Abstract
Aligned nanofibrous substrates can be created by electrospinning, but methods for creating multilamellar structures of aligned fibers are limited. Here, apposed nanofibrous scaffolds with pendant β-cyclodextrin (CD) were adhered together by adamantane (Ad) modified hyaluronic acid, exploiting the guest-host interactions of CD and Ad for macroscopic assembly. Stable user-defined multi-layered scaffolds were formed for cell culture or tissue engineering.
The unique functions of a tissue are dependent on how the cells and extracellular matrix (ECM) from which it is comprised are structured. In engineered approaches to repair lost or damaged tissues, scaffolds are used to recapitulate features of the tissue and to present cues that direct tissue regeneration by endogenous cells or those implanted with the scaffold.1 Spatial features of tissue engineering substrates, whether they are biochemical or biophysical,2 can be used to influence cell fate within engineered constructs. Nanofibrous scaffolds are often used to mimic aspects of the native ECM, and spatial features of the fibers such as alignment,3 three-dimensional variations of layer orientations,4 and physical dimensions5 can be engineered to influence cell behavior in response to the construct, as well as bulk mechanical properties. These considerations are of particular importance in tissues where the aligned layers of cells and ECM are characteristic of the native tissue and relate directly to tissue function. For example, cartilage,6 the annulus fibrosis of the intervertebral disc,7 myocardium,8 and neural tissue9 are all composed of aligned structures that vary with depth in the tissue.
Electrospinning is commonly used to create aligned nanofibrous scaffolds from a wide variety of polymers. In electrospinning, an electrical charge is applied to a polymer in a solvent as it is pumped through a needle. As a result of competing electrostatic repulsion and polymer chain entanglements, a Taylor cone forms and solvent evaporates, leading to deposition of fibers on a grounded surface. Electrospun scaffolds are readily made with control over features such as fiber alignment and fiber diameter.10 Additionally, multiple polymers can be electrospun into the same scaffold, enabling the engineering of scaffold properties such as selective fiber degradation to enhance cellular infiltration.11
Although a high degree of control over the properties of electrospun nanofibrous substrates is possible, it remains a challenge to create multilayer constructs whose fibers are aligned but oriented differently between layers. Control over fiber orientation would enable the engineering of directional properties into three-dimensional tissue constructs, a feature that is critical to approach native properties in engineered tissues.4, 12 Techniques for aligning fibers in multiple directions during the electrospinning process include repositioning the collector onto which fibers deposit13 or modifying the electric field during fiber deposition.14 However, the creation of lamellar structures of arbitrary thicknesses, alignments, and orientations by positioning and adhering a nanofibrous substrate to another after it has been collected remains a challenge. One approach is the application of force over long durations to hold mats in contact with one another while cells bind adjacent substrates.15 Likewise, fiber layers have been embedded in another matrix16 or held in place by thermoplastic microfibers extruded between nanofibrous layers.17
Supramolecular interactions, such as the guest-host interaction of adamantane (Ad) and β-cyclodextrin (CD), are non-covalent molecular interactions that are used to bind macroscale structures composed of complementary materials to each other.18 CDs are cyclic oligosaccharide host molecules with hydrophobic cores that have high affinity for a variety of small organic molecules. Ad, the guest molecule, associates strongly with CD due largely to hydrophobic interactions. We synthesized two hyaluronic acid (HA) derivatives, one modified with Ad (Ad-HA) and the other with CD (CD-HA),19 to use supramolecular interactions to adhere nanofibrous substrates together by guest-host interactions. HA is a naturally occurring polymer which is present throughout the body, including in tissues such as cartilage and neural structures, and is amenable to functionalization with many chemical moieties.20
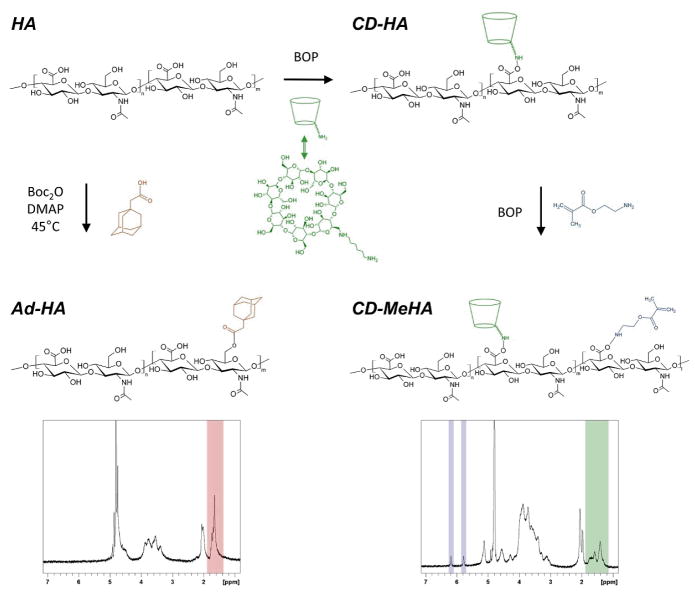
Our approach to macroscale assembly used Ad-HA as a supramolecular glue to bond apposing layers of nanofibers with pendant CD functionality to one another. The HA derivatives synthesized were each functionalized such that approximately 40% of the disaccharide repeat units were modified. In the case of Ad-HA, esterification of HA (357.4kDa) with 1-adamantane acetic acid (Fig. 1) resulted in 41% modification (by 1H-NMR) of HA. CD-HA was synthesized by coupling an amine-modified β-cyclodextrin to HA (90 kDa) via amidation, achieving 40% modification (by 1H-NMR). CD-HA was further modified with methacrylates by reacting CD-HA with 2-aminoethylmethacrylate via amidation to yield a methacrylated CD-HA (CD-MeHA). Methacrylates were added to facilitate crosslinking of fibers via radical polymerization to prevent their dissolution in an aqueous environment. With respect to the original HA backbone, 45% (by 1H-NMR) of the disaccharide groups in CD-HA were modified with methacrylates.
Fig. 1.
Chemical modifications of native hyaluronic acid (HA) to create adamantane modified HA (Ad-HA, lower left), β-cyclodextrin modified HA (CD-HA, upper right), and β-cyclodextrin and methacrylate modified HA (CD-MeHA, lower right). Representative 1H-NMR spectra highlight the peaks used to determine degree of modification with adamantane (red), β-cyclodextrin (green), or methacrylates (blue).
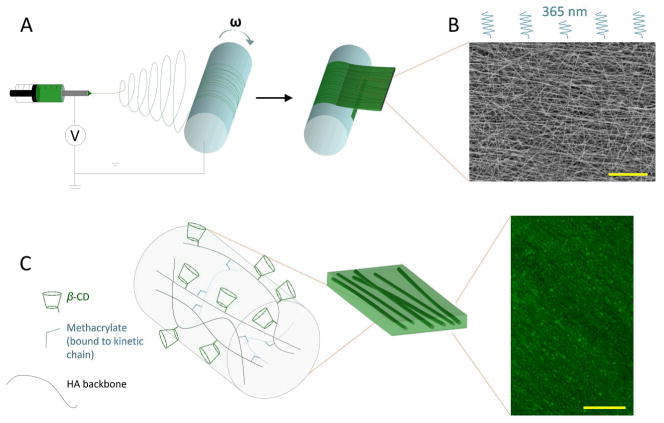
To create nanofibrous substrates, CD-MeHA was electrospun with a photoinitiator, which allowed for subsequent stabilization of the fibers by radical crosslinking with UV exposure (Fig. 2A and C). Dry fiber diameters were 191nm ± 48 nm, and they were aligned parallel to the direction of the mandrel’s rotation ± 30° (Fig. 2B). Depending on the length of time over which fibers were collected, the substrate thickness ranged from tens to hundreds of microns. To create multilamellar structures, aligned substrates were first placed on top of each other in a dry state, with the desired respective orientation of layers. These layered constructs were lightly clamped between two pieces of glass, placed into a 0.5% (w/v) aqueous solution of Ad-HA, and held together for about 15 minutes to allow complete hydration (Fig. 3A). The Ad-HA solution completely penetrated these samples of approximately 1 cm by 1 cm size. Upon separating the pieces of glass and moving the constructs into deionized water or PBS, the layers of the construct remained adhered to one another.
Fig. 2.
Schematic of nanofibrous scaffold fabrication. A) Electrospinning was used to deposit CD-MeHA as aligned nanofibers on a rotating mandrel. The fibrous, mat-like structure was then cut and removed from the mandrel. B) After crosslinking under UV light, the fibers were imaged by SEM to assess alignment (scale bar: 20 μm). C) Within the fiber, HA polymers (black) modified with β-CD (green) are crosslinked through methacrylate groups (blue) which polymerize forming kinetic chains (dashed blue). Aligned fibers, labeled with fluorescein, were swollen with PBS and imaged with multiphoton microscopy (scale bar: 100 μm).
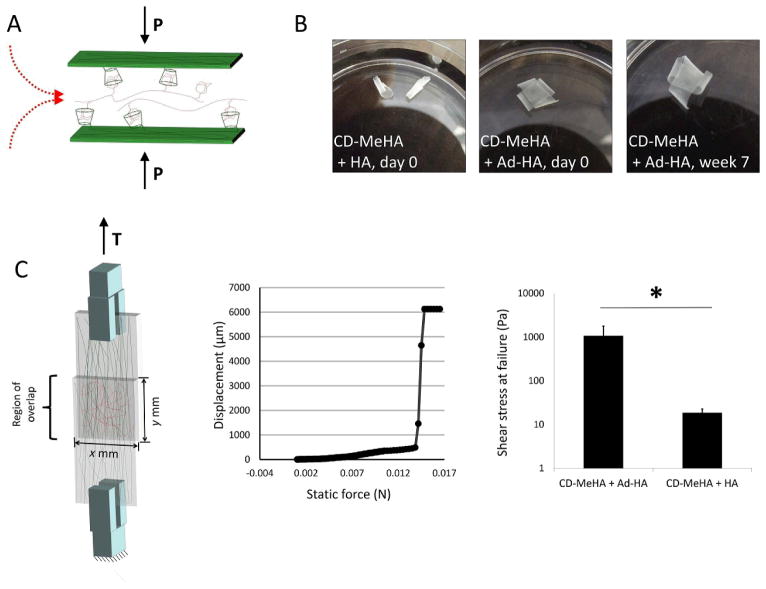
Fig. 3.
Adhesion of nanofiber layers with guest-host interactions. A) Layers of nanofibers were adhered to one another through physical bonds formed between β-CD host molecules in the nanofibers to Ad-HA, which was applied as a solution while layers were held together. B) Unmodified HA was not able to support adhesion, whereas AD-HA led to adherent structures that were stable for weeks in PBS at 37 °C. C) The force required to separate two layers of CD-HA nanofibers joined by Ad-HA was measured in a lap test. Failure of the bonds between the layers was determined from displacement during a force ramp. The force per unit area required to separate layers joined by Ad-HA was found to be two orders of magnitude greater than the force required to separate layers joined by unmodified HA.
To examine the stability of the interaction, two-layer lamellar constructs were incubated in PBS at 37°C over a period of seven weeks. At time points over the seven week period, the constructs were inspected for delamination. Among the eight samples studied, no delamination was observed over the entire seven weeks (Fig. 3C). The layered substrates remained bound to one another and could be manipulated with forceps without layer separation. However, when unmodified HA was used in place of Ad-HA the layered constructs fell apart immediately after removal of the slides.
Tensile testing was used to assess the strength of the interactions between two hydrated layers of CD-MeHA nanofibers held together by Ad-HA. In this case, nanofibrous substrates were directly electrospun onto methacrylated glass coverslips, which led to covalent bonds between the scaffolds and glass with UV exposure. Tensile testing was then used to measure the displacement of one coverslip relative to the other as a function of increasing force. When the bonds between the substrates failed, a rapid increase in the displacement of one slide was observed and denoted as the force at failure of the construct. An average shear stress of 1065±708Pa (mean±SD) at failure was observed for this system (Fig. 3B). This shear stress was significantly greater (p < 0.02) than that of controls where unmodified HA was used in place of Ad-HA (19±4Pa). These results indicate that the guest-host interactions are holding the lamellar constructs together through avidity, as the net forces are far greater than possible through individual non-covalent bonds. Adhesion strength might be tuned through an increased modification of HA with CD. This should result in an increased density of the supramolecular host moiety at the nanofiber surfaces, allowing for more bonds with Ad-HA. Strength of adherence as well as the stability of the layered structure over time might also be tuned through properties of Ad-HA, including degree of modification and molecular weight. This follows from modifying these properties, as this would affect the number of interactions between guest moieties on a polymer and host moieties on a nanofiber’s surface, where roughness and surface effects must be overcome to bind macroscopic elements assembled by supramolecular interactions.21
Multiphoton microscopy and scanning electron microscopy (SEM) allowed visualization of the aligned and oriented nanofibrous structures (Fig. 4). For multiphoton imaging, CD-MeHA was labeled with a carboxyfluorescein molecule. Z-stacks of the adherent, aligned mats, as well as SEM images, indicated that the alignment of each layer persisted after the nanofibrous substrates were positioned and bound by supramolecular interactions and that the layers were in immediate apposition to one another. To prepare adherent, layered constructs for SEM, we performed a stepwise dehydration, through which the layers remained adhered. After the final step, the substrates were cut parallel to one layer, sputter coated for SEM and imaged. The resulting images offer a high resolution view of the features in a hydrated state via fluorescent imaging as well as a higher resolution perspective of construct hierarchy—from fibers to single layers to layered constructs.
Fig. 4.
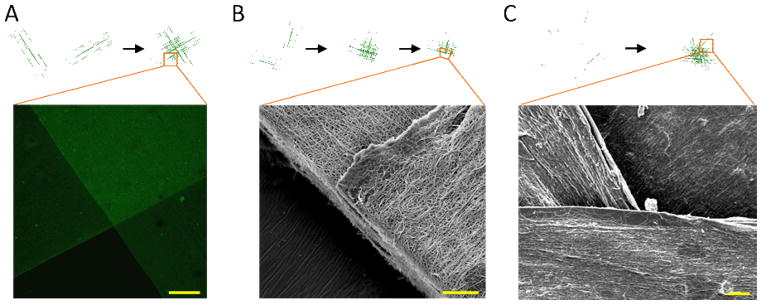
Images of aligned and adhered layers of nanofibers. A) Multiphoton microscopic image of CD-MeHA fibers labeled with fluorescein (scale bar: 200 μm). B) SEM image of CD-MeHA fibers at a cut along the midline of one layer (scale bar: 20 μm). C) SEM image of the surface of a four layer construct (scale bar: 20 μm).
This approach of adhering layers of aligned fibers together has inherent flexibility for numerous applications, including in the number and orientation of fiber layers. While the current work has focused on the development of acellular constructs that might ultimately direct tissue regeneration by native cells in vivo, the methods could be easily adapted to use with cells and cell culture components after construct fabrication for in vitro studies for tissue regeneration. Because the process of orienting the layers occurs prior to hydration, the layers can easily be repositioned as necessary until construct structures are correct. Many polymers are used in electrospinning, and the adamantane and β-cyclodextrin modifications used here could be introduced through standard conjugation techniques to a variety of systems. Also, the bonds formed are highly specific to the guest-host pair involved,18 which offers some orthogonality, should other functional groups also be needed in a construct to impart some chemical or biological functionality.
Experimental
Materials synthesis
Guest and host modified macromers were prepared as previously described19 with HA (Lifecore) having molecular weights of 357.4 kDa and 90 kDa for Ad-HA and CD-HA, respectively. Briefly, the tetrabutylammonium salt of HA was prepared by ion exchange against Dowex-100 resin and neutralization with tetrabutylammonium hydroxide to give HA-TBA after lyophilization. HA-TBA was dissolved in anhydrous dimethylsulfoxide (DMSO) under nitrogen for further modification. HA was functionalized with adamantane through a di-tert-butyl dicarbonate (BOC2O)-mediated esterification with 1-adamantane acetic acid. The percent of HA repeat units modified was determined to be 41% from integration of the ethyl multiplet of adamantane (δ=1.50–1.85, 12 H, shown red in 1H-NMR Fig. 1) relative to the HA backbone (δ=3.20–4.20, 10 H). Synthesizing the CD-HA derivative first required synthesis of an aminated cyclodextrin to enable coupling, 6-(6-aminohexyl)amino-6-deoxy-β-cyclodextrin (β-CD-HDA).19 HA was then functionalized through a (benzotriazol-1-yloxy)tris-(dimethylamino)phosphonium hexafluorophosphate (BOP)-mediated amidation reaction with β-CD-HDA. The percent of HA repeat units modified was determined to be 40% from integration of the hexane linker (δ=1.35–1.85, 12 H, shown green in 1H-NMR Fig. 1) relative to the methyl singlet of HA (δ=2.1, 3 H). To synthesize CD-MeHA, CD-HA was converted to its TBA salt, dissolved in DMSO, and reacted with 2-aminoethylmethacrylate through a second BOP-amidation reaction and purified with dialysis and lyophilization. 1H-NMR was used to assess HA modification. The percent of HA repeat units modified was determined to be 45% from integration of the vinyl protons (δ=6.20, 1 H and 5.78, 1H shown blue in 1H-NMR Fig. 1) relative to the methyl singlet of HA (δ=2.1, 3 H). To fluorescently label CD-MeHA, the thiol-containing peptide sequence GCDDD-5(6)-carboxyfluorescein was prepared by standard solid-phase Fmoc-protected peptide synthesis. CD-MeHA was then dissolved in an aqueous solution of 0.2 M triethanolamine at pH 8 and the fluorescently-labeled peptide was added for coupling between the thiols on the peptide and a small fraction of methacrylates via Michael-addition. All macromers were purified by dialysis and recovered by lyophilization before use.
Electrospinning
CD-MeHA or fluorescently-labeled CD-MeHA was dissolved at 4% (w/v) in deionized water containing 1.5% (w/v) poly(ethylene oxide) (PEO, 900 kDa) and 0.5% (w/v) Irgacure 2959. PEO serves as a carrier polymer which enables the electrospinning of a viscous CD-MeHA solution.22 The solution was electrospun using a power source of 21 kV from a 12″ long 18G blunt-ended needle driven by a syringe pump (0.75 mL/h) and positioned such that the end of the needle was 22 cm from a grounded rotating mandrel. After the nanofibers were collected on the mandrel, the substrates were cut, placed into a nitrogen purged chamber, and then crosslinked with UV light (320–390 collimated source) for 15 minutes.
Construct fabrication
The aligned nanofibrous substrates were trimmed to the desired size and then layered upon one another in the desired orientations. Here, layers were cut to be between 5 and 10 mm, and layers with thicknesses in the range 50 to 100 μm were used. The resulting, dry constructs were then gently held together between two glass coverslips. A solution of 0.5% (w/v) Ad-HA was then introduced into the space between the coverslips via capillary action and the nanofibrous constructs were held between the glass coverslips for 30 min. The construct could then be retrieved for analysis.
Mechanical analysis
For mechanical testing of adhesion strengths, fibers were electrospun directly onto glass coverslips. Prior to use, surfaces of the coverslips were methacrylated by first treating them with 10 M sodium hydroxide, then coating the surface with 3-(triethoxysilyl)propyl methacrylate and heating for 1 h at 100°C. After electrospinning, the nanofibers covered the methacrylated surfaces and were bound to coverslips with UV exposure. Two coverslips were then placed in apposition to create a lap joint where the fibers were in contact over a known area and the end of each coverslip was clamped into a dynamic mechanical analyzer (Q800 TA Instruments). A force ramp was applied and the force at failure measured from the displacement curve.
Microscopy
Constructs prepared for fluorescent microscopy were imaged on a Leica TCS SP5 confocal microscope equipped with a Coherent Chameleon Ti:Sapphire laser. To prepare samples for SEM, the constructs were first dehydrated using a series of increasingly concentrated ethanol washes. After the final wash, the construct was placed on aluminum foil and dried under vacuum. The construct was then cut along the long axis of one of the layers to expose an overlapping region, sputter-coated with gold and mounted for SEM (JOEL 7500F HRSEM).
Conclusions
In engineering biomaterial constructs for tissue regenerative therapies, whether for development in vitro or for direct implantation in vivo, success depends on the ability to engineer construct properties on a range of length scales. Layering aligned nanofibers while choosing fiber orientations from one layer to the next according to a tissue specific design may aid in the engineering of constructs for tissue types whose function is dependent on aligned, fibrous ECM structures. Supramolecular guest-host materials offer a means of assembling materials and macroscale structures, and we used them here to join together aligned layers of electrospun, HA-based nanofibers. The resulting structures were stable over time, could bear loads before separating, and may ultimately be applied in engineering constructs for the regeneration of organized tissues such as cartilage, nervous tissue, or cardiac muscle.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (R01EB008722) and the National Science Foundation through a MRSEC grant (DMR-1120901) and Graduate Research Fellowships to ILK and RJW.
Footnotes
Electronic Supplementary Information (ESI) available: Video demonstrating the adherence of two nanofiber layers in moving water is available online. See DOI: 10.1039/c000000x/
References
- 1.Burdick JA, Mauck RL, Gorman JH, Gorman RC. Sci Transl Med. 2013:5. doi: 10.1126/scitranslmed.3003997. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jia X, Kiick KL. Macromol Biosci. 2009;9:140. doi: 10.1002/mabi.200800284. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yannas IV. Biomaterials. 2013;34:321. doi: 10.1016/j.biomaterials.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Mitragotri S, Lahann J. Nat Mater. 2009;8:15. doi: 10.1038/nmat2344. [DOI] [PMC free article] [PubMed] [Google Scholar]; Marklein RA, Burdick JA. Adv Mater. 2010;22:175. doi: 10.1002/adma.200901055. [DOI] [PubMed] [Google Scholar]; Burdick JA, Murphy WL. Nat Commun. 2012:3. doi: 10.1038/ncomms2271. [DOI] [PubMed] [Google Scholar]
- 3.Li WJ, Mauck RL, Cooper JA, Yuan X, Tuan RS. J Biomech. 2007;40:1686. doi: 10.1016/j.jbiomech.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lim SH, Liu XY, Song HJ, Yarema KJ, Mao HQ. Biomaterials. 2010;31:9031. doi: 10.1016/j.biomaterials.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tulloch NL, Muskheli V, Razumova MV, Korte FS, Regnier M, Hauch KD, Pabon L, Reinecke H, Murry CE. Circ Res. 2011;109:47. doi: 10.1161/CIRCRESAHA.110.237206. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sundararaghavan HG, Saunders RL, Hammer DA, Burdick JA. Biotechnol Bioeng. 2013;110:1249. doi: 10.1002/bit.24788. [DOI] [PubMed] [Google Scholar]
- 4.Nerurkar NL, Baker BM, Sen S, Wible EE, Elliott DM, Mauck RL. Nat Mater. 2009;8:986. doi: 10.1038/nmat2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christopherson GT, Song H, Mao HQ. Biomaterials. 2009;30:556. doi: 10.1016/j.biomaterials.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Fithian DC, Kelly MA, Mow VC. Clin Orthop Relat Res. 1990:19. [PubMed] [Google Scholar]
- 7.Cassidy JJ, Hiltner A, Baer E. Connect Tissue Res. 1989;23:75. doi: 10.3109/03008208909103905. [DOI] [PubMed] [Google Scholar]
- 8.Buckberg G, Hoffman JI, Mahajan A, Saleh S, Coghlan C. Circulation. 2008;118:2571. doi: 10.1161/CIRCULATIONAHA.107.754424. [DOI] [PubMed] [Google Scholar]
- 9.Wedeen VJ, Rosene DL, Wang RP, Dai GP, Mortazavi F, Hagmann P, Kaas JH, Tseng WYI. Science. 2012;335:1628. doi: 10.1126/science.1215280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu WY, Thomopoulos S, Xia YN. Adv Healthcare Mater. 2012;1:10. doi: 10.1002/adhm.201100021. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hong JK, Madihally SV. Tissue Eng Part B. 2011;17:125. doi: 10.1089/ten.teb.2010.0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker BM, Gee AO, Metter RB, Nathan AS, Marklein RA, Burdick JA, Mauck RL. Biomaterials. 2008;29:2348. doi: 10.1016/j.biomaterials.2008.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]; Baker BM, Shah RP, Silverstein AM, Esterhai JL, Burdick JA, Mauck RL. Proc Natl Acad Sci U S A. 2012;109:14176. doi: 10.1073/pnas.1206962109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moutos FT, Freed LE, Guilak F. Nat Mater. 2007;6:162. doi: 10.1038/nmat1822. [DOI] [PubMed] [Google Scholar]; Engelmayr GC, Cheng MY, Bettinger CJ, Borenstein JT, Langer R, Freed LE. Nat Mater. 2008;7:1003. doi: 10.1038/nmat2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beachley V, Katsanevakis E, Zhang N, Wen XJ. Adv Healthcare Mater. 2013;2:343. doi: 10.1002/adhm.201200125. [DOI] [PubMed] [Google Scholar]; Laudenslager MJ, Sigmund WM. J Nanopart Res. 2013:15. [Google Scholar]
- 14.Li D, Wang YL, Xia YN. Adv Mater. 2004;16:361. [Google Scholar]; Garrigues NW, Little D, O’Conor CJ, Guilak F. J Mater Chem. 2010;20:8962. doi: 10.1039/c0jm01880e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Driscoll TP, Nakasone RH, Szczesny SE, Elliott DM, Mauck RL. J Orthop Res. 2013;31:864. doi: 10.1002/jor.22312. [DOI] [PubMed] [Google Scholar]
- 16.Coburn J, Gibson M, Bandalini PA, Laird C, Mao HQ, Moroni L, Seliktar D, Elisseeff J. Smart Struct Syst. 2011;7:213. doi: 10.12989/sss.2011.7.3.213. [DOI] [PMC free article] [PubMed] [Google Scholar]; McCullen SD, Miller PR, Gittard SD, Gorga RE, Pourdeyhimi B, Narayan RJ, Loboa EG. Tissue Eng Part C. 2010;16:1095. doi: 10.1089/ten.tec.2009.0753. [DOI] [PubMed] [Google Scholar]; Ekaputra AK, Prestwich GD, Cool SM, Hutmacher DW. Biomacromolecules. 2008;9:2097. doi: 10.1021/bm800565u. [DOI] [PubMed] [Google Scholar]
- 17.Kang R, Le DQS, Li HS, Lysdahl H, Chen ML, Besenbacher F, Bunger C. J Mater Chem B. 2013;1:5462. doi: 10.1039/c3tb20562b. [DOI] [PubMed] [Google Scholar]
- 18.Harada A, Kobayashi R, Takashima Y, Hashidzume A, Yamaguchi H. Nat Chem. 2011;3:34. doi: 10.1038/nchem.893. [DOI] [PubMed] [Google Scholar]
- 19.Rodell CB, Kaminski AL, Burdick JA. Biomacromolecules. 2013;14:4125. doi: 10.1021/bm401280z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burdick JA, Prestwich GD. Adv Mater. 2011;23:H41. doi: 10.1002/adma.201003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng M, Shi F, Li J, Lin Z, Jiang C, Xiao M, Zhang L, Yang W, Nishi T. Adv Mater. 2014;26:3009. doi: 10.1002/adma.201305177. [DOI] [PubMed] [Google Scholar]
- 22.Sundararaghavan HG, Metter RB, Burdick JA. Macromol Biosci. 2010;10:265. doi: 10.1002/mabi.200900363. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kim IL, Khetan S, Baker BM, Chen CS, Burdick JA. Biomaterials. 2013;34:5571. doi: 10.1016/j.biomaterials.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.