Abstract
Background
The pathogenesis of multiple myeloma involves complex genetic and epigenetic events. This study aimed to investigate the role and clinical relevance of the long non-coding RNA (lncRNA), metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) in multiple myeloma.
Methods
Bone marrow mononuclear cells were collected for analysis. The samples of multiple myeloma were taken from 45 patients at diagnosis, 61 post-treatment, and 18 who relapsed or had progression. Control samples were collected from 20 healthy individuals. Real-time quantitative reverse transcription polymerase chain reactions were performed to evaluate the expression of MALAT1. The clinical relevance of MALAT1 expression was also explored.
Results
MALAT1 was overexpressed in the newly diagnosed patients compared with post-treatment patients (mean ∆CT: -5.54 ± 0.16 vs. -3.84 ± 0.09, 3.25-fold change; p < 0.001) and healthy individuals (mean ∆CT: -5.54 ± 0.16 vs. -3.95 ± 0.21, 3.01-fold change; p < 0.001). The expression of MALAT1 strongly correlated with disease status, and the magnitude of change in MALAT1 post-treatment had prognostic relevance. The patients with early progression had a significantly smaller change in MALAT1 after treatment (mean ∆CT change: 1.26 ± 1.06 vs. 2.09 ± 0.79, p = 0.011). A cut-off value of the change in MALAT1 (∆CT change: 1.5) was obtained, and the patients with a greater decrease in MALAT1 (difference in ∆CT >1.5) had significantly longer progression-free survival compared with the patients with a smaller MALAT1 change (24 months vs. 11 months; p = 0.001). For the post-treatment patients, the risk of early progression could be predicted using this cut-off value.
Conclusions
MALAT1 was overexpressed in patients with myeloma and may play a role in its pathogenesis. In addition, MALAT1 may serve as a molecular predictor of early progression.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2407-14-809) contains supplementary material, which is available to authorized users.
Keywords: Multiple myeloma, Long non-coding RNA, Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1)
Background
Multiple myeloma is a hematological malignancy characterized by abnormal proliferation of monoclonal plasma cells in bone marrow leading to various end-organ damage including anemia, hypercalcemia, renal insufficiency and osteolytic bone disease [1]. The development of multiple myeloma is thought to result from monoclonal gammopathy of undetermined clinical significance [2, 3]. With the progression from monoclonal gammopathy of undetermined clinical significance to myeloma, several complex genetic events are involved including cytogenetic abnormalities, primary or secondary chromosomal translocation, and activation of oncogenes. These oncogenetic events include dysregulation of the cyclin D gene, mutation of KRAS or NRAS, and constitutively activated nuclear factor κB (NFκB) pathway [4–7]. In addition, the bone marrow microenvironment has also been reported to play an important role in the pathogenesis of this disease [8–10].
The human genome project revealed that at least 90% of the human genome is actively transcribed to RNA, but less than 2% of RNA encodes proteins [11, 12]. Non-coding RNAs (ncRNAs) are a class of RNA with little or no capacity for protein synthesis that includes small ncRNAs and long ncRNAs (lncRNAs), which have a length of more than 200 nucleotides. The lncRNAs have been highly conserved throughout mammalian evolution including in humans, and they have been shown to be aberrantly expressed in cancer tissue and to be involved in oncogenic or tumor suppressive processes [13].
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is one of the few biologically well-studied lncRNAs, and is located on chromosome 11 (11q13.1). This lncRNA is highly conserved in mammals and is more than 8000 nucleotides in length [14–16]. MALAT1 has been shown to expressed in numerous tissues including the central nervous, endocrine, immune, reproductive and lymphoid systems [17, 18]. With respect to its function, MALAT1 is localized to nuclear speckles and has been associated with regulation of gene expressions [19, 20]. In addition, MALAT1 may play a role in the regulation of alternative splicing and cell cycle [21–23]. In terms of its association with cancer, MALAT1 has been shown to be oncogenic and to be overexpressed in several solid tumors including lung, colorectal, bladder and laryngeal cancers [24–27].
The association between lncRNAs and multiple myeloma remains undetermined, and related studies are lacking. It has been reported that deregulation of the cell cycle is an important event during carcinogenesis, and that this event is also associated with MALAT1 [23]. MALAT1 has also been reported to be expressed broadly in human tissues including lymphoid tissues, bone marrow and B lymphocytes [28, 29]. Taken together, we hypothesized that MALAT1 may play a role in multiple myeloma. Therefore, the aim of the present study was to evaluate the expression of MALAT1 in bone marrow mononuclear cells from patients with multiple myeloma and with different disease status and healthy individuals.
Methods
Multiple myeloma patients and samples
The study cohort included adult patients (aged 20 years and older) with multiple myeloma diagnosed at Kaohsiung Medical University Hospital from 2007 to 2012 who were free from other coexisting malignant diseases. The diagnosis of multiple myeloma was confirmed by bone marrow analysis which revealed a monoclonal plasma cell count over 10% by definition and related laboratory tests. The patients of extramedullary myeloma were not enrolled to this study. The diagnostic criteria, disease status and response to treatment were based on the criteria of the International Myeloma Working Group [17–19]. Forty-five samples were collected from newly diagnosed patients (29 males, 16 females; median age 62.3 years, range 49 to 79 years) with different subtypes (IgG: 21, IgA: 13, light chain: 11) and clinical stages (Durie-Salmon stage 1: 1, stage 2: 6, stage 3: 38 or international staging system stage 1: 7, stage 2: 17, stage 3: 21). In addition, 61 samples were collected from patients after myeloma treatment, and 18 samples from patients who had experienced disease progression or relapse. The disease status of the post-treatment patients was mainly a complete response (CR) and very good partial response (VGPR) based on the criteria of International Myeloma Working Group. In addition, the percentage of plasma cells in the patients achieving VGPR or CR after treatment was less than 5%.
We also enrolled 20 healthy and genetically unrelated Taiwanese volunteers (healthy individuals) as the control group. These healthy individuals had undergone bone marrow analysis to investigate cytopenia that had been noted in blood tests, but whose bone marrow examinations revealed no abnormalities. All patients and healthy individuals signed informed consent forms after the study had been thoroughly explained.
The research protocol was created in accordance with the Declaration of Helsinki, and it was reviewed, approved and registered by the Ethics Committee of Kaohsiung Medical University Hospital (KMUHIRB-2012-01-08(II)).
RNA extraction and reverse transcription
Bone marrow mononuclear cells were isolated for this study. First, the bone marrow samples were collected in tubes containing ethylenediaminetetraacetic acid (EDTA), preserved at 4°C and processed within 4 hours of collection. The bone marrow samples were then centrifuged at 12,000 × g for 15 minutes, after which ammonium chloride lysis buffer (10 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA) was used to clear the red blood cells and effectively isolate the fraction of mononuclear cells.
The isolated bone marrow samples were stored at -80°C until RNA extraction. Isolation of RNA from 200 μL of cell suspension was carried out using the TRIzol protocol (Invitrogen). The extracted RNA was then treated with DNase (Promega) and the concentration was determined by spectrophotometric OD260 measurement. The integrity of the RNA was examined by 1.2% RNA denaturing agarose gel electrophoresis.
Reverse transcription was performed to generate complementary DNA in a final volume of 20 μL, containing 2 μg RNA, 25X dNTP mix (100 mM), 10X random primer (0.5 μM), RNase inhibiter, reverse transcriptase, reverse transcriptase buffer (10X) and diethylpyrocarbonate (DEPC)-treated water. The procedure was performed according to the manufacturer’s protocol (Applied Biosystems).
Real-time quantitative reverse transcription polymerase chain reaction (RT-PCR) analysis of MALAT1expression
Real-time quantitative RT-PCR was performed in a final volume of 10 μL containing 1 μL of RT product, 0.6 μL of primer (Roche), 1.2 μL of probe (Roche, cat. no. 04688945001), 2.2 μL of DEPC H2O and 5 μL of qPCR Master Mix (2X) (KAPA Biosystem, KK 4600). Analysis of the human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as the internal control.
The primer sequences of MALAT1 were as follows: forward, 5’-GACCCTTCACCCCTCACC-3’; reverse, 5’-TTATGGATCATGCCCACAAG-3’, and the primer sequences of GAPDH were as follows: forward, 5’-AAAGTCCGCCATTTTGCCACT-3’; and reverse, 5’-CCAAATCGTTAGCGCTCCTT-3’.
Real-time quantitative RT-PCR was performed in a LightCycler 480 Real-Time PCR System (Roche). The PCR cycling program consisted of incubation for enzyme activation at 95°C for 10 minutes, followed by melting at 95°C for 10 seconds, annealing at 60°C for 30 seconds, and then extension at 72°C for 1 second, for a total of 50 cycles.
The expression levels of MALAT1 were normalized to the internal control GAPDH reference to obtain the relative threshold cycle (∆CT). The relative expression levels were calculated by the comparative CT (∆∆CT) method, and relative expression folds (2-ΔΔCT) were calculated.
Statistical analysis
The independent two samples t-test was used to compare the expression levels of MALAT1 in the different subgroups. The frequency between each categorical variable was compared by the chi-square test (χ2 test), with Yates correction or Fisher’s exact test. Analysis of correlation was performed using Pearson correlations or Spearman correlation coefficients. Receiver operating characteristic (ROC) analysis was used to evaluate the cut-off value. Survival curves were plotted using the Kaplan–Meier method and compared using the log-rank test. Relative risk analysis was performed by calculating the odds ratio (OR) and 95% confidence interval (CI) by Cox regression analysis.
All statistical analyses were based on two-sided hypothesis tests with a significance level of p < 0.05. The analyses were performed using SPSS version 17.0 (SPSS, Chicago, IL, USA).
Results
Correlation of MALAT1expression with disease status in multiple myeloma
The expression of MALAT1 was significantly higher in the patients at diagnosis compared with the patients post-treatment (mean ∆CT: -5.54 ± 0.16 vs. -3.84 ± 0.09, 3.25-fold change; p < 0.001) or the healthy individuals (mean ∆CT: -5.54 ± 0.16 vs. -3.95 ± 0.21, 3.01-fold change; p < 0.001) (Table 1). This suggests that MALAT1 may be deregulated and overexpressed in patients with multiple myeloma.
Table 1.
Expression of MALAT1 in patients with multiple myeloma and healthy individuals
| Population | No. | Expression of MALAT1(Mean ΔC T) | |
|---|---|---|---|
| Newly diagnosed | 45 | -5.54 ± 0.16 | |
| Post-treatment | 61 | -3.84 ± 0.09 | |
| Relapse or progression | 18 | -4.92 ± 0.23 | |
| Healthy individuals | 20 | -3.95 ± 0.21 | |
| ΔΔCT | Fold change | P value | |
| Newly diagnosed vs. Post-treatment | -1.70 | 3.25 | <0.001 |
| Newly diagnosed vs. Healthy individuals | -1.59 | 3.01 | <0.001 |
| Relapse or progression vs. Post-treatment | -0.92 | 1.89 | <0.001 |
| Post-treatment vs. Healthy individuals | 0.11 | 1.08 | 0.614 |
The 61 post-treatment samples were composed of 58 samples collected in disease status of VGPR or CR from 42 patients. The percentages of plasma cells were all less than 5%.
The 3 samples collected at a disease status of partial response came from 3 patients.
Note:
ΔCT = CT (MALAT1 - GAPDH).
Increased expression (fold change) was calculated as 2-ΔΔCT.
The association of MALAT1 expression pattern with disease status was further analyzed. The expression of MALAT1 was found to be significantly decreased in the post-treatment patients to a level that was similar to that of the healthy individuals (mean ∆CT: -3.84 ± 0.09 vs. -3.95 ± 0.21, p = 0.614). In addition, in the patients in whom the disease had progressed or relapsed, the expression of MALAT1 was significantly increased compared with the post-treatment patients (mean ∆CT: -4.92 ± 0.23 vs. -3.84 ± 0.09, 1.89-fold change; p < 0.001) (Table 1). For the patients who underwent multiple bone marrow examinations during treatment and follow-up, the expression of MALAT1 changed dynamically and was correlated with disease status (Figure 1).
Figure 1.
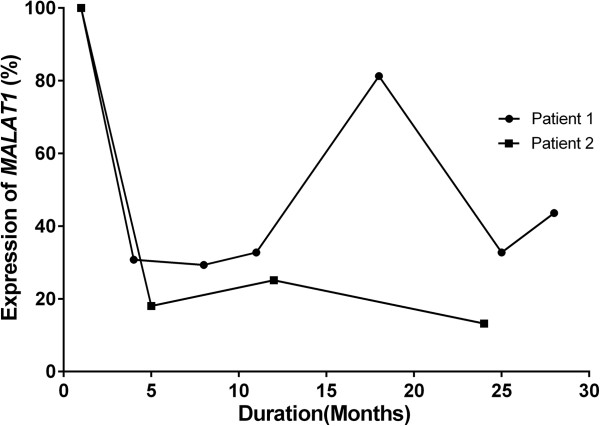
Expression of MALAT1 during treatment and follow-up in two representative patients. The first patient (Patient 1) had a high expression of MALAT1 initially, which then decreased after successful induction chemotherapy and autologous peripheral blood stem cell transplantation, 9 months after the time of diagnosis. At 18 months, disease relapse was accompanied by an increase in MALAT1 expression. After salvage treatment, the disease was controlled and the expression of MALAT1 decreased. Eventually, the disease progressed and the expression of MALAT1 increased. The second patient (Patient 2) received induction chemotherapy followed by peripheral blood stem cell transplantation. The expression of MALAT1 decreased markedly after the treatment achieved complete remission. The disease status remained in remission during the follow-up period and was accompanied by a low expression of MALAT1.
Association between MALAT1expression and clinical outcome
The clinical relevance of MALAT1 was analyzed. The expressions of MALAT1 in the 45 newly diagnosed patients with different clinical characteristics were listed in Additional file 1: Table S1. We noticed that the expression of MALAT1 was not associated with the percentage of plasma cells in the bone marrow (r = -0.037, p = 0.808) (Additional file 2: Table S2). With regards to the association between MALAT1 expression and prognosis, the results showed that the initial higher MALAT1 expression level (Cut-off value: ∆CT = -5.30) determined by ROC analysis was not associated with inferior prognosis including progression-free survival (PFS) (median PFS: 21.0 ± 9.9 vs. 15.0 ± 6.0 months, p = 0.390) or overall survival (OS) (median OS: Not reached; mean OS: 31.9 ± 4.3 vs. 37.8 ± 3.6 months, p = 0.172) (Additional file 3: Figure S1). However, we hypothesized the magnitude of the change (decrease) after myeloma-related therapy may have been related to the degree of treatment response and prognosis, because the expression of MALAT1 changed after treatment.
Among the 45 patients, 36 (including 21 men) with a mean age of 61.3 years received bone marrow examinations at least twice including at diagnosis and after treatment, and hence the expression of MALAT1 was available for further analysis. The median PFS of these 36 patients was 18 months (95% CI 8.95-25.35), and we used this median PFS as a cut-off value to divide the patients into two groups of early (PFS ≤18 months) or late (PFS >18 months) progression, with 18 patients in each group. We found that the only parameter which showed a significant difference between these two groups was the magnitude of MALAT1 change after treatment. The patients with early progression had a significantly smaller magnitude of MALAT1 change after treatment compared with the patients with late progression (mean difference of ∆CT: 1.26 ± 1.06 vs. 2.09 ± 0.79; p = 0.011) (Table 2).
Table 2.
The clinical characteristics of patients with early (PFS ≤18 months) or late (PFS >18 months) progression
| All patients (N = 36) | PFS ≤ 18 months (N = 18) | PFS > 18 months (N = 18) | Pvalue | |
|---|---|---|---|---|
| Age (years, mean(SD)) | 61.3(8.3) | 61.9 ± 7.8 | 60.6 ± 8.7 | 0.633 |
| Male, n (%) | 21(58.3%) | 10(55.6%) | 11(61.1%) | 1.000 |
| M protein | 0.210 | |||
| IgG, n (%) | 19(52.8%) | 7(38.9%) | 12(66.6%) | |
| IgA, n (%) | 10(27.8%) | 7(38.9%) | 3(16.7%) | |
| Light chain, n (%) | 7(19.4%) | 4(22.2%) | 3(16.7%) | |
| International staging system | 0.881 | |||
| Stage 1, n (%) | 5(13.9%) | 2(11.1%) | 3(16.7%) | |
| Stage 2, n (%) | 12(33.3%) | 6(33.3%) | 6(33.3%) | |
| Stage 3, n (%) | 19(52.8%) | 10(55.6%) | 9(50%) | |
| Durie-Salmon stage | 1.000 | |||
| Stage 1, n (%) | 0 | 0 | 0 | |
| Stage 2, n (%) | 5(13.9%) | 2(11.1%) | 3(16.7%) | |
| Stage 3, n (%) | 31(86.1%) | 16(88.9%) | 15(83.3%) | |
| Percentage of plasma cell in bone marrow (%, mean (SD)) | 50.8 ± 25.3 | 54.3 ± 26.8 | 47.3 ± 23.8 | 0.740 |
| Anemia, n (%) | 27(75%) | 14(77.8%) | 13(72.2%) | 1.000 |
| Renal insufficiency, n (%) | 9(25%) | 5(27.8%) | 4(22.2%) | 1.000 |
| Hypercalcemia, n (%) | 14(38.9%) | 8(44.4%) | 6(33.3%) | 0.733 |
| Bone disease, n (%) | 25(69.4%) | 14(77.8%) | 11(61.1%) | 0.471 |
| Cytogenetic abnormality, n (%) | 7(19.4%) | 3(16.7%) | 4(22.2%) | 1.000 |
| Bortezomib-containing induction Tx, n (%) | 11(30.6%) | 4(22.2%) | 7(38.9%) | 0.471 |
| Auto-HSCT in 1st fine Tx, n (%) | 9(25%) | 2(11.1%) | 7(38.9%) | 0.121 |
| Treatment response: | ||||
| CR, n (%) | 7(19.4%) | 1(5.6%) | 6(33.3%) | 0.088 |
| VGPR, n (%) | 26(72.2%) | 14(77.8%) | 12(66.7%) | 0.711 |
| PR, n (%) | 3(8.3%) | 3(16.7%) | 0 | 0.229 |
| Expression of MALAT1 at diagnosis (Mean ΔCT ± SD) | -5.52 ± 1.15 | -5.77 ± 0.89 | 0.353 | |
| Magnitude of MALAT1 change after treatment (Difference in ΔCT) | 1.26 ± 1.06 | 2.09 ± 0.79 | 0.011 |
Difference in ΔCT = ΔCT (Post-treatment - newly diagnosed).
Auto-HSCT, autologous hematopoietic stem cell transplantation; CR, complete response; VGPR, very good partial response; PFS, progression-free survival; Tx, treatment.
Role of MALAT1in predicting early progression
The previous results showed that the magnitude of MALAT1 change (as quantified by the difference in ∆CT) was the only parameter associated with PFS. We then used ROC analysis and obtained a cut-off expression change value of 1.5 (post-treatment ∆CT – pre-treatment ∆CT; approximately a 2.8-fold change) with an estimated area under the ROC curve of 0.79 (p = 0.003). The proportion of patients with a lower MALAT1 change (difference in ∆CT ≤1.5) was significantly higher in those who displayed early progression (n = 13, 72.2%) compared with those who displayed late progression (n = 4, 22.2%; p = 0.007).
In terms of an association between clinical characteristics and magnitude of MALAT1 change, the patients with greater changes (difference in ∆CT >1.5) may have had a better treatment response (Additional file 4: Table S3). The PFS and OS rates were also analyzed using the cut-off value from ROC analysis. With a minimum follow-up period of 12 months (range: 12 to 48 months), the patients with a greater MALAT1 decrease (difference in ∆CT >1.5) had a significantly prolonged median PFS (24 months, range 11-48 months) compared with the patients with a smaller MALAT1 change (11 months, range: 6-21 months; p = 0.001). There was no significant difference in OS between the two groups (median OS: Not reached; mean OS: 39.2 ± 3.6 months, range: 12-48 months vs. 32.8 ± 4.2 months, range: 12-48 months; p = 0.313) (Figure 2).
Figure 2.
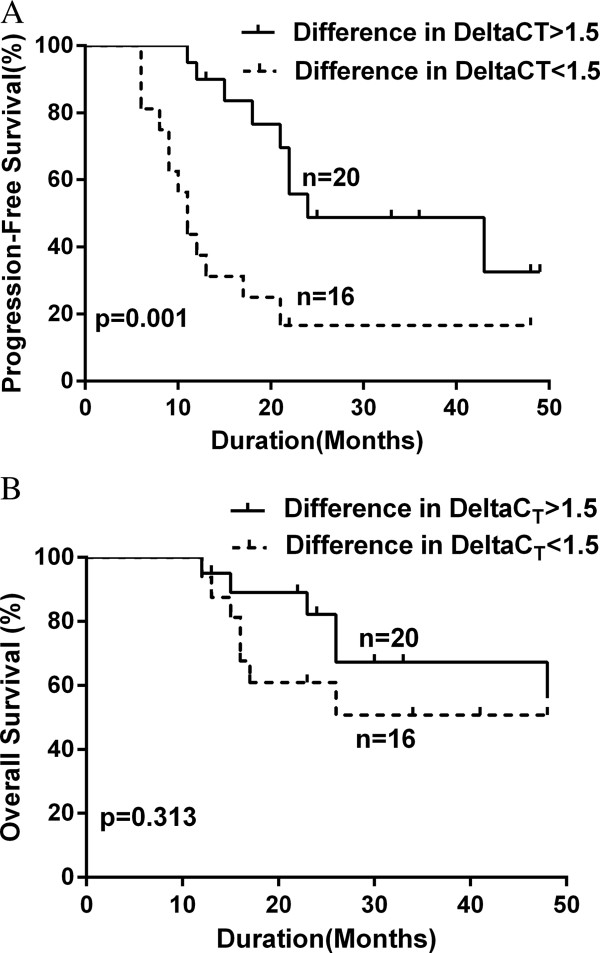
Kaplan-Meier estimates of the probability of progression-free survival (PFS, A) and overall survival (OS, B) are shown according to the magnitude in the change of MALAT1 expression after treatment. The patients were divided into two groups by a cut-off value (difference in ΔCT: 1.5).
Cox regression analysis was used to identify the relative risk of early progression, which revealed that autologous hematopoietic stem cell transplantation (Auto-HSCT) and the magnitude of MALAT1 change were significantly associated with the prognosis. For all post-treatment patients (n = 36), those with a smaller MALAT1 change (difference in ∆CT ≤1.5) had a significantly higher risk of early progression of disease (OR 4.89, 95% CI 1.73-13.86; p = 0.003), while auto-HSCT reduced the risk of early progression (OR 0.22, 95% CI 0.05-0.97; p = 0.046). For the post-treatment patients with a VGPR or CR (n = 33), a smaller MALAT1 change (difference in ∆CT ≤1.5) remained the single factor predictive of early progression of multiple myeloma (OR 4.38, 95% CI 1.48-12.99; p = 0.008) (Table 3). Using the cut-off value to predict the patients who would show early progression (PFS ≤18 months), the estimated accuracy was 75% with a sensitivity of 72.2%, a specificity of 77.8%, a positive predictive value of 76.5%, and a negative predictive value of 73.7%.
Table 3.
Multivariate Cox regression analysis for all post-treatment patients and post-treatment patients with a treatment response of VGPR/CR
| PFS ≤18 months (N = 18) | PFS >18 months (N = 18) | Cox regression analysis | |||||
|---|---|---|---|---|---|---|---|
| OR | 95% CI | Pvalue | |||||
| All patients (N = 36) | Auto-HSCT in 1st line treatment, n | 9 | 2(11.1%) | 7(38.9%) | 0.22 | 0.05-0.97 | 0.046 |
| Difference in △CT ≤1.5, n | 17 | 13(72.2%) | 4(22.2%) | 4.89 | 1.73-13.86 | 0.003 | |
| PFS ≤18 months (N = 15) | PFS >18 months (N = 18) | ||||||
| Patients with VGPR/CR (N = 33) | Auto-HSCT in 1st line treatment, n | 9 | 2(13.3%) | 7(38.9%) | 0.24 | 0.05-1.09 | 0.066 |
| Difference in △CT ≤1.5, n | 14 | 10(66.7%) | 4(22.2%) | 4.38 | 1.48-12.99 | 0.008 | |
OR, Odds ratio; CI, confidential interval; Auto-HSCT, autologous hematopoietic stem-cell transplantation; CR, complete response; VGPR, very good partial response; PFS, progression-free survival.
Discussion
In the current study, we demonstrated that MALAT1 was overexpressed in the patients with newly diagnosed multiple myeloma. This finding indicates that MALAT1 may play a role in multiple myeloma.
The results of the present study are in contrast with the study by Isin et al., in which the expression of MALAT1 was found to be significantly lower in patients with multiple myeloma [30]. A possible explanation for this discrepancy may be due to different sample sources. Our study analyzed the expression of MALAT1 in bone marrow mononuclear cells rather than plasma samples, because the pathogenesis of myeloma is closely related to bone marrow. Another possible explanation for the higher expression of MALAT1 in the current study may be associated with the bone marrow microenvironment which supports the proliferation of myeloma cells. In addition, our analysis revealed that expression of MALAT1 in newly diagnosed myeloma patients is not associated with the total percentage of plasma cells in the bone marrow. This finding indicated that the expression of MALAT1 may be associated with interactions between myeloma cells and the bone marrow microenvironment. The detailed mechanism needs further studies to elucidate.
The current study also investigated the clinical relevance of MALAT1 in patients with multiple myeloma. We found that the expression of MALAT1 changed dynamically when stratified by disease status. In addition, the major clinical significance was the magnitude of change in expression after treatment rather than the initial expression. This finding is different from previous studies of solid tumors which have reported that a higher expression is related to poorer prognosis. We observed that the patients with a greater decrease in MALAT1 after initial treatment had a significantly prolonged PFS, which is consistent with the current consensus that therapeutic intervention to achieve a maximal response is beneficial for patients with multiple myeloma [31, 32]. In terms of OS, we did not find a significant benefit in the post-treatment patients with a greater decrease in MALAT1. A possible explanation may be the incorporation of potent and effective salvage treatment in the patients who experienced a relapse or progression of disease, as well as the fact that some patients received auto-HSCT after salvage treatment.
We also found that MALAT1 may serve as a marker to predict early progression. Because the duration of response decreases with an increasing number of salvage regimens after progression, identification of patients at risk of early progression after first-line treatment is an important issue. More intensive treatment may improve the prognosis in this subgroup. We also found that patients with a smaller MALAT1 change after treatment had a significantly higher risk for early progression, even in those with a VGPR, CR and normal percentage of plasma cells in bone marrow. This finding suggests that the expression of MALAT1 can be used to identify the patients at risk of early progression. Accordingly, the therapeutic strategy may be adjusted to be initially more aggressive, as more potent treatment may reduce the risk of early progression and prolong PFS.
Our findings may provide a new insight into the pathogenesis of multiple myeloma. However, there are some limitations to this study. First, the cytogenetic examinations were done by conventional G-band metaphase chromosome analysis, and the percentage of cytogenetic abnormalities was relative low. Therefore, the association between MALAT1 and specific cytogenetic abnormalities remains to be determined. Further analysis by fluorescent in-situ hybridization with larger cohort may provide more impactful insight on the clinical relevance of MALAT1 expression in multiple myeloma. Second, we didn’t evaluate the expression of MALAT1 in patients resistant to myeloma therapy due to no available samples. Third, the number of cases to evaluate the clinical relevance of MALAT1 was limited, which was likely due to the stringency of the enrollment criteria.
Conclusions
In conclusion, this study revealed that MALAT1 was overexpressed in patients with multiple myeloma, and this lncRNA may play a role in the pathogenesis of the disease. In addition, the change in MALAT1 expression after treatment was clinically significant and may serve as a molecular predictor of the patients at risk of early progression of multiple myeloma.
Electronic supplementary material
Additional file 1: Table S1: The clinical characteristics and expression of MALAT1 in 45 newly diagnosed patients with multiple myeloma. (DOC 56 KB)
Additional file 2: Table S2: Expression of MALAT1 and plasma cell percentage in the bone marrow in 45 newly diagnosed patients. (DOC 58 KB)
Additional file 3: Figure S1: Kaplan-Meier estimates of the probability of progression-free survival (PFS, 1A) and overall survival (OS, 1B) are based on the initial level of MALAT1 expression. The patients were divided into two groups by a cut-off value (△CT: -5.30). (ZIP 137 KB)
Additional file 4: Table S3: The clinical characteristics of the patients with different cut-off values. (DOC 58 KB)
Acknowledgments
The authors thank the Statistical Analysis Laboratory, Department of Medical Research, Kaohsiung Medical University Hospital, Kaohsiung Medical University for their help.
Footnotes
Competing interests
The authors declare that they have no competing interest.
Authors’ contributions
SFC, JGC and TCL designed the study. SFC, YCC, CSC, SFL, YCL, HHH, TCL contributed to collection and review of clinical data. SFC and YCC performed molecular examination. S-FC and CSC performed statistical analysis. SFC wrote the manuscript. SFC, TCL, JGC critically revised the manuscript. TCL and JGC approved the final version of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Shih-Feng Cho, Email: sifong96@gmail.com.
Yuli Christine Chang, Email: yuli33@ms28.hinet.net.
Chao-Sung Chang, Email: ccschang@gmail.com.
Sheng-Fung Lin, Email: shlin@kmu.edu.tw.
Yi-Chang Liu, Email: ycliu@kmu.edu.tw.
Hui-Hua Hsiao, Email: huhuhs@cc.kmu.edu.tw.
Jan-Gowth Chang, Email: d6781@mail.cmuh.org.tw.
Ta-Chih Liu, Email: d730093@cc.kmu.edu.tw.
References
- 1.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364:1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 2.Weiss BM, Abadie J, Verma P, Howard RS, Kuehl WM. A monoclonal gammopathy precedes multiple myeloma in most patients. Blood. 2009;113:5418–5422. doi: 10.1182/blood-2008-12-195008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landgren O, Kyle RA, Pfeiffer RM, Katzmann JA, Caporaso NE, Hayes RB, Dispenzieri A, Kumar S, Clark RJ, Baris D, Hoover R, Rajkumar SV. Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study. Blood. 2009;113:5412–5417. doi: 10.1182/blood-2008-12-194241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuehl WM, Bergsagel PL. Multiple myeloma: evolving genetic events and host interactions. Nat Rev Cancer. 2002;2:175–187. doi: 10.1038/nrc746. [DOI] [PubMed] [Google Scholar]
- 5.Seidl S, Kaufmann H, Drach J. New insights into the pathophysiology of multiple myeloma. Lancet Oncol. 2003;4:557–564. doi: 10.1016/S1470-2045(03)01195-1. [DOI] [PubMed] [Google Scholar]
- 6.Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol. 2005;23:6333–6338. doi: 10.1200/JCO.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 7.Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12:335–348. doi: 10.1038/nrc3257. [DOI] [PubMed] [Google Scholar]
- 8.Kumar S, Witzig TE, Timm M, Haug J, Wellik L, Fonseca R, Greipp PR, Rajkumar SV. Expression of VEGF and its receptors by myeloma cells. Leukemia. 2003;17:2025–2031. doi: 10.1038/sj.leu.2403084. [DOI] [PubMed] [Google Scholar]
- 9.Nefedova Y, Cheng P, Alsina M, Dalton WS, Gabrilovich DI. Involvement of Notch-1 signaling in bone marrow stroma-mediated de novo drug resistance of myeloma and other malignant lymphoid cell lines. Blood. 2004;103:3503–3510. doi: 10.1182/blood-2003-07-2340. [DOI] [PubMed] [Google Scholar]
- 10.Hideshima T, Mitsiades C, Tonon G, Richardson PG, Anderson KC. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer. 2007;7:585–598. doi: 10.1038/nrc2189. [DOI] [PubMed] [Google Scholar]
- 11.Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, Gerstein M, Snyder M. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 12.International Human Genome Sequencing C Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 13.Hauptman N, Glavac D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14:4655–4669. doi: 10.3390/ijms14034655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Muller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 15.Guru SC, Agarwal SK, Manickam P, Olufemi SE, Crabtree JS, Weisemann JM, Kester MB, Kim YS, Wang Y, Emmert-Buck MR, Liotta LA, Spiegel AM, Boguski MS, Roe BA, Collins FS, Marx SJ, Burns L, Chandrasekharappa SC. A transcript map for the 2.8-Mb region containing the multiple endocrine neoplasia type 1 locus. Genome Res. 1997;7:725–735. doi: 10.1101/gr.7.7.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Asseldonk M, Schepens M, de Bruijn D, Janssen B, Merkx G, Geurts van Kessel A. Construction of a 350-kb sequence-ready 11q13 cosmid contig encompassing the markers D11S4933 and D11S546: mapping of 11 genes and 3 tumor-associated translocation breakpoints. Genomics. 2000;66:35–42. doi: 10.1006/geno.2000.6194. [DOI] [PubMed] [Google Scholar]
- 17.Wilusz JE, Freier SM, Spector DL. 3' end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedel CC, Dolken L, Ruzsics Z, Koszinowski UH, Zimmer R. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res. 2009;37:e115. doi: 10.1093/nar/gkp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyagawa R, Tano K, Mizuno R, Nakamura Y, Ijiri K, Rakwal R, Shibato J, Masuo Y, Mayeda A, Hirose T, Akimitsu N. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA. 2012;18:738–751. doi: 10.1261/rna.028639.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang F, Yi F, Han X, Du Q, Liang Z. MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett. 2013;587:3175–3181. doi: 10.1016/j.febslet.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 23.Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, Prasanth KV. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt LH, Spieker T, Koschmieder S, Schaffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, Hillejan L, Wiebe K, Berdel WE, Wiewrodt R, Muller-Tidow C. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 25.Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3' end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39:169–175. doi: 10.3892/ijo.2011.1007. [DOI] [PubMed] [Google Scholar]
- 26.Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8:2289–2294. doi: 10.1039/c2mb25070e. [DOI] [PubMed] [Google Scholar]
- 27.Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, Zornig M, MacLeod AR, Spector DL, Diederichs S. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lane L, Argoud-Puy G, Britan A, Cusin I, Duek PD, Evalet O, Gateau A, Gaudet P, Gleizes A, Masselot A, Zwahlen C, Bairoch A. neXtProt: a knowledge platform for human proteins. Nucleic Acids Res. 2012;40:D76–83. doi: 10.1093/nar/gkr1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu C, Macleod I, Su AI. BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic Acids Res. 2013;41:D561–565. doi: 10.1093/nar/gks1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isin M, Ozgur E, Cetin G, Erten N, Aktan M, Gezer U, Dalay N. Investigation of circulating lncRNAs in B-cell neoplasms. Clin Chim Acta. 2014;431:255–259. doi: 10.1016/j.cca.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 31.Chanan-Khan AA, Giralt S. Importance of achieving a complete response in multiple myeloma, and the impact of novel agents. J Clin Oncol. 2010;28:2612–2624. doi: 10.1200/JCO.2009.25.4250. [DOI] [PubMed] [Google Scholar]
- 32.Moreau P, Attal M, Pegourie B, Planche L, Hulin C, Facon T, Stoppa AM, Fuzibet JG, Grosbois B, Doyen C, Ketterer N, Sebban C, Kolb B, Chaleteix C, Dib M, Voillat L, Fontan J, Garderet L, Jaubert J, Mathiot C, Esseltine D, Avet-Loiseau H, Harousseau JL, investigators IFMs Achievement of VGPR to induction therapy is an important prognostic factor for longer PFS in the IFM 2005-01 trial. Blood. 2011;117:3041–3044. doi: 10.1182/blood-2010-08-300863. [DOI] [PubMed] [Google Scholar]
Pre-publication history
- The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2407/14/809/prepub
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1: The clinical characteristics and expression of MALAT1 in 45 newly diagnosed patients with multiple myeloma. (DOC 56 KB)
Additional file 2: Table S2: Expression of MALAT1 and plasma cell percentage in the bone marrow in 45 newly diagnosed patients. (DOC 58 KB)
Additional file 3: Figure S1: Kaplan-Meier estimates of the probability of progression-free survival (PFS, 1A) and overall survival (OS, 1B) are based on the initial level of MALAT1 expression. The patients were divided into two groups by a cut-off value (△CT: -5.30). (ZIP 137 KB)
Additional file 4: Table S3: The clinical characteristics of the patients with different cut-off values. (DOC 58 KB)


