Abstract
Microsatellites are simple tandem repeats that are present at millions of loci in the human genome. Microsatellite instability (MSI) refers to DNA slippage events on microsatellites that occur frequently in cancer genomes when there is a defect in the DNA mismatch repair system. These somatic mutations can result in inactivation of tumor suppressor genes or disrupt other non-coding regulatory sequences, thereby playing a role in carcinogenesis. Here, we will discuss the ways in which high-throughput sequencing data can facilitate a genome- or exome-wide discovery and more detailed investigation of MSI events in microsatellite-unstable cancer genomes. We will address the methodological aspects of this approach and highlight insights from recent analyses of colorectal and endometrial cancer genomes from The Cancer Genome Atlas project. These include identification of novel MSI targets within and across tumor types and the relationship between the likelihood of MSI events to chromatin structure. Given the increasing popularity of exome and genome sequencing of cancer genomes, a comprehensive characterization of MSI may serve as a valuable marker of cancer evolution and aid in a search for therapeutic targets.
Introduction
Since its discovery in sporadic and familial colorectal cancer genomes in the early nineties (1-3), microsatellite instability (MSI) has been well studied as a unique type of somatic mutation that can occur in cancer genomes with a dysfunctional DNA mismatch repair (MMR) system (4, 5). MSI has been used mainly to discriminate cancer genomes into microsatellite-stable vs -unstable cases, based on PCR analysis of microsatellite markers in the Bethesda panel (6, 7) or on immunohistochemistry of MMR proteins (8). Their clinical utilities in the prediction of chemosensitivity of colorectal cancers (9) and the screening of individuals or families with inherited syndromes of hereditary nonpolyposis colorectal cancers (HNPCC), also called Lynch syndrome (10), have been well established. However, the use of MSI beyond screening with a pre-determined set markers has been limited due to low throughput of traditional MSI screening methods. For example, exactly which loci are mutated in individual genomes, which genomic or epigenomic features are associated with their occurrences, and what the functional consequences are in terms of mRNA expression and perturbed pathways have not been fully examined.
The advent of high-throughput sequencing technology has enabled detailed examination of various types of somatic alterations in cancer genomes. Whole-genome or exome-wide interrogation of genomic aberrations has identified recurrent mutations across various cancer types as potential driver events, leading to a widespread effort in cataloguing actionable changes with potential applications to individualized therapeutics. The popularity of cancer genome sequencing has also raised the question of whether this technology can be exploited for a more thorough characterization of MSI. In a recent article (11), we addressed this issue by developing a method to identify tumor genome-specific DNA slippage events from whole-genome and exome sequencing data and applying it to the colorectal and endometrial cancer genomes from The Cancer Genome Atlas (TCGA) consortium (12, 13). Here, we will describe this method and our findings on the genomic distributions and characteristics of MSI events as well as their functional consequences. We will also address its potential utility as a cancer marker and discuss open questions related to MSI.
Molecular basis of MSI
Microsatellites are abundant, simple repetitive sequences distributed across the human genome. During DNA replication, these tandem repeats are prone to DNA polymerase slippage events, which lead to variations in the repeat length. Most of these errors are corrected by the MMR system, but loss-of-function mutations in MMR genes can render the repair machinery ineffective, leaving the DNA slippage events unfixed and resulting in an elevated mutation rate overall across the genome as shown in knock-out experiments (14). MMR genes whose mutations may lead to a mutator phenotype include MSH2 (MutS homolog 2), MSH3, MSH6, MLH1 (MutL homolog 1), MLH3, PMS1 (post-meiotic segregation-1), and PMS2. In Lynch syndrome, germline mutations of MLH1, MSH2, MSH6, or PMS2 increases the lifetime risk of a number of cancers, especially colorectal cancer, although their recessive nature requires a second hit or somatic inactivation of the wild-type allele to be functional inactivation (15).
Detection of MSI events using high-throughput sequencing
Traditional MSI detection methods are based on PCR analysis of specific loci. For each microsatellite marker, primers are designed so that PCR amplicons span the entire repeat and the length of the microsatellites can be measured by a fragment assay. The autoradiography using radiolabeled PCR primers by gel eletrophoresis was used until the late 90s, and then replaced by fluoresceinated primers with automatic sequencers (16). In distinguishing the microsatellite-unstable and -stable genomes, the frequently-used Bethesda panel consists of three dinucleotide (D2S123, D5S346 and D17S250) and two mononucleotide repeats (BAT-25 and BAT-26) (6, 7). Unlike common genetic tests utilizing markers that are highly polymorphic in the population, these MSI markers are monomorphic or quasimonomorphic in terms of the repeat length (e.g., BAT26 is composed of (A)26, which is observed in >99% of the European population). Based on the calls from these markers, cancer genomes are classified into microsatellite-unstable (MSI-high or “MSI-H”; two or more out of the five markers) or microsatellite-stable (“MSS”; no positive marker) cases, with MSI-low (“MSI-L”; positive for one marker) for intermediates. For panels with more than five markers, MSI-H genome is typically called when ≥ 30-40% of the markers are positive, while the remaining cases are classified into MSI-L (some but <30-40% of the markers) or MSS genomes (no positive markers).
To identify the MSI targets in a genome-wide manner, some studies used in silico screening of coding mononucleotide microsatellites (several hundred markers discovered per study) followed by extensive PCR validations to measure the frequency of instability of each marker (17, 18). Although some novel MSI targets in colorectal cancer genomes were identified, this approach cannot be widely adopted because it is laborious, time-consuming, and low-throughput. Thus, the current use of MSI markers is to distinguish the mutator phenotype of microsatellite-unstable cancers from the stable genomes, rather than to examine individual mutations on microsatellite loci.
To overcome these limitations, we recently developed a method to identify MSI events from whole-genome or exome sequencing data (Figure 1). First, we scanned the human reference sequence to obtain a reference set of ~8 million microsatellites (~140,000 for an exome-wide reference set). To identify microsatellites, we used a repeat-searching algorithm called Sputnik (source code at http://wheat.pw.usda.gov/ITMI/EST-SSR/LaRota/); the repeats identified by the algorithm is variable depending on the criteria employed (e.g., the number of base pairs or the unit length) as well as the degree of degeneracy allowed in defining a microsatellite. From whole-genome or exome sequencing data, we collected a set of sequencing reads that fully contain (i.e., spanning the entire length of the repeat with enough flanking sequences to map the reads) each of the microsatellites in the reference set. With 100-bp reads, we can capture >99% of the microsatellites. Due to both biological variations and minor errors in measurement, these reads result in a distribution of lengths for each microsatellite. With sequencing data from tumor and matched normal genomes, we can compare the distributions of observed lengths at each locus to call MSI events. We used the Kolmogorov-Smirnov test to compare the two distributions and adjusted for multiple testing to determine the significance threshold.
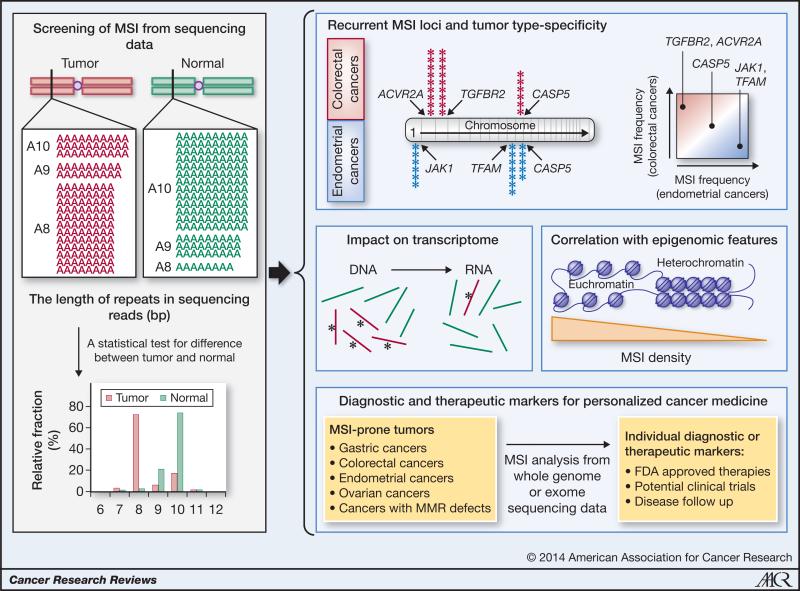
Figure 1. Comprehensive MSI analysis of cancer genomes.
An example of MSI in TGFBR2 (A10 mononucleotide repeat) is shown (Left). Estimates for the microsatellite length are obtained from the sequencing data of the tumor and matched normal genomes. When the difference between the length distributions is significant, the locus is said to have an MSI event. This MSI screening can be extended to all repeats covered by the sequencing data, with more than 8 million microsatellites for whole-genome sequencing data. The panels on the right illustrate biological and clinical applications. For example, recurrent MSI targets in different tumor types (e.g., colorectal and endometrial cancers) can be identified as cancer drivers (top). The mutant allele frequencies (red bars with asterisks and green bars representing mutant and wildtype alleles) can be compared between the genome and transcriptome (of the same individual); significant biases in frequencies may indicate cis-impact of the mutation on expression levels. The genome-wide MSI occupancy profiles can be examined for epigenetic configurations that favor MSI (middle). Finally, MSI screening of a patient's genome may reveal potentially druggable targets for individualized treatment.
To examine the performance of the method, we compared the sequencing-based MSI calls made on the TGFBR2 A10 microsatellite with those from capillary sequencing-based fragment length assay (12, 13). Across from 147 colorectal and 130 endometrial cancer genomes, the sequenced-based calls replicated the results from the fragment length assay with 91% sensitivity and 100% specificity (11). We note that a list of indels typically does not include microsatellites; a previous investigation of MSI in TCGA colorectal cancer genomes required a manual examination of the 30 selected microsatellite loci (13). Although conventional indel-calling algorithms have been adopted to call MSI events in some studies (19, 20), we suspect that such approaches may be less sensitive than a direct test we employed. A further investigation will be required to compare different statistical methods and to optimize the testing procedure, including the use of non-parametric tests.
Other technical issues involve uneven genome coverage and artifacts in sequencing technology. The sequencing coverage is variable across the genome due to the uneven target capture process in exome sequencing as well as GC bias in general. Determining the minimum sequencing coverage for making MSI calls and characterizing the loss of power to detect in low coverage regions will be helpful. With respect to sequencing technology, measuring the correct size of homopolymer runs is challenging for any platforms, with experimental errors (sometimes called “stutter”) increasing with the length of the homopolymer. The Illumina platform is reliable in this regard compared to other platforms (21), but its accuracy for longer homopolymers has not been carefully examined. Our work utilized the available sequencing data from matched normal to minimize the shortcomings of the sequencing technology and to account for possible polymorphism of microsatellite length across individuals. However, a technique for more accurate measurement of the absolute length of a homopolymer and a better characterization of population-wide length distribution at each microsatellite loci from a normal population may enable a single sample-based (without the matched normal) testing.
Enlarging the repertoire of somatic mutations in MS-unstable cancer genomes
An MSI event may occur in a coding sequence and produce a frameshift. Such a frameshift MSI (not in-frame) can potentially disrupt the structure and consequently the function of the encoded proteins, thus serving as a potent mechanism of inactivation for tumor suppressor genes. The discovery of MSI as a cancer driver was based on a frameshift MSI on the TGFBR2 A10 homopolymer in colorectal cancers (22), followed by identification of additional genes recurrently targeted by MSI in other cancers including gastric and endometrial cancers (23). The frequency of MSI on known loci such as TGBFR2 and ACVR2A in 30 microsatellite-unstable colorectal cancers from TCGA (11) was largely concordant with previous estimates in colorectal cancers (23).
Although distinct MSI profiles have been observed in different cancer types (23, 24), those studies were based on small number of microsatellite markers established mainly in colorectal cancers. Thus, they were limited in scope and few novel MSI targets were identified in other cancer types. In contrast, our exome-based examination of microsatellite-unstable endometrial cancers revealed many novel targets, such as JAK1 (Janus kinase 1) and TFAM1 (11) which were not found in colorectal cancers and thus not in the list of potential MSI targets previously studied (17, 18, 23). Although the concept of tumor type-specificity for MSI is not new, a comprehensive identification of differential MSI targets enabled a more detailed view of MSI targeting in a tumor type-specific and pathway-dependent manner. For example, the most frequent targets TGFBR2 and ACVR2A in colorectal cancers are associated with TGF-beta signaling pathway, whereas this pathway is not affected in endometrial cancers. It will be valuable to identify recurrent targets of MSI in other cancer types that have not been extensively studied for MSI occurrences, such as ovarian cancers, as those targets may function as drivers of tumorigenesis.
An early study proposed that a microsatellite-unstable colorectal cancer genome can harbor >100,000 MSI events (2). Our genome-wide survey of MSI using whole-genome sequencing revealed that as much as 300,000 loci can be affected (11). This large number of MSI events can facilitate genome-wide correlative analyses with various genetic features. For example, we observed that the local density of MSI is inversely correlated with that of point mutations, with overrepresentation in euchromatic regions (11). The relative depletion of MSI at stable nucleosome positions also supports the notation that chromatin configuration is a major determinant of genomic distribution of MSI events (11). Our findings also clearly suggest that factors involved in occurrence of MSI in the contexts of an MMR deficiency are different from those of point mutations in cancer genomes.
Combined analyses with transcriptome sequencing data revealed that the alleles harboring coding MSI often showed repressed transcript levels (11). This finding is consistent with earlier observations in cell lines that harbor MSI on TGFBR2 (22) and in recent MSI studies in gastric cancer cell lines (20). Altered transcript stability due to nonsense-mediated decay is a potential mechanism for this phenomenon (25), but other mechanisms including altered promoter usage and splicing may be involved. The observation that the MSI events in 3’ untranslated regions are often associated with the increased transcript levels may be explained by the disruption of microRNA binding sites by MSI, but it also should be further examined along with the potential impact of MSI located on other regulatory sequences such as promoters.
Beyond the Bethesda panel
A genome-wide approach delivers a more complete description of MSI. One issue that arises with using the Bethesda panel is whether a sufficient number of markers are present for robust discrimination of the cases. It is easy to infer from the extensive tumor type-specificity of MSI targets described above that using the same small subset of markers would be inadequate for tumor types other than colorectal cancers. With an increasingly large amount of exome and whole-genome data available from TCGA and the International Cancer Genome Consortium (ICGC) as well as other published genomic profiling studies, it will be possible to develop a specific set of markers for each tumor type in the near future.
Another example is on the interpretation of MSI-L genomes. As an intermediate between the MSI-H and MSS cases (e.g., positive for one out of five Bethesda markers; varying criteria are applied when more markers are used) (26), the MSI-L tumors were thought to comprise 3-10% of colorectal cancers. But whether these genomes represent a unique disease entity that can be clearly separated from MSI-H and MSS genomes with distinct clinical or genetic features has been debated (26). Our exome-wide analyses revealed that MSI-L and MSS colorectal cancers (n = 23 and 97) do not show significant difference in the number of MSI events (5 and 4 for the median count of MSI events, respectively, in MSI-L and MSS, compared to 290 in MSI-H genomes) and the majority of the cases (95.6% and 87.6% of MSI-L and MSS genomes) showed at least one MSI call in the exome-wide screening (11). These findings support a view that MSI-L arises largely due to the basal instability level of cancer genomes (27) rather than representing a distinct disease category. Although some previous studies proposed the use of gene expression signatures or specific markers (e.g., KRAS mutations) in evaluating MSI-L cases, we observed that MSI-L and MSS genomes did not show a significant difference in the frequency of KRAS mutations (31.8% and 37.8% for MSI-L and MSS genomes, respectively) or in the level of MSH3 expression (11).
The unique clinicopathological features of MSI-H colorectal genomes—such as a favorable prognosis, overrepresentation in old females (sporadic cases) and proximal location in the colon and a relative lack of copy number changes—have served as a rationale for MSI testing in clinical settings. Besides discrimination of cancer genomes into microsatellite unstable or stable ones, our study has revealed a wide range in the number of MSI events (e.g., 79 - 647, 0 - 49 and 0 - 50 events identified, respectively, in 27 MSI-H, 23 MSI-L and 97 MSS colorectal cancers using exome data). This quantitative view will enable more sensitive correlative analyses for previously unrecognized associations between the clinical covariates and the extent of MSI. Also, the use of an extended set of microsatellite markers may facilitate further investigation into some unresolved issues related to the MSI-L category, e.g., whether approximately 10 - 25% of MSI-H genomes are misannotated as MSI-L (28) or some MSI-L cases may represent the pre-MSI-H phase during the cancer progression (26).
The mutational landscape associated with loss of MMR may be different depending on the type of affected MMR genes. But whether this genetic information can be used to distinguish Lynch syndrome cases from sporadic microsatellite-unstable colorectal cancers is not clear. Although the underlying genetic constitutions and initial genetic events may be different, the resulting genomic profiles might be similar in the two groups, especially as the elevated mutation rate may help incur multiple passenger mutations in other MMR genes and makes it difficult to infer the initial cancer drivers (11).
As mentioned above, it is possible that the MMR mutations are merely consequences of elevated mutations rates and whether those mutations are functionally relevant needs to be examined more carefully. For colorectal cancers, germline mutations in POLD1 and POLE were identified in individuals with familial colorectal cancers as predisposing genetic variants (29). The proteins encoded by these genes are DNA polymerases with catalytic and proofreading activities and a notable association of POLE mutation and hypermutated phenotype has been shown in TCGA cases (13). Given the availability of sequencing data in a wide range of tumor types, a study of MSI in cancer types beyond those commonly recognized as MSI-associated may reveal its infrequent but nonetheless significant presence.
MSI as a marker of tumor evolution
A mutational landscape can provide insights into the evolution of a cancer genome. The presence of point mutations and their allele frequencies are frequently used as evolutionary markers, but repeat length polymorphisms in MSI can potentially serve as quantitative evolutionary markers as well. In a model that utilizes the accumulation of repeat length variation (30), the genotype (i.e., the fixed length of microsatellite repeats) of the last common ancestor responsible for the terminal clonal amplification in the cancer evolution can be inferred from the distribution of repeat lengths. The differences between this and the germline genotypes are then summarized into evolutionary ages or time interval between the initiating events (loss of MMR) in the founder cell and the emergence of the last common ancestor. The variability of repeat length accumulated in the last common ancestor can also be measured to derive the time interval required for the terminal clonal amplification. (30). The repeat length distribution obtained from high-throughput sequencing data can be easily incorporated into this model to examine the evolutionary history of these microsatellite-unstable genomes.
Future perspectives
The advent of high-throughput sequencing has allowed for a substantial progress in the identification of somatic alterations in cancer genomes. Although much of the attention so far has gone to the more common aberrations (e.g., point mutations, copy number changes, and translocations), sequencing-based analysis of MSI has yielded many new insights on its pattern of occurrence, variation across patients, and functional impact (11). MSI analysis also holds promise as a tool for identifying novel cancer drivers and as key factors in clinical correlative analyses. Using the framework developed in our analysis of colorectal and endometrial cancers, gastric and ovarian genomes as well as others currently not associated with MSI should be examined, especially if the genomes are hypermutated. Our analysis utilized the matched normal genomes as a control in tumor genome analysis; but with improved algorithms and sequencing platforms for detection of homopolymer in the future, it should be possible to carry out a similar analysis using only the tumor genomes. With the ever-increasing popularity of exome and whole-genome sequencing, we will be able to learn a great deal more on the nature of MSI and its clinical implications in the near future.
Acknowledgments
Grant Support
This study was supported by the National Research Foundation of Korea (NRF) (2013R1A1A2060959) and the US National Institutes of Health (U24CA144025).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkanen L, Mecklin JP, et al. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–16. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 2.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–61. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 3.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–9. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 4.Hemminki A, Peltomaki P, Mecklin JP, Jarvinen H, Salovaara R, Nystrom-Lahti M, et al. Loss of the wild type MLH1 gene is a feature of hereditary nonpolyposis colorectal cancer. Nat Genet. 1994;8:405–10. doi: 10.1038/ng1294-405. [DOI] [PubMed] [Google Scholar]
- 5.Parsons R, Li GM, Longley MJ, Fang WH, Papadopoulos N, Jen J, et al. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993;75:1227–36. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 6.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–57. [PubMed] [Google Scholar]
- 7.Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–8. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thibodeau SN, French AJ, Roche PC, Cunningham JM, Tester DJ, Lindor NM, et al. Altered expression of hMSH2 and hMLH1 in tumors with microsatellite instability and genetic alterations in mismatch repair genes. Cancer Res. 1996;56:4836–40. [PubMed] [Google Scholar]
- 9.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–57. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453–6. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 11.Kim TM, Laird PW, Park PJ. The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell. 2013;155:858–68. doi: 10.1016/j.cell.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegan DC, Narayanan L, Jirik FR, Edelmann W, Liskay RM, Glazer PM. Differing patterns of genetic instability in mice deficient in the mismatch repair genes Pms2, Mlh1, Msh2, Msh3 and Msh6. Carcinogenesis. 2006;27:2402–8. doi: 10.1093/carcin/bgl079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peltomaki P. Lynch syndrome genes. Fam Cancer. 2005;4:227–32. doi: 10.1007/s10689-004-7993-0. [DOI] [PubMed] [Google Scholar]
- 16.Cawkwell L, Li D, Lewis FA, Martin I, Dixon MF, Quirke P. Microsatellite instability in colorectal cancer: improved assessment using fluorescent polymerase chain reaction. Gastroenterology. 1995;109:465–71. doi: 10.1016/0016-5085(95)90334-8. [DOI] [PubMed] [Google Scholar]
- 17.Alhopuro P, Sammalkorpi H, Niittymaki I, Bistrom M, Raitila A, Saharinen J, et al. Candidate driver genes in microsatellite-unstable colorectal cancer. Int J Cancer. 2012;130:1558–66. doi: 10.1002/ijc.26167. [DOI] [PubMed] [Google Scholar]
- 18.Mori Y, Yin J, Rashid A, Leggett BA, Young J, Simms L, et al. Instabilotyping: comprehensive identification of frameshift mutations caused by coding region microsatellite instability. Cancer Res. 2001;61:6046–9. [PubMed] [Google Scholar]
- 19.Wang L, Tsutsumi S, Kawaguchi T, Nagasaki K, Tatsuno K, Yamamoto S, et al. Whole-exome sequencing of human pancreatic cancers and characterization of genomic instability caused by MLH1 haploinsufficiency and complete deficiency. Genome Res. 2012;22:208–19. doi: 10.1101/gr.123109.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoon K, Lee S, Han TS, Moon SY, Yun SM, Kong SH, et al. Comprehensive genome- and transcriptome-wide analyses of mutations associated with microsatellite instability in Korean gastric cancers. Genome Res. 2013;23:1109–17. doi: 10.1101/gr.145706.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–8. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 23.Duval A, Hamelin R. Mutations at coding repeat sequences in mismatch repair-deficient human cancers: toward a new concept of target genes for instability. Cancer Res. 2002;62:2447–54. [PubMed] [Google Scholar]
- 24.Kuismanen SA, Moisio AL, Schweizer P, Truninger K, Salovaara R, Arola J, et al. Endometrial and colorectal tumors from patients with hereditary nonpolyposis colon cancer display different patterns of microsatellite instability. Am J Pathol. 2002;160:1953–8. doi: 10.1016/S0002-9440(10)61144-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 26.Tomlinson I, Halford S, Aaltonen LA, Hawkins N, Ward R. Does MSI-low exist? J Pathol. 2002;197:6–13. doi: 10.1002/path.1071. [DOI] [PubMed] [Google Scholar]
- 27.Laiho P, Launonen V, Lahermo P, Esteller M, Guo M, Herman JG, et al. Low-level microsatellite instability in most colorectal carcinomas. Cancer Res. 2002;62:1166–70. [PubMed] [Google Scholar]
- 28.Halford S, Sasieni P, Rowan A, Wasan H, Bodmer W, Talbot I, et al. Low-level microsatellite instability occurs in most colorectal cancers and is a nonrandomly distributed quantitative trait. Cancer Res. 2002;62:53–7. [PubMed] [Google Scholar]
- 29.Palles C, Cazier JB, Howarth KM, Domingo E, Jones AM, Broderick P, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45:136–44. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsao JL, Yatabe Y, Salovaara R, Jarvinen HJ, Mecklin JP, Aaltonen LA, et al. Genetic reconstruction of individual colorectal tumor histories. Proc Natl Acad Sci USA. 2000;97:1236–41. doi: 10.1073/pnas.97.3.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]