Abstract
HLA-DRB1*0401 has been associated with predisposition to develop rheumatoid arthritis (RA) and collagen-induced arthritis (CIA) while *0402 is not associated with susceptibility. In this study, we determined if *0401 mice have CD4 T cell repertoire that is predetermined to produce proinflammatory cytokines. The data presented here shows that both *0401 and *0402 mice can produce TH1/TH17 cytokines although kinetics of response may be different. However, in the context of antigen-specific response in CIA, *0402 mice generate a TH2 response that may explain its resistance to develop arthritis. In addition, a significant subset of naïve CD4 T cells when activated in polarizing conditions differentiate into T regulatory cells in *0402 mice that produce IFNγ. *0401 mice harbor memory CD4 T cells that differentiate in to IL-17+ cells in various conditions. We hypothesize that *0401 has been evolutionarily selected due to its ability to clear infection. Our data suggests that *0401 generates a strong immune response to LPS and may be efficient in clearing infection. Autoimmunity is a bystander effect of the cytokine storm that along with the presence of low number of T regulatory cells in *0401 mice ensues immune dysregulation.
Keywords: T regulatory, T cell polarization, immune dysregulation, Rheumatoid arthritis, humanized mice
Introduction
Most autoimmune diseases, including rheumatoid arthritis (RA), have a strong genetic risk demonstrated by the presence of certain genes in the HLA region that account for around 50% of the risk [1]. Association of HLA-DR4 with susceptibility to RA has been studied extensively ever since the initial observations made by Stastny [2]. Among the HLA-DR4 genes, DRB1*0401 (Dw4), DRB1*0404/0408 (Dw14), and DRB1*0405 (Dw15) alleles confer genetic predisposition to RA while DRB1*0402 (Dw10) does not. Genetic susceptibility to RA in most populations has been explained by the presence of an amino acid motif in the 3rd hypervariable region comprising residues 67-74 known as ‘shared epitope’ (SE) [3, 4]. On the other hand, motif expressed by the sequences at this region in DRB1*0402 confers resistance to RA. Positive selection of potentially autoreactive T cells and antigen presentation by susceptible class II alleles have been suggested as plausible mechanisms for the association of SE with RA [5, 6]. Presentation of naturally processed peptides derived from endogenous class II molecules [7, 8] suggests that self-peptide presentation could be one mechanism for selection of autoreactive/proinflammatory T cells. However, it is unknown if HLA polymorphism is associated with presence of autoreactive T cells.
In order to understand the role of SE in arthritis, we generated transgenic mice expressing DRB1*0401 and *0402 gene that lack all four endogenous class II murine chains, Aα, Aβ, Eα, and Eβ due to a deletion of the entire class II region. These mice were tested for susceptibility to collagen-induced arthritis (CIA), an animal model for rheumatoid arthritis. Our previous studies with this model showed that while *0401.AEo mice develop Type II collagen (CII) induced arthritis (CIA) that mimics human disease in sex bias, autoantibodies and histopathology, *0402.AEo mice were resistant to CIA [6, 9]. Our data suggested that DRB1*0401 predisposes to develop arthritis by proliferation of autoreactive antigen-specific T cells while the DRB1*0402 causes deletion of T cells in the thymus that may be autoreactive providing protection from arthritis [10]
In this study we addressed our hypothesis that T cell selection by shared epitope has a predetermined immune response profile. We investigated this hypothesis by determining the role of SE in driving specific immune response that could generate an environment for susceptibility/resistance to develop arthritis. Even though both HLA-DRB1*0401 and *0402 can present type II collagen and its derived peptide to generate immune response, only *0401 mice produce proinflammatory response. Here we show that *0401 mice have memory T cells that, when activated, generate CD4+IL-17+ T cells while *0402 mice do not. Under polarizing conditions for TH17, TGFb+IL-6, *0402 mice generated more T regulatory (Treg) cells than *0401 mice. However, *0402 mice can produce TH17 response to arthritis-unrelated antigen which suggests the presence of and activation of TH17 cells occurs in certain conditions. Thus “shared epitope” can impact pathogenesis of RA by activating existing autoreactive cells following infection or due to external/environmental factors
RESULTS
Shared epitope polarizes production of pro-inflammatory cytokines by CD4 T cells
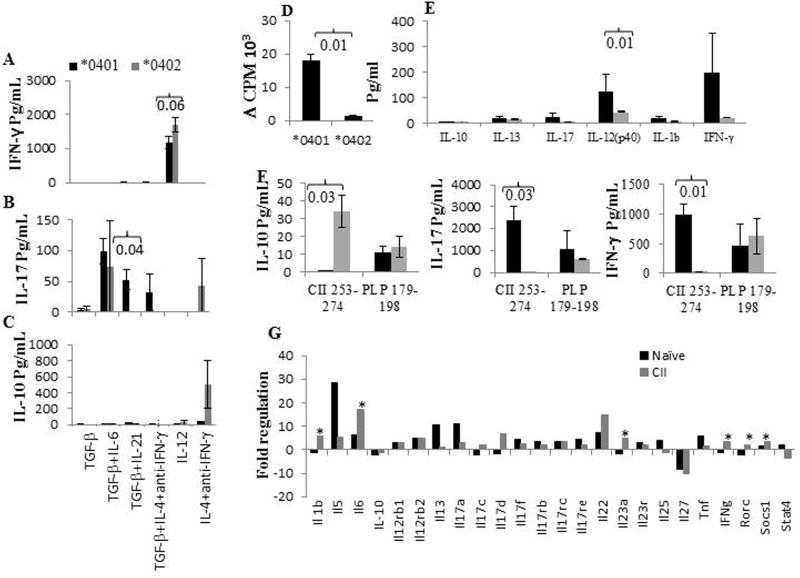
A recent study has suggested that in vitro generated TH cells maintain cytokine expression in vivo [11]. We addressed the hypothesis that T cells selected by the shared epitope are predetermined to produce cytokines that generate pro-inflammatory conditions, while *0402-selected T cells produce pro- and anti-inflammatory cytokine profile. We tested this by determining if T cells from naïve *0401 and *0402 mice can be driven to produce a certain TH profile in vitro. CD4+CD25-CD62Lhi T cells isolated from naïve Tg mice were cultured in the presence of CD3/CD28 with or without polarizing conditions to drive them to produce IL-17, IL-10, or IFNγ (Fig 1A-C). CD4 T cells from *0401 mice produced significantly higher IL-17 levels in the presence of TH17 polarizing conditions compared to *0402 mice, p=0.04. DRB1*0402 mice, on the other hand, showed a trend towards higher levels of IL-10 and IFNγ than *0401 mice, though differences between the strains were not significant. Next we evaluated the antigen-specific immune response; CII- immunized mice were challenged in vitro with CII or DR4-restricted CII-peptide 254-273 [10]. The *0402 mice generated a much lower response to CII and also produced significantly lower levels of pro-inflammatory IL-12(p40) compared to *0401 mice (Fig 1D, 1E). Both *0401 and *0402 can bind CII 254-273 [12]. To make sure that T cells selected by *0402 are not defective in producing proinflammatory cytokines, both strains were immunized with CII- 254-273 or arthritis-irrelevant PLP 179-189 and production of IL-17, IFNγ and IL-10 was measured in response to in vitro challenge with the antigen (Fig 1F). *0402 mice produced significantly higher amounts of IL-10 with lower IL-17 compared to *0401 mice in response to CII-254-273. Both strains produced similar amounts of cytokines in response to PLP-179-189 suggesting that *0402 mice can produce TH17 response in an antigen-specific manner.
Figure 1.
Shared epitope determines cytokine profile. Naïve CD4+CD25-CD62Lhi cells derived from transgenic mice were cultured with anti-CD3/CD28 in polarizing conditions, and production of cytokines, A) IFNγ, B) IL-17 and C) IL-10 from the supernatants were measured by ELISA (N= 4/group). The values are normalized with the media controls. D) In vitro T cell response to CII in transgenic mice, E) Cytokines measured in the supernatants of the culture in 1D, F) Mice immunized with CII-254-273 or arthritis-unrelated peptide PLP 179-198 were cultured with the immunizing peptide and culture supernatants measured for production of TH1 (IFNγ), TH2 (IL-10), and TH17 (IL-17) cytokines. G) Comparison of the expression levels of cytokines transcripts, in the spleens of *0401 and *0402 mice (n=3 in each group). The bar graphs depict comparison of mean fold change in gene transcripts of *0401 mice compared to *0402 mice in naïve mice and CII-immunized mice. * P<0.05. Black boxes indicate *0401 mice and gray boxes represent *0402 mice in all figures.
SE is associated with higher gene expression of TH17 cytokines
Next we determined if the presence of *0401 is associated with increased gene expression of TH17 regulatory network that can cause immune dysregulation (Fig 1G). Naïve *0401 mice expressed TH17, IL-6 and, IL-17a, transcripts at 7 fold or higher compared to *0402 mice while transcripts for IL-27, a cytokine associated with suppression of TH17 response, was expressed 8 fold lower in the former (Fig 1G). In addition, TNF transcripts were also expressed at 6 fold higher levels in *0401 compared to *0402 mice. Although naïve mice did not show significant differences between the 2 strains in gene expression of TH1/TH17 cytokines, the differences in some of the TH1/Th17 cytokines were significant in splenic cells isolated from CII-immunized mice (Fig 1G). After CII immunization, *0401 mice expressed TH17 suppressors, IL-27 and STAT4, at 5 or more fold lower levels and cytokines involved in TH17 network, IL-6, Rorc and IL-23, at significantly higher levels than *0402 mice. Among chemokines, transcripts for CXCL5, a potential target for arthritis, were expressed at significantly increased levels in naïve *0401 mice compared to *0402 mice, P<0.01 (data not shown).
TH17 differentiation requires different polarizing conditions for *0401 and *0402 mice
We further ascertained if CD4 T cells from *0401 mice are predetermined to differentiate into TH17 subset which might explain the association of SE and RA. Since in vivo conditions involve activation of T cells via APCs, we addressed the kinetics of response of naïve CD4 T cells in the presence of irradiated APCs. Naïve CD4 T cells (5×105) were cultured with irradiated APCs and CD3/CD28 (Fig 2A). In the absence of any polarizing conditions, *0401 mice showed a trend for higher levels of pro-inflammatory cytokines, IFNγ and IL-17a, compared to *0402 mice. Over 6 days of culture, *0401 mice showed a shift in cytokine response even though the changes were not significant except for IL-10 (P<0.05). However, no significant differences were observed between the 2 strains. Next we addressed the kinetics of response generated in polarizing conditions. Both strains produced similar levels of IFNγ in TH1 polarizing condition (Fig 2B, 2C). When cultured in TH2 or TH17 polarizing conditions for 3 days, production of IL-10 and IL-17 showed the same trend as observed in Figure 3A. However, the differences were not significant due to variability with in the strains which is reflective of the variability in responses generally observed in human studies. After day 6, CD4 T cells from *0402 mice produced very low levels of most of the cytokines except IL-10 and IFNγ. Although IL-10 has been suggested to be protective for autoimmune diseases, CD4+ T cells from *0401 mice can be driven to produce IL-10 under polarizing conditions as well as in response to CII (Fig 2B, 2C and Fig 1F). From this data one can speculate that kinetics of immune response may differ in the presence of SE and non-SE alleles.
Figure 2.
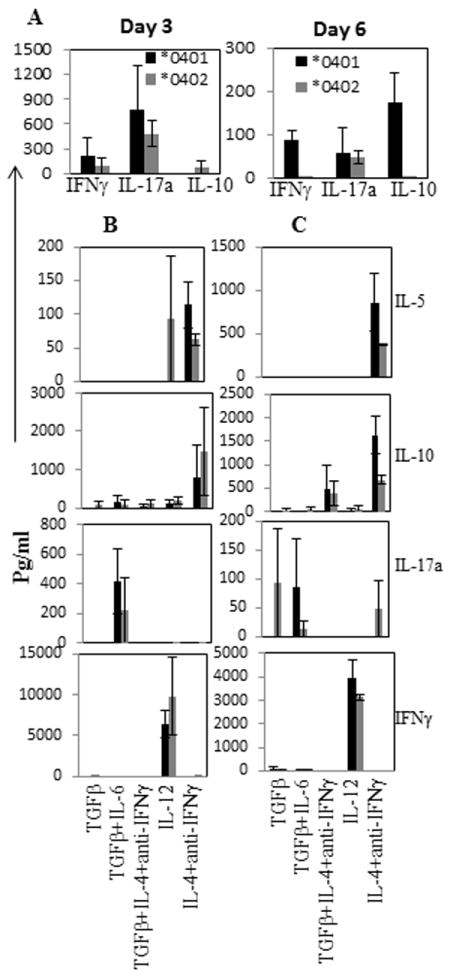
Kinetics of cytokine production, day 3 and day 6, in the supernatant by naïve CD4+CD25-CD62Lhi cells when cultured in the presence of matured BMDCs and anti-CD3/CD28 without polarizing conditions A), and with polarizing conditions B) day 3 and C) day 6. Fig depicts mean and SEM (N=3-5 mice in each group)
Figure 3.
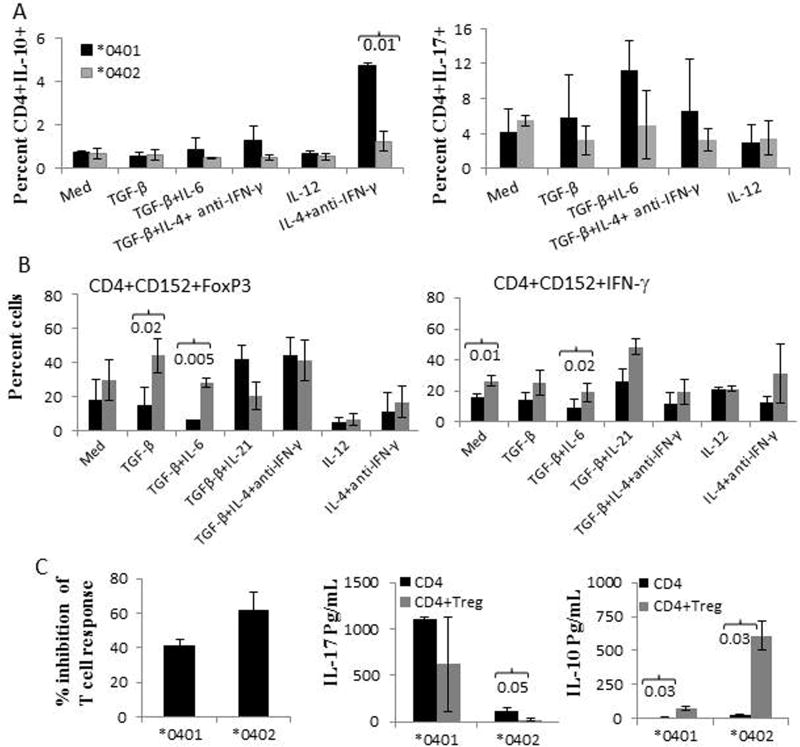
*0402 promotes generation of T regulatory cells. Naïve T cells from transgenic mice were cultured in the presence of BMDCs and various polarizing conditions for 6 days, (N=4 mice in each group) A) CD4+IL-10+ and CD4+IL-17+ cells. B) Generation of T regulatory, CD4+CD152+FoxP3+ cells and T regulatory cells producing IFNγ from naive CD4+CD25-CD62L cells in the presence of BMDCs and different polarizing conditions C) CD4+CD25- T cells from *0401 and *0402 mice immunized with CII peptide 254-273 were cultured in the absence or presence of CD4+CD25hi T cells (ratio 1:1), Fig depicts mean and SEM, N=4-6 mice/group. Supernatants were used to measure IL-17 and IL-10. Gating of various cells is depicted in supplementary Fig.
*0402 leads to differentiation of CD4 T cells to Treg cells in all polarizing conditions
The above observations were confirmed by enumerating the IL-10 or IL-17 producing CD4 T cells. Bone marrow derived dendritic cells (BMDCs) were cultured with sorted naïve CD4 T cells in the presence of various polarizing conditions. *0401 mice showed a significant increase in CD4+IL-10+ T cells compared to *0402 mice, P<0.01, in TH2 polarizing conditions (Fig 3A). In TH17 polarizing conditions, CD4+ T cells differentiated into CD4+IL-17+ T cells in *0401 mice, while in *0402 mice no change in the numbers of CD4+IL-17+ T cells was observed between TH17 polarizing condition and medium control. None of the differences were significant between the 2 strains (Fig 3A). Since previous studies have shown that *0402 mice harbor higher number of Tregs [10], we ascertained if CD4+ T cells are being differentiated into Treg cells in them.
Naïve CD4 T cells were cultured with CD11c+ BMDCs in polarizing conditions. Differentiation of CD4 T cells to Tregs was determined by the expression of CD152 and FOXP3 (Fig 3B). CD4 T cells differentiated into Treg cells in *0402 mice in various polarizing conditions, with significant differences between the 2 strains in the presence of TGFβ and TGFβ+IL-6. More than 25% of CD4+CD152 Treg cells express IFNγ in various polarizing conditions in *0402 mice and around 5-20% of CD4+CD152+FoxP3+ cells expressed IFNγ (not shown), confirming the above results and suggesting that IFNγ may be involved in protection from arthritis. Our observations suggest that the presence of *0402 gene leads to selection of TCR that is predetermined to differentiate into Treg cells via DCs in certain conditions. We next determined if Tregs suppress antigen-specific immune response and inflammatory cytokines (Figure 4C). Peptide CII 254-273 activated CD4+CD25- sorted T cells were cultured in the absence or presence of CD4+CD25hi cells and the peptide. T cell response was suppressed in both strains in the presence of CD4+CD25hi (40% suppression of T cell response in *0401 and 62% in *0402). Cytokines measured from the supernatants of the cultures showed a significant suppression of IL-17 in *0402 mice (P<0.05) when Tregs were cultured with the activated CD4 T cells. Both strains showed significant increase in IL-10 production after culturing in the presence of Tregs.
Figure 4.
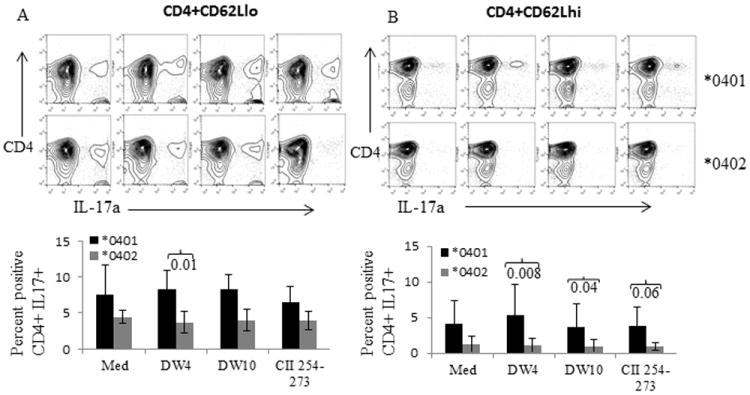
Genetically susceptible *0401 mice have autoreactive T cells. Naïve CD4+CD62Lhi and memory CD4+CD62Llo T cells from *0401 and *0402 mice were cultured with BMDCs primed overnight with *0401 derived peptide 65-79 (DW4), *0402 derived peptide 65-79 (DW10) and CII 254-273. Cultured cells were analyzed by FACS for the expression of CD4+IL-17A+ cells. A representative example of FACS analysis depicting CD4+ IL-17A+ cells and histogram showing mean ±S.E. (N=3-4 mice per group) for percent CD4+CD17+ T cells in both strains A) CD4+CD62Llo cells and B) CD4+CD62Lhi cells.
Shared epitope activated DCs facilitate proliferation of IL-17 producing autoreactive memory T cells
Next we determined the role of peptide loaded DCs in differentiation of CD4 T cells. We hypothesized that SE and CII-peptide activated DCs should facilitate differentiation of CD4 T cells to TH17+ cells in *0401 mice only. Bone marrow derived DCs were cultured overnight with SE peptides DW4-65-79, DW10-65-79 or CII-254-273. After 24 hours, naïve (CD4+CD62Lhi) and memory (CD4+CD62Llo) T cells were added to the culture with polarizing cytokines and TH17 CD4 T cells were analyzed. Activated DCs in media as well as shared epitope loaded DCs led to differentiation of memory as well as naïve CD4 T cells to CD4+IL-17+ T cells in *0401 mice (Fig 4). Naive T cells also showed a robust SE and CII 254-273 induced TH17 polarization in *0401 mice as compared to *0402 mice (Fig 4B). These data support the above observations and suggest that *0401 mice harbor autoreactive cells that can proliferate to become proinflammatory cells and produce TH17 cytokines.
*0402 mice produce antigen-specific TH2/TH17 cytokines but do not develop arthritis
To determine the significance of the in vitro data shown here, we evaluated the effect of SE in vivo in arthritis by measuring the cytokine profile in the sera of CII-immunized mice (Fig 5A). The *0402 mice produced significantly higher levels of TH17 cytokines (IL-9 and IL-17) compared to *0401 mice. On the other hand, *0401 mice had higher levels of IL-13 and IL-23. Together with previous data [13], our observations suggest that IL-13 and IL-23 may be crucial in the progression of CIA. High Eotaxin and granulocyte macrophage colony stimulating factor (GMCSF) in *0401 mice suggest that recruitment of inflammatory cells may be critical for pathogenesis.
Figure 5.
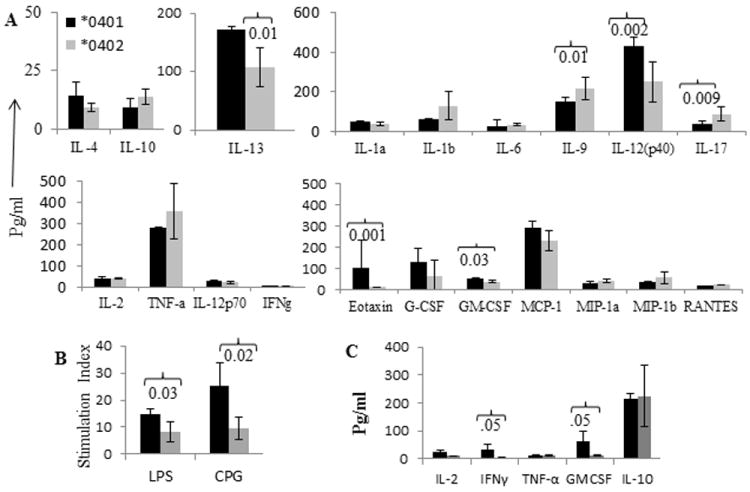
Antigen-specific TH17 related cytokine production is delayed in *0402 mice. A) Cytokine profile from sera collected after 5 weeks of CII immunization of transgenic mice (Mean± SEM, N=3 per group). *0401 mice produce more robust innate immune response compared to *0402 mice. B) In vitro immune response to LPS and CPG in naïve mice and C) Cytokines produced in response to LPS in both strains (Mean± SEM, N=4 per group).
*0401 mice mount a much robust innate immune response
Both LPS and CPG are bacterial products that generate response via TLR4 and TLR9 respectively. We tested if both strains differ in CII-specific response but generate similar innate immune response. Our data showed that *0401 mice generated significantly higher response to both (LPS and CPG P<0.03 and P<0.02 respectively) and produced higher pro-inflammatory cytokines to LPS as compared to *0402 mice (Fig 5B, 5C).
DISCUSSION
DRB1*0401 and *0402 molecules differ in the peptide binding region by 3 amino acids but depict a complete opposite association with RA. However, the mechanism of association has not been clearly elucidated. Using transgenic mice expressing the *0401 and *0402 gene constructs, we have previously shown that *0401 renders susceptibility to develop arthritis while *0402 mice do not develop arthritis [9, 14]. We hypothesized that HLA molecules influence susceptibility/protection to inflammatory disease by predetermining the cytokine profile. In this study we have addressed this hypothesis and shown that naïve CD4 T cells from *0401 mice are driven to produce TH1/TH17 cytokines in response to TCR dependent proliferation, while CD4 T cells from *0402 mice produce a TH1/TH2 response. Further, our observations suggest that the T cell repertoire selected by *0401 may harbor autoreactive T cells that are predetermined to produce a proinflammatory immune response.
However, in vitro conditions may not recapitulate in vivo conditions. We simulated in vivo conditions by culturing naïve CD4 cells with CD11c+ BMDCs. Even though we observed strong trends, there was no significant difference between *0401 and *0402 mice under TH1/TH17 polarizing conditions. Interestingly, under the TH2 polarizing conditions, increased number of naïve CD4+ T cells differentiated into IL-10+ cells in *0401 mice compared to *0402 mice (Fig 4). Although IL-10 has been suggested to be protective in most of the models of arthritis, it is important for isotype switching and has been shown to be increased in RA patients [15, 16]. We believe that production of IL-10 later during effector phase may be associated with CD4 T cells and B cells interaction for antibody production as suggested before [17]. However, in response to CII 254-273 peptide, *0402 mice produced a very robust IL-10 in confirmation with our previous findings [10] while *0401 mice produced IL-17. Peptide CII-254-273 is a DR4-restricted peptide and has been shown to be important in the development of CIA [10, 18]. Binding studies have shown that both *0401 and *0402 can bind this peptide although in different registries [12] and may activate different T cells. This data suggests that while *0401 mice produce IL-10 to CII during late phase of disease, processing of CII by APCs may lead to availability of T cell epitope comprising amino acids at 254-273 position that generates IL-17 response. Both strains produced similar cytokine profile to an arthritis irrelevant peptide. This suggests that while SE augments proliferation and differentiation of autoreactive memory cells and naïve T cells to CD4+IL-17+ cells in arthritis, *0402 leads to generation of Tregs that may be involved in protection from arthritis.
We hypothesized that HLA alleles have been selected by nature to clear infection via TCR selection, that may be predetermined to produce certain cytokines. However, some of these HLA alleles, under certain conditions, can generate autoimmune response [19]. Thus autoimmunity could be a bystander effect of the enormous immune response generated to clear infection or due to epitope mimicry. To test this hypothesis, we investigated the immune response generated by LPS and CPG by *0401 and *0402 mice. Our data showed that *0401 mice generated much more robust response and produced higher pro-inflammatory cytokines to LPS compared to *0402 mice. IL-17 is required for clearing infections [20]. A recent study has suggested that MHC class II molecules may regulate innate immune response [20]. Although the initial response to LPS is generated by innate immunity, it suggests indirectly that *0401 mice produce high immune response and may be able to clear infection but the cytokine profile favors autoimmunity. Our recent observations support this contention as the gut microbiome of naïve *0401 mice is associated with TH17 profile while *0402 mice is not [21]. IL-17 is required for clearing infections [22].
An important role of IL-17 has been suggested in the pathogenesis of RA [23] which is supported by the recent phase II trial with IL-17 neutralizing antibodies in RA patients [24]. We confirmed the significance of TH17 response in pathogenesis of CIA. In vivo studies with the cytokine profile in sera of CII-immunized mice and in vitro data suggest that IL-13 and IL-12(p40), a chain for IL-23, may be critical in progression of disease as they were produced at significantly higher levels in *0401 mice than in *0402 mice. IL-13 is involved in activation and differentiation of B cells into antibody producing cells [25, 26]. Our previous studies with *0401 mice have shown a significant involvement of B cells and IL-13 in CIA [27-29]. Similar observations have been reported in RA patients where higher levels of IL-13 in sera were reported compared to normal sera [30]. Interestingly, *0402 mice produce IL-9 and IL-17 cytokines in response to CII suggesting that the TH17 response to CII is delayed in them. IL-9 is a pleiotropic cytokine that enhances function of FOXP3 Treg cells as well as induction of TH17 differentiation [31]. It is also possible that IL-17 production is required early during the onset of disease and IL-23 is required later for progression. This notion is supported by the in vitro observations in this study.
DRB1*0401 mice produced much higher levels of chemokines Eotaxin and GMCSF, both known to exacerbate inflammatory immune response in RA [32]. In humans, Eotaxin1 is involved in increased expression of matrix metalloproteinase (MMP) -3 and MMP 13 that causes cartilage degradation [33]. These data suggest that cytokines and chemokines milieu produced by CD4 T cells and APCs are involved in disease pathogenesis or drive generation of Tregs. Our observations support this contention as in certain polarizing conditions, *0402 generated Treg cells that produce IFNγ and probably control the immune response. However, these data do not directly prove that IFNγ produced by Treg cells is protective as suggested by a previous study [34] and further studies are needed to prove this hypothesis.
In conclusion our study provides an insight into the role of shared epitope and suggests that *0401 and *0402 molecules can produce all types of cytokine though the requirement for producing proinflammatory cytokines like TH1/TH17 may differ. One can speculate that an initial event during the onset of disease may involve differentiation of CD4 T cells to TH17 producing cells or activation of memory autoreactive T cells in certain conditions in individuals with SE alleles while non-associated HLA alleles can generate Treg cells in association with TH1 profile thus controlling immune dysregulation. These speculations need to be further investigated.
Materials and Methods
Transgenic mice
The generation of DRB1*0401 and DRB1*0402 transgenic (Tg) mice has been described previously [6, 35]. Aβ°.DRB1*0401 and Aβ°.DRB1*0402 mice were mated with MHCII Δ/Δ (AE°) mice [36] to generate AE°.DRB1*0401 and AE°.DRB1*0402 mice [6, 9]. Thus both strains have the same background except for the HLA gene construct. Mice of both sexes (8-12 weeks of age) used in this study were bred and maintained in the pathogen-free Immunogenetics Mouse Colony at the Mayo Clinic, Rochester, MN in accordance with the Animal Use and Care Committee. All the experiments included littermate controls and were carried out with the approval of the Institute’s Animal use and care committee.
For convenience, DRB1*0402.AEo Tg mice will be referred to as *0402 and DRB1*0401 Tg mice as *0401 in the manuscript.
Flow Cytometry
The expression of DR, was analyzed by flow cytometry using mAbs L227 (anti-DR), as previously described [9]. Conjugated specific anti-CD3, CD4, CD11c, CD62L, CD152, (BD biosciences) were also utilized. All cell surface markers were analyzed with cells pooled from 2 mice/ strain and repeated 2-3 times.
Intracellular staining for FoxP3 was performed using commercial antibodies (eBioscience) as per the manufacturer’s instructions. Phycoerythrin-conjugated rat IGg2a (BD biosciences) was used as the isotype control for FoxP3 staining. For CD4+IL10+ and CD4+IL-17+ T cells, CD4 T cells were isolated using anti mouse CD4 magnetic beads and stained with anti-CD4 FITC conjugated (BD biosciences). Cells were fixed, and permeabilized using a cytofix kit and stained with PE-conjugated anti-mouse IL-17A or IL-10 (eBioscience). Flow cytometry data was analyzed with Flo Jo software. Strategy for gating cells positive for various markers is depicted in supplementary fig.
Generation of and culture with CD11c DCs
Bone marrow cells harvested from transgenic mice were cultured for 5 days in medium (RPMI1640 containing 1% penicillin-streptomycin and supplements) with GM-CSF (10ng/ml) and IL-4 (1 ng/ml) for maturation of dendritic cells (DCs). After 5 day culture, DCs were harvested and checked by FACS for CD11c. Mature CD11c DC were cultured with splenic CD4 cells isolated by positive selection using CD11c microbeads (Miltenyi Biotech). In some experiments, CD4 cells were sorted by FAC Sorter for the CD4+CD62Lhi and CD4+CD62lo cells before culturing with DCs.
Proliferation assay
Mice were immunized with 200 μg of CII emulsified 1:1 in CFA (Difco) intradermally at the base of the tail and one hind footpad and in vitro T cell proliferation was done as described [9]. Sera was collected 35 days after CII-immunization in some mice. In some experiments peptide derived from type II collagen, CII- 254-273, and proteolipid protein, PLP 179-198, were tested for T cell proliferation. Both peptides were synthesized and purified at Mayo Clinic Peptide Facility. Mice were primed with 200 μg of peptide and challenged in vitro with 100 μg/ml of the peptide. For presentation of CII and its derived peptide 254-273, CD4+ cells were sorted by FAC sorter from LNCs harvested from primed mice. CD4+ cells (5×105) were cultured in vitro alone or in the presence or absence of irradiated spleen cells as antigen presenting cells (APCs) and CII (50 μg/ml). The other cultures utilized CD3/CD28 coated plates. Lipopolysaccharide (LPS) (5ug/ml) and CPG (1ug/ml) were also used.
T cell proliferation was determined by thymidine incorporation. Stimulation index of 2 or more was taken as positive response. All experiments were reproduced 2 or 3 times. Each experiment was done twice with cells pooled from 2 mice. Lipopolysaccharide (LPS) (5μg/ml) and cytosine connected to guanine through phosphodiester bonds (CPG) (1μg/ml) were also used in some experiments.
TH1/TH2/TH17 differentiation
Naïve CD4 T cells, 50,000 CD4+CD25-CD62L+ cells, sorted from spleen after staining with conjugated antibodies were plated on anti CD3 Abs (5ug/ml) and anti-CD28 Abs (1ug/ml) coated plates. For differentiation, following cytokines were added to the cultures- for TH17 differentiation TGFβ (5ng/ml) and a mixture of TGFβ (5ng/ml) and IL-6 (20 ng/ml), for TH2 differentiation a mixture of TGFβ (5ng/ml) +IL-4 (2μg/ml) + anti IFNγ (2 μg/ml) and, a mixture of IL4+anti IFNγ; and for Th1 differentiation, IL-12 (2μg/ml). In some cases TGFβ+IL-21 were also used for TH17 differentiation. The supernatant from the cultures were collected on day 5 and measured for IFNγ, IL-17, IL10 and IL-5. For enumerating CD4+IL10+ and CD4+IL-17+ T cells, cells were permeabilized using a cytofix kit and intracellular staining for IL-17 and IL-10 using flourochrome labeled anti-IL17 and IL-10 Abs (eBioscience).
For determining if presentation of self-DRB1 derived peptide 65-79 can lead to differentiation of CD4 T cells to CD4+IL-17+ T cells, BM-derived DCs (10,000) were cultured with or without the peptides, DRB1*0401 derived peptide DW4-65-79 and DRB1*0402 derived peptide DW10-65-79 overnight at a concentration of 50ug/ml. CD4+CD25-CD62L+ cells (5×104) were added to the culture with a mixture of polarizing cytokines (TGFβ, TGFβ+IL-6, TGFβ+IL-4+ anti IFNγ, IL4+anti IFNγ, IL12) in anti-CD3 Abs and anti-CD28 coated plates. Cultures were stimulated with phorbol myristate acetate (PMA)/ionomycin/Brefeldin (lymphocyte activation cocktail with BD glogiplug) during the last 6 hours of culture. Cells were harvested and stained for CD4+IL-17+ T cells as above.
T regulatory cell differentiation assay
Bone marrow DCs were cultured with 5×105 CD4+CD25-CD62L+ naïve T cells at a ratio of 1:5 of DC: CD4 cells in the presence of anti-CD3 Abs (5ug/ml) and anti-CD28 Abs (1ug/ml) for 5 days with polarizing cytokines as described. Lymphocyte activation cocktail with BD glogiplug was added to the cultures 6 hours before harvesting. T regulatory, CD4+CD152+Foxp3, and CD4+CD152+ IFNγ cells were analyzed by Flow-cytometry after staining with conjugated specific antibodies and gating on CD4+ cells.
T cell suppression assay
Mice were immunized with type II collagen (CII)-derived peptide 254-273. Ten days post-immunization, CD4+CD25- and CD4+CD25hi (Tregs) T cells were sorted from spleen. CD4+CD25- T cells were cultured in the presence of CII-254-273 and in the absence or presence of Tregs in a ratio of 1:1 (1×105 CD4+CD25- and 1×105 CD4+CD25hi). Cells were pooled from 2 mice in some experiments and experiment was done thrice. T cell response was measured as described above. Culture supernatants were used to measure IL-17 and IL-10 using available kits (eBioscience).
Cytokines
Cytokines from sera and supernatants were measured using the Bio-Plex array system with the mouse cytokine 23-plex panel as per manufacturer’s instructions and analyzed with Bio-Plex manager 2.0 software (Bio-Rad laboratories, Hercules, CA). Some cytokines were measured by capture ELISA using kits according to manufacturer’s instructions (BD Biosciences). Cytokines in sera were tested in mice immunized with CII for 6 weeks.
RNA isolation and real- time polymerase chain reaction
Total RNA from cells was extracted using the RNeasy kit and protocol (Qiagen). cDNA was prepared using RT2 First Strand Kit cDNA Synthesis Kit and Primer Mixes (SA Biosciences). The quantification of gene expression related to the TH1/TH17 Regulatory Network was performed using the RT2 Profiler PCR Array PAMM-0773 (SA Biosciences) and the HT7900 Fast Real-Time PCR System (ABI). Product amplification was measured and analyzed according to the manufacturer’s instructions.
Statistical analysis
Difference in levels of cytokines and various was calculated using Student’s t test or T test with unequal variance where ever mentioned. Level of p≤0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Julie Hanson and her staff in the Mayo Immunogenetics mouse colony for breeding and taking care of the mice. The study was supported by the NIH grants AR30752 and AI 075262.
Footnotes
Conflict of Interest
The authors declare no commercial or financial conflict of interest.
References
- 1.Wordsworth BP, Lanchbury JS, Sakkas LI, Welsh KI, Panayi GS, Bell JI. HLA-DR4 subtype frequencies in rheumatoid arthritis indicate that DRB1 is the major susceptibility locus within the HLA class II region. Proc Natl Acad Sci U S A. 1989;86:10049–10053. doi: 10.1073/pnas.86.24.10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stastny P. Association of the B-cell alloantigen DRw4 with rheumatoid arthritis. N Engl J Med. 1978;298:869–871. doi: 10.1056/NEJM197804202981602. [DOI] [PubMed] [Google Scholar]
- 3.Gregersen PK, Shen M, Song QL, Merryman P, Degar S, Seki T, Maccari J, Goldberg D, Murphy H, Schwenzer J, et al. Molecular diversity of HLA-DR4 haplotypes. Proc Natl Acad Sci U S A. 1986;83:2642–2646. doi: 10.1073/pnas.83.8.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 5.Moller E, Bohme J, Valugerdi MA, Ridderstad A, Olerup O. Speculations on mechanisms of HLA associations with autoimmune diseases and the specificity of “autoreactive” T lymphocytes. Immunol Rev. 1990;118:5–19. doi: 10.1111/j.1600-065x.1990.tb00811.x. [DOI] [PubMed] [Google Scholar]
- 6.Taneja V, Taneja N, Behrens M, Pan S, Trejo T, Griffiths M, Luthra H, David CS. HLA-DRB1*0402 (DW10) transgene protects collagen-induced arthritis-susceptible H2Aq and DRB1*0401 (DW4) transgenic mice from arthritis. J Immunol. 2003;171:4431–4438. doi: 10.4049/jimmunol.171.8.4431. [DOI] [PubMed] [Google Scholar]
- 7.Snijders A, Elferink DG, Geluk A, van Der Zanden AL, Vos K, Schreuder GM, Breedveld FC, de Vries RR, Zanelli EH. An HLA-DRB1-derived peptide associated with protection against rheumatoid arthritis is naturally processed by human APCs. J Immunol. 2001;166:4987–4993. doi: 10.4049/jimmunol.166.8.4987. [DOI] [PubMed] [Google Scholar]
- 8.Kirschmann DA, Duffin KL, Smith CE, Welply JK, Howard SC, Schwartz BD, Woulfe SL. Naturally processed peptides from rheumatoid arthritis associated and non-associated HLA-DR alleles. Journal of immunology. 1995;155:5655–5662. [PubMed] [Google Scholar]
- 9.Taneja V, Behrens M, Mangalam A, Griffiths MM, Luthra HS, David CS. New humanized HLA-DR4-transgenic mice that mimic the sex bias of rheumatoid arthritis. Arthritis Rheum. 2007;56:69–78. doi: 10.1002/art.22213. [DOI] [PubMed] [Google Scholar]
- 10.Taneja V, Behrens M, Basal E, Sparks J, Griffiths MM, Luthra H, David CS. Delineating the role of the HLA-DR4 “shared epitope” in susceptibility versus resistance to develop arthritis. J Immunol. 2008;181:2869–2877. doi: 10.4049/jimmunol.181.4.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nurieva R, Yang XO, Chung Y, Dong C. Cutting edge: in vitro generated Th17 cells maintain their cytokine expression program in normal but not lymphopenic hosts. Journal of immunology. 2009;182:2565–2568. doi: 10.4049/jimmunol.0803931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diab BY, Lambert NC, L’Faqihi FE, Loubet-Lescoulie P, de Preval C, Coppin H. Human collagen II peptide 256-271 preferentially binds to HLA-DR molecules associated with susceptibility to rheumatoid arthritis. Immunogenetics. 1999;49:36–44. doi: 10.1007/s002510050461. [DOI] [PubMed] [Google Scholar]
- 13.Behrens M, Trejo T, Luthra H, Griffiths M, David CS, Taneja V. Mechanism by which HLA-DR4 regulates sex-bias of arthritis in humanized mice. Journal of autoimmunity. 2010;35:1–9. doi: 10.1016/j.jaut.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taneja V, Griffiths MM, Luthra H, David CS. Modulation of HLA-DQ-restricted collagen-induced arthritis by HLA-DRB1 polymorphism. International immunology. 1998;10:1449–1457. doi: 10.1093/intimm/10.10.1449. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Li J, Gao H, Wang C, Luo J, Lv Z, Li X. Comprehensive evaluation of different T-helper cell subsets differentiation and function in rheumatoid arthritis. Journal of biomedicine & biotechnology. 2012;2012 doi: 10.1155/2012/535361. 535361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maini R, St Clair EW, Breedveld F, Furst D, Kalden J, Weisman M, Smolen J, Emery P, Harriman G, Feldmann M, Lipsky P. Infliximab (chimeric anti-tumour necrosis factor alpha monoclonal antibody) versus placebo in rheumatoid arthritis patients receiving concomitant methotrexate: a randomised phase III trial. ATTRACT Study Group. Lancet. 1999;354:1932–1939. doi: 10.1016/s0140-6736(99)05246-0. [DOI] [PubMed] [Google Scholar]
- 17.Hummelshoj L, Ryder LP, Poulsen LK. The role of the interleukin-10 subfamily members in immunoglobulin production by human B cells. Scandinavian journal of immunology. 2006;64:40–47. doi: 10.1111/j.1365-3083.2006.01773.x. [DOI] [PubMed] [Google Scholar]
- 18.Rosloniec EF, Brand DD, Myers LK, Esaki Y, Whittington KB, Zaller DM, Woods A, Stuart JM, Kang AH. Induction of autoimmune arthritis in HLA-DR4 (DRB1*0401) transgenic mice by immunization with human and bovine type II collagen. Journal of immunology. 1998;160:2573–2578. [PubMed] [Google Scholar]
- 19.Mangalam AK, Taneja V, David CS. HLA class II molecules influence susceptibility versus protection in inflammatory diseases by determining the cytokine profile. Journal of immunology. 2013;190:513–518. doi: 10.4049/jimmunol.1201891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X, Zhan Z, Li D, Xu L, Ma F, Zhang P, Yao H, Cao X. Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining activation of the kinase Btk. Nature immunology. 2011;12:416–424. doi: 10.1038/ni.2015. [DOI] [PubMed] [Google Scholar]
- 21.Gomez A, Luckey D, Yeoman CJ, Marietta EV, Berg Miller ME, Murray JA, White BA, Taneja V. Loss of sex and age driven differences in the gut microbiome characterize arthritis-susceptible 0401 mice but not arthritis-resistant 0402 mice. PloS one. 2012;7:e36095. doi: 10.1371/journal.pone.0036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cua DJ, Tato CM. Innate IL-17-producing cells: the sentinels of the immune system. Nature reviews Immunology. 2010;10:479–489. doi: 10.1038/nri2800. [DOI] [PubMed] [Google Scholar]
- 23.van den Berg WB, Miossec P. IL-17 as a future therapeutic target for rheumatoid arthritis. Nature reviews Rheumatology. 2009;5:549–553. doi: 10.1038/nrrheum.2009.179. [DOI] [PubMed] [Google Scholar]
- 24.Patel DD, Lee DM, Kolbinger F, Antoni C. Effect of IL-17A blockade with secukinumab in autoimmune diseases. Annals of the rheumatic diseases. 2013;72(Suppl 2):iii116–iii123. doi: 10.1136/annrheumdis-2012-202371. [DOI] [PubMed] [Google Scholar]
- 25.Minty A, Chalon P, Derocq JM, Dumont X, Guillemot JC, Kaghad M, Labit C, Leplatois P, Liauzun P, Miloux B, et al. Interleukin-13 is a new human lymphokine regulating inflammatory and immune responses. Nature. 1993;362:248–250. doi: 10.1038/362248a0. [DOI] [PubMed] [Google Scholar]
- 26.Defrance T, Carayon P, Billian G, Guillemot JC, Minty A, Caput D, Ferrara P. Interleukin 13 is a B cell stimulating factor. The Journal of experimental medicine. 1994;179:135–143. doi: 10.1084/jem.179.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Behrens M, Smart M, Luckey D, Luthra H, Taneja V. To B or not to B: role of B cells in pathogenesis of arthritis in HLA transgenic mice. Journal of autoimmunity. 2011;37:95–103. doi: 10.1016/j.jaut.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taneja V, Krco CJ, Behrens MD, Luthra HS, Griffiths MM, David CS. B cells are important as antigen presenting cells for induction of MHC-restricted arthritis in transgenic mice. Molecular immunology. 2007;44:2988–2996. doi: 10.1016/j.molimm.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luckey D, Medina K, Taneja V. B cells as effectors and regulators of sex-biased arthritis. Autoimmunity. 2012;45:364–376. doi: 10.3109/08916934.2012.665528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tokayer A, Carsons SE, Chokshi B, Santiago-Schwarz F. High levels of interleukin 13 in rheumatoid arthritis sera are modulated by tumor necrosis factor antagonist therapy: association with dendritic cell growth activity. The Journal of rheumatology. 2002;29:454–461. [PubMed] [Google Scholar]
- 31.Elyaman W, Bradshaw EM, Uyttenhove C, Dardalhon V, Awasthi A, Imitola J, Bettelli E, Oukka M, van Snick J, Renauld JC, Kuchroo VK, Khoury SJ. IL-9 induces differentiation of TH17 cells and enhances function of FoxP3+ natural regulatory T cells. Proc Natl Acad Sci U S A. 2009;106:12885–12890. doi: 10.1073/pnas.0812530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cornish AL, Campbell IK, McKenzie BS, Chatfield S, Wicks IP. G-CSF and GM-CSF as therapeutic targets in rheumatoid arthritis. Nature reviews Rheumatology. 2009;5:554–559. doi: 10.1038/nrrheum.2009.178. [DOI] [PubMed] [Google Scholar]
- 33.Hsu YH, Hsieh MS, Liang YC, Li CY, Sheu MT, Chou DT, Chen TF, Chen CH. Production of the chemokine eotaxin-1 in osteoarthritis and its role in cartilage degradation. Journal of cellular biochemistry. 2004;93:929–939. doi: 10.1002/jcb.20239. [DOI] [PubMed] [Google Scholar]
- 34.Page CE, Smale S, Carty SM, Amos N, Lauder SN, Goodfellow RM, Richards PJ, Jones SA, Topley N, Williams AS. Interferon-gamma inhibits interleukin-1beta-induced matrix metalloproteinase production by synovial fibroblasts and protects articular cartilage in early arthritis. Arthritis research & therapy. 2010;12:R49. doi: 10.1186/ar2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan S, Trejo T, Hansen J, Smart M, David CS. HLA-DR4 (DRB1*0401) transgenic mice expressing an altered CD4-binding site: specificity and magnitude of DR4-restricted T cell response. Journal of immunology. 1998;161:2925–2929. [PubMed] [Google Scholar]
- 36.Kouskoff V, Fehling HJ, Lemeur M, Benoist C, Mathis D. A vector driving the expression of foreign cDNAs in the MHC class II-positive cells of transgenic mice. Journal of immunological methods. 1993;166:287–291. doi: 10.1016/0022-1759(93)90370-m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.