Abstract
Proton pump inhibitors (PPI) have been associated with infectious complications in cirrhosis, but their impact on distal gut microbiota composition and function is unclear. We aimed to evaluate changes in stool microbiota composition and function in patients with cirrhosis and healthy controls after omeprazole therapy. Both 15 compensated cirrhotic patients and 15 age-matched controls underwent serum gastrin measurement, stool microbiota profiling with multitagged pyrosequencing, and urinary metabolic profiling with NMR spectroscopy to assess microbial cometabolites before/after a 14-day course of 40 mg/day omeprazole under constant diet conditions. Results before (pre) and after PPI were compared in both groups, compared with baseline by systems biology techniques. Adherence was >95% without changes in diet or MELD (model for end-stage liver disease) score during the study. Serum gastrin concentrations significantly increased after PPI in cirrhosis (pre 38.3 ± 35.8 vs. 115.6 ± 79.3 pg/ml P < 0.0001) and controls (pre 29.9 ± 14.5 vs. 116.0 ± 74.0 pg/ml, P = 0.001). A significant microbiota change was seen in both controls and cirrhosis after omeprazole (QIIME P < 0.0001). Relative Streptococcaceae abundance, normally abundant in saliva, significantly increased postomeprazole in controls (1 vs. 5%) and cirrhosis (0 vs. 9%) and was correlated with serum gastrin levels (r = 0.4, P = 0.005). We found significantly reduced hippurate in cirrhosis vs. controls both pre- and postomeprazole and increased lactate in both groups post vs. preomeprazole, whereas dimethylamine (DMA) decreased in cirrhosis only. On correlation network analysis, significant changes in linkages of bacteria with metabolites (hippurate/DMA/lactate) were found postomeprazole, compared with pre-PPI in cirrhosis patients. In conclusion, omeprazole is associated with a microbiota shift and functional change in the distal gut in patients with compensated cirrhosis that could set the stage for bacterial overgrowth.
Keywords: gut barrier, infection, metabolomics, microbiome, proton pump inhibitor, gastrin
in cirrhosis, alteration in gut microbiota may predispose to several complications, such as hepatic encephalopathy, acute-on-chronic liver failure and infections, such as spontaneous bacterial peritonitis (SBP), presumably through bacterial translocation and bacterial overgrowth (3, 8, 24). Proton pump inhibitors (PPI), by reducing gastric acidity, have been implicated in small intestinal bacterial overgrowth and these infectious complications (6, 18, 28). Most studies evaluating this association have found an indiscriminate use of these medications in patients with cirrhosis (7). The impact of these medications on the composition and functionality of distal gut microbiota in cirrhosis is not fully understood.
We hypothesized that PPI therapy can significantly change gut microbiota composition and function in patients with cirrhosis and healthy controls. The aim was to analyze systematically the stool microbiota composition and urinary metabolic profile in patients with compensated cirrhosis and healthy controls before and after a 2-wk course of omeprazole therapy.
MATERIALS AND METHODS
We recruited patients with cirrhosis (diagnosed with biopsy or evidence of portal hypertension in patients with chronic liver disease) without decompensation (Child A, based on Child-Turcotte-Pugh scoring system), who were not currently on PPI therapy. Patients who had Child B/C cirrhosis, unclear diagnosis of cirrhosis, or Helicobacter pylori positivity on serology, were already on PPI therapy, or were unable to give consent were excluded. We also included age-matched healthy controls without chronic disease who were also not already on PPI therapy. This protocol was approved by the McGuire VA Medical Center Institutional Review Board.
At the baseline visit, all subjects underwent a detailed 24-h food recall, blood was drawn for baseline gastrin (for all subjects) and model for end-stage liver disease (MELD) score (for patients with cirrhosis), fresh stool was collected for microbiota, and urine was collected for urinary nuclear magnetic resonance (NMR) metabolomics.
Subjects were given instructions regarding maintaining the dietary pattern and omeprazole 40 mg to be taken before breakfast every day for 14 days.
After 14 days, subjects were reevaluated and all procedures performed at baseline visit were repeated. Adherence was assessed by use of pill return, and adverse effects related to omeprazole were also investigated.
Sample Analysis
Gastrin.
Gastrin concentrations were measured by radioimmunoassay at Quest Diagnostics (Chantilly, VA).
Microbiota.
Stool was collected and DNA was extracted by published techniques (4). We first used Length Heterogeneity PCR (LH-PCR) fingerprinting of the 16S rRNA to rapidly survey our samples and standardize the community amplification. We then interrogated the microbial taxa associated using Multitag Pyrosequencing (MTPS) (11). This technique allows the rapid sequencing of multiple samples at one time.
Microbiome community fingerprinting.
About 10 ng of extracted DNA was amplified by PCR by use of a fluorescently labeled forward primer 27F [5′-(6FAM) AGAGTTTGATCCTGGCTCA G-3′] and unlabeled reverse primer 355R′ (5′-GCTGCCTCCCGTAGGAGT-3′). Both primers are universal primers for bacteria. The LH-PCR products were diluted according to their intensity on agarose gel electrophoresis and mixed with ILS-600 size standards (Promega) and HiDi Formamide (Applied Biosystems, Foster City, CA). The diluted samples were then separated on a ABI 3130xl fluorescent capillary sequencer (Applied Biosystems) and processed with the GeneMapper software package (Applied Biosystems). Normalized peak areas were calculated via a custom PERL script and operational taxonomic units (OTUs) constituting less than 1% of the total community from each sample were eliminated from the analysis to remove the variable low-abundance components within the communities.
MTPS.
Specifically, using MTPS (11) we have generated a set of 96 emulsion PCR fusion primers that contain the 454 emulsion PCR linkers on the 27F and 355R primers and a different eight-base “barcode” between the A adapter and 27F primer. Thus each fecal sample was amplified with unique bar-coded forward 16S rRNA primers, and then up to 96 samples were pooled and subjected to emulsion PCR and pyrosequenced by use of a GS-FLX pyrosequencer (Roche). Data from each pooled sample were “deconvoluted” by sorting the sequences into bins based on the barcodes by use of custom PERL scripts. Thus we were able to normalize each sample by the total number of reads from each barcode. We have noted that ligating tagged primers to PCR amplicons distorts the abundances of the communities, and thus it is critical to incorporate the tags during the original amplification step.
Microbiome community analysis.
We identified the taxa present in each sample using the Bayesian analysis tool in Version 10 of the Ribosomal Database Project (RDP10). The abundances of the bacterial identifications were then normalized with a custom PERL script, and genera present at >1% of the community were tabulated. We chose this cutoff because of our a priori assumption that genera present in <1% of the community vary between individuals and have minimal contribution to the functionality of that community, and 2,000 reads per sample will only reliably identify community components that are greater than 1% in abundance.
Urinary NMR metabolic profiling.
Urinary metabolic profiling was undertaken by NMR, since metabolites measured by NMR have been shown to provide information about the gut microbial-mammalian metabolic axis and the contribution of the gut microbiota to drug metabolism (10, 17).
Urinary NMR spectra were acquired at 298 K from 400 μl of urine, buffered to pH 7.4 with 200 μl of phosphate buffer, by use of a JEOL ECP 500 MHz NMR spectrometer (JEOL, Tokyo, Japan). A standard pulse-collect sequence with water presaturation was used to acquire the NMR data (90° pulse length, relaxation delay 2 s, acquisition time 4.36 s, spectral width 15 ppm, 32 K data points, 64 data collects). Chemical shifts were referenced to added 3-trimethylsilyl-(2,2,3,3-2H4)-1-propionate (TSP) at 0.00 pm.
Systems Biology Analysis
Analysis of microbiota.
Quantitative Insights Into Microbial Ecology (QIIME) analysis was used to evaluate changes in overall microbial abundance before and after PPI, and principal component analysis (PCA) was performed before and after PPI to gauge microbiota clustering between groups (http://qiime.org/tutorials/tutorial.html). We also performed Metastats to evaluate changes in relative abundance between groups before and after PPI therapy with correction for the false discovery rate (FDR) (29).
Analysis of urinary NMR spectra.
NMR peaks were assigned using publically available databases (31). The resonances assigned to residual water and urea (δ 4.6–6.4 ppm) were excluded from further analysis. Prior to multivariate analysis the NMR spectra were subdivided into smaller regions, some representing specific metabolites, by use of the Intelligent Bucketing algorithm within KnowItAll (KnowItAll v9.0, Bio-Rad, Philadelphia, PA), and the data were mean centered. PCA was used as an unsupervised method for data visualization. Univariate analysis was used to determine the significance of differences in specific metabolite levels, compared with the total spectral integral, between groups (IBM SPSS Statistics 21, IBM 2012). A Mann-Whitney U-test was used to compare the control and cirrhosis groups both pre- and postomeprazole. A Wilcoxon signed-rank test was used to compare individual subjects in the control group pre- and postomeprazole and also in the cirrhosis group pre- and postomeprazole.
Correlation network analysis.
We created correlation networks using tools in the Galaxy Portal at the Microbiome Analysis Center and only included nodes consisting of microbiota and urinary NMR metabolites at baseline and post-PPI for controls and patients with cirrhosis separately (19). The correlation differences between baseline and post-PPI networks were visualized in Cytoscape and then analyzed separately for controls and patients with cirrhosis (22).
RESULTS
We enrolled 15 patients with cirrhosis and 15 age-matched healthy controls. The MELD score was 8.4 ± 2.7 and the etiology of cirrhosis was predominantly hepatitis C (n = 8), alcohol (n = 2) and hepatitis C + alcohol (n = 5). All subjects were negative for H. pylori on serology.
Subjects returned 14 ± 3 days later with >95% adherence on omeprazole intake documented by pill bottle return. No adverse events, including infections, occurred during the study period. There was no significant change in daily caloric intake in the control group (2,354 vs. 2,431 kcal/day, P = 0.4) or patients with cirrhosis (2,214 vs. 2,245 kcal/day, P = 0.6, Table 1) over the study period. The MELD score in patients with cirrhosis remained stable (8.4 vs. 8.0, P = 0.5) and no complications of cirrhosis occurred during the study.
Table 1.
Baseline features of the groups
| Cirrhotic Patients (n = 15) | Controls (n = 15) | |
|---|---|---|
| Age, years ± SD | 55 ± 4 | 54 ± 3 |
| Sex, Men/Women | 10/5 | 8/7 |
| Race, Caucasian/African-American/Hispanic | 7/6/2 | 8/4/3 |
| Caloric intake in the last 24 h, kcal ± SD | 2,214 ± 457 | 2,354 ± 342 |
None of the comparisons were statistically significant.
Gastrin Results
There was a significant increase in serum gastrin after omeprazole compared with baseline in patients with cirrhosis (values expressed as means ± SD; pre 38.3 ± 35.8 vs. 115.6 ± 79.3 pg/ml P < 0.0001) and controls (pre 29.9 ± 14.5 vs. 116.0 ± 74.0 pg/ml, P = 0.001) using paired t-tests. However, there was no significant difference in gastrin concentrations when controls were compared with patients with cirrhosis, either at baseline (P = 0.43) or after omeprazole (P = 0.98) by use of unpaired t-tests.
Urinary NMR Metabolic Profiling Results
Urine samples were archived from 15 control subjects and 14 patients with cirrhosis, since one NMR data set, in a subject with cirrhosis before omeprazole, was of inadequate quality to be included for further analysis. All the NMR spectra were normalized to the total spectral integral in the range δ = 0.2–10 ppm, excluding the water, urea, glucose, acetaminophen, and ethanol regions, since these resonances complicated interpretation of the metabolites of interest. PCA plots from controls (n = 15) vs. cirrhosis (n = 14) showed separate clustering of controls and patients with cirrhosis, both preomeprazole and postomeprazole (Fig. 1). Metabolites that were found to be different on NMR (relative signal level in a metabolite region compared with total NMR signal levels with confounding regions excluded) were hippurate, dimethylamine (DMA), and lactate. Relative hippurate levels were significantly reduced in patients with cirrhosis, compared with controls both at baseline (P = 0.007) and postomeprazole (P = 0.003). Considering metabolite changes on an individual basis, lactate levels were increased in both controls (P = 0.001) and patients with cirrhosis (P = 0.034), postomeprazole compared with pre-PPI values. Additionally there was a significant reduction in DMA levels in individuals in the cirrhosis group post-PPI compared with their baseline state (P = 0.034)
Fig. 1.
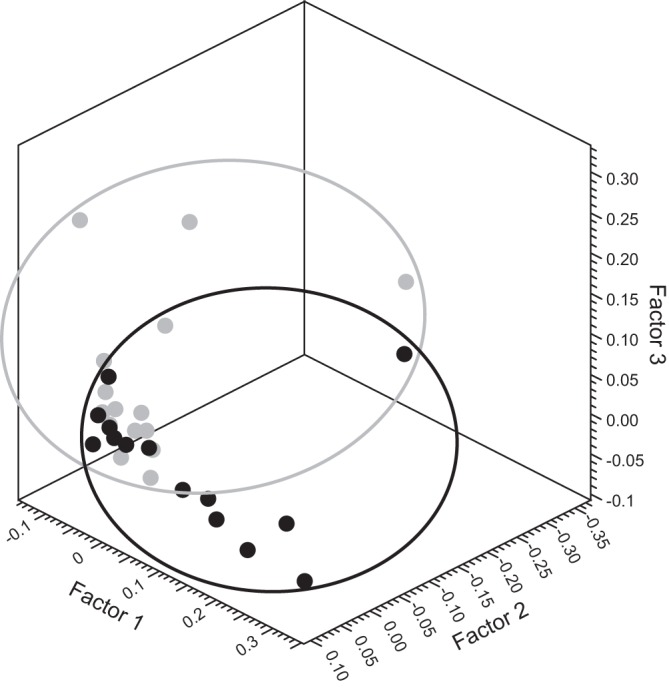
Urinary NMR metabolic profiling. Principal component analysis of urinary NMR data sets postomeprazole from controls (gray circles) and cirrhotic patients (black circles) showing clear separation of the groups.
Microbiota Results
The QIIME analysis indicated that there was a significant change in the overall microbiota composition after PPI therapy using the weighted UNIFRAC significant test in controls as well as in patients with cirrhosis (P < 0.0001).
The PCA analysis indicated that the pre-PPI patients with cirrhosis were clustered together compared with a significant change after PPI therapy (Fig. 2A). However, the change in controls was not pronounced in PCA (Fig. 2B).
Fig. 2.
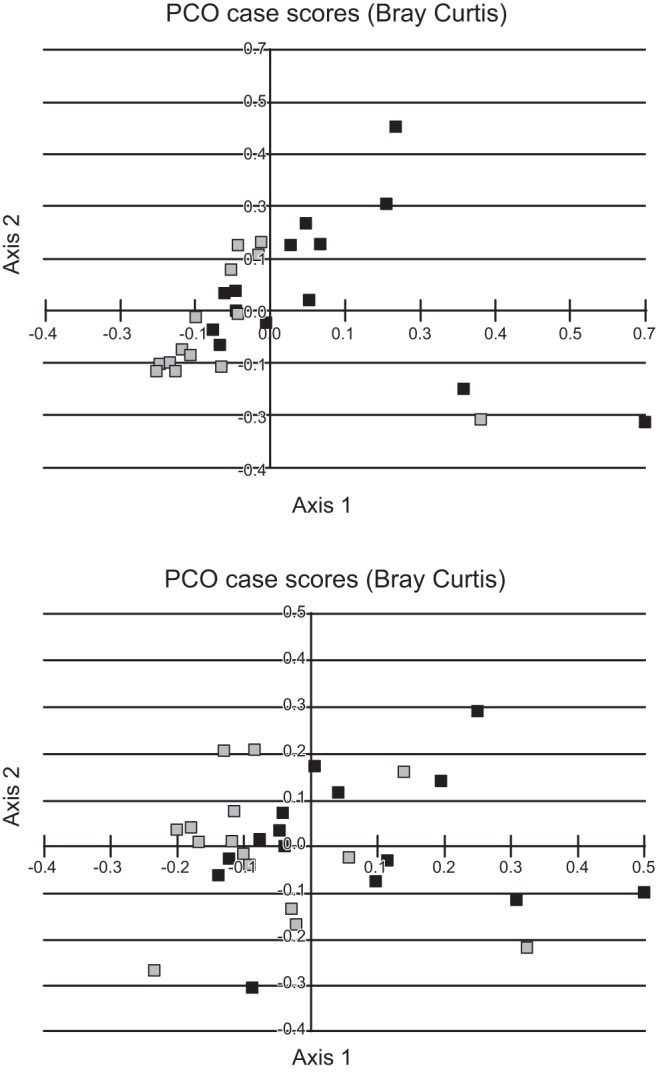
Principal component analysis of microbiota before and after omeprazole. Light gray squares, microbiota at baseline; dark gray squares, postomeprazole microbiota. A: significant clustering before omeprazole that changed after omeprazole. B: no clustering before or after omeprazole therapy.
The Metastats analysis indicated a significant increase in the relative abundance of Streptococcaceae after PPI therapy, compared with baseline in both patients with cirrhosis (baseline 0.0 vs. 8.9%, P = 0.0008, FDR = 0.008) and controls (baseline 0.2 vs. 5.7%, P = 0.0009, FDR = 0.007). The relative change in Streptococcaceae after PPI was statistically similar between the two groups (P = 0.16). In addition, there was a reduction of autochthonous bacterial abundance in patients with cirrhosis (pre-PPI Lachnospiraceae 13.9 vs. 9.9%, P = 0.04, FDR = 0.26, pre-PPI Ruminococcaceae 6.2 vs. 3.7%, P = 0.02, FDR = 0.12).
There was no significant change in median relative abundances of other major families in patients with cirrhosis before/after omeprazole (Bacteroidaceae 14 vs. 9.9%, P = 0.59; Enterobacteriaceae 0 vs. 0%, P = 0.85; Clostridiales incertae sedis XIV 4.8 vs. 3.8%, P = 0.29; Peptostreptococcaceae 0 vs. 0%, P = 0.23; Porphyromonadaceae 17.9 vs. 18.4%, P = 0.87; Prevotellaceae 0 vs. 0%, P = 0.76; Veillonellaceae 0 vs. 1%, P = 0.84; and Rikenellaceae 0 vs. 0%, P = 0.68). Similarly, no significant change in other major bacterial taxa was found in controls before/after omeprazole (Bacteroidaceae 22.3 vs. 11.1%, P = 0.08; Enterobacteriaceae 0 vs. 0%, P = 0.92; Clostridiales incertae sedis XIV 8.1 vs. 7.8%, P = 0.81; Lachnospiraceae 19.2 vs. 23.7%, P = 0.88; Ruminococcaceae 7.6 vs. 8.9%, P = 0.17; Peptostreptococcaceae 0 vs. 0%, P = 0.23; Porphyromonadaceae 1.9 vs. 1.4%, P = 0.22; Prevotellaceae 0 vs. 0%, P = 0.88; Veillonellaceae 0 vs. 0%, P = 0.23; and Rikenellaceae 1.5 vs. 2.3%, P = 0.88).
There was a significant positive correlation between serum gastrin and Streptococcaceae (r = 0.4, P = 0.005) but not with other taxa.
Correlation Network Difference Analysis
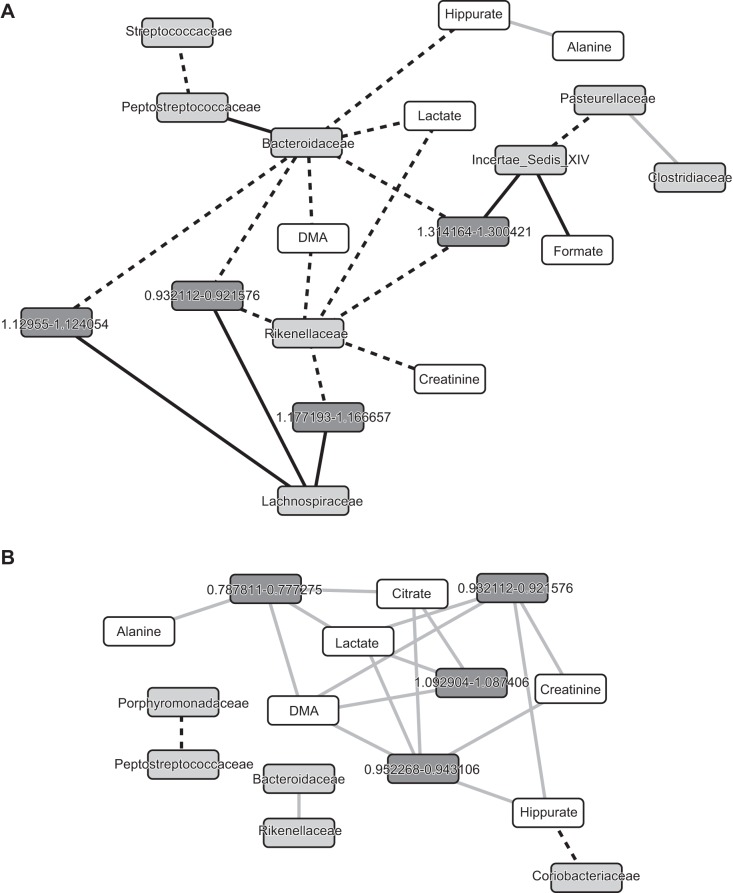
In patients with cirrhosis (Fig. 3A), there was a significant change between Bacteroidaceae and the metabolites DMA, lactate, and hippurate, which were positive before omeprazole but became negatively correlated afterward. After omeprazole, Bacteroidaceae also changed from positive to negative correlation with several unnamed metabolites linking it to autochthonous taxa (Lachnospiraceae and Clostridiales incertae sedis XIV) and Rikenellaceae. Hippurate remained negatively correlated with alanine whereas Streptococcaceae changed their linkage from positive to negative with Peptostreptococcacae.
Fig. 3.
Correlation network differences before and after omeprazole. Node colors: light gray, bacterial families; dark gray, unnamed urinary metabolites; white, named urinary metabolites. Edge colors: black dashed, was positive and became negative; black solid, was negative became positive; gray, remained negative but significant change. A: correlation network difference in cirrhotic patients. B: correlation network difference in controls.
In the control group (Fig. 3B), no significant changes in correlations were seen. The correlations between several unnamed metabolites and urinary lactate, hippurate, citrate, DMA, and alanine remained negative before and after omeprazole. The only significant change between urinary metabolites and microbiota was that the hippurate linkage changed from positive to negative with Coriobacteriaceae.
DISCUSSION
The present study shows that a 14-day course of 40 mg/day of omeprazole is associated with a significant shift in gut microbiota composition and function in patients with cirrhosis as well as healthy controls. The use of PPIs has been exponentially increasing in the general population and in patients with cirrhosis (7). The use of PPIs is being increasingly associated with potentially life-threatening infections, such as SBP and Clostridium difficile, both of which are infections with a distal gut origin (6, 7, 12, 18, 24). These infections are potentially life threatening despite treatment, and therefore the impact of PPI therapy on distal gut microbiota composition and function is important to analyze to delineate future preventive measures (5, 24).
The significant increase in the relative abundance of Streptococaceae after omeprazole in both controls and patients with cirrhosis is intriguing. Streptococci are associated with pneumonia in the setting of PPI use because of their presumed overgrowth in the upper gut (13). There is a high abundance of Streptococcacae in the oral cavity and throat that should normally be destroyed by intact gastric acid (9, 20). Corroborating this potential mechanism is the correlation between Streptococcaceae abundance and serum gastrin in our population. The relationship with serum gastrin is important because with reduced luminal acid after PPI the somatostatin-associated inhibition of gastrin is decreased. As shown previously and in the present study, PPIs induce a two- to fourfold increase in serum gastrin, which acts as a corollary to acid suppression and reconfirms adherence on the drug (16). Although studies of omeprazole therapy in patients without cirrhosis have shown a significant change in microbiota after PPI in the gastric fluid and esophageal mucosa and dysbiosis in rodent studies in the small bowel, our findings extend it to the distal gut, which is a key location for initiation of cirrhosis-associated complications (1, 28). Although Streptococcaceae is not a predominant family that is associated with SBP or spontaneous bacteremia, the striking increase in its relative abundance points toward a potential failure of quorum-sensing mechanisms that would usually keep it in check after the gastric acid barrier is decreased by omeprazole. Given the relatively early stage of cirrhosis, the short therapy duration, and the focus on the entire microbiota rather than specific species, we did not find significantly increased abundances of traditional gram-negative pathogens or C. difficile in our population.
As noted in several studies, the microbiota functionality, and not just altered composition, is critically important in influencing the course in cirrhosis. We found that the change in microbiota composition after omeprazole was also accompanied by increased lactate in both controls and cirrhotic patients and reduced DMA in patients with cirrhosis. Hippurate levels were reduced in the cirrhosis group compared with controls, both at baseline and also post-PPI. Hippurate is an intriguing molecule that is a culmination of human and bacterial metabolism of dietary polyphenols (30). Gut microbiota convert these polyphenols to benzoic acid that is in turn converted to hippurate after glycine conjugation in the human liver and kidney. DMA can be a product of gut bacterial metabolism of dietary choline, although it can also originate from the N-methylation of methylamines or from the breakdown of creatine (14). Since the diet and liver function were stable in each subject during the study, it is likely that the changes in both hippurate and DMA were due to altered microbiota function after omeprazole. Interestingly, there was also a significant increase in lactate levels after omeprazole treatment, which could be related to the relative increase in Streptococcaceae that can produce lactate through fermentation (26). This is consistent with a prior study of distal microbiota change after acid suppression in rodents, which demonstrated an overgrowth of lactate-producing bacteria (15). Although the significance of this finding is unclear, lactate production is also a mode for certain taxa to inhibit growth of potentially harmful microbiota (25). These functional changes were correlated with microbiota composition. Correlations between Bacteroidaceae and DMA, lactate, and hippurate dramatically shifted from being positive at baseline to a negative linkage after omeprazole in patients with cirrhosis. Although it is not clear whether Bacteroidaceae itself was responsible for the reduction, since its relative abundance did not change, it is likely that the metabolic behavior of this taxon could have been affected. Linkages between Streptococcaceae and Peptostreptococcaceae and Bacteroidaceae were also significantly changed after omeprazole in the cirrhosis group. Autochthonous bacterial taxa are associated with a higher urinary DMA in patients without cirrhosis, and the reduced Lachnospiraceae and Ruminococcaceae with reduced DMA in our population extends this on to cirrhosis (23). In a prior longitudinal study in patients with more advanced cirrhosis, a decrease in choline conversion to DMA was seen in patients with worse outcomes with a reduction in autochthonous gut microbiota (2). Therefore, our results indicate that acid suppression can impact even the distal gut microbiota composition and function within a short duration. The exponential increase in Streptococcaceae abundance over this short duration indicates that further gut ecosystem changes impacting bacterial translocation could occur in patients with more advanced cirrhosis in whom typically PPI therapy is given for several months.
The omeprazole dose used is associated with adequate acid suppression and indeed resulted in significantly higher serum gastrin levels compared with predrug values (http://www.accessdata.fda.gov/drugsatfda_docs/label/2008/022056s001019810s087lbl.pdf). We used a 14-day drug period since short course are not associated with potential rebound acid hypersecretion in subjects who do not otherwise require this medication (21, 27). To avoid confounding effects due to antibiotics, treatments for hepatic encephalopathy, and advanced liver disease on microbiota itself, we only studied patients with compensated cirrhosis. Additionally, in a prior cross-sectional study including patients with advanced cirrhosis, there was no significant change in those on PPI compared with the rest, probably because of the severe inherent dysbiosis (3). However, longitudinal studies in patients with advanced cirrhosis on PPI are required but will probably be difficult to implement since a large majority are already on PPI for unclear reasons or have confounders related to medications that can affect the microbiota independently. We also relied on stool results, which may be different from mucosal microbiota analyses (4). The results show that patients with cirrhosis and healthy controls can have changes in microbiota due to PPI as a proof of concept; further studies are needed to link these changes definitively to infectious and other complications in patients with cirrhosis. PPIs are effective medications for the recommended indications and for prescribed durations (http://www.accessdata.fda.gov/drugsatfda_docs/label/2008/022056s001019810s087lbl.pdf). However, given their potential to change the gut microbiota function and composition as far as the distal gut and their association with infectious complications in cirrhosis, their indiscriminate use should be discouraged.
We conclude that a short course of omeprazole therapy in healthy controls and compensated cirrhotic patients can significantly shift the urinary NMR metabolic profile and also the stool microbiota function and composition, which are linked to the gastric acid suppression and overgrowth of bacteria normally present in the oral cavity. Further studies to link these changes with complications of cirrhosis are required.
GRANTS
This work was partly supported by grant RO1AA020203 from the National Institute on Alcohol Abuse and Alcoholism, grant RO1DK087913 from the National Institute of Diabetes and Digestive and Kidney Diseases, and the McGuire Research Institute (J. S. Bajaj). S. D. Taylor-Robinson, M. M. E. Crossey, and M. Ratneswaran are grateful to the British National Institute for Health Research (NIHR) Biomedical facility at Imperial College London for infrastructure support. M. M. E. Crossey is supported by a personal fellowship form the Sir Halley Stewart Trust (Cambridge, UK).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author (s).
AUTHOR CONTRIBUTIONS
J.S.B., M.L.S., P.B.H., S.D.T.-R., and P.M.G. conception and design of research; J.S.B., I.J.C., N.S.B., M.R., M.S., R.W., M.M.C., S.D.T.-R., and P.M.G. analyzed data; J.S.B., I.J.C., N.S.B., M.L.S., M.R., P.B.H., K.D., M.S., R.W., M.M.C., S.D.T.-R., and P.M.G. interpreted results of experiments; J.S.B., N.S.B., and P.M.G. prepared figures; J.S.B., I.J.C., D.M.H., P.B.H., M.S., R.W., S.D.T.-R., and P.M.G. drafted manuscript; J.S.B., I.J.C., N.S.B., D.M.H., M.L.S., M.R., P.B.H., M.B.W., M.S., R.W., S.D.T.-R., and P.M.G. edited and revised manuscript; J.S.B., I.J.C., N.S.B., D.M.H., M.L.S., M.R., P.B.H., K.D., N.A.N., M.S., R.W., M.M.C., S.D.T.-R., and P.M.G. approved final version of manuscript; I.J.C., N.S.B., M.R., M.B.W., K.D., N.A.N., M.S., M.M.C., and P.M.G. performed experiments.
ACKNOWLEDGMENTS
We thank the Pathology Group at the Mary Lyon Centre, MRC Harwell for access to the NMR facility. We also thank Dr A. Riva, Foundation for Liver Research, for statistical advice.
REFERENCES
- 1.Amir I, Konikoff FM, Oppenheim M, Gophna U, Half EE. Gastric microbiota is altered in oesophagitis and Barrett's oesophagus and further modified by proton pump inhibitors. Environ Microbiol 16: 2905–2914, 2014. [DOI] [PubMed] [Google Scholar]
- 2.Bajaj JS, Gillevet PM, Patel NR, Ahluwalia V, Ridlon JM, Kettenmann B, Schubert CM, Sikaroodi M, Heuman DM, Crossey MM, Bell DE, Hylemon PB, Fatouros PP, Taylor-Robinson SD. A longitudinal systems biology analysis of lactulose withdrawal in hepatic encephalopathy. Metab Brain Dis 27: 205–215, 2012. [DOI] [PubMed] [Google Scholar]
- 3.Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, Noble NA, Unser AB, Daita K, Fisher AR, Sikaroodi M, Gillevet PM. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol 60: 940–947, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bajaj JS, Hylemon PB, Ridlon JM, Heuman DM, Daita K, White MB, Monteith P, Noble NA, Sikaroodi M, Gillevet PM. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol 303: G675–G685, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bajaj JS, O'Leary JG, Reddy KR, Wong F, Olson JC, Subramanian RM, Brown G, Noble NA, Thacker LR, Kamath PS, Nacseld Second infections independently increase mortality in hospitalized patients with cirrhosis: the North American consortium for the study of end-stage liver disease (NACSELD) experience. Hepatology 56: 2328–2335, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bajaj JS, Ratliff SM, Heuman DM, Lapane KL. Proton pump inhibitors are associated with a high rate of serious infections in veterans with decompensated cirrhosis. Aliment Pharmacol Ther 36: 866–874, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chavez-Tapia NC, Tellez-Avila FI, Garcia-Leiva J, Valdovinos MA. Use and overuse of proton pump inhibitors in cirrhotic patients. Med Sci Monitor 14: CR468–CR472, 2008. [PubMed] [Google Scholar]
- 8.Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, Wang Y, Zhu B, Li L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology 54: 562–572, 2011. [DOI] [PubMed] [Google Scholar]
- 9.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. The human oral microbiome. J Bacteriol 192: 5002–5017, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duarte IF, Diaz SO, Gil AM. NMR metabolomics of human blood and urine in disease research. J Pharm Biomed Anal 93: 17–26, 2014. [DOI] [PubMed] [Google Scholar]
- 11.Gillevet P, Sikaroodi M, Keshavarzian A, Mutlu EA. Quantitative assessment of the human gut microbiome using multitag pyrosequencing. Chem Biodivers 7: 1065–1075, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goel GA, Deshpande A, Lopez R, Hall GS, van Duin D, Carey WD. Increased rate of spontaneous bacterial peritonitis among cirrhotic patients receiving pharmacologic acid suppression. Clin Gastroenterol Hepatol 10: 422–427, 2012. [DOI] [PubMed] [Google Scholar]
- 13.Gulmez SE, Holm A, Frederiksen H, Jensen TG, Pedersen C, Hallas J. Use of proton pump inhibitors and the risk of community-acquired pneumonia: a population-based case-control study. Arch Intern Med 167: 950–955, 2007. [DOI] [PubMed] [Google Scholar]
- 14.Heinzmann SS, Merrifield CA, Rezzi S, Kochhar S, Lindon JC, Holmes E, Nicholson JK. Stability and robustness of human metabolic phenotypes in response to sequential food challenges. J Proteome Res 11: 643–655, 2012. [DOI] [PubMed] [Google Scholar]
- 15.Kanno T, Matsuki T, Oka M, Utsunomiya H, Inada K, Magari H, Inoue I, Maekita T, Ueda K, Enomoto S, Iguchi M, Yanaoka K, Tamai H, Akimoto S, Nomoto K, Tanaka R, Ichinose M. Gastric acid reduction leads to an alteration in lower intestinal microflora. Biochem Biophys Res Commun 381: 666–670, 2009. [DOI] [PubMed] [Google Scholar]
- 16.Kuipers EJ. Proton pump inhibitors and gastric neoplasia. Gut 55: 1217–1221, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Jia W. Cometabolism of microbes and host: implications for drug metabolism and drug-induced toxicity. Clin Pharmacol Ther 94: 574–581, 2013. [DOI] [PubMed] [Google Scholar]
- 18.Merli M, Lucidi C, Di Gregorio V, Giannelli V, Giusto M, Ceccarelli G, Riggio O, Venditti M. The chronic use of beta-blockers and proton pump inhibitors may affect the rate of bacterial infections in cirrhosis. Liver Int 2014 May 17. 10.1111/liv.12593 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 19.Naqvi A, Rangwala H, Keshavarzian A, Gillevet P. Network-based modeling of the human gut microbiome. Chem Biodivers 7: 1040–1050, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piombino P, Genovese A, Esposito S, Moio L, Cutolo PP, Chambery A, Severino V, Moneta E, Smith DP, Owens SM, Gilbert JA, Ercolini D. Saliva from obese individuals suppresses the release of aroma compounds from wine. PLoS One 9: e85611, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reimer C, Sondergaard B, Hilsted L, Bytzer P. Proton-pump inhibitor therapy induces acid-related symptoms in healthy volunteers after withdrawal of therapy. Gastroenterology 137: 80–87, 87.e1, 2009. [DOI] [PubMed] [Google Scholar]
- 22.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottiere HM, Dore J, Marteau P, Seksik P, Langella P. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA 105: 16731–16736, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tandon P, Garcia-Tsao G. Bacterial infections, sepsis, and multiorgan failure in cirrhosis. Semin Liver Dis 28: 26–42, 2008. [DOI] [PubMed] [Google Scholar]
- 25.Tsai YT, Cheng PC, Pan TM. The immunomodulatory effects of lactic acid bacteria for improving immune functions and benefits. Appl Microbiol Biotechnol 96: 853–862, 2012. [DOI] [PubMed] [Google Scholar]
- 26.Vesper BJ, Jawdi A, Altman KW, Haines GK, 3rd, Tao L, Radosevich JA. The effect of proton pump inhibitors on the human microbiota. Curr Drug Metab 10: 84–89, 2009. [DOI] [PubMed] [Google Scholar]
- 27.Waldum HL, Qvigstad G, Fossmark R, Kleveland PM, Sandvik AK. Rebound acid hypersecretion from a physiological, pathophysiological and clinical viewpoint. Scand J Gastroenterol 45: 389–394, 2010. [DOI] [PubMed] [Google Scholar]
- 28.Wallace JL, Syer S, Denou E, de Palma G, Vong L, McKnight W, Jury J, Bolla M, Bercik P, Collins SM, Verdu E, Ongini E. Proton pump inhibitors exacerbate NSAID-induced small intestinal injury by inducing dysbiosis. Gastroenterology 141: 1314–1322, 1322.e1–5, 2011. [DOI] [PubMed] [Google Scholar]
- 29.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol 5: e1000352, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams HR, Cox IJ, Walker DG, Cobbold JF, Taylor-Robinson SD, Marshall SE, Orchard TR. Differences in gut microbial metabolism are responsible for reduced hippurate synthesis in Crohn's disease. BMC Gastroenterol 10: 108, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res 41: D801–D807, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]