepatology. 2011;53:1333–1341. doi: 10.1002/hep.24178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawnsley DR, Xiao J, Lee J, Liu X, Mericko-Ishizuka P, Kumar V, He J, Basu A, Lu M, Lynn FC, Pack M, Gasa R, Kahn ML. The transcription factor Atonal homolog 8 regulates Gata4 and Friend of Gata-2 during vertebrate development. Journal of Biological Chemistry. 2013;288:24429–24440. doi: 10.1074/jbc.M113.463083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb A, Wessling-Resnick M. Regulation of transferrin receptor 2 protein levels by transferrin. Blood. 2004;104:4294–4299. doi: 10.1182/blood-2004-06-2481. [DOI] [PubMed] [Google Scholar]
- Schmidt PJ, Toran PT, Giannetti AM, Bjorkman PJ, Andrews NC. The transferrin receptor modulates Hfe-dependent regulation of HAMP expression. Cell Metabolism. 2008;7:205–214. doi: 10.1016/j.cmet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RJ, Lombard M, Raja KB, Thatcher R, Peters TJ. Iron absorption by hypotransferrinaemic mice. British Journal of Haematology. 1991;78:565–570. doi: 10.1111/j.1365-2141.1991.tb04490.x. [DOI] [PubMed] [Google Scholar]
- Talbot NP, Lakhal S, Smith TG, Privat C, Nickol AH, Rivera-Ch M, León-Velarde F, Dorrington KL, Mole DR, Robbins PA. Regulation of HAMP expression at high altitude. Blood. 2012;119:857–860. doi: 10.1182/blood-2011-03-341776. [DOI] [PubMed] [Google Scholar]
- Tanno T, Bhanu NV, Oneal PA, Goh SH, Staker P, Lee YT, Moroney JW, Reed CH, Luban NL, Wang RH, Eling TE, Childs R, Ganz T, Leitman SF, Fucharoen S, Miller JL. High levels of GDF15 in thalassemia suppress expression of the iron regulatory protein HAMP. Nature Medicine. 2007;13:1096–1101. doi: 10.1038/nm1629. [DOI] [PubMed] [Google Scholar]
- Tanno T, Porayette P, Sripichai O, Noh SJ, Byrnes C, Bhupatiraju A, Lee YT, Goodnough JB, Harandi O, Ganz T, Paulson RF, Miller JL. Identification of TWSG1 as a second novel erythroid regulator of HAMP expression in murine and human cells. Blood. 2009;114:181–186. doi: 10.1182/blood-2008-12-195503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein DA, Roy CN, Fleming MD, Loda MF, Wolfsdorf JI, Andrews NC. Inappropriate expression of HAMP is associated with iron refractory anemia: implications for the anemia of chronic disease. Blood. 2002;100:3776–3781. doi: 10.1182/blood-2002-04-1260. [DOI] [PubMed] [Google Scholar]
- Wilkins SJ, Frazer DM, Millard KN, McLaren GD, Anderson GJ. Iron metabolism in the hemoglobin-deficit mouse: correlation of diferric transferrin with HAMP expression. Blood. 2006;107:1659–1664. doi: 10.1182/blood-2005-07-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighting DM, Andrews NC. Interleukin-6 induces HAMP expression through STAT3. Blood. 2006;108:3204–3209. doi: 10.1182/blood-2006-06-027631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Zhou J, Liu Q, Lu D, Wang L, Qiao X, Jia W. Atoh8, a bHLH transcription factor, is required for the development of retina and skeletal muscle in zebrafish. PLoS ONE. 2010;5:e10945. doi: 10.1371/journal.pone.0010945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1. Hepatic Id1 and Smad7 mRNA levels in control and Hamp1−/− mice with induced haemolytic anemia.
Fig S2. Effect of modulators of erythropoiesis and changes in transferrin levels on hepatic Hamp1 mRNA expression.
Fig S3. Effect of modulators of erythropoiesis and changes in transferrin levels on liver non-heme iron levels.
Fig S4. Effect of modulators of erythropoiesis and changes in transferrin levels on hepatic Id1 and Smad7 expression.
Table SI. Primer sequences.