Abstract
While it is currently estimated that 40–50% of eukaryotic proteins are phosphorylated, little is known about the frequency and local effects of phosphorylation near pharmaceutical inhibitor binding sites. In this study, we investigated how frequently phosphorylation may affect the binding of drug inhibitors to target proteins. We examined the 453 non-redundant structures of soluble mammalian drug target proteins bound to inhibitors currently available in the Protein Data Bank (PDB). We cross-referenced these structures with phosphorylation data available from the PhosphoSitePlus database. 322/453 (71%) of drug targets have evidence of phosphorylation that has been validated by multiple methods or labs. For 132/453 (29%) of those, the phosphorylation site is within 12Å of the small molecule-binding site, where it would likely alter small molecule binding affinity. We propose a framework for distinguishing between drug-phosphorylation site interactions that are likely to alter the efficacy of drugs vs. those that are not. In addition we highlight examples of well-established drug targets, such as estrogen receptor alpha, for which phosphorylation may affect drug affinity and clinical efficacy. Our data suggest that phosphorylation may affect drug binding and efficacy for a significant fraction of drug target proteins.
Keywords: drug target, data mining, crystal structure, PhosphoSitePlus, binding affinity, ligand, inhibitor
INTRODUCTION
Current estimates indicate that approximately 40–50% of human proteins are phosphorylated. 20,266 non-redundant human proteins have been reported and reviewed in the UniProt database (http://www.uniprot.org)1. PHOSIDA reports 8283 (www.phosida.com)2 non-redundant phosphorylated human proteins, Phospho.ELM reports 8698 (phospho.elm.eu.org)3, and a search of phosphorylation sites identified in either multiple high throughput reports or in low throughput reports in PhosphoSitePlus returns 10,062 human proteins (www.phosphosite.org)4. Two recent advances have greatly accelerated the pace of progress in our understanding of protein phosphorylation. First, whole-cell mass spectrometry efforts have identified large numbers of phosphorylated proteins 5. Secondly, phosphorylation sites in mammalian proteins that have been reported in the literature are maintained in a curated database, PhosphoSitePlus 4. The number of reported protein phosphorylation sites has increased from ~2,000 in 2003, when the database was created, to 20,000 in 2007 and over 200,000 currently 4,6.
Protein kinases, which regulate other proteins through phosphorylation, are one of the most important classes of drug targets. Kinases are commonly turned on through phosphorylation of the activation loop near the active site 7. Small molecule inhibition of an unphosphorylated kinase can cause feedback phosphorylation of the available kinase pool 8. In some cases where phosphorylation is known to reduce drug affinity, strategies have been developed to either lock the drug target in an inactive conformation or retain drug affinity in the phosphorylated state 9,10. Recent studies in MEK inhibiton even suggests different upstream activators and MEK phosphorylation states have dramatic effects on clinical drug candidate efficacy 11,12. Given that a large fraction of proteins in general, and kinases in particular are phosphorylated, it is reasonable to suggest that changes in protein phosphorylation could affect small molecule drug binding and efficacy for a significant fraction of drug target proteins. Yet, to our knowledge, no group has investigated the frequency of with which phosphorylation may affect drug binding, nor its occurrence outside of kinases. Both target and computational-based screening methods do not typically consider phosphorylation given the limited number of available phosphorylated protein structures. As a result, these screening methods may discount significant local structural effects of phosphorylation on the drug target protein 13. Here we investigate the effects and frequency of phosphorylation near the site of drug binding for 453 drug targets in the PDB.
METHODS
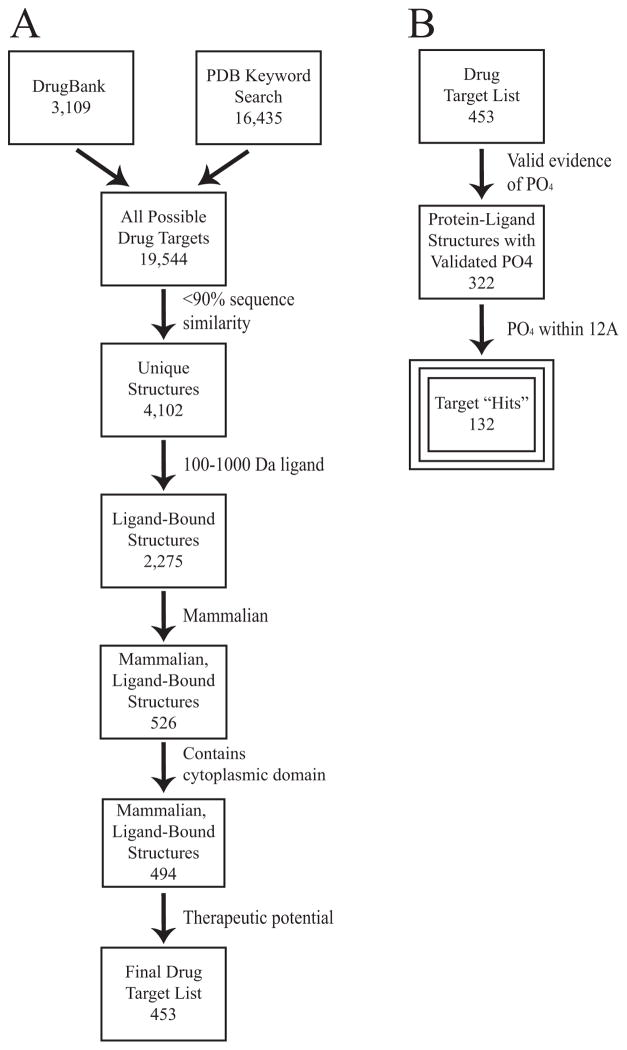
We first generated a comprehensive list of experimental drug targets in both preclinical and clinical studies, by combining those in the DrugBank database with all available PDB structures containing keywords “drug”, “inhibitor”, “agonist”, “antagonist” in the PDB. From this dataset, we removed (1) redundant targets at ≥90% sequence identity, (2) all structures that did not contain a 100–1000 Da ligand, a lenient range for drug-like molecules, (3) all non-mammalian targets, whose species is not covered in PhosphoSitePlus, (4) transmembrane protein structures lacking a cytoplasmic domain, whose extracellular domains may bind to inhibitors but are rarely phosphorylated and (5) off-target ligand-structure complexes and other proteins that do not have any known therapeutic potential based on a literature search [Fig. 1(a)]. 15,442 (79%) of the available drug target structures were redundant with other available structures, due to targets for which multiple ligand structures are available in the PDB. A ligand-bound structure was available for 2275 (55%) of unique targets. By investigating exclusively mammalian targets, we eliminated 1749 drug-bound structures, notably including a number of antibacterial agents. These drug-target interactions certainly may be affected by phosphorylation, but far fewer bacterial phosphorylation sites have been reported as the bacterial Phosphorylation Site Database contains only 2250 entries 14.
Figure 1.
Flow diagrams. Each criterion is listed along with the number of targets meeting it. A: Dataset preparation. B: Identification of the 132 validated hits, as defined in text.
We next cross-referenced the PDB entries for the 453 non-redundant mammalian ligand-bound drug target structures with reported phosphorylation sites in PhosphoSitePlus 4. We investigated serine, threonine, and tyrosine phosphorylation exclusively as histidine phosphorylation is rare and other post-translational modifications are rarely validated beyond mass spectrometry (MS) studies 15,16. Data are discussed in terms of the number of phosphorylated “hits” out of 453.
In the absence of additional data, identification of a phosphorylated site by mass spectrometry alone is generally considered to be insufficient to demonstrate phosphorylation. We consider a phosphorylated site to be “valid” if multiple groups have published evidence of endogenous phosphorylation of the protein target in cells, either by mass spectrometry, biochemical and cell biological studies, or combination of those. By this criterion, 322/453 (71%) of drug targets with structures have valid evidence of phosphorylation, while 131/453 (29%) have no known validated phosphorylation sites [Figure 1(b)].
We define “hits” as ligand-bound structures with validated phosphorylated residues within 12Å of any atom in the small molecule (Table I, Supporting Information Table S1). A 12Å distance cutoff is based on past studies of several phospho-proteins, in which phosphorylation within 12Å of the site of small molecule binding could induce local conformational rearrangements that would affect ligand affinity. For example, Hsp90α is phosphorylated at Thr90, where the hydroxyl is 11.8Å away from a bound inhibitor 17. This phosphorylation causes a decrease in affinity for ATP 18. In cyclin-dependent kinase 2 (Cdk2), phosphorylation 9.5Å away from ATP reduces substrate peptide affinity 2.5 fold 19. Peptide phosphorylation can similarly induce conformational changes on the order of 10Å 20. A much more stringent cutoff is phosphorylation within 3Å of the drug binding site, which would directly affect drug binding through steric and electrostatic interactions, as shown in the case of E. coli isocitrate dehydrogenase and its natural substrate 21. While phosphorylations greater than 12Å away from a drug binding site may induce more global conformational changes within proteins that significantly affect affinity, we cannot easily predict these effects without phosphorylated or phospho-mimetic structures, which are rarely available. As a result, our estimate of effects is likely conservative considering the changes to activity long-range phosphorylation produce 22. The resolution of the crystal structure data for the 453 ligand-bound drug targets in our study ranges from ~1.5Å to ~3Å. Crystal structure resolution does not directly correlate with proper ligand geometry and fit to the electron density 23, therefore, differences in structural resolution were ignored in determining distances between the ligand and phosphorylation site.
TABLE I.
Target “hits” with cellular, biochemical, or clinical evidence of phosphorylation that can be classified by function. Entries are listed in order of class, validation, and stage of development. For validation, an entry of 1 indicates validation of phosphorylation by biochemical studies only, 2 indicates that validation by cellular functional studies, 3 indicates that both biochemical and cellular functional studies have been done to validate the phosphorylation sites, 4 indicates that there is a clinical correlation between phosphorylation of the drug target at a particular site and patient prognosis, and a (*) indicates there are less than 2 MS studies listed on PhosphoSitePlus. Evidence for kinesin-5 phosphorylation is based on our laboratory’s unpublished data.
| Target Protein | Uniprot Sequence ID | Stage of Drug Development | PDB ID | PDB Ligand ID | Phosphorylated Residue(s) | Distance (Å) | Validation, References | Hit Class |
|---|---|---|---|---|---|---|---|---|
| Androgen Receptor (AR) | P10275 | Approved | 1Z95 | 198 | Ser791 | 10.0 | 3, 4* 26,29,30 | Class 1 |
| Liver X receptor beta (LXRβ) | P55055 | Preclinical | 1UPV | 444 | Ser426 | 9.5 | 3 63 | Class 1 |
| SH2 domain containing protein tyrosine phosphatase-2 (SHP2) | Q06124 | Phase I | 3O5X | JZG | Tyr279 | 3.9 | 3 64 | Class 1 |
| Angiopoietin-1 receptor (Tek/Tie2) | Q02763 | Phase II | 3L8P | 0CE | Tyr879 | 9.0 | 3 65 | Class 1 |
| Cyclin-dependent kinase 2 (Cdk2) | P24941 | Phase II | 3LFQ | A28 | Thr14 | 5.8 | 3 66 | Class 1 |
| Cyclin-Dependent Kinase 6 (Cdk6) | Q00534 | Phase III | 4AUA | 4AU | Tyr24 | 9.1 | 3 67 | Class 1 |
| Retinoid X Receptor Alpha (RXRα) | P19793 | Approved | 1G5Y | REA | Tyr249 | 9.4 | 3 68,69 | Class 1 |
| Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (Pin1) | Q13526 | Preclinical | 3TC5 | 3T5 | Ser71 | 5.3 | 3* 70,71 | Class 1 |
| Ectonucleotide pyrophosphatase/phosphodiesterase 2 (ENPP2) | Q9R1E6 | Preclinical | 3WAY | DWY | Thr210 | 2.4 | 3* 72,73 | Class 1 |
| Tyrosine-protein phosphatase non-receptor type 22 (PTPN22) | Q9Y2R2 | Preclinical | 4J51 | N75 | Ser35 | 10.6 | 3* 74 | Class 1 |
| Endothelial PAS domain-containing protein 1 (HIF2a/EPAS1) | Q99814 | Preclinical | 4GHI | 0X3 | Thr324 | 7.6 | 3* 75 | Class 1 |
| Glyoxylase 1 (GLO1) | Q04760 | Preclinical | 3W0T | HPU | Thr107 | 10.5 | 3* 76 | Class 1 |
| Farnesoid X receptor (FXR) | Q96RI1 | Phase II | 3P89 | 89P | Thr456 | 10.6 | 3* 77 | Class 1 |
| Phosphatidylinositol 3-kinase catalytic subunit type 3 (PIK3C3) | Q8NEB9 | Phase II | 3LS8 | AJZ | Ser71 | 6.3 | 3* 78 | Class 1 |
| Neuronal nitricoxide synthase (nNOS) | P29476 | Phase II | 3N5W | XFJ | Tyr609 | 12.0 | 3* 79 | Class 1 |
| Heat Shock Protein 90-alpha (HSP90α) | P07900 | Phase III | 4BQJ | XKL | Ser52, Thr90 | 3.9, 12.7 | 3* 18,80 | Class 1 |
| Janus Kinase 1 (Jak1) | P23458 | Approved | 3EYG | MI1 | Tyr940 | 8.7 | 3* 81 | Class 1 |
| Janus Kinase 2 (Jak2) | O60674 | Approved | 3UGC | 046 | Tyr913 | 7.9 | 3* 81 | Class 1 |
| Mineralocorticoid Receptor (MCR) | P08235 | Approved | 3VHV | LD1 | Ser843 | 6.4 | 3* 28 | Class 1 |
| Cytochrome P450 19A1 (CYP19A1) | P11511 | Approved | 3S7S | EXM | Ser118 | 11.8 | 3* 82 | Class 1 |
| Retinoic acid receptor alpha (RARα) | P10276 | Approved | 4DQM | LUF | Ser219 | 9.9 | 3* 83 | Class 1 |
| Deoxyribonucleoside 5′-monophosphate N-glycosidase (Rcl) | O35820 | Preclinical | 4FYH | TR5 | Ser28 | 3.5 | 1 84 | Class 1 |
| Kinesin-5 (Eg5) | P52732 | Phase II | 3KEN | ZZD | Tyr231 | 3.5, 9.3 | 1 85 | Class 1 |
| Calcium/calmodulin-dependent protein kinase type 4 (CAMK4) | Q16566 | Preclinical | 2W4O | DKI | Tyr940 | 9.3 | 1* 86 | Class 1 |
| Protein Mdm4 (MDMX) | O15151 | Preclinical | 3LBJ | WW8 | Tyr99 | 4.4 | 2 87 | Class 1 |
| Ribosomal protein S6 kinase alpha-1 (Rsk1) | Q15418 | Preclinical | 2Z7R | STU | Ser154 | 9.4 | 2* 88 | Class 1 |
| BCR-Abelson murine leukemia viral oncogene homolog 1 Thr→Ala (BCR-Abl) | P00519 | Approved | 3CS9 | NIL | Tyr253 | 3.7 | 2* 89 | Class 1 |
| Proliferating Cell Nuclear Antigen (PCNA) | P12004 | Preclinical | 3VKX | T3 | Tyr211 | 9.6 | 2, 4* 24,90 | Class 2 |
| Estrogen Receptor α (ERα) | P03372 | Approved | 3ERT | OHT | Tyr537 | 9.9 | 3, 4* 25,35,36 | Class 2 |
| L-Lactate Dehydrogenase A-Chain (LDHA) | P00338 | Preclinical | 4AJJ | 88R | Tyr83 | 4.4 | 3 91 | Class 2 |
| Tyosine Protein Phosphotase Non-Receptor Type 1 (PTP1B Phosphotase) | P18031 | Preclinical | 2CM7 | IZD | Tyr46 | 3.5 | 3 92 | Class 2 |
| Eukaryotic Translation Initiation Factor 4E (eIF4E) | Q13541 | Preclinical | 2V8Y | MGV | Ser209 | 11.1 | 3 93 | Class 2 |
| p-21 Activated Kinase 1 (PAK1) | Q13153 | Preclinical | 4EQC | XR1 | Tyr285 | 5.5 | 3 94 | Class 2 |
| Deoxycytidine kinase (dCK) | P27707 | Preclinical | 4JLK | 1NO | Ser74 | 11.9 | 3 95 | Class 2 |
| Mitogen-activated protein kinase kinase 6 (MAP2K6/Mek6) | P52564 | Preclinical | 3VN9 | ANK | Ser207 | 6.6 | 3 96 | Class 2 |
| Interleukin-2 Inducible Kinase (Itk) | Q08881 | Preclinical | 4HCU | 13L | Tyr512 | 8.9 | 3 97,98 | Class 2 |
| Mitogen-activated kinase 8 (MAPK8/JNK1) | P45983 | Preclinical | 3ELJ | GS7 | Tyr185 | 6.4 | 3 99 | Class 2 |
| Activated CDC42 kinase 1 (TNK2/ACK1) | Q07912 | Preclinical | 3EQR | T74 | Tyr284 | 9.7 | 3 100 | Class 2 |
| Glutathione S-transferase P (GSTP1) | P09211 | Phase II | 3GUS | N11 | Tyr8 | 4.0 | 3 101 | Class 2 |
| Macrophage colony-stimulating factor 1 receptor (CSF1R) | P077333 | Phase II | 3DPK | 8C5 | Tyr809 | 8.9 | 3 102,103 | Class 2 |
| Mitogen-Activated Protein Kinase Kinase (MEK1) | Q02750 | Phase II | 4LMN | EUI | Ser218, Ser222 | 5.9, 10.5 | 3 104,105 | Class 2 |
| BDNF/NT-3 growth factors receptor (TrkB) | Q16620 | Phase II | 4AT5 | MUJ | Tyr706 | 10.3 | 3 106,107 | Class 2 |
| Insulin-like growth factor 1 receptor (IGF1R) | P08069 | Phase II | 3NW7 | LGV | Tyr1161 | 7.1 | 3 108,109 | Class 2 |
| Dual specificity tyrosine-phosphorylation-regulated kinase 1A (DYRK1A) | Q13627 | Phase II | 4MQ1 | 2C3 | Tyr321 | 6.5 | 3 110,111 | Class 2 |
| Hepatocyte growth factor receptor (c-Met) | P08581 | Approved | 3F66 | IHX | Tyr1234 | 10.3 | 3 112 | Class 2 |
| Mast/stem cell growth factor kit (c-Kit) | P10721 | Approved | 1T46 | STI | Tyr823 | 10.8 | 3 113,114 | Class 2 |
| Bruton’s tyrosine kinase (Btk) | Q06187 | Approved | 3OCS | 746 | Tyr551 | 3.7 | 3 115,116 | Class 2 |
| Kelch-like ECH-associated protein 1 (Keap1) | Q14145 | Preclinical | 4IQK | IQK | Ser602 | 2.5 | 3* 117 | Class 2 |
| Co-activator-associated arginine methyltransferase 1 (CARM1) | Q86X55 | Preclinical | 2Y1W | 849 | Ser216 | 3.9 | 3* 118 | Class 2 |
| Mitogen-activated protein kinase 1 (MAPK1/Erk2) | P28482 | Preclinical | 3QYZ | Z8B | Ser29 | 8.4 | 3* 119 | Class 2 |
| Receptor Interacting Protein 1 (RIP1) Kinase | Q13546 | Preclinical | 4NEU | Q1A | Ser161 | 9.5 | 3* 120 | Class 2 |
| PRK-like Endoplasmic Reticulum Kinase (PERK) | Q9NZJ5 | Preclinical | 4G31 | 0WH | Tyr619 | 6.4 | 3* 121 | Class 2 |
| Transforming growth factor-β activated kinase-1 (MAP3K7/TAK1) | O43318 | Preclinical | 4GS6 | 1FM | Ser192 | 9.8 | 3* 122 | Class 2 |
| NF-κβ-inducing Kinase (MAP3K14/NIK) | Q99558 | Preclinical | 4IDT | T28 | Thr559 | 10.2 | 3* 123 | Class 2 |
| Choline Kinase alpha (CHKA) | P35790 | Phase 1 | 4DA5 | 0H7 | Tyr333 | 4.4 | 3* 124 | Class 2 |
| Maternal Embryonic Zipper Kinase (MELK) | Q14680 | Phase 1 | 4BKY | 82B | Ser171 | 7.1 | 3* 125 | Class 2 |
| Polo-like kinase 1 (PLK1) | P53350 | Phase I | 4J52 | 1J3 | Ser137 | 6.5 | 3* 126 | Class 2 |
| 3-Phosphoinositide-dependent protein kinase 1 (PDK1) | O15530 | Phase I | 4AW0 | MJF | Ser160 | 3.0 | 3* 127 | Class 2 |
| Serine/Threonine kinase B-Raf (BRAF) | P15056 | Approved | 4JVG | B96 | Thr599 | 3.6 | 3* 128 | Class 2 |
| Janus Kinase 3 (Jak3) | P52333 | Approved | 3LXL | IZA | Tyr904 | 3.4 | 3* 129 | Class 2 |
| Mitogen-activated protein kinase 11 (MAPK11) | Q15759 | Approved | 3GP0 | NIL | Tyr323 | 8.1 | 3* 130,131 | Class 2 |
| Protein Kinase C iota type (PKCI) | P41743 | Approved | 3ZH8 | C58 | Tyr265 | 2.9 | 3* 132 | Class 2 |
| Insulin Receptor (IR) | P06213 | Approved | 4IBM | IR1 | Ser1033 | 7.1 | 3* 133 | Class 2 |
| Fatty acid-binding protein, adipocyte (FABP4) | P15090 | Preclinical | 3HK1 | B64 | Tyr20 | 3.4 | 1 134 | Class 2 |
| Cell Division Cycle 7-related Kinase (Cdc7) | O00311 | Phase II | 4F9C | 0SX | Thr376 | 9.0 | 1 135 | Class 2 |
| Caspase 1 (Casp1) | P29466 | Preclinical | 1RWX | YBH | Ser376 | 7.0 | 2 136 | Class 2 |
| Mammalian STE20-like Kinase 4 (Mst4) | Q9P289 | Preclinical | 4FZF | DKI | Thr178 | 11.3 | 2* 137 | Class 2 |
| MEK Kinase 5 (MAP3K5/ASK1) | Q99683 | Preclinical | 4BF2 | STU | Thr842 | 9.8 | 2* 138 | Class 2 |
| Protein Kinase B (Akt1) | P31749 | Phase II | 4GV1 | 0XZ | Tyr176 | 6.5 | 2* 139 | Class 2 |
| Proto-oncogene tyrosine-protein kinase Src (c-Src) | P12931 | Approved | 3G5D | 1N1 | Tyr338 | 3.3 | 2 140 | Class 2 |
RESULTS AND DISCUSSION
Nearly one-third of known drug targets are phosphorylated near the drug binding site
Our results show that of the 453 small-molecule bound drug targets in the PDB, 322 (71%) have at least one validated phosphorylation site. It is interesting that the frequency of phosphorylation for drug target proteins appears to significantly exceed the estimated frequency of phosphorylation for all human proteins. This may be because highly phosphorylated proteins in signaling cascades and also make good drug targets, as with kinases. It could also be the case that proteins with measurable signaling activity, such as kinases, are more likely to be selected as drug targets. Those drug target proteins are likely to be more well-studied and validated when compared to other phospho-proteins in general.
132 of the 453 small-molecule bound drug targets in the PDB (29%) contained a validated phosphorylation site within 12Å of the drug binding site. These are the “hits” of our study, which are discussed in detail below (Table I, Supporting Information Table S1). In these tables the highest resolution drug target structures available in the PDB were used, and reported distance is the shortest from the small molecule to the phosphorylated residue. The ligand in the crystal structure is rarely the drug candidate in used clinical trials. Target “hits” are listed in Table I and Supporting Information Table S1 in order of class, validation, and stage of drug development as described below. Table I contains all proteins for which phosphorylation has known effects on target function, and Supporting Information Table S1 contains all other “hits”.
The distribution of the distances between the bound ligand and the phosphorylated residue are shown in Figure 2(a). 9/132 hits contained a site within 3Å of the ligand binding site. There are large bottlenecks between MS verification of phosphorylation by multiple groups and more detailed data such as biochemical and cell based studies, as well as data establishing physiological effects of phosphorylation for the 132 hits [Fig. 2(b)]. Phosphorylation sites reported for 70/132 hits have been accompanied by functional biochemical or cellular studies, with 57/132 having demonstrated both biochemical and cellular functional relevance. To date, only 3 of the 132 hits have established clinical correlation of drug treatment outcome with target protein phosphorylation 24–26. All of these are described in detail below.
Figure 2.
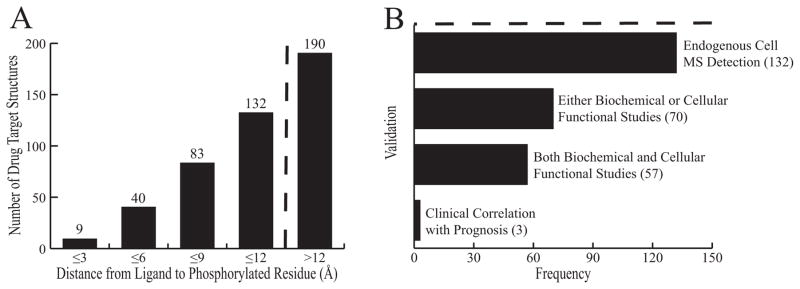
A: Distribution of distance from phosphorylated residue to the target. B: Validation of the 132 phosphorylation sites within 12Å of drug binding sites. The dashed line indicates the cutoff for investigation in our study. Each criterion is listed along with the number of targets meeting it. Each level is inclusive of the one below.
Not surprisingly, protein kinases comprised the largest fraction of our hits. Of the 453 drug target structures, 130 are kinases. 95% (123/130) of those kinase targets are phosphorylated, with 77/130 (59%) of those phosphorylation sites occurring within 12Å of the inhibitor. The fact that phosphorylation occurs in the vast majority of kinase targets is expected given activation loop phosphorylation and involvement in signaling cascades. In addition to protein kinases, nuclear receptors were an interesting group, comprising 25 of the 453 drug target structures. 9 of those 25 had phosphorylation within 12Å of drug binding sites (36%), as phosphorylation has been established as a common mechanism of modulating the activity of these proteins 27, (Supporting Information Fig. S1).
Classification of hits
For 70/453 of the hits, it is known whether phosphorylation activates the target, has little effect, or whether it is inhibitory [Fig. 2(b)]. These 70 hits fell into one of two classes as shown in Figure 3. Class 1 hits (27/70, 39%) have an inhibitory phosphorylation site that is close to the site of drug binding. In this situation, if phosphorylation reduces drug efficacy, it will also inactivate the target by modifying the same site. Regardless of whether the drug binds or the target is phosphorylated such that the drug cannot bind, the target is inactivated. Mineralocorticoid receptor is an example of a Class 1 hit, in which phosphorylation at Ser843 reduces the affinity for the natural agonist and inactivates the receptor 28 [(Fig. 4(a)]. Because phosphorylation occurs at the binding site for both the agonist and inhibitor of mineralcorticoid receptor, we would predict that phosphorylation of Ser843 results in reduced drug affinity. This would not translate to a loss in efficacy, in fact one could expect the opposite effect. If a larger fraction of targets are phosphorylated, then a larger fraction of targets will be inactivated for a particular dose of a drug compound.
Figure 3.
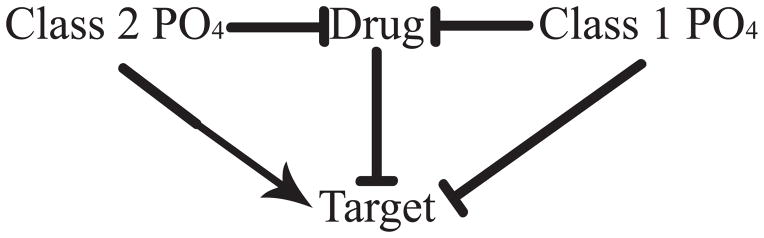
Mechanisms by which phosphorylation may affect drug efficacy. For Class 1 hits, phosphorylation inhibits (⊤) both drug binding and target activity, whereas for Class 2 hits, phosphorylation inhibits drug binding while activating (→) the target. While this schematic depicts phosphorylation inhibiting drug binding, it may in some cases enhance binding as discussed in the text.
Figure 4.
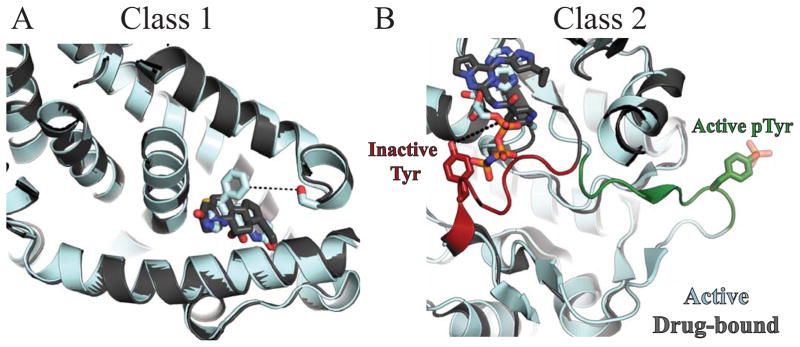
Structural views example Class 1 and Class 2 hits. Active targets are shown in cyan, inactive targets in grey. Small molecule ligands are shown in sticks. A: Overlapped structures of active (2AA2) and inactive (3VHV) forms of Class 1 target mineralocorticoid receptor. When phosphorylated, Ser843 (red) inactivates the receptor by preventing agonist binding. As shown, the inhibitor binds at the same site as the agonist. Inhibitor affinity would likely be reduced by phosphorylation of Ser843 6.4Å away (dashed lines). B: Overlapped structures of active, phopshorylated insulin receptor (1IR3) and inactive insulin-like growth factor 1 receptor (3NW7), a Class 2 target. The two proteins share 91.3% sequence similarity within the crystallized constructs and 100% sequence identity within the activation loop. In the inactive conformation, the activation loop is red. Tyr1161 (sticks) is 7.1Å away from the “DFG-out” inhibitor (dashed lines). In the active, phosphorylated conformation, the activation loop is green. Phosphorylation simultaneously activates insulin-like growth factor 1 receptor and can reduce inhibitor affinity.
Another example of a Class 1 hit is the androgen receptor (AR, Supporting Information Table S1). Approved antiandrogens like bicalutamide act as antagonists to the androgen receptor in hormonal dysfunction diseases such as prostate cancer. AR Ser790 is phosphorylated in vitro and in vivo by Akt 29. Phosphomimetic substitution of Ser790 to aspartate nearly abolishes the ability of AR to bind androgen and localize to the nucleus 30. While some research suggests Akt inhibition/activation of AR is not critical for progression of prostate cancer 31,32, clinical studies indicate that high phosphorylation of Ser790 is associated to a longer time to death from recurrence in castration-resistant prostate cancer 26.
For Class 2 hits (43/70, 61%), phosphorylation does not significantly inhibit target function or in some cases it may actually increase activity. As phosphorylation can reduce drug affinity without inhibiting activity, Class 2 proteins may avoid inhibition by small molecule drugs when phosphorylated. Nearly all kinase hits fall into this category (33/41), including insulin-like growth factor 1 receptor [IGF-1R, Fig. 4(b)]. One study on IGF-1R showed that despite high membrane permeability, lack of efflux transporters, and nanomolar affinity to the unphosphorylated state, inhibitors failed to show efficacy in cell-based assays 33. The inhibitors used in this study were “DFG-out” inhibitors that bind only to the inactive conformation when the DFG loop is out, away from the active site. This causes the activation loop to occlude the nucleotide pocket and prevent ATP binding [Fig. 4(b), dark gray/red structure]. While “DFG-out” inhibitors can keep the protein in an inactive conformation, they do not bind well to the active, phosphorylated enzyme. The phosphorylated crystal structure of IGF-1R shows large rearrangements of the activation loop. The DFG sequence is “in”, while the activation loop itself is out and away from the nucleotide pocket, enabling ATP binding [Fig. 4(b), cyan/green structure, 34]. Therefore it is plausible that in vivo phosphorylation causes a reduction in drug affinity. In the case of the BCR-Abl “DFG-out” inhibitor imatinib, there is a 200-fold reduction in affinity when the kinase is phosphorylated 9.
Among the examples of Class 2 proteins listed in Table I is Estrogen Receptor Alpha (ERα), a well-established drug target in the treatment of hormone-responsive breast cancer. The alpha carbon of tyrosine 537 is 11.3 Å away from the binding site for 4-hydroxytamoxifen. Tyr537 makes several key interactions that stabilize the inactive conformation 35, and is phosphorylated both in vitro and in vivo by Src-family kinases 36. Phosphorylation of Tyr537 activates the ERα receptor, possibly by facilitating dimerization and interactions with other binding partner proteins37,38, although the precise mechanism is disputed 39,40. Phosphomimetic mutation of Tyr537 to glutamate reduces the affinity of estradiol to ERα by 10-fold 41 and results in ligand-independent activation of the receptor 42. Recent clinical evidence associated high levels of Tyr537 phosphorylation, as studied in breast cancer, with poor overall survival of patients treated with tamoxifen 25. Furthermore, it is intriguing that in the vast majority of tamoxifen-resistant, ERα-positive cancers, ERα does not contain any mutations 43,44. These data together suggest that aberrant phosphoregulation of ERα, rather than mutation of the receptor, can cause poor drug response.
The emerging cancer drug target Proliferating Cell Nuclear Antigen (PCNA) also falls under Class 2. When Tyr211 is phosphorylated, PCNA is activated and localized to the nucleus where it is involved in DNA replication/repair and cell cycle progression 45. Because of its function in these proliferative processes, it is being investigated as a drug target and biomarker in cancer 46,47. The hydroxyl group of Tyr211 is 11.4Å away from the inhibitor and makes several interactions with a flexible loop 48. This loop moves to accommodate the inhibitor and phosphorylation would likely alter PCNA inhibitor binding. Clinically, phosphorylation at Tyr211 is correlated with poor survival in breast cancer patients 24, and blocking Tyr211 phosphorylation using PCNA peptide mimetics reduces tumor growth in vivo 49. These data together suggest that for Class 2 hits, including kinase targets of “DFG-out” inhibitors, ERα, and PNCA, phosphorylation of the target protein may block the action and efficacy of drug inhibitors.
CONCLUSIONS
In this comprehensive bioinformatics study, we found that 132/453, or 29% of proteins analyzed in this study are known to have phosphorylation occurring within 12 Å of a drug binding site. For 70 of the 132 phosphorylated targets, it is known whether phosphorylation activates or inhibits the target. 39% (27/70) of these were classified as Class 1 hits, for which the drug and phosphorylation have similar effects on activity, while 61% (43/70) were Class 2 hits, for which the drug and phosphorylation have opposing effects on activity. These results suggest that phosphorylation can alter drug efficacy for a rather large fraction of target proteins. Kinases and nuclear receptors represented a large fraction of the hits in our study, and some of the reason for their high representation is likely selection bias in the available data. For example numerous studies of kinase activation by phosphorylation in or near the active site have led to inhibitor design, as in the case of the “DFG-out” inhibitors described above.
Clinical studies examining the effects of specific target phosphorylation sites on specific therapies were only performed for 3 drug target proteins in our study. For the Class 1 hit AR, phosphorylation near the drug binding site was correlated with good outcome/drug sensitivity. In contrast, for the Class 2 hits ERα and PCNA, phosphorylation near the drug binding site was correlated with poor sensitivity to the drug. Our speculation is that in the case of Class 1 hits, a large fraction of the target proteins may be inactivated via phosphorylation, such that a lower concentration of drug may be required to cause sufficient inhibition of the remaining active targets. In Class 2 hits, phosphorylation of the target protein can directly cause drug resistance in the absence of any target protein mutations.
Our model in Figure 3 depicts phosphorylation either directly or allosterically inhibiting drug binding. Phosphorylation should inhibit drug binding if it causes a direct structural clash with the drug in its binding site or causes unfavorable structural rearrangements. For the three proteins with known clinical effects of phosphorylation described above, phosphorylation does appear to reduce drug efficacy, consistent with inhibition of drug binding. However, for the majority of our hits (127/132), it is not known whether phosphorylation inhibits or enhances drug binding, and examples of phosphorylation enhancing drug binding do exist. In phosphodiesterase-5, phosphorylation of Ser102 increases the enzymatic activity 1.6-fold 50. Through allosteric rearrangements, phosphorylation increases the affinity of cGMP to PDE5, causing activation50,51. The inhibitor tadalafil is able to overcome this by showing a 3-fold increase in binding affinity when Ser102 is phosphorylated 52. We did not identify PDE5 in our screen because the domain containing Ser102 has not been crystallized in an inhibitor-bound structure. In another example, phosphorylation of phenylalanine dehydroxylase (PAH) at Ser16 increased substrate-dependent enzymatic activation 53. The site is in a disordered region of the protein and there is no electron density present in crystal structure. Small-molecule activators of PAH are being pursued for treatment of phenylketonuria (PKU). While both phosphorylation54 and drug candidates55 stabilize protein folding and maintain the activity of PAH, the combined effects have yet to be investigated and are difficult to determine through crystallography.
Our study is likely to have under-reported the phenomenon of phosphorylation affecting drug inhibition of targets, for several reasons. First, because drug-bound structures are often kept confidential during drug development while phosphorylation sites are continually being identified and verified, we are under-reporting the actual occurrence of phosphorylated, drug-bound proteins. Second, other post-translational modifications like acetylation are reported to have critical roles in both upregulating56 and downregulating57 activity, but are rarely investigated to the extent that phosphorylation is. Third, we were unable to assess targets with phosphorylation in unstructured domains. Extremely flexible protein regions are commonly phosphorylated but are rarely seen in crystal structures. Fourth, given the difficulties of predicting long-distance allosteric effects of phosphorylation on protein activity or drug binding, we did not assess the 131/453 drug target proteins with phosphorylation sites that were not within 12 Å of the drug binding site in the crystal structures. Some of these long-range phosphorylation sites even have known physiological effects. For example, phosphorylation of PPAR-γ at Ser112 reduces drug agonist binding by 10-fold despite being nearly 40 Å away 58. In addition to the site at Tyr537 described above, ERα also contains a long distance phosphorylation site that affects antagonist binding. When ERα is phosphorylated at Ser305, tamoxifen still binds, but fails to induce an inactive conformation. Based on intramolecular FRET experiments, it appears phosphorylation of Ser305 causes tamoxifen to exhibit agonist behavior towards ERα, leading to tamoxifen resistance 59. This critical conformational rearrangement is not reflected in published crystal structures, most of which contain only the ligand-binding domain 60. Lastly, it should also be noted that phosphorylation sites are enriched for mutations in cancer cells 61. The effects of these mutations on target protein phospho-regulation are highly clinically relevant, but they are not reported in PhosphoSitePlus, and hence are not included in our study.
In target-based drug discovery, target proteins are often initially produced and assayed in vitro or in cultured cells where the set of post-translational modifications occurring on the target protein could be vastly different than in vivo. Similarly, structure-based in silico efforts can only seldom investigate the structural changes induced by phosphorylation because phosphorylated structures are rarely available (1,475/97,180 structures in the PDB, 2%). Except in the case of kinase auto-phosphorylation, structural information about these phospho-proteins is extremely limited and can require partial chemical synthesis to achieve a high percentage of phosphorylated protein 62. Given the findings of this study, we encourage researchers and clinicians alike to consider the possibility of protein post-translational modification as they approach rational drug design and evaluate potential reasons for drug treatment outcomes that are either much better or worse than expected.
Supplementary Material
Acknowledgments
We thank J. Klosowiak for discussions and assistance. K.P.S. is funded by NIGMS T32GM008382, J.S.W. is funded by the Myhrvold Family Fellowship from the Fannie and John Hertz Foundation, K.G.M. and S.E.R. are funded by NIH R01GM107209.
Footnotes
Research Conducted at:
Department of Cell & Molecular Biology, Northwestern University Feinberg School of Medicine
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
References
- 1.UniProt C. Activities at the Universal Protein Resource (UniProt) Nucleic acids research. 2014;42(Database issue):D191–198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gnad F, Gunawardena J, Mann M. PHOSIDA 2011: the posttranslational modification database. Nucleic acids research. 2011;39(Database issue):D253–260. doi: 10.1093/nar/gkq1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dinkel H, Chica C, Via A, Gould CM, Jensen LJ, Gibson TJ, Diella F. Phospho. ELM: a database of phosphorylation sites--update 2011. Nucleic acids research. 2011;39(Database issue):D261–267. doi: 10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic acids research. 2012;40(Database issue):D261–270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loyet KM, Stults JT, Arnott D. Mass spectrometric contributions to the practice of phosphorylation site mapping through 2003: a literature review. Molecular & cellular proteomics : MCP. 2005;4(3):235–245. doi: 10.1074/mcp.R400011-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4(6):1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 7.Johnson LN, Noble ME, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85(2):149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 8.Friday BB, Yu C, Dy GK, Smith PD, Wang L, Thibodeau SN, Adjei AA. BRAF V600E disrupts AZD6244-induced abrogation of negative feedback pathways between extracellular signal-regulated kinase and Raf proteins. Cancer Res. 2008;68(15):6145–6153. doi: 10.1158/0008-5472.CAN-08-1430. [DOI] [PubMed] [Google Scholar]
- 9.Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289(5486):1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nature reviews Cancer. 2009;9(1):28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- 11.Lee Rosen PL, Ma Wen Wee, Goldman Jonathon, Weise Amy, Dimitrios Colevas A, Adjei Alex, Yazji Salem, Shen Angela, Johnston Stuart, Gates Mary R, Jones Cheryl, Musib Luna, De Crespigny Alex, Chan Iris, Sikic Branimir. A first-in-human phase 1 study to evaluate the MEK1/2 inhibitor GDC-0973 administered daily in patients with advanced solid tumors. Cancer Research. 2011;71(8) Supplement 1 [Google Scholar]
- 12.Hatzivassiliou G, Haling JR, Chen H, Song K, Price S, Heald R, Hewitt JF, Zak M, Peck A, Orr C, Merchant M, Hoeflich KP, Chan J, Luoh SM, Anderson DJ, Ludlam MJ, Wiesmann C, Ultsch M, Friedman LS, Malek S, Belvin M. Mechanism of MEK inhibition determines efficacy in mutant KRAS- versus BRAF-driven cancers. Nature. 2013;501(7466):232–236. doi: 10.1038/nature12441. [DOI] [PubMed] [Google Scholar]
- 13.Sams-Dodd F. Target-based drug discovery: is something wrong? Drug discovery today. 2005;10(2):139–147. doi: 10.1016/S1359-6446(04)03316-1. [DOI] [PubMed] [Google Scholar]
- 14.Wurgler-Murphy SM, King DM, Kennelly PJ. The Phosphorylation Site Database: A guide to the serine-, threonine-, and/or tyrosine-phosphorylated proteins in prokaryotic organisms. Proteomics. 2004;4(6):1562–1570. doi: 10.1002/pmic.200300711. [DOI] [PubMed] [Google Scholar]
- 15.Puttick J, Baker EN, Delbaere LT. Histidine phosphorylation in biological systems. Biochimica et biophysica acta. 2008;1784(1):100–105. doi: 10.1016/j.bbapap.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 16.Seo J, Lee KJ. Post-translational modifications and their biological functions: proteomic analysis and systematic approaches. Journal of biochemistry and molecular biology. 2004;37(1):35–44. doi: 10.5483/bmbrep.2004.37.1.035. [DOI] [PubMed] [Google Scholar]
- 17.Kreusch A, Han S, Brinker A, Zhou V, Choi HS, He Y, Lesley SA, Caldwell J, Gu XJ. Crystal structures of human HSP90alpha-complexed with dihydroxyphenylpyrazoles. Bioorganic & medicinal chemistry letters. 2005;15(5):1475–1478. doi: 10.1016/j.bmcl.2004.12.087. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Lu XA, Song X, Zhuo W, Jia L, Jiang Y, Luo Y. Thr90 phosphorylation of Hsp90alpha by protein kinase A regulates its chaperone machinery. The Biochemical journal. 2012;441(1):387–397. doi: 10.1042/BJ20110855. [DOI] [PubMed] [Google Scholar]
- 19.Welburn JP, Tucker JA, Johnson T, Lindert L, Morgan M, Willis A, Noble ME, Endicott JA. How tyrosine 15 phosphorylation inhibits the activity of cyclin-dependent kinase 2-cyclin A. The Journal of biological chemistry. 2007;282(5):3173–3181. doi: 10.1074/jbc.M609151200. [DOI] [PubMed] [Google Scholar]
- 20.Sahoo H, Nau WM. Phosphorylation-induced conformational changes in short peptides probed by fluorescence resonance energy transfer in the 10A domain. Chembiochem : a European journal of chemical biology. 2007;8(5):567–573. doi: 10.1002/cbic.200600466. [DOI] [PubMed] [Google Scholar]
- 21.Hurley JH, Dean AM, Sohl JL, Koshland DE, Jr, Stroud RM. Regulation of an enzyme by phosphorylation at the active site. Science. 1990;249(4972):1012–1016. doi: 10.1126/science.2204109. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Oh H, Burton RA, Burgner JW, Geahlen RL, Post CB. Tyr130 phosphorylation triggers Syk release from antigen receptor by long-distance conformational uncoupling. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(33):11760–11765. doi: 10.1073/pnas.0708583105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liebeschuetz J, Hennemann J, Olsson T, Groom CR. The good, the bad and the twisted: a survey of ligand geometry in protein crystal structures. Journal of computer-aided molecular design. 2012;26(2):169–183. doi: 10.1007/s10822-011-9538-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang SC, Nakajima Y, Yu YL, Xia W, Chen CT, Yang CC, McIntush EW, Li LY, Hawke DH, Kobayashi R, Hung MC. Tyrosine phosphorylation controls PCNA function through protein stability. Nature cell biology. 2006;8(12):1359–1368. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- 25.Skliris GP, Nugent Z, Watson PH, Murphy LC. Estrogen receptor alpha phosphorylated at tyrosine 537 is associated with poor clinical outcome in breast cancer patients treated with tamoxifen. Hormones & cancer. 2010;1(4):215–221. doi: 10.1007/s12672-010-0049-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCall P, Adams CE, Willder JM, Bennett L, Qayyum T, Orange C, Underwood MA, Edwards J. Androgen receptor phosphorylation at serine 308 and serine 791 predicts enhanced survival in castrate resistant prostate cancer patients. International journal of molecular sciences. 2013;14(8):16656–16671. doi: 10.3390/ijms140816656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao D, Lazar MA. Modulating nuclear receptor function: may the phos be with you. The Journal of clinical investigation. 1999;103(12):1617–1618. doi: 10.1172/JCI7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shibata S, Rinehart J, Zhang J, Moeckel G, Castaneda-Bueno M, Stiegler AL, Boggon TJ, Gamba G, Lifton RP. Mineralocorticoid receptor phosphorylation regulates ligand binding and renal response to volume depletion and hyperkalemia. Cell metabolism. 2013;18(5):660–671. doi: 10.1016/j.cmet.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin HK, Yeh S, Kang HY, Chang C. Akt suppresses androgen-induced apoptosis by phosphorylating and inhibiting androgen receptor. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(13):7200–7205. doi: 10.1073/pnas.121173298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palazzolo I, Burnett BG, Young JE, Brenne PL, La Spada AR, Fischbeck KH, Howell BW, Pennuto M. Akt blocks ligand binding and protects against expanded polyglutamine androgen receptor toxicity. Human molecular genetics. 2007;16(13):1593–1603. doi: 10.1093/hmg/ddm109. [DOI] [PubMed] [Google Scholar]
- 31.Lin HK, Hu YC, Yang L, Altuwaijri S, Chen YT, Kang HY, Chang C. Suppression versus induction of androgen receptor functions by the phosphatidylinositol 3-kinase/Akt pathway in prostate cancer LNCaP cells with different passage numbers. The Journal of biological chemistry. 2003;278(51):50902–50907. doi: 10.1074/jbc.M300676200. [DOI] [PubMed] [Google Scholar]
- 32.Xin L, Teitell MA, Lawson DA, Kwon A, Mellinghoff IK, Witte ON. Progression of prostate cancer by synergy of AKT with genotropic and nongenotropic actions of the androgen receptor. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(20):7789–7794. doi: 10.1073/pnas.0602567103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sampognaro AJ, Wittman MD, Carboni JM, Chang C, Greer AF, Hurlburt WW, Sack JS, Vyas DM. Proline isosteres in a series of 2,4-disubstituted pyrrolo[1,2-f][1,2,4]triazine inhibitors of IGF-1R kinase and IR kinase. Bioorganic & medicinal chemistry letters. 2010;20(17):5027–5030. doi: 10.1016/j.bmcl.2010.07.045. [DOI] [PubMed] [Google Scholar]
- 34.Nemecek C, Metz WA, Wentzler S, Ding FX, Venot C, Souaille C, Dagallier A, Maignan S, Guilloteau JP, Bernard F, Henry A, Grapinet S, Lesuisse D. Design of potent IGF1-R inhibitors related to bis-azaindoles. Chemical biology & drug design. 2010;76(2):100–106. doi: 10.1111/j.1747-0285.2010.00991.x. [DOI] [PubMed] [Google Scholar]
- 35.Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95(7):927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 36.Arnold SF, Obourn JD, Jaffe H, Notides AC. Phosphorylation of the human estrogen receptor on tyrosine 537 in vivo and by src family tyrosine kinases in vitro. Molecular endocrinology. 1995;9(1):24–33. doi: 10.1210/mend.9.1.7539106. [DOI] [PubMed] [Google Scholar]
- 37.Migliaccio A, Di Domenico M, Castoria G, de Falco A, Bontempo P, Nola E, Auricchio F. Tyrosine kinase/p21ras/MAP-kinase pathway activation by estradiol-receptor complex in MCF-7 cells. The EMBO journal. 1996;15(6):1292–1300. [PMC free article] [PubMed] [Google Scholar]
- 38.Arnold SF, Vorojeikina DP, Notides AC. Phosphorylation of tyrosine 537 on the human estrogen receptor is required for binding to an estrogen response element. The Journal of biological chemistry. 1995;270(50):30205–30212. doi: 10.1074/jbc.270.50.30205. [DOI] [PubMed] [Google Scholar]
- 39.Weis KE, Ekena K, Thomas JA, Lazennec G, Katzenellenbogen BS. Constitutively active human estrogen receptors containing amino acid substitutions for tyrosine 537 in the receptor protein. Molecular endocrinology. 1996;10(11):1388–1398. doi: 10.1210/mend.10.11.8923465. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen HD, Phan TT, Carraz M, Brunsveld L. Estrogen receptor alpha/beta-cofactor motif interactions; interplay of tyrosine 537/488 phosphorylation and LXXLL motifs. Molecular bioSystems. 2012;8(12):3134–3141. doi: 10.1039/c2mb25257k. [DOI] [PubMed] [Google Scholar]
- 41.Zhong L, Skafar DF. Mutations of tyrosine 537 in the human estrogen receptor-alpha selectively alter the receptor’s affinity for estradiol and the kinetics of the interaction. Biochemistry. 2002;41(13):4209–4217. doi: 10.1021/bi0121095. [DOI] [PubMed] [Google Scholar]
- 42.White R, Sjoberg M, Kalkhoven E, Parker MG. Ligand-independent activation of the oestrogen receptor by mutation of a conserved tyrosine. The EMBO journal. 1997;16(6):1427–1435. doi: 10.1093/emboj/16.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karnik PS, Kulkarni S, Liu XP, Budd GT, Bukowski RM. Estrogen receptor mutations in tamoxifen-resistant breast cancer. Cancer Res. 1994;54(2):349–353. [PubMed] [Google Scholar]
- 44.Roodi N, Bailey LR, Kao WY, Verrier CS, Yee CJ, Dupont WD, Parl FF. Estrogen receptor gene analysis in estrogen receptor-positive and receptor-negative primary breast cancer. J Natl Cancer Inst. 1995;87(6):446–451. doi: 10.1093/jnci/87.6.446. [DOI] [PubMed] [Google Scholar]
- 45.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. Journal of cell science. 2003;116(Pt 15):3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 46.Stoimenov I, Helleday T. PCNA on the crossroad of cancer. Biochemical Society transactions. 2009;37(Pt 3):605–613. doi: 10.1042/BST0370605. [DOI] [PubMed] [Google Scholar]
- 47.Malkas LH, Herbert BS, Abdel-Aziz W, Dobrolecki LE, Liu Y, Agarwal B, Hoelz D, Badve S, Schnaper L, Arnold RJ, Mechref Y, Novotny MV, Loehrer P, Goulet RJ, Hickey RJ. A cancer-associated PCNA expressed in breast cancer has implications as a potential biomarker. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(51):19472–19477. doi: 10.1073/pnas.0604614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Punchihewa C, Inoue A, Hishiki A, Fujikawa Y, Connelly M, Evison B, Shao Y, Heath R, Kuraoka I, Rodrigues P, Hashimoto H, Kawanishi M, Sato M, Yagi T, Fujii N. Identification of small molecule proliferating cell nuclear antigen (PCNA) inhibitor that disrupts interactions with PIP-box proteins and inhibits DNA replication. The Journal of biological chemistry. 2012;287(17):14289–14300. doi: 10.1074/jbc.M112.353201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao H, Lo YH, Ma L, Waltz SE, Gray JK, Hung MC, Wang SC. Targeting tyrosine phosphorylation of PCNA inhibits prostate cancer growth. Molecular cancer therapeutics. 2011;10(1):29–36. doi: 10.1158/1535-7163.MCT-10-0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corbin JD, Turko IV, Beasley A, Francis SH. Phosphorylation of phosphodiesterase-5 by cyclic nucleotide-dependent protein kinase alters its catalytic and allosteric cGMP-binding activities. European journal of biochemistry / FEBS. 2000;267(9):2760–2767. doi: 10.1046/j.1432-1327.2000.01297.x. [DOI] [PubMed] [Google Scholar]
- 51.Wang H, Robinson H, Ke H. Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain. The Journal of biological chemistry. 2010;285(49):38149–38156. doi: 10.1074/jbc.M110.141614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bessay EP, Blount MA, Zoraghi R, Beasley A, Grimes KA, Francis SH, Corbin JD. Phosphorylation increases affinity of the phosphodiesterase-5 catalytic site for tadalafil. The Journal of pharmacology and experimental therapeutics. 2008;325(1):62–68. doi: 10.1124/jpet.107.133405. [DOI] [PubMed] [Google Scholar]
- 53.Doskeland AP, Martinez A, Knappskog PM, Flatmark T. Phosphorylation of recombinant human phenylalanine hydroxylase: effect on catalytic activity, substrate activation and protection against non-specific cleavage of the fusion protein by restriction protease. The Biochemical journal. 1996;313 (Pt 2):409–414. doi: 10.1042/bj3130409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miranda FF, Teigen K, Thorolfsson M, Svebak RM, Knappskog PM, Flatmark T, Martinez A. Phosphorylation and mutations of Ser(16) in human phenylalanine hydroxylase. Kinetic and structural effects. The Journal of biological chemistry. 2002;277(43):40937–40943. doi: 10.1074/jbc.M112197200. [DOI] [PubMed] [Google Scholar]
- 55.Pey AL, Ying M, Cremades N, Velazquez-Campoy A, Scherer T, Thony B, Sancho J, Martinez A. Identification of pharmacological chaperones as potential therapeutic agents to treat phenylketonuria. The Journal of clinical investigation. 2008;118(8):2858–2867. doi: 10.1172/JCI34355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133(4):612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mukherjee S, Keitany G, Li Y, Wang Y, Ball HL, Goldsmith EJ, Orth K. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312(5777):1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- 58.Shao D, Rangwala SM, Bailey ST, Krakow SL, Reginato MJ, Lazar MA. Interdomain communication regulating ligand binding by PPAR-gamma. Nature. 1998;396(6709):377–380. doi: 10.1038/24634. [DOI] [PubMed] [Google Scholar]
- 59.Michalides R, Griekspoor A, Balkenende A, Verwoerd D, Janssen L, Jalink K, Floore A, Velds A, van’t Veer L, Neefjes J. Tamoxifen resistance by a conformational arrest of the estrogen receptor alpha after PKA activation in breast cancer. Cancer cell. 2004;5(6):597–605. doi: 10.1016/j.ccr.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 60.Bapat AR, Frail DE. Full-length estrogen receptor alpha and its ligand-binding domain adopt different conformations upon binding ligand. The Journal of steroid biochemistry and molecular biology. 2003;86(2):143–149. doi: 10.1016/s0960-0760(03)00262-0. [DOI] [PubMed] [Google Scholar]
- 61.Radivojac P, Baenziger PH, Kann MG, Mort ME, Hahn MW, Mooney SD. Gain and loss of phosphorylation sites in human cancer. Bioinformatics. 2008;24(16):i241–247. doi: 10.1093/bioinformatics/btn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mocklinghoff S, Rose R, Carraz M, Visser A, Ottmann C, Brunsveld L. Synthesis and crystal structure of a phosphorylated estrogen receptor ligand binding domain. Chembiochem : a European journal of chemical biology. 2010;11(16):2251–2254. doi: 10.1002/cbic.201000532. [DOI] [PubMed] [Google Scholar]
- 63.Huang W, Ghisletti S, Saijo K, Gandhi M, Aouadi M, Tesz GJ, Zhang DX, Yao J, Czech MP, Goode BL, Rosenfeld MG, Glass CK. Coronin 2A mediates actin-dependent de-repression of inflammatory response genes. Nature. 2011;470(7334):414–418. doi: 10.1038/nature09703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mitra S, Beach C, Feng GS, Plattner R. SHP-2 is a novel target of Abl kinases during cell proliferation. Journal of cell science. 2008;121(Pt 20):3335–3346. doi: 10.1242/jcs.035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shewchuk LM, Hassell AM, Ellis B, Holmes WD, Davis R, Horne EL, Kadwell SH, McKee DD, Moore JT. Structure of the Tie2 RTK domain: self-inhibition by the nucleotide binding loop, activation loop, and C-terminal tail. Structure. 2000;8(11):1105–1113. doi: 10.1016/s0969-2126(00)00516-5. [DOI] [PubMed] [Google Scholar]
- 66.Gu Y, Rosenblatt J, Morgan DO. Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. The EMBO journal. 1992;11(11):3995–4005. doi: 10.1002/j.1460-2075.1992.tb05493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bertero T, Gastaldi C, Bourget-Ponzio I, Mari B, Meneguzzi G, Barbry P, Ponzio G, Rezzonico R. CDC25A targeting by miR-483-3p decreases CCND-CDK4/6 assembly and contributes to cell cycle arrest. Cell death and differentiation. 2013;20(6):800–811. doi: 10.1038/cdd.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee HY, Suh YA, Robinson MJ, Clifford JL, Hong WK, Woodgett JR, Cobb MH, Mangelsdorf DJ, Kurie JM. Stress pathway activation induces phosphorylation of retinoid X receptor. The Journal of biological chemistry. 2000;275(41):32193–32199. doi: 10.1074/jbc.M005490200. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Z, Kovalenko P, Cui M, Desmet M, Clinton SK, Fleet JC. Constitutive activation of the mitogen-activated protein kinase pathway impairs vitamin D signaling in human prostate epithelial cells. Journal of cellular physiology. 2010;224(2):433–442. doi: 10.1002/jcp.22139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee TH, Chen CH, Suizu F, Huang P, Schiene-Fischer C, Daum S, Zhang YJ, Goate A, Chen RH, Zhou XZ, Lu KP. Death-associated protein kinase 1 phosphorylates Pin1 and inhibits its prolyl isomerase activity and cellular function. Molecular cell. 2011;42(2):147–159. doi: 10.1016/j.molcel.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nechama M, Lin CL, Richter JD. An unusual two-step control of CPEB destruction by Pin1. Molecular and cellular biology. 2013;33(1):48–58. doi: 10.1128/MCB.00904-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Clair T, Lee HY, Liotta LA, Stracke ML. Autotaxin is an exoenzyme possessing 5′-nucleotide phosphodiesterase/ATP pyrophosphatase and ATPase activities. The Journal of biological chemistry. 1997;272(2):996–1001. doi: 10.1074/jbc.272.2.996. [DOI] [PubMed] [Google Scholar]
- 73.Lee HY, Clair T, Mulvaney PT, Woodhouse EC, Aznavoorian S, Liotta LA, Stracke ML. Stimulation of tumor cell motility linked to phosphodiesterase catalytic site of autotaxin. The Journal of biological chemistry. 1996;271(40):24408–24412. doi: 10.1074/jbc.271.40.24408. [DOI] [PubMed] [Google Scholar]
- 74.Yu X, Sun JP, He Y, Guo X, Liu S, Zhou B, Hudmon A, Zhang ZY. Structure, inhibitor, and regulatory mechanism of Lyp, a lymphoid-specific tyrosine phosphatase implicated in autoimmune diseases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(50):19767–19772. doi: 10.1073/pnas.0706233104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.To KK, Sedelnikova OA, Samons M, Bonner WM, Huang LE. The phosphorylation status of PAS-B distinguishes HIF-1alpha from HIF-2alpha in NBS1 repression. The EMBO journal. 2006;25(20):4784–4794. doi: 10.1038/sj.emboj.7601369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Hemptinne V, Rondas D, Toepoel M, Vancompernolle K. Phosphorylation on Thr-106 and NO-modification of glyoxalase I suppress the TNF-induced transcriptional activity of NF-kappaB. Molecular and cellular biochemistry. 2009;325(1–2):169–178. doi: 10.1007/s11010-009-0031-7. [DOI] [PubMed] [Google Scholar]
- 77.Frankenberg T, Miloh T, Chen FY, Ananthanarayanan M, Sun AQ, Balasubramaniyan N, Arias I, Setchell KD, Suchy FJ, Shneider BL. The membrane protein ATPase class I type 8B member 1 signals through protein kinase C zeta to activate the farnesoid X receptor. Hepatology. 2008;48(6):1896–1905. doi: 10.1002/hep.22431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Furuya T, Kim M, Lipinski M, Li J, Kim D, Lu T, Shen Y, Rameh L, Yankner B, Tsai LH, Yuan J. Negative regulation of Vps34 by Cdk mediated phosphorylation. Molecular cell. 2010;38(4):500–511. doi: 10.1016/j.molcel.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cordelier P, Esteve JP, Najib S, Moroder L, Vaysse N, Pradayrol L, Susini C, Buscail L. Regulation of neuronal nitric-oxide synthase activity by somatostatin analogs following SST5 somatostatin receptor activation. The Journal of biological chemistry. 2006;281(28):19156–19171. doi: 10.1074/jbc.M602024200. [DOI] [PubMed] [Google Scholar]
- 80.Wang X, Song X, Zhuo W, Fu Y, Shi H, Liang Y, Tong M, Chang G, Luo Y. The regulatory mechanism of Hsp90alpha secretion and its function in tumor malignancy. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(50):21288–21293. doi: 10.1073/pnas.0908151106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Funakoshi-Tago M, Tago K, Kasahara T, Parganas E, Ihle JN. Negative regulation of Jak2 by its auto-phosphorylation at tyrosine 913 via the Epo signaling pathway. Cellular signalling. 2008;20(11):1995–2001. doi: 10.1016/j.cellsig.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 82.Miller TW, Shin I, Kagawa N, Evans DB, Waterman MR, Arteaga CL. Aromatase is phosphorylated in situ at serine-118. The Journal of steroid biochemistry and molecular biology. 2008;112(1–3):95–101. doi: 10.1016/j.jsbmb.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Santos NC, Kim KH. Activity of retinoic acid receptor-alpha is directly regulated at its protein kinase A sites in response to follicle-stimulating hormone signaling. Endocrinology. 2010;151(5):2361–2372. doi: 10.1210/en.2009-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dupouy C, Zhang C, Padilla A, Pochet S, Kaminski PA. Probing the active site of the deoxynucleotide N-hydrolase Rcl encoded by the rat gene c6orf108. The Journal of biological chemistry. 2010;285(53):41806–41814. doi: 10.1074/jbc.M110.181594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gifford K, Waitzman JS, Poor TA, Mann B, Gonzalez MC, Wadsworth P, Rice SE. Src phosphorylation regulates the human kinesin-5, Eg5, and disrupts the binding of Eg5 inhibitors. Unpublished data presented at the 2014 Biophysical Society Annual Meeting; (poster #3930) [Google Scholar]
- 86.Kitani T, Okuno S, Fujisawa H. Studies on the site of phosphorylation of Ca2+/calmodulin-dependent protein kinase (CaM-kinase) IV by CaM-kinase kinase. Journal of biochemistry. 1997;121(4):804–810. doi: 10.1093/oxfordjournals.jbchem.a021656. [DOI] [PubMed] [Google Scholar]
- 87.Zuckerman V, Lenos K, Popowicz GM, Silberman I, Grossman T, Marine JC, Holak TA, Jochemsen AG, Haupt Y. c-Abl phosphorylates Hdmx and regulates its interaction with p53. The Journal of biological chemistry. 2009;284(6):4031–4039. doi: 10.1074/jbc.M809211200. [DOI] [PubMed] [Google Scholar]
- 88.Cavet ME, Lehoux S, Berk BC. 14-3-3beta is a p90 ribosomal S6 kinase (RSK) isoform 1-binding protein that negatively regulates RSK kinase activity. The Journal of biological chemistry. 2003;278(20):18376–18383. doi: 10.1074/jbc.M208475200. [DOI] [PubMed] [Google Scholar]
- 89.Skaggs BJ, Gorre ME, Ryvkin A, Burgess MR, Xie Y, Han Y, Komisopoulou E, Brown LM, Loo JA, Landaw EM, Sawyers CL, Graeber TG. Phosphorylation of the ATP-binding loop directs oncogenicity of drug-resistant BCR-ABL mutants. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(51):19466–19471. doi: 10.1073/pnas.0609239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lo YH, Ho PC, Wang SC. Epidermal growth factor receptor protects proliferating cell nuclear antigen from cullin 4A protein-mediated proteolysis. The Journal of biological chemistry. 2012;287(32):27148–27157. doi: 10.1074/jbc.M112.388843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fan J, Hitosugi T, Chung TW, Xie J, Ge Q, Gu TL, Polakiewicz RD, Chen GZ, Boggon TJ, Lonial S, Khuri FR, Kang S, Chen J. Tyrosine phosphorylation of lactate dehydrogenase A is important for NADH/NAD(+) redox homeostasis in cancer cells. Molecular and cellular biology. 2011;31(24):4938–4950. doi: 10.1128/MCB.06120-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Boubekeur S, Boute N, Pagesy P, Zilberfarb V, Christeff N, Issad T. A new highly efficient substrate-trapping mutant of protein tyrosine phosphatase 1B (PTP1B) reveals full autoactivation of the insulin receptor precursor. The Journal of biological chemistry. 2011;286(22):19373–19380. doi: 10.1074/jbc.M111.222984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Whalen SG, Gingras AC, Amankwa L, Mader S, Branton PE, Aebersold R, Sonenberg N. Phosphorylation of eIF-4E on serine 209 by protein kinase C is inhibited by the translational repressors, 4E-binding proteins. The Journal of biological chemistry. 1996;271(20):11831–11837. doi: 10.1074/jbc.271.20.11831. [DOI] [PubMed] [Google Scholar]
- 94.Rider L, Shatrova A, Feener EP, Webb L, Diakonova M. JAK2 tyrosine kinase phosphorylates PAK1 and regulates PAK1 activity and functions. The Journal of biological chemistry. 2007;282(42):30985–30996. doi: 10.1074/jbc.M701794200. [DOI] [PubMed] [Google Scholar]
- 95.Smal C, Vertommen D, Bertrand L, Ntamashimikiro S, Rider MH, Van Den Neste E, Bontemps F. Identification of in vivo phosphorylation sites on human deoxycytidine kinase. Role of Ser-74 in the control of enzyme activity. The Journal of biological chemistry. 2006;281(8):4887–4893. doi: 10.1074/jbc.M512129200. [DOI] [PubMed] [Google Scholar]
- 96.Humphreys JM, Piala AT, Akella R, He H, Goldsmith EJ. Precisely ordered phosphorylation reactions in the p38 mitogen-activated protein (MAP) kinase cascade. The Journal of biological chemistry. 2013;288(32):23322–23330. doi: 10.1074/jbc.M113.462101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Heyeck SD, Wilcox HM, Bunnell SC, Berg LJ. Lck phosphorylates the activation loop tyrosine of the Itk kinase domain and activates Itk kinase activity. The Journal of biological chemistry. 1997;272(40):25401–25408. doi: 10.1074/jbc.272.40.25401. [DOI] [PubMed] [Google Scholar]
- 98.Wilcox HM, Berg LJ. Itk phosphorylation sites are required for functional activity in primary T cells. The Journal of biological chemistry. 2003;278(39):37112–37121. doi: 10.1074/jbc.M304811200. [DOI] [PubMed] [Google Scholar]
- 99.Lu X, Nemoto S, Lin A. Identification of c-Jun NH2-terminal protein kinase (JNK)-activating kinase 2 as an activator of JNK but not p38. The Journal of biological chemistry. 1997;272(40):24751–24754. doi: 10.1074/jbc.272.40.24751. [DOI] [PubMed] [Google Scholar]
- 100.Yokoyama N, Miller WT. Biochemical properties of the Cdc42-associated tyrosine kinase ACK1. Substrate specificity, authphosphorylation, and interaction with Hck. The Journal of biological chemistry. 2003;278(48):47713–47723. doi: 10.1074/jbc.M306716200. [DOI] [PubMed] [Google Scholar]
- 101.Okamura T, Singh S, Buolamwini J, Haystead T, Friedman H, Bigner D, Ali-Osman F. Tyrosine phosphorylation of the human glutathione S-transferase P1 by epidermal growth factor receptor. The Journal of biological chemistry. 2009;284(25):16979–16989. doi: 10.1074/jbc.M808153200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Roussel MF, Shurtleff SA, Downing JR, Sherr CJ. A point mutation at tyrosine-809 in the human colony-stimulating factor 1 receptor impairs mitogenesis without abrogating tyrosine kinase activity, association with phosphatidylinositol 3-kinase, or induction of c-fos and junB genes. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(17):6738–6742. doi: 10.1073/pnas.87.17.6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.van der Geer P, Hunter T. Tyrosine 706 and 807 phosphorylation site mutants in the murine colony-stimulating factor-1 receptor are unaffected in their ability to bind or phosphorylate phosphatidylinositol-3 kinase but show differential defects in their ability to induce early response gene transcription. Molecular and cellular biology. 1991;11(9):4698–4709. doi: 10.1128/mcb.11.9.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pham CD, Arlinghaus RB, Zheng CF, Guan KL, Singh B. Characterization of MEK1 phosphorylation by the v-Mos protein. Oncogene. 1995;10(8):1683–1688. [PubMed] [Google Scholar]
- 105.Gopalbhai K, Jansen G, Beauregard G, Whiteway M, Dumas F, Wu C, Meloche S. Negative regulation of MAPKK by phosphorylation of a conserved serine residue equivalent to Ser212 of MEK1. The Journal of biological chemistry. 2003;278(10):8118–8125. doi: 10.1074/jbc.M211870200. [DOI] [PubMed] [Google Scholar]
- 106.Huang YZ, McNamara JO. Mutual regulation of Src family kinases and the neurotrophin receptor TrkB. The Journal of biological chemistry. 2010;285(11):8207–8217. doi: 10.1074/jbc.M109.091041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Middlemas DS, Meisenhelder J, Hunter T. Identification of TrkB autophosphorylation sites and evidence that phospholipase C-gamma 1 is a substrate of the TrkB receptor. The Journal of biological chemistry. 1994;269(7):5458–5466. [PubMed] [Google Scholar]
- 108.Kato H, Faria TN, Stannard B, Roberts CT, Jr, LeRoith D. Essential role of tyrosine residues 1131, 1135, and 1136 of the insulin-like growth factor-I (IGF-I) receptor in IGF-I action. Molecular endocrinology. 1994;8(1):40–50. doi: 10.1210/mend.8.1.7512194. [DOI] [PubMed] [Google Scholar]
- 109.Pautsch A, Zoephel A, Ahorn H, Spevak W, Hauptmann R, Nar H. Crystal structure of bisphosphorylated IGF-1 receptor kinase: insight into domain movements upon kinase activation. Structure. 2001;9(10):955–965. doi: 10.1016/s0969-2126(01)00655-4. [DOI] [PubMed] [Google Scholar]
- 110.Kentrup H, Becker W, Heukelbach J, Wilmes A, Schurmann A, Huppertz C, Kainulainen H, Joost HG. Dyrk, a dual specificity protein kinase with unique structural features whose activity is dependent on tyrosine residues between subdomains VII and VIII. The Journal of biological chemistry. 1996;271(7):3488–3495. doi: 10.1074/jbc.271.7.3488. [DOI] [PubMed] [Google Scholar]
- 111.Kaczmarski W, Barua M, Mazur-Kolecka B, Frackowiak J, Dowjat W, Mehta P, Bolton D, Hwang YW, Rabe A, Albertini G, Wegiel J. Intracellular distribution of differentially phosphorylated dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A) Journal of neuroscience research. 2014;92(2):162–173. doi: 10.1002/jnr.23279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Longati P, Bardelli A, Ponzetto C, Naldini L, Comoglio PM. Tyrosines1234–1235 are critical for activation of the tyrosine kinase encoded by the MET proto-oncogene (HGF receptor) Oncogene. 1994;9(1):49–57. [PubMed] [Google Scholar]
- 113.DiNitto JP, Deshmukh GD, Zhang Y, Jacques SL, Coli R, Worrall JW, Diehl W, English JM, Wu JC. Function of activation loop tyrosine phosphorylation in the mechanism of c-Kit auto-activation and its implication in sunitinib resistance. Journal of biochemistry. 2010;147(4):601–609. doi: 10.1093/jb/mvq015. [DOI] [PubMed] [Google Scholar]
- 114.Agarwal S, Kazi JU, Ronnstrand L. Phosphorylation of the activation loop tyrosine 823 in c-Kit is crucial for cell survival and proliferation. The Journal of biological chemistry. 2013;288(31):22460–22468. doi: 10.1074/jbc.M113.474072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dinh M, Grunberger D, Ho H, Tsing SY, Shaw D, Lee S, Barnett J, Hill RJ, Swinney DC, Bradshaw JM. Activation mechanism and steady state kinetics of Bruton’s tyrosine kinase. The Journal of biological chemistry. 2007;282(12):8768–8776. doi: 10.1074/jbc.M609920200. [DOI] [PubMed] [Google Scholar]
- 116.Baba Y, Hashimoto S, Matsushita M, Watanabe D, Kishimoto T, Kurosaki T, Tsukada S. BLNK mediates Syk-dependent Btk activation. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(5):2582–2586. doi: 10.1073/pnas.051626198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Niture SK, Gnatt A, Jaiswal AK. Oncogene PKCepsilon controls INrf2-Nrf2 interaction in normal and cancer cells through phosphorylation of INrf2. Journal of cell science. 2013;126(Pt 24):5657–5669. doi: 10.1242/jcs.133819. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 118.Feng Q, He B, Jung SY, Song Y, Qin J, Tsai SY, Tsai MJ, O’Malley BW. Biochemical control of CARM1 enzymatic activity by phosphorylation. The Journal of biological chemistry. 2009;284(52):36167–36174. doi: 10.1074/jbc.M109.065524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Won M, Park KA, Byun HS, Kim YR, Choi BL, Hong JH, Park J, Seok JH, Lee YH, Cho CH, Song IS, Kim YK, Shen HM, Hur GM. Protein kinase SGK1 enhances MEK/ERK complex formation through the phosphorylation of ERK2: implication for the positive regulatory role of SGK1 on the ERK function during liver regeneration. Journal of hepatology. 2009;51(1):67–76. doi: 10.1016/j.jhep.2009.02.027. [DOI] [PubMed] [Google Scholar]
- 120.Degterev A, Hitomi J, Germscheid M, Ch’en IL, Korkina O, Teng X, Abbott D, Cuny GD, Yuan C, Wagner G, Hedrick SM, Gerber SA, Lugovskoy A, Yuan J. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nature chemical biology. 2008;4(5):313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Su Q, Wang S, Gao HQ, Kazemi S, Harding HP, Ron D, Koromilas AE. Modulation of the eukaryotic initiation factor 2 alpha-subunit kinase PERK by tyrosine phosphorylation. The Journal of biological chemistry. 2008;283(1):469–475. doi: 10.1074/jbc.M704612200. [DOI] [PubMed] [Google Scholar]
- 122.Kishimoto K, Matsumoto K, Ninomiya-Tsuji J. TAK1 mitogen-activated protein kinase kinase kinase is activated by autophosphorylation within its activation loop. The Journal of biological chemistry. 2000;275(10):7359–7364. doi: 10.1074/jbc.275.10.7359. [DOI] [PubMed] [Google Scholar]
- 123.Lin X, Mu Y, Cunningham ET, Jr, Marcu KB, Geleziunas R, Greene WC. Molecular determinants of NF-kappaB-inducing kinase action. Molecular and cellular biology. 1998;18(10):5899–5907. doi: 10.1128/mcb.18.10.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Miyake T, Parsons SJ. Functional interactions between Choline kinase alpha, epidermal growth factor receptor and c-Src in breast cancer cell proliferation. Oncogene. 2012;31(11):1431–1441. doi: 10.1038/onc.2011.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Beullens M, Vancauwenbergh S, Morrice N, Derua R, Ceulemans H, Waelkens E, Bollen M. Substrate specificity and activity regulation of protein kinase MELK. The Journal of biological chemistry. 2005;280(48):40003–40011. doi: 10.1074/jbc.M507274200. [DOI] [PubMed] [Google Scholar]
- 126.Wu B, Jiang P, Mu Y, Wilmouth RC. Cancer Osaka thyroid (Cot) phosphorylates Polo-like kinase (PLK1) at Ser137 but not at Thr210. Biological chemistry. 2009;390(12):1271–1277. doi: 10.1515/BC.2009.141. [DOI] [PubMed] [Google Scholar]
- 127.Riojas RA, Kikani CK, Wang C, Mao X, Zhou L, Langlais PR, Hu D, Roberts JL, Dong LQ, Liu F. Fine tuning PDK1 activity by phosphorylation at Ser163. The Journal of biological chemistry. 2006;281(31):21588–21593. doi: 10.1074/jbc.M600393200. [DOI] [PubMed] [Google Scholar]
- 128.Zhang BH, Guan KL. Activation of B-Raf kinase requires phosphorylation of the conserved residues Thr598 and Ser601. The EMBO journal. 2000;19(20):5429–5439. doi: 10.1093/emboj/19.20.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cheng H, Ross JA, Frost JA, Kirken RA. Phosphorylation of human Jak3 at tyrosines 904 and 939 positively regulates its activity. Molecular and cellular biology. 2008;28(7):2271–2282. doi: 10.1128/MCB.01789-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Salvador JM, Mittelstadt PR, Guszczynski T, Copeland TD, Yamaguchi H, Appella E, Fornace AJ, Jr, Ashwell JD. Alternative p38 activation pathway mediated by T cell receptor-proximal tyrosine kinases. Nature immunology. 2005;6(4):390–395. doi: 10.1038/ni1177. [DOI] [PubMed] [Google Scholar]
- 131.Jirmanova L, Giardino Torchia ML, Sarma ND, Mittelstadt PR, Ashwell JD. Lack of the T cell-specific alternative p38 activation pathway reduces autoimmunity and inflammation. Blood. 2011;118(12):3280–3289. doi: 10.1182/blood-2011-01-333039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.White WO, Seibenhener ML, Wooten MW. Phosphorylation of tyrosine 256 facilitates nuclear import of atypical protein kinase C. Journal of cellular biochemistry. 2002;85(1):42–53. [PubMed] [Google Scholar]
- 133.Strack V, Hennige AM, Krutzfeldt J, Bossenmaier B, Klein HH, Kellerer M, Lammers R, Haring HU. Serine residues 994 and 1023/25 are important for insulin receptor kinase inhibition by protein kinase C isoforms beta2 and theta. Diabetologia. 2000;43(4):443–449. doi: 10.1007/s001250051327. [DOI] [PubMed] [Google Scholar]
- 134.Hresko RC, Hoffman RD, Flores-Riveros JR, Lane MD. Insulin receptor tyrosine kinase-catalyzed phosphorylation of 422(aP2) protein. Substrate activation by long-chain fatty acid. The Journal of biological chemistry. 1990;265(34):21075–21085. [PubMed] [Google Scholar]
- 135.Masai H, Matsui E, You Z, Ishimi Y, Tamai K, Arai K. Human Cdc7-related kinase complex. In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a criticial threonine residue of Cdc7 bY Cdks. The Journal of biological chemistry. 2000;275(37):29042–29052. doi: 10.1074/jbc.M002713200. [DOI] [PubMed] [Google Scholar]
- 136.Basak C, Pathak SK, Bhattacharyya A, Mandal D, Pathak S, Kundu M. NF-kappaB- and C/EBPbeta-driven interleukin-1beta gene expression and PAK1-mediated caspase-1 activation play essential roles in interleukin-1beta release from Helicobacter pylori lipopolysaccharide-stimulated macrophages. The Journal of biological chemistry. 2005;280(6):4279–4288. doi: 10.1074/jbc.M412820200. [DOI] [PubMed] [Google Scholar]
- 137.ten Klooster JP, Jansen M, Yuan J, Oorschot V, Begthel H, Di Giacomo V, Colland F, de Koning J, Maurice MM, Hornbeck P, Clevers H. Mst4 and Ezrin induce brush borders downstream of the Lkb1/Strad/Mo25 polarization complex. Developmental cell. 2009;16(4):551–562. doi: 10.1016/j.devcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 138.Tobiume K, Saitoh M, Ichijo H. Activation of apoptosis signal-regulating kinase 1 by the stress-induced activating phosphorylation of pre-formed oligomer. Journal of cellular physiology. 2002;191(1):95–104. doi: 10.1002/jcp.10080. [DOI] [PubMed] [Google Scholar]
- 139.Mahajan K, Coppola D, Challa S, Fang B, Chen YA, Zhu W, Lopez AS, Koomen J, Engelman RW, Rivera C, Muraoka-Cook RS, Cheng JQ, Schonbrunn E, Sebti SM, Earp HS, Mahajan NP. Ack1 mediated AKT/PKB tyrosine 176 phosphorylation regulates its activation. PloS one. 2010;5(3):e9646. doi: 10.1371/journal.pone.0009646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Barker SC, Kassel DB, Weigl D, Huang X, Luther MA, Knight WB. Characterization of pp60c-src tyrosine kinase activities using a continuous assay: autoactivation of the enzyme is an intermolecular autophosphorylation process. Biochemistry. 1995;34(45):14843–14851. doi: 10.1021/bi00045a027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.