Abstract
The T-cell receptor (TCR)–CD3 complex is critical for T-cell development and function, and represents one of the most complex transmembrane receptors. Models of different stoichiometry and valency have been proposed based on cellular experiments and these have important implications for the mechanisms of receptor triggering. Since determination of receptor stoichiometry in T-cells is not possible due to the presence of previously synthesized, unlabeled receptor components with different half-lives, we examined the stoichiometry of the receptor assembled in endoplasmic reticulum (ER) microsomes of B-cell origin. The stoichiometric relationship among all subunits was directly determined using intact radiolabeled TCR–CD3 complexes that were isolated with a sequential, non-denaturing immunoprecipitation method, and identical results were obtained with two detergents belonging to different structural classes. The results firmly establish that the αβ TCR–CD3 complex assembled in the ER is monovalent and composed of one copy of the TCRαβ, CD3δɛ, CD3γɛ and ζ−ζ dimers.
Keywords: assembly, endoplasmic reticulum, membrane biochemistry, stoichiometry, T-cell receptor
Introduction
The T-cell receptor (TCR)–CD3 complex is one of the most complex transmembrane (TM) receptor structures that has been identified and serves a critical function in the immune system. Signals delivered through this receptor are required for T-cell development in the thymus, the induction of T-cell-mediated immune responses against infectious agents and differentiation of T-cells into effector and memory populations with discrete functional properties. The αβ TCR–CD3 complex is composed of six different type I single-spanning TM proteins: the TCRα and TCRβ chains that form the TCR heterodimer responsible for ligand recognition, and the non-covalently associated CD3γ, CD3δ, CD3ɛ and ζ chains, which bear cytoplasmic sequence motifs that are phosphorylated upon receptor activation and recruit a large number of signaling components (Klausner et al, 1990; Exley et al, 1991; Garboczi et al, 1996; Davis et al, 1997; Sun et al, 2001). The complex is formed in the endoplasmic reticulum (ER) by an ordered assembly process driven by interactions among both extracellular and TM portions of the subunits (Alarcon et al, 1988; Bonifacino et al, 1990; Wileman et al, 1993). The signaling components form the CD3γɛ and CD3δɛ heterodimers and the ζ−ζ homodimer, which associate with the TCRαβ heterodimer through coordination of ionizable residues in the TM regions (Alcover et al, 1990; Blumberg et al, 1990a; Cosson et al, 1991; Manolios et al, 1991; Call et al, 2002).
The mechanism of receptor triggering is not understood and two major models of the TCR–CD3 complex have been proposed, each with unique implications for signal initiation. A model in which a single TCR heterodimer associates with all signaling components (Manolios et al, 1991; Punt et al, 1994; Kearse et al, 1995; Call et al, 2002) implies a triggering mechanism based on a conformational change in individual ligand-engaged receptors and/or recruitment of two or more separate TCR–CD3 complexes into close proximity. In contrast, the model in which two TCR heterodimers are present in a complex (Exley et al, 1995; Jacobs, 1997; San Jose et al, 1998; Fernandez-Miguel et al, 1999) raises the possibility that signaling is initiated by a conformational change in a pre-assembled dimer, as recently described for the erythropoietin receptor (Livnah et al, 1999; Remy et al, 1999) and other cytokine and hormone receptors (Carr et al, 2001; He et al, 2001). The two related but distinct issues of TCR valency (number of TCR heterodimers) and stoichiometry (molar ratios of the different subunits) must therefore be clarified in order to elucidate the mechanism of activation. It has been determined that at least two CD3ɛ subunits are present in the fully assembled structure, as both human and murine forms could be identified in individual TCR–CD3 complexes from murine T-cells expressing both proteins (Blumberg et al, 1990b; de la Hera et al, 1991). However, no direct assessment of the stoichiometric relationships among all of the subunits has been performed. Two studies of the composition of the TCR–CD3 complex addressed receptor valency using transgenic mice that expressed two distinct αβ TCR (Punt et al, 1994) or two different TCRβ chain sequences with the same TCRα (Fernandez-Miguel et al, 1999). However, these two studies came to opposite conclusions regarding the valency of the receptor, which reflects the experimental difficulties of examining such a complex receptor structure in the available cellular systems.
A method for direct determination of the stoichiometry of such a multicomponent receptor structure must meet a number of critical experimental requirements for the conclusions to be valid. All components of the receptor have to be homogenously labeled at a defined number of positions. The stoichiometry among all receptor components can therefore not be determined by labeling of surface receptors with biotin or 125I, since the number of modified sites cannot be determined with certainty. Metabolic labeling is adequate for homogenous labeling of all receptor components, but a number of assembly intermediates and unassembled chains are present in the ER in addition to complete receptor structures. It is thus essential that the receptor is isolated using an approach that yields a population in which each member has the same composition. Finally, analysis of a membrane protein complex requires that proteins be extracted using detergents that can efficiently solubilize the lipid bilayer while simultaneously preserving the non-covalent interactions among the subunits. It is thus critical to demonstrate that the observed stoichiometry does not change with detergent choice. For many complex protein assemblies, absolute subunit stoichiometry is only established when high-resolution structural information becomes available. However, membrane-anchored protein complexes have been generally refractory to the biophysical methods developed to study the structures of water-soluble proteins. Therefore, while complex membrane proteins represent some of the most important structures for basic cellular functions, they are also among the most difficult subjects for structural biologists and protein biochemists.
In this study, we addressed both the valency and the stoichiometry of the TCR–CD3 complex using direct biochemical approaches. We employed a method for isolating intact, radiolabeled protein complexes of known composition to first determine the TCR valency, and then to define the stoichiometric relationships among all TCR–CD3 subunits in the complete receptor structure as well as major assembly intermediates. These results define the TCR–CD3 complex as monovalent, with a stoichiometry of TCRαβ–CD3γɛ–CD3δɛ–ζζ. These conclusions are supported by a large body of data on TCR–CD3 assembly (Punt et al, 1994; Kearse et al, 1995; Call et al, 2002).
Results
Valency of the TCR–CD3 complex
To examine the valency of the TCR–CD3 complex, we assembled human αβ TCR–CD3 complexes in ER microsomes of B-cell origin. This system has been previously used for a mutational analysis of the polar TM residues whose interaction coordinates TCR–CD3 assembly (Call et al, 2002), and has been shown to reflect accurately the membrane protein interactions observed in cells using metabolic labeling techniques (Ribaudo and Margulies, 1992; Bijlmakers et al, 1994; Huppa and Ploegh, 1997; Hebert et al, 1998). The major strength of this method is that radiolabeled proteins are synthesized only from input mRNAs, and that the presence of individual subunits can therefore be controlled. We reasoned that it would be possible to discern whether one or more TCRαβ heterodimers are present in a complex by performing assembly reactions in the presence of two TCRβ chains that differed only by the sequence and molecular weight of attached affinity tags, the nine-amino-acid HA tag and the 47-amino-acid streptavidin-binding peptide (SBP). Two different affinity tags were used rather than two different TCRβ chain sequences, since TCRα and β pairing efficiency is sequence dependent and highly variable. Figure 1A shows the results of such an experiment for the human MHC class II-restricted HA 1.7 αβ TCR (Lamb et al, 1982; Hennecke et al, 2000). Streptavidin (SA) precipitation of digitonin lysate from a reaction containing an SBP-tagged HA 1.7 TCRβ chain and all other TCR–CD3 subunits precipitated TCRβSBP, the TCRαβSBP disulfide-linked heterodimer, and all four associated polypeptides (lane 1), indicating full assembly of TCR–CD3 complexes. The same result is shown for a reaction with the HA-tagged TCRβ chain (lane 2). When separated by SDS–PAGE under non-reducing conditions, the TCRαβSBP and TCRαβHA heterodimers were clearly resolved based on molecular weight (arrowheads). However, when the two TCRβ mRNAs were co-translated in the same reaction, antibodies to one affinity tag failed to co-precipitate the TCRαβ heterodimer bearing the other affinity tag (lanes 4 and 5), despite equivalent levels of assembly (lane 3).
Figure 1.

The αβ TCR–CD3 complex assembled in the ER is monovalent. (A) Translation/assembly reactions were performed with mRNAs encoding the TCRα and TCRβ chains of the human HA 1.7 TCR as well as CD3γ, CD3δ, CD3ɛ and ζ chains. In order to determine whether two TCRβ chains could be incorporated into a TCR–CD3 complex, C-terminal HA or SBP tags were placed onto the HA 1.7 TCRβ chain and either one (lanes 1 and 2) or both β chains (lanes 3–6) were used in the reactions. Reactions were incubated for 15 min at 30°C under reducing conditions to facilitate rapid translation of mRNAs and translocation of radiolabeled proteins into ER microsomes (mouse IVD12; see Materials and methods), followed by a 4-h folding and assembly period under oxidizing conditions, as described in Materials and methods. Single-step IPs were performed for lanes 1–5 as indicated under each gel using SA (SBP tag), anti-HA mAb (HA tag) or mAb UCH-T1 (CD3ɛ), while two-step snIPs were performed for lanes 6 and 7 in which SBP-tagged complexes were captured onto SA beads, released by competition with biotin and re-precipitated with anti-HA mAb. In these experiments and all others except Figure 2B, final products were digested for 1 h at 37°C with 500 U endoglycosidase H. TCRαβ heterodimers that incorporated TCRβHA or TCRβSBP could be resolved by SDS–PAGE due to the size difference between the two tags. All other components of the complex were precipitated when either TCRβ tag was targeted in the IP (lanes 1 and 2). When the two TCRβ chains were present in the same assembly reaction, both TCRβ chains and TCRαβ heterodimers were precipitated by the anti-CD3ɛ mAb (lane 3). However, precipitation directly targeting one TCRβ tag failed to co-precipitate the alternate TCRβ chain or TCRαβ heterodimer (lanes 4 and 5). When both TCRβ tags were targeted in a snIP, no radiolabeled proteins were recovered (lane 6). As a control (*) for background signal in the snIP analysis, aliquots of reactions 1 and 2 were mixed after assembly and analyzed in parallel with reaction 6 (lane 7). (B) The experiment in (A) was repeated using constructs encoding the human A6 αβ TCR. In (C, D), the experiments were performed with constructs in which the tags were placed on TCRα rather TCRβ (HA 1.7 TCR in (C), A6 TCR in (D)). In these experiments, the PC tag recognized by the protein C mAb was used in combination with the SBP tag. All experiments demonstrated that a single TCR heterodimer is incorporated into a TCR–CD3 complex.
As a second approach, both tags were targeted in a sequential non-denaturing immunoprecipitation (snIP) designed to yield intact complexes only if they contained both TCRβ chain affinity tags. Following the first precipitation step, captured radiolabeled protein complexes were eluted from SA beads by competition with biotin and re-precipitated using a mAb directed against the HA tag. No radiolabeled proteins were recovered in this procedure (lane 6), indicating that complexes containing both TCRβ chains in direct or indirect association were not present. However, the same immunoprecipitation (IP) procedure yielded fully assembled complexes when the tags were placed on TCRβ (SBP) and ζ (HA; see Figures 3 and 4), or on CD3ɛ (Call et al, 2002), which is known to occur in at least two copies in the assembled receptor (Blumberg et al, 1990b; de la Hera et al, 1991). This method is therefore suitable for addressing the question of receptor valency and does not disrupt intact radiolabeled protein complexes since elution following the first precipitation step was performed under non-denaturing conditions. We controlled for affinity tag placement and sequence effects by shifting the tags to the C-terminus of TCRα, and by using a different second-step affinity tag, which is recognized by a protein C (PC)-specific antibody (C and D). In addition, the experiments were repeated using a second human αβ TCR pair (MHC class I-restricted A6 TCR; Garboczi et al, 1996; Utz et al, 1996), with identical results (panels B and D). In all experiments, the differentially tagged TCRαβ heterodimers associated equally well with CD3 proteins, yet failed to co-precipitate, indicating that only a single TCR heterodimer is present in the TCR–CD3 complex assembled in the ER.
Figure 3.

Direct measurement of subunit stoichiometry in intact radiolabeled TCR–CD3 complexes isolated by snIP. Translation reactions were performed in the presence of [35S]methionine and ER membranes (mouse IVD12) with mRNAs encoding the A6 TCR and all other components of the TCR–CD3 complex. The SBP tag was present on the A6 TCRβ chain and the HA tag on the ζ chain so that complexes containing all subunits could be isolated by TCRβSBP → TCRζHA snIP from 1.0% digitonin lysates. Quadruplicate reactions were subjected to SDS–PAGE (12%, non-reducing conditions) and proteins were transferred to sequencing-grade PVDF membranes overnight (30 V/4°C). Radiolabeled proteins were detected using a phosphor imager and the signal from each lane was quantitated by densitometry using the Wide Line tool in ImageQuant; the histogram to the right of the gel shows the data for lane 1. Raw counts (line a) were normalized by the number of methionine residues present in each polypeptide (line b) to reflect the relative abundance of each subunit (line c). These values were expressed as a ratio with regard to TCRαβ heterodimer (d), which was present in a single copy per TCR–CD3 complex (Figures 1 and 2). The results from two independent experiments (each performed in quadruplicate) were combined and subjected to statistical analysis (line e; numbers expressed as mean±s.d.).
Figure 4.
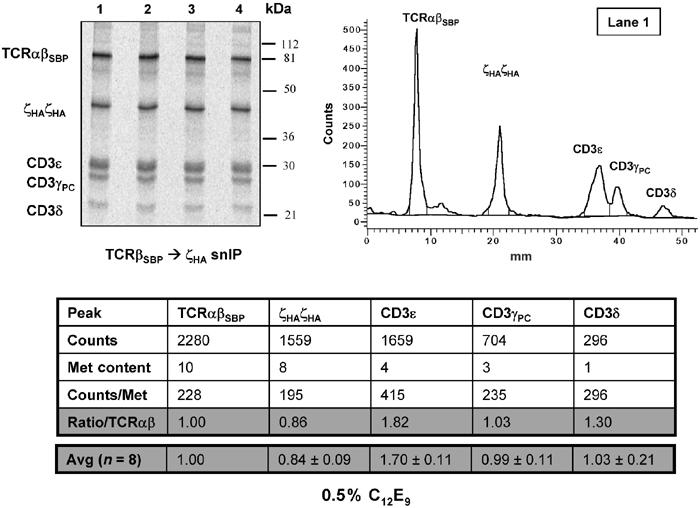
The same TCR–CD3 stoichiometry is observed when a detergent belonging to a different structural class is used for membrane solubilization. Intact radiolabeled TCR–CD3 complexes were isolated and analyzed as in Figure 3, with the exception that detergent solubilization and IPs were performed using 0.5% C12E9, a non-ionic polyoxyethylene detergent, instead of 1.0% digitonin, a deoxycholate derivative.
These experiments were performed using human B-cell-derived ER microsomes to avoid complications arising from the presence of previously synthesized, unlabeled TCR–CD3 components. Therefore, the possibility remained that additional T-cell-specific factors were required for assembly of a higher-order structure in T-cells. We therefore isolated ER microsomes from a TCRβ-deficient human Jurkat T-cell line, and repeated the analysis using these membranes (Figure 2). T-cell-specific proteins were found to be present in these microsomes and available to participate in assembly (Figure 2A), as illustrated by co-precipitation of newly translated A6 TCRαHA and TCRβSBP chains with antibodies to CD3ɛ (lane 3) and ζ (lane 4). As mRNAs encoding CD3 and ζ chains were not present, complexes were formed from newly synthesized TCR heterodimers and previously synthesized signaling chains. This result was clearly specific to the T-cell-derived microsomes, since anti-CD3ɛ and anti-ζ antibodies did not co-precipitate radiolabeled TCR from reactions containing B-cell-derived microsomes (lanes 7 and 8). We therefore tested whether the presence of these or other native T-cell-specific proteins would alter the outcome of the experiments described in Figure 1. To permit parallel analysis in both T-cell- and B-cell-derived membranes, mRNAs for two tagged TCRβ chains (TCRβSBP and TCRβHA) as well as all other components of the complex were included in the reactions, and precipitations were performed by targeting the tag on either TCRβ chain, CD3ɛ or ζ (Figure 2B). As before, the TCRαβSBP and TCRαβHA heterodimers each assembled with CD3ɛ and ζ (lanes 1 and 4), yet were not co-precipitated in the same complex (lanes 2 and 3) regardless of whether assembly reactions were performed with membranes from T-cell or B-cell origin (compare lanes 1–4 with lanes 5–8). These results validate that microsomes of B-cell origin are suitable for studying TCR–CD3 assembly, including a direct assessment of receptor stoichiometry.
Figure 2.

T-cell-derived ER membranes contain previously synthesized T-cell-specific proteins and do not induce assembly of multivalent TCR–CD3 complexes. (A) T-cell-derived ER membranes contain endogenous, assembly-competent components of the TCR–CD3 complex. HA-tagged A6 TCRα and SBP-tagged A6 TCRβ mRNAs were translated in the presence of ER microsomes isolated either from TCRβ-deficient Jurkat T-cells (lanes 1–4) or MGAR B-cells (lanes 5–8), without addition of CD3γ, CD3δ, CD3ɛ or ζ mRNAs. Radiolabeled proteins present in digitonin lysates of membrane fractions were precipitated by targeting TCRαHA (lanes 1 and 5), TCRβSBP (lanes 2 and 6), CD3ɛ (lanes 3 and 7) and ζ chain (lanes 4 and 8). Anti-CD3ɛ and anti-ζ antibodies co-precipitated TCR proteins from T-cell membranes (lanes 3 and 4), but not B-cell membranes (lanes 7 and 8). (B) The TCR–CD3 complex assembled in T-cell-derived ER microsomes is monovalent. Assembly reactions with mRNAs encoding A6 TCRα, TCRβSBP and TCRβHA, CD3γ, CD3δ, CD3ɛ and ζ were carried out in the presence of T-cell- or B-cell-derived ER microsomes. As in Figure 1, the two differentially tagged TCR heterodimers were precipitated by antibodies to TCR-associated signaling chains (lanes 1, 4, 5 and 8), but only one heterodimer was recovered when either of the two TCRβ tags was directly targeted in the precipitation (lanes 2, 3, 6 and 7). Products were not EndoH digested and are therefore observed at different positions compared to the other figures.
Stoichiometry of the fully assembled TCR–CD3 complex
The experiments presented thus far established that the ER-assembled TCR–CD3 complex contains a single TCRαβ heterodimer, but the stoichiometric relationships among all TCR–CD3 subunits has never been directly addressed using a quantitative biochemical approach. The experiment shown in Figure 2A demonstrated as to why it would be difficult to perform a quantitative analysis in T-cells or T-cell-derived membranes, due to the presence of previously synthesized, unlabeled components and partial complexes that are known to differ substantially in their half-life (Bonifacino et al, 1990, 1991). We solved this problem by assembling uniformly 35S-methionine-labeled TCR–CD3 complexes in ER microsomes derived from B-cells, a cell type that is closely related to T-cells but does not synthesize any TCR–CD3 proteins. Complexes of defined composition were isolated by two-step snIP, separated by SDS–PAGE under non-reducing conditions, transferred to polyvinylidene fluoride (PVDF) membranes and quantitated using a phosphor imager.
In the experiment shown in Figure 3, fully assembled TCR–CD3 complexes were purified from cleared digitonin lysates by snIP targeting TCRβSBP followed by HA-tagged ζ chain, which is known to be the last subunit to join during assembly (Sussman et al, 1988; Geisler et al, 1989; Weissman et al, 1989). Since the elution following the first precipitation step was performed by competition with biotin, protein complexes were not disrupted. The counts contained in each band were integrated using the Wide Line tool in the ImageQuant software package (Molecular Dynamics). A sample histogram (right panel) is shown for the gel in the left panel (lane 1) with corresponding peak assignments. The raw counts integrated from each peak (line a) were first adjusted according to the known number of methionine positions in the respective polypeptide (line b). This normalized count (line c) represents the relative amounts of each component. Finally, since the experiments described above established that the TCR heterodimer is present in only one copy per complex, we expressed the abundance of each component relative to TCRαβ (line d). The results indicated that each TCRαβ heterodimer is associated with precisely one ζζ homodimer, two CD3ɛ subunits and one copy of CD3γ as well as CD3δ (line d). These values were highly reproducible (line e), and are consistent with a model in which the complex is built from one copy of each of four distinct modules: the TCRαβ ligand-binding subunit and the non-covalently associated CD3γɛ, CD3δɛ and ζζ signaling dimers.
We took great care in our choice of detergent, since too harsh a detergent can disrupt non-covalent protein complexes, yet too weak a detergent could leave patches of incompletely solubilized lipid carrying embedded radiolabeled proteins. We initially screened a total of 16 different non-ionic detergents from a variety of structural classes for their effects on the integrity of the TCR–CD3 complex (not shown). Digitonin was clearly the best detergent choice based on three independent criteria: (1) digitonin was among the most effective in extracting proteins from the lipid; (2) digitonin produced the highest yield of non-covalently associated proteins co-precipitating with the IP target; and (3) digitonin lysates did not produce artifactual associations due to incompletely solubilized membrane patches. This last criterion was tested in an experiment where TCR–CD3 components were co-translated with two irrelevant polypeptides that assemble into the HLA-DR1αβ heterodimer. Anti-CD3ɛ IP from digitonin lysates co-precipitated the associated TCRα and TCRβ polypeptides but not HLA-DR1 α or β (not shown). To further validate our choice and to rule out detergent-specific effects, we repeated these measurements with a second detergent that belonged to a different structural class than digitonin. As shown in Figure 4, analysis of TCR–CD3 complexes extracted using the polyoxyethylene detergent C12E9 (Lubrol) rather than digitonin produced the same result. These measurements were therefore unchanged by an alternative detergent choice, and reflect the actual stoichiometry of the assembled TCR–CD3 complex.
Stoichiometry of key assembly intermediates
Translation and assembly of protein complexes in ER microsomes allow complete control over the mRNAs that are translated into radiolabeled proteins, thus providing an opportunity to determine the stoichiometry of isolated assembly intermediates that have been shown to represent key steps in the TCR–CD3 assembly process in cells. An analysis of partial complexes formed in the absence of the ζ chain indicated that the stoichiometric relationships among the remaining subunits were unchanged, and that this complex was composed of one TCRαβ heterodimer, one CD3γ, one CD3δ and two CD3ɛ subunits (Figure 5). This is consistent with the finding that a complex containing TCR and all three CD3 subunits can be recovered from an intracellular compartment in murine T-cells lacking expression of the ζ chain (Sussman et al, 1988; Weissman et al, 1989). The same product was also observed in ER microsome assembly experiments when the TCRα chain carried a point mutation of the arginine in its TM domain that prevented association of the ζζ homodimer (Call et al, 2002).
Figure 5.

Stoichiometry of a TCR–CD3 assembly intermediate lacking the ζ chain. Assembly reactions were carried out as in previous experiments using mRNAs encoding all subunits with the exception of the ζ chain. Radiolabeled protein complexes containing all five polypeptides were isolated by TCRβSBP → CD3γPC snIP and analyzed as in Figure 4. The small, broad peak between TCRαβSBP and CD3ɛ may contain non-disulfide-linked TCRα and/or TCRβSBP chains, and was not included in the calculations shown. If assumed to represent TCR, it contributes less than 10% of the total amount of TCR detected. The absence of the ζ chain did not alter the composition or stoichiometry of the remaining subcomplex.
TCR–CD3 assembly proceeds in an ordered fashion with a preferred sequence of association: interaction of CD3δɛ with the TCRα TM domain, followed by association of CD3γɛ via the TCRβ TM domain, and binding of the ζζ homodimer to the complex via a second, distinct site in the TCRα TM domain (Sussman et al, 1988; Geisler, 1992; Kearse et al, 1995; Call et al, 2002). This last step requires prior association of both CD3δɛ and CD3γɛ to TCRαβ since it does not occur when either CD3γ or CD3δ is absent. The TCRα–CD3δɛ and TCRαβ–CD3δɛ subcomplexes are formed in the absence of CD3γ and ζ chain (Figure 6; Call et al, 2002), and these early intermediates have been observed in developing thymocytes in pulse-chase experiments (Kearse et al, 1995) as well as in a Jurkat mutant cell line deficient in CD3γ (Geisler, 1992). As illustrated in Figure 6, densitometric analysis of purified TCRαβ–CD3δɛ complexes reflects the presence of only one copy of CD3δ and CD3ɛ per TCRαβ heterodimer. Interestingly, the TCRβSBP → CD3ɛ snIP strategy employed to isolate this partial complex yielded a significant amount of non-disulfide-linked TCRα and TCRβ chains (arrowheads). These two proteins were in a 1:1 stoichiometric relationship with one another, indicating that they most likely represent a non-covalent TCRαβ heterodimer associated with CD3δɛ. Indeed, when these signals were included in the calculations as additional counts deriving from TCRαβ heterodimer, the relative quantities of the subunits more closely approximated 1:1:1 (TCRαβ:CD3δ:CD3ɛ). The stoichiometric analysis of assembly intermediates thus allows us to exclude models in which a higher-order structure is formed at a particular assembly step through association of a certain signaling dimer. Rather, these results reflect an ordered assembly process in which a monovalent TCR–CD3 complex is formed by interaction of three dimeric modules with a single TCR heterodimer.
Figure 6.

The TCRαβ–CD3δɛ assembly intermediate contains only one copy of CD3ɛ. Assembly reactions lacking mRNAs for both CD3γ and ζ were carried out as before; complexes containing both TCR and CD3 components were isolated by TCRβSBP → CD3ɛ snIP. Recovered proteins were analyzed as described for previous experiments. The majority of TCR heterodimers were disulfide-linked, but non-covalently linked TCR heterodimers were also present in these reactions. The results indicate that only one copy of CD3ɛ is associated with TCR when CD3γ is absent from the reaction.
The experiments presented here clearly demonstrate that all assembly steps can occur within the ER. However, recent work from Geisler and co-workers (Dietrich et al, 1999) has shown that in Jurkat T-cell mutants lacking various TCR–CD3 components, the ζ chain also localizes to the Golgi compartment while other subunits and intermediate complexes are retained in the ER. In cells lacking ζ chain, TCRαβ–CD3δɛ–CD3γɛ complexes exit the ER and are directed from the Golgi to lysosomes for degradation (Sussman et al, 1988; Dietrich et al, 1999). Together, these observations suggest that the final step of TCR–CD3 assembly may also occur in the Golgi compartment.
Discussion
We have demonstrated that the TCR–CD3 complex formed in the ER is a monovalent structure in which a single TCRαβ heterodimer is associated with exactly one CD3γɛ heterodimer, one CD3δɛ heterodimer and one ζζ homodimer. Comparison of TCR–CD3 assembly in T-cell- and B-cell-derived ER microsomes establishes that the machinery required for the proper assembly of this receptor is present in microsomes of B-cell origin. This allowed us to perform reliable stoichiometry measurements on receptor complexes assembled in the ER of a cell type that is closely related to T-cells, but lacks expression of any TCR–CD3 components, and thus provided a solution to the problem that previously synthesized, unlabeled components make stoichiometry measurements in T-cells unreliable. An efficient procedure for snIP (Call et al, 2002) was critical for accurate stoichiometry measurements, since this approach permitted isolation of uniformly labeled, fully assembled structures despite the presence of assembly intermediates and unassembled subunits. We also drew upon knowledge of the stepwise nature of the assembly process from previous studies to isolate specifically key assembly intermediates and measure their stoichiometry. The results of these experiments are fully consistent with mutational analyses showing how critical polar residues in the TM regions of the subunits coordinate the assembly of a single TCRαβ heterodimer with each signaling dimer in discrete steps (Call et al, 2002).
These data are in agreement with the results of previous studies showing that in T-cells from transgenic mice expressing two distinct TCRαβ heterodimers, the two TCRs were not associated (Punt et al, 1994). However, models have been proposed in which two (or more) TCR heterodimers associate through CD3 subunits within a single complex (Exley et al, 1995; Jacobs, 1997; San Jose et al, 1998; Fernandez-Miguel et al, 1999). Exley et al (1995) attempted to determine the molecular weight of the TCR–CD3 complex by labeling cell surface proteins with 125I and fractionating detergent-solubilized membrane proteins on sucrose gradients. Analysis of immunoprecipitated TCR–CD3 complexes from these fractions demonstrated two broad, overlapping TCR–CD3 peaks when membranes were solubilized with 0.2% digitonin and 0.04% Triton X-100. One of these peaks appeared to have a molecular weight higher than calculated for a monovalent TCR–CD3 complex. However, accurate mass measurements are difficult to make, and the presence of aggregates or partially solubilized complexes cannot be excluded in these experiments. Complexes with an even higher apparent molecular weight were immunoprecipitated following solubilization of membranes with 100 mM octyl-β-D-glucoside. The authors hypothesized that the integrity of TCR–CD3 complexes was poorly maintained in digitonin compared to octyl-β-D-glucoside. However, we observed that the complex is preserved in digitonin, but that it is partially disrupted by solubilization with octyl-β-D-glucoside (data not shown). As a second line of experimental evidence, Exley et al (1995) examined antibody-induced downmodulation of TCR–CD3 complexes on T-cell hybridomas that expressed two distinct TCR heterodimers. An antibody directed at one of the TCR heterodimers resulted in a modest degree of downmodulation of the other TCR heterodimer, but the reciprocal experiment showed little to no effect. Large numbers of TCR–CD3 complexes are recruited to the T-cell–APC interface during the formation of immunological synapses (reviewed by Bromley et al, 2001), and antibody-induced co-modulation of distinct TCR–CD3 complexes may therefore not be indicative of a direct physical interaction between the receptors.
Fernandez-Miguel et al (1999) cite co-precipitation of two distinct TCRβ chains from transgenic murine T-cells as the major piece of experimental evidence in support of a multivalent TCR–CD3 model. These investigators used one Vβ-specific antibody for IP of the first TCRβ chain and another Vβ-specific antibody for detection of the second TCRβ chain by Western blotting, and interpreted detection of associated TCRβ chains as support for a multivalent structure. However, this conclusion cannot be reconciled with our data on TCR–CD3 assembly in ER microsomes (Call et al, 2002; present study), or with data from other cellular studies (Punt et al, 1994). A number of technical issues could explain this discrepancy. The investigators detected associated TCRβ chains using both 1% NP-40 and Brij 96, and since NP-40 is known to disrupt TCR–CD3 interactions (San Jose et al, 1998; data not shown), these structures are unlikely to represent mature, intact complexes. Importantly, cells were not surface labeled in this study, leaving open the possibility that the precipitated material could represent partially assembled intermediates, side products or aggregates from intracellular compartments. Indeed, the majority of co-precipitated TCRβ was not covalently associated with TCRα and was sensitive to endoglycosidase H digestion, indicating that it did not derive from mature TCR–CD3 complexes. In an earlier study utilizing a similar experimental design (Punt et al, 1994), structures containing two receptors were not observed when surface-labeled T-cells were analyzed, emphasizing the importance of discriminating between cell surface and intracellular sources when the analysis does not distinguish between partial and complete receptor structures.
Fernandez-Miguel et al (1999) also proposed a particular arrangement among the subunits, but this model was not supported by mutagenesis experiments that directly tested key predictions of the model. The key feature of their model was that the two TCR heterodimers were bridged by association through the ζζ homodimer. However, comparison of stoichiometry measurements for partial and fully assembled complexes in the present study demonstrated that this prediction is not correct since two copies of CD3ɛ and one copy each of CD3γ and CD3δ were present per TCR heterodimer, even when the ζ−ζ homodimer was not present (Figures 3, 4 and 5). Another prediction of the dual-TCR model is that CD3 proteins associate indiscriminately with TCRα and TCRβ chains since one TCR heterodimer can interact directly with only one CD3 heterodimer, yet the fully assembled complex contains both CD3γ and CD3δ (Hall et al, 1991; Huppa and Ploegh, 1997; Call et al, 2002). Individual TCR and CD3 components do associate in various combinations when transfected into non-T-cells in groups of two or three (Cosson et al, 1991; Manolios et al, 1991), and both CD3γ and CD3δ can be found in association with TCRβ in TCRα-deficient murine T-cells (San Jose et al, 1998). However, we have recently shown that while such products can be detected when pairs and trios of polypeptides are studied in isolation, the criteria for progression to the final structure are stringent and require an interaction of CD3δɛ with TCRα and of CD3γɛ with TCRβ, respectively (Call et al, 2002). The assignment of distinct, non-redundant roles for the three basic TM residues in the TCRαβ heterodimer as binding sites for CD3γɛ, CD3δɛ or ζζ also supports a model in which a single TCR interacts directly with all other subunits (Call et al, 2002). These findings are in agreement with cellular studies showing that CD3δɛ and TCRα, as well as CD3γɛ and TCRβ, interact specifically in murine thymocytes (Kearse et al, 1995), and the early identification of an interaction between TCRβ and CD3γ at the cell surface using chemical crosslinking techniques (Brenner et al, 1985).
The major intellectual driving force for the two-TCR models was that they appeared to provide an attractive solution to the problem that six acidic residues are present in the TM domains of the three signaling dimers while only three basic TM residues are located in the TM domains of the TCR heterodimers (Jacobs, 1997; Fernandez-Miguel et al, 1999). These models assume that the basic and acidic residues form pairwise interactions, analogous to salt bridges formed in an aqueous environment (Cosson et al, 1991). However, the basic and acidic residues do not interact as pairs since the association of each signaling dimer with the TCR requires one basic and two acidic side chains correctly positioned among three TM helices (Call et al, 2002). Depending on the protonation state of these ionizable residues, there may nevertheless not be a charge imbalance in the assembled receptor (Engelman, 2003). For example, partial protonation of the pair of acidic TM residues in a signaling dimer could reduce the resulting charge from −2 to −1.
The data of Punt et al (1994) clearly argue against the notion that higher-order structures involving more than one TCR heterodimer are constitutively present on the cell surface. It remains possible, however, that monovalent receptor complexes transiently associate at the cell surface to form higher-order structures under particular conditions. Such transient interactions may occur, for example, during the formation of immunological synapses, since a large number of TCR–CD3 complexes are highly concentrated in a small fraction of the membrane, and such interactions could involve other membrane proteins concentrated at the interface between T-cells and antigen-presenting cells. Identifying such interactions and evaluating their relevance to TCR activation will require sophisticated methods for studying the dynamics of surface-expressed complexes in live cells.
Materials and methods
Antibodies and reagents
Anti-CD3ɛ (mouse mAb UCH-T1) and anti-ζ (mouse mAb 6B10.2) were purchased from Santa Cruz (Santa Cruz, CA). High-affinity anti-HA (rat mAb 3F10) and calcium-dependent anti-PC (mouse mAb HPC4) were purchased from Roche (Indianapolis, IN). SA coupled to agarose beads was purchased from Sigma (St Louis, MO). Digitonin was purchased from Biosynth International (Naperville, IL). C12E9 detergent (Lubrol) was from Sigma.
cDNA constructs and in vitro transcription
Human CD3 γ, δ, ɛ and ζ sequences were amplified from peripheral blood by RT–PCR. A6 TCRα and TCRβ sequences were amplified from the A6 T-cell clone by RT–PCR, and HA 1.7 TCRα and TCRβ sequences were obtained from D Wiley (Hennecke et al, 2000). All sequences were cloned into a modified pSP64 vector (provided by M Kozak) with the murine H-2Kb signal sequence. Mutations were introduced by PCR using overlapping primers. Peptide tags were added as C-terminal in-frame fusions, usually with a three-amino-acid flexible linker. The SBP was sequence C4 (Wilson et al, 2001), with four methionines and one cysteine changed to serine so that radiolabeled methionine or cysteine would not be incorporated into the tag. Peptide tag sequences (single-letter code) were as follows: SBP, SDEKTTGWRGGHVVEGLAGELEQLRARLEHHPQGQ REPSSSGGSKLG; PC peptide, EDQVDPRLIDGK; HA peptide, YPYDVPDYA. In vitro transcription was performed from linearized cDNA constructs using RiboMax T7 large-scale RNA production kit and methyl-7G cap analog (Promega, Madison, WI).
Translation and assembly reactions
Each 25 μl reaction contained 17.5 μl of nuclease-treated rabbit reticulocyte lysate (Promega), 0.5 μl of amino-acid mixture minus methionine (Promega), 0.5 μl of SUPERase-In RNase inhibitor (Ambion), 1.0 μl of 35S-labeled methionine (Amersham, Piscataway, NJ), equivalent molar amounts of each RNA (60–130 ng each) and 2.0 μl of ER microsomes. These microsomes were prepared on continuous Iodixanol gradients as previously described (Call et al, 2002) from human TCRβ-deficient Jurkat T-cells (J.RT3-T3.5 from ATCC), human MGAR B-cells or mouse IVD12 hybridoma (ATCC). All translation and assembly reactions were performed at 30°C. An initial translation period of 15 min under reducing conditions was followed by a 2–4 h assembly period after addition of oxidized glutathione (4 mM). Reaction volumes were 25–100 μl as required for optimal signal with multistep snIP procedures.
Immunoprecipitation, electrophoretic analysis and densitometry
Translation and assembly reactions were stopped by dilution with 1 ml of ice-cold PBS/10 mM iodoacetamide, and microsomes were pelleted (10 min/20 000 g/4°C) and rinsed. Pellets were resuspended in 20 μl of solubilization/IP buffer (PBS+1% digitonin, 10 mM iodoacetamide, 0.1% BSA, 5 μg/ml leupeptin, 1 mM PMSF; with 1 mM CaCl2 when anti-protein C mAb was used) by vigorous pipetting, and then rotated for 30 min at 4°C in a total of 400 μl of solubilization/IP buffer. Lysates were pre-cleared for 1 h with Tris/BSA-blocked Sepharose 4 beads, and primary captures were performed overnight at 4°C. Primary IP products were washed twice in 0.5 ml of wash buffer (PBS+1% digitonin, 10 mM iodoacetamide; with 1 mM CaCl2 for anti-protein C mAb binding). Non-denaturing elution of SA-captured complexes was performed by incubation with 400 μl of solubilization/IP buffer with 100 μM free biotin for 1 h at 4°C, and eluted complexes were incubated with subsequent antibodies and protein G–Sepharose 4 beads (Amersham) for 2 h at 4°C and washed as before. In all experiments but Figure 2B, final precipitates were digested for 1 h at 37°C with 500 U endoglycosidase H (NEB), separated on 12% SDS–PAGE gels, transferred to PVDF membranes overnight at 30 V constant voltage (4°C, with stirring) to ensure complete transfer, and exposed to phosphor imager plates. Densitometry was performed using the Wide Line tool in the ImageQuant software package (Molecular Dynamics).
References
- Alarcon B, Berkhout B, Breitmeyer J, Terhorst C (1988) Assembly of the human T cell receptor–CD3 complex takes place in the endoplasmic reticulum and involves intermediary complexes between the CD3-gamma.delta.epsilon core and single T cell receptor alpha or beta chains. J Biol Chem 263: 2953–2961 [PubMed] [Google Scholar]
- Alcover A, Mariuzza RA, Ermonval M, Acuto O (1990) Lysine 271 in the transmembrane domain of the T-cell antigen receptor beta chain is necessary for its assembly with the CD3 complex but not for alpha/beta dimerization. J Biol Chem 265: 4131–4135 [PubMed] [Google Scholar]
- Bijlmakers MJ, Benaroch P, Ploegh HL (1994) Assembly of HLA DR1 molecules translated in vitro: binding of peptide in the endoplasmic reticulum precludes association with invariant chain. EMBO J 13: 2699–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg RS, Alarcon B, Sancho J, McDermott FV, Lopez P, Breitmeyer J, Terhorst C (1990a) Assembly and function of the T cell antigen receptor. Requirement of either the lysine or arginine residues in the transmembrane region of the alpha chain. J Biol Chem 265: 14036–14043 [PubMed] [Google Scholar]
- Blumberg RS, Ley S, Sancho J, Lonberg N, Lacy E, McDermott F, Schad V, Greenstein JL, Terhorst C (1990b) Structure of the T-cell antigen receptor: evidence for two CD3 epsilon subunits in the T-cell receptor–CD3 complex. Proc Natl Acad Sci USA 87: 7220–7224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Cosson P, Klausner RD (1990) Colocalized transmembrane determinants for ER degradation and subunit assembly explain the intracellular fate of TCR chains. Cell 63: 503–513 [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Cosson P, Shah N, Klausner RD (1991) Role of potentially charged transmembrane residues in targeting proteins for retention and degradation within the endoplasmic reticulum. EMBO J 10: 2783–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner MB, Trowbridge IS, Strominger JL (1985) Cross-linking of human T cell receptor proteins: association between the T cell idiotype beta subunit and the T3 glycoprotein heavy subunit. Cell 40: 183–190 [DOI] [PubMed] [Google Scholar]
- Bromley SK, Burack WR, Johnson KG, Somersalo K, Sims TN, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML (2001) The immunological synapse. Annu Rev Immunol 19: 375–396 [DOI] [PubMed] [Google Scholar]
- Call ME, Pyrdol J, Wiedmann M, Wucherpfennig KW (2002) The organizing principle in the formation of the T cell receptor–CD3 complex. Cell 111: 967–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr PD, Gustin SE, Church AP, Murphy JM, Ford SC, Mann DA, Woltring DM, Walker I, Ollis DL, Young IG (2001) Structure of the complete extracellular domain of the common beta subunit of the human GM-CSF, IL-3, and IL-5 receptors reveals a novel dimer configuration. Cell 104: 291–300 [DOI] [PubMed] [Google Scholar]
- Cosson P, Lankford SP, Bonifacino JS, Klausner RD (1991) Membrane protein association by potential intramembrane charge pairs. Nature 351: 414–416 [DOI] [PubMed] [Google Scholar]
- Davis MM, Lyons DS, Altman JD, McHeyzer-Williams M, Hampl J, Boniface JJ, Chien Y (1997) T cell receptor biochemistry, repertoire selection and general features of TCR and Ig structure. Ciba Found Symp 204: 94–100, discussion 100–104 [DOI] [PubMed] [Google Scholar]
- de la Hera A, Muller U, Olsson C, Isaaz S, Tunnacliffe A (1991) Structure of the T cell antigen receptor (TCR): two CD3 epsilon subunits in a functional TCR/CD3 complex. J Exp Med 173: 7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich J, Kastrup J, Lauritsen JP, Menne C, von Bulow F, Geisler C (1999) TCRzeta is transported to and retained in the Golgi apparatus independently of other TCR chains: implications for TCR assembly. Eur J Immunol 29: 1719–1728 [DOI] [PubMed] [Google Scholar]
- Engelman DM (2003) Electrostatic fasteners hold the T cell receptor–CD3 complex together. Mol Cell 11: 5–6 [DOI] [PubMed] [Google Scholar]
- Exley M, Terhorst C, Wileman T (1991) Structure, assembly and intracellular transport of the T cell receptor for antigen. Semin Immunol 3: 283–297 [PubMed] [Google Scholar]
- Exley M, Wileman T, Mueller B, Terhorst C (1995) Evidence for multivalent structure of T-cell antigen receptor complex. Mol Immunol 32: 829–839 [DOI] [PubMed] [Google Scholar]
- Fernandez-Miguel G, Alarcon B, Iglesias A, Bluethmann H, Alvarez-Mon M, Sanz E, de la Hera A (1999) Multivalent structure of an alphabetaT cell receptor. Proc Natl Acad Sci USA 96: 1547–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC (1996) Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature 384: 134–141 [DOI] [PubMed] [Google Scholar]
- Geisler C (1992) Failure to synthesize the CD3-gamma chain. Consequences for T cell antigen receptor assembly, processing, and expression. J Immunol 148: 2437–2445 [PubMed] [Google Scholar]
- Geisler C, Kuhlmann J, Rubin B (1989) Assembly, intracellular processing, and expression at the cell surface of the human alpha beta T cell receptor/CD3 complex. Function of the CD3-zeta chain. J Immunol 143: 4069–4077 [PubMed] [Google Scholar]
- Hall C, Berkhout B, Alarcon B, Sancho J, Wileman T, Terhorst C (1991) Requirements for cell surface expression of the human TCR/CD3 complex in non-T cells. Int Immunol 3: 359–368 [DOI] [PubMed] [Google Scholar]
- He X, Chow D, Martick MM, Garcia KC (2001) Allosteric activation of a spring-loaded natriuretic peptide receptor dimer by hormone. Science 293: 1657–1662 [DOI] [PubMed] [Google Scholar]
- Hebert DN, Zhang JX, Helenius A (1998) Protein folding and maturation in a cell-free system. Biochem Cell Biol 76: 867–873 [DOI] [PubMed] [Google Scholar]
- Hennecke J, Carfi A, Wiley DC (2000) Structure of a covalently stabilized complex of a human alphabeta T-cell receptor, influenza HA peptide and MHC class II molecule, HLA-DR1. EMBO J 19: 5611–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppa JB, Ploegh HL (1997) In vitro translation and assembly of a complete T cell receptor–CD3 complex. J Exp Med 186: 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs H (1997) Pre-TCR/CD3 and TCR/CD3 complexes: decamers with differential signalling properties? Immunol Today 18: 565–569 [PubMed] [Google Scholar]
- Kearse KP, Roberts JL, Singer A (1995) TCR alpha–CD3 delta epsilon association is the initial step in alpha beta dimer formation in murine T cells and is limiting in immature CD4+ CD8+ thymocytes. Immunity 2: 391–399 [DOI] [PubMed] [Google Scholar]
- Klausner RD, Lippincott-Schwartz J, Bonifacino JS (1990) The T cell antigen receptor: insights into organelle biology. Annu Rev Cell Biol 6: 403–431 [DOI] [PubMed] [Google Scholar]
- Lamb JR, Eckels DD, Lake P, Woody JN, Green N (1982) Human T-cell clones recognize chemically synthesized peptides of influenza haemagglutinin. Nature 300: 66–69 [DOI] [PubMed] [Google Scholar]
- Livnah O, Stura EA, Middleton SA, Johnson DL, Jolliffe LK, Wilson IA (1999) Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science 283: 987–990 [DOI] [PubMed] [Google Scholar]
- Manolios N, Letourneur F, Bonifacino JS, Klausner RD (1991) Pairwise, cooperative and inhibitory interactions describe the assembly and probable structure of the T-cell antigen receptor. EMBO J 10: 1643–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt JA, Roberts JL, Kearse KP, Singer A (1994) Stoichiometry of the T cell antigen receptor (TCR) complex: each TCR/CD3 complex contains one TCR alpha, one TCR beta, and two CD3 epsilon chains. J Exp Med 180: 587–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy I, Wilson IA, Michnick SW (1999) Erythropoietin receptor activation by a ligand-induced conformation change. Science 283: 990–993 [DOI] [PubMed] [Google Scholar]
- Ribaudo RK, Margulies DH (1992) Independent and synergistic effects of disulfide bond formation, beta 2-microglobulin, and peptides on class I MHC folding and assembly in an in vitro translation system. J Immunol 149: 2935–2944 [PubMed] [Google Scholar]
- San Jose E, Sahuquillo AG, Bragado R, Alarcon B (1998) Assembly of the TCR/CD3 complex: CD3 epsilon/delta and CD3 epsilon/gamma dimers associate indistinctly with both TCR alpha and TCR beta chains. Evidence for a double TCR heterodimer model. Eur J Immunol 28: 12–21 [DOI] [PubMed] [Google Scholar]
- Sun ZJ, Kim KS, Wagner G, Reinherz EL (2001) Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3 epsilon gamma heterodimer. Cell 105: 913–923 [DOI] [PubMed] [Google Scholar]
- Sussman JJ, Bonifacino JS, Lippincott-Schwartz J, Weissman AM, Saito T, Klausner RD, Ashwell JD (1988) Failure to synthesize the T cell CD3-zeta chain: structure and function of a partial T cell receptor complex. Cell 52: 85–95 [DOI] [PubMed] [Google Scholar]
- Utz U, Banks D, Jacobson S, Biddison WE (1996) Analysis of the T-cell receptor repertoire of human T-cell leukemia virus type 1 (HTLV-1) Tax-specific CD8+ cytotoxic T lymphocytes from patients with HTLV-1-associated disease: evidence for oligoclonal expansion. J Virol 70: 843–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman AM, Frank SJ, Orloff DG, Mercep M, Ashwell JD, Klausner RD (1989) Role of the zeta chain in the expression of the T cell antigen receptor: genetic reconstitution studies. EMBO J 8: 3651–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wileman T, Kane LP, Young J, Carson GR, Terhorst C (1993) Associations between subunit ectodomains promote T cell antigen receptor assembly and protect against degradation in the ER. J Cell Biol 122: 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DS, Keefe AD, Szostak JW (2001) The use of mRNA display to select high-affinity protein-binding peptides. Proc Natl Acad Sci USA 98: 3750–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]


