Abstract
Acquired resistance to transforming growth factor-β (TGF-β) is a key step in the early stages of tumorigenesis. Mutations in TGF-β signaling components are rare, and little is known about development of resistance in breast cancer. On the other hand, an activated Notch pathway is known to play a substantial role in promoting breast cancer development. Here, we present evidence of crosstalk between these two pathways through HEYL. HEYL, a basic helix-loop-helix (bHLH) transcription factor and a direct target of Notch signaling, is specifically overexpressed in breast cancer. HEYL represses TGF-β activity by binding to TGF-β-activated Smads. HeyL−/− mice have defective mammary gland development with fewer terminal end buds. On the other hand, HeyL transgenic mice show accelerated mammary gland epithelial proliferation and 24% of multiparous mice develop mammary gland cancer. Therefore, repression of TGF-β signaling by Notch acting through HEYL may promote initiation of breast cancer.
INTRODUCTION
Transforming growth factor-β (TGF-β) is a multifunctional cytokine that exerts pleiotropic effects on virtually all known cell types through a seemingly simple signal transduction pathway. TGF-β binds to its transmembrane receptors, which leads to phosphorylation of two Receptor-regulated Smads (R-Smads), Smad2 and Smad3. The activated R-Smads then bind to Smad4, forming a Smad complex that associates with other transcriptional cofactors to regulate gene expression (1, 2). TGF-β is able to suppress tumor initiation at early stages of cancer development. However, cancer cells usually develop various ways to evade the growth-inhibitory effect of TGF-β. Indeed, loss of TGF-β sensitivity is a hallmark of tumor initiation (3). Although mutations of TGF-β signaling components are common in gastrointestinal cancers (4), such mutations are rarely found in breast cancer. How breast cancer cells acquire resistance to TGF-β-mediated growth inhibition is largely unknown.
In contrast to the TGF-β pathway, activation of the Notch pathway has been frequently shown to promote breast cancer development (5). Upon binding of Notch ligand to its receptor, the intracellular domain of the Notch receptor is released from the cell membrane through the action of a γ-secretase complex and translocates to the nucleus, where it forms a complex with RBP-J to induce the expression of genes that promote cell growth and inhibit cellular differentiation (6, 7). Given the fact that these two pathways have opposing effects on cell growth and mammary gland tumor development, it seems plausible that activation of the Notch pathway in breast cancer can counteract the inhibitory effects of TGF-β signaling. The Notch pathway has been reported to either synergize or antagonize TGF-β signaling depending on the cellular context, but the detailed mechanism of their crosstalk is not well established (8, 9). Moreover, the significance of their crosstalk in breast cancer is not known.
HEYL, HEY1 and HEY2 are the members of the HEY (hairy/enhancer-of-split related with YRPW motif) family (10). All three of these genes and the related HES (hairy and enhancer of split) family members are basic helix-loop-helix (bHLH) transcription factors and have been shown to be the direct targets of the Notch pathway (11–14). Hey2-null mice die in the early postnatal period and have a variety of cardiovascular defects including atrioventricular valve and ventricular septal defect, tetralogy of Fallot and congestive heart failure (15–17). Hey1- or HeyL-null mice do not show detectable developmental abnormality, while combined loss of Hey1 and HeyL causes similar cardiovascular defects (18). To date, the function of HEYL in breast cancer has not been studied. Here, we show that HEYL can inhibit TGF-β signaling by binding to Smad proteins, thereby rendering the cells resistant to the effect of TGF-β.
MATERIALS AND METHODS
Cell culture and reagents
HaCaT, HS578T, Cos1, MDA-MB-231 and HepG2 were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, Penicillin and Streptomycin. SUM159 cells were grown in Ham's F-12 with 5% Fetal Bovine Serum, 0.5 µg/ml hydrocortisone, 10 µg/ml insulin, Penicillin and Streptomycin. MCF-10A cells were cultured in Dulbecco's modified Eagle's medium/nutrient mixture F-12 supplemented with 5% horse serum, 20 ng/ml human recombinant epidermal growth factor, 0.5 µg/ml hydrocortisone, 100 ng/ml cholera toxin, 10 µg/ml insulin, Penicillin and Streptomycin.
Antibodies used were anti-Myc-tag (9B11) mouse Ab (Cell Signaling), anti-Smad3 rabbit Ab (Cell Signaling), anti-Flag M2 mouse Ab (Sigma), anti-Ki67 Ab (Thermo Scientific), anti-HEYL mouse Ab (Abnova) and anti-HEYL rabbit Ab.
Details of the construction of HEYL expression vectors and infection of cells with viral supernatants are provided in Supplementary Methods.
Generation of anti-HEYL rabbit antibody and immunohistochemical (IHC) staining
A synthetic peptide corresponding to a peptide sequence (EPSGSDGESDGPID) in the N-terminus of HEYL was used to generate polyclonal antibodies in rabbits. Details of the preparation and tests to validate its specificity are provided in Supplementary Methods.
Generation and characterization of HeyL knockout and transgenic mice
HeyL knockout mice were generated as previously described (18, 28). Exons 2–4 of HeyL were deleted. The mammary glands of 13 week old HeyL knockout mice and wild type littermates were excised and processed for whole mount staining, IHC staining and frozen chunks were embedded in OCT.
To generate HeyL transgenic mice, the full-length murine HeyL cDNA was inserted into the EcoRI site of MKbpA vector (provided by Dr. Jeffrey Rosen at Baylor College of Medicine). The resulting vector was cut using BssHII restriction endonuclease, and the microinjection of the fragment containing the transgene into single-cell embryos isolated from FVB/N mice was performed by the Transgenic Mouse Core Facility at National Cancer Institute. The establishment of founder mice was confirmed by Southern blotting. The FVB/N founder mice were then crossed with FVB/N wild type mice for 4–5 generations. The genotype of HeyL transgenic mice and the wild type littermates was verified by PCR using genomic DNA isolated from mouse tails. The HeyL transgenic mice and the wild type littermates were euthanized at 13 week old, 10 days of pregnancy, 10 days of lactation and 30 days after involution. The Inguinal mammary fad pads were excised and processed for whole mount staining, IHC staining and frozen chunks embedded in OCT.
Whole mount staining, RT-PCR and immunostaining of mouse mammary glands
For whole mount staining, the mammary glands were placed between two glass slides and fixed with 10% Formalin overnight. Details are provided in Supplementary Methods.
Treatment of HeyL knockout mice with TGF- β type I receptor inhibitor, SB 535334
SB 535334 powder (Selleck) was dissolved in 100% ethanol to 18mg/ml. The solution was further diluted 1:10 in 100mg/ml cyclodextrin. 100ul final solution (10mg/kg) was intraperitoneally injected into 7–8 weeks old HeyL knockout mice daily. The treatment lasted for 4–5 weeks. The mammary glands were then removed for analysis.
Immunoprecipitation and Western blotting
Transfected Cos1 cells in T25 flasks were lysed in 250ul 1× lysis buffer (1% Triton-X, 150 nM NaCl, 10mM Tris, 1mM EDTA and EGTA and 0.5% NP-40) with protease inhibitor on ice for 30 minutes. Details are provided in Supplementary Methods.
Quantitative RT-PCR
Details of oligonucleotide primers and methods are provided in Supplementary Methods.
Luciferase assay
HepG2 or HS578T cells on 12 or 24 well plates were transfected with an equal amount of corresponding vectors using Lipofectamine 2000 (Invitrogen). 4ng/ml TGF-β (Peprotech) was added into cell media and incubated for 16 hours. The luciferase reading was measured 48 hours after transfection and normalized to β-galactosidase activities. The luciferase assays were repeated three times.
BrdU Cell Proliferation Assay
5,000 MCF-10A control or HEYL expressing cells were seeded on a 96-well plate and treated the next day with TGF-β for 12 hours. Bromodeoxyuridine (BrdU) was added into cell media and incubated for 4 hours. Additional details are provided in Supplementary Methods.
Bimolecular fluorescence complementation assay
The fusion protein expression vectors were transfected into Cos1 cells on 24 well plates. 36 hours later, the cells were fixed with formalin, and the nuclei were stained using DAPI. The fluorescence was then observed under a microscope.
In vitro binding assay
35S labeled full-length, Basic domain deletion and Helix-Loop-Helix domain deletion HEYL proteins were synthesized in vitro using Coupled Transcription/Translation System (Promega) and incubated with equal amount of GST or GST-Smad3 fusion protein bound to glutathione-Sepharose beads. The associated proteins were revealed by SDS-PAGE and autoradiography.
RESULTS
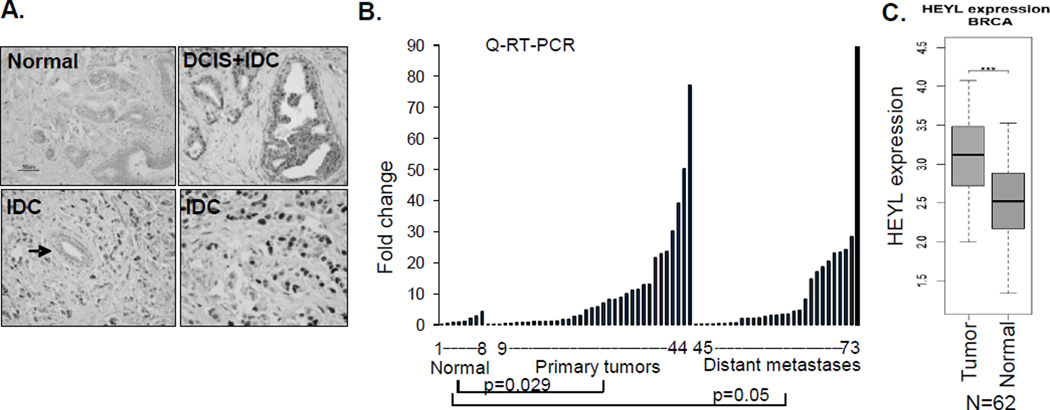
HEYL was first discovered to be highly expressed in colon and breast cancer endothelial cells. We and another group isolated endothelial cells from normal and tumor samples using beads conjugated with endothelial-specific antibody. SAGE (Serial Analysis of Gene Expression) analysis of the purified endothelial cells showed that HEYL expression was 3- and 20-fold higher in colon and breast cancer endothelial cells, respectively, compared to their normal counterparts (19, 20). To determine HEYL protein expression in cancer, we raised a polyclonal antibody targeting the peptide sequence (EPSGSDGESDGPID) at the N- terminus of HEYL that is specific to HEYL, and performed immunohistochemical (IHC) analysis on breast tumors. This antibody specifically recognized HEYL, since the staining in breast cancer tissue was blocked by the peptide that was used as an antigen for generating the HEYL antibody (Supp Figure 1A). In addition, only HS578T breast cancer cells stably expressing HEYL, but not vector controls, were positive by immunofluorescent (IF) staining (Supp. Figure 1 B). We also confirmed that in IF analysis this antibody showed positive staining with MDA-MB-435-HEYL cells, but did not crossreact with HEY1- and HES1-transfected cells (Supp. Figure 1C). IHC analysis of breast cancer tissue arrays indicated that, in addition to the positive staining in tumor endothelial cells, HEYL is also abundantly expressed in tumor epithelial cells. Compared to normal breast tissues that showed negative staining (0/13), significantly strong nuclear immunostaining was detected in 45% (50/111; p=0.002) of invasive ductal carcinomas and in 46% (6/13; p=0.015) of invasive lobular carcinomas (representative photomicrographs shown in Figure 1A). Quantitative RT-PCR showed high HEYL mRNA expression (cutoff value: 5.0 units) in 50% (18/36) of primary and in 34.5% (10/29) metastatic breast cancers but not in normal breast tissue organoids (0/8) (Figure 1B). We also compared HEYL expression in 62 matched normal and primary breast tumors (abbreviated as BRCA) in the TCGA (The Cancer Genome Atlas) database, and found that HEYL expression is significantly higher in tumors (p<0.0001) compared to normal breast tissue (Figure 1B).
Figure 1. HEYL is overexpressed in breast cancer and antagonizes TGF-β-mediated signaling.
A. HEYL IHC staining of representative sections of normal duct, ductal carcinoma in situ (DCIS), and invasive ductal carcinoma (IDC) are shown. Note the absence of staining in the normal duct located in the tumor (arrow). Difference in HEYL IHC staining was compared between the groups using two-sided Fisher’s exact T test.
B. HEYL mRNA expression in normal breast organoids (enriched epithelium following enzymatic digestion of reduction mammoplasty tissue) and malignant samples was shown. Expression difference was compared between the groups using one-sided Wilcoxon rank sum test. Differential expression of HEYL in 62 matched normal and breast cancers (BRCA) in TCGA database using two-tailed Student t test.
HEYL is a known direct target of the Notch pathway as shown by studies in cell culture and mouse models (12, 13, 21). Taking into account the cell type-specific action of the Notch pathway, we tested whether Notch can activate HEYL expression in breast epithelial cells. Expression of the Notch1 intracellular domain (N1IC), which mimics Notch pathway activation upon ligand binding, in the breast cancer cell line HS578T, resulted in increasing levels of HEYL expression (Supp. Figure 2A). Co-transfection of the N1IC vector and a HEYL promoter-driven luciferase vector resulted in a significant increase in luciferase activity (Supp. Figure 2B). Knockdown of the expression of RBP-J, a critical mediator of the canonical Notch pathway, in MDA-MB-231 cells reduced HEYL expression (Supp. Figure 2C). Consistent with our findings, it was reported that soluble Notch receptor interfering with the Notch ligand-receptor binding can repress HEYL expression in MDA-MB-231 cells, and HEYL was the only Notch target gene that was associated with the expression of the Notch ligand Jagged1 in breast cancer clinical samples (22). These data suggest that HEYL may potentially mediate part of Notch oncogenic activities in breast cancer.
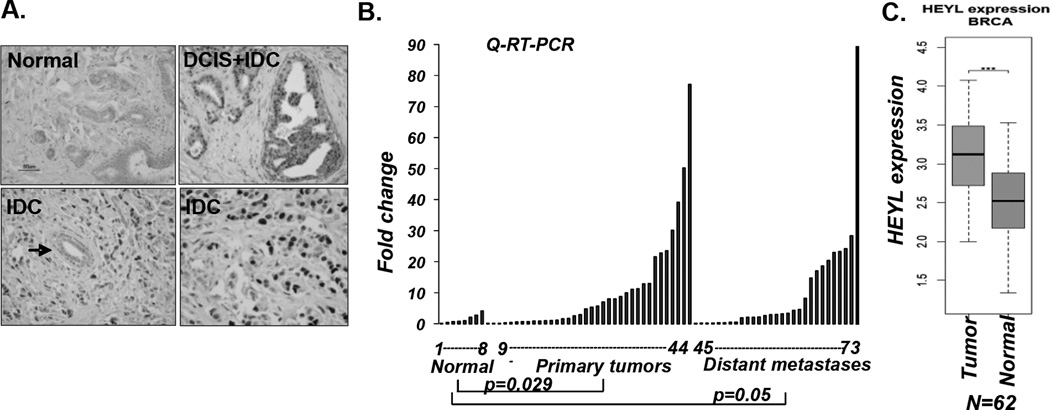
Since very few experimental investigations have been performed on the functions of HEYL, we performed bioinformatics analysis of multiple public databases to retrieve biochemical and genetic data that might provide heretofore unknown links of HEYL to key signaling pathways. A genome-wide mass spectrometry-based immune-precipitation proteomics study indicated that HEYL can bind to Smad3, a finding corroborated by a large-scale yeast-2-hybrid study (23, 24). Taking into account the contradictory growth promoting effects of Notch and growth inhibitory effects of TGF-β signaling in early breast cancer development, we examined the possibility that HEYL could inhibit the TGF-β pathway. Using a TGF-β responsive reporter vector, p3TP-Luc, as seen before, TGF-β treatment or Smad3 overexpression significantly increased the luciferase activity; however this transactivation was strongly inhibited by HEYL (Figure 2A). We observed a similar transcription repression when a second TGF-β responsive reporter vector containing the P15 (CDKN2B) gene promoter was used in the same assay (Figure 2B). Consistent with findings in the luciferase assays, TGF-β treatment of immortalized human keratinocyte cells, HaCaT, resulted in dynamic upregulation of endogenous mRNA levels of PAI-1 and P15 at different time points; similarly, this transcriptional induction was repressed in the presence of HEYL (Figure 2C). We performed the same experiment on MCF10A breast cell line and measured PAI-1 and Smad7 expression, (but not of P15, since both alleles of this gene are deleted in MCF10A cells). Again, HEYL repressed the expression of PAI-1 and Smad7 induced by TGF-β (Figure 2C). As shown in a Bromodeoxyuridine (BrdU) incorporation assay, MCF-10A human breast epithelial cells expressing HEYL proliferated faster compared to control cells upon TGF-β treatment (Figure 2D).
Figure 2. HEYL antagonizes TGF-β-mediated signaling.
A. Left panel: 0.25ug p3TP-Luc vector alone or with 0.5ug HEYL expression vector was transfected into HepG2 cells. On the next day 4ng/ml TGF-β was added for 16 hours. Right panel: Varying amounts of HEYL (0.1, 0.25, 0.5ug) were cotransfected with 0.25ug 3TP-Luc and 0.5ug Smad3 expression vectors. Luciferase activity was measured 48 hours after transfection. Error bar represents s.d. (n=3). Difference was compared between the groups using two-tailed unpaired t test; *p <0.05, ** p<0.01
B. p15-promoter luciferase vector, p15-Luc, was used in the same luciferase assay as in figure 2A.
C. HaCaT or MCF10A vector control and stably expressing -HEYL cells were treated with 4ng/ml TGF-β for 0, 4, 8 and 18 hours. mRNA levels of PAI and p15 were measured in HaCaT, and PAI and Smad7 were measured in MCF-10A by Q-RT-PCR. Error bar represents s.e.m.
D. MCF-10A control and stably expressing MCF10A-HEYL cells were treated with 0.5ng/ml TGF-β for 12 hours, and then incubated with BrdU for 4 hours. The growth of the cells was quantified by measuring the relative BrdU incorporation. Error bar represents s.e.m.
E. Measure of HEYL expression by gel-based (inset) and Q-RT-PCR in MDA-MB-231 cells expressing scramble shRNA or 2 shRNAs targeting HEYL.
F. MDA-MB-231-HEYL shRNA and vector control cells grown in 1%FBS were treated with 4 ng/ml TGF-β for 0, 2, 4 and 8 hours. mRNA levels of PAI and Smad7 were measured by Q-RT-PCR. Error bar represents s.e.m.
Next, we studied TGF-β responses in breast cancer cells that were depleted of HEYL. HEYL expression was significantly reduced in MDA-MB-231 breast cancer cells infected with retroviral vectors and stably expressing two different HEYL shRNAs (Figure 2E). MDA-MB-231 breast cancer cells have an intact TGF-β signaling pathway, but growth of these cells, for unknown reasons, is not inhibited upon TGF-β treatment. Since P15 is not expressed in MDA-MB-231 cells, we examined alterations of endogenous mRNA expression of PAI-1 and Smad7 upon TGF-β treatment. As expected, TGF-β treatment activated expression of PAI-1 and Smad7 in MDA-MB-231 cells, and their expression levels were higher in the HEYL-knockdown cells (Figure 2F). Collectively, these results support the notion that HEYL expression renders cells less sensitive to TGF-β.
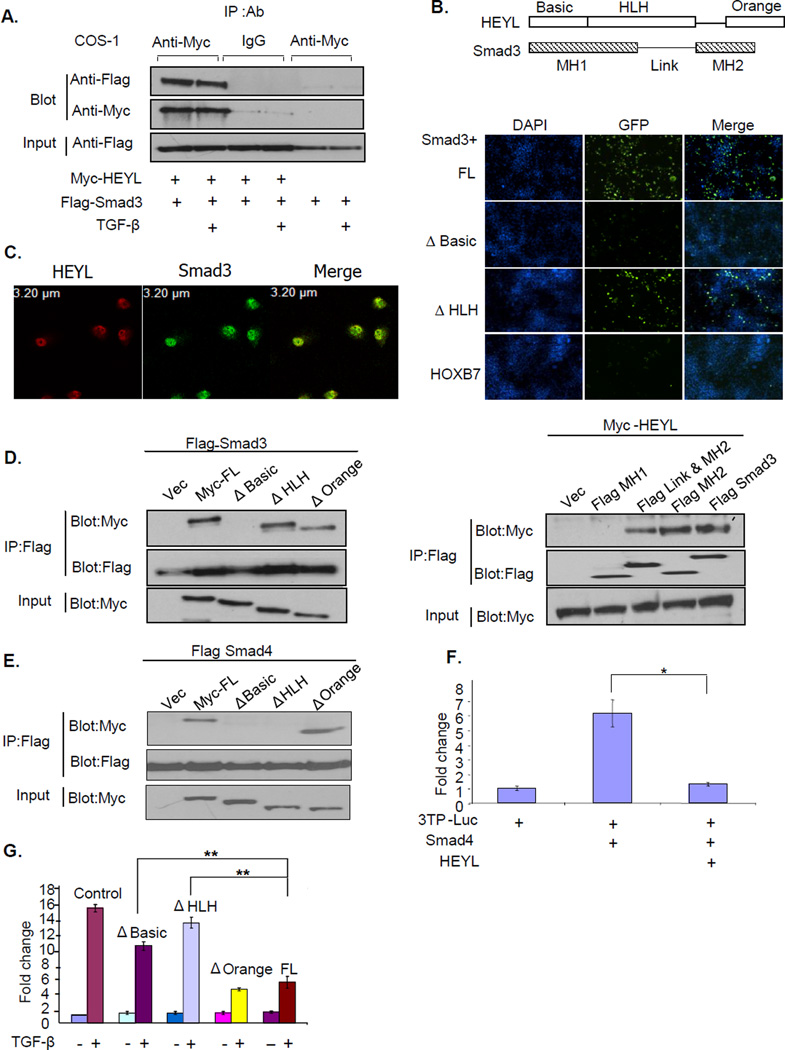
Direct interaction with Smad3 is one mechanism through which HEYL might interfere with TGF-β signaling as shown by bioinformatics analysis of databases. Using co-immunoprecipitation assays in Cos 1, we detected a strong protein-protein interaction between HEYL and Smad3; addition of TGF-β did not increase the strength of this interaction (Figure 3A). To confirm the interaction between HEYL and Smad3 in vivo, we performed the bimolecular fluorescence complementation assay in Cos 1 cells. Two fusion proteins, one containing the N-terminal half of GFP fused with HEYL and the other containing the C-terminal half of GFP fused with Smad3, were co-expressed in Cos1 cells. The emission of green fluorescence in the nuclei of these cells indicated that the interaction between HEYL and Smad3 had pulled the two halves of GFP into close proximity. On the other hand, a HOXB7 fusion protein, used as a negative control, showed very faint fluorescence when co-expressed with the C-terminal half of GFP-Smad3 fusion protein. Deleting the Basic domain of HEYL, but not the Helix-Loop-Helix domain, lowered the fluorescence intensity to the levels of the control, suggesting that the Basic domain of HEYL may mediate its interaction with Smad3 (Figure 3B and schematic of HEYL domains). We also detected the interaction of endogenous Smad3 and HEYL in MDA-MB-231 cells by co-immunofluorescent staining and confocal microscopy imaging (Figure 3C). Moreover, GST pull-down assays indicated that their interaction was direct and that deletion of the Basic domain significantly decreased the interaction (Supp. Figure 3).
Figure 3. HEYL interacts directly with Smad3.
A. Myc-tagged-HEYL alone, or along with Flag-tagged-Smad3 was transfected into Cos1 cells, and cells were treated with or without 4ng/ml TGF-β for 3 hours. The cell lysates were immunoprecipitated with the either anti-Myc antibody or control IgG, and blotted with the anti-Flag or anti-Myc antibody.
B. Bimolecular complementation assay in Cos cells showing green fluorescence at 36 hours after transfection. The N-terminal half of GFP fused with full-length HEYL (FL), Basic domain deletion of HEYL (Δ Basic), Helix-Loop-Helix domain deletion of HEYL (Δ HLH) or HoxB7 was co-expressed with the C-terminal of GFP fused with Smad3 in Cos1 cells. Cell nuclei were stained with DAPI.
C. Confocal microscopy imaging showing the co-localization of HEYL (red) and Smad3 (green) in MDA-MB-231 cells.
D. Left panel; Flag-tagged Smad3 was coexpressed with control or various Myc-tagged constructs of HEYL in Cos1 cells. Right panel; Myc-tagged full-length HEYL was coexpressed with control or various Flag-tagged constructs of Smad3 in Cos1 cells. The cell lysates were immunoprecipitated with anti-Flag antibody, and blotted with the anti-Myc or anti-Flag antibody.
E. Flag-tagged Smad4 was coexpressed with control or various Myc-tagged constructs of HEYL in Cos1 cells. The immunoprecipitation and blotting were performed as in Figure 3D.
F. Smad4 expression vector was used in a luciferase assay as described in Figure 2A.
G. HepG2 cells were transfected with 0.25ug 3TP-Luc construct and 0.5ug various constructs of HEYL. The cells were treated with 4ng/ml TGF-β for 16 hours. Luciferase activity was measured described in Figure 2A.
To map the domains of HEYL and Smad3 that are involved in the interaction, immunoprecipitation assays were performed in Cos1 cells using a series of constructs with various domain deletions of HEYL and Smad3 (domains of HEYL and Smad3 shown in Figure 3B). We found that the deletion of the Basic domain of HEYL abolished its interaction with Smad3, while the MH2 domain of Smad3 alone can mediated the interaction. Therefore, the Basic domain of HEYL can interact with the MH2 domain of Smad3 (Figure 3D). The MH2 domain is highly conserved in the Smad protein family. This raises the possibility that HEYL can interact with other Smads as well. Immunoprecipitation assays showed that HEYL, in fact, did interact with Smad4. While HEYL interacted with Smad3 through its Basic domain only, HEYL interaction with Smad4 required both the Basic and Helix-Loop-Helix domain (Figure 3E). In addition, HEYL was able to repress the transactivation of p3TP-Luc induced by Smad4, suggesting that the binding of HEYL to Smad4 also resulted in the inhibition of Smad4 activity (Figure 3F).
To test whether the interaction between HEYL and Smad3 was necessary and sufficient for the inhibition of TGF-β signaling, we used (SBE) 4-Luc, another TGF-β responsive reporter vector consisting of tandem synthetic Smad-binding elements transfected into HepG2 cells. Unlike other TGF-β responsive luciferase vectors, (SBE)4-Luc can be activated by Smad3/4 complex in the absence of additional transcriptional cofactors (25). TGF-β treatment or Smad3 overexpression also transactivates its luciferase activity. However, HEYL repressed the transactivation of (SBE)4-Luc induced by either TGF-β or Smad3 (Supp Figure 4). Therefore, the direct interaction of HEYL with Smad3 alone appears to be sufficient to inhibit TGF-β signaling. In line with our finding that the Basic and Helix-Loop-Helix domain of HEYL mediated interaction with either Smad3 or Smad4, the repressive effect of HEYL on the TGF-β pathway was abrogated when either domain was deleted, strongly suggesting that the binding of HEYL to Smad proteins was necessary for HEYL to inhibit the TGF-β pathway (Figure 3G).
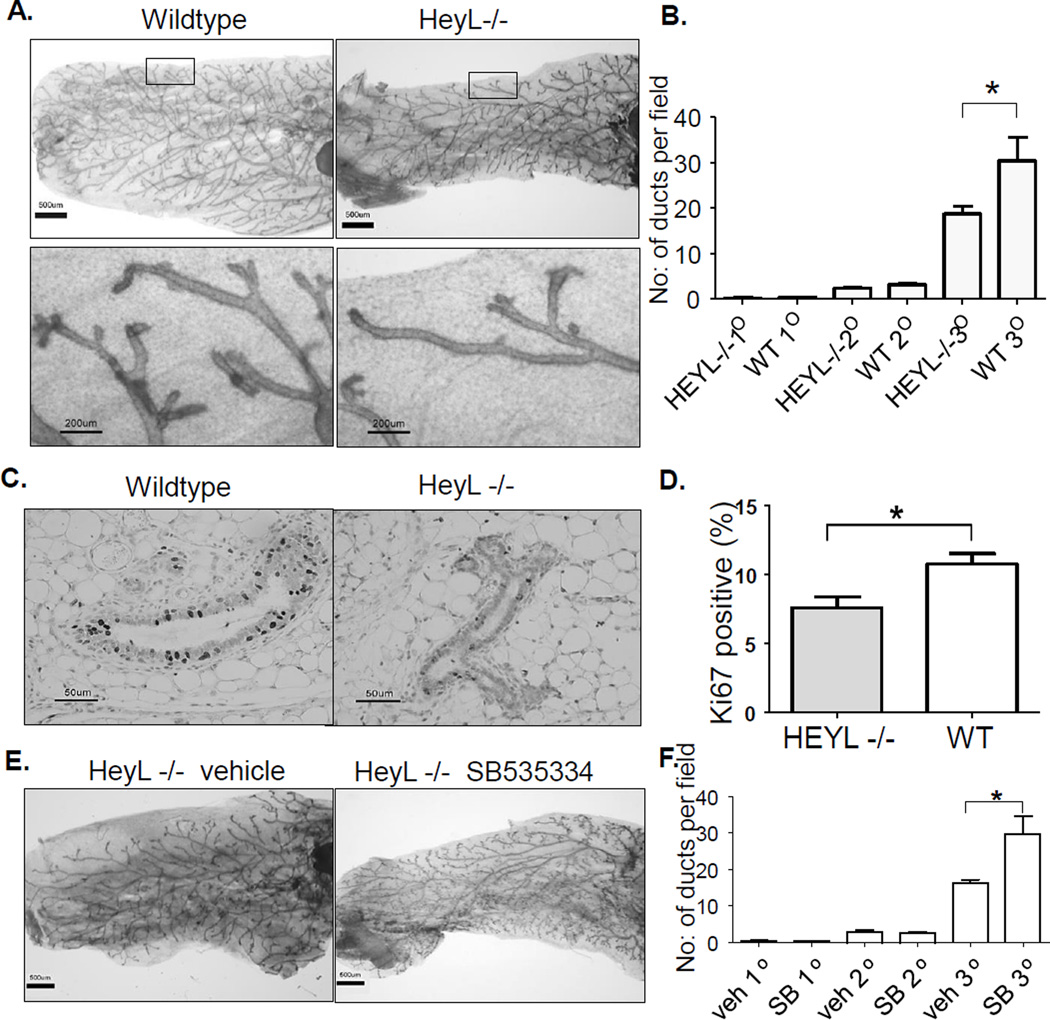
Our data indicate a counteractive effect of Notch and TGF-β on breast epithelial cell growth. In fact, in both Notch2 knockout mice and TGF-β mammary gland-specific transgenic mice the phenotypes of mammary gland development are remarkably similar- reduced epithelial cell growth and fewer terminal end buds (26, 27) were observed. If HeyL dampens TGF-β signaling, HeyL knockout mice (18, 28) are predicted to show mammary gland defects similar to TGF-β transgenic mice. To test this hypothesis, we examined the mammary glands of virgin wild-type and HeyL knockout littermates at 13 weeks old. We found that the mammary glands of HeyL knockout mice have much less side branching and very few terminal ducts (Figure 4A and B). The epithelial cells of HeyL knockout mice show reduced cell proliferation with significantly lower Ki67-positive stained cells (Figure 4C and D). To test the involvement of TGF-β in the generation of this phenotype, we treated HeyL knockout mice with a specific TGF-β type I receptor inhibitor, SB535334. In response to the treatment, the mammary gland ducts developed more side branches and terminal ducts(Figure 4E and F), suggesting that enhanced TGF-β signaling inhibits mammary gland development in HeyL knockout mice.
Figure 4. Abnormal mammary gland development in HeyL knockout mouse.
A. Whole-mount analysis of wildtype and HeyL knockout mice at 13 weeks old
B. Quantitative measurement of mammary gland side branching. Significant differences between the groups was determined using two-tailed paired t test; * p<0.05, N=6
C. Ki67 staining of the mammary glands.
D. Quantitative measurement of Ki67 staining. Significant differences between the groups was determined using two-tailed paired t test; * p<0.05, N=6
E. Whole-mount analysis of HeyL knockout mice treated with either vehicle or SB 535334
F. Quantitative measurement of mammary gland side branching of treated mice. Significant differences between the groups was determined using two-tailed paired t test; * p<0.05, N=4
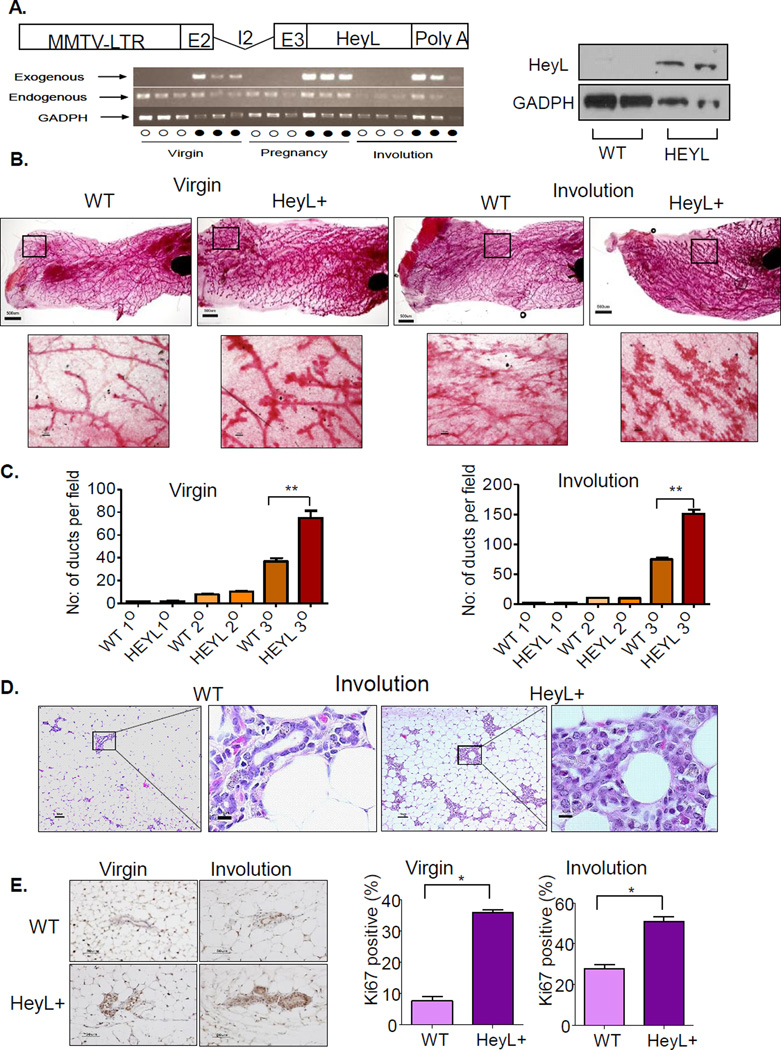
To further probe HeyL’s function in vivo, we generated transgenic mice that specifically express HeyL in the mammary gland under the control of the MMTV-LTR promoter (Figure 5A). HEYL transgenic mice showed robust transgene HeyL expression in mammary glands at different development stages, and low level of endogenous HeyL expression in mammary glands in both wild-type littermates and transgenic mice (Figure 5A). While endogenous HeyL expression did not change significantly at different stages of mammary gland development, the HeyL transgene expression level increased at the pregnancy and involution stages, consistent with the fact that MMTV-LTR promoter can be activated by progestins and corticosteroids. Increased HeyL protein expression level in pregnant transgenic mice was also confirmed by western blotting (Figure 5A). Comparing mammary gland development between wild-type littermates and HeyL transgenic virgin mice at 13 weeks old, HeyL transgenic virgin mice showed more mammary gland duct side-branching in the whole-mount analysis (Figure 5B and C), which was also confirmed by histological examination (Supplementary Figure 5). Strong ductal cell proliferation in HeyL transgenic mice was evidenced by increased KI67 staining (Figure 5E). During pregnancy, wild-type mice developed well-differentiated secretory epithelium containing lipid filled secretory vesicles, while HeyL transgenic mice showed limited epithelial differentiation and very few secreted vesicles (Supplementary Figure 6). Thirty days after weaning, the wild-type mouse mammary gland returned back to normal, leaving mainly simple ductal structures. However, the HeyL transgenic mice showed delayed and incomplete involution with numerous clusters of cells with disorganized structure, and ductal structures were poorly defined and highly proliferative (Figure 5B–E).
Figure 5. Abnormal mammary gland development in HeyL transgenic mouse.
A. The MMTV-HeyL transgenic construct contains the MMTV-LTR promoter and murine HeyL cDNA that are linked by the rabbit β-globin exon II (E2), intron II (I2) and exon III (E3). Poly A: polyadenylation signal; HeyL transgene and endogenous HeyL expression in mouse mammary glands of wild-type (○) and HeyL transgenic mice (●) at different stages were measured by RT-PCR. Western blotting confirmed higher HeyL expression in pregnant transgenic mice.
B. Whole-mount analysis of wildtype and HeyL transgenic mice at 13 weeks old or 30 days after involution.
C. Quantitative measurement of mammary gland side branching of wildtype and HeyL transgenic mice at 13 weeks old or 30 days after involution.
D. H&E staining of mammary glands at 30 days after involution
E. Ki67 staining of the mammary glands. Significant differences between the groups was determined using two-tailed paired t test; * p<0.05, N=4
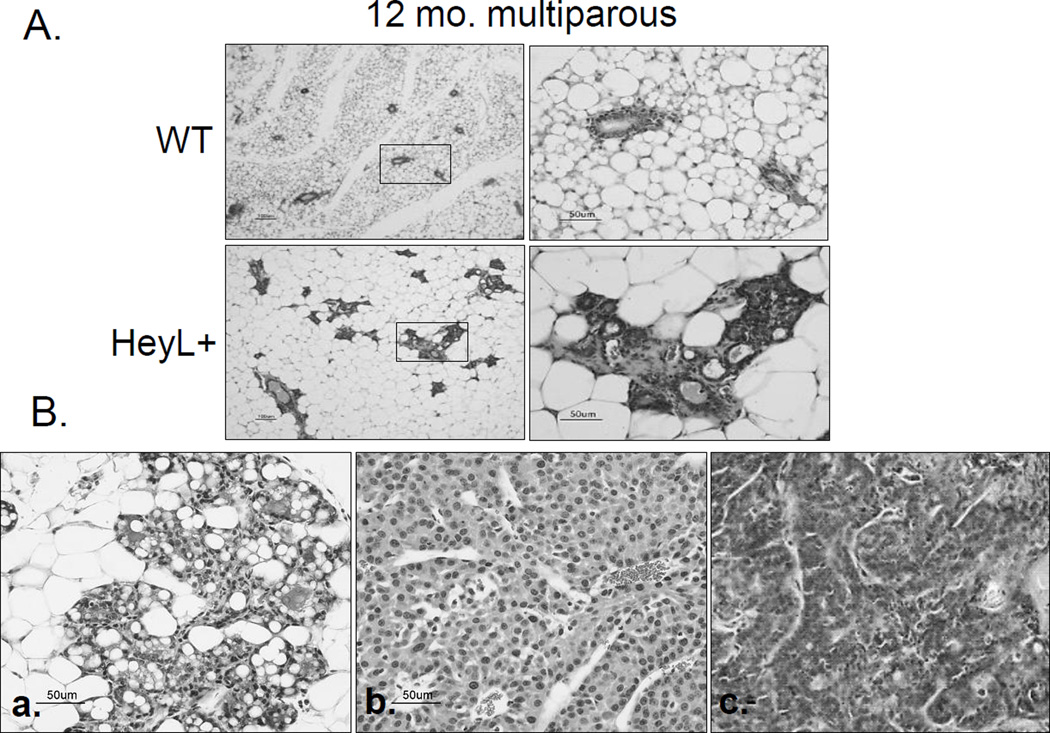
We also monitored mammary tumor development in adult HeyL transgenic mice from at least 3 different founders. Frequent mammary gland hyperplasia, not observed in age-and parity-matched wildtype littermates, was found in multiparous HeyL transgenic mice (32%, 8/25, Figure 6A and Supplementary Figure 7), and 6 out of 25 (24%) multiparous transgenic mice (range from 13 months to 20 months) developed mammary tumors (Figure 6B, a–c). Wild-type multiparous littermates showed low frequency of hyperplasia (19%, 4/21), and no tumors were detected. A low frequency of mammary gland hyperplasia (8%, 2/25) was also found in virgin transgenic mice. Compared to the wild-type group, mammary tumor and hyperplasia incidence was significantly higher in HeyL transgenic mice (Fisher’s exact test, p=0.016). Histopathologically, these tumors were adenocarcinomas or adenosquamous carcinomas, and adjacent mammary glands showed extensive hyperplasia, suggesting that some hyperplastic regions may evolve into cancers. High levels of HeyL mRNA and protein expression were detected in these tumors (Supp. Figure 8). In addition, the expression of PAI-1 and P15, two TGF-β target genes, was lower in HEYL+ transgenic mammary tumors than in normal mammary glands (Supp. Figure 9), providing evidence for low TGFβ function in high HEYL-expressing mammary tumors.
Figure 6. mammary gland hyperplasia and tumor development in HeyL transgenic mouse.
A. H&E staining of mammary glands from wild-type and HeyL transgenic mice at 12 months old.
B. (a,b) H&E staining of one mammary gland tumor and its adjacent hyperplasic lesion. (c) another mammary tumor of HeyL transgenic mouse.
DISCUSSION
In this paper, we provide the first comprehensive report of the role of HEYL in breast cancer. We report that HEYL, a direct target gene of the Notch pathway, is found overexpressed in breast cancer. HEYL inhibits TGF-β signaling through direct binding to Smads. The basic and Helix-loop-Helix domains that mediate HEYL’s interaction with Smads are highly conserved among the members of HERP family. In fact, our unpublished data indicate that HEY1 also binds to Smads. Thus, the Notch pathway tightly represses TGF-β signaling through the binding of several of its targets to Smad proteins, indicating a closely interactive, but functionally antagonistic, network between the two pathways (model in Supp. Figure 10). Consistent with the in vitro data, HeyL knockout mice have enhanced TGF-β signaling and less mammary gland development. In addition, our in vivo mouse model shows that HeyL transgenic mice have enhanced cell proliferation during mammary gland development and develop mammary gland hyperplasia and mammary tumors. Similar phenotypes are seen between the abnormal mammary gland development in type II TGF-β receptor knockout mice or transgenic mice expressing dominant negative type II TGF-β receptor (27, 29, 30). Thus, HeyL transgenic mice can recapitulate most of the phenotypes of mice with reduced TGF-β signaling.
The early protection provided by the TGF-β pathway plays a central role in suppressing the formation of most types of cancers. Previous studies have identified an oncogene SnoN that interferes with TGF-β signaling in breast cancer in a manner similar to HEYL. However, HEYL and SnoN are regulated in a dissimilar fashion. SnoN expression is upregulated by TGF-β, while HEYL expression is induced by the Notch pathway (31). Moreover, HEYL is frequently overexpressed in human breast cancer but not in normal human breast tissues, while SnoN expression is present in normal breast epithelial cells, but is variable in breast cancer in terms of subcellular localization and expression level (32).
Extensive reports indicate that the Notch pathway promotes breast cancer development. However, the interplay between the Notch and TGF-β pathways in breast cancer is unclear. In this paper, we have provided several lines of evidence to show that HEYL is a novel negative regulator of the TGF-β pathway. The identification of HEYL as a direct target of the Notch pathway that associates with Smad proteins and inhibits TGF-β signaling provides a new insight into the tumor promoting capabilities of the Notch pathway.
Supplementary Material
Acknowledgments
We would like to thank the following for providing valuable reagents: Joan Massagué and Addgene for the Smad1, 2 3, 4, Notch1 intracellular domain, p3TP-Luc and various Smad3 deletion constructs, Bert Vogelstein for the (SBE)4-Luc vector, Jeffery Rosen for MMTV transgenic vector, Xiao-Fan Wang for the p15 luciferase vector, and Osamu Nakagawa for the HEYL-luciferase vector. We are grateful to Xinyan Wu and Mary Jo Fackler for their thoughtful discussions and to Kenneth Kinzler for his critical review.
Funding: This work was supported by the Department of Defense predoctoral fellowship W81XWH-04-1-0382 to L. Han, the DOD Center of Excellence Grant W81XWH-04-1-0595 and Susan G Komen Grant SAC110050 to S. Sukumar. This work was also supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute (NCI) Center for Cancer Research.
Footnotes
Potential conflicts of interest: No potential conflicts of interest were disclosed.
Author Contributions
L.H and S.S designed experiments and wrote the manuscript. L.H. performed the experiments and S.S. supervised the research. A.D., N.N. and P.K. performed animal experiments. A.R. and L.F. generated the HeyL transgenic mice. W.T provided technical assistance, S.C. analyzed the TCGA database and S.K. helped perform IHC staining. D.H provided animal pathology expertise and reporting, G.L. and P.A. provided human histopathology analysis and M.G. generated the HeyL knockout mice.
References
- 1.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19(23):2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 2.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Levy L, Hill CS. Alterations in components of the TGF-beta superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 2006;17(1–2):41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nature reviews. 11(5):338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 6.De Strooper B, Annaert W, Cupers P, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398(6727):518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 7.Mumm JS, Schroeter EH, Saxena MT, et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5(2):197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 8.Masuda S, Kumano K, Shimizu K, et al. Notch1 oncoprotein antagonizes TGF-beta/Smad-mediated cell growth suppression via sequestration of coactivator p300. Cancer Sci. 2005;96(5):274–282. doi: 10.1111/j.1349-7006.2005.00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niimi H, Pardali K, Vanlandewijck M, Heldin CH, Moustakas A. Notch signaling is necessary for epithelial growth arrest by TGF-beta. J Cell Biol. 2007;176(5):695–707. doi: 10.1083/jcb.200612129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steidl C, Leimeister C, Klamt B, et al. Characterization of the human and mouse HEY1, HEY2, and HEYL genes: cloning, mapping, and mutation screening of a new bHLH gene family. Genomics. 2000;66(2):195–203. doi: 10.1006/geno.2000.6200. [DOI] [PubMed] [Google Scholar]
- 11.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194(3):237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 12.Nakagawa O, McFadden DG, Nakagawa M, et al. Members of the HRT family of basic helix-loop-helix proteins act as transcriptional repressors downstream of Notch signaling. Proc Natl Acad Sci U S A. 2000;97(25):13655–13660. doi: 10.1073/pnas.250485597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin MH, Leimeister C, Gessler M, Kopan R. Activation of the Notch pathway in the hair cortex leads to aberrant differentiation of the adjacent hair-shaft layers. Development. 2000;127(11):2421–2432. doi: 10.1242/dev.127.11.2421. [DOI] [PubMed] [Google Scholar]
- 14.Maier MM, Gessler M. Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochemical and biophysical research communications. 2000;275(2):652–660. doi: 10.1006/bbrc.2000.3354. [DOI] [PubMed] [Google Scholar]
- 15.Donovan J, Kordylewska A, Jan YN, Utset MF. Tetralogy of fallot and other congenital heart defects in Hey2 mutant mice. Curr Biol. 2002;12(18):1605–1610. doi: 10.1016/s0960-9822(02)01149-1. [DOI] [PubMed] [Google Scholar]
- 16.Sakata Y, Kamei CN, Nakagami H, Bronson R, Liao JK, Chin MT. Ventricular septal defect and cardiomyopathy in mice lacking the transcription factor CHF1/Hey2. Proc Natl Acad Sci U S A. 2002;99(25):16197–16202. doi: 10.1073/pnas.252648999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gessler M, Knobeloch KP, Helisch A, et al. Mouse gridlock: no aortic coarctation or deficiency, but fatal cardiac defects in Hey2−/− mice. Curr Biol. 2002;12(18):1601–1604. doi: 10.1016/s0960-9822(02)01150-8. [DOI] [PubMed] [Google Scholar]
- 18.Fischer A, Steidl C, Wagner TU, et al. Combined loss of Hey1 and HeyL causes congenital heart defects because of impaired epithelial to mesenchymal transition. Circ Res. 2007;100(6):856–863. doi: 10.1161/01.RES.0000260913.95642.3b. [DOI] [PubMed] [Google Scholar]
- 19.Parker BS, Argani P, Cook BP, et al. Alterations in vascular gene expression in invasive breast carcinoma. Cancer research. 2004;64(21):7857–7866. doi: 10.1158/0008-5472.CAN-04-1976. [DOI] [PubMed] [Google Scholar]
- 20.St Croix B, Rago C, Velculescu V, et al. Genes expressed in human tumor endothelium. Science (New York, NY. 2000;289(5482):1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- 21.Leimeister C, Schumacher N, Steidl C, Gessler M. Analysis of HeyL expression in wild-type and Notch pathway mutant mouse embryos. Mech Dev. 2000;98(1–2):175–178. doi: 10.1016/s0925-4773(00)00459-7. [DOI] [PubMed] [Google Scholar]
- 22.Leong KG, Niessen K, Kulic I, et al. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J Exp Med. 2007;204(12):2935–2948. doi: 10.1084/jem.20071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colland F, Jacq X, Trouplin V, et al. Functional proteomics mapping of a human signaling pathway. Genome research. 2004;14(7):1324–1332. doi: 10.1101/gr.2334104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rual JF, Venkatesan K, Hao T, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437(7062):1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 25.Alliston T, Ko TC, Cao Y, et al. Repression of bone morphogenetic protein and activin-inducible transcription by Evi-1. J Biol Chem. 2005;280(25):24227–24237. doi: 10.1074/jbc.M414305200. [DOI] [PubMed] [Google Scholar]
- 26.Barcellos-Hoff MH, Ewan KB. Transforming growth factor-beta and breast cancer: Mammary gland development. Breast Cancer Res. 2000;2(2):92–99. doi: 10.1186/bcr40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sale S, Lafkas D, Artavanis-Tsakonas S. Notch2 genetic fate mapping reveals two previously unrecognized mammary epithelial lineages. Nature cell biology. 15(5):451–460. doi: 10.1038/ncb2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer A, Schumacher N, Maier M, Sendtner M, Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004;18(8):901–911. doi: 10.1101/gad.291004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forrester E, Chytil A, Bierie B, et al. Effect of conditional knockout of the type II TGF-beta receptor gene in mammary epithelia on mammary gland development and polyomavirus middle T antigen induced tumor formation and metastasis. Cancer research. 2005;65(6):2296–2302. doi: 10.1158/0008-5472.CAN-04-3272. [DOI] [PubMed] [Google Scholar]
- 30.Gorska AE, Joseph H, Derynck R, Moses HL, Serra R. Dominant-negative interference of the transforming growth factor beta type II receptor in mammary gland epithelium results in alveolar hyperplasia and differentiation in virgin mice. Cell Growth Differ. 1998;9(3):229–238. [PubMed] [Google Scholar]
- 31.Stroschein SL, Wang W, Zhou S, Zhou Q, Luo K. Negative feedback regulation of TGF-beta signaling by the SnoN oncoprotein. Science (New York, NY. 1999;286(5440):771–774. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]
- 32.Zhang F, Lundin M, Ristimaki A, et al. Ski-related novel protein N (SnoN), a negative controller of transforming growth factor-beta signaling, is a prognostic marker in estrogen receptor-positive breast carcinomas. Cancer research. 2003;63(16):5005–5010. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.