Abstract
Based on extensive pre-clinical studies, the oncolytic parvovirus H-1 (H-1PV) is currently applied to patients with recurrent glioblastoma in a phase I/IIa clinical trial (ParvOryx01, NCT01301430). Cure rates of about 40% in pediatric high-risk medulloblastoma (MB) patients also indicate the need of new therapeutic approaches. In order to prepare a future application of oncolytic parvovirotherapy to MB, the present study preclinically evaluates the cytotoxic efficacy of H-1PV on MB cells in vitro and characterizes cellular target genes involved in this effect. Six MB cell lines were analyzed by whole genome oligonucleotide microarrays after treatment and the results were matched to known molecular and cytogenetic risk factors. In contrast to non-transformed infant astrocytes and neurons, in five out of six MB cell lines lytic H-1PV infection and efficient viral replication could be demonstrated. The cytotoxic effects induced by H-1PV were observed at LD50s below 0.05 p. f. u. per cell indicating high susceptibility. Gene expression patterns in the responsive MB cell lines allowed the identification of candidate target genes mediating the cytotoxic effects of H-1PV. H-1PV induced down-regulation of key regulators of early neurogenesis shown to confer poor prognosis in MB such as ZIC1, FOXG1B, MYC, and NFIA. In MB cell lines with genomic amplification of MYC, expression of MYC was the single gene most significantly repressed after H-1PV infection. H-1PV virotherapy may be a promising treatment approach for MB since it targets genes of functional relevance and induces cell death at very low titers of input virus.
Keywords: medulloblastoma, oncolytic virus, parvovirus H-1PV, cellular targets, MYC, master regulators of neurogenesis
Introduction
Medulloblastoma is the most frequent malignant brain tumor in children accounting for about 10% of the mortality in pediatric cancer patients.1 The introduction of multimodal treatment concepts including surgery, local irradiation, and high-dose chemotherapy according to risk group and therapy response has improved the outcome of high-risk medulloblastoma patients. However, about 60%2 of the high-risk patients do not survive the disease and even children surviving medulloblastoma suffer from severe long-term neurologic, endocrinological, and cognitive sequelae resulting in relevant morbidity.
High-risk medulloblastoma can either be classified clinically (metastatic disease at the time of initial diagnosis) or histopathologically (large cell or anaplastic subtype), or by molecular subtypes. Chromosomal aberration patterns (genomic amplification of the oncogenes MYC and/or MYCN, gain of 17q or 6q) and gene expression profiles (group 3 and 4) identifying medulloblastoma patients with extremely poor prognosis have been characterized.3–6
What’s new? —
Medulloblastoma, the most frequent pediatric brain cancer, causes death in about 60 percent of high-risk patients, and so there is a major need for novel, highly effective therapies. One therapy of interest is parvovirus H-1 (H-1PV), which was found in this study to produce marked cytotoxic effects in six medulloblastoma cell lines. Gene expression profiling revealed that H-1PV infection causes down-regulation of key regulatory genes involved in early neurogenesis, with significant repression of MYC. The master regulators affected may represent putative direct or indirect H-1PV target genes.
In search of promising therapeutic approaches for high-risk medulloblastoma patients, oncolytic virotherapy has been pre-clinically evaluated in a variety of cell culture systems and animal models.7–9 Medulloblastoma could be shown to be a promising target for the application of self-replicating oncolytic viruses, such as reovirus, Seneca Valley virus and recombinant measles virus, inducing significant increase of survival in medulloblastoma xenograft-bearing nude mice.10–13 The rodent parvovirus H-1 (H-1PV) is a non-recombinant oncolytic virus naturally occurring in rats. Although the virus is apathogenic in humans viremia and subsequent seroconversion has been reported under experimental conditions.14 For high-grade glioma, the oncolytic efficacy of H-1PV has been demonstrated in vitro and in rat models. Long time survival has been observed after intratumoral, intravenous or intranasal virus application in both orthotopic allograft and xenograft bearing rats.15,16 Recently, a clinical phase I/IIa trial on adult patients with primary progressive or recurrent glioblastoma has been initiated. The present publication preclinically addressed the applicability of H-1PV to the treatment of medullo-blastoma as the most frequent malignant brain tumor in children. Subsequently, we characterized the transcriptional response to H-1PV infection in MB cells in order to identify target genes associated with virus-induced cytotoxicity.
Viral entry into human medulloblastoma cells, efficient virus replication and cellular lysis induced by the virus could be demonstrated. As a first step in the investigation of the mechanisms of selective cytotoxicity in MB, we used gene expression profiling in responsive cell lines during H-1PV treatment to identify target genes or pathways modulated by H-1PV infection and preceding cellular death. Surprisingly, among the genes we found to be most significantly repressed, several had been previously identified as overexpressed in primary medulloblastoma samples and to be associated with poor prognosis. These direct or indirect H-1PV target genes are well characterized morphogens of early embryonic CNS development and potent mitogens in medulloblastoma formation or metastatic spreading such as MYC,17,18 NFIA and NFIB.19
Material and Methods
Cell culture
The human medulloblastoma cell lines D425, D458 and DAOY, were purchased from the ATCC. MED8A were a friendly gift from Michael D. Taylor (Toronto, ON, Canada). ONS76 was obtained from the Institute for Fermentation (Osaka, Japan), and UW228-2 cells were provided by John Silber (Seattle, WA). All cell lines were cultured at 37°C, 5% CO2 in different media containing 100 units of penicillin and 100 μg of streptomycin per ml, and 10% FCS. The cell lines DAOY, MED8A, ONS76 and UW228-2 were kept in DMEM, the cell lines D425 and D458 in improved MEM. The culture conditions for the MYCN over expressing neuroblastoma cell line WAC-2 were published previously.20 Primary human infant astrocytes were obtained in 2003 during routine neurosurgical treatment by Marta Herrero y Calle, Department of Neurosurgery, University Hospital Freiburg, as published previously. Informed consent was obtained from all parents/patients in this study before the neurosurgical procedure.20 They were cultured in DMEM supplemented with astrocyte growth supplement (ScienCell, Carlsbad, CA). HCN1A infant cortical neurons were obtained from the ATCC and cultured in neuronal growth medium (ScienCell, Carlsbad, CA).
Virus production and infection
Wild-type H-1PV was produced by infecting NBK-324K human embryonic kidney cells. The recombinant, replication-deficient parvovirus H-1 expressing EGFP (H-1EGFP) was produced by co-transfection of 293T cells with the recombinant vector DNA and a helper plasmid expressing the viral capsid genes as previously described.21 Virus purification was performed by filtration (maximal diameter of particles 0.2 μm) and subsequent iodixanol gradient centrifugation (Visipaqueμ, Amersham Biosciences Europe, Freiburg, Germany). The contamination of virus stocks with endotoxins was below 2.5 EU/ml.
Detection of infectious H-1PV particles by dot blot assay
Virus titers were determined as described previously.22 Briefly, NB-324K cells were seeded in 96-well plate 24 h before the assay. Cells were infected with 10-fold serial dilutions of the virus preparation and incubated for 72 h at 37°C, 5% CO2. After alkaline lysis applying 0.75 M NaOH, DNA was transferred to a nylon membrane, cross-linked, and hybridized with a NS-1 specific, 32P-labeled probe and detected by autoradiography on X-ray films. All titration experiments were performed in triplicates.
Immunofluorescence staining and microscopy
In a 96-well plate cells were seeded in a density of 2,500 cells per well and either infected with MOIs of 1 or 10 p. f. u per cell of the wild type H-1PV virus or mock-infected with the same volume of 40% iodixanol (Visipaqueμ, GE Healthcare, Chalfont St Giles, U.K.) in Ringer solution. After 24 h infection cells were fixed in precooled methanol and acetone, and immunofluorescence for NS1 and Lamin β was performed as published previously.23 Immunofluorescence was documented by confocal microscopy and the OLYMPUS FLUOVIEW 3.0 Viewer software and counted subsequently. Phase contrast images were generated by inverted phase contrast microscopy (Olympus; Model CKX41) using Cell B software (Olympus, Hamburg, Germany) or an Olympus digital camera.
Assessment of cell viability and lysis
After treatment the metabolic activity of the cells was tested using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and cell lysis was determined by measuring the release of lactate dehydrogenase (LDH) into the culture medium. Cells were seeded in 96-well plates and either mock-infected as described above or infected with H-1PV at increasing MOIs. Cells in 4 out of 11 wells infected with a defined virus dose were lysed with detergent 72 h after treatment. Subsequently, 50 μl of the supernatant from all wells were transferred into a new plate to perform the LDH-release assay using the Cytotox96 cytotoxicity assay kitμ (Promega, Mannheim, Germany) as described by the manufacturer. Cells were further incubated with 0.8 μg/ml MTT for up to 1 h. The supernatant was discarded and cells were allowed to dry. Subsequently the dye was redissolved in 100 μl of 2-propanol. Absorbance was measured photometrically (Multiscan Plusμ, Titertek Instruments Inc., Huntsville, AL) at 495 nm for the LDH-release assays and 570 nm for the MTT-tests.
Real-time proliferation measurement
Proliferation of the cells was measured based on real-time impedance measurement in intervals of 30 min using the xCELLigence system for real-time proliferation measurement (xCELLigence SP and MP, Roche Applied Science, Mannheim, Germany). For medulloblastoma cell lines, 3,000 cells per well were seeded in a 96-well plate (E-plate 96, Roche Applied Science, Mannheim, Germany) and the proliferation index was recorded in multiplicates of 10 wells infected with a defined virus dose until the non-treated control cells reached confluence and eventually died. Dose–response curves and resulting LD50s were calculated by analysis of 10 wells per dose according to the manufacturer’s advice. Due to the low cell proliferation rate in HCN1A culture of non-transformed infant neurons, 10,000 cells per well were seeded and only triplicates were analyzed.
Total RNA preparation of cell lines
RNA from cell lines was extracted by phenolic extraction adding Trizol (Invitrogen, Carlsbad) solution, and further purified using RNeasy mini columns (Qiagen, Hilden, Germany) according the manufacturers’ instructions. Quality and quantity of RNA were determined by absorption measurement in an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA) and NanoDrop ND-100 (NanoDrop Technologies, Wilmington, DE). Only high quality RNA (RNA Integrity Number of minimum 7.0) was used for array analysis.
Array-comparative genomic hybridization
As previously described, array-CGH analyses were carried out for the six medulloblastoma cell lines at an average probe spacing of 0.4 Mb.3
Microarray analysis
RNA was hybridized to the 4x44K feature Agilent Whole Human Genome Oligo Microarray following the manufacturer’s instructions, as described previously.24 All hybridization experiments were investigated by labeling tumor sample against commercial reference probes (normal brain pool (cerebellum), Clonetech, Mountain View) and were scanned in a two-color Agilent Scanner G25505B (Agilent Technologies, Santa Clara, CA) according to the manufacturer’s specification. Normalization of raw signals was performed using vsn.25 Based on BLASTing the probes’ sequence information against the genome, biological annotations were retrieved from EnsEMBL database (version 54, NCBI Build 36 of the human genome reference sequence).
Biostatistics and bioinformatics
Missing Agilent transcript data were imputed using k-nearest neighbor imputation, as described previously.26 The first objective was to identify clones that are regulated in cells showing significant oncolysis in response to H-1PV (cell lines DAOY, D425, D458, MED8A, and UW228-2) at any time point compared to baseline reference, which allows statistical testing. In consequence, only clones with similar up- or down-regulation in all five cell lines were selected. The five cell lines were treated as replicates of the same condition in order to allow for statistical testing. The empirical Bayes approach was used to identify differentially expressed genes.24 The selection of genes was based on the moderated F statistics. Benjamini-Hochberg correction was applied in order to keep the false discovery rate (FDR) at 5%. p Values of 0.05 or below were considered significant. All biostatistical and statistical analyses were performed with the R/Bioconductor software environment, Version 2.11/2.6 using add-on R packages as previously described.24
Quantitative real-time PCR analysis
Quantification of mRNA expression for selected candidate genes was performed by real-time quantitative PCR (QRT-PCR) using the ABI PRISM® 7700HT Sequence Detection System Instrument (Applied Biosystems, Applera Deutschland GmbH, Darmstadt, Germany). Total RNA was isolated and reversely transcribed as described previously.24 Primers were obtained from Sigma-Aldrich. QRT-PCR reactions were performed with the power SYBR® green PCR master mix according to the manufacturer’s instructions in a MicroAmp optical 96-well reaction plate with an ABI PRISM® 7700HT sequence detector (Applied Biosystems). Gene expression levels were normalized to HPRT1 and SDHA expression, which was used as a control housekeeping gene.
Western Blot Analysis
Cells (106) were either mock-treated with 10% iodixanol (Visipaqueμ, Amersham Biosciences) or infected with 1 p. f. u. of H-1PV per cell. At indicated time points, cells were harvested with a cell scraper, pelleted, and washed with PBS, and cell pellets were kept at –80°C until further analysis. Cells were allowed to lyse in RIPA lysis buffer for 1 h on ice. Lysates were centrifuged at 15,000g for 30 min at 4°C. Protein concentrations in the cell lysates were determined photometrically according to the manufacturer’s advice (BIO-RAD protein assay, Bio-Rad GmbH, München, Germany). A total of 10 μg of extracted proteins were fractionated by 9% SDS-PAGE, and transferred to nitrocellulose membranes (Schleicher & Schüll, Kassel, Germany). For the detection of viral proteins, polyclonal rabbit antisera (kindly provided by Dr. N. Salomé) were used: MK-3 raised against the viral NS1 protein,27 and anti-VP1 and anti-VP2 against the viral proteins VP1 and VP2 respectively.20,28,29 For the detection of putative cellular protein targets of H-1PV, the following commercially available antibodies were applied: monoclonal mouse anti-human c-myc (clone 9E10), polyclonal rabbit anti-human NF1A (Sigma-Aldrich), and polyclonal rabbit anti-human NFIB (Sigma-Aldrich, Hamburg, Germany). Bound first antibodies were detected using horseradish peroxidase-conjugated IgGs (goat anti-mouse IgG-HRP, clone sc-2005, and goat anti-rabbit, clone W401B, Santa Cruz Biotechnology, Santa Cruz (CA)) in dilutions from 1:2,500 to 1:5,000. Chemoluminescence was assayed as described previously.30
Results
Medulloblastoma cells are competent for parvovirus transduction
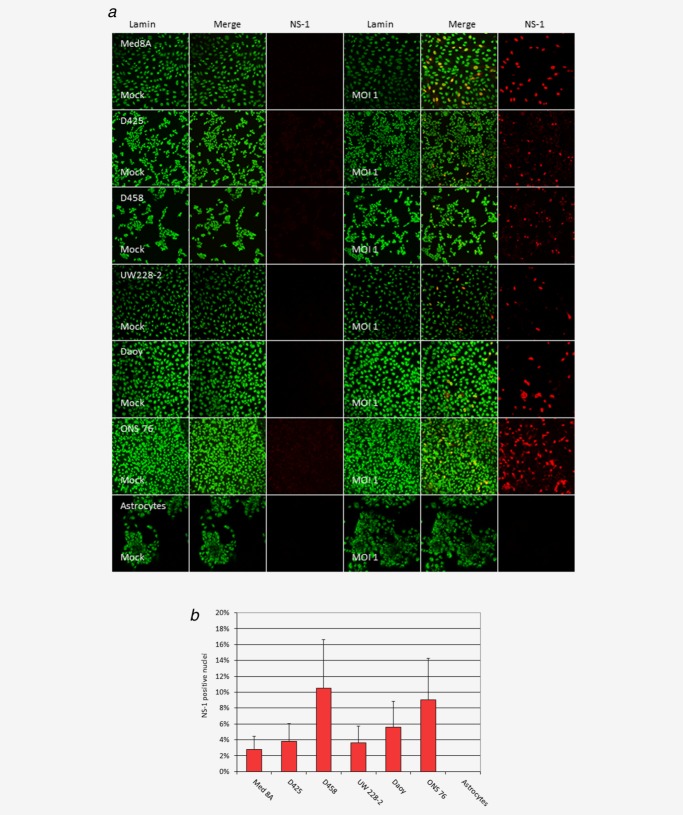
Using recombinant H-1EGFP, virus-mediated transduction was confirmed for all six MB cell lines (data not shown). Entry of wild-type H-1PV virus and onset of replication could be demonstrated by immunofluorescence detection of NS-1 expression in all six MB cell lines tested 24 h after infection (Fig. 1a). The fraction of NS-1 expressing cells varied between the different MB lines, however not being of high statistical significance (p ≥ 0.04) (Fig. 1b). In contrast, no relevant NS-1 expression was detected in cultures of pediatric astrocytes with the same dose of wild-type H-1PV (Figs. 1a and 1b). Quantification of the proportion of NS-1 positive cells (Fig. 1b) significantly differed between each of the MB cell lines and infant astrocytes, which were analyzed as non-transformed control cells (determined by t-test, p < 0.001).
Figure 1.
Permissiveness of medulloblastoma cells for parvovirus H-1 (H-1PV) infection and onset of replication Medulloblastoma cells or infant non-transformed astrocytes were infected with 1 plaque forming unit per cell of wild type H-1PV, fixed after 24 h, and examined by confocal microscopy after immunofluorescence staining for the nuclear membrane protein Lamin beta (green) and for the viral protein NS-1 (red). (a) Representative pictures of indicated cell cultures after mock (three left panels) or H-1PV infection (three right panels). (b) The proportion of NS1-expressing medulloblastoma cells in a field was determined in triplicates by manual counting. Error bars indicate standard errors of the mean.
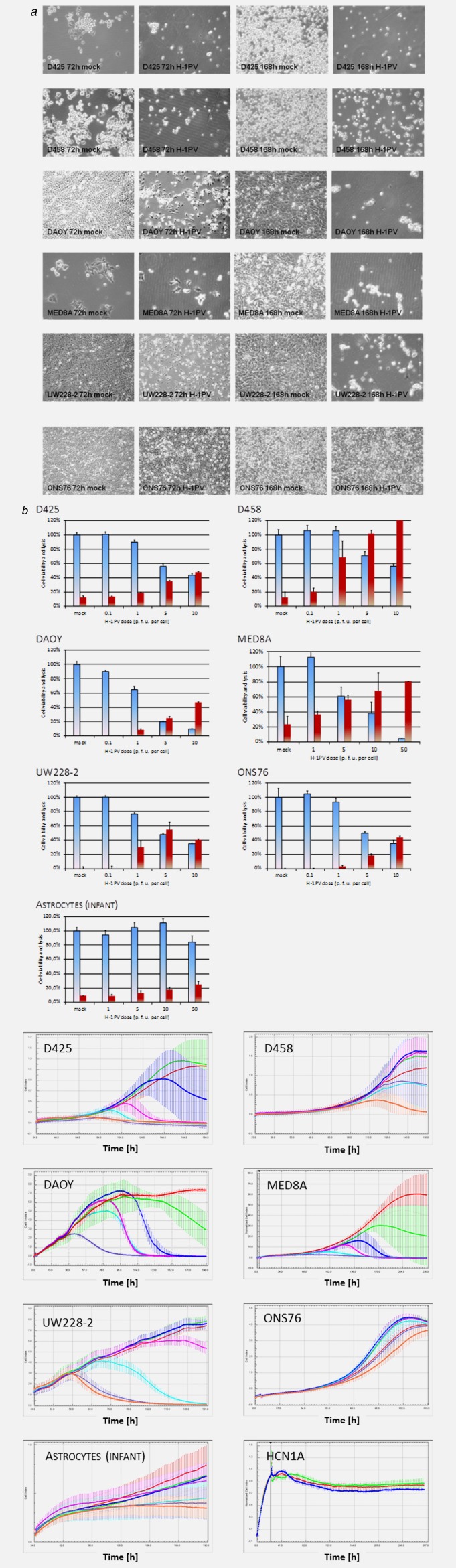
H-1PV infection of MB cells was accompanied by cytomorphological changes (Fig. 2a). Under the assay conditions used, these cytopathic effects were detected at low multiplicities of infection with IC50 doses lower than 1 p. f. u. per cell for all MB cell lines except for one (ONS76), which only showed these effects at high multiplicities of infection (Supporting Information Table 2). In contrast, cultures of non-transformed infant brain cells, such as astrocytes or neurons, remained unaffected by H-1PV infection with doses below 50 p. f. u. per cell, with TCID50s that were 5,000 fold higher than in the five responsive medulloblastoma cell lines (Supporting Information Table 2). These in vitro findings provided first indications of the therapeutic index for oncolytic virus treatment of pediatric brain tumors with H-1PV and for the clinical safety of the application to children.
Figure 2.
Cytotoxicity of parvovirus H-1 (H-1PV) for medulloblastoma cell lines. Indicated medulloblastoma (MB) cells were mock-treated or infected with one plaque forming unit per cell of H-1PV and analyzed for cytomorphological changes by conventional phase contrast microscopy at 72 h and 168 h after infection (a). Seventy-two hours postinfection cell viability and lysis was measured by MTT- (blue columns) and LDH release (red columns) assays (b). Real-time proliferation measurement of H-1PV treated cell lines and short term cultures of nontransformed neuroepithelial cells. The cell index correlating to the number of living cells was determined in intervals of 15 minutes. For each time point average values with standard deviation bars are displayed. In MB cell lines in multiplicates of 10 per cell line and dose of input virus applied were analyzed and the respective graphs are given in a colour code: For the MB cell lines: mock infected (red), 0.001 p. f. u. per cell (green), 0.01 p. f. u. per cell (royal blue), 0.1 p. f .u. per cell (magenta), 1 p. f. u. per cell (cyan), 5 p. f. u. per cell (violet), and 10 p. f. u. per cell (orange). For infant astrocytes: mock infected (red), 0.01 p.f. u. per cell (green), 0.1 p.f. u. per cell (royal blue), 1 p.f.u. (magenta), 10 p. f. u. per cell (cyan), 50 p. f. u. per cell (violet), and 100 p.f. u. per cell (orange); and for HCN1A cortical neurons: Untreated (red), mock infected (green), and 100 p. f. u. per cell (royal blue). A comparative analysis was performed in cultures of non-transformed infant astrocytes (multiplicates of ten) and cortical neurons (triplicates) (C).
H-1PV infection of medulloblastoma cell lines is lytic
The viability of infected medulloblastoma cells was quantified by MTT assay at 72 h and 168 h after infection with H-1PV in a dose range from 0.001 p. f. u. to 50 p. f. u. per cell. In all six medulloblastoma cell lines, a dose-dependent reduction of cellular viability was observed at day 3 after infection (Fig. 2b). In order to confirm that H-1PV had a lytic infectious cycle in medulloblastoma cells, cell lysis was quantified by determining extracellular LDH activity in the supernatant. At 72 h after infection, lytic infection was observed in all MB cell lines, and dependent on the dose of input virus administered (Fig. 2c). In five medulloblastoma cell lines (D425, D458, DAOY, MED8A, and UW228-2) this dose-dependent cytotoxic effect increased over time (Fig. 2a), suggesting that H-1PV is able to replicate and spread in these lines.
The oncolytic effects of H-1PV on medulloblastoma cells were confirmed by real-time proliferation measurement. H-1PV infection resulted in the time-dependent suppression of growth and eventual cell death in the five responsive MB lines (Fig. 2b, Supporting Information Table 2). In agreement with their above-mentioned greater resistance to H-1PV infection, the medulloblastoma cell line ONS76 and normal infant astrocytes were not eradicated in the dose-range tested.
H-1PV replicates in medulloblastoma cells
Extensive cell lysis was induced by H-1PV in the five responsive MB cell cultures over time after infection at low MOIs of 0.05 p. f. u. per cell or less, arguing for efficient virus replication and spreading in these cell cultures. In order to quantify virus production in medulloblastoma cell lines, we first determined the increase in titers of DNA-containing viral particles (QPCR-assay) in the culture supernatant 72 h after infection (Fig. 3a). To confirm the release of fully infectious of H-1PV progeny virions, supernatants were then tested over time for virus titers as measured by dot blot assay (Fig. 3b). In the five responsive medulloblastoma cell lines (DAOY, D425, D458, MED8A, and UW228-2), a significant production of infectious H-1PV virions was detected. Virus multiplication was especially striking in two of the three MYC-amplified MB cell lines (D425 and D458) which sustained 100- to 7,000-fold amplification of input virus within 72 h after infection. Consistent with the greater resistance to H-1PV cytotoxic and cytostatic effects described above, the nonresponsive cell line ONS76 supported no or little virus production.
Figure 3.
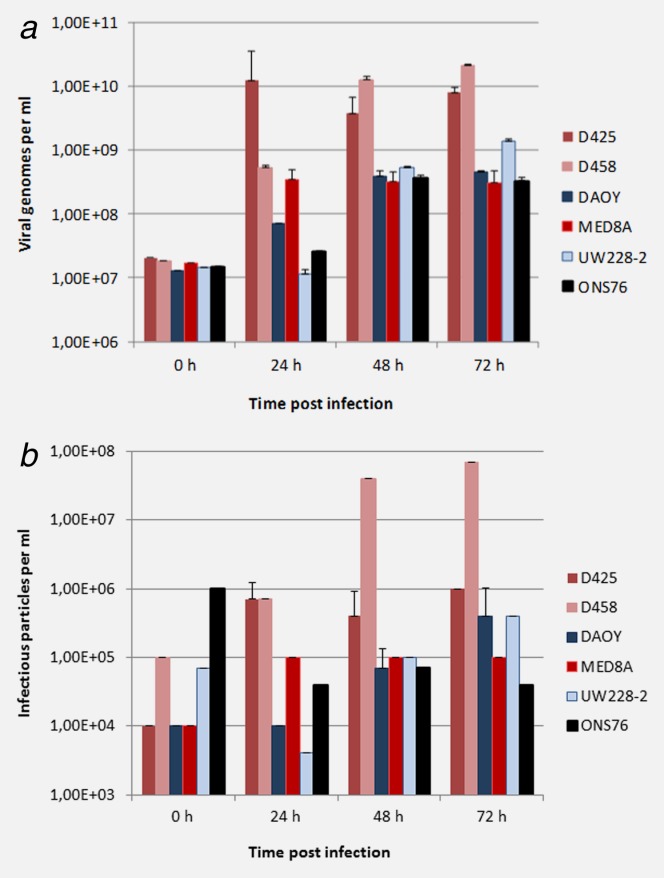
h-1pv replication in medulloblastoma cell lines. Cells were infected with one plaque forming unit per cell of wild type Parvovirus H-1 (H-1PV). At given intervals after infection, supernatants from triplicate cultures were harvested, and the virus titers were determined by quantitative RT-PCR (genomic viral DNA titers, a) and infectious center assay (infectious titers, b). Average values are shown with standard deviation bars.
Parvovirus-responsive medulloblastoma cell lines harbor cytogenetic features of high-risk medulloblastoma
The panel of six pediatric medulloblastoma cell lines tested was categorized with regard to genomic aberrations associated with high-risk in medulloblastoma patients such as genomic amplification of MYC or MYCN, and chromosomal gain of 6q or 17q3 by array Comparative Genomic Hybridization (aCGH) and p53 mutation status as shown in Supporting Information Table 1. According to the present aCGH data, MED8A, D425 and D458 cells constitute a subset of MB cell lines characterized by genomic amplification of MYC (Fig. S1, Supporting Information Tables 1 and 3) with an additional genomic amplification at the MYCN locus in D425 cells (Supporting Information Fig. S1). All changes were detected in the panel, namely gain of 6q in UW228-2, and gain of 17q in D425, D458, MED8A, and ONS76 cells. In a whole genome expression array analysis comparing expression profiles of the MB cell lines to that of samples of non-transformed cerebellum, overexpression of MYC could be quantified and was observed in all cell lines except for DAOY (Supporting Information Table 3). Thus, except for DAOY, all MB cell lines tested harbor molecular or cytogenetic features of high-risk medulloblastoma.
Table 1.
Pathway analysis of potential host cell genes targeted by parvovirus h-1
| p Value | Pathway |
|---|---|
| A | |
| 0.0004 | Pathogenic Escherichia coli infection |
| 0.0005 | Spliceosome |
| 0.0023 | Wnt signaling pathway |
| B | |
| 0.000026 | Steroid biosynthesis |
| 0.0013 | Ether lipid metabolism |
| 0.0018 | TGF-beta signaling pathway |
| 0.0056 | Wnt signaling pathway |
| 0.0095 | GnRH signaling pathway |
| 0.0102 | Jak-STAT signaling pathway |
a kegg pathway analysis was independently performed for the three MYC/MYCN amplified medulloblastoma (MB) cell lines (A) and for the five responsive MB cell lines with cytogenetic features of high-risk medulloblastoma (B).
Expression of distinct cellular target genes is transcriptionally dysregulated upon H-1PV infection of responsive medulloblastoma cells
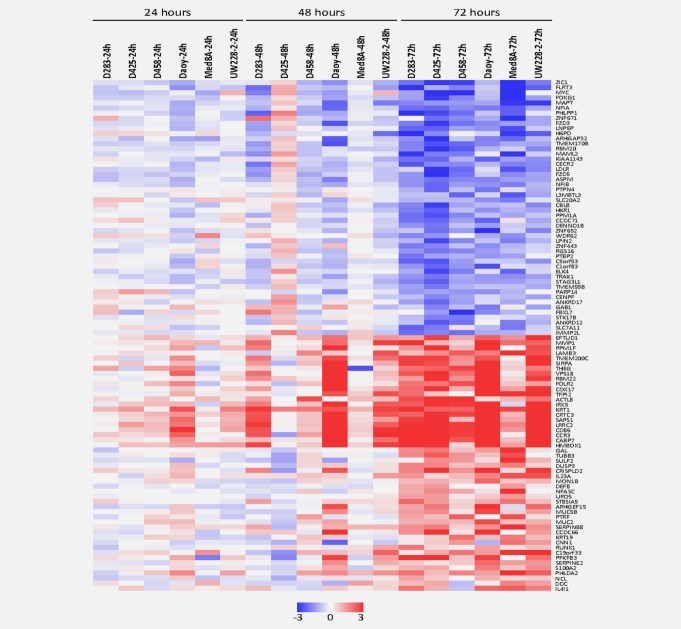
As a first step in the investigation of the molecular mechanisms underlying H-1PV toxicity in MB cells, we identified the sets of cellular genes which are either transcriptionally up- or down-regulated in all five responsive MB cell lines (D425, D458, MED8A, DAOY, and UW228-2) at time points post infection preceding cell death, and whose expression is not affected or inversely modulated in the poorly responsive MB cell line ONS76. Figure 4 lists the cellular genes which are up-regulated or down-regulated most significantly in course of H-1PV infection. These gene subsets could be assumed to comprise candidate genes whose expression dysregulation contributes to H-1PV toxicity for MB cells.
Figure 4.
Parvovirus h-1 induced changes in the transcriptional profiles of medulloblastoma cells. Effects of Parvovirus h-1 (h-1pv) on mRNA expression patterns in responsive medulloblastoma cell lines were determined by whole genome oligonucleotide microarray analysis at 24 h intervals after H-1PV infection with one plaque forming unit per cell in five responsive medulloblastoma cell lines (MED8A, D425, D458, UW228-2, and DAOY) versus the resistant cell line ONS76. Genes showing continuous differential expression after H-1PV treatment at the time points analyzed in all responsive medulloblastoma cell lines were included in the statistical analysis. Genes with most significant differential expression in course of H-1PV infection were determined by LIMMA (FDR < 5). Thereof expression levels of the 25 genes most significantly repressed (upper panel) and the 25 genes most significantly induced (lower panel) at 72 h after infection are shown.
Among the genes consistently up-regulated in course of H-1PV infection in MB cells some may play relevant roles in virus-host cell interactions, such as ILR4I1 and IL23A possibly involved in the innate anti-viral immune response, MMP1 and PPM1F related to the cells’ capability of migration and genes inducing neuronal differentiation such as RUNX1 or DUSP3 (Fig. 4). Although these candidates remain to be confirmed by functional experiments two interesting clues were provided by the present analysis.
Repression of regulator genes involved in early stages of CNS development and in medulloblastoma formation precedes H1-PV cytotoxic effects in H-1PV responsive MB cell lines
The only gene found to be significantly repressed as early as 24 h after H-1PV infection in all responsive MB cells encodes LAMA1, a potent mitogen in granule cell precursor cells.31 Furthermore, the 20 cellular genes whose transcription was continuously further repressed within the first 72 h after H-1PV infection of responsive MB cells and showed most significant repression include a number of other genes known to control the neural progenitor state in early embryonic development, such as ZIC1, MYC, FOXG1, FLRT3, NFIA and NFIB (Fig. 4a).
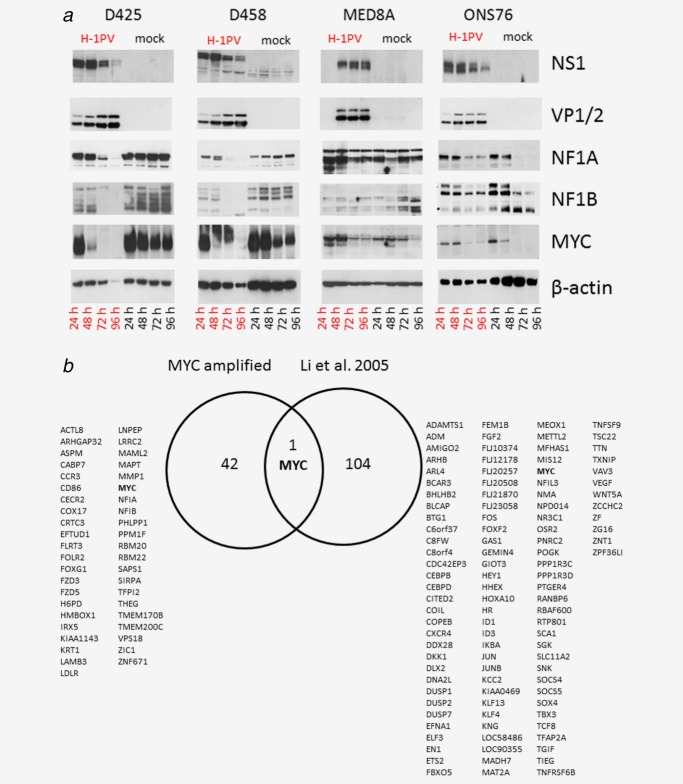
In a comparative analysis in all five responsive MB cell lines 72 h post H-1PV infection (MOI = 1 p. f. u. per cell) repression was most significant for ZIC1. In a separate data analysis in the subset of MYC amplified MB cell lines MYC is the gene most significantly repressed (p < 0.0001) and repression of MYC was strongest in these cells (Supporting Information Table 3). The H-1PV induced transcriptional repression of these master regulator genes of neurogenesis was exemplarily confirmed for MYC, NFIA and NFIB at the transcriptional level by qRT-PCR (data not shown) and at the protein level by Western Blot of all responsive cell lines, as shown in Figure 5. On the transcriptional level, MYC is the only common cellular target gene to H-1PV infection identified by expression profiling in two independent studies, the present one and previously published work on hepatocellular carcinoma cells,32 see Figure 5b.
Figure 5.
(a) Validation of cellular targets for parvovirus h-1 (h-1pv) dependent repression in all responsive medulloblastoma cell lines at protein level. Steady state levels of the putative H-1PV target proteins MYC, NFIA and NFIB were determined by Western blot analysis in the course of H-1PV infection. In parallel, expression of the viral proteins NS1 and VP1/2 was confirmed in the medullobastoma (MB) cell cultures infected. Results are exemplarily shown for the MYC amplified cell lines D425, D458, and MED8A and compared to the partially resistant cell line ONS76. (b) Venn-diagram displaying common cellular targets of h-1pv in all responsive MB cell lines (left panel) compared to those in the hepatocellular carcinoma cell line QGY-7703 (right panel). The expression profiling data characterizing the transcriptional response to oncolytic H-1PV infection has already been published.35 [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Pathway analysis of the major H-1PV genes induced or repressed in high-risk MB cell lines responsive to H-1PV infection
These findings prompted us to perform a systematic pathway analysis of putative H-1PV target genes identified (see Fig. 4). This analysis was focused on the genes found to be transcriptionally dysregulated by infection in the completely responsive H-1PV MB cell lines showing the molecular and cytogenetic features of high risk medulloblastoma (D425, D458, MED8A, and UW 228-2, Supporting Information Tables 1 and 3).
Interestingly, the Wnt signaling pathway was the third most significantly disturbed pathway upon infection of these cells with H-1PV (p = 0.0023). Indeed, genes encoding the Wnt receptors FZD3 and FZD5 were among the genes most significantly repressed in H-1PV infected responsive MB cell lines (Fig. 4a). Furthermore, expression of the signal regulatory protein α1 encoding gene (SIRPA) is one of the genes most strongly induced in these cells. Both subsets of KEGG pathway analyses, either focusing on MB cell lines harboring genomic amplification of MYC (Table 1; A) or on MB cell lines harboring cytogenetic features of high-risk MB (Table 1; B) showed highly significant dysregulation in the expression of genes related to Wnt signaling (p < 0.01). Thus, H-1PV infection is likely to result in the impairment of Wnt signaling in responsive medulloblastoma cell lines prior to the onset of cell death.
Discussion
Compared to other antineoplastic agents, self-replicating oncolytic viruses offer striking advantages, including the ability to establish an effective virus titer by replicating in initially infected malignant cells. In five out of six human medulloblastoma cell lines, efficient H-1PV multiplication was confirmed. In vitro these cell lines undergo H-1PV induced cell lysis after infection with much lower titers of input virus than cell culture models of adult brain tumors such as glioblastoma.33 A selective toxicity of H-1PV to a panel of different medulloblastoma cell lines harboring molecular and genomic features high-risk medulloblastoma could be demonstrated. The present data on human brain cells confirm that recombinant H-1PV is able to enter non-transformed infant brain cells—but with low efficacy. However, viral transduction by wild type H-1PV and expression of its main cytotoxic viral protein NS-1 is restricted to the medulloblastoma cells analyzed.
In contrast, transplacental H-1PV infection in hamsters and rats has been shown to be pathogenic indicating a strong tropism to embryonic tissues derived from all three germinal layers. After pre- or perinatal infection rats being the natural host organism seem to be especially prone to H-1PV induced neurotoxicity. H-1PV had been reported to induce hypoplasia of the cerebellum in newborn rats after late embryonic and early neonatal infection. Histopathologically, this developmental disorder corresponded to a virus induced selective induction of apoptosis in granule cell neurons.34 Published preclinical data obtained in adult glioblastoma-bearing rodent models suggest clinical safety of H-1PV. Even the intracranial application of the virus into rats after the neonatal period could not be associated with any neuropathogenicity in the natural host of the virus.33
In conclusion, the observation of a selective neurotropism of H-1PV in rats after perinatal or early postnatal infection to granule cell precursor cells raised concerns with respect to the clinical safety of an intracranial application of H-1PV to infants. This well-defined neurotoxicity has been described for H-1PV infections occurring during the first postnatal week in the natural host organism. In contrast, no infection by wild type H-1PV has been observed in mice. Thus, the conclusions to be drawn from neonate rodent models for the safety issues of H-1PV application in infant human patients remain limited.
The present in vitro data on human infant brain cells do not show significant effects of H-1PV infection on the viability of non below a titer of 50 p. f. u. per cell, which confirms previously published data.20 However, human granule cell precursor cells have not been available for an in vitro analysis of cytotoxic effects induced by H-1PV. The present data on human infant brain cells imply clinical safety of H-1PV in children. Based on the present and previously published data, however, adverse effects of H-1PV infection on the developing brain cannot entirely be excluded.
The selective cytotoxicity of H-1PV to embryonic granule cell precursor (GCP) cells in the natural host organism may be mediated by repression of the key regulator genes indicating a virus induced disruption of main embryonic signaling pathways. The parallels between the molecular control of GCP proliferation and the formation of medulloblastoma are one of the best characterized examples of the crucial role of developmental molecular pathways in oncogenesis.35 In course of H-1PV infection the repression of regulator genes maintaining proliferation of rodent granule cell precursor cells may contribute to its selective toxicity to the granule cell layer of the cerebellum in embryonic and newborn rats.
We hypothesized that an analogous transcriptional response to H-1PV infection mediates its selective cytotoxic effects in human medulloblastoma cells. Among the genes induced in course of H-1PV infection IL4I1 and IL23A have been shown to play key roles in innate immune response. MMP1 and PPM1F over expression have been demonstrated to be related to increased cell migration in cancer cells, but have not been studied in MB yet.36 Other genes induced have shown to play relevant roles in the induction of neuronal differentiation such as DUSP337,38 or RUNX1, which induces differentiation in early dorsal root precursor cells.39 However, the role of these genes in the embryonic development of the cerebellum or in MB formation remains unclear.
Among the genes most significantly repressed in course of H-1PV infection of responsive medulloblastoma cell lines we identified a cluster of genes highly enriched for functional association to the control of a neural progenitor state in cerebellar neurogenesis. These included members of the Wnt signaling pathway, such as FRZ3, FRZ5, and SIRPA, and known Wnt-target genes exerting key regulatory functions on early neurogenesis such as NFIA and MYC. Moreover, other, unrelated key regulator genes of early neurogenesis such as ZIC1, FLRT3, or FOXG1 could be identified.
ZIC1 is one of the earliest regulators of neural induction during embryonic development.40 Heterozygous deletions of ZIC1 in mice have been shown to induce decreased postnatal granule cell progenitor proliferation and to lead, thereby, to reduction of cerebellar size in affected animals. The functional role of ZIC1 promoting the proliferation of granule cell progenitor cells could be demonstrated to be dependent on SHH signaling.41 Overexpression of ZIC1 has been shown to play a crucial role in the development of Wnt signaling dependent MB in a beta-catenin mutant mouse model.42
FOXG1 also is a master regulator of neural differentiation that has been demonstrated to be able to induce differentiation into neuronal cells by reprogramming of embryonic fibroblasts. FOXG1 is a transcriptional repressor that protects neuroepithelial progenitor cells from cytostatic and differentiation inducing signals. Excess FOXG1 expression in vivo is associated with neural progenitor cell overgrowth43 and repression of FOXG1 is able to induce neuronal death.44 Recently, FOXG1 could be shown to promote the survival of cerebellar granule neurons. FOXG1 has been shown to be frequently over expressed in MB, mainly due to genomic amplification of the FOXG1 locus.45 Over expression of the MB oncogene FOXG1B has been consistently found in Group 3 and Group 4 tumors representing the subgroups with poorest prognosis in pediatric MB.6
NFIA and NFIB are members of the transcription factor I family, that encode a transcription factors which play an important role in granule cell precursor development46 and medulloblastoma formation, especially in metastatic models.4 Recently, NFIA has been shown to be a direct Wnt target gene with TCF/Lef binding sites in its promoter region,47 thus suggesting that NFIA repression might also be mediated by down-regulation of Wnt signaling in response to H-1PV infection.
Amplification of the MYC oncogene is one of the hallmarks of malignant transformation in medulloblastoma. In childhood medulloblastoma MYC/MYCN amplification is associated with very poor prognosis.6 The Wnt-group of MB characterized by the over expression of inhibitors of Wnt-signaling such as DKK1, DKK2, and WIF1 associated with the best prognosis. Over expression of MYC without over expression of Wnt-signaling inhibitors characterizes MB tumors associated with the worst prognosis referred to as Group 3.4 MYC is a known Wnt target gene and has been hypothesized to play a key role in mediating the effects of SHH, WNT and PI3K/AKT in medulloblastoma.48,49 In transplant-bearing mouse models with orthotopically implanted neural stem cells harboring p53 mutations and inducible MYC expression the functional role of MYC over expression in conferring aggressive and metastatic medulloblastoma phenotypes has recently been demonstrated.17,18
In all responsive MB cell lines H-1PV was able to repress MYC expression on the transcript level. In MYC amplified MB cell lines MYC repression could also be confirmed on protein level indicating a potential functional role of MYC repression in preventing the survival of H-1PV infected MB cells. In a data analysis focused on MB cell lines with genomic amplification of MYC, MYC was the gene being most significantly repressed in course of H-1PV infection.
The three medulloblastoma cell lines harboring genomic amplifications of MYC and thus most likely modeling group 3 MB tumors (D425, D458, and MED8A) were among the most susceptible cell lines analyzed. In these cell lines efficacy of replication was highest, which is in good correspondence with published data describing susceptibility to rodent parvovirus induced cytotoxic effects to be dependent on the over expression of the oncogenes H-ras or MYC.50 Repression of MYC preceded induction of apoptosis by H-1PV, which implicates its possible functional relevance for the antineoplastic effects of the virus. This hypothesis is supported by previous data showing down-regulation of MYC expression to be among the most significant features of H-1PV infection in promonocytic leukemia and hepatocellular carcinoma cell lines.28,32
Wnt/b-catenin pathway activation in Myc-immortalized cerebellar progenitor cells has been shown to inhibit neuronal differentiation and to induce tumors resembling medulloblastoma in an orthotopic rat model.50 The virus-induced repression of genes involved in Wnt signaling prior to cell death (e.g. FZD3, FZD5, SIRPA) lead us to hypothesize a potential role of abrogation of Wnt signaling in virus induced death of MB cells. However, the mechanism by which H-1PV infection results in the repression of the cellular target genes described and subsequently induces cytotoxic effects to MB cells remains to be clarified by functional experiments.
In conclusion, the present expression data demonstrate that H-1PV infection results in repression of known genes and pathways involved in the regulation of early neurogenesis. We propose that the repression of genes responsible for the maintenance of the neural progenitor state is a key feature of H-1PV induced effects to embryonic cancer cells of neuroectodermal origin and may contribute to the ability of H-1PV induce cytotoxic effects in these cells.
Focusing on medulloblastoma as a pre-clinical model, the present study is the first to demonstrate the cytotoxic potency of H-1PV in pediatric brain tumor cells. Medulloblastoma cells prove to be very sensitive to the cytotoxic effects of H-1PV which induced cell death with an efficacy comparable to the most promising other oncolytic viruses.7,9 These data strongly suggest that H-1PV virotherapy may be an attractive novel treatment option for very high-risk medulloblastoma.
Acknowledgments
The authors thank Dr. Nathalie Salomé and Michèle Klein for providing the polyclonal antisera against NS-1 and VP1/VP2. Rafael Josupeit is acknowledged for taking the fluorescence pictures. The authors are indebted to Marta Herrero y Calle, Department of Neurosurgery, Freiburg University Hospital, who established the short term cultures of infant astrocytes. The authors are grateful to Dr. Antonio Marchini and Dr. Jürg Nüsch for helpful discussions concerning molecular pathways mediating viral oncolysis.
Glossary
- CGH
comparative genomic hybridization
- GPC
granule cell precursor
- H-1PV
parvovirus H-1
- HSV-1
herpes simplex virus type 1
- p. f. u.
plaque forming unit(s)
- wt
wild type.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
References
- Smith MA, Seibel NL, Altekruse SF, et al. Outcomes for children and adolescents with cancer: challenges for the twenty-first century. J Clin Oncol. 2010;28:2625–34. doi: 10.1200/JCO.2009.27.0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski S, von HK, Emser A, et al. Survival and prognostic factors of early childhood medulloblastoma: an international meta-analysis. J Clin Oncol. 2010;28:4961–8. doi: 10.1200/JCO.2010.30.2299. [DOI] [PubMed] [Google Scholar]
- Pfister S, Remke M, Benner A, et al. Outcome prediction in pediatric medulloblastoma based on DNA copy-number aberrations of chromosomes 6q and 17q and the MYC and MYCN loci. J Clin Oncol. 2009;27:1627–36. doi: 10.1200/JCO.2008.17.9432. [DOI] [PubMed] [Google Scholar]
- Northcott PA, Korshunov A, Witt H, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–14. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YJ, Tsherniak A, Tamayo P, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29:1424–30. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Korshunov A, Pfister SM, et al. The clinical implications of medulloblastoma subgroups. Nat Rev Neurol. 2012;8:340–51. doi: 10.1038/nrneurol.2012.78. [DOI] [PubMed] [Google Scholar]
- Hammill AM, Conner J, Cripe TP. Oncolytic virotherapy reaches adolescence. Pediatr Blood Cancer. 2010;55:1253–63. doi: 10.1002/pbc.22724. [DOI] [PubMed] [Google Scholar]
- Friedman GK, Pressey JG, Reddy AT, et al. Herpes simplex virus oncolytic therapy for pediatric malignancies. Mol Ther. 2009;17:1125–35. doi: 10.1038/mt.2009.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix J, Witt O, Schlehofer JR, et al. Therapeutic exploitation of onclytic viruses in pediatric oncology. Drugs Future. 2010;35:1015–1027. [Google Scholar]
- Lun XQ, Zhou H, Alain T, et al. Targeting human medulloblastoma: oncolytic virotherapy with myxoma virus is enhanced by rapamycin. Cancer Res. 2007;67:8818–27. doi: 10.1158/0008-5472.CAN-07-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studebaker AW, Kreofsky CR, Pierson CR, et al. Treatment of medulloblastoma with a modified measles virus. Neuro Oncol. 2010;12:1034–42. doi: 10.1093/neuonc/noq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Baxter PA, Zhao X, et al. A single intravenous injection of oncolytic picornavirus SVV-001 eliminates medulloblastomas in primary tumor-based orthotopic xenograft mouse models. Neuro Oncol. 2011;13:14–27. doi: 10.1093/neuonc/noq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studebaker AW, Hutzen B, Pierson CR, et al. Oncolytic measles virus prolongs survival in a murine model of cerebral spinal fluid-disseminated medulloblastoma. Neuro Oncol. 2012;14:459–70. doi: 10.1093/neuonc/nor231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommelaere J, Geletneky K, Angelova AL, et al. Oncolytic parvoviruses as cancer therapeutics. Cytokine Growth Factor Rev. 2010;21:185–95. doi: 10.1016/j.cytogfr.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Geletneky K, Kiprianova I, Ayache A, et al. Regression of advanced rat and human gliomas by local or systemic treatment with oncolytic parvovirus H-1 in rat models. Neuro Oncol. 2010;12:804–14. doi: 10.1093/neuonc/noq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiprianova I, Thomas N, Ayache A, et al. Regression of glioma in rat models by intranasal application of parvovirus H-1. Clin Cancer Res. 2011;17:5333–42. doi: 10.1158/1078-0432.CCR-10-3124. [DOI] [PubMed] [Google Scholar]
- Pei Y, Moore CE, Wang J, et al. An animal model of MYC-driven medulloblastoma. Cancer Cell. 2012;21:155–67. doi: 10.1016/j.ccr.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi D, Robinson G, Uziel T, et al. A mouse model of the most aggressive subgroup of human medulloblastoma. Cancer Cell. 2012;21:168–80. doi: 10.1016/j.ccr.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Northcott PA, Dubuc A, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482:529–33. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix J, Leuchs B, Li J, et al. Parvovirus H1 selectively induces cytotoxic effects on human neuroblastoma cells. Int J Cancer. 2010;127:1230–9. doi: 10.1002/ijc.25168. [DOI] [PubMed] [Google Scholar]
- Wrzesinski C, Tesfay L, Salome N, et al. Chimeric and pseudotyped parvoviruses minimize the contamination of recombinant stocks with replication-competent viruses and identify a DNA sequence that restricts parvovirus H-1 in mouse cells. J Virol. 2003;77:3851–8. doi: 10.1128/JVI.77.6.3851-3858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Andaloussi N, Endele M, Leuchs B, et al. Novel adenovirus-based helper system to support production of recombinant parvovirus. Cancer Gene Ther. 2011;18:240–9. doi: 10.1038/cgt.2010.73. [DOI] [PubMed] [Google Scholar]
- Bar S, Daeffler L, Rommelaere J, Nuesch JP. Vesicular egress of non-enveloped lytic parvoviruses depends on gelsolin functioning. PLoS Pathog. 2008;4:e1000126. doi: 10.1371/journal.ppat.1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt H, Mack SC, Ryzhova M, et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell. 2011;20:143–57. doi: 10.1016/j.ccr.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, von HA, Sultmann H, et al. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–6. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- Faisst S, Faisst SR, Dupressoir T, et al. Isolation of a fully infectious variant of parvovirus H-1 supplanting the standard strain in human cells. J Virol. 1995;69:4538–43. doi: 10.1128/jvi.69.7.4538-4543.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayet B, Lopez-Guerrero JA, Rommelaere J, et al. Induction of programmed cell death by parvovirus H-1 in U937 cells: connection with the tumor necrosis factor alpha signalling pathway. J Virol. 1998;72:8893–903. doi: 10.1128/jvi.72.11.8893-8903.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YQ, de FF, Hertoghs J, et al. Selective killing of simian virus 40-transformed human fibroblasts by parvovirus H-1. Cancer Res. 1986;46:3574–9. [PubMed] [Google Scholar]
- Herrero y Calle M, Cornelis JJ, Herold-Mende C, et al. Parvovirus H-1 infection of human glioma cells leads to complete viral replication and efficient cell killing. Int J Cancer. 2004;109:76–84. doi: 10.1002/ijc.11626. [DOI] [PubMed] [Google Scholar]
- Heng C, Lefebvre O, Klein A, et al. Functional role of laminin alpha1 chain during cerebellum development. Cell Adh Migr. 2011;5:480–9. doi: 10.4161/cam.5.6.19191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Werner E, Hergenhahn M, et al. Expression profiling of human hepatoma cells reveals global repression of genes involved in cell proliferation, growth, and apoptosis upon infection with parvovirus H-1. J Virol. 2005;79:2274–86. doi: 10.1128/JVI.79.4.2274-2286.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommelaere J, Geletneky K, Angelova AL, et al. Oncolytic parvoviruses as cancer therapeutics. Cytokine Growth Factor Rev. 2010;21:185–95. doi: 10.1016/j.cytogfr.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Ohshima T, Iwama M, Ueno Y, et al. Induction of apoptosis in vitro and in vivo by H-1 parvovirus infection. J Gen Virol. 1998;79:3067–71. doi: 10.1099/0022-1317-79-12-3067. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Roussel MF. Development and cancer of the cerebellum. Trends Neurosci. 2011;34:134–42. doi: 10.1016/j.tins.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel A, Gutierrez-Fernandez A, Sounni NE, et al. New and paradoxical roles of matrix metalloproteinases in the tumor microenvironment. Front Pharmacol. 2012;3:140. doi: 10.3389/fphar.2012.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang TH, Kim KT. VRK3-mediated inactivation of ERK signaling in adult and embryonic rodent tissues. Biochim Biophys Acta. 2008;1783:49–58. doi: 10.1016/j.bbamcr.2007.10.011. [DOI] [PubMed] [Google Scholar]
- Patterson KI, Brummer T, O’Brien PM, et al. Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem J. 2009;418:475–89. doi: 10.1042/bj20082234. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Senzaki K, Ozaki S, et al. Runx1 promotes neuronal differentiation in dorsal root ganglion. Mol Cell Neurosci. 2012;49:23–31. doi: 10.1016/j.mcn.2011.08.009. [DOI] [PubMed] [Google Scholar]
- Marchal L, Luxardi G, Thome V, et al. BMP inhibition initiates neural induction via FGF signaling and Zic genes. Proc Natl Acad Sci USA. 2009;106:17437–42. doi: 10.1073/pnas.0906352106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank MC, Grinberg I, Aryee E, et al. Multiple developmental programs are altered by loss of Zic1 and Zic4 to cause Dandy-Walker malformation cerebellar pathogenesis. Development. 2011;138:1207–16. doi: 10.1242/dev.054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson P, Tong Y, Robinson G, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–9. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adesina AM, Nguyen Y, Mehta V, et al. FOXG1 dysregulation is a frequent event in medulloblastoma. J Neurooncol. 2007;85:111–22. doi: 10.1007/s11060-007-9394-3. [DOI] [PubMed] [Google Scholar]
- Dastidar SG, Landrieu PM, D’Mello SR. FoxG1 promotes the survival of postmitotic neurons. J Neurosci. 2011;31:402–13. doi: 10.1523/JNEUROSCI.2897-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Crandall JE, Litwack ED, et al. Targets of the nuclear factor I regulon involved in early and late development of postmitotic cerebellar granule neurons. J Neurosci Res. 2010;88:258–65. doi: 10.1002/jnr.22199. [DOI] [PubMed] [Google Scholar]
- Hasenpusch-Theil K, Magnani D, Amaniti EM, et al. Transcriptional analysis of Gli3 mutants identifies Wnt target genes in the developing hippocampus. Cereb Cortex. 2012;22:2878–9. doi: 10.1093/cercor/bhr365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. Network of WNT and other regulatory signaling cascades in pluripotent stem cells and cancer stem cells. Curr Pharm Biotechnol. 2011;12:160–70. doi: 10.2174/138920111794295710. [DOI] [PubMed] [Google Scholar]
- Baryawno N, Sveinbjornsson B, Kogner P, et al. Medulloblastoma: a disease with disorganized developmental signaling cascades. Cell Cycle. 2010;9:2548–54. doi: 10.4161/cc.9.13.12170. [DOI] [PubMed] [Google Scholar]
- Salome N, van HB, Duponchel N, et al. Sensitization of transformed rat cells to parvovirus MVMp is restricted to specific oncogenes. Oncogene. 1990;5:123–30. [PubMed] [Google Scholar]
- Rogers HA, Sousa S, Salto C, et al. WNT/beta-catenin pathway activation in Myc immortalised cerebellar progenitor cells inhibits neuronal differentiation and generates tumours resembling medulloblastoma. Br J Cancer. 2012;107:1144–52. doi: 10.1038/bjc.2012.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.