Abstract
The orphan receptor ROS1 is a human proto-oncogene, mutations of which are found in an increasing number of cancers. Little is known about the role of ROS1, however in vertebrates it has been implicated in promoting differentiation programs in specialized epithelial tissues. In this study we show that the C. elegans ortholog of ROS1, the receptor tyrosine kinase ROL-3, has an essential role in orchestrating the morphogenesis and development of specialized epidermal tissues, highlighting a potentially conserved function in coordinating crosstalk between developing epithelial cells. We also provide evidence of a direct relationship between ROL-3, the mucin SRAP-1, and BCC-1, the homolog of mRNA regulating protein Bicaudal-C. This study answers a longstanding question as to the developmental function of ROL-3, identifies three new genes that are expressed and function in the developing epithelium of C. elegans, and introduces the nematode as a potentially powerful model system for investigating the increasingly important, yet poorly understood, human oncogene ROS1. genesis 51:545–561.
Keywords: ROS1 oncogene, Caenorhabditis elegans, ROL-3, cuticle, epithelial, seam cells
INTRODUCTION
The advent of high-throughput genomic techniques for identifying mutations on a global scale is revealing the importance of specific proteins that are associated with human diseases, such as cancer. One such protein is the receptor tyrosine kinase (RTK) and proto-oncogene ROS1, the mis-expression of which has been observed in a significant number of human glioblastomas (Birchmeier et al., 1987; Sharma et al., 1989; Birchmeier et al., 1990; Charest et al., 2003). Recently, the growing importance of ROS1 has been highlighted by the discovery of transforming mutations in other cancers types, most significantly in lung (Bergethon et al., 2012; Davies et al., 2012; Janne et al., 2012; Rimkunas et al., 2012; Suehara et al., 2012), and to a lesser degree, breast (Eom et al., 2008), colon and kidney (Ruhe et al., 2007). The discovery of small molecule compounds that can inhibit ROS1, such as Crizotinib, are proving effective for treating specific tumor types (Davies et al., 2012; Komiya et al., 2012), and has led to the proposal that the ROS1 receptor tyrosine kinase is a powerful target for engineered anticancer drugs (Forde and Rudin, 2012; Ou et al., 2012). The identification of ROS1 mutations that are associated with familial heart disease (Shiffman et al., 2005) and hypertension (Yamada et al., 2008) has also implicated this protein in the development of other human diseases beyond cancer.
Although the importance of ROS1 is becoming clear, its developmental role is still poorly understood. ROS1 is conserved in a number of organisms and is one of two remaining orphan receptors, having no known ligand (Acquaviva et al., 2009). Studies in avian and rodent model systems have uncovered a complex spatio-temporal pattern of expression for ROS1 in a range of developing organs including the lung, kidney, liver, intestine, and reproductive tissues (Sonnenberg et al., 1991; Tessarollo et al., 1992; Chen et al., 1994; Sonnenberg-Riethmacher et al., 1996). Expression is highly specific to the epithelial tissues of these organs, and coincides with important morphogenic events (Kanwar et al., 1995). ROS1 −/− transgenic mice are viable, however males are infertile due to improper development of the epididymis (Sonnenberg-Riethmacher et al., 1996), a specialized epithelial structure that regulates sperm development and maturation (for review see Cooper, 2007). In humans, expression of ROS1 is reported in the tissues of a number of organs, including the lungs and the epididymis (Legare and Sullivan, 2004; Acquaviva et al., 2009), revealing potential functional conservation of ROS1 in higher organisms.
The power of model organisms, such as the nematode C. elegans, to explore the underlying biology of human disease genes is well established. The epithelium of C. elegans is a valuable model system for exploring the function of conserved genes involved in epithelial development (Kagoshima and Shigesada, 2007; Joshi et al., 2010). The outer epithelial system of C. elegans is composed of two general cell types: major hypodermal cells and the seam cells, a specialized type of hypodermal cell (reviewed by Altun and Hall, 2009a,2009b). The largest hypodermal cell (hyp7) is a single multinuclear syncytium that covers most of the main body of the animal. Several smaller hypodermal cells form the head and the tail. The major hypodermis is responsible for establishing the body structure of the animal, and for secretion of cuticle components (Johnstone and Barry, 1996; Greenwald, 1997; Michaux et al., 2001). The seam cells, a specialized group of stem cell-like epithelial cells, also secrete cuticle components and play an additional critical role in the timing of postembryonic development and co-ordination of the molt cycle (Ruaud and Bessereau, 2006; Monsalve et al., 2011; Singh et al., 2011). Seam cells progress through a highly ordered developmental program of cell division, migration, elongation and fusion that leads to the generation of a multinucleate structure running laterally along either side of the animal (Sulston and Horvitz, 1977; Podbilewicz and White, 1994). At the L1 stage, each seam is composed of 10 cells, three anterior head cells (H0–H2), six cells of the main body (V1–V6) and a single tail in the posterior (T). During the L1 stage the seam cells of the V lineage (V1–V6) undergo a symmetric cell division, separating from one another and resulting in a final complement of 16 cells in each seam. The cells of the V lineage act as stem cells, dividing asymmetrically at each of the subsequent larval stages (L2–L4). The anterior daughters of each cell pair fuse with the major hypodermis after each cell division, taking on a hypodermal cell fate. During this process the posterior seam cells elongate in an anterio-posterior direction to retain contact with their neighbors. Upon completion of the hypodermal fusion/seam cell elongation cycle the new cuticle is synthesized and the animal initiates apolysis, the process of releasing the existing cuticle. This is followed by ecdysis, the process of shedding the existing cuticle. Upon completion of ecdysis, seam cell development is re-established. At the last larval stage the seam cells fuse with one another to form a single multinucleate syncytium. This complex developmental process is under precise control of a number of developmental pathways (Ambros, 1989; Ambros, 2001; Abbott et al., 2005; Nimmo et al., 2005; Xia et al., 2007; Joshi et al., 2010; Monsalve et al., 2011; Wildwater et al. 2011). The developmental program of the major hypodermis and seam therefore represents a system in which to study conserved genes that function in the development of epithelial tissues in human diseases, such as ROS1.
The C. elegans homolog of ROS1 encoded by the ORF C16D9.2 was previously identified as ROL-3 (Simmer et al., 2003). The developmental function that ROL-3 provides is not known, however mutations in rol-3 lead to defects in gross morphology that manifest as a roller (Rol) phenotype (Brenner, 1974). Furthermore, in contrast to other Rol loci, ROL-3 also has an essential function in development, as severe alleles result in an early developmental arrest (Johnsen and Baillie, 1991). ROL-3 is of particular interest with respect to ROS1 because the Rol phenotype is known to be associated with defects in the substructure of the cuticle (Peixoto et al., 1998, 2000), structures that are intimately associated with outer epithelial tissues (Page and Johnstone, 2007). Expanding our understanding of how ROL-3 acts to control gross morphology, and confers a phenotype associated with mutation of cuticle collagens, will give insight into important processes in C. elegans development, and identify potentially conserved functions with ROS1 in higher organisms.
In this study, we describe the molecular and genetic characterization of ROL-3. We show that ROL-3 is dynamically expressed exclusively in the major outer epithelial tissues of the animal, and is closely associated with the developing seam cells. We find that animals carrying mutations in rol-3 synthesize a disorganized cuticle, and are defective in molting. Furthermore, we characterize a novel requirement for ROL-3 in the development of the seam syncytium, a tissue associated with the process of molting, and demonstrate that ROL-3 is necessary for the maintenance of the seam cell identity. Finally, we provide evidence of a direct relationship between ROL-3, the predicted mucin SRAP-1, and the Bicaudal-C homolog, BCC-1.
RESULTS
ROL-3 is Structurally Related to the Human Proto-oncogene ROS1
The rol-3 locus of C. elegans encodes a type I integral membrane protein related to the sevenless subfamily of tyrosine kinase insulin receptor genes (Fig. 1b). The structure of the 2481aa protein is typical of an RTK of this class with a single transmembrane domain separating the intracellular and extracellular regions of the protein. The large extracellular portion of the protein contains an N-terminal signal peptide sequence and six fibronectin type III (Fn3) domains. The intracellular region contains a single kinase domain (TyrK; Fig. 1c). BlastP analysis identifies strong similarity between ROL-3 and the human proto-oncogene and orphan receptor ROS1 (29% similarity, 17% identity), particularly within the kinase domain (60% similarity, 35% identity), indicating that ROL-3 is the likely C. elegans ortholog of this human proto-oncogene (Acquaviva et al., 2009).
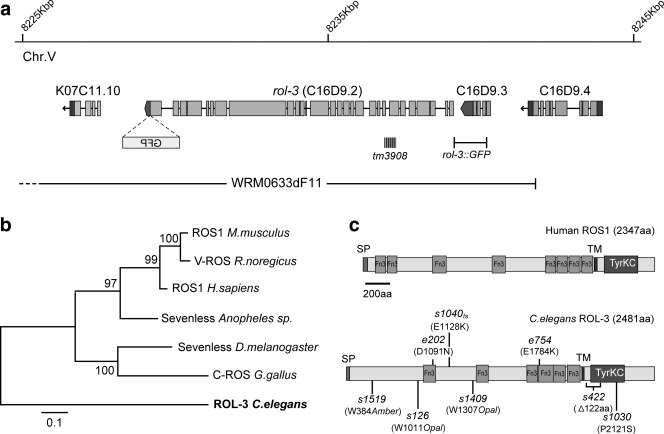
Fig 1.
ROL-3 has a conserved kinase domain and is structurally orthologous to the human proto-oncogene ROS1. a. Schematic showing the genomic region of C16D9.2. The position of a recombineered C-terminal GFP insertion into the fosmid WRM0633dF11, the genomic region used to drive ROL-3::GFP expression, and the deletion allele (tm3809) are show below the gene model. b. A closest neighbour tree of rol-3 derived from BlastP alignment with the putative kinase domain sequence. c. Protein schematics of C. elegans ROL-3 and Human ROS1. The positions of identified rol-3 alleles are shown. The alleles s1040ts and e754 have previously been reported (Simmer, Moorman et al. 2003). SP = Signal Peptide, Fn3 = Fibronectin type III repeat, TM = Transmembrane domain, TyrKC = Tyrosine Kinase Domain.
Mutations in rol-3 Define A Complex Allelic Series
The rol-3 locus was originally defined by two mutations, e202 and e754 (Brenner, 1974), both of which give rise to an adult specific left-hand roller (LRol) phenotype. A further eleven rol-3 alleles that cause an arrest during larval development were isolated in subsequent screens for essential genes (Johnsen and Baillie, 1991), including a temperature sensitive allele, s1040ts. s1040ts animals arrest as larvae when grown at 20° but develop into viable adult rollers when cultured at the permissive temperature of 15° (Table1). The molecular nature of the LRol alleles, e754 and s1040ts has been reported previously (E1822K, E1167K; Fig. 1c) (Simmer et al., 2003). We sequenced the remaining LRol allele, e202 (D1091N; Fig. 1c). All mutations that give rise to the viable rolling phenotype reside in the region encoding the extracellular portion of the protein (Fig. 1c). We also sequenced five of the remaining alleles, all of which lead to larval arrest. The alleles s422, an in-frame deletion of 366bp, and s1030, a C-T transition resulting in a Proline to Serine substitution (P2121S; Fig. 1c), are located in the putative kinase domain (Fig. 1c). The remaining three alleles s126, s1409, and s1519 are G-A transitions that introduce stop codons (W1011Opal, W1492Amber, W384Amber respectively Fig. 1c). Finally we obtained a UV-TMP generated deletion allele (tm3908; Fig. 1a,c; a kind gift from S. Mitani). This mutation is a 324bp deletion that partially deletes exons 7 and 8, resulting in a premature stop at amino acid 432. Observation of tm3908 homozygotes confirmed that these animals also arrest at an early larval stage (Table1 and data not shown). All four truncations alleles are predicted to generate proteins lacking both the kinase and transmembrane domains and are therefore likely to be null for ROL-3 function.
Table 1.
Cuticle Defects in rol-3 Mutant Animals
| Allele | Temp. | Terminal stagea | Viability (%) | Phenotype |
|---|---|---|---|---|
| N2 (wildtype) | 25° | Viable adult (n = 100) | 100% (n = 100) | WT |
| rol-3(tm3908) | 25° | L2 (100%) (n = 37) | 0% (n = 37) | Mlt |
| rol-3(s1040ts) | 15° | Viable adult (n = 148) | 99% (n = 148) | LRol |
| rol-3(s1040ts) | 20° | L2 (30%) L3 (70%) (n = 30) | 0% (n = 30) | Mlt |
| rol-3(s1040ts) | 25° | L2 (70%) L3 (30%) (n = 30) | 0% (n = 30) | Mlt |
| rol-3(s1040ts); sEx2695 | 20° | Viable adult (n = 100) | 100% (n = 100) | WT |
ND = not determined, Mlt – unshed cuticle, CC – cuticle constriction.
Terminal developmental stage assayed by elt-5::mCherry or UNC-47::GFP expression.
ROL-3 is Expressed in a Dynamic Pattern in the Major Hypodermis Throughout Development
To identify the tissues in which rol-3 is expressed, we generated a transcriptional reporter containing 3kb of the rol-3 5′ flanking region fused to the GFP sequence containing a NLS (rol-3::GFP). Animals carrying rol-3::GFP as a transgenic array (sEx1594) express GFP in all major hypodermal nuclei, with the exception of the seam cells (Fig. 2a–f). Expression is first detected during dorsal intercalation and is particularly intense throughout embryogenesis (Fig. 2a–d). Upon hatching rol-3::GFP expression persists in the major hypodermis at a relatively low-level throughout development up until the adult stage, where expression is not detected (Fig. 2e,f and data not shown).
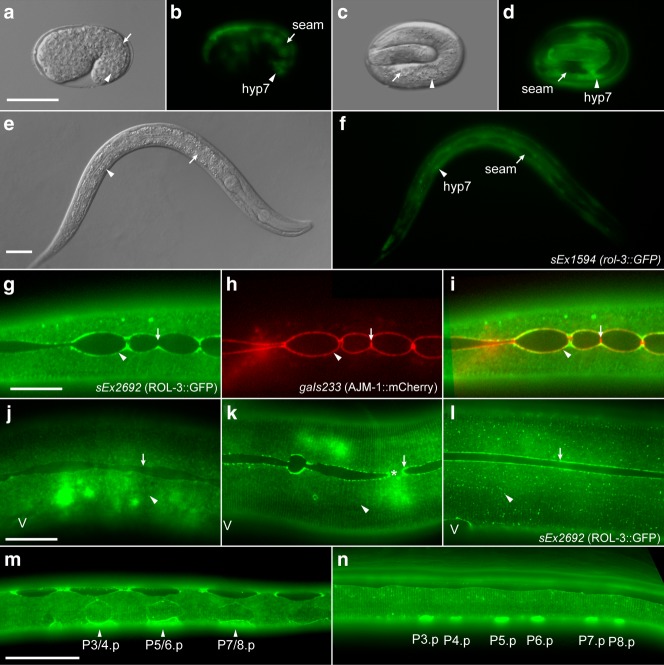
Fig 2.
ROL-3 is expressed in a dynamic pattern in the developing hypodermis. a–f, A rol-3::GFP transcriptional fusion is expressed in the major hypodermis throughout development. Expression begins at the embryonic stage in most, if not all, major hypodermal cells (arrowheads a–d) and continues throughout post-embryonic development (arrowheads e and f). Expression is not observed in the seam cells (arrows a–f). g–k, A dynamic pattern of ROL-3::GFP accumulation is observed throughout post-embryonic development. Broad expression of ROL-3::GFP is seen in the major hypodermis and accumulates in foci adjacent to seam cell boundaries (arrowheads g–i). ROL-3::GFP does not localize with the apical junctions between seam cells (arrows g–i). Prior to seam cell division broad expression of ROL-3::GFP is present in the hypoderm (arrowhead j), but is not present in foci at the seam cell boundary (arrow j). As seam cells divide asymmetrically ROL-3::GFP accumulates at the cell boundaries (arrow k). Localization at anterior daughters is lost as they migrate and fuse with the hypodermis (asterisk k). Localization at cell boundaries persists as seam cell elongation is completed (arrow l) (V = Vulva). m-n, Strong expression of ROL-3::GFP is observed in VPCs at the L1 (m) and early L2 (n) stage. Scale bars, 25 μm.
The large size of the rol-3 ORF made generating a translational reporter by traditional cloning methods technically challenging. To circumvent this we used the recombineering method (Tursun, Cochella et al., 2009) to engineer a fosmid harboring GFP fused in frame at the C-terminus of the gene (Fig. 1a). A transgenic array containing ROL-3::GFP (sEx2695) is sufficient for the rescue of all phenotypes associated with mutations in rol-3, including the lethality manifested in animals containing the null allele tm3908 (Table1 and data not shown). Consistent with the transcriptional GFP reporter ROL-3::GFP is expressed in the major hypodermal cells and excluded from seam cells (Fig. 2g–i). At the point when seam cells are quiescent ROL-3::GFP expression is generally diffuse across the surface of the major hypodermis (Fig. 2j). Prior to and during seam cell division however, ROL-3::GFP expression intensifies and accumulates in foci at the seam cell boundaries (Fig. 2k,l). ROL-3::GFP localization at the seam cell boundary is lost at the anterior seam cells as they fuse with the major hypodermis (Fig. 2k). Particularly intense ROL-3::GFP foci are observed at the leading edges of the posterior seam cells as they elongate toward one another (Fig. 2k). ROL-3 localization at the seam cell boundary persists until seam cell elongation is completed (Fig. 2l). In addition to the major hypodermal cells ROL-3::GFP expression is also detected in the anterior seam cells as they take on the major hypodermal fate at the L1 stage of development (Fig. 2m). These cells migrate to separate the vulval precursor cells (VPCs) which also express ROL-3::GFP at this stage (Fig. 2m). Expression of ROL-3::GFP in the VPC's is restricted to L1 larval stage, being rapidly reduced during the L1–L2 transition and not detected at latter stages of development (Fig. 2m and n and data not shown). The dynamic pattern of ROL-3::GFP localization is consistent with a role in orchestrating crosstalk between the developing seam cells and the hypodermal substrate over which they are developing.
rol-3 is Essential for Proper Cuticle Formation and Molting
In C. elegans, Rol phenotypes are associated with mutations that disrupt the biosynthesis, processing and assembly of cuticle collagens (Higgins and Hirsh, 1977; Cox et al., 1980; Levy et al., 1993; Bergmann et al., 1998; Peixoto et al., 1998; Yang and Kramer, 1999; Peixoto et al., 2000). These mutations result in the development of a disordered cuticle structure, arising from improper collagen secretion and assembly. To ascertain if hypomorphic mutations in rol-3 disrupt cuticle structure we visualized the adult cuticle using the COL-19::GFP reporter, the expression of which can be used as a readout for proper cuticle formation (Thein et al., 2003) (Fig. 3a–d, Supporting Information Fig. S1a,b). In rol-3(s1040ts) adults raised at permissive temperature COL-19::GFP is highly disorganized. The annuli, circumferential ridges in the hypoderm that are formed as a consequence of cuticle collagen deposition, form correctly over the dorsal and ventral major hypodermis but are absent from the cuticle in the lateral major hypodermis (Fig. 3c,d). These defects are not specific to the s1040ts allele as similar defects are also observed in animals that are mutant for the e754 allele (Supporting Information Fig. S1a,b). Additionally, him-8(e1489); rol-3(e754) animals display gross morphological defects of male specific structures, consistent with improperly formed or accumulated cuticle (Supporting Information Fig. S1c–d). Furthermore, breaks and bifurcations occur in the Alae, cuticular structures that form over the lateral seam cells (Fig. 3c,d and Supporting Information Fig. S1b). Hypomorphic mutations in rol-3 therefore give rise to cuticle defects consistent with disruption of collagen processing and assembly.
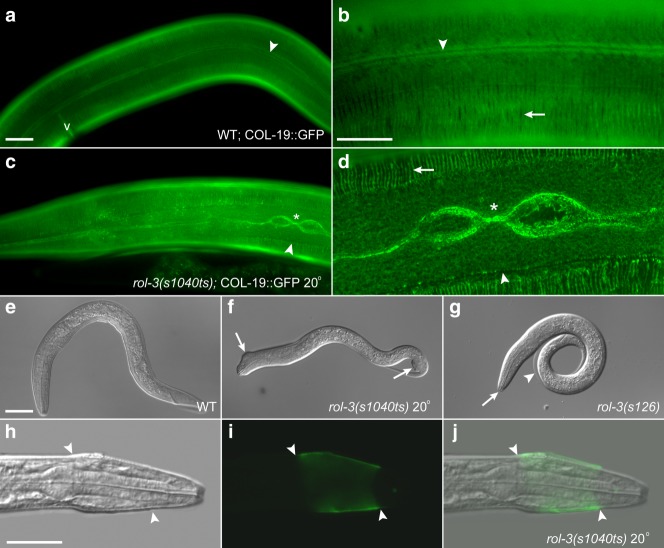
Fig 3.
Mutation of rol-3 leads to defects in cuticle structure. a-d. COL-19::GFP expression. a and b. COL-19::GFP expression reveals the ordered structure of the alae (arrowhead) and annuli (arrow) in the cuticle of adult wildtype animals. c and d. In rol-3(s1040ts) adults expressing COL-19::GFP the alae are disrupted (arrowhead) and annuli do not extend across the lateral region of the cuticle (arrow). Bifurcation of the alae is also observed (asterisks). e–j. rol-3 larval cuticle defects. E. L2 stage WT animal. F. Arrested rol-3(s1040ts) animal presenting with morphology defects (arrows). G. Arrested rol-3(s126) animal fully encased in unshed cuticle (arrowhead and arrow). h–j. FITC-WGA staining of an arrested rol-3(s1040ts) L1–L2 animal. Unshed cuticles can be seen resulting in a characteristic constriction (arrowhead). Fluorescently labelled WGA binds the exposed and unshed L1 and L2 cuticles beneath (green). Scale bars, 25 μm.
The cuticle defects associated with hypomorphic mutations of rol-3 could account for the adult specific rolling phenotype. However, the cause of the larval arrest associated with more severe alleles of rol-3 was unclear. rol-3(s1040ts) animals arrest as larvae when grown at restrictive temperature, but are superficially wildtype in appearance. Closer examination of these arrested animals revealed the presence of subtle morphological defects and a distinctive anterior constriction of the cuticle, consistent with the retention of an incompletely shed cuticle (Fig. 3f–j and Table1). To examine the integrity of the cuticle in these animals we stained arrested rol-3(s1040ts) larvae with fluorescein isothiocyanate conjugated wheat germ agglutinin (FITC-WGA). Similar probes are known to preferentially bind glycoproteins exposed by cuticle damage (Politz et al., 1990). FITC-WGA staining of arrested rol-3(s1040ts) larvae confirmed the presence of improperly shed cuticles (Fig. 3h–j). Furthermore, examination of animals homozygous for the null allele rol-3(s126) revealed that these animals appear to be fully encased in unshed cuticle (Fig. 3g). Together these observations demonstrate that both the early larval arrest and late-onset LRol phenotypes are associated with defects in cuticle formation and function.
ROL-3 is Required for the Fidelity of Seam Cell Elongation and Morphogenesis
The molting defects associated with null rol-3 alleles might be due to the inability to release an improperly formed cuticle. Molting defects are also often associated with mutations that lead to the disruption of seam cell morphology (Brooks et al., 2003; Silhankova et al., 2005; Ruaud and Bessereau, 2006; Meli et al., 2010; Monsalve et al., 2011; Singh et al., 2011). Our examination of COL-19::GFP expression in rol-3(s1040ts) adult animals revealed the presence of defects in the patterning of the alae, the formation of which are dependent on the structure of the underlying seam syncytium (Page and Johnstone, 2007; and reviewed by Altun and Hall, 2009a,2009b). This indicated that formation of seam syncytium might be compromised in these animals. To observe the developing seam syncytium we utilized the transgenic array wIs78, which expresses GFP in seam cell nuclei and adherin junctions (Li et al., 2005). In rol-3(s1040ts) and rol-3(e754) larvae raised at permissive temperature seam cell elongation progresses normally or with minor defects in seam cell contact (Fig. 4b and Supporting Information Fig. S1e). Adult animals develop a seam syncytium that can be mildly disorganized, bifurcated and/or discontinuous (Fig. 4d, Supporting Information Fig. S1f and Table2). The specification of seam cell identity is not significantly affected in rol-3(s1040) animals, based on the number of seam cells displaying robust expression of SCM::GFP (Fig. 4d and Table2). However, rol-3(s1040ts) animals arrested at the restrictive temperature of 20° exhibit severe defects in seam formation, with many cells failing to elongate correctly (Fig. 4d/e and Table2). All animals observed present with these defects, though the severity of the phenotype varies from those that complete cell contact in all but a few cells, to those where cell contact fails completely (Table2). In animals homozygous for the null allele rol-3(s126), seam cells completely fail to elongate toward one another, retaining a rounded appearance (Fig. 4g/f). Additionally, seam cell number appears to be reduced in both rol-3(s1040) and rol-3(s126) animals (Fig. 4d, g).
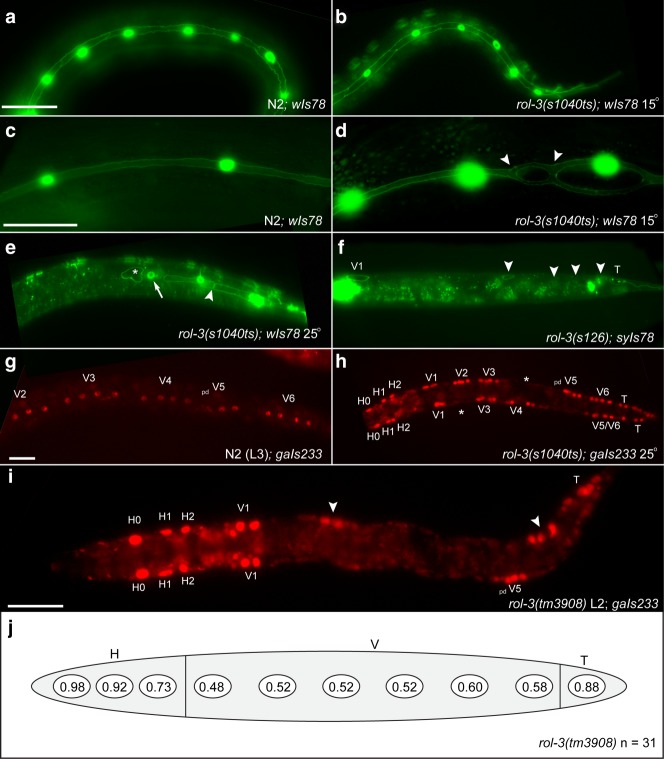
Fig 4.
Development of the seam syncytium is perturbed in animals carrying mutations in rol-3. a. Wild type L2 stage animal showing the characteristic ladder structure of the seam cells. b. rol-3(s1040ts) L2 animal raised a permissive temperature displaying a superficially wild type seam structure. c. WT adult seam syncytium is narrow and complete (arrow). d. Adult rol-3(s1040ts) animal displaying a disordered seam that is bifurcated (arrowheads). e. Arrested rol-3(s1040ts) animal. Two seam cells have elongated correctly (arrowhead) while an adjacent SCM::GFP expressing seam cell has ectopically fused with the hypodermis (arrow). A seam cell that has lost SCM::GFP expression and not elongated can be seen (asterisk). Several seam cells in the anterior of the animal have lost SCM::GFP expression and fused with the hypodermis (left image). f. Arrested rol-3(s126) animal showing a reduced number of AJM-1::GFP positive seam cells that have not elongated, note: several seam cells are not in the focal plane in this image (arrowheads). g. WT L3 animal expressing seam specific elt-5::mCherry. h. Arrested rol-3(s1040ts) animal with an L3 equivalent number of divided elt-5::mCherry positive cells. Note that the animal is much smaller than wildtype. Several cells of the V lineage have lost of elt-5::mCherry expression (asterisks). i. rol-3(tm3908) animal arrested approximately at the L3 stage showing severe loss of elt-5::mCherry specification in the V lineage J. Quantification of the loss of seam cell specification in the H, V and T seam cell lineages in arrested rol-3(tm3908) animals. Scale bars, 25μm.
Table 2.
Seam Cell Defects in rol-3 Mutants
| Allele | T (°C) | L1 seam #a | Terminal seam #b | Viability | WT seam cell elongation (%) | Seam cell bifurcation (%) | Seam cell breaks (%) | SCM expression |
|---|---|---|---|---|---|---|---|---|
| N2 | 20 | 10 (n = 40) | 15.9 (n = 37) | 100 (n = >100) | 100 (n = 37) | <1 | 0 | Robust |
| tm3908 | 20 | 10 (n = 29) | 6.6 (n = 31) | 0 (n = 76) | 0 (n = 31) | NA | NA | ND |
| s1040ts | 15 | 10 (n = 30) | 15.3 (n = 65) | 99 (n = >100) | 90 (n = 65) | 75 | 10 | Robust |
| s1040ts | 20 | 10 (n = 30) | ND | 0 (n = >100) | 0 (n = 34) | NA | NA | Variable |
| s1040ts | 25 | 10 (n = 30) | ND | 0 (n = >100) | 0 (n = 38) | NA | NA | Variable |
| bcc-1(tm3921) | 20 | ND | ND | 100 (n = >100) | 100 (n = 78) | 0 | 0 | ND |
| bcc-1(tm3821); rol-3(s1040ts) | 20 | ND | ND | 42 (d) | 0 (n = 35) | 63.5 | 13.6 | ND |
| bcc-1(tm3821); rol-3(s1040ts) | 25 | ND | ND | 0 (n = >100) | 0 (n = 59) | 0 | 100 | ND |
| bcc-1(tm3821); rol-3(s1519) | 20 | ND | ND | 0c | ND | ND | ND | ND |
| srap-1(sDp33); rol-3(s1040ts)/+ | 20 | ND | ND | 100 (54) | 100 (n = 24) | 0 | 0 | ND |
| sDp33; rol-3(s1040ts) | 20 | ND | ND | 96 (d) | 0 (n = 83) | 24.1 | 1.2 | ND |
| sDp33; rol-3(s1040ts) | 25 | ND | ND | 0 (n = >100) | 0 (n = 41) | 0 | 100 | ND |
| sDp33; rol-3(s1519) | 20 | ND | ND | 0c | ND | ND | ND | ND |
ND = not determined.
L1 stage seam cell number assayed by elt-5::mCherry expression.
Adult seam number assayed by SCM::GFP expression.
No viable double mutant animals recovered in genetic crosses (see Methods).
Jones, Rose et al. 2012.
ROL-3 is Required for the Maintenance of Seam Cell Identity in the Developing Seam Syncytium
To further explore the defects we had observed in seam cell morphology we used an elt-5::mCherry fluorescent reporter (Liu et al., 2009) to follow the fate of anterior and posterior seam cells. elt-5 encodes for a transcription factor that is expressed exclusively in seam cells (Koh and Rothman, 2001). Unlike SCM::GFP, elt-5::mCherry expression persists in the anterior daughters of the seam cells upon their fusing with the hypodermal syncytium. rol-3(s1040ts) and rol-3(tm3908) animals expressing elt-5::mCherry hatch with the correct number of elt-5::mCherry positive cells (Table2). However, after exit from the L1 stage expression of the reporter becomes erratic and is frequently lost in individual seam cells (Fig. 4h, i and Table2). Loss of elt-5::mCherry expression is more common in rol-3(tm3908) null animals, but is not eliminated entirely (Fig. 4i). Seam cells that retain elt-5::mCherry expression continue to divide normally (Fig. 4h, i). In rol-3(tm3908) animals, loss of elt-5::mCherry expression is observed in all P V and T seam cell lineages. However, losses occur more frequently in the V lineage seam cells of the central body (Fig. 4j). Together these observations indicate that ROL-3 is not required for establishing seam cell identity per se, but rather is needed for the maintenance of seam cell identity.
The Suppressors of rol-3, bcc-1, and srap-1, are Expressed in Epithelial Tissues
We have previously reported the identification of deletions of bcc-1, and duplications of srap-1 that act to suppress the lethality associated with rol-3(s1040ts) animals (Jones et al., 2012). bcc-1 encodes a homolog of the drosophila RNA binding protein Bicaudal-C (BIC-C), which acts to negatively regulate the function of specific genes by binding to and targeting their mRNAs for degradation (Saffman et al., 1998; Chicoine et al., 2007). srap-1 encodes a predicted mucin, a family of high molecular weight extracellular matrix (ECM) proteins that form a protective barrier secreted by many cell types, including epithelial cells (Gendler and Spicer, 1995; Chakraborty et al., 2011). It is unclear how the mutations in bcc-1 and srap-1 elicit their suppressive effect. As an ortholog of Bic-C, BCC-1 might provide a post-translational regulatory function, and its loss of function would therefore be expected to lead to the overexpression of specific genes through persistence of their mRNA species. Similarly duplications of srap-1 might result in an overexpression of the protein, also resulting in a gain of function phenotype. It is possible that potential overexpression phenotypes might compensate for the loss of ROL-3 function in hypodermal tissues.
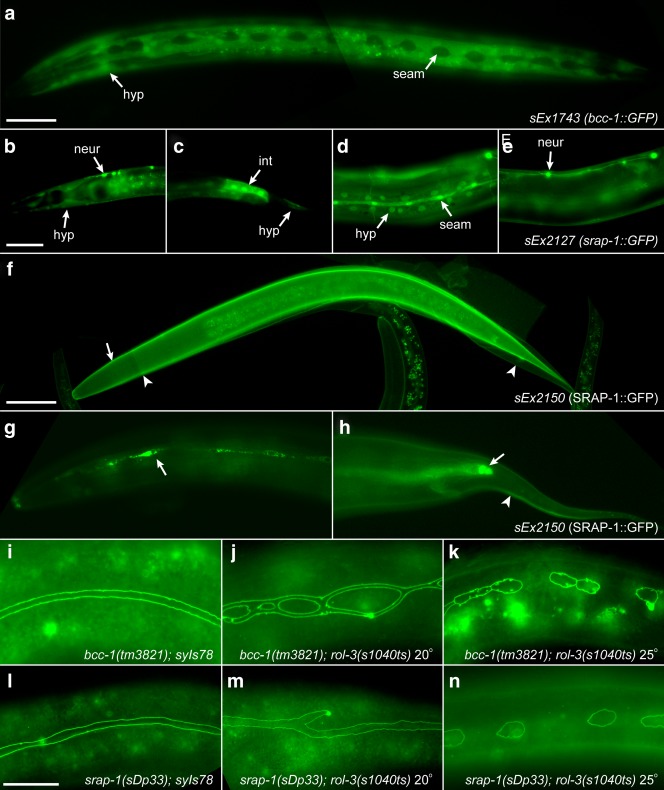
To ascertain if BCC-1 and SRAP-1 might function in the epithelium we first determined their expression patterns. A transcriptional GFP reporter driven by the putative promoter region of bcc-1 is expressed exclusively in the major hypodermis throughout development (Fig. 5a). This pattern of expression is similar to the expression pattern of rol-3 and is consistent with a potential role for BCC-1 in modulating mRNA species in the developing epithelium. A transcriptional GFP reporter driven by the putative srap-1 promoter is detected in a variety of tissues including the major hypodermis and seam cells (Fig. 5b,c). Notable expression of this reporter is also observed in the tissues of the developing vulva and central nervous system (Fig. 5d,e). To gain further insight into the expression of srap-1 we used recombineering to generate a fosmid containing GFP fused in-frame at the C-terminus of the srap-1 ORF. Expression of SRAP-1::GFP is detected at the surface of the newly synthesized cuticle coincident with the molt (Fig. 5f). Significant accumulation of SRAP-1::GFP is also detected at the seam syncytium in the adult stage (Fig. 5g,h). Given that both bcc-1 and srap-1 are expressed in epithelial tissues we asked if this expression is dependent on ROL-3 function. rol-3(s1040) mutant animals raised at the restrictive temperature of 25° display robust expression of both the bcc-1::GFP and SRAP-1::GFP reporters (Supporting Information Fig. S2a–d). Together this expression pattern analysis reveals that both bcc-1 and srap-1 are expressed in epithelial tissues associated with ROL-3 function or localization, and that this expression does not appear to be dependent on the expression of rol-3.
Fig 5.
Suppressors of ROL-3 are expressed in epithelial tissues and modify seam cell defects. a. bcc-1::GFP is expressed exclusively in the major hypodermis (hyp) being excluded from the seam cells (seam). b–e. srap-1::GFP is expressed dynamically in many developing tissues throughout development including the larval (b and c) and adult (d and e) hypodermis, seam cells, intestine (d), and neurons. Note that the same animal in different focal planes is shown in panels d and e. h–j. SRAP-1::GFP, is secreted at the newly synthesized cuticle (arrow) coincident with the release of the previous cuticle (arrowheads) (f) and accumulates at the seam cells in adults (arrows g and h). I–n. bcc-1(tm3821) and srap-1(sDp33) promote seam cell elongation in rol-3(s1040ts) animals. bcc-1(tm3821) and srap-1(sDp33) animals develop a wildtype seam syncytium (i and l). bcc-1(tm3821); rol-3(s1040ts) and srap-1(sdp33); rol-3(s1040ts) double mutant animals display disorganised and ectopic seam cell development at 20° (j and m). In double mutants at 25° seam cell elongation is severely abrogated (k and n). Scale bars, 25 μm.
Suppression of rol-3-Associated Lethality by the Mutations bcc-1(tm2831) and srap-1(sDp33) is Dependent on the Presence of Partial ROL-3 Function
The mutations bcc-1(tm3821) and srap-1(sDp33) suppress the lethality associated with the temperature sensitive allele rol-3(s1040ts) when grown at 20°. We investigated seam cell development in the double mutant animals to determine if this suppression might arise from a restoration of seam cell development. Animals mutant for either bcc-1(tm2831) or srap-1(sDp33) alone do not display any obvious seam cell defects, indicating that they are not required for the development of this tissue (Fig. 5i,l and Table2). However, rol-3(s1040ts) animals doubly mutant for the suppressor mutations develop a highly disorganised and ectopically formed seam syncytium when grown at 20° (Fig. 5j,m and Table2). This observation suggested that either the suppressor mutations are promoting seam cell development in the absence of ROL-3 function, or alternatively, they are directly or indirectly promoting the function of a hypomorphic rol-3 product. To test between these two possibilities we determined whether the suppressor mutations would rescue lethality in animals that are null for rol-3 function. We find that bcc-1(tm3921) and srap-1(sDp33) mutations are unable to suppress lethality in animals carrying the null rol-3(s1519) or rol-3(tm3908) alleles (Table2). Furthermore, we find that suppression is abolished when animals doubly mutant with the rol-3(s1040ts) allele are cultured at a higher temperature of 25°. Seam cell elongation is completely abrogated in these animals, leading to a developmental arrest and severe molting defects (Fig. 5k,n, Table2). Together these observations indicate that the suppression of lethality is dependent on partial ROL-3 function, and also implies that rol-3(s1040) animals cultured at 25° are essentially null for ROL-3 function.
DISCUSSION
In this study we have investigated the developmental role of the essential receptor tyrosine kinase ROL-3. We have shown that mutations in rol-3 lead to improper molting and the development of a compromised adult cuticle, phenotypes that are indicative of the LRol phenotype. We have also established that ROL-3 is expressed in the major hypodermis in a dynamic manner, where it forms foci at the boundaries of the developing seam cells. Furthermore, we have demonstrated that ROL-3 is required for seam cell development and the maintenance of seam cell identity. Finally we provide evidence that the suppressor mutations bcc-1(tm3821) and srap-1(sDp33) act to modify seam cell development in a manner that is dependent on partial ROL-3 function.
We find that animals carrying hypomorphic mutations in rol-3 have a compromised cuticle. Viable adult rol-3(s1040ts) animals display defects in collagen disposition specifically over the lateral hypodermis. This might account for the observed adult specific rolling phenotype as, with the exception of mutations in rol-3, all known mutations that manifest as a LRol phenotype reside in cuticle collagens (Kramer et al., 1988, 1990; Levy et al., 1993; van der Keyl et al., 1994; Novelli et al., 2004). Additionally we have demonstrated that ROL-3 is expressed in the major hypodermis where many cuticle collagens are synthesized (Page and Johnstone, 2007). Together these observations highlight a potential link between ROL-3 function and the biogenesis of cuticle collagens in the major hypodermis. The LRol phenotype has been shown to arise due to specific mutations that affect the processing and trimerization of pro-collagens (Yang and Kramer, 1999). As a putative signaling molecule ROL-3 could potentially function to directly or indirectly co-ordinate the biogenesis and incorporation of collagens into the cuticle.
In addition to the LRol phenotype, we have demonstrated that null alleles of rol-3 result in developmental arrest, associated with an inability to molt. Molting is distinctly associated with the integrity of the seam syncytium as disruption of seam cell development is associated with molting defects (Koh and Rothman, 2001; Brooks et al., 2003; Silhankova et al., 2005; Hao et al., 2006; Ruaud and Bessereau, 2006; Fritz and Behm, 2009; Monsalve et al., 2011; Singh et al., 2011). Furthermore, molting requires numerous proteases, steroid hormones, and nuclear hormone receptors (Yochem et al., 1999; Gissendanner and Sluder, 2000; Kostrouchova et al., 2001; Brooks et al., 2003; Kuervers et al., 2003; Davis et al., 2004; Frand et al., 2005; Kim et al., 2005; Hayes et al., 2006; Thein et al., 2009; Stepek et al., 2010; Kim et al., 2011), many of which are produced specifically in the seam cells. We observe defects in seam cell formation in animals carrying mutations in rol-3. In these animals the seam cells appear to be competent for developmental processes such as cell division and fusion with the surrounding hypodermal tissue. However, defects in elongation of posterior seam cells are observed as the cells attempt to retain contact with one another. The severity of these defects is directly related with the severity of the rol-3 mutation; subtle defects in seam cell morphology are observed in animals carrying the hypomorphic mutations rol-3(e754) and rol-3(s1040ts), while elongation is completely abrogated in animals carrying the null mutations rol-3(tm3908) and rol-3(s126). Together these observations indicate that ROL-3 is required to promote elongation of seam cells as they form connections with one another.
The pattern of expression and localization that we have described indicates that ROL-3 acts in a noncell-autonomous manner to influence the development of the seam syncytium. This might be surprising given that as a signaling molecule ROL-3 is expected to elicit an effect within the major hypodermis where is it expressed. As the seam syncytium is embedded within the major hypodermis its formation likely requires that the developing seam cells navigate the surface of this cell in order to establish and maintain contact with one another. A signaling mechanism mediating cross-talk between the seam cells and the major hypodermis might therefore be required to establish or modify the substrate over which the seam cells can develop. The activation of ROL-3 by a signal from the developing seam cells might represent the first step in a signal cascade leading to the establishment of a permissive substrate for seam cell development. It has been noted that no major signaling molecules that could facilitate crosstalk between the major hypodermis and the seam syncytium have been identified in large-scale screens for genes involved in molting (Frand et al., 2005). It is tempting to speculate that such a role could be provided by ROL-3. This hypothesis is supported by the observation that ROL-3 localizes dynamically at the boundary between the major hypodermis and the developing seam cells. A notable example of another gene in C. elegans that is expressed in the major hypodermis but disrupts alae formation, and by extension seam cell development or function, is cut-6, which encodes a novel cuticulin important for body morphology (Muriel et al., 2003). However we cannot rule out the possibility that ROL-3 also functions within the seam cells themselves eliciting a direct effect on seam cell morphology and development. Further work is needed to determine the precise mechanism of ROL-3 function.
In addition to seam cell morphological and developmental defects, we also describe the loss of seam specific markers in animals carrying mutations in rol-3. rol-3 mutant animals, including those harboring null alleles, hatch with the correct number of seam cells, demonstrating that ROL-3 function is not required to establish seam cell identity. However, during subsequent postembryonic development a gradual loss of seam cell identity is observed. The loss of seam cell identity is more severe in animals carrying the null allele rol-3(tm3908) but not eliminated entirely. The loss of seam cell specification therefore appears to occur stochastically. A hypothesis for these observations is that reestablishing a physical connection after cell division is a requirement for continued maintenance of seam cell identity. Seam cell identity is dependent upon a number of distinct developmental mechanisms including; miRNA mechanisms of post transcriptional control, Runx, and engrailed-type transcriptional regulation, and both canonical and noncanonical Wnt signaling (reviewed by Joshi et al., 2010). In the absence of ROL-3 mediated seam cell development these cues might be uncoupled, leading to loss of fate and ectopic fusion with the major hypodermis. In support of this hypothesis seam cell ablation studies have shown that formation for the posterior deirid from the V5 seam cell lineage is dependent up contact being made between adjacent seam cells (Austin and Kenyon, 1994). Additional studies to characterize the relationship between cell contact and the maintenance of seam cell identity using mutant alleles of rol-3 are likely to be informative.
We have shown that the suppressors of rol-3-associated lethality, BCC-1 and SRAP-1, exacerbate seam cell defects in a ROL-3 dependent manner. Mutations in bcc-1 and srap-1 were identified in a suppressor screen for mutations that promote survival of rol-3(s1040ts) animals at the restrictive temperature of 20° (Jones et al., 2012). When cultured at 25° however this suppression is completely abrogated. We also find that suppressor mutations are unable to supress lethality in null animals. Given that animals carrying rol-3 mutations arrest trapped within unshed cuticles it is possible that the suppressor mutations promote cuticle release, allowing animals with minimal ROL-3 function to progress in development. Consistent with this hypothesis we have shown that SRAP-1 is secreted into the cuticle during molting. Furthermore suppression by srap-1 is facilitated by duplications of the gene, suggesting a gain of function phenotype. Similarly, as a putative regulator of specific mRNAs, loss of BCC-1 in the major hypodermis might act to increase molting cues or the production of enzymes that promote cuticle release. An alternative model is that suppressors might act directly or indirectly to promote increased function of a disabled rol-3 product. Further investigation is needed to confirm the suppressive relationship between BCC-1, SRAP-1 and ROL-3.
ROL-3 is the closest homologue in the C. elegans genome to the human proto-oncogene ROS1. The endogenous function of ROS1 is not well known, however in the murine model ROS1 is specifically expressed in epithelial tissues where it is required for the development of the epididymis. Expression of ROL-3 in epidermal tissues is consistent with the murine model, demonstrating the potential for a conserved function between these proteins. It has been demonstrated that ROS1 directly binds SH2 domain containing tyrosine phosphatase SHP-1 (Keilhack et al., 2001), and has been shown to activate both SHP-1 and SHP-2, the mitogen-activated protein kinase ERK1/2, insulin receptor substrate 1 (IRS-1), phosphatidylinositol 3-kinase (PI3K), protein kinase B (AKT) and the STAT3 and VAV3 signaling pathways (Zong et al., 1993; Xiong et al., 1996; Zeng et al., 2000; Charest et al., 2006). Orthologs of several of these proteins are present in the C. elegans genome, leading to potential future studies to test the conserved nature of these interactions. Furthermore, like ROS1, ROL-3 is an orphan receptor having no known ligand. Given the analysis we have presented here, the seam syncytium is the likely source of the ROL-3 ligand.
Finally ROS1 is a proto-oncogene whose transforming effect is primarily manifested due to miss-expression and gain of function mutations, such as the FIG-ROS fusion, which targets the intracellular region of ROS1 to the golgi apparatus in glioblastomas (Charest et al., 2003, 2003), and the recently identified SLC34A2-ROS and CD74-ROS fusions in lung carcinomas (Rikova et al., 2007). With currently available techniques, construction of molecules that mimic ROS1 activating mutations in C. elegans are easily attainable, and would be a potentially informative approach to increase our understanding of the biology of this oncogene.
The receptor tyrosine kinase ROL-3 is at a hub of C. elegans hypodermal development, and provides an essential developmental role in epithelial development. Lack of ROL-3's function to promote seam cell elongation potentially uncouples crosstalk between specialized epithelial tissues, leading to severe pleiotropic defects in epithelial development, cuticle formation and molting. Further investigation will elucidate the precise molecular mechanisms involved. Finally, research of ROL-3 function in C. elegans development has potential utility as a model system for investigation of both the oncogenic and endogenous function of the Human proto-oncogene ROS1.
MATERIALS AND METHODS
Maintenance and handling of C. elegans were performed as previously described (Brenner, 1974). Worms were cultured at 20°, unless otherwise stated. Bristol N2 was used as Wild Type.
Mutant strains: LGI: dpy-5(e907), LGII: srap-1(sDp33), LGIV: bcc-1(tm3821), him-8(e1489), LGV: rol-3(e202, e754, s126, s422, s1030, s1040ts, s1409, s1519, tm3908)
Transgenic arrays: kaIs12 (COL-19::GFP), syIs78 (AJM-1::GFP), wIs78 (SCM::GFP; AJM-1::GFP), gaIs233 (elt-5::HIS-24::mCherry), ouEx123 (AJM-1::mCherry, RNT-1::GFP), oxIs12 (unc-47::GFP), sEX1594 (rol-3::GFP), sEx2692 (ROL-3::GFP), sEx1743 (bcc-1::GFP), sEx2127 (srap-1::GFP), sEx2150 (SRAP-1::GFP).
Fosmid selection and Preparation: C. elegans genomic fosmid clones containing C16D9.2 (rol-3) and T06D8.1 (srap-1) were identified via the web-based searching facility at the Michael Smith Genome Sciences Centre (http://elegans.bcgsc.bc.ca/perl/fosmid/CloneSearch). Fosmid DNAs were isolated as previously described (Dolphin and Hope, 2006).
Generation of fosmid subclones: A subclone of fosmid WRM067dH05 was generated by restriction enzyme digestion of 1 mg of fosmid with 10U SpeI (NEB) followed by gel extraction of the 18 kb fragment predicted to contain C16D9.2. The 18 kb fragment was subsequently injected at 10 µg/ml together with 80 µg/ml of pCeh361 and 10 µg/ml of pTG96 of into dpy-5 animals.
Generation of fluorescent reporters by PCR stitching: To generate transcriptional reporters putative promoters regions were amplified by PCR and fused to GFP using PCR stitching as previously described (Hobert, 2002). Primer sequences for rol-3::GFP, bcc-1::GFP, srap-1::GFP are available upon request.
Generation of translational fluorescent reporters by recombineering: Translational fluorescent reporters were constructed by recombineering as previously described (Tursun et al., 2009). Primer sequences for ROL-3::GFP and SRAP-1::GFP are available upon request.
Generation of transgenic strains: Transgenic animals were generated as previously described (Hunt-Newbury et al., 2007). DNA constructs were co-injected at an initial concentration of 10 µg/ml with 80µg/ml of pCeh361 (a dpy-5 rescuing construct; Thacker et al., 2006) and 10 µg/ml of pTG96 (a myo-2 promoter green fluorescent protein reporter plasmid, which has strong expression in the pharynx) (Gu et al., 1998). To generate transgenic animals containing C16D9.2, a fosmid DNA concentration of between 1 and 2 µg/ml was necessary. To confirm that each transgenic line contained fosmid DNA, a 600 bp PCR product targeting the fosmid vector sequence was amplified (Primer sequences available upon request).
Rescue of rol-3 lethal alleles: Hermaphrodites carrying a transgenic array containing the wild type rol-3 ORF and the myo-2::GFP pharyngeal fluorescent reporter mated to N2 males. myo-2::GFP positive males were isolated from the F1 progeny and mated to dpy-18(e364)/eT1(III); rol-3(x)(where x is any lethal allele of rol-3), unc-46(e177)/eT1(V) hermaphrodites. Rescue of rol-3 was confirmed by the presence of viable myo-2::GFP positive Unc animals in the F2. rol-3(s833) is linked to dpy-11(e224), in this case rescue was assayed by the presence of viable myo-2::GFP positive F2 Dpy animals.
Sequencing of mutant alleles: 50 animals of the appropriate genotype were picked into 50µl of lysis buffer (50 mM KCl, 10 mM Tris pH 8.3, 2.5 mM MgCl2, 0.45% Tween-20, 0.01% Gelatin, 0.1 mg/ml Proteinase K) and freeze cracked in liquid nitrogen. Samples were lysed and treated with proteinase K, (1 h at 60° followed by 15 min at 95°) and 1µl subsequently used in a 25 µl PCR reaction with appropriate primers (available on request). Samples were sent for sequencing at Macrogen (https://dna.macrogen.com/english/login/join.jsp).
Lectin staining of rol-3(s1040) animals: Fluorescein isothiocyanate (FITC) coupled wheat germ agglutinin (WGA; Sigma; Cat. No. L4895) was used to test for exposure of specific antigens on the surface of rol-3(s1040) animals raised at 20°. Mixed stage worm cultures were washed in sterile M9 buffer three times followed by incubation in M9 buffer for another hour to remove residual bacteria from the gut and body surface. Worms were incubated in 100 µl FITC conjugated WGA (20–25 µg/mL) for 1 h, washed in 1 ml M9 buffer four times before being mounted on slides for observation with Nomarski and fluorescent microscopy.
Microscopy and Image Processing: Analysis of mutant and GFP transgenic animals was performed using a ZEISS Stemi SVC11 dissecting microscope with GFP filters and a ZEISS Axioskop with 10×/0.25, 40×, 0.65 and 60×/0.85 Objective Lenses. All pictures were taken using QCapture software (QImaging) with a QIMAGING digital camera mounted on a ZEISS Axioskop. Images were processed using Photoshop CS5 (Adobe).
Acknowledgments
The authors wish to thank Dr. Nigel O'Neil for helpful comments on the manuscript, Dr. Shohei Mitani for the rol-3(tm3908) knock out strain, Dr. Allison Woollard and Dr. Jeff Simske for transgenic animals carrying the AJM-1::mCherry reporter and Dr. Anthony Page for the COL-19::GFP reporter strain. Funding for this research was provided by CIHR operating grants to DLB and AMR and CIHR fellowship to MRJ.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Mutant phenotypes associated with rol-3(e754). A. WT animal expressing COL-19::GFP. Uniform expression across the hypodermal surface can be seen (arrow) and formation of alae is complete (arrowhead). B. rol-3(e754) animal expressing COL-19::GFP. Annuli do not form across the lateral hypodermal surface (arrow) and the alae are irregular and improperly formed (arrowhead). C. DIC image of an adult male him-8(e1489) animal showing the WT structure of the male tail. D. Adult rol-3(e754; him- 8(e1489) animal showing the male tail structure encased in excess or improperly formed cuticle. E. Larval rol-3(e754) animal expressing syIs78 (AJM-1::GFP)[Bhagwati, Wang et al. 2003]. Subtle seam cell elongation defects can be seen (asterisks). F. Adult rol-3(e754) animal expressing AJM-1::GFP. Seam syncytium formation is complete but irregular in shape (arrow)(Compare to figure c). Scale bars, 25μm.
Expression of bcc-1 and SRAP-1 does not require functional ROL-3. A. A WT animal expressing bcc-1::GFP in the major hypodermis (arrow). B. A rol-3(s1040) animal raised at the restrictive temperature of 25o expressing bcc-1::GFP in the major hypodermis (arrow). C. A mid-larval stage WT animal expressing SRAP-1::GFP, this animal is in the process of molting (arrows). D. A rol-3(s1040) animal expressing SRAP- 1::GFP raised at the restrictive temperature of 25o. Animal has arrested encased in an improperly shed cuticle. Note the significant accumulation of SRAP-1::GFP at the constriction caused by the unshed cuticle (arrow). Scale bar for all panels, 25μm.
LITERATURE CITED
- Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acquaviva J, Wong R, Charest A. The multifaceted roles of the receptor tyrosine kinase ROS in development and cancer. Biochim Biophys Acta. 2009;1795:37–52. doi: 10.1016/j.bbcan.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Altun ZF, Hall DH. Epithelial System: Hypodermis. WormAtlas. 2009a [Google Scholar]
- Altun ZF, Hall DH. Epithelial System: Seam Cells. WormAtlas. 2009b [Google Scholar]
- Ambros V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 1989;57:49–57. doi: 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- Ambros V. The temporal control of cell cycle and cell fate in Caenorhabditis elegans. Novartis Found Symp. 2001;237:203–214. doi: 10.1002/0470846666.ch16. discussion 214–220. [DOI] [PubMed] [Google Scholar]
- Austin J, Kenyon C. Cell contact regulates neuroblast formation in the Caenorhabditis elegans lateral epidermis. Development. 1994;120:313–323. doi: 10.1242/dev.120.2.313. [DOI] [PubMed] [Google Scholar]
- Bergethon K, Shaw AT, Ou SH, Katayama R, Lovly CM, McDonald NT, Massion PP, Siwak-Tapp C, Gonzalez A, Fang R. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann DC, Crew JR, Kramer JM, Wood WB. Cuticle chirality and body handedness in Caenorhabditis elegans. Dev Genet. 1998;233:164–174. doi: 10.1002/(SICI)1520-6408(1998)23:3<164::AID-DVG2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Birchmeier C, O'Neill K, Riggs M, Wigler M. Characterization of ROS1 cDNA from a human glioblastoma cell line. Proc Natl Acad Sci USA. 1990;87:4799–4803. doi: 10.1073/pnas.87.12.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchmeier C, Sharma S, Wigler M. Expression and rearrangement of the ROS1 gene in human glioblastoma cells. Proc Natl Acad Sci USA. 1987;84:9270–9274. doi: 10.1073/pnas.84.24.9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:1–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DR, Appleford PJ, Murray L, Isaac RE. An essential role in molting and morphogenesis of Caenorhabditis elegans for ACN-1, a novel member of the angiotensin-converting enzyme family that lacks a metallopeptidase active site. J Biol Chem. 2003;278:52340–52346. doi: 10.1074/jbc.M308858200. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Bonthu N, Swanson BJ, Batra SK. Role of mucins in the skin during benign and malignant conditions. Cancer Lett. 2011;301:127–141. doi: 10.1016/j.canlet.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest A, Kheifets V, Park J, Lane K, McMahon K, Nutt CL, Housman D. Oncogenic targeting of an activated tyrosine kinase to the Golgi apparatus in a glioblastoma. Proc Natl Acad Sci USA. 2003;100:916–921. doi: 10.1073/pnas.242741799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest A, Lane K, McMahon K, Park J, Preisinger E, Conroy H, Housman D. Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma with an interstitial del(6)(q21q21) Genes Chromosomes Cancer. 2003;37:58–71. doi: 10.1002/gcc.10207. [DOI] [PubMed] [Google Scholar]
- Charest A, Wilker EW, McLaughlin ME, Lane K, Gowda R, Coven S, McMahon K, Kovach S, Feng Y, Yaffe MB. ROS fusion tyrosine kinase activates a SH2 domain-containing phosphatase-2/phosphatidylinositol 3-kinase/mammalian target of rapamycin signaling axis to form glioblastoma in mice. Cancer Res. 2006;66:7473–7481. doi: 10.1158/0008-5472.CAN-06-1193. [DOI] [PubMed] [Google Scholar]
- Chen J, Zong CS, Wang LH. Tissue and epithelial cell-specific expression of chicken proto-oncogene c-ros in several organs suggests that it may play roles in their development and mature functions. Oncogene. 1994;9:773–780. [PubMed] [Google Scholar]
- Chicoine J, Benoit P, Gamberi C, Paliouras M, Simonelig M, Lasko P. Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Dev Cell. 2007;13:691–704. doi: 10.1016/j.devcel.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Cooper TG. Sperm maturation in the epididymis: A new look at an old problem. Asian J Androl. 2007;9:533–539. doi: 10.1111/j.1745-7262.2007.00285.x. [DOI] [PubMed] [Google Scholar]
- Cox GN, Laufer JS, Kusch M, Edgar RS. Genetic and phenotypic characterization of roller mutants of CAENORHABDITIS ELEGANS. Genetics. 1980;95:317–339. doi: 10.1093/genetics/95.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KD, Le AT, Theodoro MF, Skokan MC, Aisner DL, Berge EM, Terracciano LM, Cappuzzo F, Incarbone M, Roncalli M. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin Cancer Res. 2012;18:4570–4579. doi: 10.1158/1078-0432.CCR-12-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MW, Birnie AJ, Chan AC, Page AP, Jorgensen EM. A conserved metalloprotease mediates ecdysis in Caenorhabditis elegans. Development. 2004;131:6001–6008. doi: 10.1242/dev.01454. [DOI] [PubMed] [Google Scholar]
- Dolphin CT, Hope IA. Caenorhabditis elegans reporter fusion genes generated by seamless modification of large genomic DNA clones. Nucleic Acids Res. 2006;34:e72. doi: 10.1093/nar/gkl352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom M, Han A, Yi SY, Shin JJ, Cui Y, Park KH. RHEB expression in fibroadenomas of the breast. Pathol Int. 2008;58:226–232. doi: 10.1111/j.1440-1827.2008.02215.x. [DOI] [PubMed] [Google Scholar]
- Forde PM, Rudin CM. Crizotinib in the treatment of non-small-cell lung cancer. Expert Opin Pharmacother. 2012;13:1195–1201. doi: 10.1517/14656566.2012.688029. [DOI] [PubMed] [Google Scholar]
- Frand AR, Russel S, Ruvkun G. Functional genomic analysis of C. elegans molting. PLoS Biol. 2005;3:e312. doi: 10.1371/journal.pbio.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz JA, Behm CA. CUTI-1: A novel tetraspan protein involved in C. elegans CUTicle formation and epithelial integrity. PLoS One. 2009;4:e5117. doi: 10.1371/journal.pone.0005117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendler SJ, Spicer AP. Epithelial mucin genes. Annu Rev Physiol. 1995;57:607–634. doi: 10.1146/annurev.ph.57.030195.003135. [DOI] [PubMed] [Google Scholar]
- Gissendanner CR, Sluder AE. nhr-25, the Caenorhabditis elegans ortholog of ftz-f1, is required for epidermal and somatic gonad development. Dev Biol. 2000;221:259–272. doi: 10.1006/dbio.2000.9679. [DOI] [PubMed] [Google Scholar]
- Greenwald I. Development of the Vulva. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. 2nd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1997. Chapter 19. [PubMed] [Google Scholar]
- Gu T, Orita S, Han M. Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol Cell Biol. 1998;18:4556–4564. doi: 10.1128/mcb.18.8.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Aspock G, Burglin TR. The hedgehog-related gene wrt-5 is essential for hypodermal development in Caenorhabditis elegans. Dev Biol. 2006;290:323–336. doi: 10.1016/j.ydbio.2005.11.028. [DOI] [PubMed] [Google Scholar]
- Hayes GD, Frand AR, Ruvkun G. The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development. 2006;133:4631–4641. doi: 10.1242/dev.02655. [DOI] [PubMed] [Google Scholar]
- Higgins BJ, Hirsh D. Roller mutants of the nematode Caenorhabditis elegans. Mol Gen Genet. 1977;150:63–72. doi: 10.1007/BF02425326. [DOI] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007;5:e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janne PA, Meyerson M. ROS1 rearrangements in lung cancer: A new genomic subset of lung adenocarcinoma. J Clin Oncol. 2012;30:878–879. doi: 10.1200/JCO.2011.39.4197. [DOI] [PubMed] [Google Scholar]
- Johnsen RC, Baillie DL. Genetic analysis of a major segment [LGV(left)] of the genome of Caenorhabditis elegans. Genetics. 1991;129:735–752. doi: 10.1093/genetics/129.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone IL, Barry JD. Temporal reiteration of a precise gene expression pattern during nematode development. EMBO J. 1996;15:3633–3639. [PMC free article] [PubMed] [Google Scholar]
- Jones MR, Rose AM, Baillie DL. Oligoarray comparative genomic hybridization-mediated mapping of suppressor mutations generated in a deletion-biased mutagenesis screen. G3 (Bethesda) 2012;2:657–663. doi: 10.1534/g3.112.002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi PM, Riddle MR, Djabrayan NJ, Rothman JH. Caenorhabditis elegans as a model for stem cell biology. Dev Dyn. 2010;239:1539–1554. doi: 10.1002/dvdy.22296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagoshima H, Shigesada K, Kohara Y. RUNX regulates stem cell proliferation and differentiation: Insights from studies of C. elegans. J Cell Biochem. 2007;100:1119–1130. doi: 10.1002/jcb.21174. [DOI] [PubMed] [Google Scholar]
- Kanwar YS, Liu ZZ, Kumar A, Wada J, Carone FA. Cloning of mouse c-ros renal cDNA, its role in development and relationship to extracellular matrix glycoproteins. Kidney Int. 1995;48:1646–1659. doi: 10.1038/ki.1995.460. [DOI] [PubMed] [Google Scholar]
- Keilhack H, Muller M, Bohmer SA, Frank C, Weidner KM, Birchmeier W, Ligensa T, Berndt A, Kosmehl H, Gunther B. Negative regulation of Ros receptor tyrosine kinase signaling. An epithelial function of the SH2 domain protein tyrosine phosphatase SHP-1. J Cell Biol. 2001;152:325–334. doi: 10.1083/jcb.152.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Hwang SB, Jeong PY, Lee J, Cho JW. Requirement of tyrosylprotein sulfotransferase-A for proper cuticle formation in the nematode C. elegans. FEBS Lett. 2005;579:53–58. doi: 10.1016/j.febslet.2004.11.044. [DOI] [PubMed] [Google Scholar]
- Kim TH, Kim YJ, Cho JW, Shim J. A novel zinc-carboxypeptidase SURO-1 regulates cuticle formation and body morphogenesis in Caenorhabditis elegans. FEBS Lett. 2011;585:121–127. doi: 10.1016/j.febslet.2010.11.020. [DOI] [PubMed] [Google Scholar]
- Koh K, Rothman JH. ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development. 2001;128:2867–2880. doi: 10.1242/dev.128.15.2867. [DOI] [PubMed] [Google Scholar]
- Komiya T, Thomas A, Khozin S, Rajan A, Wang Y, Giaccone G. Response to crizotinib in ROS1-rearranged non-small-cell lung cancer. J Clin Oncol. 2012;30:3425–3426. doi: 10.1200/JCO.2012.42.4556. author reply 3426. [DOI] [PubMed] [Google Scholar]
- Kostrouchova M, Krause M, Kostrouch Z, Rall JE. Nuclear hormone receptor CHR3 is a critical regulator of all four larval molts of the nematode Caenorhabditis elegans. Proc Natl Acad Sci USA. 2001;98:7360–7365. doi: 10.1073/pnas.131171898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JM, French RP, Park EC, Johnson JJ. The Caenorhabditis elegans rol-6 gene, which interacts with the sqt-1 collagen gene to determine organismal morphology, encodes a collagen. Mol Cell Biol. 1990;10:2081–2089. doi: 10.1128/mcb.10.5.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JM, Johnson JJ, Edgar RS, Basch C, Roberts S. The sqt-1 gene of C. elegans encodes a collagen critical for organismal morphogenesis. Cell. 1988;55:555–565. doi: 10.1016/0092-8674(88)90214-0. [DOI] [PubMed] [Google Scholar]
- Kuervers LM, Jones CL, O'Neil NJ, Baillie DL. The sterol modifying enzyme LET-767 is essential for growth, reproduction and development in Caenorhabditis elegans. Mol Genet Genomics. 2003;270:121–131. doi: 10.1007/s00438-003-0900-9. [DOI] [PubMed] [Google Scholar]
- Legare C, Sullivan R. Expression and localization of c-ros oncogene along the human excurrent duct. Mol Hum Reprod. 2004;10:697–703. doi: 10.1093/molehr/gah087. [DOI] [PubMed] [Google Scholar]
- Levy AD, Yang J, Kramer JM. Molecular and genetic analyses of the Caenorhabditis elegans dpy-2 and dpy-10 collagen genes: A variety of molecular alterations affect organismal morphology. Mol Biol Cell. 1993;4:803–817. doi: 10.1091/mbc.4.8.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Jones-Rhoades MW, Lau NC, Bartel DP, Rougvie AE. Regulatory mutations of mir-48, a C. elegans let-7 family MicroRNA, cause developmental timing defects. Dev Cell. 2005;9:415–422. doi: 10.1016/j.devcel.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Liu X, Long F, Peng H, Aerni SJ, Jiang M, Sanchez-Blanco A, Murray JI, Preston E, Mericle B, Batzoglou S. Analysis of cell fate from single-cell gene expression profiles in C. elegans. Cell. 2009;139:623–633. doi: 10.1016/j.cell.2009.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meli VS, Osuna B, Ruvkun G, Frand AR. MLT-10 defines a family of DUF644 and proline-rich repeat proteins involved in the molting cycle of Caenorhabditis elegans. Mol Biol Cell. 2010;21:1648–1661. doi: 10.1091/mbc.E08-07-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaux G, Legouis R, Labouesse M. Epithelial biology: Lessons from Caenorhabditis elegans. Gene. 2001;277:83–100. doi: 10.1016/s0378-1119(01)00700-4. [DOI] [PubMed] [Google Scholar]
- Monsalve GC, Van Buskirk C, Frand AR. LIN-42/PERIOD controls cyclical and developmental progression of C. elegans molts. Curr Biol. 2011;21:2033–2045. doi: 10.1016/j.cub.2011.10.054. [DOI] [PubMed] [Google Scholar]
- Muriel JM, Brannan M, Taylor K, Johnstone IL, Lithgow GJ, Tuckwell D. M142.2 (cut-6), a novel Caenorhabditis elegans matrix gene important for dauer body shape. Dev Biol. 2003;260:339–351. doi: 10.1016/s0012-1606(03)00237-9. [DOI] [PubMed] [Google Scholar]
- Nimmo R, Antebi A, Woollard A. mab-2 encodes RNT-1, a C. elegans Runx homologue essential for controlling cell proliferation in a stem cell-like developmental lineage. Development. 2005;132:5043–5054. doi: 10.1242/dev.02102. [DOI] [PubMed] [Google Scholar]
- Novelli J, Ahmed S, Hodgkin J. Gene interactions in Caenorhabditis elegans define DPY-31 as a candidate procollagen C-proteinase and SQT-3/ROL-4 as its predicted major target. Genetics. 2004;168:1259–1273. doi: 10.1534/genetics.104.027953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou SH, Tan J, Yen Y, Soo RA. ROS1 as a ‘druggable’ receptor tyrosine kinase: Lessons learned from inhibiting the ALK pathway. Expert Rev Anticancer Ther. 2012;12:447–456. doi: 10.1586/era.12.17. [DOI] [PubMed] [Google Scholar]
- Page AP, Johnstone IL. The cuticle. WormBook. 2007:1–15. doi: 10.1895/wormbook.1.138.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto CA, Alves LC, de Melo JV, de Souza W. Ultrastructural analyses of the Caenorhabditis elegans sqt-1(sc13) left roller mutant. J Parasitol. 2000;86:269–274. doi: 10.1645/0022-3395(2000)086[0269:UAOTCE]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Peixoto CA, de Melo JV, Kramer JM, de Souza W. Ultrastructural analyses of the Caenorhabditis elegans rol-6 (su1006) mutant, which produces abnormal cuticle collagen. J Parasitol. 1998;84:45–49. [PubMed] [Google Scholar]
- Podbilewicz B, White JG. Cell fusions in the developing epithelial of C. elegans. Dev Biol. 1994;161:408–424. doi: 10.1006/dbio.1994.1041. [DOI] [PubMed] [Google Scholar]
- Politz SM, Philipp M, Estevez M, O'Brien PJ, Chin KJ. Genes that can be mutated to unmask hidden antigenic determinants in the cuticle of the nematode Caenorhabditis elegans. Proc Natl Acad Sci USA. 1990;87:2901–2905. doi: 10.1073/pnas.87.8.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- Rimkunas VM, Crosby KE, Li D, Hu Y, Kelly ME, Gu TL, Mack JS, Silver MR, Zhou X, Haack H. Analysis of receptor tyrosine kinase ROS1-positive tumors in non-small cell lung cancer: Identification of a FIG-ROS1 fusion. Clin Cancer Res. 2012;18:4449–4457. doi: 10.1158/1078-0432.CCR-11-3351. [DOI] [PubMed] [Google Scholar]
- Ruaud AF, Bessereau JL. Activation of nicotinic receptors uncouples a developmental timer from the molting timer in C. elegans. Development. 2006;133:2211–2222. doi: 10.1242/dev.02392. [DOI] [PubMed] [Google Scholar]
- Ruhe JE, Streit S, Hart S, Wong CH, Specht K, Knyazev P, Knyazeva T, Tay LS, Loo HL, Foo P. Genetic alterations in the tyrosine kinase transcriptome of human cancer cell lines. Cancer Res. 2007;67:11368–11376. doi: 10.1158/0008-5472.CAN-07-2703. [DOI] [PubMed] [Google Scholar]
- Saffman EE, Styhler S, Rother K, Li W, Richard S, Lasko P. Premature translation of oskar in oocytes lacking the RNA-binding protein bicaudal-C. Mol Cell Biol. 1998;18:4855–4862. doi: 10.1128/mcb.18.8.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Birchmeier C, Nikawa J, O'Neill K, Rodgers L, Wigler M. Characterization of the ros1-gene products expressed in human glioblastoma cell lines. Oncogene Res. 1989;5:91–100. [PubMed] [Google Scholar]
- Shiffman D, Ellis SG, Rowland CM, Malloy MJ, Luke MM, Iakoubova OA, Pullinger CR, Cassano J, Aouizerat BE, Fenwick RG. Identification of four gene variants associated with myocardial infarction. Am J Hum Genet. 2005;77:596–605. doi: 10.1086/491674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhankova M, Jindra M, Asahina M. Nuclear receptor NHR-25 is required for cell-shape dynamics during epidermal differentiation in Caenorhabditis elegans. J Cell Sci. 2005;118(Pt 1):223–232. doi: 10.1242/jcs.01609. [DOI] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, Fraser AG, Ahringer J, Plasterk RH. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Chao MY, Somers GA, Komatsu H, Corkins ME, Larkins-Ford J, Tucey T, Dionne HM, Walsh MB, Beaumont EK. C. elegans. Notch signaling regulates adult chemosensory response and larval molting quiescence. Curr Biol. 2011;21:825–834. doi: 10.1016/j.cub.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg E, Godecke A, Walter B, Bladt F, Birchmeier C. Transient and locally restricted expression of the ros1 protooncogene during mouse development. EMBO J. 1991;10:3693–3702. doi: 10.1002/j.1460-2075.1991.tb04937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg-Riethmacher E, Walter B, Riethmacher D, Godecke S, Birchmeier C. The c-ros tyrosine kinase receptor controls regionalization and differentiation of epithelial cells in the epididymis. Genes Dev. 1996;10:1184–1193. doi: 10.1101/gad.10.10.1184. [DOI] [PubMed] [Google Scholar]
- Stepek G, McCormack G, Page AP. The kunitz domain protein BLI-5 plays a functionally conserved role in cuticle formation in a diverse range of nematodes. Mol Biochem Parasitol. 2010;169:1–11. doi: 10.1016/j.molbiopara.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Suehara Y, Arcila M, Wang L, Hasanovic A, Ang D, Ito T, Kimura Y, Drilon A, Guha U, Rusch V. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin Cancer Res. 2012;18:6599–6608. doi: 10.1158/1078-0432.CCR-12-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Tessarollo L, Nagarajan L, Parada LF. c-ros: The vertebrate homolog of the sevenless tyrosine kinase receptor is tightly regulated during organogenesis in mouse embryonic development. Development. 1992;115:11–20. doi: 10.1242/dev.115.1.11. [DOI] [PubMed] [Google Scholar]
- Thacker C, Sheps JA, Rose AM. Caenorhabditis elegans dpy-5 is a cuticle procollagen processed by a proprotein convertase. Cell Mol Life Sci. 2006;63:1193–1204. doi: 10.1007/s00018-006-6012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein MC, McCormack G, Winter AD, Johnstone IL, Shoemaker CB, Page AP. Caenorhabditis elegans exoskeleton collagen COL-19: An adult-specific marker for collagen modification and assembly, and the analysis of organismal morphology. Dev Dyn. 2003;226:523–539. doi: 10.1002/dvdy.10259. [DOI] [PubMed] [Google Scholar]
- Thein MC, Winter AD, Stepek G, McCormack G, Stapleton G, Johnstone IL, Page AP. Combined extracellular matrix cross-linking activity of the peroxidase MLT-7 and the dual oxidase BLI-3 is critical for post-embryonic viability in Caenorhabditis elegans. J Biol Chem. 2009;284:17549–17563. doi: 10.1074/jbc.M900831200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursun B, Cochella L, Carrera I, Hobert O. A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS One. 2009;4:e4625. doi: 10.1371/journal.pone.0004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Keyl H, Kim H, Espey R, Oke CV, Edwards MK. Caenorhabditis elegans sqt-3 mutants have mutations in the col-1 collagen gene. Dev Dyn. 1994;201:86–94. doi: 10.1002/aja.1002010109. [DOI] [PubMed] [Google Scholar]
- Wildwater M, Sander N, de Vreede G, van den Heuvel S. Cell shape and Wnt signaling redundantly control the division axis of C. elegans epithelial stem cells. Development. 2011;138:4375–4385. doi: 10.1242/dev.066431. [DOI] [PubMed] [Google Scholar]
- Xia D, Zhang Y, Huang X, Sun Y, Zhang H. The C. elegans CBFbeta homolog, BRO-1, regulates the proliferation, differentiation and specification of the stem cell-like seam cell lineages. Dev Biol. 2007;309:259–272. doi: 10.1016/j.ydbio.2007.07.020. [DOI] [PubMed] [Google Scholar]
- Xiong Q, Chan JL, Zong CS, Wang LH. Two chimeric receptors of epidermal growth factor receptor and c-Ros that differ in their transmembrane domains have opposite effects on cell growth. Mol Cell Biol. 1996;16:1509–1518. doi: 10.1128/mcb.16.4.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Kato K, Yoshida T, Yokoi K, Matsuo H, Watanabe S, Ichihara S, Metoki N, Yoshida H, Satoh K. Association of polymorphisms of ABCA1 and ROS1 with hypertension in Japanese individuals. Int J Mol Med. 2008;21:83–89. [PubMed] [Google Scholar]
- Yang J, Kramer JM. Proteolytic processing of Caenorhabditis elegans SQT-1 cuticle collagen is inhibited in right roller mutants whereas cross-linking is inhibited in left roller mutants. J Biol Chem. 1999;274:32744–32749. doi: 10.1074/jbc.274.46.32744. [DOI] [PubMed] [Google Scholar]
- Yochem J, Tuck S, Greenwald I, Han M. A gp330/megalin-related protein is required in the major epidermis of Caenorhabditis elegans for completion of molting. Development. 1999;126:597–606. doi: 10.1242/dev.126.3.597. [DOI] [PubMed] [Google Scholar]
- Zeng L, Sachdev P, Yan L, Chan JL, Trenkle T, McClelland M, Welsh J, Wang LH. Vav3 mediates receptor protein tyrosine kinase signaling, regulates GTPase activity, modulates cell morphology, and induces cell transformation. Mol Cell Biol. 2000;20:9212–9224. doi: 10.1128/mcb.20.24.9212-9224.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong CS, Poon B, Chen J, Wang LH. Molecular and biochemical bases for activation of the transforming potential of the proto-oncogene c-ros. J Virol. 1993;67:6453–6462. doi: 10.1128/jvi.67.11.6453-6462.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mutant phenotypes associated with rol-3(e754). A. WT animal expressing COL-19::GFP. Uniform expression across the hypodermal surface can be seen (arrow) and formation of alae is complete (arrowhead). B. rol-3(e754) animal expressing COL-19::GFP. Annuli do not form across the lateral hypodermal surface (arrow) and the alae are irregular and improperly formed (arrowhead). C. DIC image of an adult male him-8(e1489) animal showing the WT structure of the male tail. D. Adult rol-3(e754; him- 8(e1489) animal showing the male tail structure encased in excess or improperly formed cuticle. E. Larval rol-3(e754) animal expressing syIs78 (AJM-1::GFP)[Bhagwati, Wang et al. 2003]. Subtle seam cell elongation defects can be seen (asterisks). F. Adult rol-3(e754) animal expressing AJM-1::GFP. Seam syncytium formation is complete but irregular in shape (arrow)(Compare to figure c). Scale bars, 25μm.
Expression of bcc-1 and SRAP-1 does not require functional ROL-3. A. A WT animal expressing bcc-1::GFP in the major hypodermis (arrow). B. A rol-3(s1040) animal raised at the restrictive temperature of 25o expressing bcc-1::GFP in the major hypodermis (arrow). C. A mid-larval stage WT animal expressing SRAP-1::GFP, this animal is in the process of molting (arrows). D. A rol-3(s1040) animal expressing SRAP- 1::GFP raised at the restrictive temperature of 25o. Animal has arrested encased in an improperly shed cuticle. Note the significant accumulation of SRAP-1::GFP at the constriction caused by the unshed cuticle (arrow). Scale bar for all panels, 25μm.