Abstract
It is important to search the biomarker to predict the development and prognosis of autoimmune thyroid diseases (AITDs) such as Hashimoto's disease (HD) and Graves' disease (GD). MicroRNA (miR) bind directly to the 3′ untranslated region of specific target mRNAs to suppress the expression of proteins, promote the degradation of target mRNAs and regulate immune response. miR-125a is known to be a negative regulator of regulated upon activation normal T cell expressed and secreted (RANTES), interleukin (IL)-6 and transforming growth factor (TGF)-β; however, its association with AITDs remains unknown. To clarify the association between AITDs and miR-125a, we genotyped the rs12976445 C/T, rs10404453 A/G and rs12975333 G/T polymorphisms in the MIR125A gene, which encodes miR-125a, using direct sequencing and polymerase chain reaction–restriction fragment length polymorphism (PCR–RFLP) methods in 155 patients with GD, 151 patients with HD and 118 healthy volunteers. We also examined the expression of miR-125a in peripheral blood mononuclear cells (PBMCs) from 55 patients with GD, 79 patients with HD and 38 healthy volunteers using quantitative real-time PCR methods. We determined that the CC genotype and C allele of the rs12976445 C/T polymorphism were significantly more frequent in patients with HD compared with control subjects (P < 0·05) and in intractable GD compared with GD in remission (P < 0·05). The expression of miR-125a was correlated negatively with age (P = 0·0010) and down-regulated in patients with GD compared with control subjects (P = 0.0249). In conclusion, miR-125a expression in PBMCs and the rs12976445 C/T polymorphism were associated with AITD development and prognosis.
Keywords: autoimmune thyroid disease, intractability, miR-125a, polymorphism, severity
Introduction
Autoimmune thyroid diseases (AITDs) such as Graves' disease (GD) and Hashimoto's disease (HD) are typical organ-specific autoimmune diseases [1,2]. The severity of HD and the intractability (inducibility to remission) of GD varies among patients. Some patients with HD develop hypothyroidism early in life, whereas some maintain a euthyroid state up to old age. Some patients with GD achieve remission through medical treatment, whereas others do not [3,4]. However, GD intractability and HD severity are difficult to predict at diagnosis.
MicroRNAs (miR) are endogenously encoded single-stranded RNAs approximately 22 nucleotides in length, and they have been known to play essential roles in a variety of pathological conditions [5]. miR suppress post-transcriptional gene expression by binding specifically to their target messenger RNAs (mRNAs) and inducing either translational repression or mRNA degradation [6]. miR regulate various biological processes such as cell proliferation, differentiation, metabolism, apoptosis, development, inflammation and immunity [5–15]. Recently, strong associations between miR and the immune system have been reported, and it has been suggested that miR also have regulatory roles in the immune response [8,10,13]. miR are initially transcribed as long primary transcripts (pri-miRNA), and they are processed into a ∼65 nt hairpin-shaped precursor miR (pre-miRNA) [16]. The pri-miRNA processing is a critical step in miRNA biogenesis because it regulates the expression level of mature miRNA [17,18]. Pre-miRNA are subsequently exported to the cytoplasm and cleaved to generate a 18∼25 nt mature miRNA [13].
The MiR125A gene, which encodes miR-125a, is located on chromosome 19q13.41 in a gene cluster containing MIR99B and MIR7E. miR-125a is known to be a negative regulator of Kruppel-like factor 13 (KLF13) and the tumour necrosis factor α-induced protein 3 (TNFAIP3), inhibits the production of regulated on activation normal T cell expressed and secreted (RANTES) and promotes the nuclear factor kappa B (NF-κB) pathway [12,15]. It has been reported that miR-125a is down-regulated in systemic lupus erythematosus (SLE) [15,19], breast cancer [20], gastric cancer [21], ovarian cancer [22] and verrucous carcinoma [14], although the roles of miR-125a in AITD still remain unclear. Therefore, we performed a quantification of miR-125a expression in peripheral blood mononuclear cells (PBMCs).
A G/T single-nucleotide polymorphism, named rs12975333, has been identified in the MIR125A gene, and its T allele blocks the pri- to pre-miR-125a processing step [17]. There are also two polymorphisms in this gene, rs10404453 A/G and rs12976445 C/T, which may be correlated with the expression of mature miR-125a in patients with AITDs. In this study, we genotyped these three MIR125A SNPs in patients with AITD.
Materials and methods
Subjects for genotyping
We screened the study polymorphisms in 155 patients with GD, 151 patients with HD and 118 healthy volunteers. Among GD patients, 60 GD patients had been treated with methimazole for at least 5 years and were still positive for anti-thyrotrophin receptor antibody (TRAb) (intractable GD), 45 GD patients had maintained a euthyroid state and were negative for TRAb for more than 2 years without medication (GD in remission), and 50 patients who could not be categorized to intractable GD or GD in remission groups at the time of analysis. All patients with GD had clinical histories of positive TRAb and thyrotoxicosis. Among HD patients, 59 HD patients had developed moderate to severe hypothyroidism before 50 years of age and been treated with thyroxine (severe HD), 41 untreated euthyroid HD patients were aged more than 50 years (mild HD), and 51 patients who could not be categorized to severe HD or mild HD groups at the time of analysis. All patients with HD were positive for anti-thyroid microsomal antibody (McAb) and/or anti-thyroglobulin antibody (TgAb) and all patients with mild HD had a palpable diffuse goitre. All healthy volunteers were euthyroid and negative for thyroid-specific autoantibodies (control subjects). All patients and control subjects were Japanese and unrelated. Written informed consent was obtained from all patients and controls and the study protocol was approved by the Ethics Committee of Osaka University. Clinical characteristics of the subjects selected for genotyping are given in Table 1.
Table 1.
Clinical characteristics of the subjects for genotyping
| GD | HD | ||||
|---|---|---|---|---|---|
| Controls | Intractable | In remission | Severe | Mild | |
| n (female/male) | 118 (78/40) | 60 (50/10) | 45 (39/6) | 59 (47/12) | 41 (35/6) |
| Age of onset (years) (range) | 37·0 ± 14·6‡ (21∼66) | 34·5 ± 14·7 (11∼71) | 33·4 ± 14·3 (10∼67) | 37·1 ± 11·1 (10∼49) | 62·2 ± 10·9‡ (50∼92) |
| Goitre size (cm) | n.d. | 4·8 ± 1·3 | 4·2 ± 0·7 | 4·1 ± 1·0 | 4·5 ± 1·1 |
| Free T4 (ng/dl) | n.d. | 1·2 ± 0·3 | 1·2 ± 0·1 | 1·3 ± 0·3 | 1·2 ± 0·2 |
| Free T3 (pg/dl) | n.d. | 2·7 ± 0·5 | 2·7 ± 0·4 | 2·6 ± 0·6 | 2·8 ± 0·4 |
| TSH (μU/ml) | n.d. | 1·8 ± 1·4 | 1·9 ± 1·3 | 4·8 ± 16·9 | 2·7 ± 1·6 |
| TRAb (IU/l) (range) | <2·0 | 6·1 ± 11 (2∼71)* | <2·0 | <2·0 | <2·0 |
| TgAb (2n × 100) | Negative | 3·6 ± 3·0 | 2·9 ± 3·2 | 6·8 ± 3·4** | 3·6 ± 4·4 |
| McAb (2n × 100) | Negative | 5·1 ± 2·3 | 5·4 ± 2·1 | 5·0 ± 3·2 | 3·3 ± 3·0 |
| Current treatment | None | Methimazole or PTU | None | L-thyroxine | None |
| Duration of treatment (years) | None | 12·1 ± 6·7 | 3·0 ± 1·2§ | 10·0 ± 8·5 | None |
| Current dose of anti-thyroid drug (mg/day) (range)† | None | 12·4 ± 11·5 (2·5∼50) | None | None | None |
| Current dose of L-thyroxine drug (μg/day) (range) | None | None | None | 88·7 ± 39·0 (25∼250) | None |
Data are means ± standard deviations.
P < 0·01 [versus Graves' disease (GD) in remission].
P < 0·01 [versus mild Hashimoto's disease (HD)].
Doses were expressed as comparable doses of methimazole [50 mg of propylthiouracil (PTU) was converted to 5 mg of methimazole].
Age at the time of sampling.
Duration of treatment with anti-thyroid drug before remission.
T4 = thyroxine; T3 = triiodothyronine; TSH = thyrotrophin; TRAb = anti-thyrotrophin receptor antibody; McAb = anti-thyroid microsomal antibody; n.d. = not determined; TgAb = anti-thyroglobulin antibody.
Genotyping of polymorphisms
Genotyping was performed using the PCR–restriction fragment length polymorphism (RFLP) method and direct sequencing. Briefly, target sequences of DNA were amplified using PCR. The PCR products were then digested with BaeGI (New England Biolabs, Ipswich, MA, USA). DNA fragment length polymorphisms were visualized by ultraviolet transillumination after electrophoretic separation on 10% polyacrylamide gels. The PCR primers used for MIR125A sequencing are presented in Table 2.
Table 2.
The primers, polymerase chain reaction (PCR) conditions and a restriction enzyme used in this study
| Primer pairs | PCR condition | Restriction enzymes | |
|---|---|---|---|
| PCR–RFLP | |||
| rs12976445 C/T | 5′-TTTTGGTCTTTCTGTCTCTGG -3′ | 96°C for 5 min | BaeGI |
| 5′-TGGAGGAAGGGTATGAGGAGT-3′ | (94°C for 30 s, 58·4°C for 30 s, 72°C for 30 s) × 35 cycles | ||
| 72°C for 7 min | |||
| Direct sequence | |||
| rs10404453 A/G | 5′-CTGACTCCCTCTTATTCTGG-3′ | 96°C for 5 min | |
| 5′-TAGAGACTGGCAACATGG-3′ | (94°C for 30 s, 54·9°C for 30 s, 72°C for 30 s) × 35 cycles | ||
| 72°C for 7 min | |||
| Direct sequence | |||
| rs12975333 G/T | 5′-GAATGTCTCTGTGCCTATCTCC-3′ | 96°C for 5 min | |
| 5′-TCAGGCCAGCAATTCCC-3′ | (94°C for 30 s, 58·5°C for 30 s, 72°C for 30 s) × 35 cycles | ||
| 72°C for 7 min |
PCR–RFLP = polymerase chain reaction–restriction fragment length polymorphism.
miRNA extraction from PBMCs
We collected blood samples in ethylenediamine tetraacetic acid (EDTA)-treated tubes and isolated PBMCs from each AITD patient and healthy volunteer with Lymphoprep (Axis-Shield PoC AS, Oslo, Norway). PBMCs were washed once in sterile phosphate-buffered saline (PBS) and preserved in RNAlater® solution (Ambion, Austin, TX, USA) at −80°C until required. Total RNA was isolated from preserved PBMCs with the mirVana™ PARIS™ Kit (Ambion), in accordance with the manufacturer's protocol.
Quantitative RT–PCR
Reverse transcription (TR) of miR-125a and U6 was performed using a TaqMan® MicroRNA RT kit and target-specific stem loop primers provided in TaqMan® MicroRNA assays (Applied Biosystems, Foster City, CA, USA), following the manufacturer's protocol. Quantitative RT–PCR was performed using TaqMan® MicroRNA assays (Applied Biosystems), in accordance with the manufacturer's protocol. The following TaqMan MicroRNA assays were used: hsa-miR-125a and RNU6B. Detection and quantification were performed on a StepOnePlus™ Real-Time PCR System (Applied Biosystems). Expression levels of miRNA were normalized to U6. All reactions were performed in triplicate. The relative expression levels of each miRNA were calculated using the ΔΔCt method.
Subjects for quantification of miR expression
We examined the expression level of miRNAs in PBMCs from 55 patients with GD, 79 patients with HD and 38 healthy volunteers. Among these, 15 patients had intractable GD, 17 patients had GD in remission, 22 patients who could not be categorized to intractable GD or GD in remission groups at the time of analysis, 32 patients had severe HD and 19 had mild HD, and 28 patients who could not be categorized to severe HD or mild HD groups at the time of analysis. The clinical characteristics of the subjects used for miR quantification are given in Table 3.
Table 3.
Clinical characteristics of the subjects for quantification of microRNA (miR)-125a expression
| GD | HD | ||||
|---|---|---|---|---|---|
| Controls | Intractable | In remission | Severe | Mild | |
| n (female/male) | 38 (24/14) | 15 (14/1) | 17 (15/2) | 32 (25/7) | 19 (18/1) |
| Age of onset (years) (range) | – | 34·7 ± 14·6 (16∼64) | 28·3 ± 8·7 (15∼46) | 39·0 ± 9·8 (23∼49) | – |
| Age of sampling (years) (range) | 46·6 ± 12·7 (21∼66) | 53·6 ± 11·0 (38∼83) | 52·6 ± 15·4 (26∼87) | 53·0 ± 15·5 (26∼83) | 62·2 ± 9·7 (50∼79) |
| Goitre size (cm) | n.d | 4·6 ± 1·1 | 4·4 ± 0·5 | 4·2 ± 1·2 | 4·7 ± 1·3 |
| Free T4 (ng/dl) | 1·1 ± 0·2 | 1·1 ± 0·2 | 1·3 ± 0·2 | 1·2 ± 0·2 | 1·2 ± 0·4 |
| Free T3 (pg/dl) | 2·8 ± 0·2 | 2·6 ± 1·1 | 2·8 ± 0·3 | 2·8 ± 0·8 | 2·8 ± 0·3 |
| TSH (μU/ml) | 2·1 ± 1·3 | 1·9 ± 1·3 | 2·1 ± 1·7 | 4·8 ± 4·9 | 2·7 ± 2·1 |
| TRAb (IU/l) (range) | <2·0 | 11·6 ± 27·8 (2∼99) | <2·0 | <2·0 | <2·0 |
| TgAb (2n × 100) | Negative | 5·0 ± 2·6 | 6·0 ± 2·8 | 9·0 ± 2·6* | 2·15 ± 2·4 |
| McAb (2n × 100) | Negative | 7·0 ± 3·8 | 7·3 ± 3·0 | 9·5 ± 4·2 | 6·3 ± 2·9 |
| Current treatment | None | Methimazole or PTU | None | L-thyroxine | None |
| Current dose of anti-thyroid drug (mg/day) (range) | None | 37·5 ± 59·8 (5∼200) | None | None | None |
| Current dose of L-thyroxine drug (μg/day) (range) | None | None | None | 79·6 ± 32·1 (25∼150) | None |
Data are means ± standard deviations.
P < 0·01 [versus mild Hashimoto's disease (HD)].
Doses were expressed as comparable doses of methimazole [50 mg of propylthiouracil (PTU) was converted to 5 mg of methimazole]. T4 = thyroxine; T3 = triiodothyronine; TSH = thyrotrophin; TRAb = anti-thyrotrophin receptor antibody; McAb = anti-thyroid microsomal antibody; n.d. = not determined; TgAb = anti-thyroglobulin antibody.
Statistical analysis
We used Student's t-test for two-group comparisons to analyse the significance of differences in the expression level of miRNAs. For multiple group comparisons, parametric comparisons used analysis of variance (anova). If anova was significant, significances of patients with GD or HD compared with control subjects were evaluated by Dunnett's test. A χ2 test was used to evaluate differences in genotype frequencies and alleles among the subject groups. Pearson's correlation coefficient was used to analyse the associations between the expression of miR125a and age. Data were analysed using jmp9 software (SAS Institute, Inc., Tokyo, Japan). Probability values of less than 0·05 were considered significant.
Results
MIR125A rs12976445 C/T polymorphism
Initially, we performed direct sequencing for three SNPs (rs12976445, rs10404453, rs12976333) in a randomly selected group of 16 patients with HD and determined that rs12976445 was the most suitable polymorphism for further examination (Table 4). We found a significant difference in the genotype frequency between control subjects and patients with HD (P = 0·037) for this polymorphism. The C allele and CC genotype of this polymorphism were significantly more frequent in patients with HD compared with control subjects (P = 0·049 and P = 0·036, respectively) (Table 5). Furthermore, the C allele and CC genotype were significantly more frequent in patients with intractable GD compared with those with GD in remission (P = 0·040 and P = 0·027, respectively) (Table 6).
Table 4.
Genotype frequencies of the MIR125A polymorphisms (preliminary investigation)
| Severe Hashimoto's disease (HD) | Mild HD | ||
|---|---|---|---|
| rs12976445 | CC | 6 (75·0%) | 7 (87·5%) |
| C/T | CT | 2 (25·0%) | 1 (12·5%) |
| TT | 0 (0%) | 0 (0%) | |
| rs10404453 | GG | 8 (100%) | 8 (100%) |
| G/A | GA | 0 (0%) | 0 (0%) |
| AA | 0 (0%) | 0 (0%) | |
| rs12975333 | GG | 8 (100%) | 8 (100%) |
| G/T | GT | 0 (0%) | 0 (0%) |
| TT | 0 (0%) | 0 (0%) |
Table 5.
Genotype and allele frequencies of the miR-125a polymorphism in patients with Graves' disease (GD), Hashimoto's disease (HD) and in control subjects
| Control | All patients with GD | All patients with HD | ||||
|---|---|---|---|---|---|---|
| miR-125a | CC | 87 (73·73%) | 122 (78·71%) | n.s. | 127 (84·11%) | 0·037 |
| rs12976445 | CT | 31 (26·27%) | 31 (20%) | 24 (15·89%) | ||
| TT | 0 (0%) | 2 (1·29%) | 0 (0%) | |||
| CC + CT | 118 (100%) | 153 (98·71%) | n.s. | 151 (100%) | n.s. | |
| TT | 0 (0%) | 2 (1·29%) | 0 (0%) | |||
| CC | 87 (73·73%) | 122 (78·71%) | n.s. | 127 (84·11%) | 0·036 | |
| CT + TT | 31 (26·27%) | 33 (21·29%) | 24 (15·89%) | |||
| C allele | 205 (86·86%) | 275 (88·71%) | n.s. | 278 (92·05%) | 0·049 | |
| T allele | 31 (13·14%) | 35 (11·29%) | 24 (7·95%) | |||
n.s. = not significant.
Table 6.
Genotype and allele frequencies of the miR-125a polymorphism in patients with Graves' disease (GD), Hashimoto's disease (HD)
| GD | HD | ||||||
|---|---|---|---|---|---|---|---|
| Intractable | In remission | Severe | Mild | ||||
| miR-125a | CC | 51 (85%) | 30 (66·67%) | n.s. | 48 (81·36%) | 36 (87·80%) | n.s. |
| rs12976445 | CT | 8 (13·33%) | 14 (31·11%) | 11 (18·64%) | 5 (12·20%) | ||
| TT | 1 (1·67%) | 1 (2·22%) | 0 (0%) | 0 (0%) | |||
| CC + CT | 59 (98·33%) | 44 (97·78%) | n.s. | 59 (100%) | 41 (100%) | n.s. | |
| TT | 1 (1·67%) | 1 (2·22%) | 0 (0%) | 0 (0%) | |||
| CC | 51 (85%) | 30 (66·67%) | 0·027 | 48 (81·36%) | 36 (87·80%) | n.s. | |
| CT + TT | 9 (15%) | 15 (33·33%) | 11 (18·64%) | 5 (12·20%) | |||
| C allele | 110 (91·67%) | 74 (82·22%) | 0·040 | 107 (90·68%) | 77 (93·90%) | n.s. | |
| T allele | 10 (8·33%) | 16 (17·78%) | 11 (9·32%) | 5 (6·10%) | |||
n.s. = not significant.
Expression of miR-125a in PBMCs and clinical characteristics
The expression of miR-125a in control subject PBMCs was correlated negatively with age (Fig. 1, r = −0·52, P = 0·0010). However, in GD and HD patients, the expression of miR125a was not correlated with any clinical characteristics.
Fig. 1.
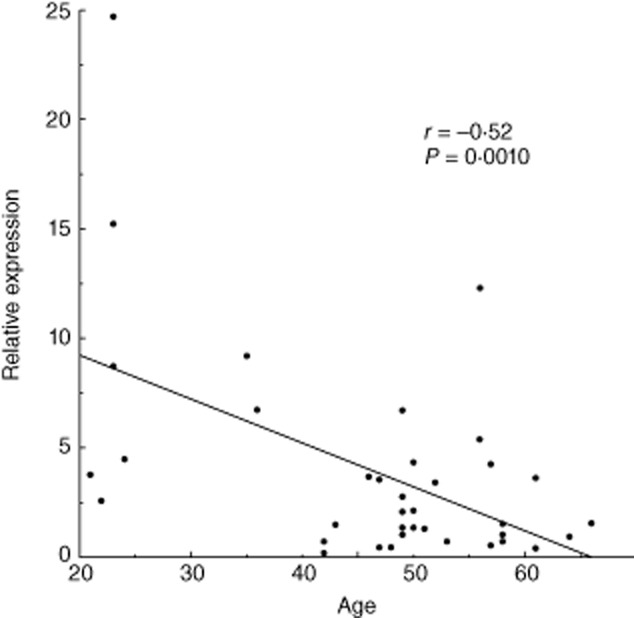
The correlation between microRNA (miR)-125a expression in peripheral blood mononuclear cells (PBMCs) and age. Pearson's correlation coefficient was used to analyse the associations between the PBMC miR125a expression level and age at sampling in control subjects.
PBMC miR-125a expression
The expression of miR-125a in GD patient PBMCs was decreased significantly compared with control subjects (P = 0·0249, Fig. 2a). We found no significant differences in miR-125a expression in PBMCs among the patients with different prognoses of AITDs (Fig. 2b,c).
Fig. 2.
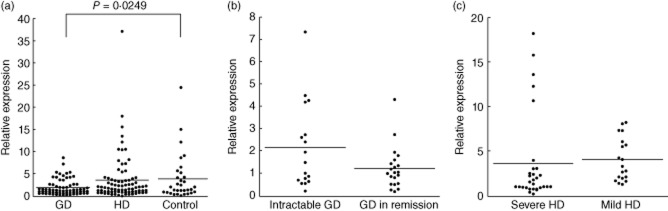
Peripheral blood mononuclear cell (PBMCs) microRNA (miR)-125a expression in study groups. The PBMC miR-125a expression level in patients with autoimmune thyroid diseases (AITDs) and control subjects (a), with different prognoses of Graves' disease (GD) (b) and Hashimoto's disease (HD) (c). A Student's t-test was used to compare the expression level of miR-125a between two groups and Dunnett's test was used between three groups.
PBMC miR-125a expression stratified by genotype
PBMC miR-125a expression did not differ significantly among patients with each genotype of the MIR125A rs12976445C/T polymorphism (Fig. 3).
Fig. 3.
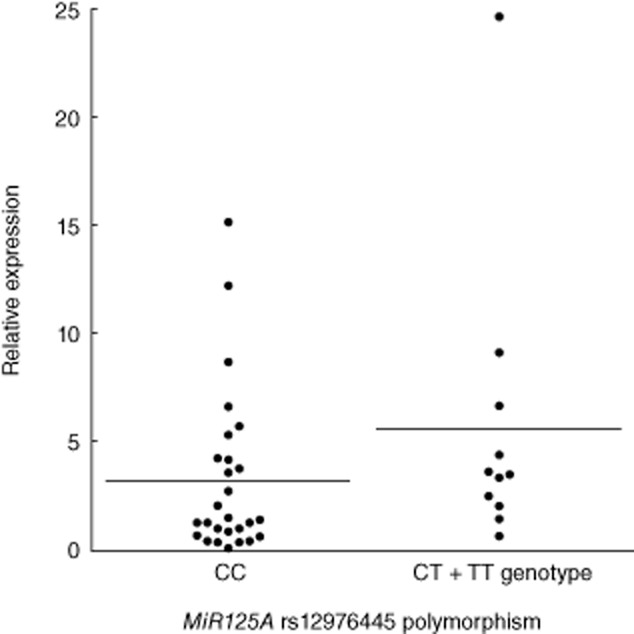
The expression of microRNA (miRNA) in patients stratified by genotype. miR-125a expression in peripheral blood mononuclear cells (PBMCs) from control subjects with each genotype of MIR125A rs12976445 C/T polymorphism. A Student's t-test was used to compare the expression levels of miR-125a among each group.
Discussion
Initially, our data indicated that miR-125a expression decreased with age (Fig. 1). Therefore, prior to the analysis of miR-125a expression, we confirmed that our subject groups were age- and sex-matched (Table 3). To our knowledge, this is the first report demonstrating the correlation between miR expression and age.
We determined in this study that the MIR125A rs12976445 C/T polymorphism was associated with the pathogenesis and prognosis of AITDs. Our data indicate that the C allele and CC genotype are correlated with HD development and GD intractability (Tables 5 and 6). A previous study reported that C allele carriers (CC + CT genotypes) correlated with low miR-125a expression compared with the TT genotype in a Polish population [23]. However, in the present study, the association between genotype and miR-125a expression level was not observed in control subject PBMCs (Fig. 3). We suggest that this polymorphism may be associated with the development and prognosis of AITDs through a role in pri-miRNA processing, because this polymorphism is known to be related to pri-miR processing rather than through variations in miR-125a expression levels [17].
Our data also demonstrated that miR-125a expression in PBMCs was decreased significantly in patients with GD (Fig. 2a). This indicates that miR-125a expression in PBMCs may suppress GD development. It has been demonstrated that miR-125a acts as a negative regulator of RANTES, interleukin (IL)-6 and transforming growth factor (TGF)-β [9,15]. Additionally, we have reported previously that the high genetic producibilities of IL-6 and TGF-β, which promote the differentiation of T helper type 17 (Th17) cells, were associated with the GD development [24,25]. Therefore we speculate that, in GD patients, the down-regulation of miR-125a may indirectly promote the differentiation of Th17 cells and cause GD development. This is supported by our previous study, where we demonstrated that Th17 cells were increased in GD patients [26]. Conversely, in some patients with severe HD, a higher miR-125a expression was observed (Fig. 2b). In these subjects, high miR-125a expression may play some role to cause a severe condition in HD through some factors such as Th1 cells, which are predominant in severe HD [26]. However, Yamada et al. reported that serum miRNA levels (miR-16, miR-22, miR-375 and miR-451) were associated with the development of AITD [27]. Therefore, further studies are necessary to clarify the association between circulating miRNA levels and the development and prognosis of AITD.
In conclusion, miR-125a expression in PBMCs was down-regulated in patients with GD. The rs12976445 C/T polymorphism was associated with HD development and GD intractability.
Disclosure
The authors declare that they have no conflicts of interest.
References
- 1.Volpe R. The immune system and its role in endocrine function. In: Becker KL, editor. Principles and practice of endocrinology and metabolism. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 1770–1781. [Google Scholar]
- 2.Davies T. Pathogenesis of Graves' disease. In: Braverman LE, Utiger RD, editors. The thyroid – a fundamental and clinical text. Philadelphia: Lippincott Williams & Wilkins; 2000. pp. 518–531. [Google Scholar]
- 3.Yoshida H, Amino N, Yagawa K, et al. Association of serum antithyroid antibodies with lymphocytic infiltration of the thyroid gland: studies of seventy autopsied cases. J Clin Endocrinol Metab. 1978;46:859–862. doi: 10.1210/jcem-46-6-859. [DOI] [PubMed] [Google Scholar]
- 4.Amino N, Hagen SR, Yamada N, Refetoff S. Measurement of circulating thyroid microsomal antibodies by the tanned red cell haemagglutination technique: its usefulness in the diagnosis of autoimmune thyroid diseases. Clin Endocrinol (Oxf) 1976;5:115–125. doi: 10.1111/j.1365-2265.1976.tb02822.x. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4:1179–1184. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 8.Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 9.Chen T, Huang Z, Wang L, et al. MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovasc Res. 2009;83:131–139. doi: 10.1093/cvr/cvp121. [DOI] [PubMed] [Google Scholar]
- 10.Graff JW, Dickson AM, Clay G, McCaffrey AP, Wilson ME. Identifying functional microRNAs in macrophages with polarized phenotypes. J Biol Chem. 2012;287:21816–21825. doi: 10.1074/jbc.M111.327031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang L, Huang Q, Chang J, Wang E, Qiu X. MicroRNA HSA-miR-125a-5p induces apoptosis by activating p53 in lung cancer cells. Exp Lung Res. 2011;37:387–398. doi: 10.3109/01902148.2010.492068. [DOI] [PubMed] [Google Scholar]
- 12.Kim SW, Ramasamy K, Bouamar H, Lin AP, Jiang D, Aguiar RC. MicroRNAs miR-125a and miR-125b constitutively activate the NF-kappaB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20) Proc Natl Acad Sci USA. 2012;109:7865–7870. doi: 10.1073/pnas.1200081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 14.Odar K, Bostjancic E, Gale N, Glavac D, Zidar N. Differential expression of microRNAs miR-21, miR-31, miR-203, miR-125a-5p and miR-125b and proteins PTEN and p63 in verrucous carcinoma of the head and neck. Histopathology. 2012;61:257–265. doi: 10.1111/j.1365-2559.2012.04242.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhao X, Tang Y, Qu B, et al. MicroRNA-125a contributes to elevated inflammatory chemokine RANTES levels via targeting KLF13 in systemic lupus erythematosus. Arthritis Rheum. 2010;62:3425–3435. doi: 10.1002/art.27632. [DOI] [PubMed] [Google Scholar]
- 16.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 17.Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet. 2007;16:1124–1131. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- 18.Hu Y, Liu CM, Qi L, et al. Two common SNPs in pri-miR-125a alter the mature miRNA expression and associate with recurrent pregnancy loss in a Han-Chinese population. RNA Biol. 2011;8:861–872. doi: 10.4161/rna.8.5.16034. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, Peng W, Ouyang X, Li W, Dai Y. Circulating microRNAs as candidate biomarkers in patients with systemic lupus erythematosus. Transl Res. 2012;160:198–206. doi: 10.1016/j.trsl.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 21.Nishida N, Mimori K, Fabbri M, et al. MicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin Cancer Res. 2011;17:2725–2733. doi: 10.1158/1078-0432.CCR-10-2132. [DOI] [PubMed] [Google Scholar]
- 22.Cowden Dahl KD, Dahl R, Kruichak JN, Hudson LG. The epidermal growth factor receptor responsive miR-125a represses mesenchymal morphology in ovarian cancer cells. Neoplasia. 2009;11:1208–1215. doi: 10.1593/neo.09942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehmann TP, Korski K, Ibbs M, Zawierucha P, Grodecka-Gazdecka S, Jagodzinski PP. rs12976445 variant in the pri-miR-125a correlates with a lower level of hsa-miR-125a and ERBB2 overexpression in breast cancer patients. Oncol Lett. 2013;5:569–573. doi: 10.3892/ol.2012.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inoue N, Watanabe M, Morita M, et al. Association of functional polymorphisms in promoter regions of IL5, IL6 and IL13 genes with development and prognosis of autoimmune thyroid diseases. Clin Exp Immunol. 2011;163:318–323. doi: 10.1111/j.1365-2249.2010.04306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamada H, Watanabe M, Nanba T, Akamizu T, Iwatani Y. The +869T/C polymorphism in the transforming growth factor-beta1 gene is associated with the severity and intractability of autoimmune thyroid disease. Clin Exp Immunol. 2008;151:379–382. doi: 10.1111/j.1365-2249.2007.03575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nanba T, Watanabe M, Inoue N, Iwatani Y. Increases of the Th1/Th2 cell ratio in severe Hashimoto's disease and in the proportion of Th17 cells in intractable Graves' disease. Thyroid. 2009;19:495–501. doi: 10.1089/thy.2008.0423. [DOI] [PubMed] [Google Scholar]
- 27.Yamada H, Itoh M, Hiratsuka I, Hashimoto S. Circulating microRNAs in autoimmune thyroid diseases. Clin Endocrinol (Oxf) 2014;81:276–281. doi: 10.1111/cen.12432. [DOI] [PubMed] [Google Scholar]


