Eukaryotic RNAs with premature termination codons (PTCs) are eliminated by nonsense-mediated decay (NMD). Lykke-Andersen et al. discover that SMG6-catalyzed endonucleolysis widely initiates the degradation of human nonsense RNAs, whereas decapping is used to a lesser extent. A large proportion of genes hosting snoRNAs in their introns produce considerable amounts of NMD-sensitive splice variants, indicating that these RNAs are merely by-products of a primary snoRNA production process.
Keywords: RNA decapping, endonucleolytic RNA cleavage, intron-encoded snoRNA, nonsense-mediated decay, regulation of snoRNA expression, snoRNA host genes
Abstract
Eukaryotic RNAs with premature termination codons (PTCs) are eliminated by nonsense-mediated decay (NMD). While human nonsense RNA degradation can be initiated either by an endonucleolytic cleavage event near the PTC or through decapping, the individual contribution of these activities on endogenous substrates has remained unresolved. Here we used concurrent transcriptome-wide identification of NMD substrates and their 5′–3′ decay intermediates to establish that SMG6-catalyzed endonucleolysis widely initiates the degradation of human nonsense RNAs, whereas decapping is used to a lesser extent. We also show that a large proportion of genes hosting snoRNAs in their introns produce considerable amounts of NMD-sensitive splice variants, indicating that these RNAs are merely by-products of a primary snoRNA production process. Additionally, transcripts from genes encoding multiple snoRNAs often yield alternative transcript isoforms that allow for differential expression of individual coencoded snoRNAs. Based on our findings, we hypothesize that snoRNA host genes need to be highly transcribed to accommodate high levels of snoRNA production and that the expression of individual snoRNAs and their cognate spliced RNA can be uncoupled via alternative splicing and NMD.
All functional transcripts, whether they are protein-coding (mRNA) or noncoding (ncRNA), are produced as precursor molecules that undergo various processing steps before they take on their final forms. Eukaryotic RNA polymerase II transcribed genes often encode more than one mature RNA species, as exemplified by the alternative splicing of exonic sequences into a variety of transcript isoforms (Braunschweig et al. 2013; Kornblihtt et al. 2013), usage of alternative promoters (Carninci et al. 2006), and the hosting of smaller RNAs, like miRNAs and snoRNAs, within introns (Brown et al. 2008). Regulation of such alternative RNA production confers great plasticity to eukaryotic gene expression because parameters such as expression specificity, stability, localization, and protein-coding potential can be altered between transcript isoforms (McGlincy and Smith 2008; Valen et al. 2009; Kelemen et al. 2013). However, many alternative processing options also increase the likelihood of mistakes, and, throughout the life span of a transcript, its integrity and functionality is continuously being monitored, which ensures that nonfunctional processing by-products and erroneously processed or outworn molecules are degraded by decay machineries residing in either the nucleus or the cytoplasm (Doma and Parker 2007; Muhlemann and Jensen 2012; Arraiano et al. 2013). For cytoplasmic RNAs with mRNA-like characteristics, the distinction between functional and nonfunctional is carried out by means of translation-dependent RNA surveillance systems (Shoemaker and Green 2012; Inada 2013). One such system is the nonsense-mediated RNA decay (NMD) pathway that recognizes and eliminates RNAs containing premature termination codons (PTCs) (Kervestin and Jacobson 2012, Schweingruber et al. 2013). NMD establishes a quality control system by filtering away PTC-containing (nonsense) RNAs arising by random errors during early steps of gene expression or genomic mutations and rearrangements. Moreover, NMD plays a prominent role in gene expression homeostasis and as part of regulatory responses during, for example, the execution of differentiation programs (Kervestin and Jacobson 2012; Schweingruber et al. 2013; Ge and Porse 2014).
Two decades of research has shed light on many aspects of NMD, and a current working model can be summarized as follows: Prior to its degradation, a human nonsense RNA is marked near the PTC by an NMD-eliciting protein complex nucleated around the general NMD factor UPF1 (Kervestin and Jacobson 2012; Schweingruber et al. 2013). A stop codon is recognized as being premature when an eRF1/eRF3-bound ribosome stalled at the stop codon interacts with UPF1 instead of with the cytoplasmic polyA tail-binding protein PABPC1, which normally “identifies” the termination codon as proper and allows for productive translation termination (Amrani et al. 2004; Behm-Ansmant et al. 2007; Ivanov et al. 2008; Silva et al. 2008; Singh et al. 2008). UPF1 recruitment is favored when the physical distance between the terminating ribosome and the polyA tail is sufficiently long (Behm-Ansmant et al. 2007; Eberle et al. 2008; Singh et al. 2008) and/or when the stop codon is situated more than ∼50 nucleotides (nt) upstream of the last splice junction (Nagy and Maquat 1998). The latter mechanism is stimulated as a result of splice junctions being marked by the exon junction complex (EJC), which includes the UPF1-interacting proteins UPF3 and UPF2 (Kunz et al. 2006; Chamieh et al. 2008; Ivanov et al. 2008; Melero et al. 2012). UPF1 is recruited together with the kinase complex SMG1C, and subsequent interactions with UPF2 and UPF3 allow SMG1C-mediated phosphorylation of UPF1 (Yamashita et al. 2009). This in turn attracts various factors that can initiate degradation of the RNA body: (1) The SMG6 endonuclease binds to phosphorylated residues in the N-terminal part of UPF1 (Okada-Katsuhata et al. 2012) and catalyzes a cleavage in the vicinity of the PTC (Gatfield and Izaurralde 2004; Huntzinger et al. 2008; Eberle et al. 2009); (2) the SMG5/SMG7 heterodimer binds to phosphorylated residues in the C-terminal part of UPF1 (Okada-Katsuhata et al. 2012) and recruits the CCR4–NOT deadenylase complex, which catalyzes polyA tail shortening that stimulates decapping by the general decapping complex (DCP1/DCP2) (Lejeune et al. 2003; Couttet and Grange 2004; Yamashita et al. 2005; Loh et al. 2013); or (3) the decapping complex is recruited to UPF1 either directly or in a PNRC2-dependent manner, leading to deadenylation-independent decapping (Lykke-Andersen 2002; Fenger-Gron et al. 2005; Cho et al. 2009, 2013; Lai et al. 2012; Loh et al. 2013). After execution of the initial deprotection step, the NMD-eliciting complex most likely dissociates and releases the RNA substrate for exonucleolytic degradation (Franks et al. 2010). The cytoplasmic 5′–3′ exonuclease XRN1 is responsible for the rapid removal of decay intermediates in all of the described pathways. Specifically, XRN1 degrades the 3′ fragment derived from the endonucleolytically cleaved as well as the decapped full-length nonsense RNA (Gatfield and Izaurralde 2004; Unterholzner and Izaurralde 2004; Huntzinger et al. 2008; Eberle et al. 2009; Arraiano et al. 2013; Nagarajan et al. 2013). In Saccharomyces cerevisiae and Drosophila melanogaster, the cytoplasmic RNA exosome, a 3′–5′ exonuclease, has been implicated in the elimination of fully deadenylated RNA species and the endocleavage-derived 5′ fragment, respectively (Mitchell and Tollervey 2003; Gatfield and Izaurralde 2004).
Early reports suggested that human nonsense RNA degradation takes place in a way similar to that of the yeast S. cerevisiae; i.e., via accelerated decapping and/or deadenylation followed by exonucleolysis (Muhlrad and Parker 1994; Cao and Parker 2003; Chen and Shyu 2003; Lejeune et al. 2003; Mitchell and Tollervey 2003; Takahashi et al. 2003; Couttet and Grange 2004; Yamashita et al. 2005). However, later studies inspired by the observation that nonsense RNAs are exclusively degraded via endocleavage in D. melanogaster (Gatfield and Izaurralde 2004) revealed that SMG6-catalyzed endocleavage can also occur during human NMD (Huntzinger et al. 2008; Eberle et al. 2009). However, the extent to which this contributes to the overall degradation of endogenous nonsense RNAs has been questioned (Yamashita 2013).
Here we establish SMG6-catalyzed endocleavage as a commonly occurring initiating step in human nonsense RNA decay. Our data suggest that decapping generally serves as a backup option, although it is the preferred pathway for a minor subset of substrates. By combining global identification of nonsense RNAs and their corresponding decay intermediates, we identified primary NMD-responsive isoforms from up to 12% of all expressed genes. Among these, spliced RNAs derived from both protein-coding and “noncoding” snoRNA host genes are highly enriched. More than 90% of human snoRNA-coding units are situated inside the intronic sequence of conventional genes, and the corresponding snoRNA production is dependent on the expression of the host gene and the productive splicing of its precursor RNA (Kiss et al. 2006; Brown et al. 2008; Dieci et al. 2009). Our findings highlight that spliced host gene RNAs are often mere by-products of the snoRNA production process. Notably, this is also the case for many snoRNA host gene-encoded spliced ncRNA and mRNA species with documented functions. The sensitivity of these species to NMD illustrates a widespread usage of translation to regulate the levels of functional RNA. Finally, our data strongly imply that genes encoding multiple snoRNAs use extensive alternative splicing events to facilitate the differential expression of individual snoRNAs.
Results
Global discovery of NMD-specific endonucleolytic cleavage events
To investigate the generality of endocleavage in NMD, we devised a massive parallel sequencing approach, “5′ end-seq,” in which siRNA-mediated depletion of XRN1 was used to identify endocleavage and decapping sites in polyadenylated cytoplasmic RNAs from HEK293 Flp-In T-Rex cells expressing the β-globin PTC39 (β-39) nonsense reporter transcript (Fig. 1A; Supplemental Fig. S1A–C; Supplemental Table S1; Eberle et al. 2009). We used polyadenylated RNA, as the β-39 3′ fragment produced by SMG6-catalyzed endocleavage harbors a polyA tail (Eberle et al. 2009) and because NMD-triggered decapping can take place either independent of deadenylation or after an initial polyA tail-shortening step that leaves some of the tail intact. Additionally, analyses of selected transcripts indicated an enrichment for both endocleaved and decapped species by oligo-dT capture (Supplemental Fig. S1B,D; Supplemental Material). 5′ end-seq exploits that an XRN1 substrate contains a monophosphate moiety at its 5′ end (Arraiano et al. 2013; Nagarajan et al. 2013) and therefore can be selectively ligated to an RNA adapter molecule within a pool of diverse RNAs (Supplemental Fig. S1A). Putative decapping and endocleavage events were distinguished through comparison with cap-selected 5′ ends of RNAs as detected by cap analysis of gene expression (CAGE) tag sequencing of RNA obtained from control HEK293 Flp-In T-Rex cells (Takahashi et al. 2012). Furthermore, NMD-specific endocleavage events were identified via codepletion of XRN1 with either SMG6 or UPF1 (Supplemental Fig. S1C). All of the samples were also subjected to standard RNA sequencing (RNA-seq) (Fig. 1A; Supplemental Fig. S1; Supplemental Table S1), and the Cufflinks2 software (Trapnell et al. 2010) was applied to conduct an annotation-guided de novo transcript assembly of the data, allowing us to estimate the transcriptomes and their isoform-specific expression levels.
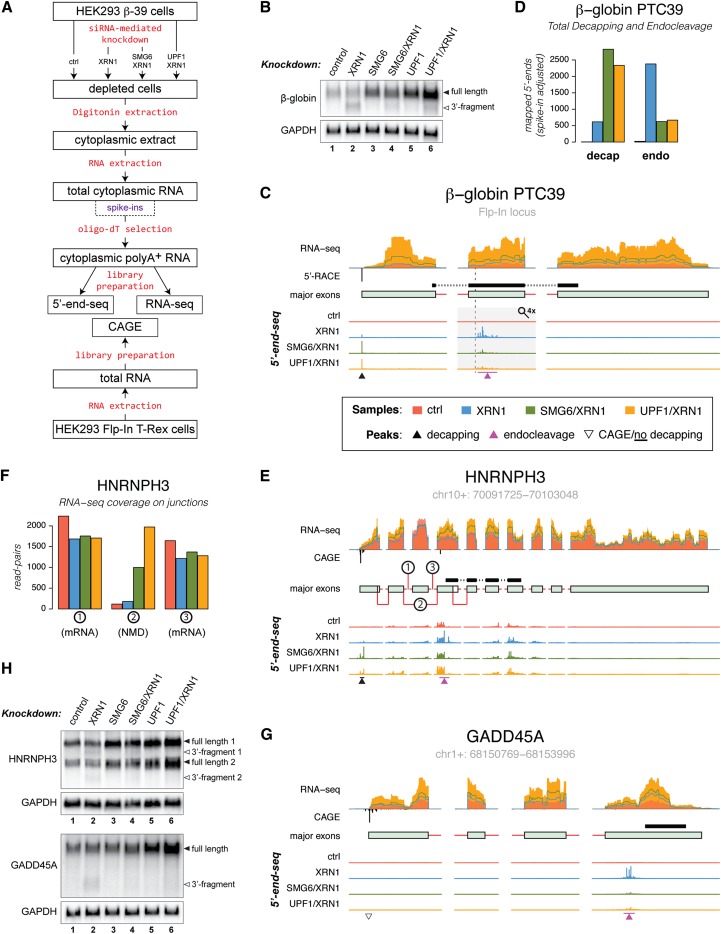
Figure 1.
NMD-specific endonucleolytic cleavage sites are revealed by 5′ end-seq. (A) Schematic outline of the sample preparation steps for the massive parallel sequencing procedures used in this study (see the text and Supplemental Fig. S1 for details). (B) Northern blotting analysis of total RNA isolated from HEK293-β-39 cells depleted for the indicated factors. The Northern membrane was hybridized with a probe directed against the region shown in C. GAPDH levels were detected as an internal loading standard. (C) Overview of the stably integrated β-39 gene illustrating the sequencing data used in the analyses. From top to bottom, the tracks display (1) RNA-seq coverage over the major exons as well as leading and trailing intronic sequences determined from control, XRN1-depleted, SMG6/XRN1-depleted, and UPF1/XRN1-depleted samples (in all figures, data from the control, XRN1-depleted, SMG6/XRN1-depleted, and UPF1/XRN1-depleted samples are depicted in red, blue, green, and orange, respectively). (2) 5′-RACE-mapped 5′ end of the transcript (black; below RNA-seq axis). For all endogenous genes, 5′ ends were determined by CAGE. (3) Schematic representation of the major exons expressed from the gene (exons and intronic sequences are represented as light-green boxes and red lines, respectively; see the Materials and Methods for details). The position of the probe used for Northern blotting is shown as a black bar above the transcript model. The PTC is indicated by a vertical dashed line. (4) 5′ end-seq-determined 5′ end signals displayed in individual tracks but on the same scale for control, XRN1-depleted, SMG6/XRN1-depleted, and UPF1/XRN1-depleted samples. To allow simultaneous visualization of decapping and endocleavage peaks (indicated by black and purple arrowheads, respectively), the 5′ end-seq tracks were scaled up by a factor of 4 in the region covering exon 2 of the β-39 RNA (indicated by a gray rectangle and a magnifying glass). (D) Histogram showing total signal in the decapping peaks (left) and NMD-specific endocleavage sites (right). (E,G) As in C, but for the HNRNPH3 and GADD45A loci. The open triangle in G indicates a transcript 5′ end determined by CAGE for which no decapping signal was detected. (F) Histogram showing the RNA-seq coverage over the exon–exon junctions indicated in E. (H) As in B, but the membranes were hybridized with probes directed against HNRNPH3 (top panel) and GADD45A (bottom panel) RNA species (see E and G for positions of probes). See also Supplemental Figure S2.
As an initial exploration of the data, we analyzed the behavior of the β-39 nonsense reporter transcript. In accordance with previous results (Eberle et al. 2009), an endocleavage-derived 3′ fragment was observed by Northern blotting analysis upon depletion of XRN1 (Fig. 1B, lane 2; Supplemental Fig. 1C, bottom panel for corresponding Western blotting analysis). Accumulation of this fragment was substantially reduced when XRN1 was codepleted with either SMG6 or UPF1 (Fig. 1B, lanes 4,6), whereas the levels of full-length spliced RNA were considerably increased, similar to when SMG6 and UPF1 were depleted individually (Fig. 1B, lanes 3–6). These effects were quantitatively reproduced by RNA-seq analyses of samples from control, XRN1-depleted, SMG6/XRN1-depleted, and UPF1/XRN1-depleted cells (the latter two will be jointly referred to as double-depleted samples) (Fig. 1C, RNA-seq track; Supplemental Fig. 1C, top panel, for corresponding Western blotting analysis). In the vicinity of and primarily downstream from the β-39 PTC (Fig. 1C, dashed vertical line), the 5′ end-seq data from the XRN1-depleted sample displayed a number of peaks (Fig. 1C, indicated by a purple arrowhead), which represent NMD-specific endocleavage events, since they were absent or at considerably reduced intensities in the control and double-depleted samples (Fig. 1D, right, endo). Additionally, there was a distinct “decapping peak” (Fig. 1C, indicated by a black arrowhead) in the XRN1-depleted sample at the 5′-most nucleotide of the β-39 RNA, which was absent in the control sample and increased in the double-depleted samples (Fig. 1D, left, decap). The peak “signatures” are summarized in Figure 1D, which shows the total number of mapped 5′ ends corresponding to decapping (decap) and endonucleolytic cleavage (endo). The particular appearance of the decapping peak is described further in the next section.
Peak types similar to the ones described for the β-39 RNA could be detected in endogenous nonsense RNAs. In general, we found examples of transcripts that displayed both NMD-specific endocleavage and the described decapping profile (for example, from the HNRNPH3 gene) (Fig. 1E,H, top panel), but, in many cases, we could only detect one of the two peak types, as exemplified by the GADD45A transcript, where only a cluster of NMD-specific endocleavage events was identified (Fig. 1G,H, bottom panel; see Supplemental Fig. S2 for additional examples).
The RNA-seq data were used to identify the specific NMD-responsive isoforms, and evidently only one major transcript variant was produced from the GADD45A gene (Fig. 1G, major exons track). This variant was highly responsive to depletion of NMD factors (Fig. 1G, RNA-seq track) and displayed a decreased decay rate upon depletion of UPF1 (Supplemental Fig. S2G). In contrast, the HNRNPH3 gene produced several splice variants, of which only those arising from an exon-skipping event are NMD substrates, which makes the NMD responsiveness less obvious from the RNA-seq data (Fig. 1E,F, the major alternative splicing events that differentiate non-NMD and NMD transcripts are indicated by encircled numbers in E, and the corresponding RNA-seq coverage on the exon–exon junctions is displayed in F).
We previously determined that NMD-specific endocleavage sites cluster in the vicinity of the PTC in model substrates (Eberle et al. 2009). To investigate whether this observation was supported by our global data, we characterized NMD-specific endocleavage events in a reference set of annotated NMD substrates, consisting of 538 transcripts with a total of 782 potential endocleavage sites (“NMD reference set” derived from annotation) (see the Materials and Methods; Supplemental Table S2; Harrow et al. 2012), by mapping their positions relative to the annotated termination codons. Although these transcripts were only computationally determined as NMD substrates, the distribution of endocleavage positions was indeed concentrated around the putative PTC (Supplemental Fig. S2H). These observations convinced us that the experimental setup is useful to reliably identify NMD-specific endocleavage events.
NMD-specific decapping is increased upon depletion of SMG6
In addition to the increased intensity of the β-39 decapping peak in the double-depleted samples compared with the XRN1-depleted sample, the signal was also slightly higher upon depletion of SMG6/XRN1 compared with UPF1/XRN1 (Fig. 1D, decap). Since levels of full-length β-39 RNA was ∼2.5-fold higher in the latter versus the former sample (Fig. 1B,C), relative decapping of the β-39 RNA was increased upon SMG6 depletion. This was confirmed by reverse transcription followed by quantitative PCR (RT-qPCR) on adapter-ligated total RNA (Fig. 2A, top panel). Furthermore, increased relative decapping in the SMG6/XRN1-depleted sample was specific for the β-39 nonsense RNA and not detectable when assaying the β-globin wild-type (β-wt) construct (Eberle et al. 2009) under similar conditions (Fig. 2A, bottom panel). The same phenomenon was detected for nonsense RNA lacking exon 3 (ex3−) produced from the HNRNPH3 gene (Fig. 2B, top panel), whereas relative decapping at an equivalent position in NMD-insensitive mRNA from the same locus (ex3+) was unchanged between the XRN1-depleted and double-depleted samples (Fig. 2B, bottom panel; Supplemental Fig. S3 for further examples).
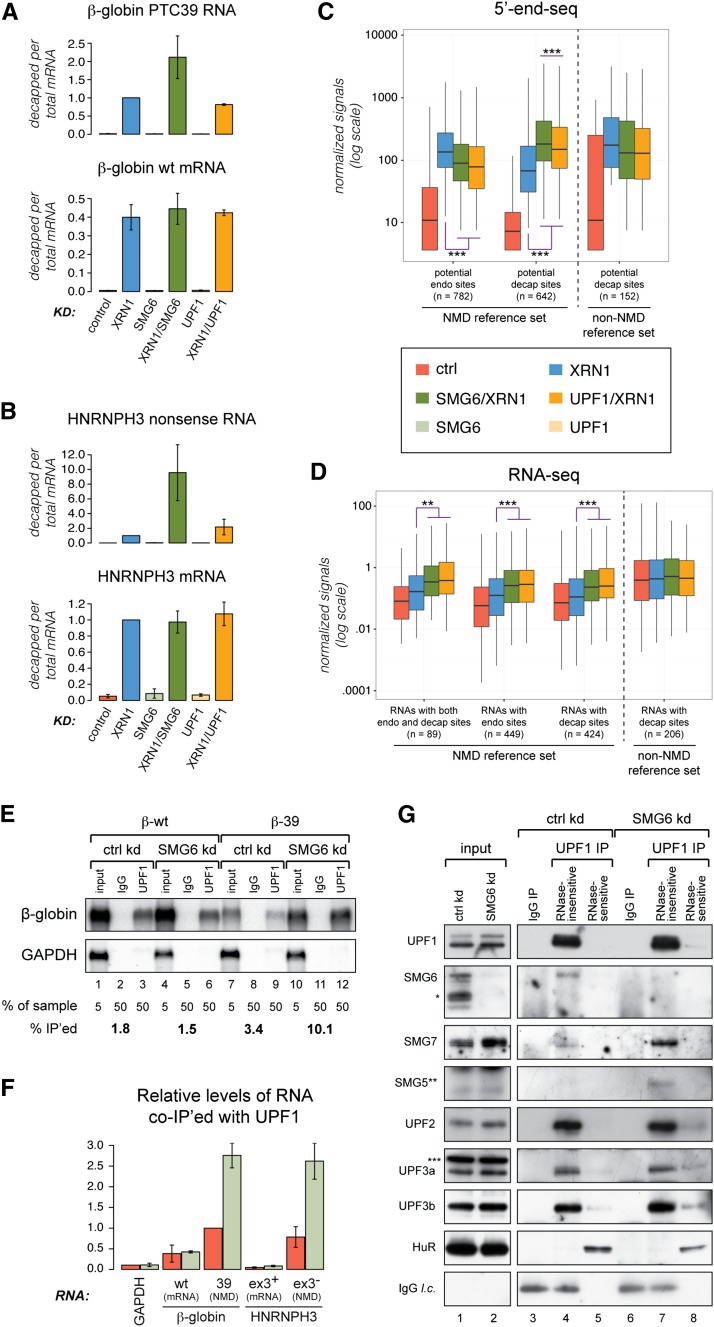
Figure 2.
NMD-specific decapping increases upon depletion of SMG6. (A,B) Relative decapping (ratio between levels of transcript-specific decapping and levels of full-length mRNA as measured by RT-qPCR on adapter-ligated RNA samples) of β-39 (top panel) and β-wt (bottom panel) transcripts under the indicated conditions (all levels are relative to the value measured for β-39 upon XRN1 depletion) (A) and HNRNPH3 nonsense RNA (exon3−; top panel) and HNRNPH3 mRNA (NMD-insensitive variant, exon3+; bottom panel) variants under the indicated conditions (for each variant, the levels are relative to the value measured upon XRN1 depletion) (B). The histograms represent data from at least three independent experiments (nA = 3 and nB = 4). Error bars depict standard deviations. (C, two left clusters) Box plots illustrating the distribution of 5′ end-seq peak intensities from potential endonucleolytic cleavage (endo) and decapping (decap) sites detected in a reference set of annotated NMD-responsive transcripts. The right cluster of box plots represents decapping sites in a non-NMD reference set of transcripts. Boxes span the second and third quartile, and the horizontal line indicates the median. Whiskers extend to the most extreme data point that is no more than 1.5 times the height of the box away from the top and bottom edge of the box. (***) P-value ≤ 0.001. (D) Box plots illustrating the distribution of RNA-seq-based expression levels of the transcripts corresponding to the 5′ end-seq signals plotted in C. (**) P-value ≤ 0.01; (***) P-value ≤ 0.05. (E) Northern blotting analysis of RNA coimmunoprecipitated by endogenous UPF1 from HEK293 Flp-In T-Rex cells stably expressing either β-wt (lanes 1–6) or β-39 (lanes 7–12). Five percent of input RNA from the β-wt and β-39 cells subjected to control and SMG6 depletion conditions (lanes 1,4,7,10) were compared with 50% of the coimmunoprecipitated RNA (IgG control antibody in lanes 2,5,8,11; anti-UPF1 IgG in lanes 3,6,9,12). The blot was hybridized as described in Figure 1B. Relative levels of β-globin mRNA coimmunoprecipitated with UPF1 are shown below the blots (percent immunoprecipitated). (F) Quantification of two independent experiments conducted as in E. Coimmunoprecipitated HNRNPH3 transcripts and GAPDH were measured by RT-qPCR. All values are relative to coimmunoprecipitated levels of β-39 under control conditions. Error bars depict standard deviations. (G) Western blotting analyses of proteins coimmunoprecipitated with endogenous UPF1 from the HEK293-β-39 cell line. We compared 0.3% of input cell extracts from control and SMG6-depleted cells (lanes 1,2) with 25%–37.5% of the coimmunoprecipitated proteins (IgG control antibody in lanes 3,6; anti-UPF1 IgG antibody in lanes 4,5,7,8). The immunoprecipitated samples were treated with RNase A before elution, and lanes 3, 4, 6, and 7 show the RNase-insensitive coimmunoprecipitated material, whereas lanes 5 and 8 show the material that is sensitive to RNase treatment (i.e., stripped off the beads upon RNase treatment). The membranes were probed with antibodies recognizing the indicated proteins. Detection of HuR served to control for the RNase A treatment. IgG light chain (l.c.) was detected by the secondary antibody. Asterisks indicate proteins recognized by the SMG6-specific (*), SMG5-specific (**), and UPF3a-specific (***) antibodies, respectively, which are not coimmunoprecipitated with UPF1 (although, in the case of SMG6, the protein is depleted by the siRNA directed against SMG6). Similar results were obtained by immunoprecipitating UPF1 from the HEK293-β-wt cell line. See also Supplemental Figure S3.
The overall distributions of 5′ end-seq endocleavage and decapping peak intensities within the aforementioned NMD reference set were similar to the signatures seen for the β-39 and HNRNPH3 nonsense RNAs; i.e., endocleavage and decapping were significantly reduced and increased, respectively, when comparing the double-depleted sample with the XRN1-depleted sample (Fig. 2C, NMD reference set). Furthermore, codepletion of SMG6 with XRN1 increased decapping levels significantly more than when UPF1 was codepleted (Fig. 2C, NMD reference set). Consistent with these transcripts being NMD targets, the corresponding RNA-seq data revealed significantly increased levels in the two double-depleted samples compared with control and XRN1-depleted samples (Fig. 2D, NMD reference set). In contrast, a set of mRNAs that were not annotated as NMD substrates but at the same time displayed decapping in the XRN1-depleted sample and no increase in the double-depleted samples did not exhibit changed RNA-seq levels (Fig. 2C,D; Supplemental Table S2, non-NMD reference set). This illustrates that distinct traits between NMD and non-NMD substrates can be discriminated by our high-throughput data.
We hypothesized that the increased decapping of nonsense RNA upon depletion of SMG6 arises because the NMD-eliciting complex, containing UPF1, can still assemble on the RNA in the absence of SMG6. This potentially allows for an extended window of opportunity for the NMD-stimulated decapping reaction to take place. In contrast, when UPF1 is depleted, the NMD-eliciting complex is not deposited on the nonsense RNA, which then is degraded via an alternative (possibly passive) decay route also involving decapping. To test this hypothesis, we characterized RNA and proteins coimmunoprecipitated with endogenous UPF1 from either HEK293-β-39 or HEK293-β-wt cells subjected to SMG6 depletion and corresponding control conditions. In accordance with our model, we observed increased steady-state binding of UPF1 to the β-39 mRNA in the absence of SMG6, which was not seen for the β-wt transcript (Fig. 2E,F). A similar binding pattern was observed for the HNRNPH3 NMD/non-NMD transcript pair (Fig. 2F; Supplemental Fig. S3 for further examples). The less efficient and SMG6-insensitive binding of UPF1 to the non-NMD RNAs probably reflects the general translation-independent binding of inactive/uncomplexed UPF1 to the 3′ untranslated regions (UTRs) of many transcripts (Hogg and Goff 2010; Hurt et al. 2013; Zund et al. 2013; Gregersen et al. 2014). Furthermore, in accordance with previously published results (Okada-Katsuhata et al. 2012), we observed a substantial increase in the amount of coimmunoprecipitated SMG7 and SMG5 upon depletion of SMG6 (Fig. 2G, cf. lanes 4 and 7). This was not due to a general increase in the levels of the NMD-eliciting complex, as coimmunoprecipitated amounts of UPF2, UPF3a, and UPF3b remained largely unaltered between the conditions (Fig. 2G). Thus, the absence of SMG6 from the NMD-eliciting complex allows increased interaction of alternative adapter proteins known to enhance decapping with nonsense RNAs.
These results demonstrated that the described massive parallel sequencing approach is an effective way of identifying NMD-specific decapping events. Moreover, as also suggested by previous studies (Lejeune et al. 2003; Loh et al. 2013), our data imply that the NMD machinery can use alternative degradation pathways.
Endonucleolytic cleavage dominates over decapping in NMD
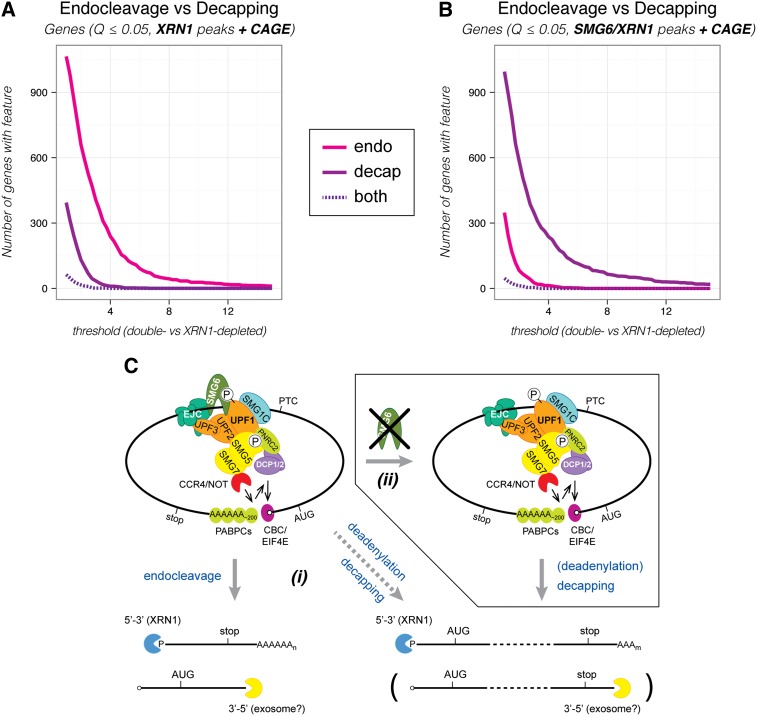
To address the global usage of endocleavage versus decapping in NMD, we first counted the extent to which these two activities occurred on a selected subset of transcripts that allowed a direct comparison (see the Materials and Methods; Supplemental Fig. S4 for details). Only transcripts supported by CAGE at their 5′ ends were included, as this allowed for an unambiguous detection of both types of peaks. Within this set, peaks were identified by a stringent set of criteria (Benjamini-Hochberg corrected P-value [Q] ≤ 0.05 based on negative binomial fitting of the 5′ end-seq data). We first identified peaks in an unbiased manner—initially not discriminating whether they arose from endocleavage or decapping—through comparison of the XRN1-depleted and control samples. Next, we asked whether the peaks were NMD-specific by requiring a reduction in peak intensities in both of the double-depleted samples for endocleavage events and, conversely, an increase for decapping events. Figure 3A shows the number of identified genes that produce one or more transcripts with NMD-specific endocleavage (magenta curve) or decapping (purple curve) events as a function of the fold change threshold in peak intensities. In contrast to the β-globin and HNRNPH3 nonsense RNA cases, we rarely detected transcripts that displayed both NMD-specific endocleavage and decapping (Fig. 3A, purple dashed curve; Supplemental Fig. S5A shows the same analysis based on the underlying numbers of transcripts). Over a broad range of possible cutoffs, the number of detected genes producing transcripts that were endocleaved by SMG6 was consistently several times higher than the number of genes yielding transcripts that displayed NMD-specific decapping (Fig. 3A). Together with the data presented in Figure 2, this indicates that SMG6-mediated endonucleolysis is the major contributor to the degradation of endogenous nonsense RNA in HEK293 cells. When conducting the initial peak identification based on a comparison of 5′ end-seq data from the SMG6/XRN1-depleted and control samples, we observed the expected reduction in the detected number of genes producing endocleaved transcripts (Fig. 3B). Importantly, we also detected a rise in the number of genes producing transcripts undergoing NMD-specific decapping, which is in agreement with the observation made on the NMD reference set that depletion of SMG6 generally leads to increased decapping of nonsense RNAs (Fig. 3B; Supplemental Fig. S5B). We hypothesize that the “local” PTC-proximal response by SMG6 is kinetically favored when the protein is available and the RNA substrate is accessible for its action. Otherwise, initiation of degradation may be relayed to one or the other terminus of the RNA (Fig. 3C; also see the Discussion).
Figure 3.
NMD-specific endocleavage dominates over decapping in HEK293 cells. (A) The number of genes producing one or more transcripts with one or more endonucleolytic cleavage sites (endo [magenta]), decapping sites (decap [purple]), or both (dashed, dark purple) as a function of the threshold used to define the given event (see the text for details). The XRN1 depletion sample was compared with the control sample to identify potential sites in transcripts with CAGE information in order to allow a “fair” comparison between endocleavage and decapping events (XRN1 peaks + CAGE). We note that the dominance of endocleavage over decapping was robust over a range of applied criteria for the initial peak detection. The NMD-specific endocleavage and decapping events were determined without taking differential expression determined by RNA-seq data into account. (B) As in A, but with potential sites initially identified in the SMG6/XRN1 depletion sample (SMG6/XRN1 peaks + CAGE). Analyses corresponding to A and B, but done for transcripts instead of genes is shown in Supplemental Figure S5, A and B. See Supplemental Figure S4 for details about peak identification. (C) Model for NMD in humans. The translation machinery stalls (not shown) when it encounters a PTC. The termination codon is marked as premature by a protein complex that includes UPF1, UPF2, and UPF3. Phosphorylation of UPF1 by SMG1C commits the RNA to NMD. (Left top panel) Subsequently, a set of proteins (SMG5/SMG7, PNRC2, and/or SMG6) that deprotects and prepares the RNA for exonucleolytic degradation is recruited to phosphorylated UPF1. SMG6 catalyzes a local PTC-proximal endocleavage (left bottom panel), whereas SMG5/SMG7 interacts with the CCR4/NOT deadenylation complex that catalyzes polyA tail shortening, which in turn stimulates decapping (right bottom panel). Furthermore, UPF1 can interact with the decapping complex (DCP1/2) and stimulate decapping either directly or indirectly via binding to PNRC2. (i) We suggest that SMG6-mediated endocleavage is the first and fastest response, whereas decapping is kinetically less favored. (ii) However, decapping can partially substitute for endocleavage if SMG6 function is somehow hindered (as seen upon depletion of SMG6). See the Discussion for further details.
Identification of hundreds of primary NMD-responsive genes
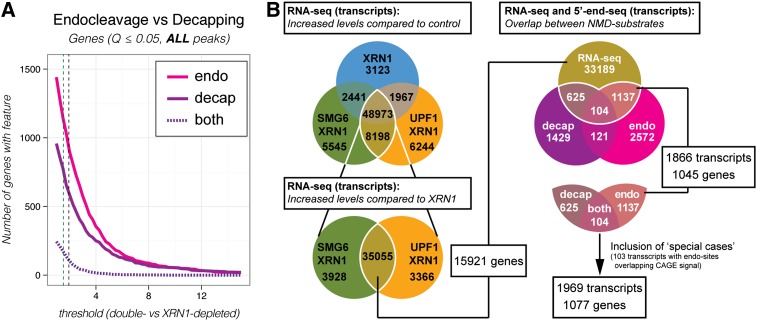
We next wanted to combine the 5′ end-seq and RNA-seq approaches to identify human endogenous nonsense RNAs. A transcript isoform expressed at higher levels in the double-depleted versus control and XRN1-depleted samples and additionally displaying NMD-specific endocleavage and/or decapping events is highly likely to be a direct target of NMD. Hence, we used the NMD reference set to determine reasonable thresholds for identifying NMD substrates in the 5′ end-seq and RNA-seq data sets. Based on mean fold changes in 5′ end-seq peak signals and RNA-seq-based expression levels, we empirically derived lower boundaries for defining a transcript as an NMD target (Fig. 4A, thresholds indicated by dashed vertical lines for decapping and endocleavages; Supplemental Fig. S5C; for details see also the Materials and Methods; Supplemental Table S2). We note that, compared with the analysis presented in Figure 3, A and B, more NMD-specific decapping and endocleavage events were included because the restrictions applied to be able to compare the extent of usage of the two activities were no longer necessary (Fig. 4A; Supplemental Fig. S5C). The thresholds determined for the RNA-seq-based expression levels were first applied to identify transcripts enriched in the three depletion samples compared with the control (Fig. 4B, top left panel). Subsequently, the double-depleted samples were compared with the XRN1-depleted sample to filter away transcripts whose increased expression levels were an effect of XRN1 depletion alone (Fig. 4B, bottom left panel). Finally, the NMD-sensitive transcripts identified independently by 5′ end-seq and RNA-seq (Supplemental Table S3) were compared, which yielded 1969 NMD-sensitive transcripts arising from 1077 assembled genes (Fig. 4B, right panel; Supplemental Table S4). When applying a less stringent (“relaxed”) cutoff for the initial identification of peaks in the 5′ end-seq data (noncorrected P-value ≤ 0.005 based on negative binomial fitting), we could detect 7267 transcripts corresponding to 3564 genes in the overlap with the RNA-seq data (Supplemental Fig. S5D; Supplemental Table S4). The large amount of detected primary substrates strengthens the notion that the NMD pathway, besides eliminating aberrant transcripts, is part of functional gene expression circuits affecting many genes. Previous attempts to identify such circuits led to the discovery of 11 SRSF protein-coding genes that produce nonsense RNA isoforms, possibly through autoregulatory feedback loops (Lareau et al. 2007). Using the present data, we confirmed that 14 of 53 SRSF protein-coding genes (Supplemental Table S5; total number of SRSF genes based on Long and Caceres 2009) could be detected in our most stringent NMD substrate set based on the combined analysis of RNA-seq and 5′end-seq. Even more genes could be included by using the “relaxed” criteria (39 out of 53) or taking the maximum union of independent RNA-seq and 5′ end-seq approaches (51 out of 53) (Supplemental Fig. S5G). However, this only explained a minor fraction of the observed NMD targets, which motivated us to find additional explanations for why certain genes are more NMD-sensitive.
Figure 4.
Global overview of NMD-specific endocleavage and decapping events and genes that produce NMD-sensitive transcript isoforms. (A) As in Figure 3A, but XRN1-depleted, SMG6/XRN1-depleted, and UPF1/XRN1-depleted samples were compared with the control sample for the initial identification of potential sites, and transcripts without CAGE information were allowed in the analysis (ALL peaks). The applied thresholds for NMD-specific endocleavage and decapping are indicated by dashed lines in the plots (endocleavage to the left and decapping to the right; for details, see Supplemental Table S2). A corresponding analysis, but done for transcripts instead of genes, is shown in Supplemental Figure S5C. (B, left panel) Venn diagrams showing the number of transcripts that display increased expression compared with control (top) and the XRN1-depleted sample (bottom) based on RNA-seq data (for thresholds, see Supplemental Table S2). (Right panel) Venn diagram showing the number of transcripts identified as NMD substrates in the combined analysis of 5′ end-seq and RNA-seq. Stringent criteria were used for 5′ end-seq peak calling. A similar diagram based on “relaxed” criteria is shown in Supplemental Figure S5D. The area encircled by a white line (magnified below the Venn diagram) indicates the interception between RNA-seq and 5′ end-seq data. A set of “special case” transcripts that contained endocleavages overlapping with CAGE signal were added to this set to produce a set of NMD-responsive transcripts/genes that was used for the next steps in the analysis. This NMD substrate set is listed in Supplemental Table S4. See also Supplemental Figure S4 for a description of the pipeline for the combined analysis of RNA-seq and 5′ end-seq data.
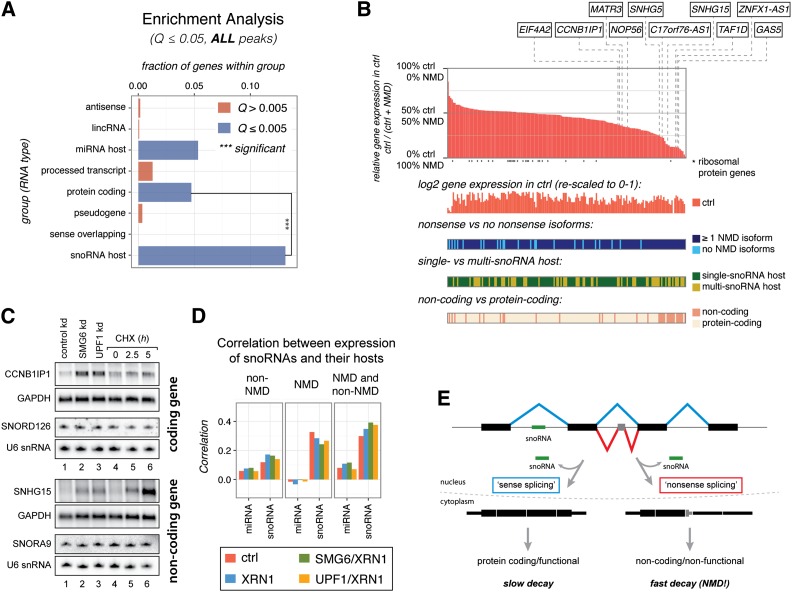
snoRNA host genes are highly enriched among NMD-sensitive genes
By investigating the genes in the identified sets of NMD substrates, we found that genes hosting snoRNAs and microRNAs (miRNAs) were significantly enriched among NMD substrates (Fig. 5A; Supplemental Fig. S5E). As a large fraction of these host genes are also protein-coding, this could simply reflect the expected enrichment of protein-coding genes among NMD-sensitive genes in general (Fig. 5A; Supplemental Fig. S5E). However, while this was the case for miRNA host genes, snoRNA host genes were significantly enriched compared with other protein-coding genes (Fig. 5A; Supplemental Fig. S5E). snoRNA host genes are generally highly expressed (see Fig. 6A), and, consequently, there is a higher likelihood that these loci produce detectable nonsense isoforms compared with genes expressed at lower levels. Still, when comparing the fraction of nonsense transcripts per locus between snoRNA host genes and a set of protein-coding genes with similar overall expression based on RNA-seq data, the former yields significantly more nonsense isoforms (Supplemental Fig. S5F).
Figure 5.
snoRNA host genes are enriched among NMD substrates. (A) Enrichment analysis for the indicated RNA biotypes in the “stringent” NMD target gene set. Bar sizes indicate the fraction of genes in the given category that is NMD substrates. Blue bars indicate that there is a significant enrichment (Fisher one-sided test, Benjamini-Hochberg-corrected Q < 0.005). Three asterisks indicate that snoRNA host genes are significantly enriched compared with protein-coding genes (Fisher two-sided test, P = 2.1 × 10−10). A similar analysis based on “relaxed” criteria is presented in Supplemental Figure S5E. (B) Summarized features of human snoRNA host genes. (Top panel) The main plot represents the relative gene expression levels of the 173 analyzed human snoRNA host genes compared between the control sample and the mean value obtained from the SMG6/XRN1 and UPF1/XRN1 samples (NMD). Individual verified genes are pointed out by dashed lines. Asterisks below the X-axis indicate genes encoding ribosomal proteins. Four additional plots below the main plot show, from top to bottom, (1) the log2-transformed absolute expression levels from the control sample (rescaled to a range of 0–1), (2) whether nonsense RNA isoforms are produced from the gene, (3) whether the gene is a single- or a multi-snoRNA host, and (4) whether the gene is annotated as protein-coding or noncoding. (C) Northern blotting analyses of total RNA isolated from the HEK293-β-39 cell line depleted for the indicated factors (lanes 1–3) or incubated for 0, 2.5, and 5 h with the translation inhibitor cycloheximide (CHX) (lanes 4–6). The membranes were hybridized with probes directed against RNA from the snoRNA host genes CCNB1IP1 (protein-coding) and SNHG15 (noncoding), respectively. GAPDH levels were detected as an internal loading standard. Separate Northern blots were probed for corresponding intron-encoded snoRNAs (SNORD126 and SNORA9) and U6 snRNA as an internal small RNA loading standard. Slowed decay under UPF1 depletion conditions is demonstrated for the SNHG15 transcript in Supplemental Figure S2G. (D) Correlation coefficients between the steady-state expression levels of small RNAs and their hosts. The left cluster in each panel shows the correlation between miRNA host transcript expression determined from the four RNA-seq libraries and the encoded miRNAs, and the right cluster shows the correlation between the snoRNA hosts and their encoded snoRNAs. The expression of small RNAs was correlated to a group of NMD-insensitive transcripts (left panel), NMD-sensitive transcripts (middle panel), and both groups together (right panel). (E) Model illustrating that a snoRNA host gene can produce both spliced stable and nonsense RNA, which in both cases leads to production of the encoded snoRNA (see the text for details). See also Supplemental Figure S6.
Figure 6.
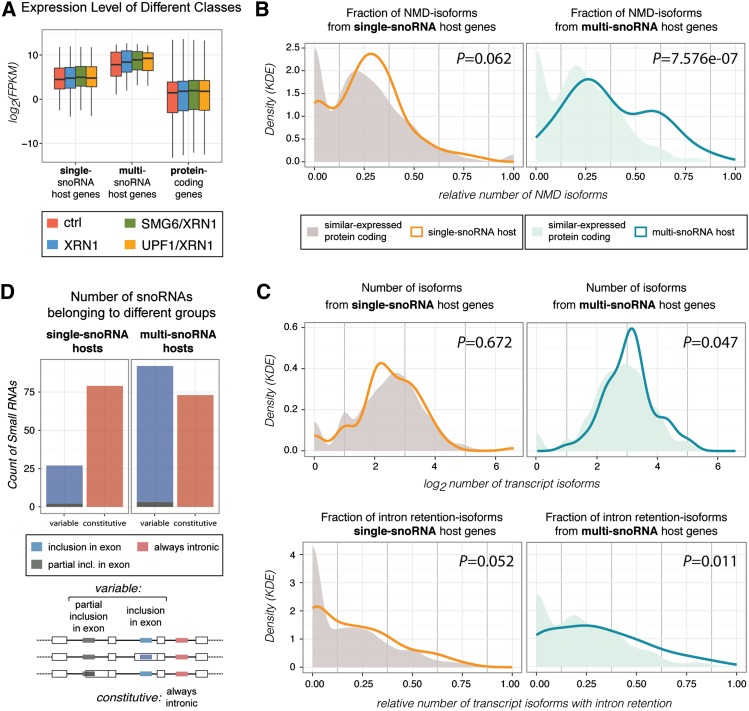
Multi-snoRNA host genes are particularly highly expressed and yield NMD-susceptible transcript isoforms. (A) Boxplot illustrating the distributions of the RNA-seq-based gene expression levels (FPKM [fragments per kilobase of exon per million mapped fragments]) of single-snoRNA and multi-snoRNA hosts and protein-coding genes in the indicated samples. (B) Density plots comparing single-snoRNA (left panel) or multi-snoRNA (right panel) host genes with similarly expressed protein-coding genes (expression distributions are shown in Supplemental Fig. S7B) in terms of the fraction of nonsense isoforms out of the total number of produced isoforms (determined by RNA-seq). Relative densities from the given groups were calculated by kernel density estimation (KDE) with Gaussian kernel. Both host gene distributions are shifted from their respective comparison group (significant for multi-snoRNA host genes). (C) As B, but here the number of produced isoforms (top panels) and the fraction of intron retention isoforms out of the total number of produced isoforms (bottom panels) are compared (data are based on RNA-seq and Cufflinks transcriptome assembly). Both single-host and multihost genes produce more intron retention transcript variants than their comparison groups (significant for multi-snoRNA host genes). (D) Count of intron-hosted snoRNAs that are exclusively intronic (constitutive [red]) or sometimes fully or partially exonic (variable [blue and dark green, respectively]) for single-snoRNA (left) and multi-snoRNA (right) host genes, respectively. The proportion of variable snoRNAs is significantly higher for multihosts compared with single hosts (Fisher two-sided test, P = 6.2 × 10−7). See also Supplemental Figure S7.
We reanalyzed a massive parallel sequencing data set of small RNAs (Kishore et al. 2013) and identified a set of 173 “active” snoRNA host genes expressing a total of 242 snoRNAs in HEK293 cells (Supplemental Table S6). For this, we applied an operational definition of a snoRNA host gene as one that produces one or more transcript isoforms with the full snoRNA sequence inside a functional intronic sequence (Kiss et al. 2006; Brown et al. 2008, Dieci et al. 2009). We found that 46 (27%) and 104 (60%) of these genes were responsive to NMD based on the “stringent” and “relaxed” NMD substrate set, respectively (Supplemental Table S4). Even more snoRNA hosts are potential targets of NMD, since the independent 5′ end-seq and RNA-seq procedures combined include up to 163 (94%) of these genes (Supplemental Fig. S5H; Supplemental Table S3). For several of the 173 snoRNA host genes, the NMD-responsiveness could be detected at the overall gene expression level, whereas others produced nonsense isoforms that could be detected only by analysis of transcript expression levels (Fig. 5B).
We used Northern blotting analyses to verify that the spliced RNA levels from a series of snoRNA host genes (indicated in Fig. 5B) were increased upon depletion of SMG6 or UPF1 and also upon short-term inhibition of translation (e.g., CCNB1IP1 and SNHG15 in Fig. 5C; see Supplemental Fig. S6A for further examples). We note that even though 33 of the 173 snoRNA host genes are annotated as noncoding, many of these are nonetheless highly responsive to NMD (Fig. 5B,C; Supplemental Figs. S6A, S7), which is in accordance with previous reports (Ideue et al. 2007; Weischenfeldt et al. 2008; Yamashita et al. 2009; Thoren et al. 2010; Beaulieu et al. 2012). In contrast to the spliced host RNAs, levels of corresponding hosted snoRNAs were essentially unaffected by UPF1 and SMG6 depletion or inhibition of translation (Fig. 5C; Supplemental Fig. S6A), which suggests that NMD only impacts the spliced host gene product, whereas the precursor RNA is unaffected. We supported this observation by expression of an artificial snoRNA from the second intron of either the β-wt or β-39 genes (Supplemental Fig. S6B). This illustrates that some snoRNA host genes give rise to an additional layer of expression that cannot be appreciated at steady state under normal conditions. A previous transcriptome-wide study reported that snoRNA host genes produce NMD substrates in UPF2-deficient tissues (Weischenfeldt et al. 2008). We reanalyzed RNA-seq data from those tissues (Weischenfeldt et al. 2012), which confirmed that snoRNA host genes are significantly more NMD-sensitive than a group of similarly or even higher-expressed genes (Supplemental Fig. S6C,D). Thus, we conclude that snoRNA host genes are considerably more likely to produce nonsense RNA than protein-coding genes are in general.
Since intron-hosted snoRNAs rely on functional splicing for their production (Kiss et al. 2006; Brown et al. 2008; Dieci et al. 2009), it is highly likely that both NMD-sensitive and NMD-insensitive spliced RNAs from a snoRNA host gene are “leftovers” of snoRNA production. In support of this, we observed an improved correlation between the expression levels of encoded snoRNAs and their corresponding spliced host RNAs when both the NMD-insensitive and NMD-sensitive transcripts were included in the analysis (Fig. 5D). In contrast, there was little correlation between miRNA expression levels and the corresponding spliced host RNAs (Fig. 5D). Based on these observations, we suggest that snoRNA host genes can use the production of nonsense isoforms—e.g., via alternative splicing—to regulate the relative expression levels of the encoded snoRNA and spliced RNA, where the snoRNA would be produced from the precursor RNA independently of whether the spliced RNA ultimately ends up as a stable (perhaps protein-coding) RNA species or a nonsense RNA (Fig. 5E). This allows host RNA and snoRNA levels to be independently regulated despite being produced from the same gene.
Host genes encoding multiple snoRNAs yield substantial numbers of nonsense isoforms
snoRNA host genes can be categorized based on whether they encode a single or multiple snoRNAs (including different snoRNA isoforms). Out of the 173 “active” examples, 108 are single hosts and 65 are multihosts encoding from two to eight snoRNAs. Additionally, 20 single-snoRNA and 13 multi-snoRNA host genes are annotated as being noncoding (Fig. 5B; Supplemental Fig. S7A; Supplemental Table S6).
We noted that multi-snoRNA host genes are expressed at significantly higher levels than their single-snoRNA host counterparts (Fig. 6A). Moreover, when comparing each subgroup with a set of similarly expressed protein-coding genes, it was also apparent that multi-snoRNA host genes produce the highest numbers of nonsense isoforms (Fig. 6B; Supplemental Fig. S7B). Finally, multi-snoRNA host genes yield large numbers of splice variants, particularly intron retention isoforms (Fig. 6C, right panels). This is especially prominent for noncoding multi-snoRNA host genes (Supplemental Fig. S7D). Given that a snoRNA needs to reside completely within an intronic sequence to be expressed (Kiss et al. 2006; Brown et al. 2008; Dieci et al. 2009), we speculated whether such sizeable numbers of alternative splice isoforms could reflect a means for regulating snoRNA expression from multi-snoRNA hosts. Indeed, we found that snoRNAs encoded by multihosts are significantly more likely to be included fully or partially within exons in one or more isoforms than when they are expressed from single hosts (Fig. 6D). This clearly reflects a potential for fine-tuned regulation of individual snoRNAs within the same host gene by post-transcriptional events.
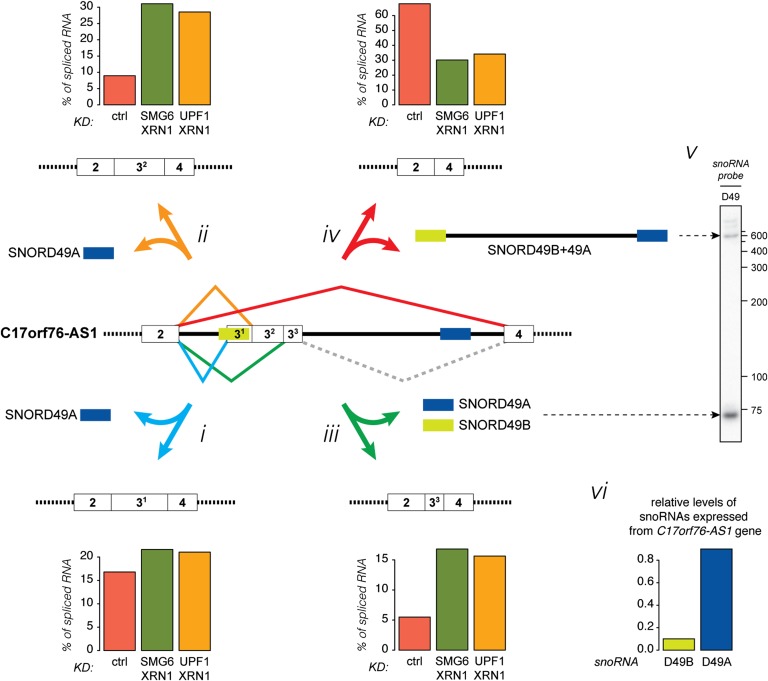
The C17orf76-AS1 gene, which encodes a putative protein as well as the snoRNAs SNORD49B, SNORD49A, and SNORD65, exemplifies this potential (Fig. 7). C17orf76-AS1 also expresses a long ncRNA from an intron (Zhang et al. 2014), which consists of SNORD49B and SNORD49A at the ncRNA termini, flanking the intervening intron sequence (Fig. 7, iv,v, SNORD49B+49A, see Northern panel at the right). The 5′ splice site downstream from exon 2 forms splice connections to four alternative 3′ splice sites, and together these alternative splicing events give rise to different snoRNA/snoRNA-like biogenesis schemes that are schematically illustrated in Figure 7: (i) splicing to exon 31, for which the 3′ splice site is situated inside SNORD49B, thus preventing SNORD49B but allowing SNORD49A expression (indicated by a blue arrow); (ii) splicing to exon 32, leading to production of SNORD49A but most likely preventing SNORD49B production due to suboptimal positioning in the intron (indicated by an orange arrow) (Hirose and Steitz 2001); (iii) splicing to exon 33, leading to expression of both SNORD49B and SNORD49A (indicated by a green arrow); and (iv) splicing to exon 4, thus skipping exon 3, giving rise to the SNORD49B+49A long ncRNA but hindering expression of any of the individual snoRNAs (indicated by a red arrow). RNAs containing the latter splice junction constitute ∼70% of the total amount of exon 2-containing RNA under control conditions (Fig. 7, histograms next to the schematics of the four isoforms show the percent of the total amount of exon 2-containing RNA within samples). However, upon inhibition of NMD, this variant, although being somewhat NMD-sensitive, only constitutes ∼30% of the total RNA. Conversely, the isoforms encoding only SNORD49A comprise approximately half of the total RNA when NMD is inhibited as opposed to a quarter under normal conditions (Fig. 7, i,ii). Similarly, the relative level of the transcript variants giving rise to both SNORD49A and SNORD49B increase from ∼5% to ∼15% upon inhibition of NMD (Fig. 7, iii). Thus, alternative splicing plays a major role in determining which intron-encoded species accumulate. Consistently, the expression of these splice variants measured in NMD-depleted samples reflects better the relative levels of the snoRNA/snoRNA-like molecules as determined by small RNA-seq and Northern blotting analysis (Fig. 7, v,vi).
Figure 7.
Multi-snoRNA host genes regulate production of individual snoRNAs from the same locus by alternative splicing. Model and data illustrating how alternative splicing leads to differential expression of intron-encoded snoRNA-related species. NMD dampens the levels of the alternative splice isoforms to a varying degree. See the text for details.
Discussion
We identified human nonsense RNAs by a transcriptome-wide approach that combines detection of the RNA substrates and their corresponding XRN1-sensitive decay intermediates. This approach has allowed us to first assess the preferred degradation pathway used by the NMD machinery and subsequently identify primary endogenous substrates of NMD.
Degradation of nonsense RNAs is preferably initiated by endocleavage in HEK293 cells
Through a global analysis of endogenous cytoplasmic polyadenylated RNAs, we show that SMG6-catalyzed endocleavages outnumber decapping events as the initial deprotection step during nonsense RNA degradation (Fig. 3). The transcriptome-wide experiment was set up to directly identify NMD-specific endocleavage events, but NMD-specific decapping events could also be detected because depletion of SMG6 leads to increased decapping of nonsense RNAs but not of regular mRNAs (Fig. 2). Although this only offers an indirect assessment of the contribution of decapping to NMD, it allows for a fair comparison of the two phenomena in the context of the same experiment, controlling for any knockdown efficiency biases. Additionally, it is most likely not feasible to design a simple experimental setup to directly detect NMD-specific decapping because NMD uses several different adapter proteins to stimulate decapping, which is furthermore carried out by the common decapping complex also involved in regular mRNA decay. Although we detect a substantial number of NMD substrates with this approach, we cannot rule out that certain nonsense RNAs fail to accumulate degradation intermediates in the polyA+ fraction either because of a general preference for 3′–5′ degradation or because this pathway is accelerated under the given conditions. Similarly, the utilization of NMD might differ between human cell types.
What does the seemingly prevalent endocleavage of endogenous nonsense RNAs tell us about the mechanism of NMD? We demonstrated that increased decapping is accompanied by increased binding of UPF1 to the RNA substrate and also to the adapter proteins SMG5 and SMG7. SMG6 is a single-strand-specific endonuclease (Arraiano et al. 2013), and we propose that the NMD-eliciting complex initially probes the substrate for available single-stranded endocleavage sites. If these or the SMG6 protein are not present, prolonged engagement of the NMD-eliciting complex on the RNA allows for the recruitment of SMG5/SMG7 or SMG5/PNRC2 heterodimers that instead stimulate deadenylation-dependent and deadenylation-independent decapping, respectively (Fig. 3C).
It was recently suggested that XRN1 can act as a cofactor for decapping (Braun et al. 2012). Our data demonstrated that this is not an obligatory requirement, and although decapping levels measured by 5′ end-seq may be slightly underestimated due to a minor effect of XRN1 depletion on decapping, we note that the suggested dominant role played by SMG6, compared with SMG5 and SMG7, is in accordance with results demonstrating that prior depletion of SMG6 is needed to uncover effects of depletion of SMG5 and SMG7 on nonsense RNA degradation but not vice versa (Loh et al. 2013). We therefore suggest that the NMD machinery preferentially uses a SMG6-catalyzed endonucleolytic cleavage as the first local response to the recognition of a PTC. Alternative means of initiation of RNA degradation can then be used if SMG6 action is hindered. It is an outstanding question as to why a subset of nonsense RNAs are preferentially decapped.
Nonsense RNA by-products derive from intron-encoded snoRNA production
We applied both stringent and relaxed criteria to identify NMD-specific endocleavage and decapping events and combined it with differential expression analysis based on RNA-seq data to identify primary targets of NMD (Fig. 4; Supplemental Fig. S5). This strategy revealed nonsense RNA isoforms produced from 4% and 12% of expressed gene loci, respectively. Supporting the validity of our approach, we confirmed that genes encoding splicing factors of the SRSF family are highly enriched among genes producing nonsense RNA isoforms. Additionally, we observed a significant enrichment of snoRNA host genes. Early studies have demonstrated that certain “noncoding” snoRNA host genes produce spliced RNAs that are associated with polysomes and highly stabilized upon inhibition of translation (Tycowski et al. 1996; Smith and Steitz 1998). Later, individual examples of snoRNA host gene-encoded nonsense RNAs from expressed pseudogenes and other “noncoding” genes were reported (Mitrovich and Anderson 2005; Ideue et al. 2007; Weischenfeldt et al. 2008; Yamashita et al. 2009; Thoren et al. 2010; Beaulieu et al. 2012). Here, we demonstrated that there is a strong and general enrichment of the combined class of protein-coding and “noncoding” snoRNA host genes among NMD substrates in HEK293 cells (Fig. 5).
An important question is why snoRNA host genes tend to produce nonsense RNAs. Expression of human intron-hosted snoRNA is dependent on splicing and subsequent liberation of the snoRNA from the excised intron. One of the by-products from snoRNA production is thus a spliced RNA, which will often have mRNA-like characteristics. An mRNA-like “by-product” can be either protein-coding or noncoding and, in the latter (and even sometimes in the former) case, will be degraded by NMD (Fig. 5E). In contrast, biogenesis of hosted miRNAs does not necessarily depend on productive splicing (Brown et al. 2008), which may be the reason that miRNA host genes are not producing NMD-sensitive isoforms to the same degree as the snoRNA host genes (Fig. 5A).
snoRNAs are often coexpressed with genes encoding proteins involved in translation, such as some ribosomal protein-coding genes (Fig. 5B), which has been suggested to be an evolutionary consequence of both molecules being engaged in the same process (Brown et al. 2008; Dieci et al. 2009). However, it is also clear that most snoRNA host gene loci are highly transcribed (Fig. 6), which likely reflects a general need for high expression of snoRNAs. Thus, while some snoRNA host genes may combine high expression of protein-coding mRNA with that of snoRNAs, we suggest that others accommodate high snoRNA expression via transcription levels that greatly exceed those sufficient for the corresponding functional spliced RNA molecules (Fig. 5B). Interestingly, snoRNA host genes come in different flavors; some are conventional protein-coding genes that may mostly produce protein from the encoded spliced RNA, whereas others, such as the SNHG genes, are more reminiscent of loci producing long spliced ncRNAs. However, other snoRNA host genes, such as TAF1D and EIF5 (Fig. 5; Supplemental Fig. S6), which clearly can produce proteins, are also expressing a large fraction of RNAs with intact ORFs that are degraded via NMD. This illustrates that a classification of both the genes and the RNAs as exclusively protein-coding or noncoding is somewhat artificial.
The existence of genes that encode functional RNAs from both exons and introns raises the question of how the expression of these individual species is coordinated. We demonstrate that multi-snoRNA host genes tend to encode large numbers of transcript variants, including many intron retention isoforms (Fig. 6). Partial or complete inclusion of the snoRNA in exonic sequence prevents its expression, and the same is obviously true when the snoRNA is excluded from the precursor transcript. We propose that the many alternative transcript isoforms produced from the multi-snoRNA host loci through usage of alternative splicing, transcription initiation, and termination serve to control differential expression of individual snoRNA species encoded from the same locus (Fig. 7).
Translate to degrade
Our analyses of snoRNA host genes revealed that the most NMD-responsive ones were those annotated as noncoding and encoding multiple snoRNAs (Figs. 5,6; Supplemental Figs. S6, S7). For example, the GAS5 and ZFAS1 (ZNFX1-AS1 in humans) ncRNAs have documented functions (Kino et al. 2010; Askarian-Amiri et al. 2011; Mourtada-Maarabouni et al. 2010; Mourtada-Maarabouni and Williams 2013), but their levels are strongly suppressed by NMD. Most snoRNA host genes are so-called 5′-TOP (terminal oligopyrimidine) genes (Dieci et al. 2009), which means that the encoded spliced RNAs contain an oligopyrimidine stretch at the 5′ end, ensuring that they are removed from the translationally active pool of cytoplasmic RNA during certain types of stress conditions (Ivanov et al. 2011). While this could be a means to up-regulate cytoplasmic levels of ncRNAs under specific conditions, it could also be a way for the cell to save energy, as ribosomes are immediately relieved of a heavy duty of both protein production and RNA degradation. Many ncRNAs are associated with polysomes, and it is debated whether they are indeed productively translated (Ingolia et al. 2011; Banfai et al. 2012; Guttman et al. 2013). Our data show that not only transcripts encoded by multi-snoRNA host genes but also other mRNA-like ncRNAs are sensitive to NMD (Fig. 5–7; Supplemental Figs. S6,S7). This underscores an important and underappreciated role for the translation machinery to not only produce protein but also facilitate cytoplasmic RNA degradation.
Materials and methods
Cell lines and siRNA-mediated depletions
The HEK293 Flp-In T-Rex β-39 and β-wt cell lines were described previously (Eberle et al. 2009). Information about the HEK293 Flp-In T-Rex cell line can be found at the manufacturer’s home page (Life Technologies). siRNA-mediated depletion of XRN1, SMG6, and UPF1 and induction of the β-globin gene variants were essentially performed as previously described (Eberle et al. 2009). We used the following siRNA sequences: control (EGFP) siRNA, GACGUAAACGGCCACAAGUdTdT/ACUUGUGGCCGUUUACGUCdTdT; XRN1 siRNA, AGAUGAACUUACCGUAGAAdTdT/UUCUACGGUAAGUUCAUCUdTdT; SMG6 siRNA, GCUGCAGGUUACUUACAAGdTdT/CUUGUAAGUAACCUGCAGCdTdT; and UPF1 siRNA, GAUGCAGUUCCGCUCCAUUdTdT/AAUGGAGCGGAACUGCAUCdTdT.
Northern blotting
For detection of RNA larger than ∼500 nt, an amount corresponding to 7.5–15 µg of total RNA per sample was separated on a 1.2% agarose gel. A [32P]-labeled DNA ladder was included on gels to verify the sizes of the detected products. For detection of snoRNAs, 2 µg of total RNA per sample was separated on a denaturing 6% polyacrylamide gel next to a [32P]-labeled 25-bp DNA ladder (Life Technologies). Subsequently, the RNA was transferred to a Hybond-N+ membrane (GE Healthcare). Hybridization was performed with [32P]-labeled riboprobes at 68°C in Rapid-Hyb buffer (GE Healthcare) or with [32P]-labeled DNA oligonucleotides at 50°C in ULTRAhyb-Oligo buffer (Life Technologies). Membranes were washed according to the manufacturers’ protocols, followed by exposure to PhosphorImager screens and analyses using Quantity One software (Bio-Rad). Riboprobes were generated by T7- or T3-driven in vitro transcription (Life Technologies) from various templates. Sequences of the ribonucleotide and oligonucleotide probes are listed in the Supplemental Material.
Generation of spike-in RNAs
A set of five spike-in RNAs containing monophosphate moieties and polyA tails (21–25 As) at their 5′ and 3′ ends, respectively, were generated to account for all experimental steps carried out after the initial RNA extraction. More or less randomly picked sense and antisense regions of EGFP, Neomycin, and Luciferase genes were PCR-amplified with primers that introduced an A stretch at one end in the PCR product. The obtained DNA fragment was inserted into the pCR4-TOPO vector (Life Technologies), which was propagated by standard procedures. Restriction enzymes were used to linearize the obtained vectors, which served as templates for T7- or T3-driven in vitro transcription (Life Technologies). The produced RNAs were gel-purified and subjected to an RNA 5′-polyphosphatase (Epicentre) reaction, which leaves a monophosphate at the 5′ end of the RNA. After an additional gel purification step, the concentrations of the spike-in RNAs were determined, and the five RNAs were mixed to a cocktail that was added to total RNA samples before library preparation procedures. Sequences of the spike-in RNAs can be found in the Supplemental Material.
RT-qPCR on adapter-ligated total RNA samples
Spike-in RNAs were added to 10 µg of purified total RNA and subjected to an RNA ligation reaction with 50 pmol of RNA adapter (sequence, ACACUCUUUCCCUACACGACGCUCUUCCGAUCU; 20 U of T4 RNA Ligase 1 [New England Biolabs], 5% [w/v] PEG-8000, 1 mM ATP, 10 U of RiboLock [Fermentas] in a total volume of 25 µL of reaction buffer). After 3 h of incubation at 37°C, the RNA was cleaned up by phenol/chloroform extraction followed by precipitation. Next, 2 µg of the adapter-ligated RNA sample was mixed with 100 pmol of a 4:1 cocktail of random hexamers and dT9 primers and subjected to reverse transcription by SuperScript III (Life Technologies). cDNA corresponding to 50 ng of the adapter-ligated RNA sample was analyzed by qPCR using amplicons detecting adapter ligation to the 5′ end (i.e., decapped) and the full-length mRNA, respectively. The relative level of ligated T7-Luc-pA spike-in RNA was used to control for ligation efficiencies. qPCR was performed with Platinum SYBR Green qPCR Supermix UDG (Life Technologies) using an Mx3005P instrument (Agilent Technologies). Primer sequences are listed in the Supplemental Material.
Coimmunoprecipitation
The Magna RIP kit (Millipore) was used for all immunoprecipitation experiments. Four 6-cm plates were used for each condition in these experiments. Immediately before harvest, the cells were rinsed in 2 mL of cold phosphate-buffered saline (PBS) and scraped in 300 µL of cold PBS per plate. Cells subjected to the same conditions were pooled and divided for later RNA and protein coimmunoprecipitations. Cells were collected by centrifugation at 500g for 5 min at 4°C. Cell pellets were lysed in 1 vol of lysis buffer prepared according to the manufacturer’s protocol, with the further addition of phosphatase inhibitor cocktail 2 (1:200 [v/v]; Sigma). This was done to prevent dephosphorylation of UPF1 and thus inhibit disassociation of the NMD-eliciting complex. To complete lysis, the resuspended cells were left for 5 min on ice followed by a complete freeze on dry ice. Cell lysates were thawed and centrifuged at 20,000g for 10 min at 4°C, and the supernatant/lysate was collected. Protein concentration was determined in the lysates by Bio-Rad protein assay dye, and concentrations were adjusted. One-hundred microliters of lysate was diluted to 1 mL in RIP buffer supplemented with RNase and phosphatase inhibitors and added to magnetic beads prebound with either RIPAb+ UPF1 or IgG control antibody (Millipore, catalog no. 03-191). Input samples were saved for both Northern and Western analyses. After 2–3 h of incubation at 4°C with rotation, the beads were washed four times, after which they were treated differently depending on whether coimmunoprecipitated RNA or protein was to be analyzed. In case of RNA coimmunoprecipitations, 10% of each reaction was used to control for immunoprecipitation of UPF1, and 90% was used for analysis of coimmunoprecipitated RNA: After the final wash, each reaction and the corresponding input samples were resuspended in 150 μL of proteinase K buffer prepared according to the manufacturer’s protocol and incubated for 30 min at 55°C with shaking. Subsequently, the RNA was cleaned up by phenol/chloroform extraction followed by precipitation. The RNA was analyzed by Northern blotting or RT-qPCR. Reverse transcription reactions were conducted as described (see “RT-qPCR on adapter-ligated total RNA samples”). Immunoprecipitated RNA was supplemented with total yeast RNA to adjust concentrations to the same level as for the input samples. For protein coimmunoprecipitation analyses, the beads were treated for 30 min at room temperature with 100 µg/mL RNase A in RIP buffer without RNase inhibitor. Subsequently, the supernatant was removed (RNase A-sensitive fraction in Fig. 2G), and proteins still bound to the beads were eluted in LDS sample buffer (RNase A-insensitive fraction in Fig. 2G).
Western blotting
Whole-cell extracts were obtained by dissolving cells in RSB100 (10 mM Tris/HCl at pH 7.4, 100 mM NaCl, 2.5 mM MgCl2) supplemented with 0.5% Triton X-100 followed by two 5-sec pulses of sonication at the lowest setting (Branson Sonifier 250). Subsequently, the cell lysates were cleared by centrifugation, and the concentrations were adjusted using Bio-Rad protein assay dye. After separation on a NuPage 4%–12% Bis/Tris (Novex/Life Technologies) (Fig. 2G) or a 10% SDS–polyacrylamide gel (Supplemental Fig. S1C), the proteins were transferred to an Immobilon-P membrane (Millipore) by standard procedures. The membranes were probed as indicated with rabbit anti-UPF1, rabbit anti-UPF2, rabbit anti-UPF3a, rabbit anti-UPF3b, rabbit anti-XRN1, and rabbit anti-SMG5 (kind gifts from J. Lykke-Andersen); rabbit anti-SMG6 (kind gift from L. Harrington or Abcam, catalog no. ab87539); rabbit anti-SMG7 (Novus Biologicals, catalog no. NBP1-22967); mouse anti- U170K (kind gift from D. Black); rabbit-anti-HuR (Santa Cruz Biotechnology, catalog no. sc-5261); and β-actin (Sigma, catalog no. A2228) antibodies followed by incubation with the appropriate HRP-conjugated secondary antibody. Bound antibodies were detected with the SuperSignal West Femto maximum sensitivity substrate (Thermo Scientific) followed by autoradiography.
Preparation of 5′ end-seq and RNA-seq libraries for massive parallel sequencing
Approximately 7 × 107 cells (HEK293 Flp-In T-Rex β-39) were seeded in 12 × 6-cm plates for each knockdown condition. After the knockdown procedure, which includes 24-h induction of the β-globin PTC39 gene, the cells were harvested (5 d after seeding). The cells were washed in PBS and subsequently collected in PBS. One-tenth of the cells was collected for analysis of total RNA (Supplemental Fig. S1B, lanes 1–4) and protein (Supplemental Fig. S1C, top panel). The remaining cells were divided into two tubes (treated identically) and pelleted by centrifugation at 500g for 5 min at 4°C, and the supernatant was removed. The cell pellet was loosened by gentle vortexing for 10 sec, and then the cells were lysed by slow addition of 5 mL of lysis buffer (10 mM Tris/HCl at pH 7.4, 10 mM NaCl, 2.5 mM MgCl2, 1% [v/v] ribonucleoside vanadyl complex, 40 µg/mL digitonin) while continuously vortexing. The cell suspension/extract was further vortexed for 10 sec and then incubated for 10 min on ice. After centrifugation at 500g for 5 min at 4°C, the supernatant was collected. The pellet was resuspended in 1 mL of lysis buffer, and the centrifugation step was repeated. The two supernatants were pooled (digitonin extract). The pellet was dissolved in 1 mL of TRIzol (Life Technologies), and the RNA was extracted (pellet) (Supplemental Fig. S1B, lanes 5–8). The digitonin extract was in essence a cytoplasmic extract where the nucleus and other membrane-bound organelles have been removed. By using this extract for RNA purification, the amounts of abundant RNA species, such as mitochondrial ribosomal RNA and snoRNA, that can find their way into the libraries were reduced. RNA was purified from the digitonin extract by phenol pH 6.6/chloroform extraction followed by ethanol precipitation (Supplemental Fig. S1B, digitonin-extracted RNA, Input, lanes 9–12). The spike-in RNA cocktail was added to 400 µg of digitonin-extracted RNA, which was subjected to oligo(dT) selection by use of the MicroPoly(A)Purist kit (Life Technologies). The obtained polyA+ and polyA− fractions were analyzed by Northern blotting analyses (Supplemental Fig. S1B, lanes 13–18). The polyA+ RNA was used to generate 5′ end-seq and RNA-seq libraries.
For 5′ end-seq libraries, 15% of the polyA+ RNA was subjected to RNA ligation to the Solexa RNA adapter essentially by the same procedure as described (in “RT-qPCR on adapter-ligated total RNA samples”). Half of the adapter-ligated polyA+ was then reverse-transcribed with SuperScript III using random hexamers with the second Solexa adapter sequence at their 5′ ends. RNA was removed from RNA/cDNA duplexes by treatment with RNase H, and, subsequently, 80% of the single-stranded cDNA was mixed with the Solexa Forward primer and used in a reaction with Herculase II fusion polymerase (Agilent Technologies) to generate double-stranded cDNA (ds-cDNA) (2 min at 95°C, 5 min at 65°C, and 10 min at 68°C). Each ds-cDNA sample was separated on a 1.5% agarose gel flanked on both sides by the Generuler low-range DNA ladder (Fermentas). ds-cDNA with sizes between 300 and 400 bp (corresponding to insertion size of ∼200–300 bp) were excised and purified using the Qiagen gel extraction kit. Next, 90% of the extracted ds-cDNA was PCR-amplified using Herculase II Fusion polymerase and primers compatible for single-end Solexa sequencing (2 min at 95°C followed by 16 cycles of 30 sec at 95°C, 30 sec at 65°C, and 60 sec at 68°C followed by 5 min at 68°C). The PCR product was purified from a 1.5% agarose gel, and the concentration was measured. The four samples were sequenced individually on an Illumina Genome Analyzer IIX instrument at the National High-Throughput DNA Sequencing Center, University of Copenhagen (the sequencing and mapping statistics can be seen in Supplemental Table S1).
For RNA-seq libraries, 50 ng of digitonin-extracted polyA+ RNA was handled according to the ScriptSeq version 2 protocol (Epicentre). ScriptSeq index PCR primers were introduced in the PCR products to allow multiplexing (indexes 1, 2, 7, and 8 for the control, XRN1-, SMG6/ XRN1-, and UPF1/XRN1-depleted samples, respectively). We used 12 PCR cycles to amplify the fragments. The obtained RNA-seq libraries were analyzed on a Bioanalyzer 2100 instrument (Agilent Technologies), mixed to equimolar levels, and sequenced on an Illumina HiSeq 2000 instrument at the Department of Molecular Medicine, Aarhus University Hospital, Skejby (the sequencing and mapping statistics can be seen in Supplemental Table S1).
Preparation of CAGE library for massive parallel sequencing
CAGE libraries were prepared from 5 μg of total RNA purified from 2 × 106 cells with the Purelink minikit (Ambion, catalog no. 12183018A), applying 1% 2-mercaptoethanol and on-column DNase I treatment (Ambion, catalog no. 12185010) as recommended by the manufacturer. Libraries were prepared according to a published protocol (Takahashi et al. 2012). The libraries were sequenced on a Illumina HiSeq 2000 instrument at the National High-Throughput DNA Sequencing Center, University of Copenhagen. To compensate for the low complexity in 5′ ends of the CAGE libraries, 30% Phi-X spike-ins were added to each sequencing lane, as recommended by Illumina.
Reference gene annotation and biotypes
We used Gencode version 17 (Harrow et al. 2012) as the reference annotation for mapping reads and for transcript assembly with de novo settings. All of the biotypes used for computational analysis, unless otherwise mentioned, were derived from the same annotation.
NMD and non-NMD reference sets
There are 12,913 transcripts arising from 6404 genes that are annotated as NMD substrates in GENCODE version 17. In order to obtain reasonable thresholds for detecting genome-wide NMD substrates, we defined an “NMD reference set” based on annotated NMD targets with detected 5′ end-seq peaks (see relevant sections below). This gave us a total set of 962 transcripts corresponding to 683 genes. For comparison, a non-NMD reference set of transcripts undergoing decapping was defined as any type of annotated transcript with decapping peaks with the highest 5′ end-seq signal in a XRN1 depletion library. This gave us a total set of 206 transcripts derived from 105 genes. Both sets are listed in Supplemental Table S2.
General data processing
The 5′ end-seq and RNA-seq data were mapped to the human genome (hg19/GRCh37, Gencode version 17) using the TopHat version 2 algorithm (Supplemental Table S1 for parameters; Kim et al. 2013). The CAGE tags were mapped to the same reference using Bowtie (Supplemental Table S1 for parameters). The mapped RNA-seq reads were used to generate a reference transcriptome by submitting them to annotation-assisted de novo transcript assembly by Cufflinks version 2 (Supplemental Table S1 for parameters; Trapnell et al. 2010).
Spike-in RNA data processing
To quantify the abundance of spike-in reads for the purpose of normalization, we used Bowtie (version 0.12.7) (Langmead et al. 2009) to map all of the RNA-seq and 5′ end-seq reads to an artificial genome concatenated by all of the spike-in sequences. The mapped spike-in reads from the four libraries (control, XRN1, SMG6/XRN1, and UPF1/XRN1) from either RNA-seq or 5′ end-seq were used to calculate normalization factors, which were then used to normalize the expression levels for RNA-seq and 5′ end signals for 5′ end-seq (Supplemental Table S1).
RNA-seq data processing and transcript assembly
RNA-seq reads were aligned to the human genome (hg19/GRCh37) with TopHat version 2 (version 2.0.6) (Kim et al. 2013). Mapping was guided by the reference annotation Gencode version 17 but allowing for novel junctions and genes. Reads that mapped to unconventional chromosomes or the mitochondrial chromosome were discarded. We then used Cufflinks version 2 (version 2.0.2) (Trapnell et al. 2010) to assemble the transcripts independently in each library, and Gencode version 17 was used as a reference genome to guide assembly. Finally, the transcripts from all of the libraries were merged by Cuffmerge and associated to the reference annotation by Cuffcompare (Trapnell et al. 2010). For the intron retention isoforms used for the analysis of snoRNA host genes, we defined retained intron isoforms as the ones fully covering at least one annotated intron sequence.
We observed some discrepancies between the gene number assembled by Cufflinks and the corresponding annotated gene numbers (official gene symbol) due to so-called conjunction genes (genes overlapping in the same transcription direction). To cope with this, the Cufflinks-assembled transcripts and genes were used for most of the presented analyses. However, official gene symbols were used for the analyses associated with gene biotypes presented in Figure 5, A and B, and Supplemental Figures S5, E, G, and H; S6, C and D; and S7A. Removal of the genes causing discrepancies between the number of Cufflinks-assembled genes and annotated genes from the analyses did not change any conclusions (data not shown). Both Cufflinks internal gene IDs and official gene IDs are provided in Supplemental Tables where relevant. The entire Cufflinks-assembled transcriptome can be found in Supplemental Data 1.
We quantified the raw read counts of Cufflinks version 2-determined transcripts and subsequently normalized them based on spike-in RNA levels. After removing the transcripts that do not have any mapped reads in any of the libraries, a set of 122,816 transcripts corresponding to 28,393 Cufflinks-assembled genes and 29,606 annotated genes (official gene symbol) were used for the downstream analysis. For gene expression, we used FPKM (fragments per kilobase of exon per million mapped fragments) values estimated by Cufflinks version 2 except for Figure 5D, where the host gene expression was calculated as the sum of spike-in normalized RNA levels of individual isoforms within the indicated categories of transcripts (non-NMD-responsive or NMD-responsive isoforms or both).
Based on mean fold changes in the RNA-seq-based transcript isoform expression levels within the NMD reference set, we empirically derived lower boundaries for defining a transcript as an NMD target (SMG6/XRN1 and UPF1/XRN1 share the same cutoff) (for the detailed calculation, see Supplemental Table S2): SMG6/XRN1 > 1.74 × CTRL and UPF1/XRN1 > 1.74 × CTRL; SMG6/XRN1 > 1.35 × XRN1, and UPF1/XRN1 > 1.35 × XRN1.
5′ end-seq data processing
TopHat version 2 was also used for the mapping of 5′ end-seq reads to the human genome. The same settings as used for RNA-seq were applied, except for the parameters specific to RNA-seq paired-end reads (see Supplemental Table S1). Only the 5′ ends of the mapped reads, including reads spanning exon–exon junctions, were finally quantified in the analysis followed by normalization based on spike-in RNA 5′ ends for each library.
CAGE data processing
By using FASTX Toolkit (http://hannonlab.cshl.edu/fastx_toolkit), we trimmed away linker sequences in the sequenced reads and subsequently filtered out the reads with >50% of the bases under a quality score of 30. Bowtie (version 0.12.7) (Langmead et al. 2009) was used to map all of the filtered reads to the human genome (hg19/GRCh37) with standard settings but allowing for multiple matches. Reads that mapped to the mitochondrial or unconventional chromosomes were discarded. Only the 5′ ends of the uniquely mapping reads were finally used in the analysis.
Neighboring CAGE signals maximally 20 bp away from each other were merged into CAGE clusters, and singleton CAGE signals were discarded. In order to exclude non-mRNA signals, we refined the CAGE clusters based on both the number of supporting reads and the distributions of the mapped 5′ end tags. This was done by filtering out the CAGE clusters with <10 tag counts and truncating the tails of the CAGE cluster by excluding the subtle signals (≤5%) from both ends. Finally, we associated the CAGE clusters with the closest assembled transcript.
Identification of endocleavage and decapping events
For identification of putative endocleavage and decapping events, we first filtered away transcripts of very low expression using the 10th percentile from the NMD reference set in both of the double-depleted libraries as an expression threshold.
All of the remaining 61,883 transcripts (18,726 Cufflinks-assembled genes corresponding to 18,526 annotated genes), including the de novo isoforms, were used for peak calling. We noted that putative decapping events could sometimes be detected slightly upstream of the assigned 5′ end of the Cufflinks-assembled transcripts. Therefore, we extended all transcripts by 100 nt upstream of the 5′ end to allow detection of such events.
For each transcript in each 5′ end-seq library, we fetched the 5′ end signals falling in the corresponding exons and fitted these signals into a negative binomial distribution using the R package MASS (Venables and Ripley 2002). We then assessed the P-values for the count in each exonic position according to the fitted distribution followed by a false discovery rate (FDR) correction (Benjamini and Hochberg 1995). Corrected P-values (Q) ≤0.05 were used as the cutoff for stringent peaks, while P-values ≤0.005 were used for the relaxed set (see also Supplemental Fig. S4).
The 5′ end-seq peaks overlapping the associated CAGE clusters were assigned as “decapping” candidates. Peaks overlapping CAGE clusters in regions that were not associated with a transcript 5′ end (further than 100 nt downstream from an assigned transcript 5′ end) were assigned as “alternative decapping” candidates. The remaining peaks were considered as “endocleavage” candidates.
Based on mean fold changes in 5′ end-seq peak signals within the NMD reference set, we empirically derived lower boundaries for defining a transcript as an NMD target (SMG6/XRN1 and UPF1/XRN1 share the same cutoff) (for the detailed calculation, see Supplemental Table S2): NMD-specific endocleavage site: XRN1 > 1.52 × SMG6/XRN1 and XRN1 > 1.52 × UPF1/XRN1; NMD-specific decapping site: SMG6/XRN1 > 1.94 × XRN1 and UPF1/XRN1 > 1.94 × XRN1.
We also considered the peak candidates from the “alternative decapping” group as an endocleavage site if the peak passed the cutoff for NMD-specific endocleavage, and the associated transcript passed the NMD-specific RNA-seq cutoff (mentioned as “special cases” in Fig. 4B; Supplemental Fig. S5D).
Graphical representation of sequencing data
All plots of sequencing data were done by use of standard packages in R (http://www.R-project.org). Alternative splice events from a given splice site were included in the plot if the major splice event constituted <75% of all possible splice events in at least one of the samples. Conversely, minor splice events were only included if they constituted >20% in at least one of the samples. To improve the representation of both coverage and differential expression between samples, the signals within each “exon window” were scaled to fit the Y-axis of the plotting window. Leading and trailing intronic sequences were included within each “exon window” to illustrate the drop in signal over exon–intron borders.
Small RNA-seq data processing
For estimating the expression of snoRNA and miRNA, we used the mapped small RNA data from a previous study (Kishore et al. 2013). Expression levels of small RNAs were calculated as the sum of the aligned reads falling within the respective annotations.
Definition of snoRNA and miRNA host genes
We defined a gene as a host gene if it produced at least one transcript (determined by RNA-seq) that fully covered an annotated snoRNA or miRNA (Gencode version 17) on the same strand. For snoRNA host genes, we excluded U3 (SNORD3A), U8 (SNORD118), and U13 (SNORD13) snoRNAs from the analysis, since they are known to be produced from independent gene loci, therefore making analysis ambiguous (Dieci et al. 2009), which led to a set of 442 snoRNA host genes. For allowing a fair comparison between snoRNA host genes and miRNA host genes, we did not apply more restrictions on this snoRNA host gene set in gene enrichment analysis (Fig. 5A; Supplemental Fig. S5E).
For the downstream analyses focusing on snoRNA host genes (from Fig. 5B and on), we removed the snoRNAs with no support in the small RNA-seq data and only allowed the top 95% expressed snoRNAs in the analyses. The same criteria were applied to miRNA host genes in expression correlation analysis (Fig. 5D). Additionally, we further restricted snoRNA host genes to the canonical understanding of snoRNA regulation; i.e., the snoRNA should be fully included in intronic sequence in at least one isoform produced from the host gene. This gave us a set of 173 “active” annotated snoRNA host genes (corresponding to 163 Cufflinks-assembled genes) expressing a total of 271 snoRNA isoforms arising from 242 snoRNAs in HEK293 cells (Supplemental Table S6).
Additional statistical analyses
Mann-Whitney two-sided tests were conducted for all statistical testing on differences between distributions unless otherwise stated.
Density plots were used to show the difference in variable distributions of interest. All densities were estimated by kernel density estimation (KDE) by use of the density function in R from the “stats” package (http://www.R-project.org). The default Gaussian kernel was used for all of the density plots.
Simulation of protein-coding genes with specified expression ranges
For comparing the different properties between snoRNA host genes and protein-coding genes, we simulated several protein-coding sets with similar expression pattern according to the corresponding snoRNA host set. This was done by breaking up the snoRNA host gene expression distribution into 25 bins. For each bin, protein-coding genes whose expression fit with the expression range of this bin were randomly sampled. In the simulation, we set the number of genes to 1000, 2000, and 3000 for comparing with multiple snoRNA host genes (65), single snoRNA host genes (108), and all snoRNA host genes (173), respectively (numbers of known snoRNA host genes are shown in parenthesis for the respective categories).
Conservation analysis in mice
For all of the analysis of mouse data, we used mapped RNA-seq libraries from a public repository (Weischenfeldt et al. 2012). In order to be consistent with the target transcriptome previously used in the mapping procedures (Weischenfeldt et al. 2012), we used University of California at Santa Cruz (UCSC) MM9 gene annotation for all of the downstream analyses, including the FPKM quantification by Cufflinks 2 and the enrichment of gene biotypes.
Accession numbers
All sequencing data described in this study have been deposited at The National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE57433 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=qhelyqishpitfqn&acc=GSE57433).
Supplementary Material
Acknowledgments
We thank Karina Hjort and Dorthe C. Riishøj for technical assistance, and Thomas B. Hansen for help with initial bioinformatics analyses. We thank Oliver Mühlemann (University of Bern, Switzerland), Jens Lykke-Andersen (University of California at San Diego), and Christian K. Damgaard (Aarhus University, Denmark) for critical reading of the manuscript. Antibodies were generously provided by Jens Lykke-Andersen (XRN1, UPF1, UPF2, UPF3a, UPF3b, and SMG5 antibodies), Lea Harrington (Wellcome Trust Centre for Cell Biology, Edinburgh; SMG6 antibody), and Douglas Black (University of California at Los Angeles; U170K antibody). The Lundbeck Foundation supported this project (S.L.-A.). Additional support was obtained from Danish National Research Foundation grant DNRF58 (T.H.J.), the Danish Natural Science Research Council (T.H.J.), the interdisciplinary sequencing (iSEQ) centre (T.H.J.), Lundbeck Foundation grants R100-A9297 and R34-A3770 (A.S.), the Novo Nordisk Foundation (A.S.), the Risk and Modifying Factors in Fronto–Temporal Dementia (RiMOD-FTD) EU joint program (A.S.), and ERC grant agreement 204135 (A.S). S.L.-A. conceived the project, designed and performed the experiments (unless otherwise stated), performed computational analyses, and drafted the manuscript. Y.C. conducted and designed computational and statistical analyses and revised the manuscript. B.R.A. carried out immunoprecipitation and RNA decay experiments. B.L. generated the CAGE library. J.W. assisted in the initial computational analyses. A.S. and T.H.J. supervised the project and revised the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.246538.114.
References
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118. [DOI] [PubMed] [Google Scholar]
- Arraiano CM, Mauxion F, Viegas SC, Matos RG, Seraphin B. 2013. Intracellular ribonucleases involved in transcript processing and decay: precision tools for RNA. Biochim Biophys Acta 1829: 491–513. [DOI] [PubMed] [Google Scholar]
- Askarian-Amiri ME, Crawford J, French JD, Smart CE, Smith MA, Clark MB, Ru K, Mercer TR, Thompson ER, Lakhani SR, et al. 2011. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA 17: 878–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE Jr, Kundaje A, Gunawardena HP, Yu Y, Xie L, et al. 2012. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res 22: 1646–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu YB, Kleinman CL, Landry-Voyer AM, Majewski J, Bachand F. 2012. Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-binding protein nuclear 1. PLoS Genet 8: e1003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. 2007. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J 26: 1591–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300. [Google Scholar]
- Braun JE, Truffault V, Boland A, Huntzinger E, Chang CT, Haas G, Weichenrieder O, Coles M, Izaurralde E. 2012. A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5′ exonucleolytic degradation. Nat Struct Mol Biol 19: 1324–1331. [DOI] [PubMed] [Google Scholar]
- Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. 2013. Dynamic integration of splicing within gene regulatory pathways. Cell 152: 1252–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JW, Marshall DF, Echeverria M. 2008. Intronic noncoding RNAs and splicing. Trends Plant Sci 13: 335–342. [DOI] [PubMed] [Google Scholar]
- Cao D, Parker R. 2003. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell 113: 533–545. [DOI] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38: 626–635. [DOI] [PubMed] [Google Scholar]
- Chamieh H, Ballut L, Bonneau F, Le Hir H. 2008. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol 15: 85–93. [DOI] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. 2003. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol Cell Biol 23: 4805–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kim KM, Kim YK. 2009. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol Cell 33: 75–86. [DOI] [PubMed] [Google Scholar]
- Cho H, Han S, Choe J, Park SG, Choi SS, Kim YK. 2013. SMG5–PNRC2 is functionally dominant compared with SMG5–SMG7 in mammalian nonsense-mediated mRNA decay. Nucleic Acids Res 41: 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couttet P, Grange T. 2004. Premature termination codons enhance mRNA decapping in human cells. Nucleic Acids Res 32: 488–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G, Preti M, Montanini B. 2009. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics 94: 83–88. [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R. 2007. RNA quality control in eukaryotes. Cell 131: 660–668. [DOI] [PubMed] [Google Scholar]
- Eberle AB, Stalder L, Mathys H, Orozco RZ, Muhlemann O. 2008. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol 6: e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55. [DOI] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell 20: 905–915. [DOI] [PubMed] [Google Scholar]
- Franks TM, Singh G, Lykke-Andersen J. 2010. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense- mediated mRNA decay. Cell 143: 938–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E. 2004. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429: 575–578. [DOI] [PubMed] [Google Scholar]
- Ge Y, Porse BT. 2014. The functional consequences of intron retention: alternative splicing coupled to NMD as a regulator of gene expression. BioEssays 35: 236–243. [DOI] [PubMed] [Google Scholar]
- Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, Dieterich C, Landthaler M. 2014. MOV10 is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol Cell 54: 573–585. [DOI] [PubMed] [Google Scholar]
- Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. 2013. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154: 240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. 2012. GENCODE: the reference human genome annotation for the ENCODE Project. Genome Res 22: 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Steitz JA. 2001. Position within the host intron is critical for efficient processing of box C/D snoRNAs in mammalian cells. Proc Natl Acad Sci 98: 12914–12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JR, Goff SP. 2010. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143: 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E. 2008. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14: 2609–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt JA, Robertson AD, Burge CB. 2013. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res 23: 1636–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideue T, Sasaki YT, Hagiwara M, Hirose T. 2007. Introns play an essential role in splicing-dependent formation of the exon junction complex. Genes Dev 21: 1993–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada T. 2013. Quality control systems for aberrant mRNAs induced by aberrant translation elongation and termination. Biochim Biophys Acta 1829: 634–642. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. 2008. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27: 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Kedersha N, Anderson P. 2011. Stress puts TIA on TOP. Genes Dev 25: 2119–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, Stamm S. 2013. Function of alternative splicing. Gene 514: 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervestin S, Jacobson A. 2012. NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol 13: 700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. 2010. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 3: ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore S, Gruber AR, Jedlinski DJ, Syed AP, Jorjani H, Zavolan M. 2013. Insights into snoRNA biogenesis and processing from PAR-CLIP of snoRNA core proteins and small RNA sequencing. Genome Biol 14: R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T, Fayet E, Jady BE, Richard P, Weber M. 2006. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb Symp Quant Biol 71: 407–417. [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR, Schor IE, Allo M, Dujardin G, Petrillo E, Munoz MJ. 2013. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol 14: 153–165. [DOI] [PubMed] [Google Scholar]
- Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH. 2006. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA 12: 1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T, Cho H, Liu Z, Bowler MW, Piao S, Parker R, Kim YK, Song H. 2012. Structural basis of the PNRC2-mediated link between mRNA surveillance and decapping. Structure 20: 2025–2037. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. 2007. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446: 926–929. [DOI] [PubMed] [Google Scholar]
- Lejeune F, Li X, Maquat LE. 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell 12: 675–687. [DOI] [PubMed] [Google Scholar]
- Loh B, Jonas S, Izaurralde E. 2013. The SMG5–SMG7 heterodimer directly recruits the CCR4–NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev 27: 2125–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Caceres JF. 2009. The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417: 15–27. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol 22: 8114–8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlincy NJ, Smith CW. 2008. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci 33: 385–393. [DOI] [PubMed] [Google Scholar]
- Melero R, Buchwald G, Castano R, Raabe M, Gil D, Lazaro M, Urlaub H, Conti E, Llorca O. 2012. The cryo-EM structure of the UPF–EJC complex shows UPF1 poised toward the RNA 3′ end. Nat Struct Mol Biol 19: 498–505. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Tollervey D. 2003. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′ → 5′ degradation. Mol Cell 11: 1405–1413. [DOI] [PubMed] [Google Scholar]
- Mitrovich QM, Anderson P. 2005. mRNA surveillance of expressed pseudogenes in C. elegans. Curr Biol 15: 963–967. [DOI] [PubMed] [Google Scholar]
- Mourtada-Maarabouni M, Williams GT. 2013. Growth arrest on inhibition of nonsense-mediated decay is mediated by noncoding RNA GAS5. Biomed Res Int 2013: 358015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourtada-Maarabouni M, Hasan AM, Farzaneh F, Williams GT. 2010. Inhibition of human T-cell proliferation by mammalian target of rapamycin (mTOR) antagonists requires noncoding RNA growth-arrest-specific transcript 5 (GAS5). Mol Pharmacol 78: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlemann O, Jensen TH. 2012. mRNP quality control goes regulatory. Trends Genet 28: 70–77. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. 1994. Premature translational termination triggers mRNA decapping. Nature 370: 578–581. [DOI] [PubMed] [Google Scholar]
- Nagarajan VK, Jones CI, Newbury SF, Green PJ. 2013. XRN 5′ → 3′ exoribonucleases: structure, mechanisms and functions. Biochim Biophys Acta 1829: 590–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E, Maquat LE. 1998. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci 23: 198–199. [DOI] [PubMed] [Google Scholar]
- Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S. 2012. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res 40: 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweingruber C, Rufener SC, Zund D, Yamashita A, Muhlemann O. 2013. Nonsense-mediated mRNA decay—mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta 1829: 612–623. [DOI] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. 2012. Translation drives mRNA quality control. Nat Struct Mol Biol 19: 594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AL, Ribeiro P, Inacio A, Liebhaber SA, Romao L. 2008. Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA 14: 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Rebbapragada I, Lykke-Andersen J. 2008. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol 6: e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Steitz JA. 1998. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol 18: 6897–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Araki Y, Sakuno T, Katada T. 2003. Interaction between Ski7p and Upf1p is required for nonsense-mediated 3′-to-5′ mRNA decay in yeast. EMBO J 22: 3951–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kato S, Murata M, Carninci P. 2012. CAGE (cap analysis of gene expression): a protocol for the detection of promoter and transcriptional networks. Methods Mol Biol 786: 181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoren LA, Norgaard GA, Weischenfeldt J, Waage J, Jakobsen JS, Damgaard I, Bergstrom FC, Blom AM, Borup R, Bisgaard HC, et al. 2010. UPF2 is a critical regulator of liver development, function and regeneration. PLoS ONE 5: e11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Steitz JA. 1996. A mammalian gene with introns instead of exons generating stable RNA products. Nature 379: 464–466. [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E. 2004. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell 16: 587–596. [DOI] [PubMed] [Google Scholar]
- Valen E, Pascarella G, Chalk A, Maeda N, Kojima M, Kawazu C, Murata M, Nishiyori H, Lazarevic D, Motti D, et al. 2009. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res 19: 255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables WN, Ripley BD. 2002. Modern applied statistics with S, Fourth edition. Springer, New York. [Google Scholar]
- Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT. 2008. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev 22: 1381–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Waage J, Tian G, Zhao J, Damgaard I, Jakobsen JS, Kristiansen K, Krogh A, Wang J, Porse BT. 2012. Mammalian tissues defective in nonsense-mediated mRNA decay display highly aberrant splicing patterns. Genome Biol 13: R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A. 2013. Role of SMG-1-mediated Upf1 phosphorylation in mammalian nonsense-mediated mRNA decay. Genes Cells 18: 161–175. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. 2005. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 12: 1054–1063. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, et al. 2009. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev 23: 1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XO, Yin QF, Wang HB, Zhang Y, Chen T, Zheng P, Lu X, Chen LL, Yang L. 2014. Species-specific alternative splicing leads to unique expression of sno-lncRNAs. BMC Genomics 15: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zund D, Gruber AR, Zavolan M, Muhlemann O. 2013. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nat Struct Mol Biol 20: 936–943. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.