Abstract
The importance of autophagy in memory CD8 T cell differentiation in vivo is not well defined. We show here that autophagy is dynamically regulated in virus-specific CD8 T cells during acute lymphocytic choriomeningitis virus infection. Autophagy decreased in activated proliferating T cells, and was then upregulated at the peak of the effector T cell response. Consistent with this model, deletion of the key autophagy genes Atg7 or Atg5 in virus-specific CD8 T cells had minimal effect on generating effector cells but greatly enhanced their death during the contraction phase resulting in compromised memory formation. These findings provide insight into when autophagy is needed during effector and memory T cell differentiation in vivo and also warrant a re-examination of our current concepts about the relationship between T cell activation and autophagy.
CD8 T cells provide protection against intracellular bacterial, parasitic, and viral infections, as well as cancers1, 2. Following antigen stimulation, naive CD8 T cells go through many rounds of proliferation, giving rise to effector T cells, which are efficiently eliminate infected cells. Upon antigen clearance, the vast majority of effector CD8 T cells undergo apoptosis, leaving only a small pool of cells to survive and differentiate into memory cells3, 4, 5. During this naive to effector to memory differentiation process, T cells undergo cellular and metabolic reprogramming shifting from anabolic processes and proliferation to catabolic processes and contraction of cell populations to generate memory. It is important to define the role of macroautophagy (herein, “autophagy”) during this process.
Autophagy is an evolutionarily conserved process involving the engulfment and delivery of cytosolic contents to the lysosome for degradation 6, 7, 8, 9, 10. This catabolic activity of autophagy is essential for cellular homeostasis and has been suggested to be inversely correlated with cell growth and proliferation11. In contrast to this paradigm, it has been reported that autophagy is up-regulated in proliferating T cells9, 12, 13. T cell receptor (TCR) stimulation promotes activation and proliferation of T cells and also induces the metabolic checkpoint kinase mTOR signaling which would be expected to inhibit rather than induce autophagy8. Thus, major questions remain related to why and how proliferating T cells up-regulate autophagy in the presence of positive mTOR signals when cells need more proteins and organelles to donate to daughter cells. Furthermore, because autophagy has been predominantly studied in vitro during T cell activation little is known about in vivo autophagy activity in antigen specific T cells during the course of effector and memory T cell differentiation after viral infections.
The in vivo functional role of autophagy in antigen specific T cells during viral infections remains unclear, but is important as pharmacologic manipulation of autophagy is being considered for many human diseases14. Conditional knockout mice in which either of the key autophagy genes Atg5 or Atg7 genes were selectively deleted during early T cell development using Lck-cre decreases mature peripheral T cells10, 15. Similarly, reduced peripheral T cells were observed in Atg5−/− chimeric mice after transplantation of fetal liver cells from Atg5−/− mice9. This study also showed that Atg5-deficient T cells exhibited a reduced proliferative capacity following in vitro TCR stimulation9. While these data indicate that autophagy plays a key role in T cell development and homeostasis, they shed less light on the function of autophagy genes in T cells responding to antigen because the cells studied had developed in the absence of autophagy genes such as Atg5 or Atg7 and exhibit abnormalities in gene expression and mitochondrial numbers and function 10, 15. Thus, a new approach using phenotypically normal naive T cells is required to study the functional role of autophagy during T cell activation in vivo.
Here, we examined two important questions; the kinetics of autophagy activity and the role of autophagy during response to lymphocytic choriomeningitis virus (LCMV). In contrast to previous observations, we found that autophagy was inversely correlated to T cell proliferation and was significantly inhibited during the early effector stage (i.e., when the cells are highly proliferative). Autophagy activity was then up-regulated at the peak of expansion (i.e., when antigen is cleared and antigen-specific cells about to undergo contraction phase). We also established the significance of autophagy proteins for the development of CD8 T cell memory in vivo by knocking out either one of the two essential autophagy genes, Atg5 and Atg7 using granzyme B cre system in which normal naive T cells were developed and autophagy genes were deleted only after T cells were activated with antigen. Our study provides important insight into the kinetics and functional role of autophagy in antigen specific CD8 T cells during effector and memory differentiation.
RESULTS
Dynamic regulation of autophagy in virus specific T cells
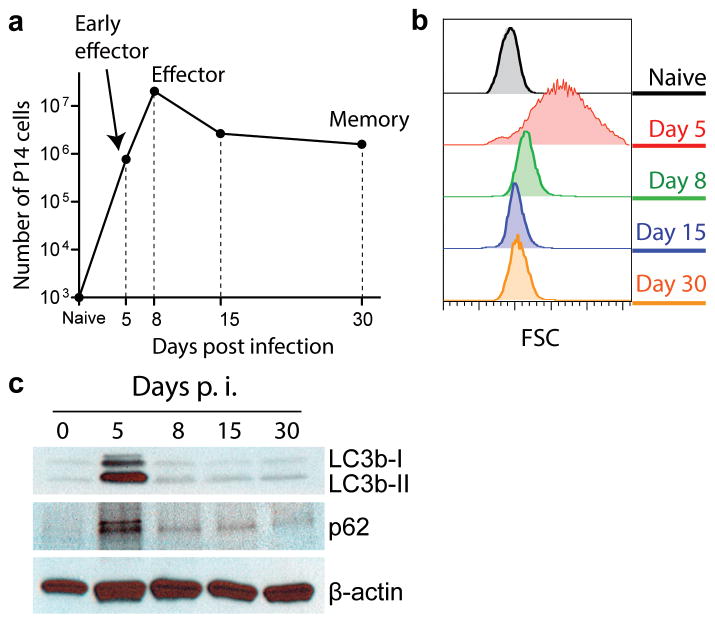
During an acute viral infection, naive CD8 T cells undergo vigorous clonal expansion, followed by contraction, in which a small percentage of effector cells survive to establish memory (Fig. 1a). To study autophagy in antigen-specific CD8 T cells through the distinct phases of the T cell response, we took several different approaches to analyze the autophagy pathway and autophagic flux in antigen-specific CD8 T cells following acute infection with lymphocytic choriomeningitis virus (LCMV) Armstrong strain. We isolated transgenic CD8 T cells that recognize the LCMV GP33-41 peptide (P14 cells) at distinct stages of the T cell response: the expansion phase; i.e., cells with a blasting phenotype (day 5 post infection (p.i.) when virus specific effector CD8 T cells are actively proliferating); the peak of expansion (day 8 p.i. when effector CD8 T cells stop proliferating and decrease in cell size); the contraction phase (day 15 p.i.); and the memory phase (day 30 p.i.) (Fig. 1a and b). To assess autophagy activity, we first examined the amount of microtubule-associated protein light chain 3 beta-I (LC3b-I) and LC3b-II in the sorted P14 cells. LC3b-II, the lipidated form of LC3b-I, is a classic marker for autophagosomes, and is incorporated into the elongating membrane that eventually forms the autophagosome and is subsequently degraded after delivery to the lysosome 7 (Supplementary Fig. 1a). The quantities of both LC3b-I and LC3b-II in antigen-specific CD8 T cells peaked at day 5 p.i. (Fig. 1c). The accumulation of LC3b proteins in P14 cells without an increase in the level of LC3b transcripts at day 5 p.i. (Supplementary Fig. 1b) would be consistent with either an induction of autophagy and increased production of these proteins or an inhibition of lysosomal degradation of these proteins.
FIGURE 1.
Analysis of autophagy in virus-specific CD8 T cells during a viral infection. (a) Kinetics of P14 cells after LCMV Armstrong infection. 10,000 P14 cells were adoptively transferred into B6 mice and were tracked in spleens over the time course of LCMV Armstrong infection. (b) Flow cytometry histograms indicate cell size by showing forward scatters of P14 cells at different stages of differentiation as indicated. (c) Immunoblot data of LC3b and p62 at different stages of P14 differentiation. (a), (b) and (c) are representative of two independent experiments of samples pooled from 5–20 mice for each time point.
To further assess autophagy activity, we therefore examined p62 (also known as Sequestosome-1) protein expression. p62 is a widely used marker for measuring autophagic activity since it is both an autophagy substrate and acts as an adaptor in targeting ubiquitinated proteins for lysosomal degradation via the autophagy pathway18 (Supplementary Fig. 1a). Similar to LC3b, p62 protein showed the highest accumulation at day 5 p.i., which was not associated with an increase in the corresponding mRNA (Fig. 1c and Supplementary Fig. 1c). In addition, we examined if LC3b and p62 proteins were accumulated at a much earlier stage of T cell activation. In these experiments, LCMV GP33-41 peptide was injected into P14 transgenic mice to activate all P14 cells in a short synchronized timeframe. Similar to what was observed in the activated P14 cells obtained from day 5 p.i., we detected higher amounts of both LC3b and p62 proteins at 20 hours post activation without an increase of their mRNAs (Supplementary Fig. 1d and e). Together these data suggested an altered flux in the autophagy pathway at the early effector stage of the T cell response, and were most consistent with a reduction of autophagy during antigen-driven T cell activation in vivo.
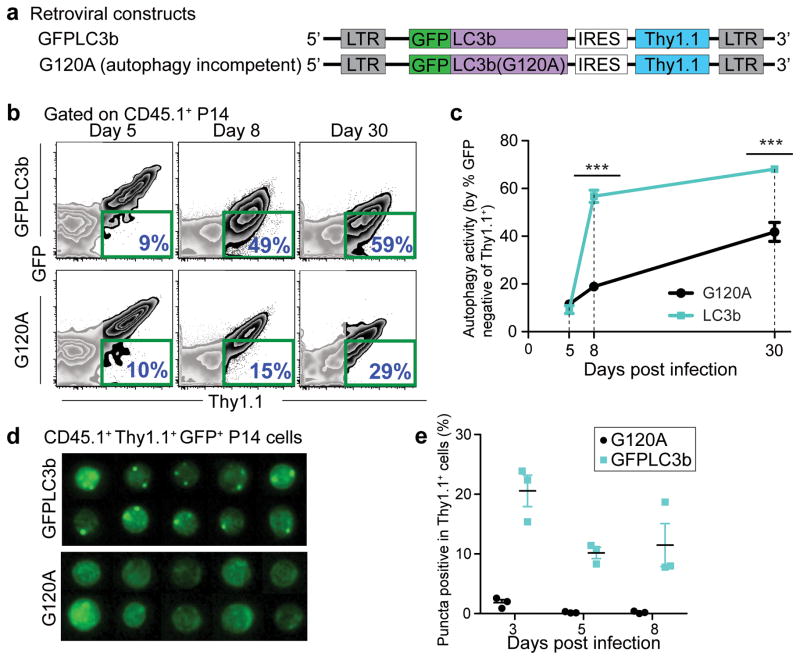
To further define how the highly dynamic autophagy pathway is regulated in vivo during the CD8 T cell response, we employed a reporter system for autophagy activity by fusing a recombinant mouse LC3b protein to green fluorescent protein (GFP) (Fig. 2a) 7. This fusion gene was introduced into P14 cells with retroviral vector system, MSCV-IRES-Thy1.1 (MIT), by which MIT-transduced P14 cells can be distinguished by the expression of Thy1.1 (Fig. 2a and Supplementary Fig. 2a). This reporter system allowed us to quantify autophagy activity by simply measuring GFP expression by flow cytometry; high autophagy activity was reflected by the reduction in GFP intensity due to either the enhanced degradation of GFP-LC3b protein through the autophagy pathway or fluorescence quenching of GFP by low pH after fusion of autophagosomes with lysosomes19. As a control we used a G120A mutant of GFP-LC3b, since this mutation removes the C-terminal amino acid to which phosphatidylethanoloamine is conjugated during the generation of LC3b-II from LC3b-I, resulting in this molecule failing to be incorporated into autophagosomes (Fig. 2a). Using this reporter system, we assessed autophagy activity at different times following LCMV infection. Consistent with our analysis of LC3b and p62, at day 5 p.i., GFP from either the WT or mutant LC3b was comparable, indicating lower autophagic flux (Fig. 2b). Similar amounts of GFP expression between WT and mutant LC3b were also observed at much earlier times post infection (days 2 and 3) (Supplementary Fig. 2b–e). However, at day 8 p.i., in contrast to days 2 to 5, there was a profound difference between the WT and mutant LC3b proteins; we observed a drastic increase in the percentage of GFP-negative P14 cells in the WT relative to the G120A mutant, suggesting a significantly elevated flux through the autophagy pathway at day 8 compared to day 5 (Fig. 2b and c). This profound difference in GFP intensity between the WT and mutant LC3b groups remained through to day 30 p.i. (Fig. 2b and c). In addition to this P14 transgenic system, we observed a similar change in the autophagy flux in endogenous LCMV specific tetramer-positive CD8 T cells (Supplemental Fig. 3). To apply the reporter system to endogenous cells, hematopoietic stem cells (HSCs) were transduced with MIT containing either WT or G120A mutant GFP-LC3b, followed by adoptive transfer of these HSCs into lethally irradiated mice (Supplementary Fig. 3a). After reconstitution, mice were infected with LCMV Armstrong and then GFP expression was monitored in MIT-transduced DbGP33 tetramer-positive CD8 T cells (Supplementary Fig. 3a). Similar to the results obtained from P14 transgenic T cells, GFP expression in DbGP33+ cells transduced with MIT carrying WT LC3b were comparable to those with MIT carrying G120A mutant at day 6 p.i., and a significant decrease in WT GFP-LC3b expression was observed at days 8, 15 and 30 p.i. (Supplementary Fig. 3b). Taken together, these data indicate that autophagy is decreased in antigen-specific CD8 T cells during the clonal expansion phase, but is significantly increased following the expansion phase and into the memory phase.
FIGURE 2.
Autophagic flux in virus-specific CD8 T cells is inversely correlated with cell proliferation status. (a) MSCV retroviral constructs showing the transgenes (GFP-LC3b and GFP-LC3b-G120A) introduced in P14 cells to probe for autophagy activity. (b) Flow cytometry plots of adoptively transferred P14 cells in spleens. P14 cells transduced with retroviral plasmid of either MIT-GFP-LC3b or MIT-G120A are positive for the congenic marker Thy1.1. Day 5, 8 and 30 p.i. splenocytes were used for the analysis. The percentage of GFP-negative cells out of the transduced P14 (Thy1.1+) cells from each group is highlighted in blue on the lower right corner of each plot and is summarized in (c). (d) Representative ImageStream® data showing images of P14 cells from either GFP-LC3b or G120A groups on day 8 p.i.. Images showing GFP signals of GFP+ MIT vector-transduced P14 cells (Ly5.1+ Thy1.1+). Summary graph of ImageStream® analysis is plotted in (e) showing percentage of GFP+ or Thy1.1+ P14 cells that exhibited more than one GFP punctum. Errors bars represent SEM. (b) and (d) are representative of three independent experiments, n≥3 mice in each group.
As an additional assessment of autophagy, we visualized GFP-LC3b puncta using multi-spectral imaging flow cytometry (ImageStream®) to quantify the number of LC3-positive autophagosomes; GFP puncta reflected the co-localization of lipidated GFP-LC3b incorporated onto the elongating phagophore membrane that subsequently forms the autophagosomes. On days 3, 5, and 8 p.i., we observed puncta in P14 cells transduced with WT GFP-LC3b, although a slightly higher frequency of puncta was detected on day 3 p.i. (Fig. 2d and e). These data indicate that antigen specific CD8 T cells can form autophagosomes during the T cell expansion phase. Collectively our results suggest that the low autophagy activity during clonal expansion is due to reduced flux through the lysosomal degradation pathway and that upon conclusion of the clonal expansion phase (day 8 p.i.) autophagic flux is significantly increased.
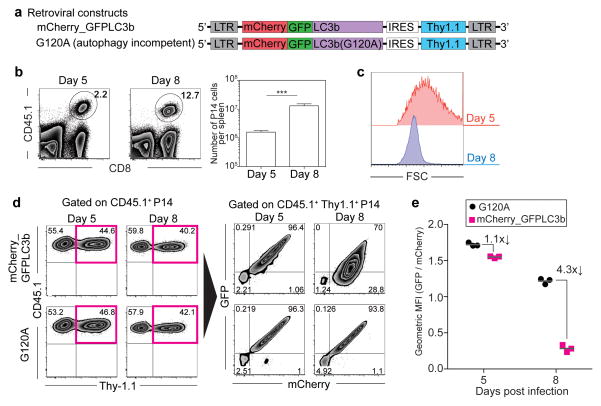
Our results concerning the in vivo kinetics of autophagy activity in virus specific CD8 T cells were unexpected because prior in vitro studies showed that T cells increased autophagy after T cell receptor (TCR) stimulation6, 7, 9, 12, 13. To further confirm our findings and examine if autophagolysosome formation is indeed inhibited during the clonal expansion phase, we made retrovirus vectors by fusing mCherry fluorescent protein into the GFP-LC3b protein used above (Fig. 3a). Since mCherry shows more stable fluorescence compared to GFP under acidic condition such as following fusion of autophagosomes to lysosomes 7, 20, 21, comparing the amount of GFP fluorescence relative to mCherry allowed us to measure whether LC3b had been delivered from autophagosomes into acidic autophagolysosomes. In this system, enhanced autophagic flux results in reduced GFP fluorescence compared to mCherry fluorescence due to delivery of the LC3b protein into an acidic environment 7, 20, 21. Using this system, we examined autophagolysosome formation in antigen specific CD8 T cells during the T cell expansion phase (day 5) and at the peak of T cell responses (day 8) (Fig. 3b). Consistent with our previous results, proliferating day 5 effector CD8 T cells were larger in size compared to effector CD8 T cells at day 8 p.i. when these cells had stopped proliferating (Fig. 3c). The GFP/mCherry fluorescence intensity ratio in retrovirus-transduced P14 cells with WT LC3b was very similar to that in P14 cells with G120A mutant LC3b at day 5 p.i. (Fig. 3d and e), indicating that autophagy decreased in proliferating virus specific CD8 T cells. In contrast, at day 8 post infection, GFP fluorescence was specifically decreased compared to mCherry fluorescence in the WT LC3b group (Fig. 3d) and the GFP/mCherry fluorescence intensity ratio showed a 4.3-fold decrease between WT and G120A groups (Fig. 3e). These results are consistent with increased delivery of LC3b into autophagolysosomes due to induction of autophagy between 5 and 8 days after infection. Taken together (Figs. 1–3, Supplementary Figs. 1–3), autophagic flux is inhibited during T cell clonal expansion phase via impairing autophagosome maturation into autophagolysosomes, followed by a rapid induction of autophagy concurrent with the peak of CD8 T cell effector responses that occurs at day 8 post infection.
FIGURE 3.
Autophagic flux is inhibited during T cell clonal expansion phase via impairing autophagosome maturation into autophagolysosomes. (a) mCherry_GFP-LC3b MSCV constructs used in experiments. Retrovirus transduction of P14 cells were performed as shown in Supplementary Fig. 2a. (b) Representative flow cytometry plots of adoptively transferred P14 cells (CD45.1+) in spleens at days 5 and 8 post infection. Percentage of P14 cells in total splenocytes is shown. The right panel represents the average number of P14 cells per spleen. n=6 mice. Error bars indicate SEM. (c) Representative flow cytometry histograms indicate cell size (forward scatters) of P14 cells at days 5 and 8 post infection. (d) Retrovirus-transduction efficiency (CD45.1+ Thy-1.1+) is shown in the left panel. GFP expression is compared with mCherry expression in the right panel in retrovirus transduced P14 cells. Ratio of geometric mean fluorescence intensity (GFP/mCherry) summarized in (e). Data are representative of two independent experiments, n≥3 mice in each group.
Atg7 and Atg5 are critical for CD8 T cell memory formation
To determine the importance of the increased autophagy activity during the transition phase from effector to memory CD8 T cells, we generated conditional knockout mice by crossing Atg7fl/fl or Atg5fl/fl mice to transgenic mice carrying the Cre recombinase gene driven by the granzyme B promoter (Gzmb-Cre)22, 23, 24. In this system, the critical autophagy genes, Atg5 or Atg7, are specifically deleted upon expression of the granzyme B gene in activated CD8 T cells. Herein, we refer to these genes as Atg7fl/flGzmb-Cre and Atg5fl/flGzmb-Cre.
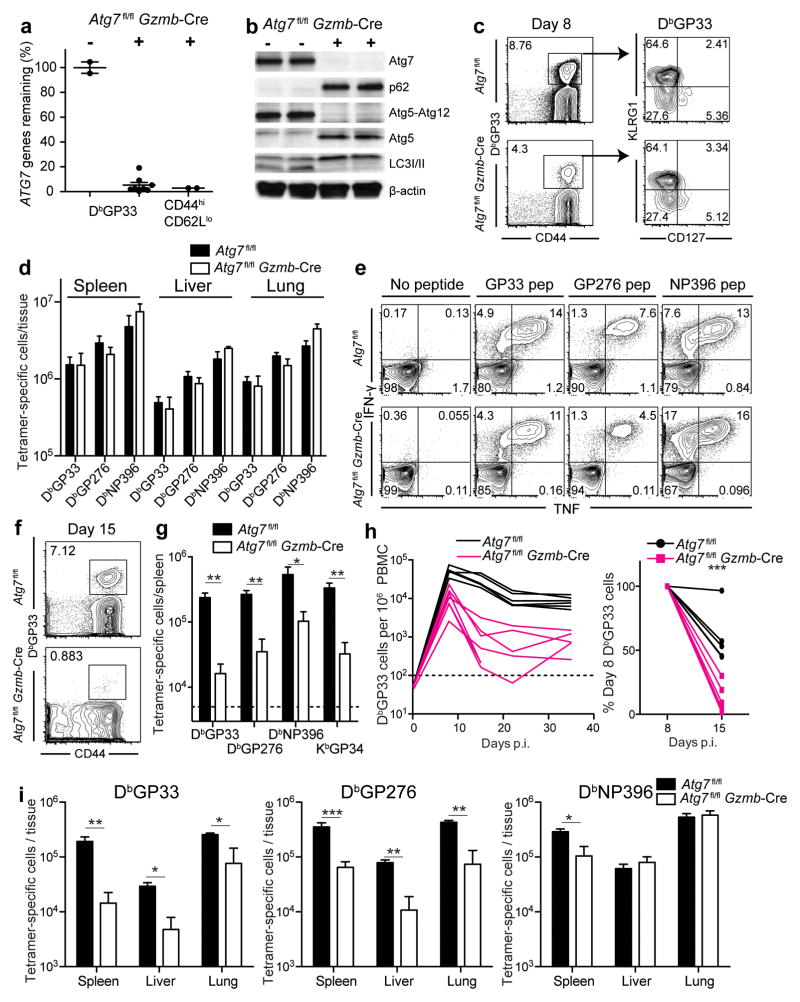
We infected Atg7fl/flGzmb-Cre mice with the LCMV Armstrong strain. In order to establish the efficiency of the Gzmb-Cre system in the context of Atg7fl/flGzmb-Cre mice, we isolated DbGP33+ CD8 T cells 8 days following infection (Supplementary Fig. 4a). Assessment of gene deletion using quantitative PCR showed a greater than 20-fold reduction in the genomic copies of the Atg7-flox allele in the DbGP33-specific CD8 T cells, demonstrating a 95% deletion of the floxed allele (Fig. 4a). A similar level of Atg7-flox gene deletion was also observed in the bulk activated CD8 T cell (CD44hiCD62Llo) population, >95% of which were LCMV-specific and expressing granzyme B at day 8 p.i.25 (Fig. 4a and Supplementary Fig. 4b). The extent of Atg7 gene deletion was also reflected by the Atg7 protein within the bulk activated CD8 T cells, whereby Atg7 protein was hardly discernible in the CD44hiCD62Llo effector CD8 T cell population from Atg7fl/flGzmb-Cre mice. We confirmed the deletion of Atg7 protein by examining its catalytic activity. Atg7 catalyzes reactions resulting in Atg5-Atg12 conjugation and lipidation of LC3-I to give rise to LC3b-II6. The enzymatic activity of Atg7 in the effector CD8 T cells was significantly diminished, as indicated by the absence of both Atg5-Atg12 conjugation, as well as lipidated LC3-II (Fig. 4b). Instead, we detected Atg5 protein in abundance as an un-conjugated species (Fig. 4b). Furthermore, p62 accumulated in the absence of Atg7, indicative of defects in autophagic activity (Fig. 4b). Together, these data indicated the high efficiency of the Cre-loxP system in deleting the target Atg7-flox allele in LCMV-specific CD8 T cells and the consequent loss of autophagy at a time point (day 8) when autophagy would normally be increased. In addition, these data validate the assays of LC3b and p62 used above to measure autophagic flux in primary T cells in vivo.
FIGURE 4.
Atg7 deficiency results in survival defects of the effector CD8 T cells during the effector to memory transition. Atg7fl/flGzmb-Cre and control mice were infected with LCMV Armstrong. (a) Genomic deletion of Atg7 in DbGP33+ T cells and in total activated CD8 T cells (CD44hiCD62Llo) day 8 p.i. Levels of Atg7 in WT mice were set to 100%. (b) Deletion of Atg7 protein and its enzymatic activities verified by Immunoblotting in CD44hiCD62Llo CD8 T cells isolated day 8 p.i. β-actin was used as a loading control. (c) DbGP33+ T cells in peripheral blood day 8 p.i.. The number represents the percentage of DbGP33+ T cells. Gated on CD8 T cells. Right, representative plots of DbGP33+ T cells showing terminal effector (KLRG1hi CD127lo) and memory precursor (KLRG1lo CD127hi) populations. (d) Total number of antigen-specific cells at day 8 p.i. (e) IFN-γ and TNF production at 8 days p.i. using ex vivo peptide stimulation. Gated on total CD8 T cells. (f) DbGP33+ T cells in peripheral blood day 15 p.i. The number indicates the DbGP33+ percentage of CD8 T cells. (g) Numbers of tetramer-positive cells in spleens day 15 p.i. (h) Kinetics of DbGP33+ CD8 T cells in peripheral blood following infection. Each line represents data from one experimental mouse. Plot on the right shows the percentage of DbGP33+ CD8 T cells remaining on day 15 relative to day 8. (i) Numbers of tetramer-positive cells at day 30 p.i. Errors bars in (a), (d), (g) and (i) indicate SEM. Data for (a) are pooled from two independent experiments and data for (c) to (i) are representative of at least two independent experiments. n=3–7 in each group. Horizontal dotted lines in (g) and (h) represent limit of detection. *, p≤0.05; **, p≤0.005; ***p≤0.0005.
To assess the importance of Atg7 in antigen-specific CD8 T cells, we infected Atg7fl/flGzmb-Cre mice with the LCMV Armstrong strain and followed the differentiation of LCMV-specific CD8 T cells over time. Eight days after infection, DbGP33+ CD8 T cells had undergone significant expansion in both Atg7fl/fl and Atg7fl/flGzmb-Cre mice (Fig. 4c). At day 8 p.i. the memory precursor population, characterized by KLRG1lo and CD127hi, was present in Atg7-deficient DbGP33-specific cells at similar proportions compared to wild-type cells (Fig. 4c)3, 4. Other phenotypic markers, including CD44 and CD62L, showed no considerable differences between the two groups (Supplementary Fig. 4c). Despite a small reduction in PBMC at day 8 p.i. (Fig. 4c left panel), antigen-specific CD8 T cells were detected at comparable quantities in all tissues examined including spleens, livers and lungs between the experimental and control groups (Fig. 4d). Supporting these data, we did not observe any differences in terms of proliferation measured by BrdU incorporation as well as the number of cells undergoing apoptosis between WT and Atg7-deficient antigen specific CD8 T cells at day 5 p.i. when T cells are rapidly proliferating (Supplementary Fig. 4d–h). An important function of CD8 T cells is the ability to produce the antiviral cytokines IFN-γ and TNF. Ex vivo stimulation of splenocytes at day 8 p.i. using peptides indicated little difference in the ability, or the number, of cells producing these cytokines (Fig. 4e). In addition, we no longer detected virus in the sera, spleen, liver or lung of wild type, nor Atg7fl/flGzmb-Cre mice by day 8 p.i. (data not shown). Collectively, our data demonstrated that virus-specific CD8 T cells deficient in Atg7 were able to proliferate and differentiate into functional effector T cells that were comparable to their wild-type counterparts. This is consistent with our findings that autophagy is actually decreased in proliferating virus specific CD8 T cells (Figs. 1–3).
Immediately following the peak of expansion, the sharpest decline in virus-specific CD8 T cells was observed in the Atg7-deficient cells between days 8 and 15 p.i. (Fig. 4f–h). T cells specific for the dominant LCMV epitopes in Atg7fl/flGzmb-Cre mice, including DbGP33, DbGP276, DbNP396 and KbGP34 were reduced approximately 5–10 fold in the spleens of Atg7fl/flGzmb-Cre mice compared to wild type mice (Fig. 4g). Few antigen-specific T cells survived the contraction phase to transition into the memory phase (day 30 p.i.) within either lymphoid or non-lymphoid tissues, or the peripheral blood (Fig. 4h and i). Overall, the data indicated that Atg7 is important for the survival of effector CD8 T cells, and further suggest that a defective autophagy pathway compromises the formation of a functional CD8 T cell memory pool. These results correlated very closely with the in vivo kinetics of autophagy. Thus, the striking survival defects of autophagy deficient effector T cells occur after day 8 p.i concurrently when autophagy is increased in virus specific T cells.
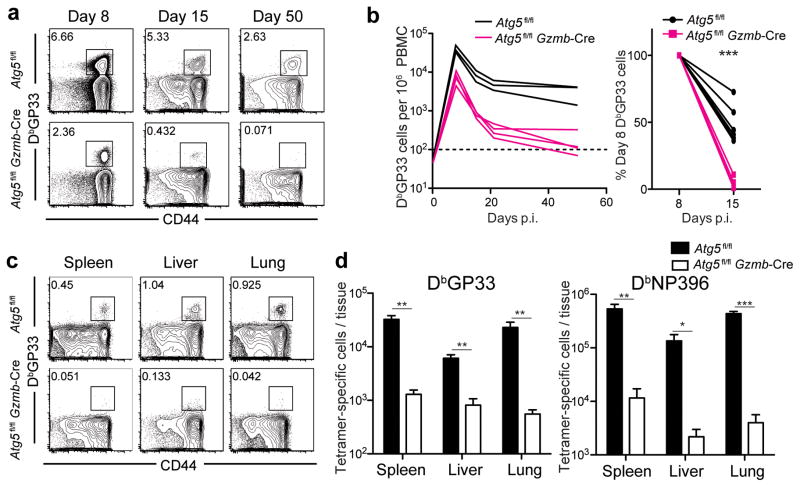
We sought to examine whether the ubiquitin-like conjugation systems necessary for autophagy were required for T cell memory formation by evaluating the importance of a second essential autophagy protein, Atg5. Similar to the Atg7fl/flGzmb-Cre phenotype, Atg5fl/flGzmb-Cre mice infected with LCMV Armstrong demonstrated an initial expansion (day 8 p.i.) followed by a drastic loss of antigen-specific CD8 T cells (Fig. 5a and b). We observed this profound loss in antigen-specific T cells in all tissues examined at the memory phase in the Atg5fl/flGzmb-Cre mice (Fig. 5c and d). These data implied a pro-survival role for proteins involved in the autophagy membrane elongation complex in antigen-specific CD8 T cells.
FIGURE 5.
Survival defects of Atg5-deficient T cells during the contraction phase following LCMV Armstrong infection. Atg5fl/flGzmb-Cre and control mice were infected with LCMV Armstrong. (a) Flow cytometric analysis of circulating LCMV-specific T cells in PBMC post-infection. The number on each plot indicates the percentage of CD8 T cells stained positive for DbGP33-tetramer from a representative mouse on days 8, 15 and 50. (b) Summary of the longitudinal kinetics of DbGP33-positive T cells in peripheral blood. Each line represents data from one experimental mouse. Plot on the right shows the percentage of DbGP33+ CD8 T cells remaining on day 15 relative to day 8. (c) DbGP33-specific T cells in tissues on day 279 post-infection. (c) The numbers on flow cytometry plots indicate the percentage of DbGP33-specific T cells of the total gated CD8 T cells. (d) The total number of DbGP33- and DbNP396-specific T cells in each tissue indicated on the graph. Errors bars in (d) indicate SEM. The horizontal dotted line in (b) represents the limit of detection. n=3–7 in each group. Data are representative of two independent experiments. *, p≤0.05; **, p≤0.005; ***p≤0.0005.
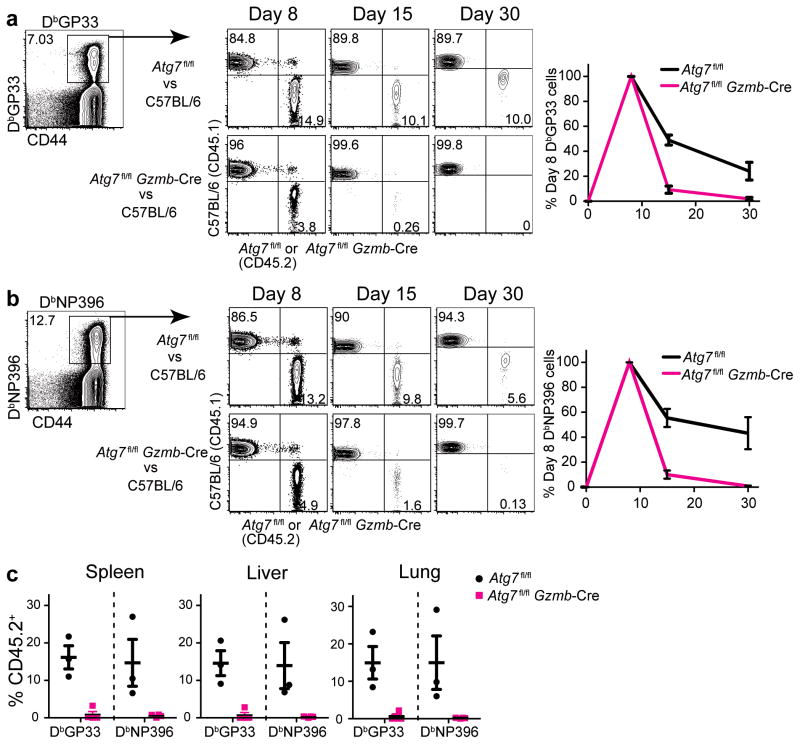
The conditional knockout system is a powerful tool to investigate functions of a gene within a specific cell population. However, one caveat of using this system is that the profound depletion of antigen-specific CD8 T cells observed in Atg7fl/flGzmb-Cre and Atg5fl/flGzmb-Cre mice during the course of LCMV infection (Figs. 4 and 5) may impact host environmental factors such as inflammatory cytokines that could exacerbate T cell survival and differentiation. Furthermore, other cell types, such as natural killer (NK) cells, also express granzyme B, introducing an additional level of complexity when comparing experimental and control mice. Recent work showed that NK cells were implicated in modulating T cell immunity26, 27. To address these issues, we generated mixed bone marrow chimeras that were reconstituted from an mixture of congenitally marked Atg7fl/flGzmb-Cre (CD45.2) and wild-type C57BL/6 (CD45.1) donor bone marrow cells into wild-type C57BL/6 (CD45.1) recipient mice (Supplementary Fig. 5a). This system allowed us to compare changes in number between wild type and Atg7fl/flGzmb-Cre antigen-specific CD8 T cells in the same environment (in the same mouse), therefore excluding factors that only act on one population of these cells. As a control, we also generated mixed bone marrow chimera mice that were reconstituted with Atg7fl/fl (CD45.2) (no cre) and wild-type (CD45.1) donor mice. After reconstitution, these animals were challenged with the LCMV Armstrong, and virus-specific CD8 T cells were examined over time (Supplementary Fig. 5a). At day 8 p.i., there was a significant expansion of antigen-specific T cells from both CD45.2 (knock-out) and CD45.1 (wild type) donor populations. The DbGP33 and DbNP396-specific CD8 T cell responses were longitudinally analyzed during the effector and memory stages (Fig. 6a and b, Supplementary Fig. 5c). Similar to the observation in the conditional knockout mice, we observed a more than 90% decline in Atg7-deficient LCMV-specific CD8 T cells (CD45.2+) from day 8 to day 15 in Atg7fl/flGzmb-Cre plus C57BL/6 chimeric mice, compared to a moderate decline of 50%–60% in the corresponding wild-type (CD45.2+) population in the Atg7fl/fl plus C57BL/6 controls. Four weeks after infection there were virtually no antigen-specific CD8 T cells of Atg7fl/flGzmb-Cre origin (CD45.2+) throughout the peripheral blood, lymphoid and non-lymphoid tissues in Atg7fl/flGzmb-Cre plus C57BL/6 chimeric mice (Fig. 6a–c). A significant proportion of naive CD8 T cells (CD44lo) from the Atg7fl/flGzmb-Cre donor persisted within the chimeric mice, indicating specific loss of the antigen-experienced effector CD8 T cells from Atg7fl/flGzmb-Cre mice throughout the progression of infection (Supplementary Fig. 5b). Since the cells from WT (CD45.1+) origin made up the majority of the reconstituted population within the chimeric mice (Supplementary Fig. 5b), the physiological and pathological conditions closely resembled those of the WT mice. Hence, in the absence of potential external variables, such as changes introduced in other cell types or any subtle variations in viral clearance, the defects rising from Atg7-deficiency were cell-intrinsic properties that affected survival of the antigen-specific effector CD8 T cells to the memory phase. Thus, these data confirmed that the autophagy pathway is critical for memory T cell formation.
FIGURE 6.
Antigen-specific CD8 T cells lacking the Atg7 gene exhibit cell-intrinsic defects in developing into long-term memory cells in bone marrow chimeric mice. Atg7fl/fl plus C57BL/6 (CD45.2/CD45.1) control and Atg7fl/flGzmb-Cre plus C57BL/6 (CD45.2/CD45.1) experimental mixed bone marrow chimera mice were generated. CD8 T cell response was evaluated following the LCMV Armstrong infection according to the scheme in Supplementary Fig. 5a. (a) and (b) Flow cytometric analysis of DbGP33- and DbNP396-specific T cells on day 8, 15 and 30 p.i. Gated on tetramer-positive CD8 T cells as indicated. Number on each quadrant represents the percentage of tetramer-positive cells that are either CD45.1+ or CD45.2+ as indicated on the axis. Average percentages of DbGP33- and DbNP396-specific T cells from day 8 to day 30 in the peripheral blood of both groups of chimeric mice are plotted to the right of the flow plots in (a) and (b). The black line represents tetramer-positive cell of Atg7fl/fl origin and the red line of Atg7fl/flGzmb-Cre origin. The number of tetramer-positive cells on day 8 p.i. from each mouse is normalized to 100%. (c) Percent of CD45.2+ antigen-specific cells in tissues on day 30 post-infection. The error bars in (a)–(c) indicate SEM. n=3–4 mice in each group. Data in (a)–(c) are representative of two independent experiments.
Distinct metabolic profiles of Atg7−/− effector T cells
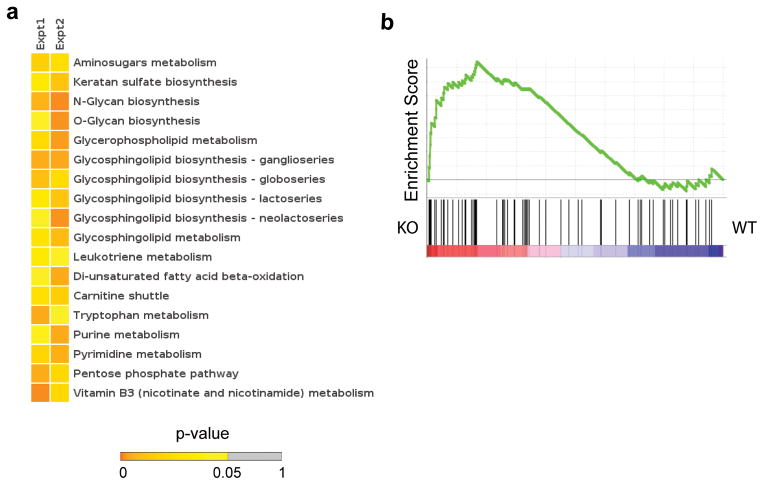
To investigate the potential mechanism underlying the survival defects in the Atg7-deficient CD8 T cells, we performed metabolomic and transcriptomic analyses on antigen-specific T cells from Atg7-deficient (Atg7fl/flGzmb-Cre) or WT (Atg7fl/fl) mice 8 days following LCMV Armstrong infection. This time point was selected as representative of a transition time point in T cell differentiation before significant loss of Atg7-deficient cells has occurred. In the DbGP33+ CD8 T cells isolated from mice 8 day p.i, the overall gene expression pattern of the Atg7-deficient CD8 T cells closely resembled that of the WT group; only a small number of genes showed significantly different expression between the two groups in the DNA microarray data, mostly associated with cell cycle (data not shown). However, these cells exhibited distinct metabolic profiles measured on a liquid chromatography coupled mass spectrometry (LC-MS) metabolomics platform. Among the metabolites that were significantly different between Atg7-sufficent and deficient cells (Supplementary Table 1), many are contained within well-defined metabolic pathways with clear links to cell survival, as well as some of which have been implicated in T cell differentiation (Supplementary Fig. 6). For example, among the pathways that were significantly different between genotypes are the carnitine shuttle and di-unsaturated fatty acid beta-oxidation, both of which are part of fatty acid metabolism in the mitochondria (Fig. 7a). T cell-intrinsic mitochondrial fatty acid oxidation has been shown to be a critical metabolic pathway for the stable development of long-lived memory T cells 16, 17. These data could indicate that autophagy contributes to the production of the lipid substrates for mitochondrial fatty acid oxidation and the fueling of oxidative phosphorylation (OXPHOS) in these cells 28. Consistent with a role for autophagy in lipid metabolism in T cells during the transition to the memory phase, our metabolite analysis showed that lipid biosynthetic pathways were dysregulated in the absence of Atg7 (Fig. 7a). Also of note, glucosamine 6 phosphate and glycan biosynthesis pathways were perturbed in Atg7-deficient T cells (Fig. 7a and Supplementary Fig. 6). The hexosamine pathway provides substrates for the glycosylation of growth factor cytokine receptors, leading to receptor stability and the maintenance of survival signals 29. Given the importance of growth factor cytokines in supporting the development of memory T cells 2, it is possible that T cells lacking autophagy machinery cannot maintain the proper growth factor signals that support memory T cell development, linking autophagy to glycosylation and the maintenance of cellular metabolism. Importantly, these metabolomics data were further validated by examining the levels of gene expression of the enzymes associated these metabolite activities. Using Gene set enrichment analysis30 and these enzymes as a gene set, the gene expression for these metabolic activities was clearly associated with the Atg7 deficient genotype (Fig. 7b and Supplementary Table 2). Overall, several metabolic pathways were altered in Atg7-deficient T cells day 8 pi, and when integrated with our transcriptomic studies suggest that autophagy supports metabolic homeostasis in CD8 T cells during the transition to memory phase.
FIGURE 7.
Metabolomic and transcriptomic analysis of Atg7-dificient CD8 T cells. Atg7fl/flGzmb-Cre (KO) and control (Atg7fl/fl, WT) mice were infected with LCMV Armstrong. DbGP33+ CD8 T cells were isolated from mice 8 day p.i for analysis. (a) Pathways differentially regulated in Day 8 Atg7-deficient antigen-specific cells. Each column represents an independent experiment of samples pooled from 3 WT or KO mice; each row a pathway colored to indicate its p-value. (b) Gene set enrichment analysis (GSEA) of genes represented in T cells, which are associated with the metabolic enzymes underlying the most differentially regulated metabolic pathways (a) in KO vs. WT. p=0.024.
Autophagy in CD8 T cells is essential for viral control
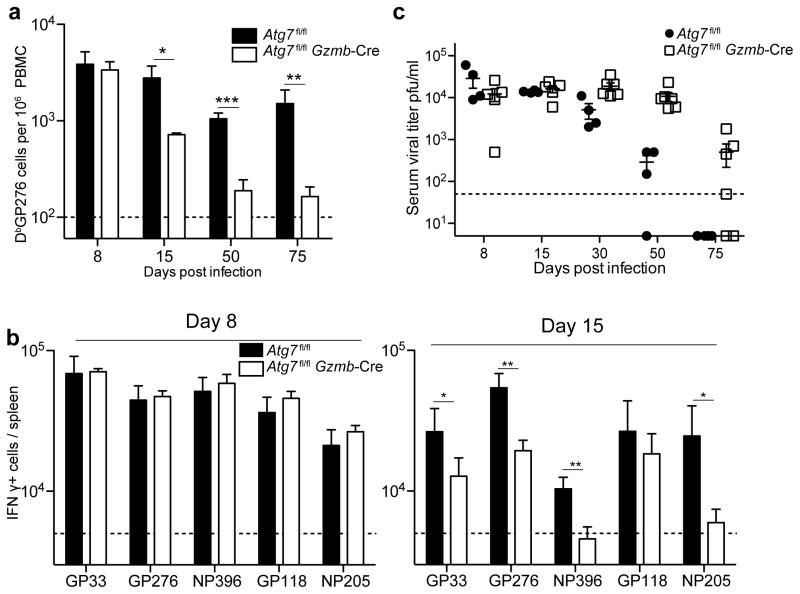
Autophagy is required for memory CD8 T cell formation during an acute viral infection as shown above. Next, we asked whether autophagy regulates CD8 T cell responses during a chronic viral infection. To address this question, the Atg7fl/flGzmb-Cre mice were challenged with the LCMV Clone 13 strain, which causes a chronic infection31. In chronic infections antigen-specific T cells gradually diminish in their effector functionality and become exhausted cells due to prolonged persistence of antigen in the host32. During a chronic infection, antigen-specific CD8 T cells in the Atg7fl/flGzmb-Cre mice again mounted a initial response to the virus in the peripheral blood, as well as in lymphoid and non-lymphoid tissues at day 8 p.i., comparable to the wild-type controls (Fig. 8a, Supplementary Fig. 7a and b). Ex vivo stimulation of splenocytes at day 8 p.i. showed little difference in the number of cells producing IFN-γ (Fig. 8b). However, by day 15 p.i., most LCMV-specific T cells were lost in Atg7fl/flGzmb-Cre mice throughout lymphoid and non-lymphoid tissues, similar to the acute infection model (Fig. 8a, Supplementary Fig. 7c and d). The observed reduction was also reflected in the number of IFN-γ-producing cells when stimulated ex vivo (Fig. 8b). As a consequence of the loss of most antigen specific cells, viral control was compromised in Atg7fl/flGzmb-Cre mice (Fig. 8c). These data further indicated a significant survival defect in the Atg7-deficient LCMV-specific effector CD8 T cells; not only were these cells compromised in memory CD8 T cell differentiation, but they also failed to survive in the chronic infection model.
FIGURE 8.
Autophagy in CD8 T cells is essential for regulating chronic LCMV infection. Atg7fl/flGzmb-Cre and control Atg7fl/fl mice were infected with LCMV Clone-13. (a) Kinetics of DbGP276-specific T cells in the peripheral blood. (b) Total number of IFN-γ secreting cells in spleens after 5-hr stimulation ex vivo with peptide indicated from days 8 and 15 p.i. (c) Viral titers in serum at various time points p.i. n=3–5 mice in each group. Data are representative of two independent experiments. Error bars indicate SEM. *, p≤0.05; **, p≤0.005; ***p≤0.0005.
DISCUSSION
Our findings that proliferating T cells show decreased autophagy in vivo were unexpected because previous in vitro studies showed that autophagy is increased in T cells after TCR stimulation9, 12, 13. Discrepancies between our current study and these previous studies may be explained by the vastly different condition of TCR stimulation in vivo versus in vitro. In addition, in some of the earlier studies only a single aspect of the autophagy pathway such as the number of autophagosomes and the amount of lipidated LC3b-II have been used to assess autophagy activity in cultured T cells. As the autophagy pathway can be blocked at any point along its progression, it has become clear that measuring a single component in the pathway, rather than measuring flux through the pathway as done here, is not reflective of whether autophagy is activated or blocked7.
Our findings of the kinetics of autophagy in vivo are totally consistent with our results in which essential autophagy genes were deleted in antigen specific CD8 T cells. The in vivo kinetics of autophagy indicate that activated T cells can proliferate with very low autophagy activity, and indeed both autophagy deficient and wild type virus-specific CD8 T cells were able to similarly proliferate in most tissues. However, this comparable proliferation of antigen specific CD8 T cells between autophagy gene conditional knockout and wild type mice was unexpected because a prior study showed that autophagy gene deletion resulted in compromised T cell proliferation after in vitro TCR stimulation9. T cells used in the prior study were autophagy defective for long periods of time before being assayed because autophagy genes were deleted during T cell development9. It is known that these autophagy gene deficient T cells are transcriptionally and phenotypically distinct from wild type T cells9, 10, 15, 33. In contrast, to avoid this complication in our studies autophagy genes were specifically deleted in granzyme B expressing cells after T cell activation, and therefore naive T cells were normal at the time of viral infection. Thus, a robust CD8 T cell expansion in the absence of autophagy genes suggest that activated T cells can proliferate and differentiate into effectors without autophagy, and this notion is consistent with the in vivo kinetics of autophagy in antigen specific CD8 T cells.
In contrast to T cell expansion phase, autophagy was required for effector T cell survival to form memory T cells during contraction phase when effector T cells undergo significant changes. In this phase, effector T cells, having just finished many rounds of vigorous cell divisions, potentially accumulate protein aggregates, reactive oxygen species (ROS) and other superfluous cellular content34, 35, 36, 37, 38. Autophagy could perform multiple pro-survival functions in these cells, including the removal of damaged mitochondria that generate ROS or misfolded proteins39, 40. Thus, upregulation of autophagy in virus-specific CD8 T cells upon conclusion of clonal expansion is important during the contraction phase.
Our results on the kinetics and the functional role of autophagy in virus specific CD8 T cells have important implications to previous studies linking the major regulator of autophagy, mTOR to the memory CD8 T cell differentiation17, 41. These studies have demonstrated that inhibition of mTOR by rapamycin improves not only the magnitude but also quality of memory CD8 T cells17, 41. These effects of rapamycin in antigen specific CD8 T cells might result from increased autophagy during the proliferation and contraction phase. Active mTOR signals directly down-regulate autophagy and inhibition of mTOR would, therefore, likely enhance autophagy activity42. Thus, it is an important future work to investigate whether rapamycin treatment up-regulates autophagy in antigen specific CD8 T cells and whether increased autophagy improves memory CD8 T cell differentiation.
Our metabolomics analysis provides insight into the metabolic pathways regulated by autophagy in virus-specific CD8 T cells. The unbiased metabolite data revealed that mitochondrial fatty acid oxidation, which has been shown to be critical for memory T cell generation 16, 17, was dysregulated in Atg7-deficient T cells at the peak of clonal expansion, right at the point Ag-specific T cells transition to the memory phase. A study of metabolomic profiles in a mouse cancer model reported that the Atg7-deficient tumor cells had defective mitochondria and reduced fatty acid oxidation43. We speculate that T cell-intrinsic autophagy could be important for generating lipid substrates for mitochondrial fatty acid oxidation (FAO). This notion is consistent with published studies showing that autophagy can regulate lipid metabolism via the breakdown of cellular lipid stores44. Precisely how the substrates for mitochondrial FAO are utilized or generated by memory T cells, and whether autophagy directly regulates mitochondrial FAO in antigen specific effector CD8 T cells is an exciting subject for future work.
In addition to an acute viral infection, our data demonstrate that autophagy in virus-specific CD8 T cells is essential for cell survival and controlling a chronic viral infection. It is well established that activated CD8 T cells during chronic infections or in tumor environments differentiate into exhausted T cells. Rejuvenating these exhausted T cells is a promising approach to treat chronic infections and cancer. Thus, our data suggest that autophagy plays a critical role in formation and survival of exhausted CD8 T cells that can be targeted for immunotherapy.
In summary, these data establish that the autophagy pathway is dynamically regulated throughout the CD8 T cell response and is critical for the formation T cell memory. Autophagy seems to play a role in the metabolic homeostasis of effector CD8 T cells during their transition into memory cells. Our studies have implications to develop effective vaccination strategies and anti-cancer immune therapy through targeting the autophagy pathway and/or other metabolic pathways.
Online Methods
Mice and infection
Atg7-floxed mice and Atg5-floxed mice were generated and characterized previously 22, 23. Conditional knockout mice were generated by crossing Atg7-floxed mice and Atg5-floxed mice to Granzyme B-Cre transgenic mice 24. To generate P14 chimeric mice bearing LCMV-specific CD8 T cells, transgenic P14 cells (Thy-1.1+) with an engineered TCR that recognizes the H-2DbGP33 epitope were adoptively transferred intravenously (1×104 antigen-specific CD8+ T cells per mouse as stated in the figure legend) to wild-type C57BL/6 mice (Jackson Laboratory). In acute infection, mice were infected with the Armstrong strain of lymphocytic choriomeningitis virus (LCMV) using an intraperitoneal (i.p.) injection of 2×105 plaque-forming unit (PFU). In chronic infection, mice were infected with the Clone 13 strain of LCMV using an intravenous injection of 2×106 PFU. All animal experiments were approved by the Institutional Animal Care and Use Committee of Emory University.
Retroviral transduction
Over-expression of transgenes in P14 was carried out using MSCV-IRES-Thy1.1 retrovirus vector (kindly provided by A. Rao, La Jolla Institute for Allergy and Immunology, La Jolla, CA; Addgene plasmid 17442). LC3b gene was cloned from mouse cDNAs and then fused N-terminal to a green fluorescent protein, Emerald GFP (Invitrogen). G120A mutant was made using Quickchange kit (Agilent technology). For mCherry constructs, the mCherry gene (Clontech Laboratories, Inc.) was fused to N-terminal of wild type and G120A mutant GFP-Lc3b. To activate P14 cells, GP33-41 peptide (200 μg) was intravenously injected into P14 transgenic mice (Ly5.1+Thy1.1−). Splenocytes were then isolated 18–24 hours post injection and spin transduced with the desired retrovirus. 1 × 104 retroviral transduced P14 cells were adoptively transferred into naive C57BL/6 mice, which were immediately infected i.p. with 2 × 105 pfu LCMV Armstrong. Retroviral transduction of HSCs were performed as described previously 45. These transduced HSCs were adoptive transferred into lethally irradiated mice.
Flow cytometry and antibodies
The following antibodies were purchased from BD Bioscience (San Diego, CA); anti-CD8 (53–6.7), anti-Thy1.1 (OX-7), anti-CD45.1 (A20), anti-CD45.2 (104), anti-CD44 (IM7), anti-IFN-γ (XMG1.2), anti-TNF (MP6-XT22). For BrdU staining, BrdU flow kit (catalog#559619) was purchased from BD Bioscience. Anti-KLRG1 (2F1) antibody was purchased from Southern Biotech. Anti-CD127 (A7R34) and CD62L (MEL-14) antibodies were purchased from eBioscience. MHC class I tetramer was prepared as previously described 46. All the analyses were done on a FACSCanto flow cytometer (BD Biosciences).
Cellular Imaging
Cells were analyzed on Image Stream Mark II (Amnis) using software INSPIRE. Images were acquired at 60x or 40x magnification with a low speed and high sensitivity setting. Single focused cells were selected for image collection. Data were analyzed using software IDEA. Spot counting analysis was used to evaluate LC3b puncta positive cells in each sample.
Quantitative PCR
To assess the genomic deletion of the Atg7 floxed allele, DbGP33-specific CD8 T cells were sorted and DNA was extracted using a DNeasy kit (Qiagen). Quantitative PCR was performed using iQSYBR green supermix (BioRad) with primers flanking one floxed site (forward: 5′-AGCTTGGCTGCTACTTCTGC-3′; reverse 5′-TGCAGGACAGAGACCATCAG-3′) and primers targeting programmed death-1 allele as a loading control (forward: 5′-GAGGTCCTTTCACTCTCCACG-3′; reverse 5′-CGACTTGTGTGCATGCATAGTACC-3′). Primers used to quantify the levels of LC3b mRNA were: 5′-CACTGCTCTGTCTTGTGTAGGTTG-3′ (forward) and 5′-TCGTTGTGCCTTTATTAGTGCATC-3′ (reverse); of p62 mRNA were 5′-AGCTGCCCTCAGCCCTCTA-3′ (forward) and 5′-GGCTTCTCTTCCCTCCATGTT-3′ (reverse). 18s rRNA was used as internal control for the qRT-PCR; 5′-GTAACCCGTTGAACCCCATT-3′ (forward) and 5′-CCATCCAATCGGTAGTAGCCG-3′ (reverse).
Cell sorting
Total activated CD8 T cells on day 8 post LCMV infection were stained with antibodies against CD8, CD44 and CD62L and were sorted using FACSAria II (BD). DbGP33-specific cells were stained with antibodies against CD8, CD44 as well as TCR using DbGP33 tetramer 46. The typical purity after cell sorting was between 95–99%.
Immunoblot analysis
Whole cell lysates of indicated cell population were resolved on 4–15% SDS-PAGE and then transferred to a PVDF (polyvinylidene difluoride) membrane (Millipore). The membrane was blotted with rabbit anti-β-actin (13E5; Cell signaling), rabbit anti-Atg7 (catalog#A2856; Sigma-aldridge), mouse anti-Atg5 (7C6; Nanotools), rabbit anti-LC3 (catalog#L7543; Sigma-aldridge) or guinea pig anti-p62 (catalog#GP62-C; Progen Biotechnic and catalog#P0067; Sigma-aldridge); secondary HRP-linked antibodies were goat anti-rabbit (catalog#7074; Cell signaling), goat anti-mouse (catalog#1034–05; Southern Biotech) and goat anti-guinea pig (catalog#106–035–003; Jackson ImmunoResearch). The blots were developed in LumiGLO reagent (Cell signaling) prior to exposure to X-ray films (Kodak).
Generation of bone marrow chimeras
Bone marrow was collected from Atg7fl/fl (CD45.2+), Atg7fl/flGzmb-Cre (CD45.2+) or wild type C57BL/6 (CD45.1+) mice. For each chimera, 20 million BM cells of 1:1 mix of Atg7fl/fl (or Atg7fl/flGzmb-Cre) and C57BL/6 were transferred intravenously into lethally irradiated (two doses of 550rad) WT C57BL/6 (CD45.1+) recipient. Recipient mice were allowed 8–10 weeks for reconstitution prior to LCMV Armstrong infections.
LC/MS metabolomic analysis
The metabolomics analysis was performed similarly as previously described (Li2013). Cell extracts were treated with acetonitrile (2:1, v/v) and centrifuged at 14,000× g for 5 min at 4°C to remove proteins. Samples were maintained at 4°C in an autosampler until injection. A Thermo Orbitrap-Velos mass spectrometer (Thermo Fisher, San Diego, CA) coupled with switching dual chromatography (an anion exchange (AE) column and a C18 reverse-phage column) was used for data collection, via positive-ion electrospray ionization (ESI). Each sample was run in 3 to 6 analytical replicates, depending on available source material. Mouse naive CD8 T cells, kidney and splenocyte tissues were also run as analytical controls. Metabolite features were extracted and quantified using apLCMS software version 5.9.847. Only features that were detected in 12 or more injection samples (including analytical replicates) were retained for further analysis. This filtering led to 3270 metabolite features from the AE column and 6300 from the C18 column. Data were log2 transformed, averaged among analytical replicates, and normalized by total ion intensity. Missing values were imputed with half mean of the feature in all detected samples. Student t-test was used to compare between KO and WT groups. Metabolic pathway analysis was performed by mummichog software 0.9.6 using default parameters48. Top 500 metabolite features in the t-test were used as input to mummichog, and the total list of features were used as reference. Mummichog tested the enrichment of input metabolites against random data resampled from the reference list, and produced an empirical p-value per pathway48. Input metabolites in the significant pathways (p < 0.05 in both experiments) were linked in a network figure by known metabolic reactions (Supplementary Fig. 6).
Microarray analysis
Day 8 DbGP33-specific CD8 T cells from acute LCMV infected mice were FACS sorted based on CD8 and the tetramer stain. RNA was isolated using RNeasy kit (Qiangen) according to manufacturer’s protocol. RNA was amplified, biotinylated and hydridized on mouse 430.2 Affymetrix microarray chips at the Microarray Core (Dana Farber Cancer Institute, MA, USA). The expression of metabolic enzymes underlying Supplementary Fig. 6 was tested for their association with KO or WT genotype. All enzyme commission (EC) numbers in the reactions in Supplementary Fig. 6 were looked up in mouse genome for their encoding genes. Only genes that are expressed in T cells are included for this analysis. The positive expression in T cells was according to the definition of ImmGen consortium, as signal intensity higher than 128 unit in any of their microarray files (total 89 microarrays on T cell populations, retrieved on Feb. 22, 2014, http://www.immgen.org/). The resulted genes (Gpd1l, Kdsr, Ado, Acox1, Gmpr2, Tkt, Alg5, Alg13, Hprt, Nampt, Gsta4, Gstk1, Gstm1, Gstm4, Gsto1, Gstp1, Gstp2, Gstt2, Hpgds, Gfpt1, Adk, Nagk, Dck, Sphk1, Sphk2, Prps1, Prps2, Cept1, Ept1, Cept1, Cdipt, Plb1, Acot2, Lpin1, Lpin2, Pde1b, Pde2a, Pde3b, Pde4a, Pde4d, Pde7a, Pde8a, Pde5a, Arsa, Gba2, Galc, Bst1, Cd38, Asah1, Asah2, Ada, Ampd1, Ampd2, Ampd3, Cant1, Enpp1, Itpa, Enpp4, Aldoa, Aldoc, Sgpl1, Npl, Acsl1, Acsl3, Acsl4, Acsl5) were written as a custom gene set in ‘.gmt’ file, and tested in GSEA program30. Preranked method in GSEA was used, where the genes were ranked by t-statistic between KO and WT samples.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5. P values were calculated based on two-tailed unpaired Student’s t-test, assuming normal distribution of data and equal sample variance. Sample size was not predetermined via statistical means, but the number of mice used in the study was consistent with previously published work of similar nature. All mice used for the study were included for a robust statistical analysis with no randomization or blinding involved.
Supplementary Material
Acknowledgments
We thank J. Jacob (Emory University) for providing Gzmb-Cre transgenic mice; R. Karaffa and S. Durham for flow sorting at the Emory Flow Cytometry Core Facility; A. Rae for assistance in the use of ImageStream®. This work was supported by grants from the National Institutes of Health (NIH) grant R01 AI030048 (to R.A.) and Mérieux Foundation (to R.A.); and R01 AI084887 as well as by the Crohn’s and Colitis Foundation (to H.W.V.).
Footnotes
Microarray
Data are deposited in the GEO database under accession number GSE57047.
Contributions
X.X., K.A., S.L., M.W.B., J.M., D.R.G, D.P.J. H.W.V., and R.A. designed the research; X.X., K.A., S.L., J.H.H., L.Y., W.G.T., B.T.K. performed experiments. X.X., K.A., S.L., M.W.B., E.L.P., D.R.G., D.P.J., H.W.V., and R.A. analyzed data; X.X., K.A., S.L., E.L.P., H.W.V., and R.A. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Williams MA, Bevan MJ. Effector and memory CTL differentiation. Annu Rev Immunol. 2007;25:171–192. doi: 10.1146/annurev.immunol.25.022106.141548. [DOI] [PubMed] [Google Scholar]
- 2.Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 2007;27(3):393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4(12):1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 4.Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med. 2008;205(3):625–640. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27(2):281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469(7330):323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh CM, Edinger AL. The complex interplay between autophagy, apoptosis, and necrotic signals promotes T-cell homeostasis. Immunol Rev. 2010;236:95–109. doi: 10.1111/j.1600-065X.2010.00919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204(1):25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephenson LM, Miller BC, Ng A, Eisenberg J, Zhao Z, Cadwell K, et al. Identification of Atg5-dependent transcriptional changes and increases in mitochondrial mass in Atg5-deficient T lymphocytes. Autophagy. 2009;5(5):625–635. doi: 10.4161/auto.5.5.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang RC, Levine B. Autophagy in cellular growth control. FEBS letters. 2010;584(7):1417–1426. doi: 10.1016/j.febslet.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hubbard VM, Valdor R, Patel B, Singh R, Cuervo AM, Macian F. Macroautophagy regulates energy metabolism during effector T cell activation. J Immunol. 2010;185(12):7349–7357. doi: 10.4049/jimmunol.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C, Capan E, Zhao Y, Zhao J, Stolz D, Watkins SC, et al. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J Immunol. 2006;177(8):5163–5168. doi: 10.4049/jimmunol.177.8.5163. [DOI] [PubMed] [Google Scholar]
- 14.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nature reviews Drug discovery. 2012;11(9):709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol. 2009;182(7):4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 16.van der Windt GJ, Everts B, Chang CH, Curtis JD, Freitas TC, Amiel E, et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36 (1):68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460(7251):103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 19.Shvets E, Fass E, Elazar Z. Utilizing flow cytometry to monitor autophagy in living mammalian cells. Autophagy. 2008;4(5):621–628. doi: 10.4161/auto.5939. [DOI] [PubMed] [Google Scholar]
- 20.Gump JM, Staskiewicz L, Morgan MJ, Bamberg A, Riches DW, Thorburn A. Autophagy variation within a cell population determines cell fate through selective degradation of Fap-1. Nat Cell Biol. 2014;16(1):47–54. doi: 10.1038/ncb2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3(5):452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 22.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432(7020):1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 23.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 24.Jacob J, Baltimore D. Modelling T-cell memory by genetic marking of memory T cells in vivo. Nature. 1999;399(6736):593–597. doi: 10.1038/21208. [DOI] [PubMed] [Google Scholar]
- 25.Masopust D, Murali-Krishna K, Ahmed R. Quantitating the magnitude of the lymphocytic choriomeningitis virus-specific CD8 T-cell response: it is even bigger than we thought. J Virol. 2007;81(4):2002–2011. doi: 10.1128/JVI.01459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waggoner SN, Cornberg M, Selin LK, Welsh RM. Natural killer cells act as rheostats modulating antiviral T cells. Nature. 2012;481(7381):394–398. doi: 10.1038/nature10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lang PA, Lang KS, Xu HC, Grusdat M, Parish IA, Recher M, et al. Natural killer cell activation enhances immune pathology and promotes chronic infection by limiting CD8+ T-cell immunity. Proc Natl Acad Sci U S A. 2012;109(4):1210–1215. doi: 10.1073/pnas.1118834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Sullivan D, van der Windt GJ, Huang SC, Curtis JD, Chang CH, Buck MD, et al. Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity. 2014;41(1):75–88. doi: 10.1016/j.immuni.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wellen KE, Lu C, Mancuso A, Lemons JM, Ryczko M, Dennis JW, et al. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev. 2010;24(24):2784–2799. doi: 10.1101/gad.1985910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol. 2003;77(8):4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, et al. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med. 1998;188(12):2205–2213. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia W, Pua HH, Li QJ, He YW. Autophagy regulates endoplasmic reticulum homeostasis and calcium mobilization in T lymphocytes. J Immunol. 2011;186(3):1564–1574. doi: 10.4049/jimmunol.1001822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111(6):837–851. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 35.Rathmell JC, Farkash EA, Gao W, Thompson CB. IL-7 enhances the survival and maintains the size of naive T cells. J Immunol. 2001;167(12):6869–6876. doi: 10.4049/jimmunol.167.12.6869. [DOI] [PubMed] [Google Scholar]
- 36.Ma A, Koka R, Burkett P. Diverse functions of IL-2, IL-15, and IL-7 in lymphoid homeostasis. Annu Rev Immunol. 2006;24:657–679. doi: 10.1146/annurev.immunol.24.021605.090727. [DOI] [PubMed] [Google Scholar]
- 37.Jones RG, Thompson CB. Revving the engine: signal transduction fuels T cell activation. Immunity. 2007;27(2):173–178. doi: 10.1016/j.immuni.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 38.Grayson JM, Laniewski NG, Lanier JG, Ahmed R. Mitochondrial potential and reactive oxygen intermediates in antigen-specific CD8+ T cells during viral infection. J Immunol. 2003;170(9):4745–4751. doi: 10.4049/jimmunol.170.9.4745. [DOI] [PubMed] [Google Scholar]
- 39.Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-kappaB. Immunity. 2012;36(6):947–958. doi: 10.1016/j.immuni.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12(3):213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 41.Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460(7251):108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27(13):1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jordan MS. Genetic reconstitution of bone marrow for the study of signal transduction ex vivo. Methods in molecular biology. 2006;332:331–342. doi: 10.1385/1-59745-048-0:331. [DOI] [PubMed] [Google Scholar]
- 46.Murali-Krishna K, Altman JD, Suresh M, Sourdive DJ, Zajac AJ, Miller JD, et al. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 1998;8(2):177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 47.Yu T, Park Y, Johnson JM, Jones DP. apLCMS--adaptive processing of high-resolution LC/MS data. Bioinformatics. 2009;25(15):1930–1936. doi: 10.1093/bioinformatics/btp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li S, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA, et al. Predicting network activity from high throughput metabolomics. PLoS Comput Biol. 2013;9(7):e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.