Abstract
Fluid Attenuated Inversion Recovery (FLAIR) is a commonly acquired pulse sequence for multiple sclerosis (MS) patients. MS white matter lesions appear hyperintense in FLAIR images and have excellent contrast with the surrounding tissue. Hence, FLAIR images are commonly used in automated lesion segmentation algorithms to easily and quickly delineate the lesions. This expedites the lesion load computation and correlation with disease progression. Unfortunately for numerous reasons the acquired FLAIR images can be of a poor quality and suffer from various artifacts. In the most extreme cases the data is absent, which poses a problem when consistently processing a large data set. We propose to fill in this gap by reconstructing a FLAIR image given the corresponding T1-weighted, T2-weighted, and PD-weighted images of the same subject using random forest regression. We show that the images we produce are similar to true high quality FLAIR images and also provide a good surrogate for tissue segmentation.
Index Terms: Image reconstruction, regression, brain
1. INTRODUCTION
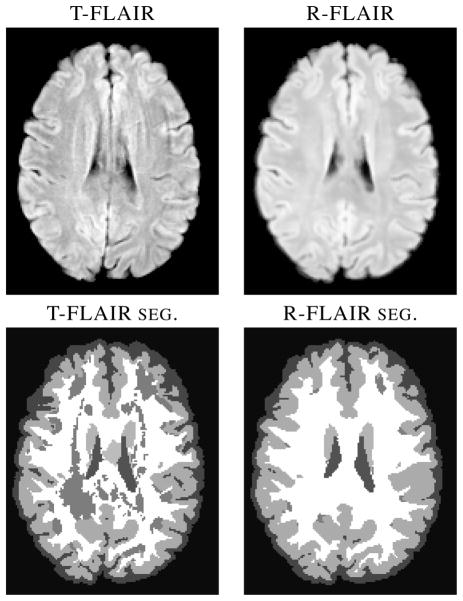
MS is a neurodegenerative disease characterized by focal lesions throughout the white matter (WM) of the brain, which can extend to the boundary of the WM and gray matter (GM). In both Alzheimer’s disease (AD) and MS, white matter lesions (WMLs) appear to be hypointense in T1-weighted (T1-w) images, similar to GM or cerebrospinal fluid (CSF). Typically other contrasts such as T2-weighted (T2-w), proton density (PD-w) weighted, and FLuid Attenuated Inversion Recovery (FLAIR) are used to help distinguish lesions from GM and CSF. Lesion volumes are known to be correlated with the progression of many sub-types of MS [1]. The contrast of magnetic resonance (MR) images is of fundamental importance for WML tissue classification. We note that it is well established that FLAIR images provide the best contrast for detection and quantification, whereas conventional T1-w, T2-w, and PD-w spin echo sequences offer diminished contrast for WML [2]. Most recent lesion segmentation algorithms therefore depend heavily on FLAIR for WML identification [3–5]. However, FLAIR images can suffer from artifacts for a variety of reasons [6]. Fig. 1 shows an artifact-ridden acquired FLAIR and the corresponding tissue segmentation result. We intend to overcome this problem by reconstructing a FLAIR image using information from the other contrasts. Our reconstructed FLAIR does not exhibit the artifacts, because each of the T1-w, T2-w, and PD-w, images used are artifact free, and together they possess enough information to recreate the FLAIR tissue intensities.
Fig. 1.
The true FLAIR (T-FLAIR) shows visible artifacts which confound LesionTOADS and other lesion segmentation algorithms (not pictured), resulting in the misclassification of healthy WM as WMLs. Our reconstructed FLAIR (R-FLAIR) does not have the artifacts, as they are not present in the T1-w, T2-w, or PD-w images.
Our work builds upon the ideas of image hallucination [7] which originally generated a high-resolution image from multiple low resolution acquisitions based on a textbook or atlas. The idea first gained traction in the medical imaging community with the work of Rousseau [8], and has since gained popularity and numerous applications from super-resolution [9, 10], contrast synthesis [9, 11–13], and inhomogeneity correction [14]. Our approach is to treat the unknown FLAIR image as dependent on other known images and model the reconstruction of the FLAIR as a nonlinear regression of image patches drawn simultaneously from the co-registered T1-w, T2-w, and PD-w contrast images. The voxel intensity in MR is a function of many nuclear magnetic resonance (NMR) parameters like proton density (PD), longitudinal and transverse relaxation times (T1 and T2) and others. We would like to frame the problem of predicting the FLAIR voxel intensities as a nonlinear function of the other contrast image intensities—which themselves are also a function of the underlying NMR parameters. The nonlinear function is learned as a nonlinear regression from data by a bagged ensemble of regression trees [15] and is significantly faster and better than the state-of-the-art.
In this paper, we build upon our previous FLAIR reconstruction work [16] which used only T1-w and T2-w data and a sparse priors model to resolve the intensity of the unknown contrast. We expand the work to include the third contrast of PD-w to help improve the FLAIR reconstruction as well as reframing the problem in terms of a random forest based reconstruction [11]. We show that the reconstructed FLAIR (R-FLAIR) image matches very closely with the original high quality FLAIR image (T-FLAIR), while also providing an important surrogate image for use in a WML segmentation algorithm.
2. METHOD
We tackle the FLAIR reconstruction with an atlas based regression approach. Our atlas consists of (
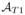 ,
,
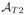 ,
,
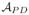 ,
,
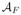 ), which have tissue contrasts of T1-w, T2-w, PD-w, and FLAIR, respectively. The atlas images have the same voxel resolution and are sampled at the same coordinates in space. The subject images are assumed to consist of the triple of (
), which have tissue contrasts of T1-w, T2-w, PD-w, and FLAIR, respectively. The atlas images have the same voxel resolution and are sampled at the same coordinates in space. The subject images are assumed to consist of the triple of (
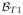 ,
,
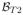 ,
,
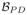 ) with the goal being to reconstruct
) with the goal being to reconstruct
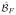 from the learned nonlinear relationships in the atlas quadruple. For several of our experiments we will have the true
from the learned nonlinear relationships in the atlas quadruple. For several of our experiments we will have the true
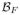 available, to which we can compare the quality of the reconstruction. At the ith voxel of the atlas images
available, to which we can compare the quality of the reconstruction. At the ith voxel of the atlas images
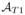 ,
,
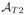 , and
, and
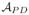 , 3D patches of size p × q × r are rasterized to form a d × 1 vector (d = pqr, p = q = r = 3, chosen empirically) for each of the three images. This triple of vectors is concatenated to form the 3d × 1 vector xi. xi and the corresponding intensity, yi, of the ith voxel in
, 3D patches of size p × q × r are rasterized to form a d × 1 vector (d = pqr, p = q = r = 3, chosen empirically) for each of the three images. This triple of vectors is concatenated to form the 3d × 1 vector xi. xi and the corresponding intensity, yi, of the ith voxel in
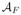 form the independent and dependent variables, respectively, for our training data denoted as 〈xi, yi〉.
form the independent and dependent variables, respectively, for our training data denoted as 〈xi, yi〉.
We solve the regression problem generated by the training data 〈xi, yi〉 using random forests (RF) [17]. A RF consists of an ensemble of regression trees [15], with each regression tree partitioning the space of xi’s into regions based on a split at each node in the tree. During training, splits at a node are performed by randomly selecting one-third of the attributes and then determining the attribute in this subset that best minimizes a least squares criterion. We ensure that nodes with less than five data values are leaf nodes, thus avoiding over-fitting of the training data. The value of the dependent variable assigned at a leaf node is the mean of the yi’s which have accumulated in that leaf during training. The bagged ensemble of regression trees allows for greater robustness in the predictive power of the RF. In our experiments, our RF consists of 60 trees each learned from bootstrapped training data. We used patches sampled from data acquired from five MS subjects, ensuring equal representation of CSF, GM, WM, and WMLs. The bootstrapping is done by choosing with replacement N times, where N is the size of the training data set (~ 105 samples). To predict the FLAIR intensity for the subject image set, we construct the rasterized 3d × 1 vector, uj, similarly to that used for the training data. The uj’s are input into each of the regression trees in the trained RF. The output of each tree is the value of the leaf node the vector uj is assigned. We aggregate these values by computing their mean to produce a final output intensity for the jth voxel of the R-FLAIR image
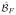 . Averaging the FLAIR intensity values at the leaf nodes of the RF lowers the peak values of the output FLAIR image. To avoid this, we linearly rescale the peak intensities of
. Averaging the FLAIR intensity values at the leaf nodes of the RF lowers the peak values of the output FLAIR image. To avoid this, we linearly rescale the peak intensities of
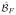 to match those of a typical FLAIR image. Training takes ten minutes and the prediction phase takes less than ten seconds, for images of size 256 × 256 × 173.
to match those of a typical FLAIR image. Training takes ten minutes and the prediction phase takes less than ten seconds, for images of size 256 × 256 × 173.
3. RESULTS
3.1. Image Similarity
Our goal in reconstructing the FLAIR from corresponding T1- w (0:83 mm3, MPRAGE), T2-w, and PD-w (Dual Spin Echo, 0:8 × 0:8 × 2:2 mm3) images is to provide an approximation to a T-FLAIR for defining lesions. Our data set consists of 49 MS subjects (44 test + 5 training), with all the data registered to the MNI space and resampled at a 1 mm3 isotropic resolution. We used the T1-w, T2-w, and PD-w images for our R-FLAIR and compare it to the T-FLAIR image in the data set. Several inferior slices of the PD-w, T2-w, and FLAIR images—covering the cerebellum–are of considerably lower quality and were not used for evaluation.
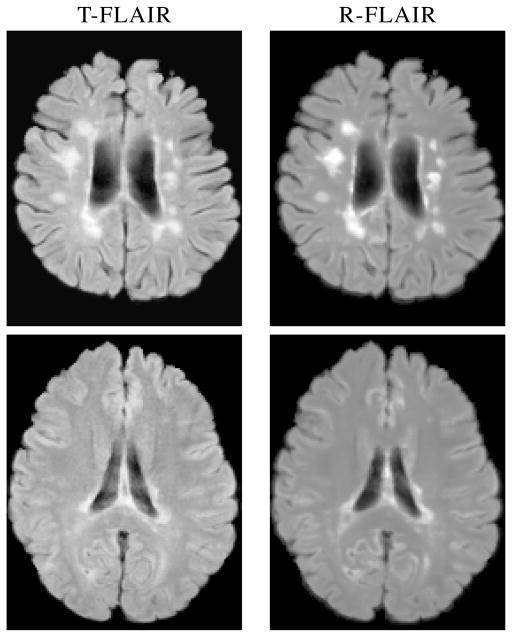
We use the image similarity metrics of peak signal to noise ratio (PSNR), the universal quality index (UQI) [18], and the structural similarity index (SSIM) [19] for evaluation (see Table 1). UQI and SSIM have a range between 0 and 1, attaining the maximum if the T-FLAIR and the R-FLAIR image are perfectly equal. Example results are shown in Fig. 2. The R-FLAIR is smoother than the T-FLAIR due to its intensities being produced by the averaging of 60 trees in the RF.
Table 1.
Mean (Std. Dev.) of PSNR (in decibels), UQI, and SSIM values over 49 subjects.
| PSNR | UQI | SSIM |
|---|---|---|
| 25.58 (1.06) | 0.88 (0.03) | 0.87 (0.02) |
Fig. 2.
Example slices of true (T-FLAIR) and reconstructed FLAIR (R-FLAIR) for a subject.
3.2. Image Segmentation
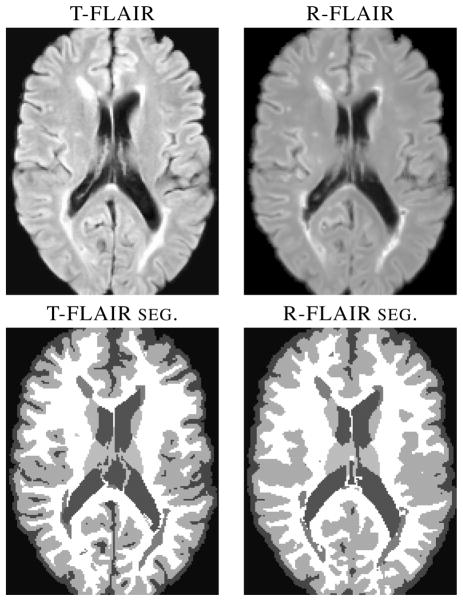
In this experiment we tested, using LesionTOADS [4], if the R-FLAIR can be used to provide reliable segmentation of brain tissues into CSF, GM, WM, and WMLs. LesionTOADS takes co-registered T1-w and FLAIR images as input and generates a segmentation. We ran LesionTOADS with the original T1-w image in conjunction with either the T-FLAIR or R-FLAIR. From the LesionTOADS segmentation, we computed the Dice coefficient to compare the stability of our R-FLAIR to T-FLAIR, the results are shown in Table 2. The Dice coefficient is low for WMLs because of algorithm errors caused by artifacts in the T-FLAIR images—see Fig. 1 for an example— which are overcome in our R-FLAIR images. For those cases where the T-FLAIR image is devoid of artifacts, the segmentation differences between the T-FLAIR and R-FLAIR are minimal (see Fig. 3).
Table 2.
Mean (Std. Dev.) of Dice coefficients based on LesionTOADS segmentation of the T-FLAIR and R-FLAIR.
| WM | GM | CSF | WML |
|---|---|---|---|
| 0.96 (0.01) | 0.99 (0.0005) | 0.95 (0.01) | 0.46 (0.22) |
Fig. 3.
LesionTOADS segmentations using true (T-FLAIR) and reconstructed FLAIRs (R-FLAIR).
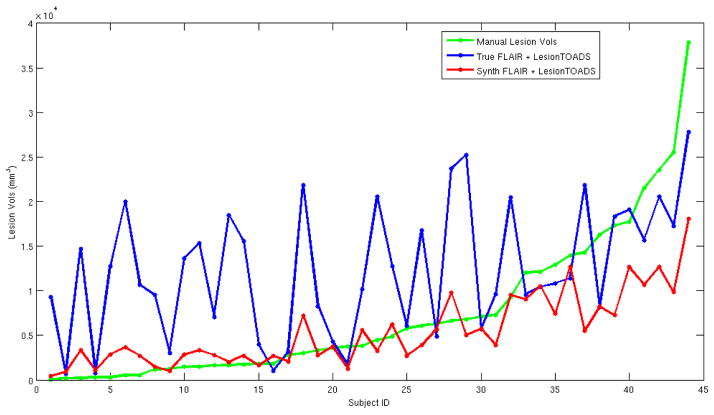
For the 44 test MS subjects, we have manual segmentations of the WMLs, from which we computed the Dice coefficient for the WMLs between the truth and the segmentations generated from each of T-FLAIR and R-FLAIR. The manual and T-FLAIR had a mean Dice score of 0.42 with a standard deviation of 0.27. Whereas, the R-FLAIR had a score of 0.38 with a standard deviation of 0.21. The Dice coefficient as a metric can be misleading because of the small diffuse nature of lesions. Thus, we also looked at the absolute relative difference of the manual WML volumes and those calculated by LesionTOADS on either T-FLAIR or the R-FLAIR. The ratio for T-FLAIR is 8.81, while for R-FLAIR it is 1.37, which is considerably less. Fig. 4 illustrates the lesion volumes for different subjects sorted in order of increasing manual lesion load. It is clear that using a R-FLAIR provides a more accurate lesion load estimation than a T-FLAIR—as the T-FLAIR has real-world artifacts that can confound any algorithm, not just LesionTOADS.
Fig. 4.
Shown are WML volumes, ordered by increasing lesion load, for 44 MS subjects. The green is the manual segmentation, while the blue is the plot of LesionTOADS based on the T-FLAIR and the red is LesionTOADS using our R-FLAIR.
4. CONCLUSION
In this work, we demonstrated a method to reconstruct a FLAIR image from the corresponding T1-w, T2-w, and PD-w images; the reconstruction is principally for image processing. Our previous work [16] could not handle WMLs, and our use of balanced training data and a third contrast has resolved this problem. The R-FLAIR is very close to the T-FLAIR in terms of image quality (see Table 1). We have shown that our R-FLAIR leads to more reasonable segmentation results with LesionTOADS than the T-FLAIR (see Fig. 4), in comparison to a human rater. In some cases, the artifacts in the T-FLAIRs produced dramatically bad segmentations (see Fig. 1), which is not the case with our R-FLAIRs.
Acknowledgments
This work is funded by the NIH/NIBIB under grant R21 EB012765 and R01 EB017743, and by the NIH/NINDS through grant R01 NS070906.
Contributor Information
Amod Jog, Email: amod@cs.jhu.edu.
Aaron Carass, Email: aaron_carass@jhu.edu.
Dzung L. Pham, Email: pham@nih.gov.
Jerry L. Prince, Email: prince@jhu.edu.
References
- 1.Fisher E, Lee JC, Rudick KNRA. Gray matter atrophy inmultiple sclerosis: a longitudinal study. Annals Neurol. 2008;64(3):255–265. doi: 10.1002/ana.21436. [DOI] [PubMed] [Google Scholar]
- 2.Filippi M, et al. Quantitative Assessment of MRI Lesion Load in Multiple Sclerosis, A comparison of conventional spin-echo with fast fluid attenuated inversion recovery. Brain. 1996;119(4):1349–1355. doi: 10.1093/brain/119.4.1349. [DOI] [PubMed] [Google Scholar]
- 3.Lecoeur J, Ferré JC, Barillot C. Optimized Supervised Segmentation of MS Lesions from Multispectral MRIs. MICCAI workshop on Med. Image Anal. on Multiple Sclerosis; 2009. [Google Scholar]
- 4.Shiee N, et al. A Topology-Preserving Approach to the Segmentation of Brain Images with Multiple Sclerosis Lesions. NeuroImage. 2010;49(2):1524–1535. doi: 10.1016/j.neuroimage.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forbes F, Doyle S, García-Lorenzo D, Barillot C, Dojat M. Adaptive weighted fusion of multiple MR sequences for brain lesion segmentation. Proc. of the Int. Symposium on Biomed. Imag. (ISBI 2011); 2010. pp. 69–72. [Google Scholar]
- 6.Stuckey S, Goh TD, Heffernan T, Rowan D. Hyperintensity in the Subarachnoid Space on FLAIR MRI. American Journal of Roentgenology. 2007;189(4):913–921. doi: 10.2214/AJR.07.2424. [DOI] [PubMed] [Google Scholar]
- 7.Hunt BR. Super-Resolution of Images: Algorithms, Principles, Performance. Intl Journal of Imag Sys and Tech. 1995;6(4):297–304. [Google Scholar]
- 8.Rousseau F. Brain hallucination. Proc. of the European Conference on Computer Vision (ECCV 2008); 2008. pp. 497–508. [Google Scholar]
- 9.Roy S, Carass A, Prince JL. Synthesizing MR Contrast and Resolution through a Patch Matching Technique. Proc. of SPIE-MI 2010; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konukoglu E, van der Kouwe A, Sabuncu MR, Fischl B. Example-Based Restoration of High- Resolution Magnetic Resonance Image Acquisitions. MICCAI. 2013;8149:131–138. doi: 10.1007/978-3-642-40811-3_17. [DOI] [PubMed] [Google Scholar]
- 11.Jog A, Roy S, Carass A, Prince JL. Magnetic Resonance Image Synthesis through Patch Regression. Proc. of the Int. Symposium on Biomed. Imag. (ISBI 2013); 2013. pp. 350–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roy S, Carass A, Prince JL. A Compressed Sensing Approach For MR Tissue Contrast Synthesis. 22nd Conf. on Inf. Proc. in Medical Imaging (IPMI); 2011. pp. 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy S, Jog A, Carass A, Prince JL. Atlas Based Intensity Transformation of Brain MR Images. Multimodal Brain Image Analysis. 2013;8159:51–62. [Google Scholar]
- 14.Roy S, Carass A, Bazin P-L, Prince JL. Intensity Inhomogeneity Correction of Magnetic Resonance Images using Patches. Proc. of SPIE-MI 2011; 2011. p. 79621F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and Regression Trees. Wadsworth Publishing Company; U.S.A: 1984. Statistics/ Probability Series. [Google Scholar]
- 16.Roy S, Carass A, Shiee N, Pham DL, Prince JL. MR contrast synthesis for lesion segmentation. Proc. of the Int. Symposium on Biomed. Imag. (ISBI 2011); 2010. pp. 932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breiman L. Bagging Predictors. Machine Learning. 1996;24(2):123–140. [Google Scholar]
- 18.Wang Z, Bovik AC. A universal image quality index. IEEE Signal Proc Letters. 2002;9(3):81–84. [Google Scholar]
- 19.Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: From error visibility to structural similarity. IEEE Trans Image Proc. 2004;13:600–612. doi: 10.1109/tip.2003.819861. [DOI] [PubMed] [Google Scholar]