Abstract
The transmembrane (TM) helices of most type II single-span membrane proteins (S-SMPs) of Escherichia coli occur near the N-terminus, where the cell’s targeting mechanisms can readily identify it as it emerges from the ribosome. But the TM helices of a few S-SMPs, such as RodZ, occur a hundred or more residues downstream from the N-terminus, which raises fundamental questions about targeting and assembly. Because of RodZ’s novelty and potential usefulness for understanding TM helix insertion in vivo, we examined its membrane targeting and assembly. We used RodZ constructs containing immuno tags before the TM domain to assess membrane insertion using Proteinase K digestion. We confirmed the Nin-Cout (Type II) topology of RodZ and established the absence of a targeting signal other than the TM domain. RodZ was not inserted into the membrane under SecA-depletion conditions or in the presence of sodium azide, which is known to inhibit SecA. Insertion failed when the transmembrane proton gradient was abolished with CCCP. Insertion also failed when RodZ was expressed in SecE-depleted E. coli, indicating that the SecYEG translocon is required for RodZ assembly. Protease accessibility assays of RodZ in other E. coli depletion strains revealed that insertion is independent of SecB, YidC, and SecD/F. Insertion was found to be only weakly dependent on the signal recognition particle (SRP) pathway: insertion was weakly dependent on the Ffh but independent of FtsY. We conclude that membrane insertion of RodZ requires only the SecYEG translocon, the SecA ATPase motor, and the transmembrane proton motive force (PMF).
Keywords: Membrane protein assembly, SecYEG translocase, SecA ATPase, single-span membrane proteins
Introduction
The mechanisms of membrane protein assembly and protein secretion of Gram-negative bacteria converge at the SecYEG translocon complex (protein-conducting channel) located in the inner membrane (Figure 1). The extensive literature on these mechanisms has been recently reviewed [1–4]. Secreted proteins and proteins destined for the outer membrane are driven post-translationally through the SecYEG channel by the ATP-powered SecA translocase. These secreted proteins are generally identified biologically by a hydrophobic N-terminal signal sequence that is cleaved by signal peptidase during secretion through the SecYEG channel. In contrast, multi-span membrane proteins (MPs) with short interhelical loops are assembled cotranslationally at SecYEG after delivery of active ribosomes by two essential components of the signal recognition particle (SRP) pathway [1–4]: Ffh, which is the protein component of the SRP protein-RNA complex, and surface-active FtsY, which partitions into the inner membrane from the cytoplasm [5] to receive the ribosome/nascent-chain (RNC) complex from the SRP and transfer it to SecYEG. In some cases, assembly is aided by another inner-membrane protein, YidC, which can directly contact SecYEG [6]. An exception to this picture occurs whenever a multi-span MP has long interhelical connecting loops: If the loops exceed about 30 amino acids in length, then SecA is required to transport the loops across the membrane, presumably through the channel [7–9].
Figure 1.

Targeting and membrane insertion of membrane proteins in E. coli and some single-span membrane proteins relevant to this study. (a) Schematic representation of the SecA protein secretion/membrane insertion pathway. (b) Schematic representation of the SRP (signal-recognition particle) pathway. The majority of secreted E. coli proteins are synthesized as preproteins with a cleavable signal peptide at their N-terminus. These preproteins are targeted to the SecYEG translocon post-translationally by the cytosolic ATPase SecA, sometimes helped by the molecular chaperone SecB [63]. The integral membrane protein YidC associates with SecYEG, but is not essential for insertion or secretion of many proteins. Many multi-span inner membrane proteins are targeted in a co-translational manner to the SecYEG translocase by the signal recognition particle (SRP, Ffh) and its receptor (FtsY) [64]. In the SRP-mediated targeting pathway, the SRP–ribosome nascent chain complex is targeted to FtsY at the membrane and then translocated via SecYEG [65]. (c) Examples of single-span membrane proteins. Momp2 [10] and LepΔH2 [8] are model type II single-span membrane proteins. FtsQ is one of the few native type-II membrane proteins whose biogenesis has been studied in much detail [13; 14]. The three proteins all have TM segments near the amino terminal end of the protein. The TM segment of RodZ, however, appears far downstream from the amino terminus. (d) RodZ constructs used in the present study for determinations of RodZ topology. Both constructs carry immuno tags on the N-terminal side of the single transmembrane segment to allow determination of the location of intact and proteolytically cleaved protein.
Much of our understanding of the long-loop SecA-intervention rules comes from extensive studies of two imitation type II single-span membrane proteins, Momp2 and LepΔH2 (Fig. 1c), which have their N-termini in the cytoplasm and their C-termini in the periplasm (Nin-Cout). A common feature of these model single-span MPs (S-SMPs) is that the hydrophobic TM segment occurs near the N-terminus, acting, in effect, as a non-cleavable signal sequence. Momp2 [10] is a chimeric proOmpA in which the cleavable signal sequence is replaced by the first 47 residues of the multi-span MtlA mannitol permease. Within these 47 residues is the first TM helix of MtlA. Because OmpA is a secreted protein driven by SecA, it was not surprising to find that Momp2 insertion required SecA [10; 11]. But in addition, targeting to SecYEG required SRP. In a much earlier study using variants of E. coli double-span leader peptidase (Lep), Andersson and von Heijne [8] found that a deletion mutant of Lep lacking TM2 (called H2) strictly required SecA for assembly based upon inhibition of SecA by sodium azide. The insertion of the native protein, FtsQ, which has an N-terminal TM segment, is also SecA dependent, but requires SRP and a transmembrane proton motive force (PMF) as well [12; 13]. In addition, FtsQ was found to interact with YidC during insertion [14]. These studies together make a strong case that SecA is required for the assembly of type II S-SMPs with TM segments near the N-terminus. But, suppose the TM segment is far downstream from the N-terminus. Could a TM segment occurring a hundred or more residues downstream affect the insertion pathway?
Single-span MPs are abundant in all branches of life [15]. Based upon a recently published database of S-SMPs [16] and a hydropathy analysis using MPEx [17], there are several S-SMPs whose TM segments occur a hundred or more residues downstream from the N-terminus. Of 94 E. coli S-SMPs identified in the database, we found six qualifying proteins by MPEx analysis (exclusive of C-tail anchored proteins). The proteins identified are CadC, RodZ, DamX, GspL, RseA, and YjhP. Two of these are of particular interest to our laboratories in the context of S-SMP assembly and stability: RodZ, which plays an important role in maintenance of the rod shape of Escherichia coli [18; 19], and CadC, which activates the CadBA operon during low-pH stress [20–22]. We report here the results of a comprehensive study of the in vivo assembly of RodZ, a 337-residue protein with a TM domain located at residues 112–133. We find that, indeed, SecA drives the insertion of RodZ via the SecYEG translocon complex. The only other requirement for insertion is the transmembrane PMF, as observed for FtsQ [13]. Unlike FtsQ, SRP is not strictly required for the assembly of RodZ. Because of the relative simplicity of its assembly requirements, RodZ is an ideal S-SMP for studies of SecA-driven insertion of TM helices.
Results
RodZ is a single-span membrane protein with Nin-Cout topology
An analysis of the RodZ sequence using the Spoctopus software [23] revealed the absence of an N-terminal signal sequence and the presence of a downstream hydrophobic segment that could serve as an internal signal/stop-transfer sequence. Membrane Protein Explorer (MPEx) [17] showed the most likely TM-segment as residues 112–133 (22 amino acids, Fig. 1c). To visualize RodZ in Western blots, we incorporated a T7 tag (MASMTGGQQMG) between amino acid Ala95 and Pro96 by mutagenesis using overlap extension (Fig. 1d). We first expressed T7-RodZ to verify its presence in the inner membrane. The cell culture was subjected to DNaseI treatment followed by several freeze-thaw cycles prior to separating different subcellular fractions [24]. RodZ appeared only in the inner membrane (IM) and total fraction (T) (Fig. 2a). We then generated a TM-deleted version of RodZ (RodZΔTM). RodZΔTM appeared in the cytoplasmic (cyt) and T fractions, but not in the IM fraction (Fig. 2b). Furthermore, we determined that RodZΔTM was not secreted into the periplasm, as judged by the absence of a Western-blot signal in the periplasmic ‘P’ fraction (Fig. 2c). These results are consistent with residues 112 – 133 forming the transmembrane domain required for stable insertion of RodZ into the inner membrane. Because targeting of both membrane and secreted proteins invariably involves recognition of a hydrophobic amino acid sequence by the targeting/assembly components of the cell (see Introduction), the TM domain is likely the targeting signal. However, further studies are required to establish unequivocally that the TM domain also serves as the targeting signal.
Figure 2.

RodZ is a type II single-span membrane protein as determined by cell fractionation and immunoblotting. In the immunoblots shown, the total fraction (T) was obtained by solubilizing whole cell pellets with SDS loading buffer. The cytoplasmic (cyt) and inner membrane (IM) fractions were obtained by subjecting the cell suspensions to ten freeze/thaw cycles in the presence of lysozyme and DNaseI. The suspensions were then centrifuged at 13,000×g in a microfuge. The supernatant, consisting of the cytoplasm and periplasm, is taken as the cyt fraction. The pellet, consisting of inner and outer membranes, was resuspended in buffer containing detergent, which solubilizes the inner membrane. The suspension was again spun at 13,000×g; the clear supernatant contained the solubilized IM proteins. Molecular weight markers (in kDa) are shown in lane 1 of all blots. (a) This Western blot for T7-RodZ shows that wild-type RodZ is found in the T and IM fractions, but is absent in the cyt fraction. Although RodZ has a molecular weight of 36 kDa, it migrates anomalously in SDS gels with an apparent molecular weight of ~55 kDa [25]. (b) The TM segment of RodZ is the insertion signal. This Western blot (T7 antibodies) shows that the expression of RodZΔTM, which is a T7-RodZ construct lacking the TM domain, is found only in the cyt and T fractions; it is absent in the IM fraction. (c) There is no signal in RodZΔTM for secretion. Secretion of RodZΔTM was tested using spheroplasts prepared by the osmotic shock method. The cells were spun at 13000×g for 15 minutes. The supernatant was used as the periplasmic fraction “P” and the pellet fraction (R) was used as non-periplasmic proteins. (d) RodZ is a TM protein oriented with a Nin-Cout topology. Spheroplasts of BL21 cells harboring pMS119-T7-RodZ were treated with or without proteinase K (protK) (lanes 2 & 3 and 4 & 5). In the presence of protK (protK+), only a fragment with the T7 tag corresponding to the cleaved N-terminal domain is observed, whereas in the absence of protK (protK−) only full-length protein is observed. In the presence of detergent, 1% CHAPS, the fragment is digested by protK (lane 6). (e) and (f) Control experiment showing the efficacy of our separation protocol for isolating the cyt and IM fractions. Western blots using YidC antibodies show that the inner-membrane protein YidC [31] is found only in the IM and T fractions (panel e). Western blots using GroEL antibodies shows cytoplasmic protein GroEL is found only in the cyt and T fractions (panel f). (g) Cartoon of the topology of RodZ.
Using a FLAG epitope, Shiomi et al. [25] demonstrated the membrane localization of RodZ and determined its likely topology using a PhoA fusion approach. We confirmed the topology of RodZ using proteinase K (protK) digestion in which spheroplasts of BL21 cells harboring the pET21-T7-RodZ were treated with protK followed by Western blotting. The cytoplasmic fragment of T7-tagged RodZ confirmed the Nin-Cout topology of RodZ (Fig. 2d). In panel d (lanes 2 & 3 and 4 & 5), the samples were loaded in different volumes to make sure of complete protK digestion of the protein and for better visibility of the digested fragment. The fragment ran at around ~17KDa, consistent with protease digestion of the RodZ periplasmic domain. RodZ is therefore a type II S-SMP. This fragment could be completely digested by protK when the detergent CHAPS was present (lane 6).
To validate the DNaseI separation protocol, we used the membrane protein YidC and the cytoplasmic protein GroEL as loading controls. YidC was found only in the inner membrane and total fractions (Fig. 2e) while GroEL was observed only in the cytoplasmic and total fractions (Fig. 2f). Fig. 2g summarizes our finding that RodZ is a type II S-SMP.
RodZ translocation is mediated by SecYEG translocon but is independent of YidC insertase
SecYEG mediates membrane insertion of most inner membrane proteins [3], but there are exceptions [26]. We therefore verified the dependence of RodZ insertion on SecYEG using a RodZ construct with an α-bungarotoxin-binding (BTX) tag (WRYYESSLLPYPD) [27; 28] inserted near the amino terminus between Gln2 and Thr3. Because SecE is required to stabilize the SecY protein, SecE depletion results in SecY depletion as well [29]. To test SecY dependence, we used the arabinose-dependent SecE depletion strain CM124 that has the secE gene under the control of araBAD promoter [30]. This cell line, harboring a pLZ1-BTX-RodZ vector, was grown under SecE expression and SecE depletion conditions in M9 minimal media supplemented with arabinose or glucose, respectively. IPTG (1 mM final concentration) was added to the CM124 cells (bearing the RodZ expression vector) 3 minutes before the cells were labeled with [35S]-methionine for 1minute. Fig. 3a demonstrates the dependence of RodZ insertion on SecYEG: ProtK treatment of spheroplasts yielded a cytoplasmic BTX-RodZ fragment for the SecE expression condition but not the SecE depletion condition. As a positive control, we demonstrated that translocation of proOmpA was inhibited under SecE-depletion conditions (Fig. S1a). Together, these data show that membrane insertion of RodZ depends upon SecYEG.
Figure 3.

Translocation of RodZ depends on SecYEG but not on YidC insertase. (a) RodZ incorporation in the membrane is blocked under SecE depletion conditions. A CM124 strain expressing pLZ1-BTX-RodZ was grown to mid-log phase under arabinose (SecE+) or glucose (SecE−) conditions. Cells were pulse labeled with [35S] methionine and analyzed by the protK assay, as described in Materials and Methods. (b) RodZ membrane translocation is independent of YidC. The YidC depletion strain JS7131 bearing the pMS119-BTX-RodZ plasmid was grown for 3h in LB media supplemented with 0.2% (m/V) arabinose or 0.2% (m/V) glucose. The membrane insertion of RodZ protein in the YidC expression and YidC depletion conditions was analyzed by proteinase accessibility assay as described in Materials and Methods. (c) Control experiment showing the effectiveness of YidC depletion. The YidC dependence of membrane insertion of YidC-dependent Pf3-p2 protein [32] was tested in JS7131, as described in Materials and Methods. Membrane translocation of Pf3-p2 was inhibited under YidC depleted conditions, as judged by the absence of a proteolytic fragment under protK+ conditions (compare lanes 1 and 3). See also Supplementary Figure S1.
The insertion of some membrane proteins requires YidC as well as SecYEG [3; 31]. To see if this is the case for RodZ, we used the YidC-depletion strain JS7131 that contains yidC gene under the control of the araBAD promoter. The depletion strain containing pMS119-BTX-RodZ vector was grown in presence of arabinose to express YidC or in glucose to deplete YidC. Following protK treatment of spheroplasts, the membrane translocation of the C-terminal domain of RodZ remained unaffected by YidC depletion conditions, as indicated by complete cleavage of full-length RodZ by protK (Fig. 3b). As a positive control, the YidC dependent membrane protein Pf3-p2 [32] was fully protected from proteolytic cleavage in spheroplasts upon YidC depletion (Fig. 3c), while mature OmpA was fully digested (Fig. S1b). We conclude from these experiments that the membrane insertion of RodZ does not require YidC but does require SecYEG.
SecA is required for membrane insertion of RodZ
Based upon the studies of the assembly of Momp2 [10; 11] and Lep-ΔH2 [8] discussed earlier, we fully expected SecA to be essential for the assembly of RodZ. As a first test of this expectation, we grew cells in the presence of sodium azide, which is known to inhibit protein translocation in E. coli by blocking SecA ATPase function [33]. We used MC1060 cells harboring the pMS119-BTX-RodZ vector at mid-logarithmic growth phase and treated with or without 3mM sodium azide (final concentration) for 5 minutes before pulse labeling with [35S] methionine for 1 minute. The spheroplasts of the cell culture were then analyzed using the protK accessibility assay. Fig. 4a shows that the sodium azide treatment blocked the membrane insertion of RodZ (lane 3), as evident from the absence of proteolytic fragment compared with cells not treated with azide. Likewise, the export of SecA-dependent proOmpA was strongly inhibited in the cells treated with sodium azide (Fig. S2a).
Figure 4.

RodZ depends on SecA and PMF for insertion into the membrane. (a) RodZ insertion is inhibited by sodium azide. MC1060 strain expressing RodZ was treated with sodium azide (3mM final concentration) for 5 minutes before pulse labeling with [35S] methionine for 1 minute. Membrane translocation of RodZ was analyzed by protK mapping as described in Materials and Methods. See also Fig. S2a. (b) RodZ membrane insertion is inhibited under SecA depletion conditions. The SecA-depletion strain EO527 containing pMS119-T7-RodZ was grown overnight in LB medium supplemented with 0.2% arabinose. The cells were then grown and Western blotted as described in Materials and Methods. The SecA-dependent maltose binding protein (MBP) was used as a control and the whole cell lysate was analyzed by Western blot using anti-His primary antibody followed by AP conjugated anti-mouse secondary antibody (Fig. S2b). (c) RodZ insertion is inhibited when a SecA dominant-negative is expressed using the SecA(R642E) cell line. RodZ carrying a T7 tag before the TM domain was coexpressed with mutant SecA(R642E) on a pET21 plasmid under control of T7 promoter in BL21 cells and induced with 40 μM IPTG at OD600 of 0.4 for 90 minutes, followed by protK treatment (lanes 2 and 4). See also Fig. S2c. (d) RodZ insertion is SecB independent. MC1060 (SecBwt) and CK1953 (SecBnull) transformed with pMS119-BTX-RodZ and the dependence of SecB for RodZ membrane insertion was examined by proteinase K mapping, as described in Materials and Methods. (e) Positive control using proOmpA expression and the protK accessibility assay, demonstrating that our depletion protocol for SecB is effective. OmpA protein was immunoprecipitated and resolved on 15% SDS-PAGE before phosphor-imaging, as described in Fig. S1a. (f) RodZ requires a proton motive force for membrane insertion (PMF). Cells were treated with CCCP (50 μM final concentration) for 45s, then pulse labeled with [35S] methionine for 1 minute and analyzed by protK mapping as described in Materials and Methods. See also Fig. S2d. (g) RodZ does not require SecD/F for membrane incorporation. JP325 strain expressing pMS119-BTX-RodZ was grown to mid-log phase under arabinose (SecD/F+) or glucose (SecD/F−) conditions before pulse labeling with [35S]-methionine for 1 minute. To test the membrane translocation of RodZ, cells expressing the RodZ protein were subjected to proteinase K mapping assay as described in Materials and Methods. (h) Positive control using proOmpA expression and the protK accessibility assay, demonstrating that our depletion protocol for SecD/F is effective. OmpA protein was immunoprecipitated and resolved on 15% SDS-PAGE before phosphor-imaging, as described in Fig. S1a.
To confirm further the involvement of SecA in the biogenesis of RodZ, we transformed the SecA depletion strain of E. coli EO527 with the plasmid (pMS119-T7-RodZ) encoding T7-tagged RodZ. The expression of the secA gene in this strain is under the control of the arabinose-inducible araBAD promoter, which causes SecA depletion when the cells are grown in the presence of glucose rather than arabinose. We grew EO527 cells in LB media supplemented with 0.2% (m/V) of arabinose or glucose and induced expression of RodZ using 1mM IPTG induction for 90 minutes. The protK-mapping assay of spheroplasts gave a clear answer for SecA+ and SecA− conditions. For SecA+, RodZ was inserted into the inner membrane with its C-terminus in the periplasm, judging by the ~17KDa proteolytic fragment (Fig. 4b, lane 1). In contrast, for SecA−, no ~17KDa proteolytic fragment was detected (Fig. 4b, lane 3). As a protocol control, we used translocation of maltose binding protein (MBP) and showed that it was impaired under SecA depletion conditions, because it accumulated in a precursor form (Fig. S2b). As a final test of the SecA requirement, we used a dominant-negative SecA system. We co-expressed RodZ with the dominant-negative SecA(R642E) under T7-promoter control by transforming BL21 cells with the pET21-T7-RodZ-SecA(R642E) vector [34]. Fig. 4c shows that membrane insertion of RodZ is inhibited, consistent with R642E abolishing SecA functionality. As a positive control, periplasmic secretion of mature MBP is shown in the presence of wild type SecA while it accumulates in the precursor form in SecA(R642E) (Fig. S2c).
SecB is a chaperone that sometimes works with SecA to keep secreted proteins in an export-competent state [35; 36] (Fig. 1a). To determine if SecB is required for RodZ insertion, we used the E. coli secB null strain CK1953 [37]. RodZ was expressed in CK1953 harboring the pMS119-BTX-RodZ vector and the spheroplasts were subjected to the protK assay. Fig. 4d shows that RodZ was digested by the externally added protK completely in the secB null strain just as in MC1060 (secB+), indicating that RodZ membrane insertion is independent of SecB. Our control experiments showed that export of SecB-dependent proOmpA was strongly inhibited in the secB null strain as expected (Fig. 4e).
Proton motive force (PMF) is required for RodZ membrane insertion
The PMF dependence of RodZ membrane insertion was examined using CCCP, a protonophore that equalizes the proton concentration across the inner membrane. MC1060 cells expressing RodZ were treated with 50μM CCCP for 45s before labeling with [35S]-methionine. The membrane insertion of RodZ was blocked upon treatment with CCCP to abolish the PMF (Fig. 4f lane 3). As a positive control, we confirmed that proOmpA, the precursor form of OmpA, accumulates when CCCP is present (Fig. S2d). This experiment shows that RodZ membrane insertion requires a transmembrane proton gradient.
The multi-span SecD/F complex, which conducts protons across the inner membrane, can stimulate SecA-driven protein secretion [4; 38]. The physiological function of SecD and SecF, however, is not entirely clear, because they are not required to maintain a proton motive force [39]. Nevertheless, we asked if SecD/F might also be required for RodZ membrane insertion. To deplete SecD/F, we used a JP325 strain in which expression of SecD/FYajC is under the control of arabinose-inducible araBAD promoter [40]. The SecD/F depletion strain JP325 containing pMS119-BTX-RodZ was grown under SecD/F expression (0.2% m/V arabinose) or SecD/F depletion (0.2% m/V glucose) conditions. To induce the expression of BTX-RodZ, we added IPTG 3 minutes before the cells were labeled with [35S]-methionine and analyzed for membrane insertion using the protease accessibility assay. We found that the membrane insertion of RodZ was unaffected by SecD/F depletion, as shown by the presence of a proteolytic fragment after protK digestion of spheroplasts (Fig. 4g, lane 3). The membrane translocation of proOmpA was inhibited under SecD/F depletion conditions, which served as a positive control for our experiments (Fig. 4h).
SRP facilitates but is not absolutely required for membrane incorporation of RodZ
Although insertion of the model S-SMP Momp2 and FtsQ requires SecA, targeting to the inner membrane also requires the SRP [10–12]. In E.coli, SRP is composed of Ffh and 4.5S RNA [41] (Fig. 1b). Thus, to determine whether SRP is required for the membrane insertion of RodZ, we used the Ffh depletion strain WAM121 with the ffh gene under control of the araBAD promoter [42]. WAM121 cells harboring the pMS119-BTX-RodZ vector were grown in the presence of arabinose or glucose, to achieve Ffh expression or Ffh depletion conditions, respectively. After IPTG induction (1mM final concentration) for 3 minutes, cells containing the RodZ expression vector were pulse labeled by [35S]-methionine for 1 minute, chased for 6 s or 2 min, and then converted to spheroplasts for protease accessibility analysis. Fig. 5a shows that 80% of the C-terminal tails of RodZ are fully digested by externally added protK at the early chase point but is nearly fully digested at the long chase time. This is in contrast to the Ffh+ conditions, in which RodZ is fully inserted at both chase points. It thus appears that Ffh is necessary for efficient insertion of RodZ. We validated our protocols by using TatC-P2 [29] (Fig. 5b) and proOmpA (Fig. S3 a, b) as controls [29].
Figure 5.
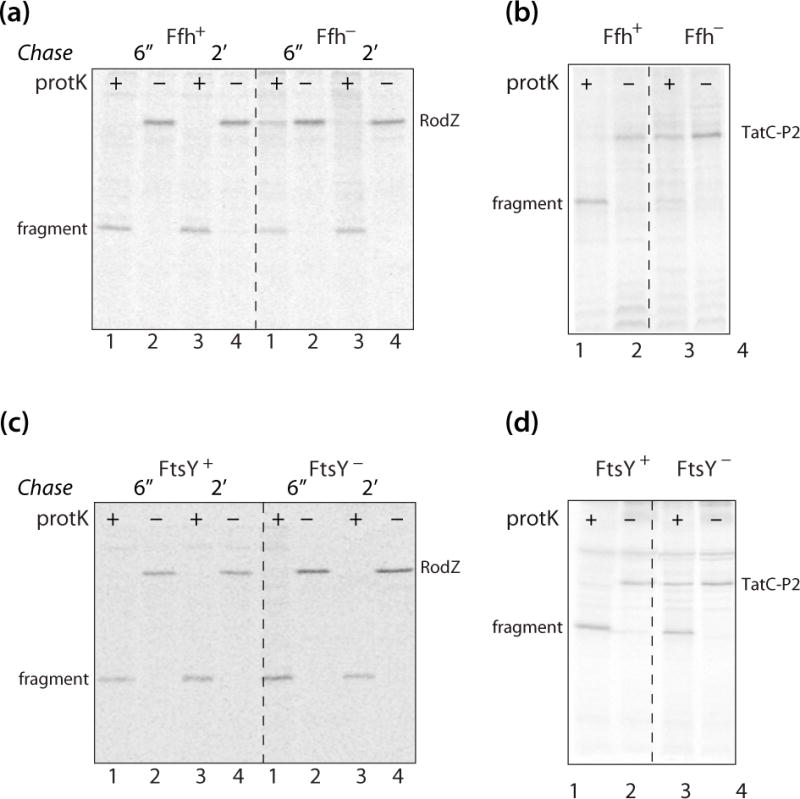
RodZ insertion does not strictly require the SRP pathway. (a) RodZ incorporation is facilitated by Ffh, but Ffh is not absolutely required. The Ffh depletion strain WAM121 bearing the RodZ expression vector pMS119-BTX-RodZ was grown to mid-log phase in the presence of arabinose (Ffh expression condition) and glucose (Ffh depletion condition), prior to pulse labeling for 1 minute and chase at 6 seconds and 2 minutes time points. The SRP pathway dependence of RodZ protein insertion was tested by proteinase accessibility assay, as described in Materials and Methods. The protK assay reveals that under Ffh depletion conditions, some uncleaved RodZ is present after 6″ and 2′ of chase, implying that Ffh affects the insertion of RodZ. (b) Control experiment showing that the Ffh-dependent TatC-P2 does not insert into the membrane under Ffh− conditions. (c) RodZ membrane insertion is independent of FtsY despite weak dependence on Ffh. To deplete FtsY, the FtsY-depletion strain IY26 with the RodZ expression vector pMS119-BTX-RodZ, was grown in the LB medium added with 0.2%(m/V) glucose. The cells expressing RodZ protein, grown in the FtsY+/FtsY− condition, were pulse labeled for 1 minute and then chased with non-radioactive methionine (40 mM final concentration) at 6 seconds and 2 minutes. The protein insertion of RodZ in FtsY+/FtsY− conditions was examined by proteinase accessibility assay as mentioned in Materials and Methods. (d) A control experiment demonstrating that TatC-P2 [29] insertion is reduced but not eliminated under FtsY− conditions. Although TatC-P2 insertion is only partially reduced under FtsY− conditions, the control demonstrates that the strain becomes depleted in the absence of arabinose. See also Fig. S3, a–d.
The insertion of multi-span membrane proteins via the SRP pathway also requires the SRP receptor, FtsY (Fig. 1b). To examine more closely the participation of the SRP pathway in RodZ assembly, we used the E. coli FtsY-depletion strain IY26 that harbored the pMS119-BTX-RodZ vector in which ftsY gene expression is under the control of an arabinose-inducible araBAD promoter. To deplete FtsY, the cells bearing the RodZ expression vector were grown in the LB medium supplemented with 0.2% (m/V) glucose. The cells were pulse labeled for 1 minute and then analyzed using the protease accessibility assay. Fig. 5c shows that FtsY depletion does not inhibit membrane insertion of RodZ (compare lanes 1 and 3). We used TatC-P2 [29] (Fig. 5d) and proOmpA expression as a positive and negative control for FtsY depletion (Fig. S3c, d). Overall, our results show that although Ffh has a small favorable effect on RodZ insertion, FtsY is not required at all. This implies that full engagement of the co-translational pathway (Fig. 1b) is not essential for RodZ insertion.
Discussion
Most studies of S-SMP assembly have used chimeric or mutant model proteins [10; 11; 43]. As far as we can establish, our examination of RodZ biogenesis presented here is the first complete exploration of the membrane insertion mechanism of a native single-span membrane protein. The study was motivated by the observation that the single TM helix of RodZ is more than a hundred residues downstream from the amino terminus, which is unusual; the TM segment of most S-SMPs occurs early in the sequence near the N-terminal. The results presented here show that membrane targeting of RodZ does not require SecB or full engagement of the SRP pathway; Ffh seems to enhance insertion but, oddly, FtsY is not required. Membrane insertion does not require YidC or SecD/F; SecA and the transmembrane PMF are sufficient for the insertion of RodZ via SecYEG. In retrospect, this might have been anticipated by the observation that SecA is required for the transmembrane transport of long interhelical loops of multi-span membrane proteins or long C-terminal domains [8–10; 12].
The partial engagement of the SRP may be related to the hydrophobicity of the RodZ TM segment. Extensive studies of the effect of N-terminal signal sequence hydrophobicity on the targeting and export of thioredoxin-1, physiologically a cytoplasmic protein, suggests that hydrophobicity is an important determinant in pathway selection [44; 45]. Thioredoxin, which folds rapidly in the cytoplasm, can nevertheless by exported to the periplasm by adding an N-terminal signal sequence that can target the protein to the SRP pathway so that secretion occurs co-translationally, thus preventing cytoplasmic folding. Schierle et al. [45] found that fusion of the MalE signal sequence did not lead to thioredoxin export whereas fusion of the DsbA signal sequence caused efficient export. Huber et al. [44] subsequently showed that the hydrophobicity of the signal sequence affects pathway selection, with high hydrophobicity favoring the SRP pathway. These results raise the possibility that the hydrophobicity of the RodZ TM segment determines the extent of engagement of the SRP pathway.
One might think of RodZ as a protein containing the first TM helix of a multi-span membrane protein with a long interhelical loop, but without the other spans. But unlike a multi-span MP, RodZ does not absolutely require the SRP pathway. Our results are similar to those of Newitt et al. [46] who showed that the insertion of a YfgA (now RodZ) chimera was affected, but not prevented, when the SRP pathway was disrupted. The differences between their study and ours are that they used a RodZ-phoA fusion protein with phoA fused after the TM segment at position 139 and they disrupted the function of the SRP pathway by inducing the dominant lethal ftsY allele (G385A). It is not clear why we did not observe that the insertion of RodZ was affected by FtsY depletion (Fig. 5c). One possibility is that induction of the FtsY G385A mutant is a more effective way of disrupting the SRP pathway. Or the different results may have to do with the fact that we used the native RodZ protein. In any case, RodZ does not appear to require the SRP pathway strictly, because Ffh depletion caused only a minor effect on its insertion while Ffh depletion caused a strong effect on the SRP-dependent TatC-P2 membrane insertion. Interestingly though, FtsY depletion had no effect on RodZ insertion while it partially inhibited TatC-P2 insertion (Fig. 5d, lane 3).
Presumably SecA mainly targets the native RodZ to SecYEG when it recognizes the late-arriving TM segment. This occurs perhaps post-translationally, but we have no evidence that bears on the question. Based upon recent structural studies showing that SecA binds to the exit tunnel of the ribosome [47; 48], it is possible that SecA interacts with the nascent chain in a co-translational manner. If insertion is post-translational, one possibility is that the TM segment first partitions into the membrane interface followed by SecA-guided TM insertion and C-terminal transport through the SecYEG translocation channel. The TM segment partitioning into the membrane may be possible because the TM segment has an extraordinarily favorable free energy of transfer from water-to-octanol of −18 kcal/mol, based on the Wimley-White whole-residue hydrophobicity scale [49]. Microsecond-scale equilibrium molecular dynamics simulations show, for example, that poly-leucine helices fold into bilayer interfaces as α-helices and never return to the aqueous phase [50]. The possibility of direct first-contact of the helix with the membrane is supported by studies of KdpD, which acts as a potassium sensor in E. coli [51; 52]. Several features of KdpD are significant: The protein is tethered to the cytoplasmic surface of the inner membrane by four TM segments (residues 401–498) with very short interhelical loops that occur far downstream from the amino terminus, it is targeted to the inner membrane by SRP-recognition of a cytoplasmic amphipathic helix (residues 22–48) [53], and membrane insertion does not require SecA, SecE, or YidC [26]. Because KdpD does not apparently engage SecA or the SecYEG channel, a reasonable explanation is spontaneous insertion following targeting by the SRP.
We have not explored the determinants of the topology of RodZ, but we note that a highly positively charge sequence (-KRRKKRD-) immediately precedes the TM segment. This should guarantee Nin-Cout topology based upon the positive-inside rule [54; 55]. Interestingly, though, the topology of leader peptidase in E. coli can be inverted simply by lengthening the normally cytoplasmic interhelical P1 loop between the two TM segments to 60–70 residues from the wild-type 39 residues [8]. This occurs despite the fact the P1 loop is heavily positively charged, suggestive of a significant role for SecA in topology determination.
The molecular mechanism of SecA transport of long periplasmic domains in multi-span MPs is not known. Although SecA-driven secretion of periplasmic and outer-membrane proteins is understood reasonably well, just how SecA engages and transports long periplasmic domains during co-translational assembly of multi-span proteins is a mystery. The insertion mechanism of type II S-SMPs by SecA is equally mysterious. Does SecA recognize the TM segment, carry it to SecYEG, and push it into SecYEG where it partitions between translocon and membrane in the course of secretion of the periplasmic domain? Or, is the TM segment delivered by SecA directly to the membrane in the vicinity of SecYEG, where it partitions spontaneously prior to ATP-dependent transport of the periplasmic domain? The molecular targeting process of the RodZ-class of proteins is also far from clear. How might targeting and insertion process be affected by the amino acid composition of the TM segment? While it seems certain that SecA is involved in targeting, are other molecules, such as trigger factor, required? We suggest that RodZ provides a starting point for finding answers to these questions.
Materials and Methods
Bacterial strains, plasmids and materials
The E. coli JS7131 (YidC depletion strain), WAM121 (Ffh depletion strain), CM124 (SecE depletion strain), JP325 (SecD/FyajC depletion strain), EO527 (SecA depletion strain) and IY26 (FtsY depletion strain) were previously described [30; 37; 42]. In these depletion strains, the araBAD promoter controls the expression of the yidC, ffh, secE, SecD/FyajC, secA and ftsY genes. SecA(R642E) dominant-negative strain was developed in our lab. In this strain, coexpression of RodZ and SecA(R642E) mutant is controlled under the T7 promoter on pET21 vector. BL21 and MC1060 were used as wild type strains. See Table 1 for more details. Media preparation and bacterial manipulations were performed according to standard methods [56].
Table 1.
Various E. coli strains and plasmids used in this study of RodZ biogenesis.
| Strains1 | Plasmids2 | Tag3 | Mutation | Reference |
|---|---|---|---|---|
| BL21 | pET21-T7-RodZ, pET16b-T7-RodZ, pET21-T7-RodZΔTM pET21-T7-SecA(R642E)RodZ, pET21-T7-SecAwt-RodZ | T7 | Novagen #69449-3 | |
| MC1060 | pMS119-BTX-RodZ | BTX | [58; 59] | |
| WAM121 | pMS119-BTX-RodZ | BTX | ffh | [42; 57] |
| IY26 | pMS119-BTX-RodZ | BTX | ftsY | [60] |
| EO527 | pMS119-T7-RodZ | T7 | secA | [61] |
| JS7131 | pMS119-BTX-RodZ | BTX | yidC | [31] |
| CM124 | pLZ1-BTX-RodZ | BTX | secE | [30] |
| CK1953 | pMS119-BTX-RodZ | BTX | secB | [37] |
| JP325 | pMS119-BTX-RodZ | BTX | SecD/F | [62] |
- ▪BL21 [E. coli B F- dcm ompT hsdS(rB- mB-) gal [malB+]K-12(λS)]
- ▪MC1060 [Δ(codB-lacI)3, galK16, galE15, λ−, relA1, rspL150, spoT1, hsdR2, ara+]
- ▪WAM121 [MC4100, ara+, ffh::kan, attB::(OriR6K, ParaBAD-ffh, tet)]
- ▪Y26 [Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ−, rph-1, Δ(rhaD-rhaB)568, hsdR514, kan-araC-PBAD-ftsY]
- ▪EO527 [λ−, IN(rrnD-rrnE)1, rph-1, λcI857, Δ(cro-bioA),SecM<>kan-araC-PBAD]
- ▪JS7131 [MC1060, ΔyidC, attB::(OriR6K, ParaBAD−yidC+, (Specr)]
- ▪M124 [secE19–111 pcnB80 zadL::(Tn10 Tcs Strr) phoAΔPvuII, lacΔX74, galE galK, rpsL, recA::cat, pCM22 (ParaBAD-secE+)]
- ▪CK1953 [MC4100, secB::Tn5]
- ▪JP325 [araΔ714, Δ(argF-lac)U169, rpsL150, relA1, thi, flb5301, deoC1, ptsF25, recA::cat, tgt::kan-araC+-PBAD::yajC-SecD/F]
- pMS119: (ColE1 origin, Tac promoter, Ampicillin resistant)
- pLZ1: (RSF origin, T7/Lac promoter, Kanamycin resistant)
- T7: MASMTGGQQMG inserted between residues Ala95 and Pro96.
- BTX: WRYYESSLLPYPD inserted between Gln2 and Thr3.
Sodium azide, CCCP, phenylmethylsulfonyl fluoride (PMSF), nitro blue tetrazolium (NBT), 5-Bromo-4-chloro-3-indolyl phosphate (BCIP), and lysozyme were obtained from Sigma. Proteinase K was purchased from Invitrogen. IPTG was purchased from Research Products International Corporation. α-Bungarotoxin, Biotin-XX Conjugate was from Invitrogen. Streptavidin agarose resin was purchased from Thermo Scientific Company. Anti-T7 antibody was from Novagen (#69999). Trans-[35S] label methionine was from Perkin Elmer. Phusion PCR kit and restriction enzymes were from New England Biolabs (NEB).
RodZ gene was amplified by PCR from E.coli K12 strain and subcloned into expression vector pET16b and pET21 using NdeI and XhoI with a stop codon. An 11 amino acid T7 tag (MASMTGGQQMG) was introduced by mutagenesis by overlap extension between Ala95 and Pro96 for immuno detection of RodZ. These constructs, named as pET16b-T7-RodZ and pET21-T7-RodZ, were was expressed in a Bl21 cell line. RodZΔTM was made by mutagenesis by overlap extension. Nucleotides from 333 to 400 were not amplified to make the RodZΔTM construct. This construct is called as pET21-T7-RodZΔTM. RodZ was subcloned into pMS119 expression vector using EcoRI and HindIII restriction sites. This construct was named as pMS119-T7-RodZ and was expressed in SecA depletion strain EO527. To make a dominant-negative strain, we generated the SecA-RodZ coexpression plasmid named pET21-SecA-T7-RodZ and pET21-SecA(R642E)-T7-RodZ plasmids using the BglII site, in which SecA was added with T7 promoter and terminator. These plasmids were expressed in BL21 cell line. We made one more construct in which we inserted a 13 amino acid α-bungarotoxin-binding (BTX) peptide (WRYYESSLLPYPD) near the amino terminus between 2Gln and 3Thr of RodZ rather than the T7 tag close to the TM domain [27; 28]. This construct was called pMS119-BTX-RodZ and was expressed in YidC depletion strain JS7131, Ffh depletion strain WAM121, FtsY depletion strain IY26, and SecD/F depletion strain JP325. It was also expressed in SecB null strain CK1953 and wild type strain MC1060. The DNA encoding BTX-RodZ was subcloned into a modified pEH1 vector pLZ1 (GenBank Accession # KF918333) to allow protein expression in the SecE depletion strain CM124. The coding regions of all constructs were verified by sequence analysis.
Growth conditions
For Ffh depletion, WAM121 [57], FtsY depletion IY26, SecD/F depletion JP325, YidC depletion JS7131, the following growth-condition protocol was followed: The cells of various E. coli strains containing pMS119-BTX-RodZ plasmid were grown overnight at 37°C in LB medium containing 0.2% (w/v) arabinose, washed in medium lacking arabinose and back-diluted 1:100 in fresh LB medium containing 0.2% (w/v) arabinose or 0.2% (w/v) glucose. The SecB null strain CK1953 and MC1060 bearing the RodZ expression vector were grown in the LB medium as SecB deletion and SecB wild type conditions, respectively. When cultures reached an OD600 of 0.6, the cells were switched into M9 minimal medium before pulse labeling with [35S]-methionine. The SecE depletion strain CM124 containing pLZ1-BTX-RodZ plasmid was grown overnight in M9 minimal medium containing 0.2% arabinose and 0.4% glucose. The cells were back diluted 1:100 and grown in fresh M9 media containing 0.4% glucose supplemented with or without 0.4% arabinose, and grown at 37°C for 9h. The SecA depletion strain EO527 containing the pMS119-T7-RodZ plasmid was grown overnight in LB medium containing 0.2% (w/v) arabinose, washed in medium lacking arabinose and back-diluted 1:10 in LB medium containing 0.4% (w/v) glucose. When cultures reached an OD600 of 0.6, the cells were induced with 1mM IPTG for 90 minutes. The BL21 cells containing pET21-SecA(R642E)-T7-RodZ and pET21-SecAwt-T7-RodZ plasmids were grown overnight in LB media. Cultures were back-diluted 1:100 in fresh LB media and were induced with 40μM IPTG at OD600 of 0.4 and grown for 90 minutes. Where appropriate, ampicillin (100 μg·ml−1, final concentration), kanamycin (50 μg·ml−1, final concentration), or chloramphenicol (50 μg·ml−1, final concentration) was added to the medium.
Cell Fractionation
Cell fraction was performed by cell lysis using freeze-thaw and DNaseI treatment. The bacterial cells (containing the over expressed protein RodZΔTM and RodZwt) were harvested and washed with PBS buffer to remove residual media. The pellet was resuspended in Lysis-Equilibration-Wash buffer (LEW buffer: 50 mM NaH2PO4, 300 mM NaCl, pH 8.0) containing DNaseI enzyme, DNaseI buffer, lysozyme and PMSF in it. Thereafter, the cell pellet was subjected to 10 cycles of freeze (liquid Nitrogen) and thaw (at 37°C water bath) followed by incubation at 37°C for 10 minutes. The cell suspension was centrifuged at 13,000×g for 10 minutes at 4°C, and the supernatant containing the soluble and periplasmic proteins (cyt fraction) was transferred to a new tube. The pellet was washed twice and resuspended in the LEW+1% Triton X-100 (detergent) to solubilize membrane proteins. The freeze and thaw cycle was repeated for 10 minutes followed by centrifugation at 13,000×g at 4°C for 15 minutes. The supernatant now contains the inner membrane fraction. Cells expressing MBP were treated by the osmotic shock method [56]. After a high-speed spin, the supernatant contains the periplasmic fraction “P” while the pellet/rest fraction “R” includes the total proteins without the periplasmic proteins. Both fractions were precipitated with 10% (m/V) TCA buffer and the MBP protein was visualized by Western blot analysis using anti-his antibody.
Protease treatment studies
For all experiments, cells were grown to mid-logarithmic phase under the various growth conditions. IPTG (1 mM final concentration) was added to cells cultured in M9 minimal media to induce RodZ protein expression, and then pulse-labeled for 1 min with trans-[35S]methionine (0.2 mCi/ml final concentration). The azide study involved treatment of cells with sodium azide (3mM final concentration) for 5 minutes prior to labeling. For the PMF dependence study, cells were treated with CCCP (50 μM final concentration) for 45s prior to [35S] labeling the cells. For non-radioactive experiment, the BL21 cells bearing the RodZ expression vector were induced with 1mM IPTG for 90 minutes before production of spheroplasts. The unlabeled or radiolabeled cells were converted to spheroplasts. To form spheroplasts, cells were resuspended in 0.25 ml of ice-cold buffer A (0.1 M Tris-acetate, pH 8.2, 0.5 M sucrose, 5 mM EDTA) then treated with lysozyme (80μg/ml final concentration) and 0.25 ml of ice-cold water for 15 minutes. 150μl of 0.2 M MgSO4 was added to stabilize the spheroplasts and cells were spun down then resuspended in buffer B (50mM Tris-acetate, 0.25 M sucrose, 10 mM MgSO4). Aliquots of the spheroplast suspension were incubated on ice for 1 h either in the presence or absence of proteinase K (0.8 mg ml−1, final concentration). After addition of phenylmethylsulfonyl fluoride (5mM final concentration) to inactivate Proteinase K, samples were precipitated with TCA (15%, final concentration), and washed with ice-cold acetone. For radiolabeled proteins, the cell pellet was solubilized in Tris-SDS buffer (10 mM Tris–HCl (pH 8.0) and 2% SDS). The TatC-P2 and Pf3-p2 proteins were immunoprecipitated using antiserum to leader peptidase while anti-OmpA antiserum was for OmpA protein immunoprecipitation. For non-radiolabeled proteins, the acetone-washed pellet was then dissolved in 50 μl Hoechst sample buffer (containing 6M urea) for Western blotting using anti-T7-AP conjugated antibody after SDS-PAGE.
Western Blots
T7-RodZ protein was detected by Western blot using the chromogenic method of detection. In this process, spheroplasts treated with protK were solubilized in SDS sample-loading buffer containing 6M urea, boiled for 10 minutes, and subjected to SDS-PAGE analysis. The gel was transferred to a nitrocellulose membrane using the iBLOT semi-dry technique (Invitrogen) and then blocked with 1% BSA in TBS buffer for 30 minutes. The membrane was then blotted with AP conjugated T7-tag antibody (1:10,000 dilution in TBS buffer) for one hour. Before developing the blot, the membrane was washed three times with TBST (TBS+Tween 20) to remove unbound antibody. For chromogenic development, 5% NBT and 5% BCIP dissolved in Buffer A (alkaline pH 9.0) were used to develop the blot. NBT and BCIP yield an intense, insoluble black-purple precipitate when reacted with alkaline phosphatase, which provides a good signal for tagged protein detection.
Pull-down protocol for BTX-tagged protein
E. coli cells were pulse labeled with 0.2mCi/ml [35S]-methionine for 1 minute. Following the protK accessibility assay, the spheroplasts were pelleted and solubilized with 100μl Tris-SDS buffer (10 mM Tris-HCl, pH 8.0, 2% (w/v) sodium dodecyl sulfate). The protein lysate was diluted with 1ml of Triton-IP buffer (10 mM Tris-HCl, pH 8.0, 5 mM EDTA, 150 mM NaCl, 2.5% (v/v) Triton X-100). After incubation with 30µl of streptavidin agarose resin, the supernatant was transferred to a new tube and incubated with 2µg/ml (final concentration) of α-Bungarotoxin (Biotin-XX Conjugate) for 45 minutes before incubation with 30µl streptavidin agarose resin for 30 minutes. The resin was then spun down and washed with Triton-IP buffer for 3 times and the BTX-tagged RodZ protein was eluted by SDS-sample buffer. The protein samples were resolved on 15% SDS-PAGE and visualized by phosphor-imaging, as described earlier [29].
Supplementary Material
Highlights.
How are single-span membrane proteins with a far downstream TM segment inserted?
One such protein, RodZ, does not require the signal recognition particle pathway.
RodZ requires only SecA, SecYEG, and a transmembrane PMF for insertion.
RodZ is an ideal tool for studying the molecular basis for TM insertion by SecA.
Acknowledgments
This work was supported in part by National Institutes of Health Grant GM074637 (to S. H. W.) and National Science Foundation Grant MCB-1052033 (to R. E. D.).
Abbreviations
- S-SMPs
single-span membrane proteins
- TM
transmembrane
- MP
membrane protein
- PMF
proton motive force
- SRP
signal recognition particle
- ProtK
proteinase K
- CCCP
carbonyl cyanide-p-chlorophenylhydrazone
- BTX
α-bungarotoxin
- HRP
horseradish peroxidase
References
- 1.Papanikou E, Karamanou S, Economou A. Bacterial protein secretion through the translocase nanomachine. Nature Rev Microbiol. 2007;5:839–851. doi: 10.1038/nrmicro1771. [DOI] [PubMed] [Google Scholar]
- 2.du Plessis DJF, Nouwen N, Driessen AJM. The Sec translocase. Biochim Biophys Acta. 2011;1808:851–865. doi: 10.1016/j.bbamem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 3.Dalbey RE, Wang P, Kuhn A. Assembly of bacterial inner membrane proteins. Annu Rev Biochem. 2011;80:161–187. doi: 10.1146/annurev-biochem-060409-092524. [DOI] [PubMed] [Google Scholar]
- 4.Park E, Rapoport T. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu Rev Biophys. 2012;41:21–40. doi: 10.1146/annurev-biophys-050511-102312. [DOI] [PubMed] [Google Scholar]
- 5.Parlitz R, Eitan A, Stjepanovic G, Bahar I, Bange G, Bibi E, Sinning I. Escherichia coli signal recognition particle receptor FtsY contains an essential and autonomous membrane-binding amphipathic helix. J Biol Chem. 2007;282:32176–32184. doi: 10.1074/jbc.M705430200. [DOI] [PubMed] [Google Scholar]
- 6.Sachelaru I, Petriman NA, Kudva R, Kuhn P, Welte T, Knapp B, Drepper F, Warscheid B, Koch H-G. YidC occupies the lateral gate of the SecYEG translocon and is sequentially displaced by a nascent membrane protein. J Biol Chem. 2013;288:16295–16307. doi: 10.1074/jbc.M112.446583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhn A. Alterations in the extracellular domain of M13 procoat protein make its membrane insertion dependent on secA and secY. Eur J Biochem. 1988;177:267–271. doi: 10.1111/j.1432-1033.1988.tb14372.x. [DOI] [PubMed] [Google Scholar]
- 8.Andersson H, von Heijne G. Sec dependent and sec independent assembly of E. coli inner membrane proteins: The topological rules depend on chain length. EMBO J. 1993;12:683–691. doi: 10.1002/j.1460-2075.1993.tb05702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi H-Y, Bernstein HD. SecA is required for the insertion of inner membrane proteins targeted by the Escherichia coli signal recognition particle. J Biol Chem. 1999;274:8993–8997. doi: 10.1074/jbc.274.13.8993. [DOI] [PubMed] [Google Scholar]
- 10.Neumann-Haefelin C, Schäfer U, Müller M, Koch H-G. SRP-dependent co-translational targeting and SecA-dependent translocation analyzed as individual steps in the export of a bacterial protein. EMBO J. 2000;19:6419–6426. doi: 10.1093/emboj/19.23.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deitermann S, Sprie GS, Koch H-G. A dual function for SecA in the assembly of single-spanning membrane proteins in Escherichia coli. J Biol Chem. 2005;280:39077–39085. doi: 10.1074/jbc.M509647200. [DOI] [PubMed] [Google Scholar]
- 12.Scotti PA, Valent QA, Manting EH, Urbanus ML, Driessen AJM, Oudega B, Luirink J. SecA is not required for signal recognition particle-mediated targeting and intial membrane insertion of a nascent inner membrane protein. J Biol Chem. 1999;274:29883–29888. doi: 10.1074/jbc.274.42.29883. [DOI] [PubMed] [Google Scholar]
- 13.van der Laan M, Nouwen N, Driessen AJM. SecYEG proteoliposomes catalyze the ΔΨ-dependent membrane insertion of FtsQ. J Biol Chem. 2004;279:1659–1664. doi: 10.1074/jbc.M306527200. [DOI] [PubMed] [Google Scholar]
- 14.van der Laan M, Houben ENG, Nouwen N, Luirink J, Driessen AJM. Reconstitution of Sec-dependent membrane protein insertion: nascent FtsQ interacts with YidC in a SecYEG-dependent manner. EMBO Rep. 2001;2:519–523. doi: 10.1093/embo-reports/kve106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Worch R, Bökel C, Höfinger S, Schwille P, Weidemann T. Focus on composition and interaction potential of single-pass transmembrane domains. Proteomics. 2010;10:4196–4208. doi: 10.1002/pmic.201000208. [DOI] [PubMed] [Google Scholar]
- 16.Bejleri O, Litou Z, Hamodrakas S. SstmpDB: a database of single-spanning transmembrane proteins. EMBnetjournal. 2013;19.B:64–65. [Google Scholar]
- 17.Snider C, Jayasinghe S, Hristova K, White SH. MPEx: A tool for exploring membrane proteins. Protein Sci. 2009;18:2624–2628. doi: 10.1002/pro.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Ent F, Johnson CM, Persons L, de Boer P, Löwe J. Bacterial actin MreB assembles in complex with cell shape protein RodZ. EMBO J. 2010;29:1081–1090. doi: 10.1038/emboj.2010.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alyahya SA, Alexander R, Costa T, Henriques AO, Emonet T, Jacob-Wagner C. RodZ, a component of the bacterial core morphogenic apparatus. Proc Natl Acad Sci USA. 2009;106:1239–1244. doi: 10.1073/pnas.0810794106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watson N, Dunyak DS, Rosey EL, Slonczewski JL, Olson ER. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J Bacteriol. 1992;174:530–540. doi: 10.1128/jb.174.2.530-540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Küper C, Jung K. CadC-mediated activation of the cadBA promoter in Escherichia coli. J Mol Microbiol Biotechnol. 2005;10:26–39. doi: 10.1159/000090346. [DOI] [PubMed] [Google Scholar]
- 22.Haneburger I, Eichinger A, Skerra A, Jung K. New insights into the signaling mechanism of the pH-responsive, membrane-integrated transcriptional activator CadC of Escherichia coli. J Biol Chem. 2011;286:10681–10689. doi: 10.1074/jbc.M110.196923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viklund H, Bernsel A, Skwark M, Elofsson A. SPOCTOPUS: A combined predictor of signal peptides and membrane protein topology. Bioinformatics. 2008;24:2928–2929. doi: 10.1093/bioinformatics/btn550. [DOI] [PubMed] [Google Scholar]
- 24.Burgess RR, Deutscher MP. Guide to protein purification. Methods Enzymol. 2009;463:1–851. doi: 10.1016/S0076-6879(09)63003-2. [DOI] [PubMed] [Google Scholar]
- 25.Shiomi D, Sakai M, Niki H. Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J. 2008;27:3081–3091. doi: 10.1038/emboj.2008.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Facey SJ, Kuhn A. The sensor protein KdpD inserts into the Escherichia coli membrane independent of the Sec translocase and YidC. Eur J Biochem. 2003;270:1724–1734. doi: 10.1046/j.1432-1033.2003.03531.x. [DOI] [PubMed] [Google Scholar]
- 27.Kasher R, Balass M, Scherf T, Fridkin M, Fuchs S, Katchalski-Katzir E. Design and synthesis of peptides that bind α-bungarotoxin with high affinity. Chem Biol. 2001;8:147–155. doi: 10.1016/s1074-5521(00)90063-2. [DOI] [PubMed] [Google Scholar]
- 28.Sanders T, Hawrot E. A novel pharmatope tag inserted into the β4 subunit confers allosteric modulation to neuronal nicotinic receptors. J Biol Chem. 2004;279:51460–51465. doi: 10.1074/jbc.M409533200. [DOI] [PubMed] [Google Scholar]
- 29.Zhu L, Klenner C, Kuhn A, Dalbey RE. Both YidC and SecYEG are required for translocation of the periplasmic loops 1 and 2 of the multispanning membrane proteinTatC. J Mol Biol. 2012;424:354–367. doi: 10.1016/j.jmb.2012.09.026. [DOI] [PubMed] [Google Scholar]
- 30.Traxler B, Murphy C. Insertion of the polytopic membrane protein MalF is dependent on the bacterial secretion machinery. J Biol Chem. 1996;271:12394–12400. doi: 10.1074/jbc.271.21.12394. [DOI] [PubMed] [Google Scholar]
- 31.Samuelson JC, Chen M, Jiang F, Möller I, Wiedmann M, Kuhn A, Phillips GJ, Dalbey RE. YidC mediates membrane protein insertion in bacteria. Nature. 2000;406:637–641. doi: 10.1038/35020586. [DOI] [PubMed] [Google Scholar]
- 32.Chen M, Samuelson JC, Jiang F, Muller M, Kuhn A, Dalbey RE. Direct interaction of YidC with the sec-independent Pf3 coat protein during its membrane protein insertion. J Biol Chem. 2002;277:7670–7675. doi: 10.1074/jbc.M110644200. [DOI] [PubMed] [Google Scholar]
- 33.Oliver DB, Cabelli RJ, Dolan KM, Jarosik GP. Azide-resistant mutants of Escherichia coli alter the SecA protein, an azide-sensitive component of the protein export machinery. Proc Natl Acad Sci USA. 1990;87:8227–8231. doi: 10.1073/pnas.87.21.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mori H, Ito K. The long α-helix of SecA is important for the ATPase coupling of translocation. J Biol Chem. 2006;281:36249–36256. doi: 10.1074/jbc.M606906200. [DOI] [PubMed] [Google Scholar]
- 35.Randall LL, Hardy SJS. Correlation of competence for export with lack of tertiary strcture of the mature species: a study in vivo of maltose-binding protein in E. coli. Cell. 1986;46:921–928. doi: 10.1016/0092-8674(86)90074-7. [DOI] [PubMed] [Google Scholar]
- 36.Weiss JB, Ray PH, Bassford PJ., Jr Purified SecB protein of Escherichia coli retards folding and promotes membrane translocation of the maltose-binding protein in vitro. Proc Natl Acad Sci USA. 1988;85:8978–8982. doi: 10.1073/pnas.85.23.8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumamoto CA, Beckwith J. Evidence for specificity at an early step in protein export in Escherichia coli. J Bacteriol. 1985;163:267–274. doi: 10.1128/jb.163.1.267-274.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsukazaki T, Mori H, Echizen Y, Ishitani R, Fukai S, Tanaka T, Perederina A, Vassylyev DG, Kohno T, Maturana AD, Ito K, Nureki O. Structure and function of a membrane component SecDF that enhances protein export. Nature. 2011;474:235–238. doi: 10.1038/nature09980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nouwen N, van der Laan M, Driessen AJM. SecDFyajC is not required for the maintenance of the proton motive force. FEBS Lett. 2001;508:103–106. doi: 10.1016/s0014-5793(01)03033-2. [DOI] [PubMed] [Google Scholar]
- 40.Pogliano JA, Beckwith J. SecD and secF facilitate protein export in Escherichia coli. EMBO J. 1994;13:554–561. doi: 10.1002/j.1460-2075.1994.tb06293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 42.Phillips GJ, Silhavy TJ. The E. coli ffh gene is necessary for viability and efficient protein export. Nature. 1992;359:744. doi: 10.1038/359744a0. [DOI] [PubMed] [Google Scholar]
- 43.Zhu L, Wasey A, White SH, Dalbey RE. Charge composition features of model single-span membrane proteins that determine selection of YidC and SecYEG translocase pathways in Eschericia coli. J Biol Chem. 2013;288:7704–7716. doi: 10.1074/jbc.M112.429431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huber D, Boyd D, Xia Y, Olma MH, Gerstein M, Beckwith J. Use of thioredoxin as a reporter to identify a subset of Escherichia coli signal sequences that promote signal recognition particle-dependent translocation. J Bacteriol. 2005;187:2983–2991. doi: 10.1128/JB.187.9.2983-2991.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schierle CF, Berkmen M, Huber D, Kumamoto C, Boyd D, Beckwith J. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J Bacteriol. 2003;185:5706–5713. doi: 10.1128/JB.185.19.5706-5713.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newitt JA, Ulbrandt ND, Bernstein HD. The structure of multiple polypeptide domains determines the signal recognition particle targeting requirment of Escherichia coli inner membrane proteins. J Bacteriol. 1999;181:4561. doi: 10.1128/jb.181.15.4561-4567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huber D, Rajagopalan N, Preissler S, Rocco MA, Merz F, Kramer G, Bukau B. SecA interacts with ribosomes in order to facilitate posttranslational translocation in bacteria. Mol Cell. 2011;41:343–353. doi: 10.1016/j.molcel.2010.12.028. [DOI] [PubMed] [Google Scholar]
- 48.Singh R, Kraft C, Jaiswal R, Sejwal K, Kasaragod VB, Kuper J, Bërger J, Mielke T, Luirink J, Bhushan S. Cryo-electron microscopic structure of SecA bound to the 70S ribosome. J Biol Chem. 2014;289:7190–7199. doi: 10.1074/jbc.M113.506634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White SH, Wimley WC. Membrane protein folding and stability: Physical principles. Annu Rev Biophys Biomol Struc. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 50.Ulmschneider JP, Smith JC, White SH, Ulmschneider MB. In silico partitioning and transmembrane insertion of hydrophobic peptides under equilibrium conditions. J Am Chem Soc. 2011;133:15487–15495. doi: 10.1021/ja204042f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zimmann P, Puppe W, Altendorf K. Membrane topology analysis of the sensor kinase KdpD of Escherichia coli. J Biol Chem. 1995;270:28282–28288. doi: 10.1074/jbc.270.47.28282. [DOI] [PubMed] [Google Scholar]
- 52.Rothenbucher MC, Facey SJ, Kiefer D, Kossmann M, Kuhn A. The cytoplasmic c-terminal domain of the Escherichia coli KdpD protein functions as a K+ sensor. J Bacteriol. 2006;188:1950–1958. doi: 10.1128/JB.188.5.1950-1958.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maier KS, Hubich S, Liebhart H, Krauss S, Kuhn A, Facey SJ. An amphiphilic region in the cystoplasmic domain of KdpD is recognized by the signal recognition particle and targeted to the Escherichia coli membrane. Molecular Microbiology. 2008:1471–1484. doi: 10.1111/j.1365-2958.2008.06246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.von Heijne G. Net N–C charge imbalance may be important for signal sequence function in bacteria. J Mol Biol. 1986;192:287–290. doi: 10.1016/0022-2836(86)90365-7. [DOI] [PubMed] [Google Scholar]
- 55.von Heijne G. Membrane protein structure prediction: Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 56.Green MR, Sambrook J. Molecular Cloning A Laboratory Manual. 4. Vol. 3. Cold Spring Harbor Press; Cold Spring Harbor: 2012. [Google Scholar]
- 57.De Gier J-W, Scotti PA, Sääf A, Valent QA, Kuhn A, Luirink J. Differential use of the signal recognition particle translocase targeting pathway for inner membrane protein assembly in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:14646–14651. doi: 10.1073/pnas.95.25.14646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meissner PS, Sisk WP, Berman ML. Bacteriophage λ cloning system for the construction of directional cDNA libraries. Proc Natl Acad Sci USA. 1987;84:4171–4175. doi: 10.1073/pnas.84.12.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 60.Seluanov A, Bibi E. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J Biol Chem. 1997;272:2053–2055. doi: 10.1074/jbc.272.4.2053. [DOI] [PubMed] [Google Scholar]
- 61.Or E, Boyd D, Gon S, Beckwith J, Rapoport T. The bacterial ATPase SecA functions as a monomer in protein translocation. J Biol Chem. 2005;280:9097–9105. doi: 10.1074/jbc.M413947200. [DOI] [PubMed] [Google Scholar]
- 62.Economou A, Pogliano JA, Beckwith J, Oliver DB, Wickner W. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell. 1995;83:1171–1181. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- 63.Driessen AJM, Manting EH, van der Does C. The structural basis of protein targeting and translocation in bacteria. Nat Struct Biol. 2001;8:492–498. doi: 10.1038/88549. [DOI] [PubMed] [Google Scholar]
- 64.Herskovits AA, Bochkareva ES, Bibi E. New prospects in studying the bacterial signal recognition particle pathway. Molecular Microbiol. 2000;38:927–939. doi: 10.1046/j.1365-2958.2000.02198.x. [DOI] [PubMed] [Google Scholar]
- 65.Akopian D, Shen K, Zhang X, Shan S-O. Signal recognition particle: an essential protein-trageting machine. Annu Rev Biochem. 2013;82:693–721. doi: 10.1146/annurev-biochem-072711-164732. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


