Abstract
Bladder cancer (BC) is a highly prevalent human disease in which retinoblastoma (Rb) pathway inactivation and epigenetic alterations are common events. However, the connection between these two processes is still poorly understood. Here we show that the in vivo inactivation of all Retinoblastoma (Rb) family genes in the mouse urothelium is sufficient to initiate BC development. The characterization of the mouse tumors revealed multiple molecular features of human BC, including the activation of E2F transcription factor and subsequent Ezh2 expression, and the activation of several signaling pathways previously identified as highly relevant in urothelial tumors. These mice represent a genetically defined model for human high-grade superficial BC. Whole transcriptional characterizations of mouse and human bladder tumors revealed a significant overlap and confirm the predominant role for Ezh2 in the downregulation of gene expression programs. Importantly, the increased tumor recurrence and progression in human superficial BC patients is associated with increased E2F and Ezh2 expression and Ezh2-mediated gene expression repression. Collectively, our studies provide a genetically defined model for human high-grade superficial BC and demonstrate the existence of an Rb-E2F-Ezh2 axis in bladder whose disruption can promote tumor development.
INTRODUCTION
Bladder cancer (BC) is a complex and heterogeneous disease caused by both genetic and environmental factors (1). Most tumors arise from the inner lining epithelial cells of the bladder wall, being over 90% transitional cell carcinomas. At diagnosis, two major types of transitional cell carcinomas can be identified according to the pathological characteristics: two thirds of patients present with superficial non-muscle invasive tumors (NMIBC), the remaining third of patients presents (or develops) a highly aggressive, muscle invasive disease (MIBC) that leads to the death of 50% of patients. In general, NMIBC have a favorable prognosis and are treated by transurethral resections and intravesical therapy. However, these tumors show a high rate of recurrence, which in some cases can progress into MIBC. This makes necessary a regular surveillance with cystoscopy and urine cytology indefinitely (EAU guidelines) (2). Therefore, NMIBC represents one of the most costly malignancies to health care systems in developed countries (3). In MIBC, surgery, radiotherapy, and chemotherapy are effective treatments for the disease but there has been little progress in survival in the last 20 years.
The interpretation of the genetic landscape of BC has largely been influenced by the current clinical (NMIBC vs. MIBC) and pathological (stage and grade; papillary vs. solid-invasive) classifications (4). Molecular taxonomy provides a distinct view of BC subphenotypes and their relationship with the disease progression (5). TP53 mutations, and RB1 inactivation are more prevalent in MIBC and may favour tumor progression and muscle invasion (6). However, there is insufficient evidence to use them as independent predictors of poor outcome in the clinical practice of BC patients (7). Low grade NMIBCs are genomically stable whereas high-grade NMIBC and MIBC display genomic instability (8). FGFR3 and PIK3CA mutations are more prevalent in NMIBC and can also be predictive of local recurrences in NMIBC (9). Bladder cancer is also characterized by significant alterations in genes involved in chromatin regulation, affecting in particular to those genes implicated in histone modification (10).
In an attempt to reproduce human tumors, multiple genetically engineered mouse models have been generated (11). In these, the expression of large TAg from SV40 (leading to the elimination of all Rb family members and p53) can induce bladder aggressive tumors in transgenic mice (11, 12). This is in contrast with the absence of bladder tumors upon Rb1 ablation in the urothelium (13). In this regard, we did not observe bladder tumor development in large cohorts of mice bearing the specific elimination of Rb gene directed by keratin K14cre expression (14), which is active in the adult mouse urothelium (15), even in the absence of p107 (16), E2F1 (17) or p130 (18). These data indicate the existence of large overlapping roles for the Rb family members in bladder urothelial cells, similar to the reported for other mouse epithelial cells (19). To circumvent this possible problem and to specifically delete Rb family function in the urothelium of adult mice, we took advantage of a mouse strain bearing floxed alleles of Rb1 and Rbl2 genes and whole deficiency in Rbl1 gene (RbF/F;p130F/F;p107−/−) (20). Conditional gene deletion in the urothelium was induced by delivering an Adeno-Cre into the bladder lumen of adult male mice (13). Here we show that the ablation of all three Rb family members in the mouse bladder urothelium leads to tumor development. The molecular characteristics of these triple knock out (TKO) mouse tumors combined with their genomic characterization provide new possible molecular mechanisms of BC development. Collectively, the present data support a molecular connection between Rb-E2F and Ezh2 that may explain human NMIBC recurrence and progression.
MATERIAL AND METHODS
Patients
Tumor samples and medical records were analyzed from 77 patients who had been consecutively evaluated at the Urology Department of the University Hospital “12 de Octubre” between October 2009 and March 2011, and diagnosed with a Ta or T1 bladder cancer. Informed consent was obtained from all patients and the study was approved by the Ethical Committee for Clinical Research of University Hospital “12 de Octubre”. The pathologic and clinical data, as well as the sample recollection and preservation procedures have been reported elsewhere (9). Samples and united data from patients included in this study were provided by the Biobanco i+12 in the Hospital 12 de Octubre integrated in the Spanish Hospital Biobanks Network (RetBioH; www.redbiobancos.es) following standard operation procedures with appropriate approval of the RETHICAL AN Scientific Committees.
Retinoblastoma family-deficient bladder tumor mouse model
All the animal experiments were approved by the Animal Ethical Committee and conducted in compliance with Centro de Investigaciones Energéticas, Medioambientales y Tecnológicas (CIEMAT) Guidelines. RbF/F;p130F/F;p107−/−mice were generated by breeding RbF/F;p107−/− (16) and p130F/F (20) mice. Adenovirus expressing Cre recombinase was obtained from University of Iowa's Vector Core Facility (www.uiowa.edu) and surgically delivered to the bladder lumen as previously described (13). At the time of sacrifice, tissues were collected and processed as previously reported (16, 17, 21).
Tissue microarray (TMA)
The construction and analysis of tissue microarray containing the human samples has been reported elsewhere (9). At least two representative duplicate cores for each case were scored.
Immunohistochemistry
Immunohistochemistry analyses of human and mouse were performed essentially as previously described (9). Antibodies used were anti Ezh2 (Abnova mAb9542 diluted 1/200), anti K5 (Covance, Princeton, NJ, USA diluted 1/500), anti K8 (TROMA, Univ of Iowa rat mAb diluted 1/10), anti laminin (Sigma L9393, diluted 1/100), anti p63 (Sta Cruz Biotech. mAb 4A4), anti pRb (Sta Cruz Biotech. sc50, diluted 1/50) and anti p130 (Sta Cruz Biotech. sc317, diluted 1/50). Signal was amplified using avidinperoxidase (ABC elite kit Vector) and peroxidase was visualized using diaminobenzidine as a substrate (DAB kit Vector). Negative control slides were obtained by replacing primary antibodies with PBS (data not shown). Scoring of the results and selection of the thresholds, internal controls for reactivity of each antibody, and tissue controls for the series were done according previously published methods (9). Mice were injected intraperitoneally with BrdU (0.1mg/g weight in 0.9% NaCl; Roche, Basel, Switzerland) 1h before sacrifice. BrdU incorporation was monitored in formalin-fixed sections using an anti-BrdU antibody (Roche) as described (16, 17, 21).
Determination of FGFR3 and PIK3CA gene mutation
The presence of mutations in the PIK3CA and FGFR3 genes was assessed in tumor genomic DNA by PCR test (QIAGEN) and/or by snapshot technique has been reported elsewhere (9).
RTqPCR
Total RNA was isolated from mouse and human samples and analyzed by RTqPCR as previously described (9, 16, 17, 21, 22). Briefly, total RNA was isolated using miRNeasy Mini Kit (Qiagen) according to the manufacturer's instructions and DNA was eliminated (Rnase-Free Dnase Set Qiagen). Reverse transcription was performed using the Omniscript RT Kit (Qiagen) and a primer mix specific for all genes of interest, in the case of human samples, and oligo dT primer for the mouse samples, using 10ng and 1μg of total RNA, respectively. PCR was performed in a 7500 Fast Real Time PCR System using Go Taq PCR master mix (Promega) and 1 μl of cDNA as a template. Melting curves were performed to verify specificity and absence of primer dimerization. Reaction efficiency was calculated for each primer combination, and TBP gene was used as reference gene for normalization (23). The sequences of the specific oligonucleotides used are listed in Supp Table 1. Discrimination between samples showing increased or decreased relative expression was made using the median.
Whole transcriptome analysis
Genome-wide transcriptome experiments were performed using the Affymetrix HuGene-1_0-st-v1 microarray or Mo Gene-1_0-st-v1 at the Genomics Facility of the Cancer Research Center (Salamanca, Spain) using standard procedures (see Supp Information). Datasets have been deposited in GEO (GSE38264). Mouse to human comparison was performed essentially as described elsewhere (22).
Immunoblot
Immunoblot was performed as described previously (16, 21). Briefly, dissected bladder tumors were disrupted by freeze-thawing cycles in lysis buffer (200mM4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid pH 7.9, 25% glycerol, 400mM NaCl, 1mM ethylenediaminetetraacetic acid, 1mM ethylene glycol tetraacetic acid, 1mg/ml aprotinin, 1mg/ml leupeptin, 1mM phenylmethanesulfonyl fluoride, 20mM NaF, 1mM NaPPi, 1mM Na3VO4, 2.5mM dithiothreitol), and centrifuged to obtain supernatant containing total protein. 35 μg protein per sample were resolved in sodium dodecyl sulfate—polyacrylamide gel electrophoresis, gels and transferred to nitrocellulose membranes (Amersham, Little Chalfont, UK). Membranes were blocked with 5% non-fat milk diluted in Tris-buffered saline (TBS) and incubated with the appropriate antibodies diluted in TBS-T 0.5% bovine serum albumin. Secondary antibodies were purchased from Jackson ImmunoResearch. Super Signal West Pico Chemiluminescence Substrate (Pierce) was used according to the manu-facturer's recommendations to visualize the bands. Antibodies used are against E2F1, E2F2, E2F3, pRb, p130 (Santa Cruz Biotechnology), Ezh2 (Abnova), phosphoSer473-Akt (Epitomics or Cell Signaling), Phospho-Erk1/2 (Thr202/Tyr204-P), p19Arf (AbCam), p53 (Novocastra) Stat3-Tyr805-P and S6-P (Cell signaling). Loading was controlled by using an anti-Actin antibody (Santa Cruz Biotechnology).
Statistical analysis
Comparisons were performed using the Wilcoxon–Mann–Whitney test (for two groups) and Student's t Test for paired samples showing normal distribution. Survival analyses (recurrence free or tumor progression in recurrence) according to various variables were performed using the Kaplan– Meyer method and differences between the patient groups were tested by the log-rank test. Discrimination between samples showing increased or decreased gene expression was made using the median. Overlapping significance was monitored by exact F Fisher test. SPSS 17.0 and Graph prism 5.0 software were used.
RESULTS
Ablation of all Rb family members causes bladder tumor development
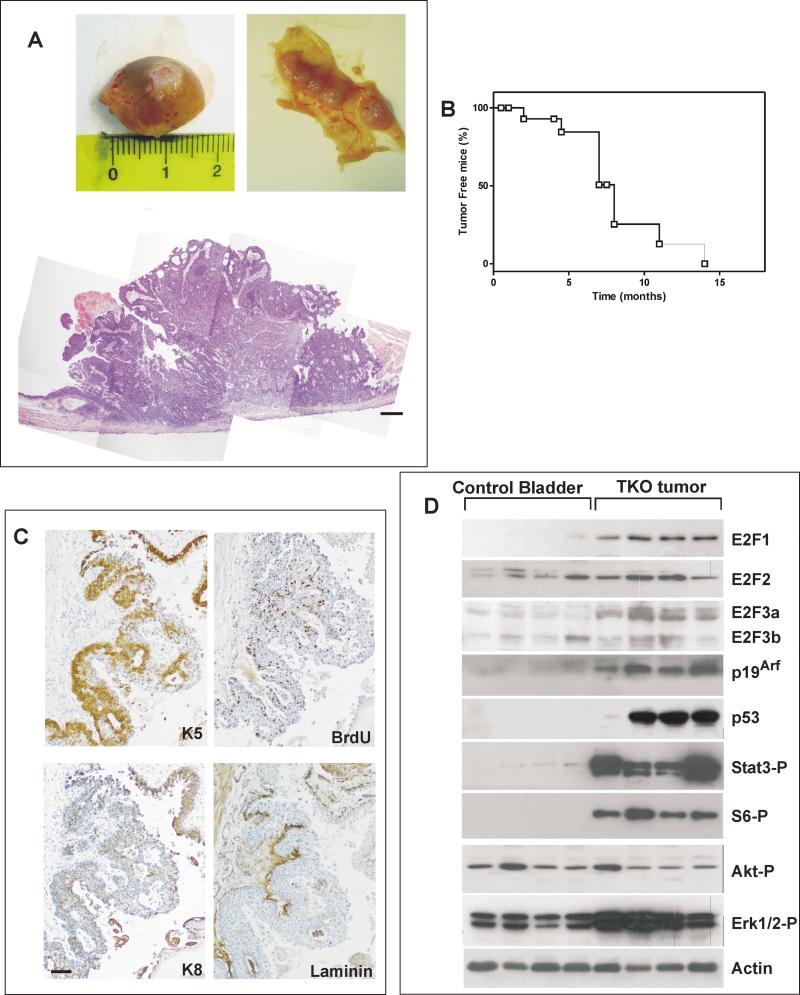
To study the functional consequences of Rb family loss in the urothelium of adult mice, conditional gene deletion mediated by delivering an Adeno-Cre into the bladder lumen (13) of adult male RbF/F;p130F/F;p107−/− mice (20) was used. We observed the development of bladder lesions in all the Adeno Cre infected mice (n=18) (Fig 1A B), but not in control mice (non-infected littermates or infected with Adeno GFP n=30). The presence of these lesions was easily monitored by computerized tomography (Supp Fig 1), and in some cases was accompanied by hematuria. The histological analysis of these lesions revealed papillary masses growing inward to the bladder lumen (Fig 1A, Supp Fig 1), displaying histological features of high-grade non-muscle invasive carcinoma. The mouse bladder tumors were keratin K5-positive, keratin K8-negative (Fig 1C), displayed high proliferation (analyzed by BrdU incorporation, Fig 1C), and invaded the basal lamina (as determined by laminin staining, Fig 1C). When mice were followed up to one year after infection (Fig 1B) no visible metastases were detected.
Fig. 1. In vivo Rb family ablation causes bladder cancer development in mice.
A) Example of external (upper left panel) or internal (upper right panel) view of a bladder from an RbF/F;p130F/F;p107-/- mouse 11 months after inducing the gene recombination by AdenoCre infection. Lower panel depicts an example of H&E stained section of the lesions shown in upper panels. Bar =500μm
B) Kaplan-Meier curve showing the kinetics of tumor development as assessed by CT scan in the adenoCre injected mice (n=18).
C). Representative immunohistochemistry of a mouse tumor stained for Keratin K5, BrdU, Keratin K8 and Laminin as indicated. Bar=150μm
D) Immunoblot analyses of control (uninfected littermates bladder samples) and mouse bladder tumors (denoted TKO: triple knock out) showing the expression of the quoted proteins, Actin was used to normalize protein loading.
We observed the ablation of all Rb family members (Supp Fig 2), and the increased expression of E2F1, 2 and 3a (Fig 1D) in these tumors, by both immunoblot and RTqPCR analyses (Supp Fig 3). In addition, tumors were characterized by the upregulation of p19Arf and p53 and the activation of Erk, S6 and Stat3 signaling pathways, as demonstrated by immunoblot analysis using phospho-specific antibodies (Fig 1D). In contrast, tumors did not show increased Akt activity (Fig 1D).
Tumors in retinoblastoma-deficient mouse resemble human BC
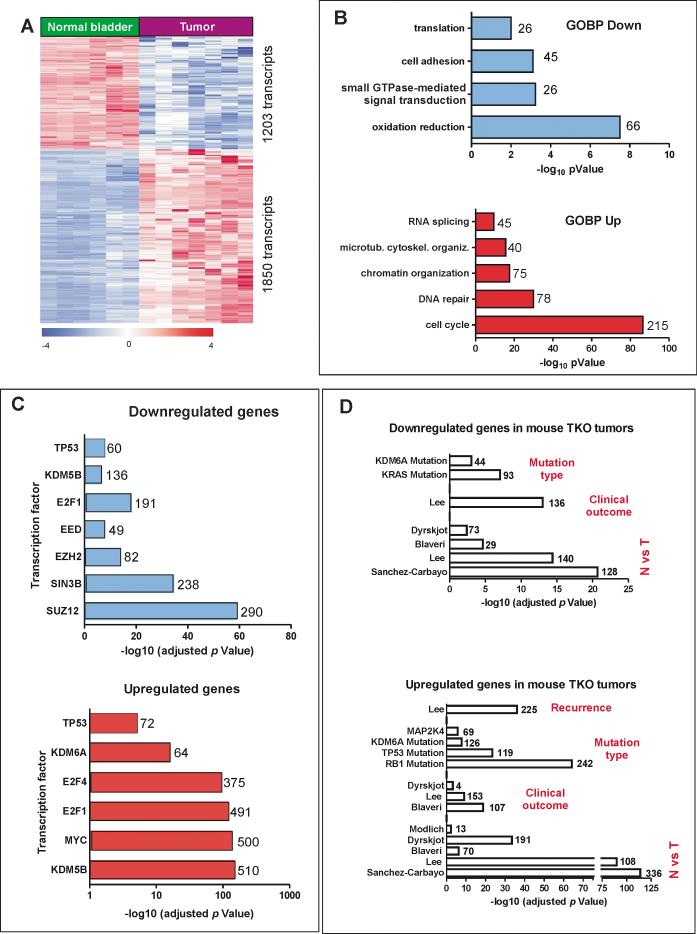
To further characterize the Rb-family mutant mouse bladder tumors and to compare with human BC, a whole transcriptome characterization of the mouse samples was performed. This analysis revealed a differential expression of 3053 transcripts (1849 downregulated and 1204 upregulated) in tumors compared to normal bladder (Fig 2A and Supp Table S2). Gene ontology characterization of these deregulated transcripts indicated that downregulated genes were involved in oxido-reduction, small GTPases transduction signaling, cell adhesion and translation processes (Fig 2B and Supp Table S3), whereas the upregulated genes were primarily involved in cell cycle and DNA repair processes, as would be expected in tumors with high E2F activity (Fig 2B and Supp Table S4). To characterize the putative transcription factors controlling these differentially expressed genes, we performed ChIP enrichment analyses (ChEA) (24). These revealed the possible involvement of Polycomb repressive group 2 (PRC2) in downregulated genes (Fig 2C and Supp Table S5), whereas the upregulated genes showed a primary involvement of E2F and Myc, together with histone demethylases (Fig 2C and Sup Table S6). Gene set enrichment analysis (GSEA) showed significant overlap of the upregulated and downregulated genes in mouse bladder tumors with various human bladder datasets and also with stem cell signatures, including genes silenced in ES cells by H3K27me3 (Supp Table S7), thus reinforcing the possible role of PRC2 in gene expression deregulation. The comparative analyses between the downregulated (Fig 2D upper panel and Supp Table S8) or upregulated (Fig 2D lower panel and Supp Table S9) mouse genes and human bladder tumors available in Oncomine database (25) showed a very significant overlapping with multiple external bladder cancer datasets comparing normal vs. tumor samples, tumors with poor clinical outcome, and the presence of specific gene mutations in human tumors, including KDM6A and RB mutations. Remarkably, this comparison (Fig 2D and Supp Table S9) also showed a statistically significant overlap between the upregulated mouse genes and upregulated genes in human recurrent bladder tumors, and also between the downregulated genes in mouse tumors and multiple datasets identifying genes silenced by PRC2 in stem cells (Supp Table S8).
Fig. 2. Transcriptome analysis of mouse bladder tumors.
A) Heatmap showing the distribution of genes (rows) identifying tumors and normal control uninfected mouse bladder samples. Each column represents a sample. A red (overexpressed) to blue (downregulated) scheme following the above scale limits (in log2 scale) is shown. Numbers on the right denote the number of transcripts of each group (upregulated or downregulated).
B) Representative Gene Ontology biological processes categories affecting the functions of the downregulated (upper panel) or upregulated (lower panel) genes in mouse bladder tumors. Numbers on the right of each bar indicate the genes on COBP category:
C) Relative relevance of transcription factors and histone modifying enzymes in the genes downregulated (upper panel) or upregulated (lower panel) in tumors as obtained by ChEA. The relevance of each factor is provided by the p-value (in –log10 scale). Numbers on the right of each bar represent genes bound by each transcription factor from the database.
D) Summary of relevant overlap between downregulated (upper panel) or upregulated (lower panel) genes in mouse bladder TKO (triple knock out) tumors with human bladder tumors from the Oncomine database. The different comparison concepts (Tumor vs Normal, Mutation Type, Clinical Outcome and Recurrence) are provided for each group. p-values were obtained by exact F Fisher test. The number of overlapping genes is shown for each dataset.
Increased Ezh2 in retinoblastoma family-deficient mouse bladder tumors
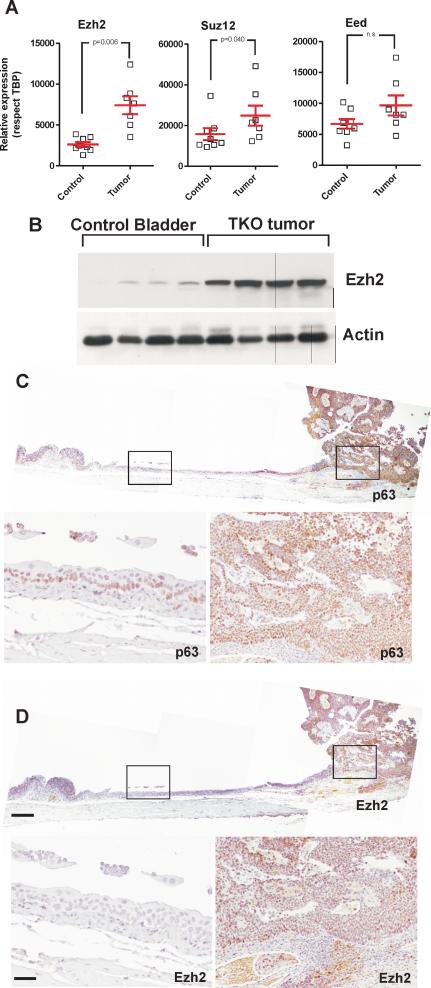
The observed similarities between downregulated genes in mouse bladder tumors and those being silenced by PRC2 prompted us to analyze possible changes in the expression of PRC2 elements, Ezh2, Suz12 and Eed, in the mouse tumors. RTqPCR analysis (Fig 3A) showed significant increased expression of Ezh2 and Suz12 genes, but not Eed in mouse tumors compared with control samples, being more relevant the increased expression of Ezh2 gene. Immunoblot analysis confirmed the increased expression of Ezh2 protein (Fig 3B), the catalytically active member of the PRC2 complex and responsible of trimethylation of H3K27. Finally, immunohistochemistry analysis of Ezh2 expression (Fig 3D) compared to that of urothelium basal layer marker p63 (Fig 3C), revealed the increased expression of Ezh2 in mouse tumors, but not in the adjacent normal urothelium.
Fig 3. Mouse bladder tumors display increased Ezh2 expression.
A) Expression of Ezh2 Suz12 and Eed genes in control (uninfected bladder samples) and mouse bladder tumors as assessed by RTqPCR (respect to Tbp). p-value was obtained by the Mann Whitney t-test, mean and SEM are denoted in red.
B) Immunoblot analyses of control (uninfected bladder samples) and mouse bladder TKO (triple knock out) tumors showing the expression of Ezh2 protein Actin was used to normalize protein loading.
C) Representative examples of the immunohistochemistry analysis of mouse bladder tumor showing the expression of p63. Lower panels are higher magnification of the corresponding areas denoted in upper panel.
D) Representative examples of the immunohistochemistry analysis of mouse bladder tumor showing the expression of Ezh2. Lower panels are higher magnification of the corresponding areas denoted in upper panel. Bar in upper panel=500μm, in lower panel =150μm
Genome-wide transcriptome analysis of human recurrent bladder tumor
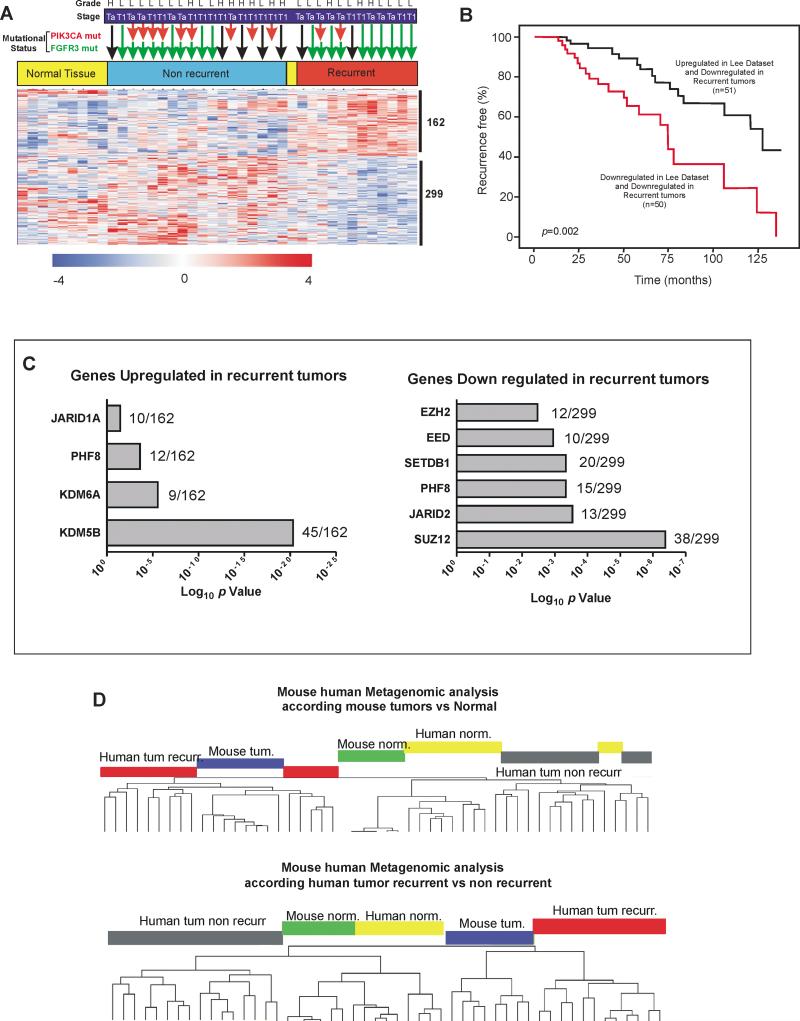
These observations indicate that the mouse Rb family mutant tumors represent a bona fide model of human BC. The pathological characteristics resemble those of human high grade NMIBC, a possibility also reinforced by the genomic comparisons. To confirm this, a whole transcriptome study was performed using a series of human NMIBC samples focusing in differential gene expression between recurrent and non-recurrent tumors. The analysis identified 351 transcripts (162 overexpressed and 299 underexpressed) differentially expressed in recurrent tumors (Fig 4A, and Supp Table S10), which also discriminated normal tissue. Remarkably, this classification did not discriminate between Ta and T1 stages, high and low grade tumors, or tumors bearing FGFR3 mutations (Fig 4A). On the contrary, tumors bearing PIK3CA gene mutation were predominantly associated with non-recurrent samples (Fig 4A). These findings may help to explain our previously reported association between PIK3CA gene alterations and reduced recurrence in superficial bladder tumors (9). Gene Ontology (GOBP Supp Table S11 and Supp Table S12) and GSEA (Supp Table S13) revealed that the upregulated genes in recurrent tumors played a major role in cell cycle control, proliferation and proteosomal protein degradation, whereas the downregulated genes displayed an association with ribosome, 3’UTR mediated translational regulation and protein translation. The GSEA also indicated similarities with stem cell transcription and PRC2 silencing (Supp table S13). The comparison of these differentially expressed genes with other human bladder cancer datasets in Oncomine database revealed a significant overlap and trend with previously reported bladder cancer datasets (Supp. Table S14). Of note, we found that the downregulated genes in recurrent samples of our study were able to identify early recurrence of NMIBC in an external dataset (26) (Fig 4B). ChEA of these deregulated genes revealed, besides the common involvement of multiple transcription factors in all datasets (Supp Table S15 and S16), a statistically significant involvement of histone modifying enzymes, affecting histone methyl transferases and demethylases, and including the PRC2 members: Ezh2, Suz12 and Eed (Fig 4C). In agreement, we also observed a significant overlap with H3K27me3 in downregulated genes in recurrent tumors by GSEA analysis (Supp Table S13).
Fig. 4. Transcriptome studies of human NMIBC (non-muscle invasive bladder cancer) recurrence.
A) Heatmap showing the distribution of genes (rows) identifying recurrent tumor samples. Each column represents a sample, the mutation of PIK3CA or FGFR3 genes is denoted by red and green arrows, respectively. The stage (Ta and T1) and grade (high: H, low: L) for each tumor sample is also provided. A red (upregulated) to blue (downregulated) scheme following the scale limits (in log2 scale) is shown. Numbers on right side denote the number of transcripts characterizing each class.
B) Kaplan-Meier distribution of patients in Lee Dataset (26) according the expression levels of the downregulated genes identified in our study. p-values were obtained by the Log rank test. n denotes the number of samples scored of each group.
C) Relative relevance of the different histone modifying enzymes in the genes of each quoted group obtained by ChEA. The relevance of each enzyme is provided by the p-value. The number on the right of each bar represents the number of specific genes bearing this type of modification and the total number of transcripts on each group: upregulated (left panel) or downregulated (right panel) in recurrent tumors.
D) Dendrograms of mouse-human transcriptome comparison upon unsupervised clustering (Pearson correlation and average linkage method) according the expression of genes differentiating mouse control and bladder tumor samples (upper panel) or genes discriminating human recurrent tumors (lower panel).
Finally, we integrated our transcriptome data from both mouse and human genes in a common dataset. The unsupervised classification of this common dataset according i) the expression of genes discriminating mouse normal and tumor bladder samples (Fig 4D upper panel), or ii) human genes discriminating human recurrent and non-recurrent NMIBC samples (Fig 4D Lower panel), invariably showed that mouse tumors were clustered with human recurrent tumors. Thus, the mouse bladder tumors initiated by the ablation of all retinoblastoma family members represent a putative model of human recurrent NMIBC.
Increased EZH2 expression in human recurrent bladder tumors
Genomic data in mouse and human bladder tumors analyzed pointed to a major involvement of PRC2, and in particular Ezh2, in the development and recurrence of NMIBC. To further support this, those genes identified by ChEA to be bound by any PRC2 element or specifically by Ezh2 (n= 38 and n=12, respectively) were loaded into Oncomine database. A statistically significant overlap was found with the Lee-bladder dataset (26) in the case of PRC2-bound genes (overlapping genes n= 4 of 38, p-value= 0.00044; Odds ratio 12.7). Moreover, we also observed a significant overlap with both PRC2- and Ezh2-bound genes in another external dataset (27) (overlapping genes n=5 of 38; p-value=0.002; Odds ratio 9.1, and n=3 of 12, p-value= 0.008; Odds ratio= 13.6, respectively). Importantly, even these limited numbers of overlapping genes were able to discriminate patients of NMIBC with high likelihood of recurrence (Supp Fig S4A, B, C). These observations point to a primordial role of Ezh2-mediated gene silencing in NMIBC early recurrence
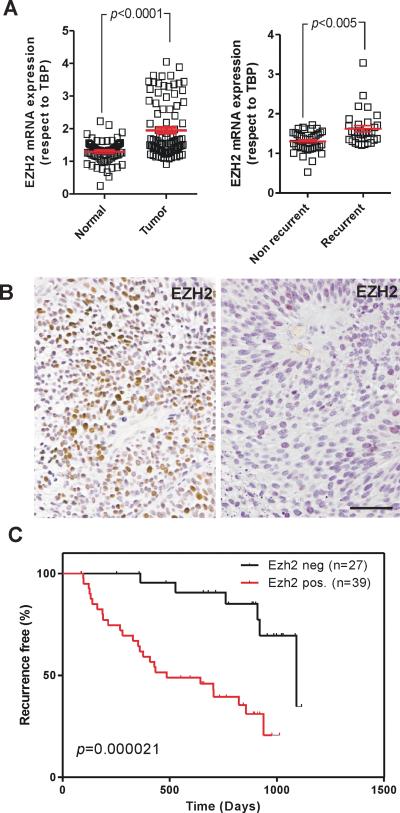
We next analyzed expression changes of PRC2 members in a series of human NMIBC samples previously described (9). RTqPCR analysis revealed a significant increase of EZH2 expression (Fig 5A), but not of SUZ12 and EED (Supp Fig S5A, B), in tumors compared with paired normal bladder samples. The EZH2 gene expression was also higher in recurrent tumors compared to non-recurrent (Fig 5A). When the patients were stratified according Ezh2 expression in a tissue microarray (Fig 5C, C’’ and data not shown), we found that Ezh2 increased expression correlated with tumor recurrence (Fig 5D; p=0.000021).
Fig. 5. EZH2 Expression in human NMIBC (non-muscle invasive bladder cancer).
A) Expression of EZH2 gene as assessed by RTqPCR (respect to TBP) in normal vs. tumor samples, and in recurrent vs non-recurrent tumors. p-values were obtained by the Mann Whitney t-test, mean and SEM are shown.
B) Representative examples of human tumors showing positive (left panel), and negative (right panel) staining for EZH2 protein. Bar= 100μm.
C) Kaplan-Meier distribution of recurrence according the expression of EZH2 protein form tissue microarrays. p-values were obtained by the Log rank test (n denotes the number of samples in each group).
A possible involvement of E2F in Ezh2 deregulated expression
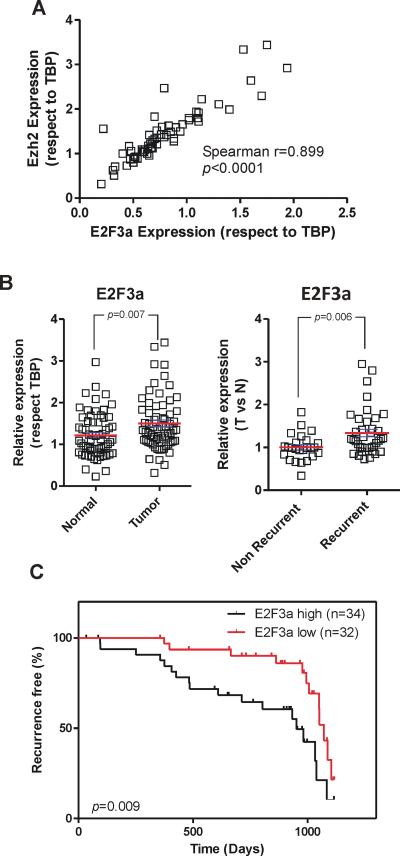
The expression of EZH2 gene is regulated by E2F activity (28). Accordingly, we monitored whether EZH2 expression correlated with the expression of different E2Fs in our series of human bladder tumor samples. This analysis revealed a striking correlation between EZH2 and E2F3a gene expression (Fig 6A). We also observed that E2F3a expression was significantly higher in tumor than in normal samples, and in recurrent than in non-recurrent tumors (Fig 6B). Importantly, increased E2F3a expression levels stratified recurrent NMIBC patients (Fig 6C). On the other hand, E2F1, 2 and 3b gene expression did not discriminate between normal and tumor samples (Supp Fig S6A), or between recurrent and non-recurrent human NMIBC (Supp Fig S6B). Further the expression of E2F1, 2 and 3b genes did not show significant correlation with EZH2 gene expression (Supp Fig S6C, E, G), and did not allow statistical stratification of recurrence in our series of NMIBC patients (Supp Fig S6D, F, H). These results indicate that the early recurrence mediated by increased EZH2 expression could be facilitated primarily by increased E2F3a expression. However, a concerted action of other E2F family members in this process could not be discarded at this point.
Fig. 6. E2F3a expression in human NMIBC (non-muscle invasive bladder cancer).
A) Expression of EZH2 gene as a function of E2F3a gene expression as assessed by RTqPCR in NMIBC samples. The r and p values are provided according the Spearmann correlation method
B) Expression of E2F3a gene in normal and tumor bladder samples (left panel) and in recurrent and non-recurrent bladder tumors (right panel). p-values were obtained by the Mann Whitney t-test, mean and SEM are denoted in red.
C) Kaplan-Meier distribution of NMIBC recurrence according increased expression of E2F3a gene p-value was obtained by the Log rank test. n denotes the number of samples scored of each group.
EZH2 affects tumor progression in recurrence
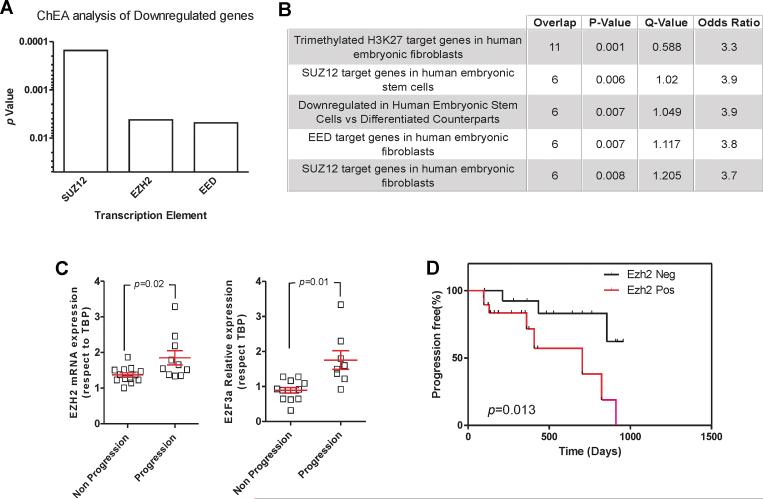
Besides the high recurrence rate, an important problem in NMIBC is the relatively high rate of recurrent tumors appearing with increased stage and/or grade compared with the primary tumors indicative that tumor progression upon recurrence occurred. During this study, 11 out of 33 patients suffering recurrence in our series also displayed tumor progression. To study if similar events to those described for recurrence were also affecting tumor progression, the differential gene expression between tumors showing or not tumor progression upon recurrence was studied. This analysis revealed a very limited number of transcripts (Supp Table S17). ChEA analysis of these transcripts showed that a significant number of under-expressed genes in tumors showing progression were bound by PRC2 complex elements (Fig 7A). In addition, these transcripts displayed a significant overlap with external datasets of gene lists regulated by PRC2, including genes with H3K27me3 marks (Fig 7B). Accordingly, samples of tumors showing progression in recurrence showed also a significant increase in EZH2 and E2F3a expression (Fig 7C). Moreover, the increased expression of EZH2 protein was an independent predictor of tumor progression in the recurrences of our series of NMIBC samples (Fig 7D).
Fig. 7. EZH2-dependent signaling in tumor recurrence and progression.
A) Relative relevance of the binding of the different PRC2 elements to the downregulated genes in tumors showing progression upon recurrence as assessed by ChEA. The relevance of each enzyme is provided by the p-value.
B) Summary of relevant overlap of the downregulated genes in tumors showing progression upon recurrence and Oncomine database in the concept “literature defined” showing the relevance of PRC2 components.
C) Expression of EZH2 and E2F3a genes as assessed by RTqPCR (respect to TBP) in tumors displaying or not progression upon recurrence. p-values were obtained by the Mann Whitney t-test, mean and SEM are denoted in red.
D) Kaplan-Meier distribution of tumor progression in recurrences according increased expression of EZH2. p-values were obtained by the Log rank test.
These data indicated that increased EZH2 expression mediates not only early recurrence, but also increased probability of tumor progression in human NMIBC.
DISCUSSION
Bladder cancer is one of the most common cancers in men worldwide making it a current problem in terms of social and medical relevance. The genomic landscape of these tumors may contribute to identify novel targets of possible therapeutic interest. These studies have revealed multiple alterations and mutations in BC, including chromatin remodeling, cell cycle and specific signal transduction pathways (5, 10, 29-33). Regarding cell cycle, RB1 mutations are relevant. However, these studies are biased by the analysis of mainly aggressive, MIBC tumors and few examples of NMIBC are included. Nonetheless, even in these few examples, RB1 alterations are present (29, 32). This, together with other possible alterations leading to functional pRb inactivation, such as CycD or E2F amplification (26, 30, 32), may suggest that Rb-dependent pathway is also of relevance in NMIBC. Our present data, showing that the complete ablation of Rb family in vivo leads to urothelial tumors supports this possibility.
It has been previously shown that the inactivation of Rb1 gene in mouse urothelium is insufficient to allow spontaneous tumor development (13). Regarding our observations of complete penetrance of BC development upon Rb-family ablation, this apparent discrepancy could be attributed to functional compensation by the other family members, as previously reported in mouse epithelia (19). Our findings are in agreement with the reported results of urothelial expression of T antigen in transgenic mice (12, 34). However, T antigen transgenic mice usually develop muscle invasive tumors; this could be attributed to impaired p53 tumor suppressive functions. Such possibility, which is also reinforced by our previous findings indicating that p53 loss in Rb-deficient stratified epithelia facilitate the development of invasive metastatic disease (21, 35), remains to be determined. The pathological characteristics of the mouse bladder tumors suggest that these mice could represent a bona fide NMIBC model. Also, the mouse tumors display activation of MAPK/Erk, Stat3 and S6 pathways, which have been previously involved in human BC (36-38).
In view of the findings in the mouse model, we aimed to use it as a tool to gain a better understanding of the molecular mechanisms responsible for the initiation, recurrence and progression of BC. This would also provide potential biomarkers. In a first approach, we performed a whole transcriptome analysis. This reinforced the similarities between mouse and human tumors, and provided a possible mechanism for the upregulation and downregulation of genes in the mouse tumors involving E2F and PRC2. We found that the removal of Rb family led to overexpression of E2F family members, and these mouse tumors invariably showed increased Ezh2 expression. Further, our GSEA and ChEA data indicated a predominant role for E2Fs, in particular E2F3a, in gene upregulation in mouse tumors. Given that i) E2F3 is frequently amplified and overexpressed in human BC (39-42), ii) the inactivation of the Rb pathway is required in addition to E2F3 overexpression in human bladder carcinogenesis (40), iii) E2F3 is a primary regulator of EZH2 expression (28), and iv) there is emerging evidence supporting the RB-E2F3-EZH2 pathway as a key oncogenic axis in cancer development and aggressiveness (43), our findings provide a new molecular mechanism for bladder tumor development. Importantly, various Ezh2 inhibitors have been developed and are being tested in various preclinical models (44-46). The possibility that these inhibitors may impair bladder tumor growth in our Rb family-deficient model will be the subject of future investigations. In addition the identification of possible overlapping mutations between human NMIBC and our mouse model will provide further evidence of the relevance of this model for preclinical studies.
In order to confirm these findings and to validate the mouse model as a suitable tool for understanding human NMIBC, we carried out a whole transcriptome analysis in human bladder cancer samples. Furthermore, as recurrence and progression are common in this type of human tumors and constitute a severe clinical problem, this study was designed to determine possible molecular factors affecting recurrence in NMIBC. The metagenomic analyses also reinforced the extreme similarities between mouse and human recurrent tumors. We also observed that recurrence is coupled to gene upregulation mediated by E2F activity, and downregulation associated with increased PRC2 activity. Importantly, we could also validate the role of PRC2- and Ezh2-mediated gene silencing in NMIBC recurrence in external datasets, indicating that increased Ezh2 expression and activity could act as a predictor of early recurrence. Further, we could also associate the increased Ezh2 expression and activity with progression upon recurrence in human NMIBC. In this regard, the increased expression of Ezh2 had been previously reported associated with increased malignancy in bladder cancer (47, 48). In agreement, KDM6A, (which catalyzes the H3K27 demethylation and thus acting in opposite manner to EZH2), had also been described as frequently mutated in bladder cancer, in some cases in association with RB1 gene mutation in high stage or grade human bladder tumors (29, 30, 33). Future research, aimed to detect possible overlapping mutations in these genes in our mouse model, will provide further data of the relevance of this model for preclinical studies. Nonetheless, to the best of our knowledge, this is the first report associating EZH2 activity with NMIBC recurrence and progression.
Collectively, these findings could be highly relevant in the clinical management and therapy of human bladder cancers. The possible therapeutic use of Ezh2 inhibitors in the management of BC is a crucial aspect that merits future research. In this regard, the mouse model here described arises as an essential and invaluable tool for such analyses.
Supplementary Material
ACKNOWLEDGEMENTS
We want to particularly acknowledge the patients enrolled in this study for their participation, and the Biobanco i+12 in the Hospital 12 de Octubre integrated in the Spanish Hospital Biobanks Network (RetBioH; www.redbiobancos.es). Biobank is supported by Instituto de Salud Carlos III. We appreciate FX Real from the CNIO for his suggestions and critical reading of the manuscript.
FUNDING. MICINN grants SAF2012-34378 and SAF2011-26122-C02-01, Comunidad Autónoma de Madrid grants S2006/BIO-0232 and S2010/BMD-2470 (Oncocycle Programs), MSyC grants ISCIIIRETIC RD06/0020/0029 and RD12/0036/0009, and from Fundación Sandra Ibarra to JMP. Grants AP99782012 and 40100017 from MMA Foundation to MD and MLM, respectively. MSyC grants ISCIIIFIS PI12/01959 to MS. M. Martínez-Fernández is funded by a ‘Juan de la Cierva’ research fellowship (JCI-2010-06167) from MICINN. Work on Rb family mutant mice in the Sage lab (JS, PV) is funded by the NIH (R01 CA114102). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflict of interest
AUTHOR CONTRIBUTIONS. MS, MMF, MD, RGE, BA, CSL, CC, MO, MAM, Performed experimental work and analyzed experimental data. FV, JD, VM, MJGR, MLM, MF, DC, JLRP and FR collected clinical samples and corresponding clinical data. JS and PV provided essential reactive and discussed the data. JMP designed and supervised the study, secured funding, analyzed the data, and wrote the manuscript. All authors discussed the results and commented on the manuscript.
REFERENCES
- 1.Wu X, Hildebrandt MA, Chang DW. Genome-wide association studies of bladder cancer risk: a field synopsis of progress and potential applications. Cancer metastasis reviews. 2009;28:269–80. doi: 10.1007/s10555-009-9190-y. [DOI] [PubMed] [Google Scholar]
- 2.Gallagher DJ, Milowsky MI. Bladder cancer. Current treatment options in oncology. 2009;10:205–15. doi: 10.1007/s11864-009-0112-6. [DOI] [PubMed] [Google Scholar]
- 3.Burger M, Oosterlinck W, Konety B, Chang S, Gudjonsson S, Pruthi R, et al. ICUD-EAU International Consultation on Bladder Cancer 2012: Non-muscle-invasive urothelial carcinoma of the bladder. European urology. 2013;63:36–44. doi: 10.1016/j.eururo.2012.08.061. [DOI] [PubMed] [Google Scholar]
- 4.Knowles MA. Molecular subtypes of bladder cancer: Jekyll and Hyde or chalk and cheese? Carcinogenesis. 2006;27:361–73. doi: 10.1093/carcin/bgi310. [DOI] [PubMed] [Google Scholar]
- 5.Iyer G, Al-Ahmadie H, Schultz N, Hanrahan AJ, Ostrovnaya I, Balar AV, et al. Prevalence and co-occurrence of actionable genomic alterations in high-grade bladder cancer. J Clin Oncol. 2013;31:3133–40. doi: 10.1200/JCO.2012.46.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rebouissou S, Herault A, Letouze E, Neuzillet Y, Laplanche A, Ofualuka K, et al. CDKN2A homozygous deletion is associated with muscle invasion in FGFR3-mutated urothelial bladder carcinoma. The Journal of pathology. 2012;227:315–24. doi: 10.1002/path.4017. [DOI] [PubMed] [Google Scholar]
- 7.van Rhijn BW. Combining molecular and pathologic data to prognosticate non-muscle-invasive bladder cancer. Urol Oncol. 2012;30:518–23. doi: 10.1016/j.urolonc.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Lindgren D, Sjodahl G, Lauss M, Staaf J, Chebil G, Lovgren K, et al. Integrated genomic and gene expression profiling identifies two major genomic circuits in urothelial carcinoma. PLoS One. 2012;7:e38863. doi: 10.1371/journal.pone.0038863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duenas M, Martinez-Fernandez M, Garcia-Escudero R, Villacampa F, Marques M, Saiz-Ladera C, et al. PIK3CA gene alterations in bladder cancer are frequent and associate with reduced recurrence in non-muscle invasive tumors. Mol Carcinog. 2013 doi: 10.1002/mc.22125. doi: 10.1002/mc.22125. [DOI] [PubMed] [Google Scholar]
- 10.Network CeAR Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014 doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmad I, Sansom OJ, Leung HY. Exploring molecular genetics of bladder cancer: lessons learned from mouse models. Disease models & mechanisms. 2012;5:323–32. doi: 10.1242/dmm.008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He F, Mo L, Zheng XY, Hu C, Lepor H, Lee EY, et al. Deficiency of pRb family proteins and p53 in invasive urothelial tumorigenesis. Cancer Res. 2009;69:9413–21. doi: 10.1158/0008-5472.CAN-09-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puzio-Kuter AM, Castillo-Martin M, Kinkade CW, Wang X, Shen TH, Matos T, et al. Inactivation of p53 and Pten promotes invasive bladder cancer. Genes Dev. 2009;23:675–80. doi: 10.1101/gad.1772909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruiz S, Santos M, Segrelles C, Leis H, Jorcano JL, Berns A, et al. Unique and overlapping functions of pRb and p107 in the control of proliferation and differentiation in epidermis. Development. 2004;131:2737–48. doi: 10.1242/dev.01148. [DOI] [PubMed] [Google Scholar]
- 15.Liang FX, Bosland MC, Huang H, Romih R, Baptiste S, Deng FM, et al. Cellular basis of urothelial squamous metaplasia: roles of lineage heterogeneity and cell replacement. The Journal of cell biology. 2005;171:835–44. doi: 10.1083/jcb.200505035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costa C, Santos M, Segrelles C, Duenas M, Lara MF, Agirre X, et al. A Novel Tumor suppressor network in squamous malignancies. Scientific reports. 2012;2:828. doi: 10.1038/srep00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costa C, Santos M, Martinez-Fernandez M, Duenas M, Lorz C, Garcia-Escudero R, et al. E2F1 loss induces spontaneous tumour development in Rb-deficient epidermis. Oncogene. 2013;32:2937–51. doi: 10.1038/onc.2012.316. [DOI] [PubMed] [Google Scholar]
- 18.Lara MF, Garcia-Escudero R, Ruiz S, Santos M, Moral M, Martinez-Cruz AB, et al. Gene profiling approaches help to define the specific functions of retinoblastoma family in epidermis. Mol Carcinog. 2008;47:209–21. doi: 10.1002/mc.20376. [DOI] [PubMed] [Google Scholar]
- 19.Costa C, Paramio JM, Santos M. Skin Tumors Rb(eing) Uncovered. Front Oncol. 2013;3:307. doi: 10.3389/fonc.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Viatour P, Somervaille TC, Venkatasubrahmanyam S, Kogan S, McLaughlin ME, Weissman IL, et al. Hematopoietic stem cell quiescence is maintained by compound contributions of the retinoblastoma gene family. Cell Stem Cell. 2008;3:416–28. doi: 10.1016/j.stem.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bornachea O, Santos M, Martinez-Cruz AB, Garcia-Escudero R, Duenas M, Costa C, et al. EMT and induction of miR-21 mediate metastasis development in Trp53-deficient tumours. Scientific reports. 2012;2:434. doi: 10.1038/srep00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duenas M, Santos M, Aranda JF, Bielza C, Martinez-Cruz AB, Lorz C, et al. Mouse p53-Deficient Cancer Models as Platforms for Obtaining Genomic Predictors of Human Cancer Clinical Outcomes. PLoS One. 2012;7:e42494. doi: 10.1371/journal.pone.0042494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohl F, Jung M, Radonic A, Sachs M, Loening SA, Jung K. Identification and validation of suitable endogenous reference genes for gene expression studies of human bladder cancer. The Journal of urology. 2006;175:1915–20. doi: 10.1016/S0022-5347(05)00919-5. [DOI] [PubMed] [Google Scholar]
- 24.Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma'ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics (Oxford, England) 2010;26:2438–44. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JS, Leem SH, Lee SY, Kim SC, Park ES, Kim SB, et al. Expression signature of E2F1 and its associated genes predict superficial to invasive progression of bladder tumors. J Clin Oncol. 2010;28:2660–7. doi: 10.1200/JCO.2009.25.0977. [DOI] [PubMed] [Google Scholar]
- 27.Dyrskjot L, Thykjaer T, Kruhoffer M, Jensen JL, Marcussen N, Hamilton-Dutoit S, et al. Identifying distinct classes of bladder carcinoma using microarrays. Nature genetics. 2003;33:90–6. doi: 10.1038/ng1061. [DOI] [PubMed] [Google Scholar]
- 28.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. Embo J. 2003;22:5323–35. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balbas-Martinez C, Sagrera A, Carrillo-de-Santa-Pau E, Earl J, Marquez M, Vazquez M, et al. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nature genetics. 2013;45:1464–9. doi: 10.1038/ng.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross JS, Wang K, Al-Rohil RN, Nazeer T, Sheehan CE, Otto GA, et al. Advanced urothelial carcinoma: next-generation sequencing reveals diverse genomic alterations and targets of therapy. Mod Pathol. 2013 doi: 10.1038/modpathol.2013.135. [DOI] [PubMed] [Google Scholar]
- 31.Guo G, Sun X, Chen C, Wu S, Huang P, Li Z, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nature genetics. 2013;45:1459–63. doi: 10.1038/ng.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cazier JB, Rao SR, McLean CM, Walker AL, Wright BJ, Jaeger EE, et al. Whole-genome sequencing of bladder cancers reveals somatic CDKN1A mutations and clinicopathological associations with mutation burden. Nature communications. 2014;5:3756. doi: 10.1038/ncomms4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nature genetics. 2011;43:875–8. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Espana A, Salazar E, Sun TT, Wu XR, Pellicer A. Differential expression of cell cycle regulators in phenotypic variants of transgenically induced bladder tumors: implications for tumor behavior. Cancer Res. 2005;65:1150–7. doi: 10.1158/0008-5472.CAN-04-2074. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Cruz AB, Santos M, Lara MF, Segrelles C, Ruiz S, Moral M, et al. Spontaneous squamous cell carcinoma induced by the somatic inactivation of retinoblastoma and Trp53 tumor suppressors. Cancer Res. 2008;68:683–92. doi: 10.1158/0008-5472.CAN-07-3049. [DOI] [PubMed] [Google Scholar]
- 36.Ho PL, Lay EJ, Jian W, Parra D, Chan KS. Stat3 activation in urothelial stem cells leads to direct progression to invasive bladder cancer. Cancer Res. 2012;72:3135–42. doi: 10.1158/0008-5472.CAN-11-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jebar AH, Hurst CD, Tomlinson DC, Johnston C, Taylor CF, Knowles MA. FGFR3 and Ras gene mutations are mutually exclusive genetic events in urothelial cell carcinoma. Oncogene. 2005;24:5218–25. doi: 10.1038/sj.onc.1208705. [DOI] [PubMed] [Google Scholar]
- 38.Korkolopoulou P, Levidou G, Trigka EA, Prekete N, Karlou M, Thymara I, et al. A comprehensive immunohistochemical and molecular approach to the PI3K/AKT/mTOR (phosphoinositide 3-kinase/v-akt murine thymoma viral oncogene/mammalian target of rapamycin) pathway in bladder urothelial carcinoma. BJU international. 2012;110:E1237–48. doi: 10.1111/j.1464-410X.2012.11569.x. [DOI] [PubMed] [Google Scholar]
- 39.Olsson AY, Feber A, Edwards S, Te Poele R, Giddings I, Merson S, et al. Role of E2F3 expression in modulating cellular proliferation rate in human bladder and prostate cancer cells. Oncogene. 2007;26:1028–37. doi: 10.1038/sj.onc.1209854. [DOI] [PubMed] [Google Scholar]
- 40.Hurst CD, Tomlinson DC, Williams SV, Platt FM, Knowles MA. Inactivation of the Rb pathway and overexpression of both isoforms of E2F3 are obligate events in bladder tumours with 6p22 amplification. Oncogene. 2008;27:2716–27. doi: 10.1038/sj.onc.1210934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oeggerli M, Tomovska S, Schraml P, Calvano-Forte D, Schafroth S, Simon R, et al. E2F3 amplification and overexpression is associated with invasive tumor growth and rapid tumor cell proliferation in urinary bladder cancer. Oncogene. 2004;23:5616–23. doi: 10.1038/sj.onc.1207749. [DOI] [PubMed] [Google Scholar]
- 42.Feber A, Clark J, Goodwin G, Dodson AR, Smith PH, Fletcher A, et al. Amplification and overexpression of E2F3 in human bladder cancer. Oncogene. 2004;23:1627–30. doi: 10.1038/sj.onc.1207274. [DOI] [PubMed] [Google Scholar]
- 43.Foster CS, Falconer A, Dodson AR, Norman AR, Dennis N, Fletcher A, et al. Transcription factor E2F3 overexpressed in prostate cancer independently predicts clinical outcome. Oncogene. 2004;23:5871–9. doi: 10.1038/sj.onc.1207800. [DOI] [PubMed] [Google Scholar]
- 44.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–63. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–12. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 46.Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890–6. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 47.Weikert S, Christoph F, Kollermann J, Muller M, Schrader M, Miller K, et al. Expression levels of the EZH2 polycomb transcriptional repressor correlate with aggressiveness and invasive potential of bladder carcinomas. International journal of molecular medicine. 2005;16:349–53. [PubMed] [Google Scholar]
- 48.Wang H, Albadine R, Magheli A, Guzzo TJ, Ball MW, Hinz S, et al. Increased EZH2 protein expression is associated with invasive urothelial carcinoma of the bladder. Urol Oncol. 2012;30:428–33. doi: 10.1016/j.urolonc.2010.09.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.