Summary
Although inland water bodies are more heterogeneous and sensitive to environmental variation than oceans, the diversity of small protists in these ecosystems is much less well-known. Some molecular surveys of lakes exist, but little information is available from smaller, shallower and often ephemeral freshwater systems, despite their global distribution and ecological importance. We carried out a comparative study based on massive pyrosequencing of amplified 18S rRNA gene fragments of protists in the 0.2-5 μm-size range in one brook and four shallow ponds located in the Natural Regional Park of the Chevreuse Valley, France. Our study revealed a wide diversity of small protists, with 812 stringently defined operational taxonomic units (OTUs) belonging to the recognized eukaryotic supergroups (SAR –Stramenopiles, Alveolata, Rhizaria–, Archaeplastida, Excavata, Amoebozoa, Opisthokonta) and to groups of unresolved phylogenetic position (Cryptophyta, Haptophyta, Centrohelida, Katablepharida, Telonemida, Apusozoa). Some OTUs represented deep-branching lineages (Cryptomycota, Aphelida, Colpodellida, Tremulida, clade-10 Cercozoa, HAP-1 Haptophyta). We identified several lineages previously thought to be marine including, in addition to MAST-2 and MAST-12, already detected in freshwater, MAST-3 and possibly MAST-6. Protist community structures were different in the five ecosystems. These differences did not correlate with geographical distances, but seemed to be influenced by environmental parameters.
Keywords: Biodiversity, 18S rRNA, protist, plankton, freshwater, cryptophyte, MAST
Introduction
Aquatic ecosystems occupy most of the Earth surface. Oceans alone cover around 71% of that surface (Costanza, 1999). Lakes, ponds and reservoirs cover more than 3% (nearly 4.5 million km2) of non-oceanic regions (Downing et al., 2006). Although comparatively smaller, these freshwater systems offer a large array and diversity of ecological niches including various trophic levels, light accessibility, temperature and oxygen concentrations. This is especially true for small and shallow lentic inland ecosystems (sizes from 0.001 to 0.1 km2), which are widespread, varied and numerous, corresponding to more than 99% of the total number of lakes on earth (Downing et al., 2006). Microbial communities dominate aquatic ecosystems and their activity has profound impact at global scales, being largely implicated in carbon fixation (Li, 1994; Jardillier et al., 2010) and climate regulation (Simó, 2001). Within these communities, microbial eukaryotes play key roles in nutrient cycling acting as photosynthesizers, heterotrophs (predators, parasites) or mixotrophs (Caron, 1994; Zubkov et al., 2008; Jardillier et al., 2010; Massana, 2011).
Small eukaryotes (<20 μm) have been known to constitute a non-negligible part of aquatic microbial communities in both freshwater and marine systems for a long time (Johnson et al., 1982; Corpe et al., 1992). Based on their cell size, small protists were initially classified in nanoeukaryotes (cells between 2 and 20 μm in diameter) and picoeukaryotes (cells ≤ 2 μm). In the last two centuries, many small eukaryotic species have been described, including phototrophs such as prasinophytes and other chlorophytes (Knight-Jones et al., 1951; Stockner, 1988; Guillou et al., 1999), which suggested a potentially significant role in primary production (Johnson et al., 1982), but also heterotrophs (Fenchel, 1982; Patterson and Larsen, 1991). Indeed, the ecological importance of heterotrophic nanoflagellates as predators has long been acknowledged (Fenchel, 1982; Wright et al., 1984). However, being too small to show easily-identifiable, unambiguous morphological differences, their true diversity remained largely inaccessible and their taxonomy poorly studied or oversimplified (many of these tiny protists were simply classed as incertae sedis).
In the past 15 years, the use of molecular methods based on 18S rRNA gene analysis has largely overcome that problem, allowing studying the phylogenetic diversity and distribution of small protists at unprecedented scales. Most such studies largely explored oceanic systems, including surface waters (Diez et al., 2001; Moon-van-der-Staay et al 2001), the deep-sea (López-García et al., 2001), and coastal regions (Massana, et al., 2004; Romari et al., 2004; Cheung et al., 2010). They revealed a huge protist diversity encompassing members of all recognized eukaryotic super-groups (SAR–Stramenopiles, Alveolata, Rhizaria–, Archaeplastida, Excavata, Amoebozoa and Opisthokonta as well as lineages of uncertain position in the eukaryotic tree, such as haptophytes, cryptophytes or picozoa (López-García and Moreira, 2008; Massana, 2011; Seenivasan et al., 2013; Moreira and López-García, 2014). Molecular surveys also allowed the discovery of lineages previously unknown in spite of their abundance, such as new clades affiliated to alveolates (e.g. Groups I and II), which turned out to be members of the classical Syndiniales (Groisillier et al., 2006; Harada et al., 2007; Guillou et al., 2008) and stramenopiles (the MAST clades, for MArine STramenopiles) (López-García et al., 2001; Moon-van der Staay et al., 2001; Diez et al., 2001; Stoeck et al., 2003; Massana et al., 2004). Thought for a long time to be exclusively marine, the MAST groups remain poorly known. In a few cases a correspondence has been found between particular organisms and their environmental sequences (e.g. the colonial protist Solenicola setigera and the MAST-3; Gómez et al., 2011). However, many other MAST lineages have not yet been linked to any cultured or described organisms (Massana et al., 2013), even though fluorescent in situ hybridization (FISH) labeling has provided some hints on their morphology, life style and ecology (Massana et al., 2002, 2006).
The molecular exploration of very small protists in freshwater began slightly later than in the oceans and also revealed a large diversity (Richards, 2005; Slapeta et al., 2005; Lefranc et al., 2005). In lakes, alveolates (especially ciliates and Perkinsozoa, by contrast to the Syndiniales-Duboscquellids-dinoflagellate dominance in marine systems), stramenopiles, cryptophytes and fungi were found to be abundant, but protists belonging to the Archaeplastida, Rhizaria, and Cercozoa or groups of uncertain affiliation were also diverse (Lefranc and Thénot, 2005; Slapeta et al., 2005; Lepère et al., 2008; Zhao et al., 2011; Mangot et al., 2012). In some cases, a significant part of the detected diversity was composed only of environmental sequences without known close relatives in databases (Lefèvre et al., 2008; Mangot et al., 2012; Triadó-Margarit and Casamayor, 2012). Some groups were observed in both oceans and freshwater systems while others seem, according to current knowledge, to be specific to marine (e.g. Syndiniales; Guillou et al., 2008) or freshwater (e.g. HAP-1 lineage of haptophytes; Slapeta et al., 2005; Shalchian-Tabrizi et al., 2011) environments. Even within lineages present both in marine and freshwater systems, 18S rRNA sequences obtained from freshwater samples often form phylogenetic clades distinct from those of oceanic sequences, suggesting a limited number of, mostly ancient, freshwater colonization events followed by radiations (Logares et al., 2009). That scarcity of marine-freshwater transitions has been observed for many small eukaryote groups such as the perkinsids (Bråte et al., 2010), haptophytes (Shalchian-Tabrizi et al., 2011; Simon et al., 2013) or MAST lineages (Massana et al., 2013), and could be explained by the difficulty of crossing the salinity barrier (Logares et al., 2009). However, freshwater aquatic systems remain largely understudied and massive high-throughput sequencing techniques have been applied to very few among them. Therefore, it might be possible that failure to detect some of these lineages is due to undersampling. Furthermore, the vast majority of protist molecular diversity studies in freshwater have been conducted in lakes (eg. Richards, 2005, Lepère et al., 2006, 2008). However, smaller systems like ponds or brooks have been overlooked, despite their abundance, distribution and ecological importance. Yet, previous analyses suggest that they host several lineages undetected in marine environments or lakes (Slapeta et al., 2005; Simon et al., 2013). In addition, because they are shallow and small, this kind of freshwater systems may display very different local physico-chemical conditions and, hence, they constitute ideal models to test whether local environmental selection is more influential than geographic distance in determining community composition, which remains controversial (Green et al., 2004; Martiny et al., 2006). A recent study on lakes suggest that geographic distance might be important to explain protist biogeography (Lepère et al., 2013), but lakes have much more buffering capacity than small shallow systems.
In this work, we have investigated the diversity of small eukaryotes (essentially the 0.2-5 μm size-fraction) in a set of shallow freshwater environments by amplification of 18S rRNA gene fragments and direct high-throughput 454-pyrosequencing. We selected one brook and four ponds located in the same geographic area (the Natural Regional Park of the Chevreuse Valley, France) but differing in size, depth and physico-chemical conditions. The objectives of our work were threefold: i) describing protist diversity at unprecedented depth in this kind of habitats, ii) checking whether increasing sequence depth leads to the discovery in freshwater systems of eukaryotes previously thought to be exclusively marine and iii) testing which parameters (geographical distance vs. physico-chemical characteristics) determine eukaryotic microbial communities.
Results
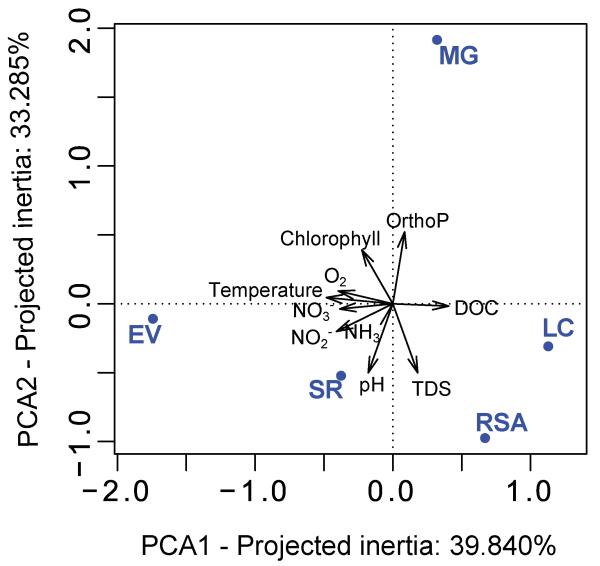
To study protist diversity in five small and shallow freshwater ecosystems, we selected one brook and four shallow ponds in the Natural Regional Park characterized by different physico-chemical parameters and trophic status (Table 1; Fig. S1). The Mare Gabard is a small pond located in the middle of the forest (Fig. S1). It smelled strongly of H2S when sediments were stirred and had high phosphate concentrations (0.15 mg.l−1) but the lowest pH, conductivity and TDS (total dissolved solids) values. Saint Robert pond is the most anthropized system selected due to its location in a hamlet (Fig. S1). It contained very high ammonia (1.22 mg.l−1) and, to a lesser extent, also nitrite concentrations (Table 1, Fig. 1), likely influenced by the permanent presence of a sizeable duck population. La Claye pond is part of a complex of several ponds on ancient peat bog substrate, with a dense population of eagle ferns in the surroundings. Lying on acidic and organic-rich soils, it was characterized by high concentrations of dissolved organic carbon (36.3 mg.l−1). The Etang des Vallées is a nearly 1.5 m deep pond and, contrary to other sampled systems, was supersaturated in oxygen (116.8%; Table 1). It also had high temperature at the time of sampling (13.1°C), and high concentrations of nitrate (5.1 mg.l−1) as compared to the other systems. The Ru Sainte Anne is the brook and had the highest amounts of total dissolved solids (746 mg. l−1) possibly due to sediment input in a very shallow (~10 cm) system of running waters (Table 1). Principal component analysis (PCA) of the different physico-chemical parameters measured showed that, despite their close proximity (distances between these systems varied from 2 to 9 km; Fig. S1), the ponds and brook were clearly distinct from each other (Fig. 1). This made of this set of shallow aquatic ecosystems a good model to study whether geographic proximity is more influential than physico-chemical parameters in determining community composition.
Table 1. Characteristics of freshwater systems studied and diversity and richness estimates for the different samples.
Replicate samples are indicated by a small case letter after the sample name. TDS, total dissolved solids; DOC, dissolved organic carbon.
| Sampled ecosystem | Gabard | Saint Robert | Etang des Vallees | Sainte Anne | La Claye | |||
|---|---|---|---|---|---|---|---|---|
| Ecosystem type | Forest pond | Village pond | Large pond | Forest brook | Pond on peaty soil | |||
| GPS coordinates | 48°39′15.83″N 1°55′20.26″E |
48°39′54.82″N 1°56′45.28″E |
48°41′23,0″N 001°54′59,2″E |
48°36′45.91″N 1°58′16.61″E |
48°36′31.72″N 1°56′17.33″E |
|||
| Approximate surface (m2) | 850 (210 × 75 m) | 495 (20 × 28 m) | 12 880 (210 × 75 m) | 1 m width at sampling point | 265 (24 × 10 m) | |||
| Approximate depth (cm) | 50 | 50 | 150 | 20 | 60 | |||
| Sampling Date | April 5, 2012 | April 5, 2012 | March 30, 2012 | April 6, 2012 | April 6, 2012 | |||
| Water temperature (°C) | 9.0 | 10.7 | 13.1 | 7.1 | 7.8 | |||
| pH | 6.6 | 7.2 | 7.31 | 7.36 | 7 | |||
| Oxygen (%) | 78.9 | 58.9 | 116.8 | 72.8 | 61.2 | |||
| Conductivity (μS/cm) | 81.4 | 531 | 288 | 746 | 520 | |||
| TDS (mg/l) | 81 | 531 | 288 | 746 | 520 | |||
| Chlorophyll (μg /l) | 74.9 | 61.9 | 44.7 | 2.6 | 10.7 | |||
| NO3− (mg/l) | 1.41 | 1.17 | 5.08 | 1.45 | 2.02 | |||
| NO2− (mg/l)2 | ~ 0 | 0.054 | 0.047 | 0.010 | 0 | |||
| NH3 (mg/) | 0.02 | 1.22 | 0.02 | 0.03 | 0.04 | |||
| DOC (mg/l) | 19.0 | 15.7 | 9.8 | 17.7 | 36.3 | |||
| PO43− (mg/l−) | 0.15 | 0.03 | 0.03 | 0.03 | 0.03 | |||
| Size fraction studied | 0.2 - 5 μm | 0.2 - 5 μm | 5 - 30 μm | 0.2 - 5 μm | 0.2 - 5 μm | 0.2 - 5 μm | ||
| Sample Name | MG25 | MG25b | SR25 | SR25b | EV34 | EV34b | EV33 | EV33c | RSA25 | RSA25b | LC25 | LC25b |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of reads before filtering |
15,020 | 5,110 | 85,803 | 6,724 | 28,418 | 14,168 | 22,314 | 7,938 | 37,766 | 7,140 | 21,283 | 14,215 |
| Number of quality- filtered reads |
10,616 | 4,197 | 42,034 | 4,506 | 10,982 | 6,906 | 17,670 | 3,947 | 19,652 | 4,243 | 11,191 | 10,605 |
| Observed number of OTUs |
76 | 55 | 15 | 37 | 147 | 132 | 177 | 87 | 427 | 198 | 66 | 68 |
| Richness1 (Standard error) |
67.9 (2.37) | 55.0 (0.15) | 11.4 (1.06) | 36.8 (0.44) | 134.4 (2.94) | 126.9 (2.07) | 148.5 (3.99) | 87.0 (0.00) | 313.7 (7.52) | 197.8 (0.44) | 57.9 (2.34) | 57.0 (2.67) |
| Diversity (Simpson Index) |
0.86 | 0.80 | 0.75 | 0.68 | 0.96 | 0.95 | 0.95 | 0.95 | 0.92 | 0.91 | 0.50 | 0.38 |
| Evenness | 0.58 | 0.56 | 0.61 | 0.45 | 0.75 | 0.72 | 0.69 | 0.78 | 0.64 | 0.7 | 0.34 | 0.27 |
Expected number of OTUs in random subsamples of the size of the smallest sequence library (3,947 reads in EV33c)
Nitrite concentrations in Gabard and La Claye ponds were under the kit detection limit of 0.006 mg/l and were set as 0 in analysis.
Fig. 1. Principal Component Analysis (PCA) plot of the measured physicochemical parameters.
Sampled ecosystems appear in blue and physico-chemical parameters in black. MG, Mare Gabard; EV, Etang des Vallees; LC, La Claye; RSA, RuSainte Anne; SR, Saint Robert. TDS, Total Dissolved Solutes; DOC, Dissolved Organic Carbon; OrthoP, orthophosphate.
Overall protist community composition
The composition of protist communities was estimated for the five selected shallow freshwater systems based on 454-pyrosequenced 18S rDNA fragments amplified from DNA of plankton of the 0.2-5 μm cell diameter fraction. In addition, protist diversity in the size fraction 5-30 μm was studied for the largest sampled ecosystem, the Etang des Vallées. Replicate samples were included in all cases. We described protist diversity in these systems from a total of 146,549 quality-filtered reads using highly stringent criteria to avoid sequence artefacts and chimeras. The reads from the 12 different samples (the 0.2-5 μm cell fraction of 5 systems with replicas plus the two replicas of the 5-30 μm for the Etang des Vallées) were treated together in order to define operational taxonomic units (OTUs) that could be fully compared among samples. All those sequences grouped in 812 OTUs defined using a cut-off value of 98% sequence identity. From these, 768 OTUs (128,661 reads) were detected at least in the 0.2-5 μm size fraction, the remaining 44 OTUs corresponding to protists found only in the larger 5-30 μm at the Etang de Vallées (Table 1). The different OTUs were then assigned to known taxonomic groups based on sequence similarity, which revealed a wide phylogenetic diversity of protists in general and of small eukaryotes in particular. Our stringently defined OTUs affiliated to the major recognized eukaryotic supergroups SAR (Stramenopiles- Alveolata- Rhizaria), Archaeplastida, Excavata, Amoebozoa and Opisthokonta (López-García et al., 2008; Adl et al., 2012) and to several lineages of unresolved phylogenetic position such as Cryptophyta, Haptophyta and Apusozoa (Fig. 2).
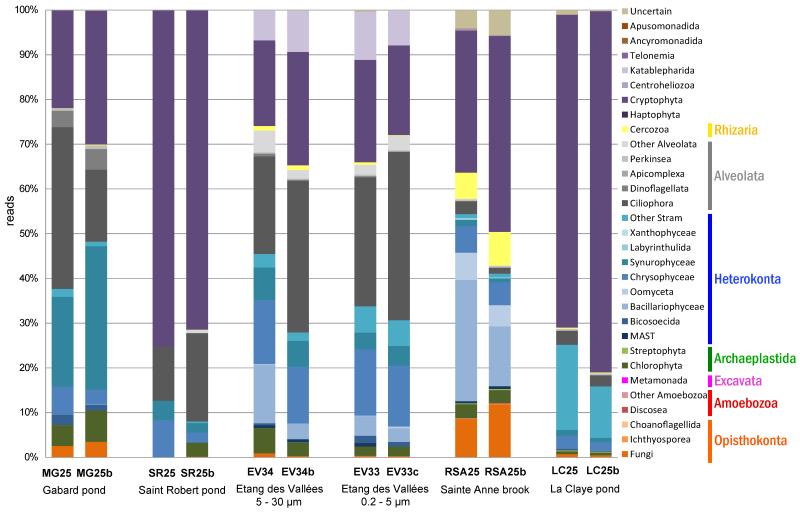
Fig. 2. Histogram showing the relative proportion of 18S rRNA gene amplicon reads assigned to high rank taxa in the five shallow ecosystems studied.
Replicate samples are labeled as ‘b’ or ‘c’. In all cases, the distribution corresponds to protists in the 0.2-5 μ size range, except in the Etang des Vallees, where the 5-30 μ size fraction was additionally analyzed.
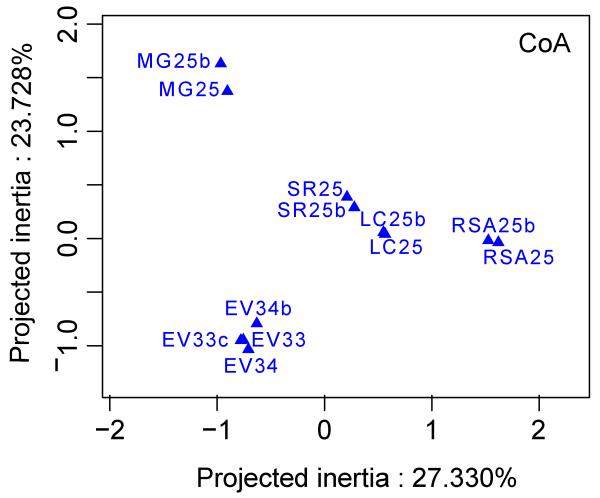
To evaluate the reliability of the community composition determined for each ecosystem at this stage, we compared the pairwise Bray-Curtis distances between the two replicates for each ecosystem. These were very small (0.26 on average, min. 0.12, max. 0.36) as compared to distances between libraries from distinct ecosystems (0.84 on average, min. 0.44, max. 0.99). Replicates grouped together in both Non-metric MultiDimensional Scaling (NMDS; Fig. S2) and Correspondence (CoA) analyses (Fig. 3), revealing their high similarity in OTU composition.
Fig. 3. Correspondence Analysis (CoA) plot showing protist community composition similarities and differences among the five ecosystems studied.
MG25 and MG25b, Mare Gabard; LC25 and LC25b, La Claye; SR25 and SR25b, Saint Robert; RSA25 and RSA25b, Ru Saint Anne; EV33 and EV33c, Etang des Vallees (0.2-5 μm size samples). EV34 and 34b, Etang des Vallees (5-30 μm size samples).
The composition of protist communities from the small size fraction samples (0.2-5 μm) differed greatly between ecosystems (Fig. 2). First, richness and diversity indexes were highly variable (Table 1). Samples from Ru Sainte Anne and Etang des Vallées were the richest and the most diverse, whereas La Claye was highly dominated by few abundant OTUs (evenness = 0.34 – 0.27 in replicates). La Claye and Saint Robert appeared relatively close in NMDS plots (Fig. S2) and clustered close together in CoA plots (Fig. 3). These two systems displayed relatively similar physico-chemical parameters, notably oxygen content and conductivity, and low diversity as compared to the other ecosystems (Table 1). The similarity between La Claye and St Robert community composition was likely influenced by the fact that they were largely dominated by the same cryptophyte OTU (Fig. 2; see below).
Second, the relative abundance of taxonomic groups varied between ecosystems, even though OTUs affiliated to Cryptophyta, Stramenopiles (or Heterokonta) and Alveolata accounted for the majority of sequences in all libraries (Figs. 2 and 4). Cryptophyte sequences were remarkably abundant, representing up to 49 and 53% of the total number of reads and reads of the 0.2-5 μm size fraction respectively. Cryptophytes were the dominant group in Saint Robert and La Claye (around 75% of reads in those samples). They were also the dominant group in the brook Sainte Anne (between 32-44% of reads), and were the second most abundant group in the two other systems (Fig. 2, Table S1). Most cryptophyte reads belonged to a unique OTU affiliated to Cryptomonas curvata (OTU_52; Figs. 4 and S3). Stramenopiles constituted the second most abundant group in our systems and represented 25% of all reads and nearly 24% of the reads from the 0.2 -5 μm size fraction samples (Fig. 2, Table S1). Within this supergroup, Chrysophyceae were abundant in all samples. However, the most abundant stramenopile groups in Mare Gabard and Ru Sainte Anne were, respectively, Synurophyceae and Bacillariophyceae, two lineages producing silica skeletons or scales. Oomycetes were also relatively abundant in the brook Sainte Anne. Alveolates constituted the third most abundant supergroup in our study. It was the dominant group in Etang des Vallées (32-41% of reads in replicates of the 0.2-5 μm size fraction, and similar values for the larger size fraction analyzed) and one of the dominant groups in Mare Gabard (22-40% of reads in duplicate samples). It was also a major component in Saint Robert pond (12-20%). However, they represented only a small proportion of the taxa detected in the brook Sainte Anne and La Claye pond (less than 5 % of reads; Fig. 2, Table S1). Among alveolates, ciliates were the most represented in all samples; though OTUs affiliated to dinoflagellates and other alveolates had occasionally significant proportions.
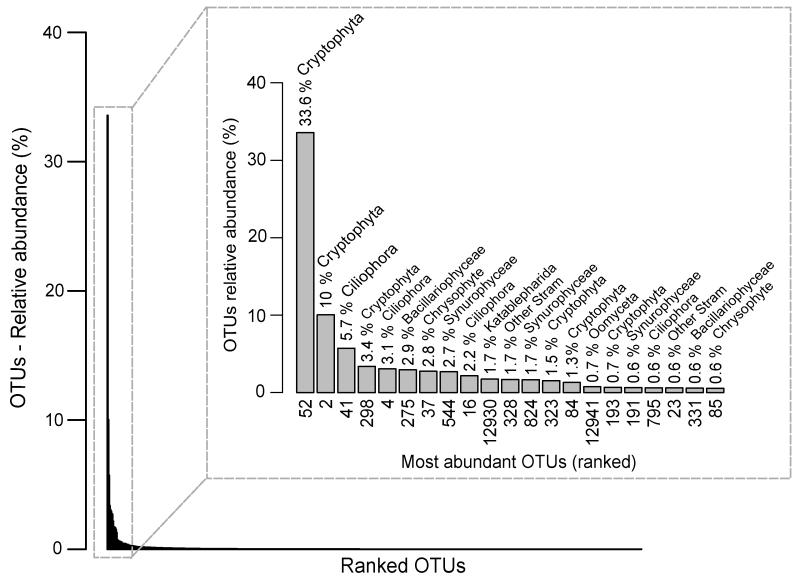
Fig. 4. Rank abundance curve for the total 768 OTUs detected collectively in the 0.2-5 μm size fraction of the five shallow freshwater systems studied.
Sequence data from all samples were pooled to define OTUs with high stringency. The relative abundance of protist OTUs representing more than 0.5% of the total number of reads are shown in the inset. The identity number of the respective OTUs and their taxonomic affiliation are shown below and above the histogram bars.
Along with these three abundant supergroups, members of the Opisthokonta, Amoebozoa, Excavata, Archaeplastida, Rhizaria and of groups of uncertain position in the eukaryotic tree were also detected. Since we purposefully used general eukaryotic primers biasing against Metazoa, opisthokonts were essentially represented by fungi, though choanoflagellates and ichthyosporeans were also detected. The highest abundance of fungi was observed in Mare Gabard and Ru Sainte Anne, but they generally represented less than 10% of reads. Rhizarian OTUs were retrieved in all ecosystems and were only represented by cercozoans. They were not abundant except for the Ru Saint Anne, where they reached 6-8% of the reads. Katablepharid sequences represented as much as 8-11% of reads from the 0.2-5 μm fraction in the Etang des Vallées but were less abundant elsewhere (0.03 % on average in each small size fraction sample). Archaeplastida (streptophytes and, mainly, chlorophytes) were detected in all ecosystems. Haptophytes were also identified, though in low proportions, and only in the Etang des Vallées and the Ru Sainte Anne. OTUs affiliated to Excavata, Labyrinthulida, Xanthophyceae, Apicomplexa, Centroheliozoa, Telonemida, Amoebozoa and Apusomonadida (Apusozoa) were detected only in the highly diverse Sainte Anne brook (Fig. 2, Table 1). 3 additional apusozoan OTUs were detected in Sainte Anne brook and the Etang des Vallées.
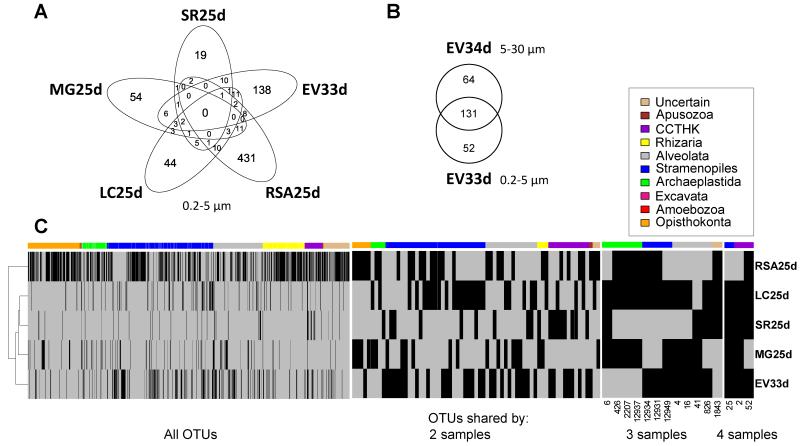
Despite some similarity in the distribution of large phylogenetic groups, with cryptophytes, stramenopiles and alveolates dominating the different shallow water systems, protist communities were very different at the phylotype scale. Indeed, no OTU was shared by all the systems and the vast majority of the 768 OTUs detected in the small size fraction was specific to the libraries of a single ecosystem (Fig. 5A). 67 and 12 OTUs were shared by 2 and 3 different ecosystems, respectively (Fig. 5C). Only three OTUs were shared by four systems (Fig. 5C), two of which affiliated to cryptophytes and corresponded to the most abundant OTUs (Fig. 4). Remarkably, OTU_52, affiliated to Cryptomonas curvata, was particularly abundant in St Robert and La Claye samples where it represented as much as 43-52% and 69-78% of reads respectively. However, OTU_52 dropped to 4-7% of reads in the 0.2-5 μm fraction of the Etang des Vallées and was not detected in Mare Gabard. OTU_25, affiliated to stramenopiles, was also shared by four systems, but represented only 0.13% of all reads in 0.2-5 μm fractions.
Fig. 5. Distribution of protist OTUs in the five shallow freshwater systems studied.
Sequence data from replicates were pooled. A, five-set Venn diagram showing the number of specific and shared OTUs in the different freshwater systems (0.2-5 μm fraction size). B, Venn diagram showing OTUs shared by the two fraction sizes analyzed in Etang des Vallees. C, clustering analysis of the five ecosystems based on the presence of shared OTUs, as shown by the heatmap. Only 0.2-5 μm size fractions are considered. Heatmaps show all OTUs and OTUs shared by 2, 3 or 4 ecosystems. No OTU is shared by all the 5 ecosystems. Each row represents an ecosystem and each vertical bar an OTU. Black: OTU present, Grey: OTU absent. The colored sidebar indicates the taxonomic affiliation of the OTU represented by the bar below. Color codes representing different phylogenetic affiliation are indicated in the box. CCTHK: Cryptophyta, Centroheliozoa, Telonemia, Haptophyta and Katablepharida. For presentation reasons, the width of OTU bars is not the same in all the heatmaps. 29 RSA25d, Ru Sainte Anne; LC25d, La Claye; SR25d, Saint Robert; MG25d, Mare Gabard; EV33d, EV34d, Etang des Vallées.
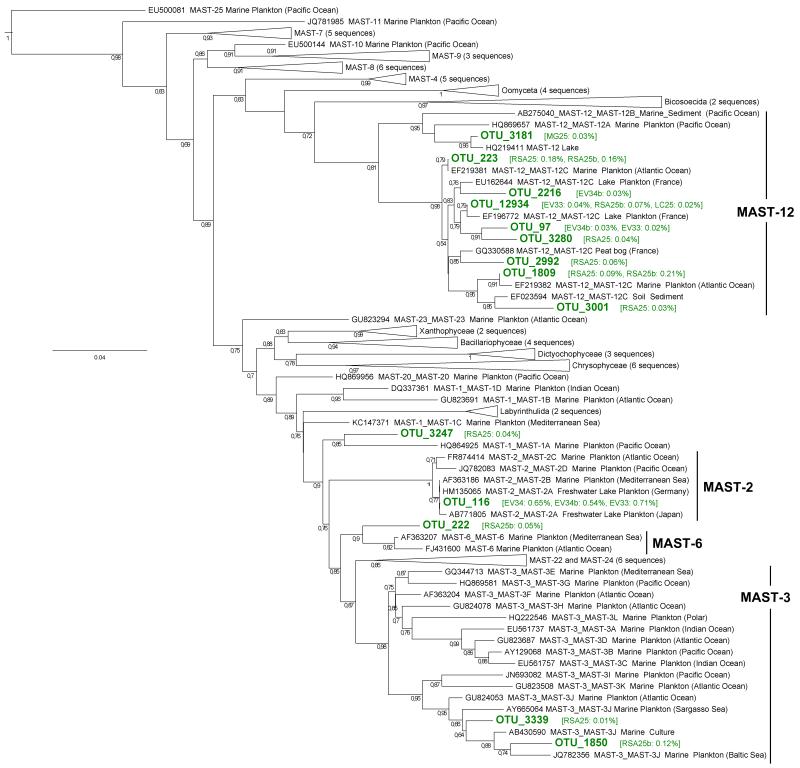
The composition of protist communities within the two different size fractions (0.2-5 μm and 5-30 μm) of the Etang des Vallées was very similar, as revealed by the NMDS and CoA analyses (Figs. 3 and S2) as well as by low Bray-Curtis distances (0.35 on average, min: 0.29, max: 0.43). Moreover, the community structure was similar in both size fractions from the Etang des Vallées, which displayed the highest diversity and evenness recorded of all samples (Table 1). The taxonomic composition was also similar at high-rank taxa level, although diatoms and green algae were in slightly higher proportions in the largest size fraction (Fig. 2). At a finer level of resolution, more OTUs were shared by both size fractions (131 OTUs) than specific to each of them (Fig. 5B).
Phylogenetic diversity
In order to get more detailed information on the different OTUs identified, we carried out phylogenetic analyses with partial 18S rRNA gene sequences representative of the different OTUs. Although cryptophytes were the most abundant group in our samples, they were not very diverse. Only 27 OTUs were affiliated to that group (3.3% of all OTUs). Most of these OTUs were closely related to existing sequences, affiliating to photosynthetic genera, especially Cryptomonas (containing the highly overrepresented OTU_52 corresponding to C. curvata mentioned above) and Chroomonas, but also to the phagocytic plastid-lacking Goniomonas (Fig. S2). However, several OTUs appeared to be quite divergent; for instance representative sequences of OTUs 838 and 1812 shared no more than 93% of identity with their first BLAST hit in the NCBI database, the sequences HM135076 from freshwater (Luo et al., 2011) and AM901364 from the cultured Cryptomonas commutata strain M1975 respectively (Fig. S3).
A total of 268 OTUs (33% of all OTUs) were affiliated with stramenopiles, the second most abundant group in our samples. Nearly half of them were related to Chrysophyta-Synurophyta (125 OTUs) while the remaining ones affiliated to Eustigmatophyta, Dictyochophyta, Bicosoecida, Oomycetes, Bacillariophyta, Xanthophyta and Labyrinthulida (Fig. S4). Surprisingly, 14 OTUs seemed to belong to various MAST groups (Fig. 6), originally thought to be exclusively marine. Nine OTUs affiliated to the group MAST-12, being related to sequences previously detected in a wide variety of ecosystems, such as a suboxic Norwegian estuary (Kolodziej et al., 2007), freshwater lakes (Lefèvre et al., 2008; Monchy et al., 2011) and a peat bog (Lara et al., 2011). OTU_116 was closely related (99% identity) to MAST-2 sequences detected in a freshwater lake (Luo et al., 2011) and the Mediterranean (Diez et al., 2001). OTUs_1850 and 3339 affiliated to MAST-3, which so far was known to contain only sequences from marine environments. Finally, OTU_222 and OTU_3247 had similarity by BLAST to MAST-6 sequences retrieved from the Mediterranean (AF363207, Diez et al., 2001) and other MAST-6 sequences, with 94% and 91% identity, respectively. However, from the phylogenetic analysis, their affiliation to this clade is unclear and will require a more in-depth exploration of these groups.
Fig. 6. Approximate Maximum Likelihood (ML) phylogenetic tree of partial 18S rRNA gene stramenopile sequences showing the presence of several MAST clades in shallow freshwater systems.
Non-MAST stramenopile lineages have been collapsed. A total of 392 unambiguously aligned positions were used to reconstruct the tree. Two alveolate sequences were used to root the tree. Representative sequences ofOTUs from this work are shown in bold green. The name of samples where MAST OTUs were detected and the respective proportion of reads are shown within brackets. The scale bar represents the number of estimated substitutions per position for a unit branch length. RSA25/RSA25b, Ru Saint Anne; MG25/MG25b, Mare Gabard; EV33/ EV33b (0.2-5 μm) and EV34/EV34b (5-30 μm), Etang des Vallées.
Alveolates were also found to be diverse (Figs. S5 and S6), but the vast majority of OTUs were assigned to ciliates (88 out of 125 alveolate OTUs). Dinoflagellates were also represented in our samples, along with 17 OTUs related to sequences of putative freshwater (Amaral-Zettler et al., 2008; Monchy et al., 2011) or marine (Scheckenbach et al., 2010; Behnke et al., 2010) perkinsid parasites. Interestingly, several OTUs grouped with Apicomplexa and Colpodellida and clustered into 2 distinct groups (Fig. S5). Eight OTUs clustered with Colpodella edax (Leander et al., 2003) and many environmental sequences from freshwater lakes or ponds (Lefèvre et al., 2007; Nakai et al., 2012; Oikonomou et al., 2012; Richards, 2005). Three other OTUs clustered with sequences affiliated to apicomplexans (mostly Cryptosporidium) coming from more diverse environments, e.g. peat bog (Lara et al., 2011), marine sediment (Dawson et al., 2002), humans (Yuan et al., 2012) or ostrich fecal samples (Martinez-Diaz, R, unpublished).
From the 131 OTUs affiliated with opisthokonts, the great majority (125 OTUs) related to fungi, mostly to chytrids but also to ascomycetes, basidiomycetes or to the basal Rozellida/Cryptomycota lineage (Fig. S7). In addition 11 OTUs were related to the aphelids, the sister group to fungi, which so far mostly contains highly divergent 18S rRNA gene environmental sequences (Karpov et al., 2014). Several OTUs clustered with ichtyosporeans and choanoflagellates (Fig. S7).
All rhizarian OTUs branched with cercozoan sequences. Even though cercozoan OTUs were not very abundant in our samples and represent only 1.2% of the total number of reads, they were composed of numerous phylotypes (106 OTUs) that represented altogether 13% of all OTUs. The vast majority of these OTUs affiliated to the Filosa (Fig. S8). Several OTUs branched with the Endomyxa, although they shared only 93.6% identity on average with their first BLAST hits in GenBank (calculated on the 13 OTU representative sequences affiliated to Endomyxa, min: 89% between OTU_12985 and AB526843, max: 99% between OTU_1810 and EU910610). Furthermore, 5 OTUs clustered with sequences from the Novel Clade 10, and OTUs_2162 and 608 shared 92% and 91% identity respectively with sequence EU567287 from Novel Clade 11 – Tremulida, two recently defined deep-branching cercozoan lineages (Bass et al., 2009; Howe et al., 2011).
65 OTUs (8% of all OTUs) belonged to Archaeplastida (Fig. S9). Five of them strongly affiliated to Embryophyta. Given the pre-filtration steps, these OTUs must correspond to pollen grains, free DNA or cells from dead leaves. Strikingly, they represented altogether only 0.04% of all reads, in spite of the dense vegetation around most sampled systems. Four additional OTUs most likely corresponded to unicellular Streptophyta, affiliated to Clausteriaceae and Desmidiaceae. The remaining archaeplastid OTUs affiliated to the chlorophyte classes Mamiellophyceae, Trebouxiophyceae, Nephroselmidophyceae and, mainly, Chlorophyceae. In addition, we detected a few OTUs belonging to other, less abundant eukaryotic groups. Thus, 4, 1 and 6 OTUs affiliated to katablepharids, telonemids and haptophytes, respectively (Fig. S3). Most haptophyte OTUs were very close to other freshwater sequences, but the OTU_3295 had a sequence nearly identical to Jomonlithus littoralis (AM490979; 99% identity), a coastal species, and to two environmental sequences (JX680345, JX680344) from a brackish pond (Simon et al., 2013). Sequences belonging to the basal haptophyte group HAP-1 (Slapeta et al., 2005) were detected in the Ru Sainte Anne (OTU_3063; Fig. S3). Six additional OTUs belonged to centroheliozoans (Fig. S3). Their closest relatives were sequences from soil or freshwater sediment, and were only retrieved in Sainte Anne brook, the most narrow and less deep ecosystem (Table 1). That observation, along with the higher proportions and diversity in Sainte Anne brook of cercozoa, fungi and diatoms, known to be usually composed of large and/or benthic cells, could be explained by a higher influence or contribution of benthic communities in this system. One excavate OTU (OTU_1857; 99% identical to Trimastix marina) and one apusomonad OTU were also detected in Sainte Anne brook. Three amoebozoan OTUs from the brook and 2 ancyromonad OTUs could also be detected in our small freshwater systems (Fig. S9). Finally, 66 of the total 812 OTUs determined could not be affiliated with confidence to any group.
Effects of physicochemical parameters and distance on community structure and composition
The five sampled ecosystems were separated by distances between 2 and 9.5 km (Fig. S1). To see whether the differences in community composition increased with geographical distance, we pooled sequence data from replicate samples for the 0.2-5 μm size fraction, calculated Bray-Curtis distances between all systems based on the pooled data, and performed a Mantel analysis to test the correlation between Bray-Curtis and geographical distance matrices. The Mantel test showed no correlation between differences in protist community composition and distance between ecosystems (r=0.2151, P-value=0.236). As an illustration, OTU_52 (Cryptomonas curvata) reached 44% of all reads (43-52 % in replicates) in the Saint Robert pond while it could not be detected in the Gabard pond (Fig. 5), its nearest system (2.1 km away). However, this OTU was detected in all other systems, up to 9.5 km away.
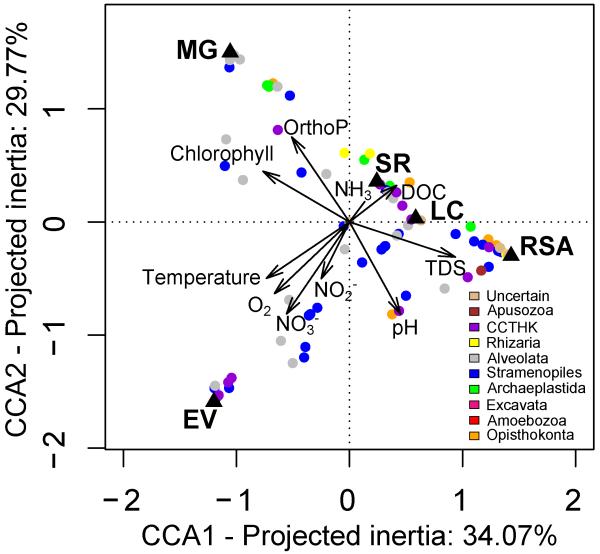
As mentioned above, each of our five ecosystems was characterized by a set of specific environmental variables, with the Mare Gabard and the Ru Saint Anne being, respectively, the less and most charged in total dissolved solids and Saint Robert being highly enriched in ammonia (Table 1; Fig. 1). All these differences in environmental parameters correlated to the structure of small protists in these ecosystems as revealed by a Mantel test linking environmental and community pairwise distances between samples (r=0.7323, P-value=0.014). However, additional canonical correspondence analyses (CCA) did not reveal any clear relationship between any of the detected eukaryotic supergroups and one or several environmental variables, although some OTUs appeared to be associated with particular samples according to their specific presence (Fig. 5) and distribution in CCA (Fig. 7). At the phylum or class level (Fig. S10), haptophytes, katablepharids, choanoflagellates, cercozoans and cryptophytes appeared to reach higher proportions where pH was the highest and phosphate concentration the lowest, the two latter variables being tightly correlated in principal component analysis (PCA) and CCA (Figs. 1 and 7). Streptophytes were associated with high values of dissolved organic carbon while bacillariophytes and fungi were more abundant where total dissolved solutes were the highest. These features could thus partially explain the differences in composition of the small eukaryotes among our contrasting ecosystems.
Fig. 7. Canonical Correspondence Analysis (CCA) plot.
Only 0.2-5 μm size fractions are considered. Each dot represents an OTU. The colorsindicate the taxonomic affiliation. Black triangles indicate samples. Duplicate samples appear superimposed. CCTHK: Cryptophyta, Centroheliozoa, Telonemia, Haptophyta and Katablepharida. TDS: Total Dissolved Solutes, DOC: Dissolved Organic Carbon, OrthoP: orthophosphate.
Discussion
Shallow freshwater systems, reservoirs of protist diversity
Compared with oceans, molecular protist diversity surveys in freshwater ecosystems are still scarce. Paradoxically, inland water bodies are collectively much more heterogeneous than oceans, and much more sensitive to environmental variation. Despite so, only a handful of studies on freshwater protist communities exist, mainly from a variety of lakes, from temperate regions and mountains to polar areas (e.g. Charvet et al., 2012; Lepère et al., 2013; Taib et al., 2013). However, nearly nothing is known from smaller, shallower and often ephemeral freshwater systems, despite their global distribution and ecological importance (Downing et al., 2006). Recent approaches based on massive sequence analysis, though prone to a variety of methodological errors and biases, are called to facilitate comparative studies among those ecologically relevant ecosystems. This is especially true for tiny protists, since morphology-based exploration very often miss their phylogenetic diversity (Massana et al., 2002; Moreira and López-García, 2002; Slapeta et al., 2006). To contribute to that task, we carried out a study based on massive pyrosequencing of amplified 18S rRNA gene fragments of protists in the 0.2-5 μm size range in five shallow freshwater ecosystems from a temperate area. Many of the high-rank taxa detected occur, although with variable relative abundances, in other freshwater systems, such as lakes. Cryptophytes, stramenopiles and alveolates were by far the most abundant supergroups detected, though their internal diversity varied greatly among ecosystems (Fig. 2). Although less abundant in the open sea (Kirkham et al., 2013; Shi et al., 2009), small cryptophytes seem to be common in freshwater (Slapeta et al., 2005; Lefranc et al., 2005; Lepère et al., 2008; Mangot et al., 2012; Taib et al., 2013). Stramenopiles, as well as ciliates, which dominate the alveolate diversity in our samples, are also common in protist communities from freshwater lakes (Lefranc et al., 2005; Lepère et al., 2008; Taib et al., 2013). Fungi, Ichthyosporea and choanoflagellates within the Opisthokonta, cercozoans within the Rhizaria, and members of the Amoebozoa, Archaeplastida, Excavata as well as members of the phylogenetically unresolved Haptophyta, Centrohelida, Katablepharida, Telonemidaand Apusozoa were also detected.
Surprisingly, fungi, which are often detected in very large proportions in some lakes (Lefranc et al., 2005; Lepère et al., 2008), accounting for up to 94% of all the 454-pyrosequences in some of them (Taib et al., 2013), were in very low proportions (generally much less than 10%; Fig. 2) in our systems. This may reveal a real difference between shallow ecosystems and deeper lakes, but it could also reflect varying population dynamics along the year (Nolte et al., 2010). Our samples were collected in early spring and many of the fungal OTUs detected correspond to potential parasitic fungi or related lineages, such as chytrids, rozellids/cryptomycota or aphelids (Fig. S7), which might experience fluctuation depending on host population dynamics along the year. Temporal surveys would be needed to test such a hypothesis. Several OTUs in our freshwater systems branched deeply within known eukaryotic supergroups. Thus, the Rozellida-Cryptomycota and the aphelids have been considered the deepest lineages of fungi (Lara et al., 2010; Jones et al., 2011), although recently, they have been re-classified as forming part of the superphylum Opisthosporidia together with Microsporidia and would be the sister group to fungi (Karpov et al., 2014). Other detected deep-branching lineages include the Colpodellida (Leander et al., 2003) within the alveolates, the poorly known novel Clade-10 Cercozoa and Tremulida (Bass et al., 2009; Howe et al., 2011) or the early diverging haptophyte lineage HAP-1, so far only detected in freshwater systems (Slapeta et al., 2005; Shalchian-Tabrizi et al., 2011).
Although protist community composition was different in the 5 analyzed systems, the comparison of protist diversity in two cell fraction sizes (0.2-5 μm and 5-30 μm) in the most diverse site, the Etang des Vallées, did not differ much in terms of high-rank taxa (Figs. 2 and 3). Indeed, despite the presence of OTUs apparently confined to each cell-size fraction (21 and 25%, for the smaller and larger size ranges, respectively), a considerable proportion of OTUs (53%) were shared between the two cell fractions (Fig. 5B). Several explanations are possible and non-mutually exclusive. One is that many small organisms may be retained in filters of larger pore-diameter if the filtered biomass is high, since they might be entangled with larger organisms. On the opposite, flexible organisms of relatively bigger sizes than those of filter pores may pass through them under the filtration pressure applied. Finally, many organisms may encompass a size range that spans the 5 μm diameter barrier imposed by our filters. This may be due to either differential sizes during their life cycle (e.g. gametes or spores and vegetative forms) or to size variability under a given life stage.
The broad protist diversity unveiled in the five freshwater systems also reflects a wide ecological diversity of functions. At a very general level, typical photosynthetic, heterotrophic and parasitic groups were detected. Among photosynthesizers, cryptophytes followed by photosynthetic stramenopiles (diatoms, chrysophytes, synurophytes, xantophytes) largely exceeded green algae. Many of these lineages may also contribute to heterotrophic activities, since many photosynthetic protists are mixotrophs (Zubkov et al., 2008; Massana, 2011; Hartmann et al., 2012). Typical predators span most of the eukaryotic diversity identified, from choanoflagellates and amoeba to bicosoecids, ciliates, cercozoa, centrohelids, katablepharids or apusozoa (Fig. 2). However, heterotrophic activities also encompass the degrading activity of osmotrophic taxa, including several fungi and labyrinthulids, and that of parasitic or parasitoid protists (many chytrids, cryptomycota, aphelids, oomycetes, apicomplexa, perkinsids).
Altogether, our extensive 18S rDNA-based survey of small protists suggests that shallow freshwater systems are important reservoirs of eukaryotic diversity. Not only members of all supergroups are present, but also several members of uncertain, poorly known or deep-branching lineages. Given the heterogeneity of this kind of systems on Earth, it can be advanced that further studies on shallow freshwater systems will uncover yet-to-characterize protists, the study of which will be of relevance to understand the ecology of these ecosystems and, from an evolutionary perspective, to reconstruct poorly resolved areas in the eukaryotic tree.
Marine-freshwater barriers transgressed
One of the potential surprises that the study of protist diversity in varied freshwater systems may bring is the increasing awareness that salinity barriers can be overcome more easily than previously thought. Although lineages with members in both freshwater and marine systems exist, such as dinoflagellates, haptophytes, perkinsids or stramenopiles (Logares et al., 2007; Bråte et al., 2010; Shalchian-Tabrizi et al., 2011; Simon et al., 2013), the marine-freshwater transition is thought to be rare (Logares et al., 2009). However, this view may be biased by current undersampling of freshwater systems. Indeed, in our study we have detected protist lineages thought to be exclusively marine in the past, notably several MAST lineages. Thus, we did not only identify members of MAST-2 and MAST-12 lineages, which have recently been detected in other freshwater systems (Massana et al., 2013), but also members of MAST lineages never identified in freshwater systems before. This was the case of MAST-3, for which we identified two bona fide OTUs and, potentially, of MAST-6 and even MAST-1 lineages, although OTUs related to the latter are of much more uncertain affiliation (Fig. 6).
Moreover, in addition to the occurrence of the emblematic MAST groups in these shallow freshwater systems, we also identified many OTUs widespread in the eukaryotic tree that share 99% identity or more with sequences retrieved from marine environments. Examples are the haptophyte OTU_3295, practically identical to Jomonlithus littoralis (Fig. S3), the stramenopile OTU_116 (Fig. S4), the ciliate OTU_255 (Fig. S6) or the excavate OTU_1857 affiliating to Trimastix marina (Fig. S9). But there are many other OTUs whose closest relatives are sequences from marine systems, even if similarity is slightly lower.
In fact, even if salinity seems to be a relevant ecological determinant structuring microbial communities (Lozupone et al., 2007), the marine-freshwater barrier seems to be relatively easy to cross. On the one hand, several protists, notably cryptophytes, are osmotolerant and can cope with various salinity concentrations by the means of contractile vacuole regulation (Hoef-Emden, 2014). On the other hand, many freshwater systems contain relatively high levels of dissolved solutes (organic and/or inorganic) requiring similar adaptations as those require for life in seawater salts. In this sense, it is interesting to note that most of the “typically marine” lineages detected in our freshwater systems, were identified in the Ru Sainte Anne, which has the highest TDS content and seems greatly influenced by this parameter (Table 1; Fig. 7).
Elements of protist biogeography
There are two essentially opposed views with regard to microbial and, more specifically, protist biogeography. Most classical views posit that small free-living protists would find little barriers to dispersal and, hence, be widely distributed. Differences in protist community structure would then be essentially explained by local environmental parameters. This “everything is everywhere, but the environment selects” view seems supported by the occurrence of cosmopolitan protist species (Baas-Becking, 1934; Finlay, 2002; Slapeta et al., 2006). On the opposite extreme, a variety of studies seem to suggest that endemic protists (Foissner, 2006) and taxa-area relationships exist for microbial eukaryotes (Green et al., 2004). Comparing community compositions sidesteps undersampling and the nearly impossible task of demonstrating true microbial endemisms (Martiny et al., 2006). A recent study suggested that the beta-diversity of the small eukaryotes between lakes was linked to the geographic distances between ecosystems (Lepère et al., 2013). However, it is extremely difficult to disentangle the effect of local environmental parameters from physical distance. Furthermore, other factors, such as the temporal (Nolte et al., 2010) and the phylogenetic scale (Ragon et al., 2012) need to be taken into account. In our case, we sampled the five systems at the same or correlative dates to limit temporal effects and, although the distances involved were different from those in the study of Lepère et al (2013) (10 km versus 100 km scale), our study clearly rejects geographic distance as a driver of community composition. On the contrary, Mantel tests show significant correlations between differences in community structure and physico-chemical parameters. However, clear associations between high taxon levels and environmental parameters were not identified (Fig. 7 and S10). Differences at finer, OTU scale might provide hints about the role of environmental selection in determining community structure. At any rate, testing which factors more greatly influence the composition of these communities would require the inclusion of biotic parameters, notably the diversity and relative abundance of bacteria, archaea and viruses from the same systems.
Experimental procedures
Sampling and measurement of physicochemical parameters
Samples were collected in spring 2012 from five small and shallow freshwater ecosystems at the Natural Regional Park of the Chevreuse Valley (France, South of Paris) (Table 1 and Fig. S1). The systems were chosen to represent a variety of conditions at local scale, from forest ponds rich in organic matter (Mare Gabard) to more urban and agricultural-influenced systems (Saint Robert). Surface water was collected using sterile plastic carboys and processed immediately back in the laboratory. Water samples were serially filtered through 30 μm pore-size nylon filters (Millipore), and through 5 and 0.2 μm pore-size diameter Nucleopore membranes (Whatman). Filters were stored frozen at −20°C until DNA extraction. The water temperature, pH, the concentration of total dissolved solutes and the level of dissolved oxygen were measured in situ using a multiparameter probe (Multi 350i, WTW). The concentrations of dissolved nitrate (NO3−), nitrite (NO2−), ammonia (NH3), orthophosphate (PO43−) and dissolved organic carbon (DOC) were measured in water samples filtered through 0.2 μm pore-size diameter Nucleopore membranes on the same day of sampling using manufactured colorimetric tests (Hach-Lange). Chlorophyll a concentrations were determined after harvesting plankton biomass on glass microfiber filters (GF/F, Whatman) that were stored at −20°C until ethanol pigment extraction. For chlorophyll extraction, filters were dried then ground in 7 ml absolute ethanol, and heated at 70°C for 20 min. After centrifugation, 1 ml of supernatant was collected and optical densities at 665 and 750 nm were measured (spectrophotometer DR5000 Hach-Lange). Chlorophyll a concentration was determined on pigment extract by spectrophotometry, as follows: [Chla] = 11.9 * (OD665 – OD750)/w* Ve/Vf with [Chla]: chlorophyll a concentration (μg.l−1), 11.95: Reciprocal specific absorbance coefficient of chlorophyll a at 665 nm (μg.cm.ml−1), OD665: optical density at 665 nm, OD750: optical density at 750 nm, w: width of the spectroscopic cuvette (1 cm), Ve: volume of the pigment extract (in ethanol, ml) and Vf: volume of water filtrated on the glass microfiber filter (l). The protocol was adapted from Ritchie (2006).
DNA extraction, amplification and sequencing of 18S rRNA genes
DNA was extracted from cells collected onto filters that were cut into pieces using the PowerSoil DNA extraction kit (MoBio) according to the manufacturer’s instructions. DNA was eluted in 80 μl of 10 mM Tris, pH 8.0. 18S rRNA gene fragments of ca. 550 bp, encompassing the V4 hypervariable region, were amplified using the newly designed primer EK-565F (5′-GCAGTTAAAAAGCTCGTAGT) and primer 18s-EUK- 1134-R - UNonMet (5′-TTTAAGTTTCAGCCTTGCG) biased against Metazoa (Bower et al., 2004). Both forward and reverse primers were tagged with 20 different 10 bp molecular identifiers (MIDs) to allow pooling and later differentiation of PCR products from 20 distinct samples. PCR amplifications were conducted in a total reaction volume of 25 μl using 1.5 mM MgCl2, 0.2 mM of each dNTP (PCR Nucleotide Mix, Promega), 0.3 μM of each primer, 0.3 to 2 μl of DNA sample and 0.5 U HotStart Taq polymerase (Taq Platinum, Invitrogen). The amplification conditions consisted of 25 cycles (94°C for 30 s, 58°C for 45 s and 72°C for 90 s), preceded by 3 min of denaturation at 94 °C and ending with a 10 min final extension step at 72°C. Amplicons from 5-7 independent PCR products for each sample were pooled together and then purified using the QIAquick PCR purification kit (Qiagen), according to the manufacturer’s instructions. The same amounts (around 200 ng) of purified amplicons from 20 samples were pooled. Amplicons were pyrosequenced using the 454 GS FLX Titanium technology from Roche (Beckman Coulter Genomics).
454 pyrosequence analysis
We obtained a total of 265,899 pyrosequences (Table 1). A series of filters were applied in order to retain only high-quality sequences. First, pyrosequences containing errors in the primer region and positions with undetermined bases were eliminated using a local pipeline (Bachy et al., 2013). The remaining sequences were analyzed with AmpliconNoise (Quince et al., 2011) to further eliminate errors introduced during PCR reactions or 454 sequencing, and build Operational Taxonomic Units (OTUs). Filtered reads were clustered by pairwise alignment and average linkage into OTUs with a 98% similarity cut-off using AmpliconNoise integrated in our local pipeline (Bachy et al., 2013). Singletons, i.e. OTUs composed of only one read, were eliminated for precaution. The most abundant sequence in each OTU was used as reference. OTU reference sequences were blasted against the Silva SSU111 database (Pruesse et al., 2007) and assigned to taxonomic groups based on sequence similarity. The sequences in all OTUs were then attributed to the different samples according to their MIDs. Chimerical OTUs were eliminated by a stringent procedure combining automated and manual steps. OTUs including sequences from at least two different samples were considered to be real. OTUs composed of sequences from only one sample were checked for chimeras using KeyDNATool (http://KeyDNAtools.com). The sequences considered suspect by the software were double checked by comparing BLAST hits recovered from independent sequence fragments (sequences were split in 2 and 3 fragments). Finally, OTUs whose representative sequence had a coverage of less than 90% with its first BLAST hit were eliminated if they were present in only one sample; they were kept but with their taxonomic affiliation changed to “uncertain” if they were present in at least two samples. OTUs affiliated to cryptophyte nucleomorphs were excluded from our analysis. After filtering, we kept 146,549 correct reads (Table 1).
Phylogenetic analyses
Phylogenetic trees were built for each eukaryotic super-group or for several high-rank taxonomic lineages if they comprised only a few OTUs. Analyses included representative sequences of OTUs, their first BLAST hit and sequences from the closest cultured members. Sequences were aligned using Probcons (Do et al., 2005). Positions retained to build trees were selected from the multiple alignments using Gblocks (Castresana, 2000) with the less stringent parameters. Phylogenetic reconstructions were then carried out by maximum likelihood approximation using FastTree (Price et al., 2010). Trees were visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). Sequences with particularly interesting positions in trees were then blasted against the NCBI database (http://blast.ncbi.nlm.nih.gov) to have an insight of their similarity with additionnal sequences in databases. The taxonomic indications given in trees are based on the taxonomic affiliation proposed in the PR2 database (http://ssu-rrna.org/index.html).
Statistical analyses
Statistical analyses were conducted using the R software (http://cran.r-project.org) (R Development Core Team 2011). To assess overall differences between microbial community compositions, we calculated pair-wise Bray-Curtis distances between all samples on the basis of OTU proportions (number of reads from each OTU for each sample normalized to the total number of reads in the corresponding sample) among replicates or sampling sites (Bray et al., 1957). This would avoid heterogeneity due to different numbers of sequences generated per sample. Non-metric multidimensional scaling (NMDS) ordination analyses were conducted based on Bray-Curtis distances (after applying a Wisconsin standardization to balance the influence of the most and least abundant OTUs) using the “Vegan” R package (Oksanen et al., 2013). Correspondence analysis (CoA) on all OTU frequencies for all 12 samples were done using the “Ade4” R package (Dray et al., 2007). Diversity and richness indices were determined using the “Vegan” package. Richness was estimated by rarefaction analysis as the estimated number of OTUs in a random subsample of each sequence library (raw counts of OTUs), of the same size as the smallest one (Hurlbert, 1971). Simpson index is defined as (Simpson, 1949) and evenness was calculated as (Pielou, 1966) with S being the observed number of OTUs and fi the frequency of each OTUi in the sample. Venn diagrams showing the number of OTUs shared by, or exclusive to, the different samples, and heatmaps showing the presence/absence of OTUs were built using the “gplots” package (Bolker et al., 2012). To test whether community composition correlated with environmental parameters in the different samples, we constructed a matrix of Bray-Curtis dissimilarities based on OTU frequencies (sequences from replicate samples were pooled given that they clustered together in previous analyses) and a matrix of Euclidean distances based on physico-chemical parameters for all ecosystems using the “Vegan” package. Both matrices were compared using a Mantel test. The Bray-Curtis matrix was also compared to a matrix of geographical distances between ecosystems. Geographical distances were estimated based on coordinates (http://biodiversityinformatics.amnh.org/open_source/gdmg/, using the mean earth radius (Moritz, 2000) as spheroid. Principal Component Analysis (PCA) and Canonical Correspondence Analysis (CCA) were conducted on centered and scaled physico-chemical parameters and OTU frequencies in replicate samples (0.2-5 μm size fractions) using the “Ade4” package.
Supplementary Material
Acknowledgements
We are thankful to F. Hardy, the Parc Naturel Régional de la Haute Vallée de Chevreuse and the Office National des Forêts du Parc de Rambouillet. We also thank Giselle Walker for critical reading of the manuscript. The research leading to these results received funding from the CNRS EC2CO program and the European Research Council under the European Union’s Seventh Framework Program ERC Grant Agreement 322669 ‘ProtistWorld’.
References
- Adl SM, Simpson AGB, Lane CE, Lukes J, Bass D, Bowser SS, et al. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral-Zettler LA, Rocca JD, Lamontagne MG, Dennett MR, Gast RJ. Changes in microbial community structure in the wake of Hurricanes Katrina and Rita. Environ. Sci. Technol. 2008;42:9072–9078. doi: 10.1021/es801904z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas-Becking L. Geobiologie of inleiding tot de milieukunde. W.P. Van Stock; Zoon, Hague, Netherlands: 1934. [Google Scholar]
- Bachy C, Dolan JR, López-García P, Deschamps P, Moreira D. Accuracy of protist diversity assessments: morphology compared with cloning and direct pyrosequencing of 18S rRNA genes and ITS regions using the conspicuous tintinnid ciliates as a case study. ISME J. 2013;7:244–255. doi: 10.1038/ismej.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass D, Chao EE-Y, Nikolaev S, Yabuki A, Ishida K, Berney C, et al. Phylogeny of Novel Naked Filose and Reticulose Cercozoa: Granofilosea cl. n. and Proteomyxidea Revised. Protist. 2009;160:75–109. doi: 10.1016/j.protis.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Behnke A, Barger KJ, Bunge J, Stoeck T. Spatio-temporal variations in protistan communities along an O/HS gradient in the anoxic Framvaren Fjord (Norway) FEMS Microbiol. Ecol. 2010;72:89–102. doi: 10.1111/j.1574-6941.2010.00836.x. [DOI] [PubMed] [Google Scholar]
- Bolker B, Lodewijk B, Gentleman R, Liaw WHA, Lumley T, Maechler M, et al. gplots: Various R programming tools for plotting data., v2.12.1. 2012 http://cran.r-project.org/web/packages/gplots/index.html.
- Bower SM, Carnegie RB, Goh B, Jones SRM, Lowe GJ, Mak Michelle, S., W. Preferential PCR Amplification of Parasitic Protistan Small Subunit rDNA from Metazoan Tissues. J. Eukaryot. Microbiol. 2004;51:325–332. doi: 10.1111/j.1550-7408.2004.tb00574.x. [DOI] [PubMed] [Google Scholar]
- Bråte J, Logares R, Berney C, Ree DK, Klaveness D, Jakobsen KS, Shalchian-Tabrizi K. Freshwater Perkinsea and marine-freshwater colonizations revealed by pyrosequencing and phylogeny of environmental rDNA. ISME J. 2010;4:1144–1153. doi: 10.1038/ismej.2010.39. [DOI] [PubMed] [Google Scholar]
- Bray JR, Curtis JT. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957;27:325. [Google Scholar]
- Caron DA. Inorganic nutrients, bacteria, and the microbial loop. Microb. Ecol. 1994;28:295–8. doi: 10.1007/BF00166820. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Charvet S, Vincent WF, Comeau A, Lovejoy C. Pyrosequencing analysis of the protist communities in a High Arctic meromictic lake: DNA preservation and change. Front. Microbiol. 2012;3:422. doi: 10.3389/fmicb.2012.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung MK, Au CH, Chu KH, Kwan HS, Wong CK. Composition and genetic diversity of picoeukaryotes in subtropical coastal waters as revealed by 454 pyrosequencing. ISME J. 2010;4:1053–1059. doi: 10.1038/ismej.2010.26. [DOI] [PubMed] [Google Scholar]
- Corpe WA, Jensen TE. An electron microscopic study of picoplanktonic organisms from a Small Lake. Microb. Ecol. 1992;24:181–197. doi: 10.1007/BF00174454. [DOI] [PubMed] [Google Scholar]
- Costanza R. The ecological, economic, and social importance of the oceans. Ecol. Econ. 1999;31:199–213. [Google Scholar]
- Dawson SC, Pace NR. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8324–8329. doi: 10.1073/pnas.062169599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez B, Pedrós-alió C, Massana R. Study of Genetic Diversity of Eukaryotic Picoplankton in Different Oceanic Regions by Small-Subunit rRNA Gene Cloning and Sequencing Study of Genetic Diversity of Eukaryotic Picoplankton in Different Oceanic Regions by Small-Subunit rRNA Gene Cloning and. Appl. Environ. Microbiol. 2001;67:2932–2941. doi: 10.1128/AEM.67.7.2932-2941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do CB, Mahabhashyam MSP, Brudno M, Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing JA, Duarte CM, Tranvik LJ, Striegl RG, McDowell WH, Kortelainen P, et al. The global abundance and size distribution of lakes, ponds, and impoundments. Limnol. Oceanogr. 2006;51:2388–2397. [Google Scholar]
- Dray S, Dufour A-B. The ade4 Package: Implementing the Duality Diagram for Ecologists. J. Stat. Softw. 2007;22:1–20. [Google Scholar]
- Fenchel T. Ecology of Heterotrophic Microflagellates. I. Some Important Forms and Their Functional Morphology. Mar. Ecol. Prog. Ser. 1982;8:211–223. [Google Scholar]
- Finlay B. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- Foissner W. Biogeography and dispersal of micro-organisms: a review emphasizing protists. Acta Protozool. 2006;45:339–366. [Google Scholar]
- Gómez F, Moreira D, Benzerara K, López-García P. Solenicola setigera is the first characterized member of the abundant and cosmopolitan uncultured marine stramenopile group MAST-3. Environ. Microbiol. 2011;13:193–202. doi: 10.1111/j.1462-2920.2010.02320.x. [DOI] [PubMed] [Google Scholar]
- Green JL, Holmes AJ, Westoby M, Oliver I, Briscoe D, Dangerfield M, et al. Spatial scaling of microbial eukaryote diversity. Nature. 2004;432:747–750. doi: 10.1038/nature03034. [DOI] [PubMed] [Google Scholar]
- Groisillier A, Massana R, Valentin K, Vaulot D, Guillou L. Genetic diversity and habitats of two enigmatic marine alveolate lineages. Aquat. Microb. Ecol. 2006;42:277–291. [Google Scholar]
- Guillou L, Chrétiennot-Dinet M-J, Medlin LK, Claustre H, Goër SL, Vaulot D. Bolidomonas: A new genus with two species belonging to a new algal class, the Bolidophyceae (Heterokonta) J. Phycol. 1999;35:368–381. [Google Scholar]
- Guillou L, Viprey M, Chambouvet A, Welsh RM, Kirkham AR, Massana R, et al. Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata) Environ. Microbiol. 2008;10:3349–3365. doi: 10.1111/j.1462-2920.2008.01731.x. [DOI] [PubMed] [Google Scholar]
- Harada A, Ohtsuka S, Horiguchi T. Species of the Parasitic Genus Duboscquella are Members of the Enigmatic Marine Alveolate Group I. Protist. 2007;158:337–347. doi: 10.1016/j.protis.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Hartmann M, Grob C, Tarran GA, Martin AP, Burkill PH, Scanlan DJ, Zubkov MV. Mixotrophic basis of Atlantic oligotrophic ecosystems. Proc. Natl. Acad. Sci. U. S. A. 2012;109:5756–5760. doi: 10.1073/pnas.1118179109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoef-Emden K. Osmotolerance in the Cryptophyceae: Jacks-of-all-trades in the Chroomonas Clade. Protist. 2014;165:123–143. doi: 10.1016/j.protis.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Howe AT, Bass D, Scoble JM, Lewis R, Vickerman K, Arndt H, Cavalier-Smith T. Novel Cultured Protists Identify Deep-branching Environmental DNA Clades of Cercozoa: New Genera Tremula, Micrometopion, Minimassisteria, Nudifila, Peregrinia. Protist. 2011;162:332–372. doi: 10.1016/j.protis.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Hurlbert SH. The nonconcept of species diversity: a critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- Jardillier L, Zubkov MV, Pearman J, Scanlan DJ. Significant CO2 fixation by small prymnesiophytes in the subtropical and tropical northeast Atlantic Ocean. ISME J. 2010;4:1180–1192. doi: 10.1038/ismej.2010.36. [DOI] [PubMed] [Google Scholar]
- Johnson PW, Sieburth JM. In-situ morphology and occurrence of eukaryotic phototrophs of bacterial size in the picoplankton of estuarine and oceanic waters. J. Phycol. 1982;18:318–327. [Google Scholar]
- Jones MDM, Forn I, Gadelha C, Egan MJ, Bass D, Massana R, Richards T. a. Discovery of novel intermediate forms redefines the fungal tree of life. Nature. 2011;474:200–203. doi: 10.1038/nature09984. [DOI] [PubMed] [Google Scholar]
- Karpov SA, Mamkaeva MA, Aleoshin VV, Nassonova E, Lilje O, Gleason FH. Morphology, phylogeny, and ecology of the aphelids (Aphelidea, Opisthokonta) and proposal for the new superphylum Opisthosporidia. Front. Microbiol. 2014;5:112. doi: 10.3389/fmicb.2014.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham AR, Lepère C, Jardillier LE, Not F, Bouman H, Mead A, Scanlan DJ. A global perspective on marine photosynthetic picoeukaryote community structure. ISME J. 2013;7:922–936. doi: 10.1038/ismej.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight-Jones EW, Walne PR. Chromulina pusilla Butcher, a Dominant Member of the Ultraplankton. Nature. 1951;167:445–446. doi: 10.1038/167445a0. [DOI] [PubMed] [Google Scholar]
- Kolodziej K, Stoeck T. Cellular identification of a novel uncultured marine stramenopile (MAST-12 Clade) small-subunit rRNA gene sequence from a norwegian estuary by use of fluorescence in situ hybridization-scanning electron microscopy. Appl. Environ. Microbiol. 2007;73:2718–2726. doi: 10.1128/AEM.02158-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara E, Mitchell EAD, Moreira D, López-García P. Highly Diverse and Seasonally Dynamic Protist Community in a Pristine Peat Bog. Protist. 2011;162:14–32. doi: 10.1016/j.protis.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Lara E, Moreira D, López-García P. The Environmental Clade LKM11 and Rozella Form the Deepest Branching Clade of Fungi. Protist. 2010;161:116–121. doi: 10.1016/j.protis.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Leander BS, Kuvardina ON, Aleshin VV, Mylkinov AP, Keeling PJ. Molecular Phylogeny and Surface Morphology of Colpodella edax (Alveolata): Insights into the Phagotrophic Ancestry of Apicomplexans. J. Eukaryot. Microbiol. 2003;50:334–340. doi: 10.1111/j.1550-7408.2003.tb00145.x. [DOI] [PubMed] [Google Scholar]
- Lefèvre E, Bardot C, Noël C, Carrias J-F, Viscogliosi E, Amblard C, Sime-Ngando T. Unveiling fungal zooflagellates as members of freshwater picoeukaryotes: evidence from a molecular diversity study in a deep meromictic lake. Environ. Microbiol. 2007;9:61–71. doi: 10.1111/j.1462-2920.2006.01111.x. [DOI] [PubMed] [Google Scholar]
- Lefèvre E, Roussel B, Amblard C, Sime-Ngando T. The molecular diversity of freshwater picoeukaryotes reveals high occurrence of putative parasitoids in the plankton. PLoS One. 2008;3:e2324. doi: 10.1371/journal.pone.0002324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc M, Thénot A. Genetic diversity of small eukaryotes in lakes differing by their trophic status. Appl. Environ. Microbiol. 2005;71:5935–5942. doi: 10.1128/AEM.71.10.5935-5942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepère C, Boucher D. Succession and regulation factors of small eukaryote community composition in a lacustrine ecosystem (Lake Pavin) Appl. Environ. Microbiol. 2006;72:2971–2981. doi: 10.1128/AEM.72.4.2971-2981.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepère C, Domaizon I, Debroas D. Unexpected importance of potential parasites in the composition of the freshwater small-eukaryote community. Appl. Environ. Microbiol. 2008;74:2940–2949. doi: 10.1128/AEM.01156-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepère C, Domaizon I, Taib N, Mangot J-F, Bronner G, Boucher D, Debroas D. Geographic distance and ecosystem size determine the distribution of smallest protists in lacustrine ecosystems. FEMS Microbiol. Ecol. 2013;85:85–94. doi: 10.1111/1574-6941.12100. [DOI] [PubMed] [Google Scholar]
- Li WKW. Primary production of prochlorophytes, cyanobacteria, and eucaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnol. Oceanogr. 1994;39:169–175. [Google Scholar]
- Logares R, Brate J, Bertilsson S, Clasen JL, Shalchian-Tabrizi K, Rengefors K. Infrequent marine-freshwater transitions in the microbial world. Trends Microbiol. 2009;17:414–422. doi: 10.1016/j.tim.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Logares R, Shalchian-Tabrizi K, Boltovskoy A, Rengefors K. Extensive dinoflagellate phylogenies indicate infrequent marine-freshwater transitions. Mol. Phylogenet. Evol. 2007;45:887–903. doi: 10.1016/j.ympev.2007.08.005. [DOI] [PubMed] [Google Scholar]
- López-García P, Moreira D. Tracking microbial biodiversity through molecular and genomic ecology. Res. Microbiol. 2008;159:67–73. doi: 10.1016/j.resmic.2007.11.019. [DOI] [PubMed] [Google Scholar]
- López-García P, Rodríguez-Valera F, Pedrós-Alió C, Moreira D. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature. 2001;409:603–607. doi: 10.1038/35054537. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11436–40. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Bock C, Li H, Padisák J, Krienitz L. Molecular and microscopic diversity of planktonic eukaryotes in the oligotrophic Lake Stechlin (Germany) Hydrobiologia. 2011;661:133–143. [Google Scholar]
- Mangot J-F, Domaizon I, Taib N, Marouni N, Duffaud E, Bronner G, Debroas D. Short-term dynamics of diversity patterns: evidence of continual reassembly within lacustrine small eukaryotes. Environ. Microbiol. 2013;15:1745–1758. doi: 10.1111/1462-2920.12065. [DOI] [PubMed] [Google Scholar]
- Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, et al. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 2006;4:102–12. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- Massana R. Eukaryotic picoplankton in surface oceans. Annu. Rev. Microbiol. 2011;65:91–110. doi: 10.1146/annurev-micro-090110-102903. [DOI] [PubMed] [Google Scholar]
- Massana R, Balagué V, Guillou L, Pedrós-Alió C. Picoeukaryotic diversity in an oligotrophic coastal site studied by molecular and culturing approaches. FEMS Microbiol. Ecol. 2004;50:231–243. doi: 10.1016/j.femsec.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Massana R, Del Campo J, Sieracki ME, Audic S, Logares R. Exploring the uncultured microeukaryote majority in the oceans: reevaluation of ribogroups within stramenopiles. ISME J. 2014;8:854–66. doi: 10.1038/ismej.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R, Castresana J, Balagué V, Romari K, Groisillier A, Valentin K, et al. Phylogenetic and ecological analysis of novel marine stramenopiles. Appl. Environ. Microbiol. 2004;70:3528–3234. doi: 10.1128/AEM.70.6.3528-3534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R, Guillou L, Diez B, Pedrós-Alió C. Unveiling the Organisms behind Novel Eukaryotic Ribosomal DNA Sequences from the Ocean. Appl. Environ. Microbiol. 2002;68:4554–4558. doi: 10.1128/AEM.68.9.4554-4558.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R, Terrado R, Forn I, Lovejoy C, Pedrós-Alió C. Distribution and abundance of uncultured heterotrophic flagellates in the world oceans. Environ. Microbiol. 2006;8:1515–1522. doi: 10.1111/j.1462-2920.2006.01042.x. [DOI] [PubMed] [Google Scholar]
- Monchy S, Sanciu G, Jobard M, Rasconi S, Gerphagnon M, Chabé M, et al. Exploring and quantifying fungal diversity in freshwater lake ecosystems using rDNA cloning/sequencing and SSU tag pyrosequencing. Environ. Microbiol. 2011;13:1433–1453. doi: 10.1111/j.1462-2920.2011.02444.x. [DOI] [PubMed] [Google Scholar]
- Moon-van der Staay SY, Wachter R. De, Vaulot D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature. 2001;409:607–610. doi: 10.1038/35054541. [DOI] [PubMed] [Google Scholar]
- Moreira D, López-García P. The molecular ecology of microbial eukaryotes unveils a hidden world. Trends Microbiol. 2002;10:31–38. doi: 10.1016/s0966-842x(01)02257-0. [DOI] [PubMed] [Google Scholar]
- Moreira D, López-García P. The rise and fall of Picobiliphytes: How assumed autotrophs turned out to be heterotrophs. Bioessays. 2014;36:468–474. doi: 10.1002/bies.201300176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz H. Geodetic Reference System 1980. J. Geod. 2000;74:128–133. [Google Scholar]
- Nakai R, Abe T, Baba T, Imura S, Kagoshima H, Kanda H, et al. Eukaryotic phylotypes in aquatic moss pillars inhabiting a freshwater lake in East Antarctica, based on 18S rRNA gene analysis. Polar Biol. 2012;35:1495–1504. [Google Scholar]
- Nolte V, Pandey R, Jost S. Contrasting seasonal niche separation between rare and abundant taxa conceals the extent of protist diversity. Mol. Ecol. 2010;19:2908–2915. doi: 10.1111/j.1365-294X.2010.04669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Not F, Gausling R. Vertical distribution of picoeukaryotic diversity in the Sargasso Sea. Environ. 2007;9:1233–1252. doi: 10.1111/j.1462-2920.2007.01247.x. [DOI] [PubMed] [Google Scholar]
- Oikonomou A, Katsiapi M, Karayanni H, Moustaka-Gouni M, Kormas KA. Plankton microorganisms coinciding with two consecutive mass fish kills in a newly reconstructed lake. Scientific World Journal. 2012;2012:504135. doi: 10.1100/2012/504135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin R,P, O’Hara RB, et al. Community Ecology Package; vegan: 2013. [Google Scholar]
- Patterson DJ, Larsen J. Biology of free-living heterotrophic flagellates. Clarendon Press; Oxford: 1991. (Systematics Association Special volume 45). [Google Scholar]
- Pielou EC. The measurement of diversity in different types of biological collections. J. Theor. Biol. 1966;13:131–144. [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R: A language and environment for statistical computing. 2013. [Google Scholar]
- Ragon M, Fontaine MC, Moreira D, López-García P. Different biogeographic patterns of prokaryotes and microbial eukaryotes in epilithic biofilms. Mol. Ecol. 2012;21:3852–3868. doi: 10.1111/j.1365-294X.2012.05659.x. [DOI] [PubMed] [Google Scholar]
- Richards TA. The molecular diversity of freshwater picoeukaryotes from an oligotrophic lake reveals diverse, distinctive and globally dispersed lineages. Environ Microbiol. 2005;7:1413–1425. doi: 10.1111/j.1462-2920.2005.00828.x. [DOI] [PubMed] [Google Scholar]
- Ritchie RJ. Consistent sets of spectrophotometric chlorophyll equations for acetone, methanol and ethanol solvents. Photosynth. Res. 2006;89:27–41. doi: 10.1007/s11120-006-9065-9. [DOI] [PubMed] [Google Scholar]
- Romari K, Vaulot D. Composition and temporal variability of picoeukaryote communities at a coastal site of the English Channel from 18S rDNA sequences. Limnol. Oceanogr. 2004;49:784–798. [Google Scholar]
- Scheckenbach F, Hausmann K, Wylezich C, Weitere M, Arndt H. Large-scale patterns in biodiversity of microbial eukaryotes from the abyssal sea floor. Proc. Natl. Acad. Sci. U. S. A. 2010;107:115–120. doi: 10.1073/pnas.0908816106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seenivasan R, Sausen N, Medlin LK, Melkonian M. Picomonas judraskeda gen. et sp. nov.: the first identified member of the Picozoa phylum nov., a widespread group of picoeukaryotes, formerly known as “picobiliphytes”. PLoS One. 2013;8:e59565. doi: 10.1371/journal.pone.0059565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, Reier-Røberg K, Ree DK, Klaveness D, Bråte J. Marine-freshwater colonizations of haptophytes inferred from phylogeny of environmental 18S rDNA sequences. J. Eukaryot. Microbiol. 2011;58:315–318. doi: 10.1111/j.1550-7408.2011.00547.x. [DOI] [PubMed] [Google Scholar]
- Shi X, Marie D, Jardillier L, Scanlan D, Vaulot D. Groups without cultured representatives dominate eukaryotic picophytoplankton in the oligotrophic South East Pacific Ocean. PLoS One. 2009;4:e7657. doi: 10.1371/journal.pone.0007657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simó R. Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol. Evol. 2001;16:287–294. doi: 10.1016/s0169-5347(01)02152-8. [DOI] [PubMed] [Google Scholar]
- Simon M, López-García P, Moreira D, Jardillier L. New haptophyte lineages and multiple independent colonizations of freshwater ecosystems. Environ. Microbiol. Rep. 2013;5:322–332. doi: 10.1111/1758-2229.12023. [DOI] [PubMed] [Google Scholar]
- Simpson EH. Measurement of Diversity. Nature. 1949;163:688–688. [Google Scholar]
- Slapeta J, López-García P, Moreira D. Global dispersal and ancient cryptic species in the smallest marine eukaryotes. Mol. Biol. Evol. 2006;23:23–29. doi: 10.1093/molbev/msj001. [DOI] [PubMed] [Google Scholar]
- Slapeta J, Moreira D, López-García P. The extent of protist diversity: insights from molecular ecology of freshwater eukaryotes. Proc. Biol. Sci. 2005;272:2073–2081. doi: 10.1098/rspb.2005.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockner JG. Phototrophic picoplankton: an overview from marine and freshwater ecosystems. Limnol. Oceanogr. 1988;33:765–775. [Google Scholar]
- Stoeck T, Taylor GT, Epstein SS. Novel Eukaryotes from the Permanently Anoxic Cariaco Basin (Caribbean Sea) Appl. Environ. Microbiol. 2003;69:5656–5663. doi: 10.1128/AEM.69.9.5656-5663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taib N, Mangot J-F, Domaizon I, Bronner G, Debroas D. Phylogenetic affiliation of SSU rRNA genes generated by massively parallel sequencing: new insights into the freshwater protist diversity. PLoS One. 2013;8:e58950. doi: 10.1371/journal.pone.0058950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triadó-Margarit X, Casamayor EO. Genetic diversity of planktonic eukaryotes in high mountain lakes (Central Pyrenees, Spain) Environ Microbiol. 2012;14:2445–2456. doi: 10.1111/j.1462-2920.2012.02797.x. [DOI] [PubMed] [Google Scholar]
- Wright RT, Coffin RB. Measuring microzooplankton grazing on planktonic marine bacteria by its impact on bacterial production. Microb. Ecol. 1984;10:137–149. doi: 10.1007/BF02011421. [DOI] [PubMed] [Google Scholar]
- Yuan CL, Keeling PJ, Krause PJ, Horak A, Bent S, Rollend L, Hua XG. Colpodella spp.-like parasite infection in woman, China. Emerg. Infect. Dis. 2012;18:125–127. doi: 10.3201/eid1801.110716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Chen M, Sun Y, Yang J, Chen F. Genetic diversity of picoeukaryotes in eight lakes differing in trophic status. Can. J. Microbiol. 2011;57:115–126. doi: 10.1139/w10-107. [DOI] [PubMed] [Google Scholar]
- Zubkov MV, Tarran GA. High bacterivory by the smallest phytoplankton in the North Atlantic Ocean. Nature. 2008;455:224–226. doi: 10.1038/nature07236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.