Abstract
We report that 10% of melanoma tumors and cell lines harbor mutations in the fibroblast growth factor receptor 2 (FGFR2) gene. These novel mutations include three truncating mutations and 20 missense mutations occurring at evolutionary conserved residues in FGFR2 as well as among all four FGFRs. The mutation spectrum is characteristic of those induced by UV radiation. Mapping of these mutations onto the known crystal structures of FGFR2 followed by in vitro and in vivo studies show that these mutations result in receptor loss of function through several distinct mechanisms, including loss of ligand binding affinity, impaired receptor dimerization, destabilization of the extracellular domains, and reduced kinase activity. To our knowledge, this is the first demonstration of loss-of-function mutations in a class IV receptor tyrosine kinase in cancer. Taken into account with our recent discovery of activating FGFR2 mutations in endometrial cancer, we suggest that FGFR2 may join the list of genes that play context-dependent opposing roles in cancer.
Introduction
Melanoma is the most lethal of all skin cancers. The American Cancer Society estimated that there would be 62,480 new cases of melanoma and about 8,420 deaths due to melanoma in 2008. Previous studies in human melanocytes and melanoma cells have revealed that one of the hallmarks of melanocytic transformation is a change from paracrine growth factor stimulation by the surrounding keratinocytes in the skin to autocrine growth factor stimulation via ectopic expression of growth factors and/or receptors by melanoma cells (1, 2). Fibroblast growth factor (FGF) signaling plays a prominent role in tissue homeostasis by providing bidirectional paracrine communication between mesenchymal and epithelial cells of parenchymal organs including the skin. The FGF family comprises 18 ligands (FGF1-FGF10 and FGF16-FGF23), which signal through four transmembrane receptor tyrosine kinases (FGFR1-FGFR4) and their tissue-specific alternatively spliced b isoform (epithelial) and c isoform (mesenchymal; ref. 3). Previous studies have shown that survival and proliferation of melanocytes depends on FGF2, which is provided by the surrounding keratinocytes in the skin. A number of functional studies have implicated both FGF2 and FGFR1 signaling in melanoma progression. Adenoviral transduction of melanocytes with FGF2 resulted in phenotypic changes consistent with increased tumorigenicity (4, 5), and down-regulation of FGF2 via antisense oligonucleotides in metastatic melanoma inhibited proliferation and colony formation in soft agar (6, 7). Introduction of antisense oligonucleotides targeted toward FGFR1 into melanocytes and metastatic melanoma cell lines resulted in decreased proliferation and signs of differentiation (8, 9). Furthermore, injection of an antisense FGFR1 construct into primary and metastatic melanomas grown in nude mice has also been shown to result in inhibition of tumor growth and induction of apoptosis (10, 11).
Constitutive activation of FGFRs through chromosomal translocation, aberrant splicing, or missense mutations has been reported in several cancers. The Catalog of Somatic Mutations in Cancer (COSMIC)7 provides a repository of all somatic changes reported to date in this receptor family. To date, FGFR3 is mutated at highest frequency in benign seborrheic keratoses and bladder cancer and to a lesser extent in multiple myeloma and cervical cancer. We have recently reported the presence of activating mutations in FGFR2 in 16% of endometrioid endometrial cancers (12).
We report here the identification of novel FGFR2 mutations in 15 of 113 (13%) of melanoma cell lines and 8 of 100 (8%) of uncultured melanoma tumors. Mapping of these mutations onto the known crystal structures of FGFR2 together with in vitro and in vivo functional analyses shows that these mutations result in receptor loss of function.
Results
Identification of FGFR2 Mutations in Melanoma Cell Lines and Uncultured Primary and Metastatic Melanoma Tumors
We screened an initial panel of 47 melanoma cell lines for the presence of mutations in the FGFR1 to FGFR4 genes by sequencing. Following the identification of mutations in FGFR2, 66 additional melanoma cell lines were screened for FGFR2 mutations using a combination of denaturing high-performance liquid chromatography and sequencing leading to the identification of fifteen different FGFR2 mutations (Table 1). None of these mutations were observed in a panel of lymphocyte DNA from 150 Caucasian controls (data not shown), suggesting that these mutations arose somatically. Of the 15 cell lines carrying FGFR2 mutations, 11 also carried mutations in either BRAF or NRAS (Table 1). No mutations were identified in either FGFR1 or FGFR3, whereas one mutation at a nonconserved codon was found in FGFR4, P716R.
Table 1.
Summary of Melanoma Samples Carrying FGFR2 Mutations
| Samples | Nucleotide Change | Codon Change | BRAF | NRAS |
|---|---|---|---|---|
| Cell Lines | ||||
| MM540 | G812A | G271E | V600E | wt |
| UACC1940 | G913A | G305R | wt | wt |
| WSB | C1109G | T370R* | V600E | wt |
| M92-001† | G1423A | E475K* | wt | G13R |
| ME10538† | G1423A | E475K | V600E | wt |
| M91-054 | T2308G | L770V | V600E | wt |
| UACC2534 | G752A | R251Q | wt | wt |
| A375 | G1906A | E636K* | V600E | wt |
| UACC3093 | GG2100-1AA | G701S | wt | Q61L |
| D11 | G1920A | M640I | wt | Q61L |
| MM369 | G655A | E219K | V600E | wt |
| D22 | G1588A | D530N | wt | wt |
| D22 | C2275T | R759X | wt | wt |
| MM229 | G2276A | R759Q | L597S | wt |
| A04 | G1942A | A648T* | wt | wt |
| A05 | A479C | E160A | V600E | wt |
| Metastatic Tumors | ||||
| Met19 | G680A | G227E | wt | nd |
| Met27 | C71T | S24F | nd | nd |
| Met15 | G229A G1720A C2063T C2122T | V77M E574K S688F P708S | V600E | nd |
| Primary Tumors | ||||
| 81‡ | IVS15-2A>T§ | FS | V600E | wt |
| AM169‖ | T743A | V248D | wt | wt |
| AM146¶ | C637T | H213Y | wt | Q61L |
| 53** | G1422A | W474X | wt | wt |
| 71** | G1924A | I642V | wt | wt |
Abbreviations: wt, wild type; nd, not done.
Samples were homozygous for this mutation.
Microsatellite genotyping confirmed that these cell lines were independent.
Superficial spreading primary melanoma, no chronic sun damage evident.
The IVS15_2A>T is predicted to result in the skipping of exon 16, subsequent frameshift for 27 amino acids, and a premature truncation at codon 690 in the kinase domain.
Acral melanoma, histologically superficial spreading melanoma.
Mucosal primary melanoma.
Primary Lentigo Maligna Melanoma, with histologic evidence of chronic sun damage.
Next, we screened a panel of 28 metastatic melanoma tumors and a panel of 72 vertical growth phase primary melanomas representing samples from different melanoma subtypes for the presence of mutations in FGFR2 gene. We identified mutations in 3 of 28 metastatic samples and 5 of 72 primary tumors (Table 1). Subset analysis revealed FGFR2 mutations in 1 of 17 (6%) superficial spreading melanomas, 2 of 10 (20%) lentigo maligna melanomas, 1 of 28 (4%) acral melanomas, and 1 of 17 (6%) mucosal melanomas. For three of these tumors, we were able to extract DNA from surrounding normal tissue, and in all cases the mutation was only present in the tumor, confirming the somatic origin of these mutations. Reminiscent of the “mutator phenotype” seen in the breast cancer kinome screen (13), one metastatic tumor was found to carry four different FGFR2 mutations (V77M, E574K, S688F, and P708S), which were not further analyzed because there was a high probability that these mutations represented “passenger” mutations rather than “driver” mutations. Notably, two primary tumors carried nonsense mutations (Table 1). The majority (20 of 22) of FGFR2 mutations occurred at residues that are conserved across FGFR1 to FGFR4 or at residues that are conserved in three of four of the FGF receptors as well as conserved in FGFR2 across evolution (data not shown). Mutations in both NRAS and BRAF were observed in tumors with FGFR2 mutations at similar frequencies to that reported in the literature.
Mapping of FGFR2 Mutations within Known FGFR2 Structures
Extracellular Mutations
To gain insights into how the different FGFR2 mutations might affect receptor function, we analyzed them in the light of our crystallographic data on FGFRs. The E219K, G227E, V248D, R251Q, and G271E mutations, which affect residues in the extracellular region of FGFR2, were mapped onto the crystal structure of the 2:2:2 FGF2-FGFR1c-heparin dimer (PDB entry 1FQ9; ref. 14; Fig. 1). We intentionally chose this structure instead of the crystal structure of the 1:1 FGF2-FGFR2c monomer to evaluate the effect of the mutations on heparin/heparan sulfate– and FGF-induced FGFR dimerization. In the dimer, two centrally located FGFRs interact with each other directly and each receptor makes contacts with both ligands as well.
FIGURE 1.
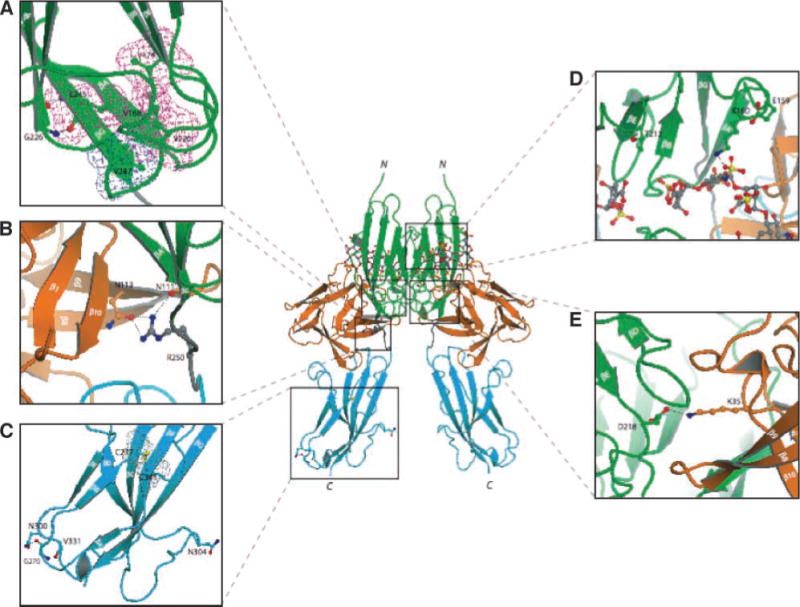
Mapping of FGFR2c extracellular mutations identified in melanoma tumors/cell lines onto the known FGF-FGFR crystal structures suggests that these mutations should impair FGFR2c activity. The locations of mutated residues in the extracellular region of FGFR2 are mapped onto the ribbon diagram of 2:2:2 FGF2-FGFR1c-heparin complex(PDB ID: 1FQ9; ref. 14). FGF is colored orange and the extracellular ligand binding region of FGFR is colored as follows: D2 in green, D3 in cyan, and D2-D3 linker in gray. The two heparin oligosaccharides in the dimer are rendered in sticks. To assist the viewer, certain β strands of D2 and D3 domains are labeled. A to E. Close-up view of the region where the mutated receptor residues are located. In each image, in addition to the mutated residue, other relevant receptor residues are shown as ball-and-sticks. A. The molecular interactions of G226 and V247, which correspond to the mutated G227 and V248 in FGFR2. G226 in βF strand makes hydrogen bonds with L245 in βG strand and contributes to proper βF–βG strand pair formation and, hence, to the overall stability of D2. V247 engages in hydrophobic contacts with other residues in the interior core of D2 and thus plays a critical role in folding of D2 as well. To emphasize these hydrophobic contacts, the surface of V247 and neighboring residues are shown in cyan (V247) and purple meshes, respectively. B. R250, homologous to R251 in FGFR2, makes three hydrogen bonds with FGF ligand. Mutation of R251 to Q should substantially reduce the binding affinity of mutated FGFR2 toward FGF ligand. C. The molecular environments of G270 and N304 in FGFR1, which correspond to the mutated G271 and G305 in FGFR2. G270 plays a role in formation of turn between βA′ and βB by making hydrogen bonds with other residues in D3. Therefore, mutation of G271 in FGFR2 should negatively affect the tertiary folding of D3. N304, on the other hand, maps to the large loop region between strand βC and βC′, which protrudes from D3 and does not play any apparent role in the structural integrity of D3. The surfaces of the conserved cysteines (C277 and C341) in the interior core of D3 are shown as gray mesh. D. The proximity of E159 and T212 to the heparin oligosaccharide chains in the dimer is emphasized. E159 and T212 correspond to E160 and H213 in FGFR2, respectively, and mutation of these residues in melanoma could alter the interaction of mutated FGFR2s with heparin/heparan sulfate. E. The important role of D218 is highlighted in the binding of the second FGF in the 2:2 FGF-FGFR dimer and hence dimerization. Mutation of E219 in FGFR2, which corresponds to D218 in FGFR1, should negatively affect the ability of the receptor to undergo ligand- and heparan sulfate–induced dimerization. Atom coloring is as follows: nitrogen in blue, oxygen in red, and sulfur in yellow. Hydrogen bonds are shown as dashed lines. Letters N and C denote the NH2 and COOH termini of FGFR1c, respectively.
Ligand interacts with the second and third immunoglobulin domains (hereafter referred to as D2 and D3) and the interconnecting D2–D3 linker. The R251Q mutation maps to the highly conserved D2–D3 linker region (Fig. 1B). In the structure, R250, the residue homologous to R251 of FGFR2, makes three hydrogen bonds with its primary FGF2, suggesting that the R251Q mutation should diminish the binding affinity of FGFR2 toward FGF2 (Fig. 1B). By contrast, the E219K mutation maps to the secondary ligand binding site on D2. In the dimer, D218, which corresponds to E219 of FGFR2, makes a hydrogen bond with K35 of the secondary FGF2 ligand (Fig. 1E). Therefore, based on the structure, E219K should not affect the ability of FGFR2c to bind FGF2, but it should reduce the ability of FGFR2 to undergo FGF- and heparin-induced dimerization.
Other extracellular mutations including V248D, G227E, and G271E are predicted to destabilize the tertiary fold of either D2 (V248D and G227E) or D3 (G271E). V247, which corresponds to V248 of FGFR2, is situated at the COOH-terminal end of the βG strand in D2 where its side chain is surrounded by other hydrophobic residues inside the hydrophobic core of D2 (Fig. 1A). Substitution of V248 with the charged aspartic acid should be detrimental to the tertiary folding of D2. G226, which corresponds to G227 in FGFR2c, maps to the start of the βF strand in D2 and is spatially in close vicinity to V247 (Fig. 1A). G270, which corresponds to G271 in FGFR2, maps to the loop between the βA′ and βB strands in D3 (Fig. 1C). Each glycine residue provides hydrogen bonds that contribute to the proper β-strand formation in D2 and D3, respectively. Therefore, based on the structure, these substitutions should cause local structural perturbations, which could ultimately affect the folding of the whole D2 and D3, respectively. Destabilization of D2 and D3 is predicted to impair the correct intramolecular disulfide bridge formation in D2 or D3, leading to exposure of the conserved cysteines and allowing them to form intermolecular disulfide-bridged receptor dimers. This destabilization can also interfere with the correct processing/maturation and trafficking of the mutated receptor through the endoplasmic reticulum (ER)/Golgi, resulting in receptor mislocalization.
Based on the structure, the E160A and H213Y mutations are likely to affect the interaction of FGFR2 with heparan sulfate. In the structure, E159 of FGFR1, which corresponds to E160 of FGFR2, facilitates the conformation of gA helix in D2, on which a key lysine residue involved in heparan sulfate binding resides (Fig. 1D). Therefore, destabilization of the gA helix due to the E160A mutation should negatively affect FGFR2-heparan sulfate interaction and thereby attenuate FGF- and heparan sulfate–mediated FGFR dimerization. T212, the residue corresponding to H213 of FGFR2, points into the heparin binding canyon of the 2:2:2 FGF2-FGFR1-heparan sulfate dimer (Fig. 1D), and the bulkier tyrosine side chain at this location could cause steric clashes with heparan sulfate oligosaccharide molecules, leading to inefficient receptor dimerization.
Kinase Domain Mutations
More than half of the melanoma mutations affect the conserved tyrosine kinase domain of FGFR2, and mapping of these mutations onto unphosphorylated and phosphorylated FGFR2 kinase domain structures (PDB ID: 2PSQ and 2PVF; ref. 15) suggests that they also represent loss-of-function mutations (Fig. 2). Notably, M640 and I642 localize onto either end of the β8 strand (Fig. 2), a secondary structure element that precedes the activation loop (A-loop) of the kinase domain. The hydrophobic side chains of these residues are in contact with residues in the αE helix in the innermost core of the C-lobe of the kinase domain. Substitutions of either residues with the smaller hydrophobic residues isoleucine or valine, respectively, would cause a cavity within the core of the kinase that should induce local structural perturbations that ultimately affect the conformation of the activation loop. The A648T mutation maps to the activation loop of the kinase domain (Fig. 2E) and is also expected to lead to loss of function. This is because in the phosphorylated activated kinase domain of FGFR2 (PDB ID: 2PVF; ref. 15), the methyl group of A648 makes hydrophobic contact with M537 in the αC helix, which contributes to the correct positioning/orientation of the αC helix relative to the COOH-terminal lobe of the kinase and, hence, to the productive alignment of the catalytic residues for the phosphotransfer reaction (15). A threonine is unfavorable at this location because its larger side chain is predicted to sterically hinder the αC helix from approaching sufficiently close to the COOH-terminal lobe, and consequently, the catalytic residues would not be aligned for the phosphotransfer reaction.
FIGURE 2.
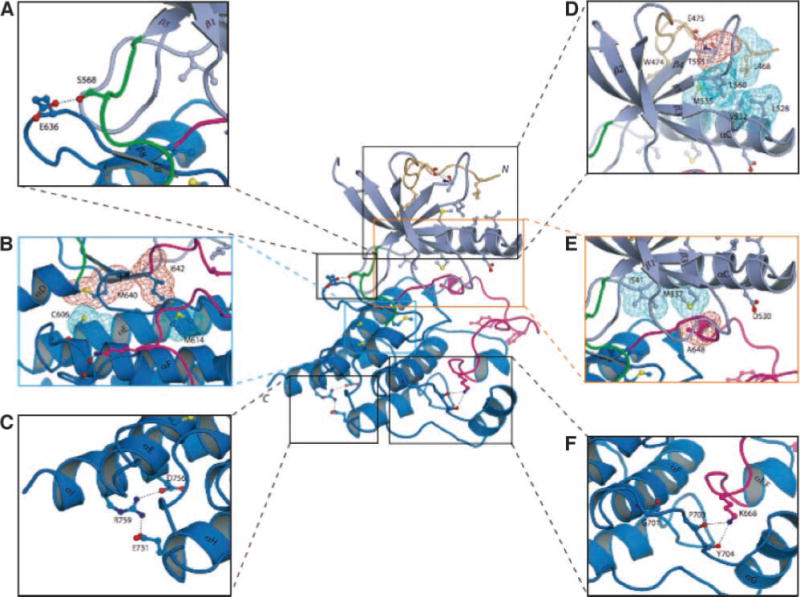
FGFR2c kinase domain mutations identified in melanoma tumors/cell lines mapped onto the crystal structures of the FGFR2 kinase domain suggest that these mutations should impair the kinase activity of FGFR2. The positions of the mutated residues are mapped onto the crystal structure of unphosphorylated wild-type FGFR2 kinase domain (PDB entry: 2PSQ; ref. 15). The coloring of the intracellular tyrosine kinase domain is as follows: the NH2-terminal lobe of kinase is in light blue; the COOH-terminal lobe is in bright blue; the activation loop is in magenta; the kinase hinge region is in green; and the NH2-terminal tail of the kinase is colored wheat. Note that ATP (not shown) binds in the cleft between the N-lobe and C-lobe of the kinase domain. To assist the viewer, certain β strands and α helices, which are relevant for explanation of the effects of the melanoma mutations, are labeled. A. E636, which maps to the sharp turn between β7 and β8 strands, makes hydrogen bond with S563 in the kinase hinge region. Mutation of this residue to lysine could influence the relative disposition of the N-lobe to the C-lobe, and hence alter the tyrosine kinase activity of FGFR2. B. The M640 and I642 side chains (red mesh) point into the inner hydrophobic core of the kinase domain and engage C606 and M614 (cyan mesh). Mutation of either M640 or I642 to smaller hydrophobic residues should weaken the extent of these core hydrophobic contacts and lead to a reduction in kinase activity. C. The hydrogen bonds between R759 and side chains of negatively charged E731 (in helix αH) and D756 (in helix αI) are shown. In addition to the loss of these hydrogen bonds, introduction of a stop codon at this location will also truncate the helix αI by one helical turn. These structural changes should reduce the stability of the kinase domain, specifically making the kinase domain temperature sensitive and ultimately leading to receptor loss of function. D. Close-up view of the region where E475 is located. The side chain of E475 engages in two strong hydrogen bonds with the side chain and backbone atom of T555 (located in the loop between β4 and β5 strands), which facilitate the conformation of the β4–β5 loop as well of the region preceding E475 itself. This in turn promotes the hydrophobic contacts between L468, T555 (the methyl group), and L560 from these regions and L528, V532, and M535 in the catalytically important αC helix. To emphasize this point, the side chains of these hydrophobic residues are shown as colored mesh. E. Close-up view of the region where D530 and A648 are located. In unphosphorylated wild-type FGFR2K structure, D530 does not play any role. However, in the A-loop phosphorylated activated FGFR2K structure (PDB entry: 2PVF; ref. 15), the side chain of D530 engages in salt bridge with R664 in the A-loop and contributes to A-loop conformation in the active state (not shown). The side chain of A648, located at the beginning of the A-loop, packs against the hydrophobic side chains of M537 in the αC helix. Replacement of A648 with threonine is expected to lead to loss of function because the larger side chain of threonine will impede αC helix from nearing sufficiently close enough to the A-loop in the active FGFR2K. F. Close-up view of the kinase region where G701 is located. G701 plays a role in stabilizing the conformation of the loop between the helices αF and αG, which is in hydrogen-bonding contacts with K668 in the A-loop. The G701S mutation should destabilize the αF-αG loop and indirectly affect the A-loop conformation.
The E636K mutation maps to the loop region between the β7 and β8 strands (Fig. 2A). In the structure, the side chain of E636 makes a solvent-exposed hydrogen bond with S568, which is located in the hinge region of the kinase domain, suggesting that it may play a role in regulation of the spatial positioning of the NH2-terminal and COOH-terminal lobes of the kinase relative to each other. Based on the structure, the E636K mutation should influence the kinase activity of the FGFR2 kinase domain. The E475 is part of the WE consensus motif that defines approximately the NH2-terminal boundary of the conserved kinase domain in many receptor tyrosine kinases. E475 engages in two strong hydrogen bonds with T555 whose methyl group is in hydrophobic contact with residues in the αC helix (Fig. 2D). Hence, these interactions hold the αC helix in an orientation found in unphosphorylated inhibited kinases. In the phosphorylated activated kinase domains, however, these hydrogen bonds are weaker and, concomitantly, the C-helix moves closer toward the C-lobe. Loss of these hydrogen bonds due to the E475K mutation could relieve this inhibition and potentially lead to activation of the kinase domain. Alternatively, the E475K mutation could introduce structural instability in the N-lobe of the kinase and reduce the half life of the mutant FGFR2. The G701S mutation maps to the loop between αF and αG helices (Fig. 2F) and facilitates the conformation of this loop region, which makes several hydrogen bonds with K668 in the activation loop. Therefore, the structural perturbations induced by the G701S mutation could indirectly lead to reduced kinase activity. The R759Q mutation maps to the COOH-terminal end of the αI helix, the last secondary structure element of the kinase domain. In the structure, R759 makes hydrogen bonds with two other residues in its vicinity (Fig. 2C), and loss of these hydrogen bonds due to the R759Q mutation could also induce subtle structural changes leading to a loss in kinase activity.
The L770V mutation should reduce the ability of FGFR2 to activate the phospholipase Cγ (PLCγ) signaling pathway. L770 follows Y769, the major phosphorylation site of FGFR2 that serves as the docking site for SH2 domains of PLCγ and thus is required for PLCγ phosphorylation and activation by the activated FGFR2. Nuclear magnetic resonance solution structure of PLCγ SH2 domain in complex with phosphopeptide indicates that the SH2 domain of PLCγ interacts with pTyr and residues at +1 to +6 positions relative to the pTyr. Hence, the L770V mutation should reduce the affinity of SH2 domains to the activated FGFR2. The L770V mutation could also reduce Y769 phosphorylation because the substrate binding pocket of the kinase domain prefers a leucine over a valine next to the tyrosine phosphorylation site (16). The reduced phosphorylation of Y769 could, in turn, reduce the ability of FGFR2 to activate the PLCγ pathway. Table 2 summarizes the different mechanisms of loss of function by the melanoma mutations inferred from crystal structures. Consistent with these analyses predicting that the FGFR2 mutations are loss-of-function mutations, two melanoma samples harbored nonsense mutations and a third sample carried a splicing mutation that would result in premature termination of FGFR2 protein translation (Table 1).
Table 2.
Summary of In silico Analysis of FGFR2 Mutations
| Novel FGFR2 Mutation | Predicted Effect Based on Molecular Modeling |
|---|---|
| E160A, H213Y | Negatively affect interaction with heparan sulfate |
| E219K | Reduced FGF- and heparin-mediated receptor dimerization |
| G227E, V248D | Destabilize the D2 domain |
| R251Q | Abrogate ligand binding |
| G271E | Destabilize the D3 domain |
| E475K | Possible increased kinase activity but less stable kinase domain |
| M640I, I642V | Local structural perturbations resulting in altered conformation of the activation loop and decreased kinase activity |
| A648T | Markedly reduced kinase activity due to misalignment of catalytic residues for phosphotransfer reaction |
| L770V | Impaired phosphorylation of Y769 and reduced recruitment of PLCγ |
Analysis of FGFR2 Melanoma Mutations In vitro
To test our structural predictions for the extracellular mutations on FGFR2 function, we expressed recombinant wild-type and mutant ectodomains in E. coli, and then isolated the inclusion bodies enriched in the proteins and subjected them to in vitro refolding. As a positive control, we also used the S252W mutant ectodomain. We have previously shown that the S252W mutation has no adverse effect on folding of FGFR ectodomain (17). We were unable to refold in vitro the ectodomain proteins containing the G227E, V248D, and G271E mutations, consistent with structural predictions that these mutations are detrimental to the tertiary fold of D2 and D3 domains. By contrast, the R251Q mutant ectodomain folded in vitro with yields similar to those of the wild-type and S252W mutant receptors. We then used surface plasmon resonance to analyze the effect of the R251Q mutation on ligand binding affinity of FGFR2c. Consistent with structural predictions, the R251Q mutation severely impaired the ability of FGFR2c to bind multiple FGF ligands including FGF2 (Fig. 3) and FGF1, FGF4, FGF7, FGF8, and FGF10 (data not shown). On the other hand, as expected, the S252W mutation enhanced the affinity of FGFR2c toward FGF2 and other FGFs (refs. 17–19; data not shown).
FIGURE 3.
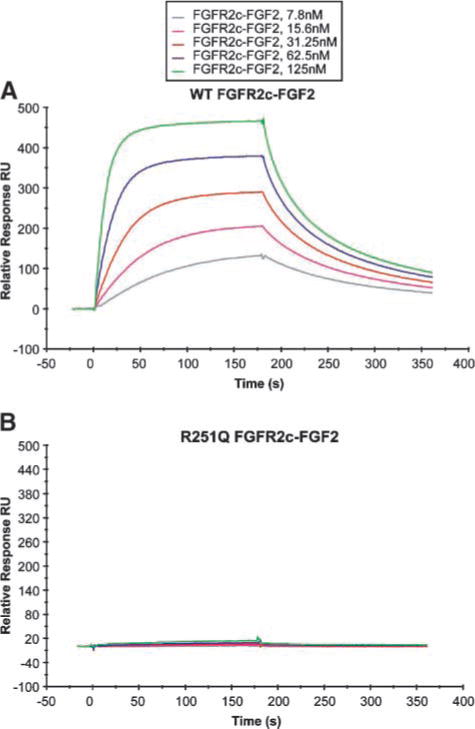
Surface plasmon resonance analysis shows that the R251Q mutation abrogates ligand binding. A and B. Increasing concentrations of wild-type (WT; A) or R251Q FGFR2c mutant ectodomain (B) were injected over a CM5 chip onto which full-length human FGF2 was immobilized. The biosensor chip response is plotted as a function of time. Surface plasmon resonance analysis was done as previously described (19).
To test the effect of kinase domain mutations, we prepared recombinant wild-type and mutant kinase domain proteins harboring the E475K, D530N, I642V, and A648T melanoma mutations and subjected them to in vitro kinase assays. Consistent with structural predictions, the D530N, I642V, and A648T mutant kinase domains exhibited reduced kinase activity relative to the wild-type kinase domain (Fig. 4). Interestingly, the E475K mutant exhibited a slight increase in kinase activity relative to the wild-type kinase domain. Notably, the protein expression yield for the E475K mutant kinase was severalfold less than that of wild-type kinase, suggesting that this mutation was partially destabilizing the kinase domain.
FIGURE 4.
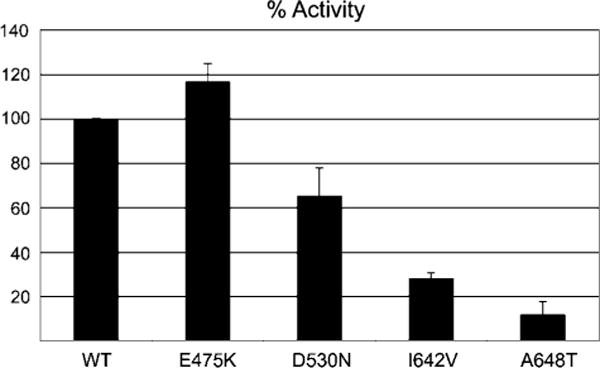
FGFR2 kinase domains harboring melanoma mutations have reduced tyrosine autophosphorylation activity. Wild-type FGFR2K and four mutant FGFR2Ks, each harboring E475K, D530N, I642V, or A648T single mutation, are subjected to in vitro kinase assays. Columns, average of three independent experiments; bars, SD. The tyrosine autophosphorylation activities of these wild-type and mutant FGFR2Ks were determined using a continuous spectrophotometric assay as previously described (43).
Analysis of FGFR2 Mutations in Cultured Cells
We next analyzed the effect of several mutations (G227E, R251Q, G271E, E475K, D530N, I642V, and A648T) on the receptor trafficking and maturation of the full-length FGFR2c. Three different FGFR2 mutants harboring C278F, S252W, or K517R served as controls. The C278F activating mutation associated with Crouzon syndrome was used to assess the effect of destabilizing D3 on both receptor maturation and activation. This mutation has been shown to result in impaired receptor processing, constitutive dimerization, and increased signaling from intracellular compartments (20). In contrast, the S252W mutation observed in patients with Apert syndrome has no effect on receptor folding (19). The K517R mutant receptor served as a kinase-deficient receptor. K517 coordinates the phosphate groups of ATP, and its mutation to arginine in the homologous FGFR3 has been previously shown to abolish kinase activity (21).
Following transfection of wild-type FGFR2 receptor into HEK293 cells, two major FGFR2-immunoreactive bands (110 and 130 kDa) are observed. PNGase digestion resulted in the reduction in size of both bands to a single 98-kDa band (open arrow), confirming that these two bands are differently glycosylated receptor populations (Fig. 5A). EndoHf digestion, which is known to remove N-linked high-mannose moieties but not complex carbohydrates from glycosylated proteins, resulted in the reduction of the 110-kDa band to 98 kDa but had no effect on the mobility of the 130-kDa band. These data show that the 110-kDa form represents immature, partially processed receptor present in the ER and early Golgi, whereas the EndoHf-resistant 130-kDa band represents the fully glycosylated mature receptor. Densitometric analysis indicates that the EndoHf-resistant 130-kDa mature form comprises ~60% of the wild-type FGFR2 (Fig. 5A and B). The R251Q and S252W mutations did not reduce the relative proportion of the EndoH-resistant 130-kDa mature form, which is fully consistent with our structural predictions of no adverse effects on protein stability by these mutations. In contrast, only 30% of G227E, V248D, G271E, and C278F mutants were EndoHf resistant, suggesting impaired receptor processing of these FGFR2 mutants. This is also consistent with our structural predictions that these mutations will adversely affect the structural integrity of the D2 or D3 domain of FGFR2 (Fig. 5B, top). There was a small increase in the relative proportion of the 110-kDa band for the E475K mutant (Fig. 5B, bottom), consistent with the decreased protein yield observed in the in vitro kinase assays.
FIGURE 5.
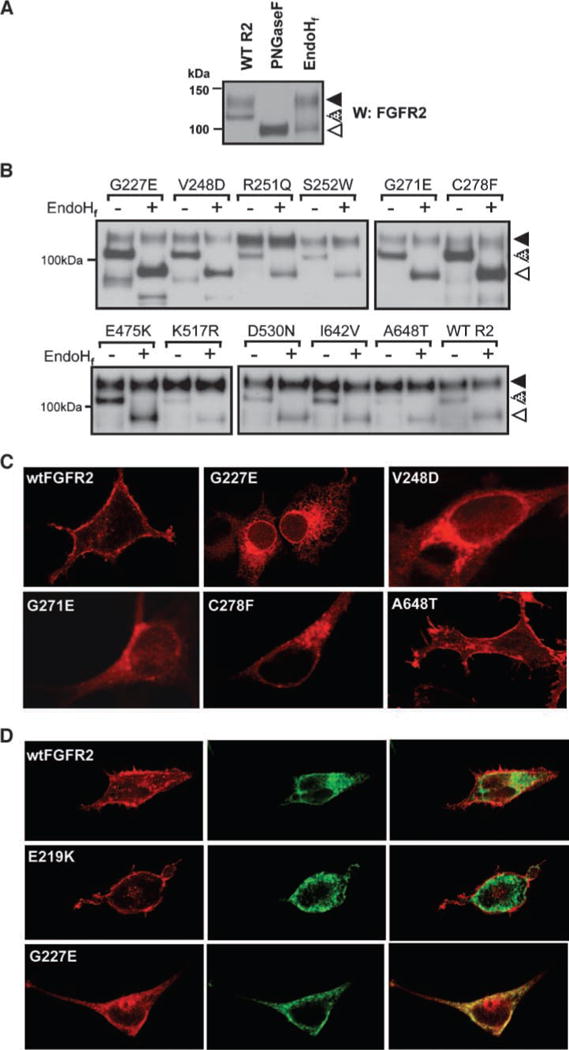
Mutations in FGFR2 impair receptor processing and localization. A. Confirmation of posttranslational glycosylation of wild-type FGFR2 by PNGase digestion of both the 130-kDa band (solid arrow) and 110-kDa band (stippled arrow) to a single 98-kDa band (open arrow) in HEK293 cells. Sensitivity to digestion with EndoHf of only the 110-kDa band indicates this 110-kDa band to be an immature, partially processed receptor predominantly present in the ER and early Golgi. B. EndoHf digestion of FGFR2 in HEK293 lysates transiently transfected with wild-type FGFR2 or mutant FGFR2. Densitometry analysis revealed that the EndoH-resistant 130-kDa band comprises ~60% of the wild-type FGFR2 but only 30% of G227E, V248D, G271E, and C278F mutants. C. Localization of wild-type and mutated FGFR2 following transient transfection into HEK293 cells and immunofluorescent staining. Impaired receptor trafficking is evident for G227E, V248D, G271E, and C278F compared with trafficking of the wild-type receptor to the cell surface. D. Immunofluorescent analysis of the colocalization of FGFR2 (red, left) and the ER-resident marker protein disulfide isomerase (green, middle) confirms the overall retention of the G227E mutant receptor in the ER compartment in HEK293 cells (yellow, right) when compared with the cell surface localization of wildtype FGFR2 and E219K (mapping onto the known extracellular crystal structure predicted the latter mutation to have no effect on misfolding).
To confirm altered cellular localization of the mutants, we performed immunofluorescence studies in transiently transfected HEK293 cells. In cells transfected with wild-type FGFR2, positive immunofluorescence signal was distributed primarily in the plasma membrane and also in the rough ER and Golgi (Fig. 5C). In cells transfected with the G227E mutant (Fig. 5C), however, there was positive perinuclear staining, particularly evident around the nuclear membrane, as well as a dispersed reticular staining indicative of retention in the ER. Immunofluorescence also showed mislocalization to intracellular membranes and decreased cell surface expression for FGFR2 mutants V248D, G271E, and C278F. In contrast, immunofluorescence of the A648T mutant shows clear cell surface localization in keeping with the structural predictions and EndoHf analyses. To confirm the ER localization of the G227E mutant, we then performed similar immunofluorescence studies looking for colocalization with the ER-resident protein, protein disulfide isomerase, which revealed a high degree of colocalization of the two proteins, confirming impaired receptor localization (Fig. 5D). In contrast, marked cell surface localization and limited colocalization with protein disulfide isomerase were observed for both the wild-type FGFR2 and the E219K mutation, predicted to have no detrimental effect on receptor folding.
BaF3 Proliferation Assay and p44/42 Mitogen-Activated Protein Kinase Phosphorylation in Response to FGF2 as Readout of Receptor Function
We next analyzed the effects of the melanoma mutations on FGFR2c activity. Given that melanocytes and melanoma cell lines express other FGFRs whose activation may mask the ability to specifically evaluate FGFR2 signaling and function, we used the murine interleukin-3 (IL-3)–dependent pro-B BaF3 cell line to determine receptor activity. The BaF3 cell line is routinely used as a model system for the evaluation of FGFR function because it does not express endogenous FGF ligands or receptors. Although BaF3 cell proliferation and survival is normally dependent on IL-3, activated tyrosine kinase signaling can substitute for IL-3 to maintain cell viability and proliferation. We included the N549K FGFR2 receptor as a constitutively activated positive control because this activating mutation has been reported in endometrial cancer (12) and an identical mutation at the paralogous positions in FGFR3 has been associated with hypochondroplasia (N540K; ref. 22). Polyclonal stable lines were generated following lentiviral transduction and selection in geneticin. Proliferation assays were then done in the absence of IL-3 and in the presence of 1 nmol/L FGF2 and 10 μg/mL heparin, and cell viability was assayed after 5 days. As anticipated, the constitutively active N549K mutant resulted in levels of proliferation above that achieved by the wild-type receptor, whereas expression of all mutant constructs including G227E, G271E, E475K, I642V, and A648T FGFR2 resulted in reduced cell proliferation compared with wild-type FGFR2 (Fig. 6A), verifying that these mutations impaired or abrogated the ability of FGFR2 to stimulate cell proliferation in response to ligand in an in vivo setting. Surprisingly, the N549K mutation did not cause constitutive BaF3 proliferation in the absence of ligand stimulation.
FIGURE 6.
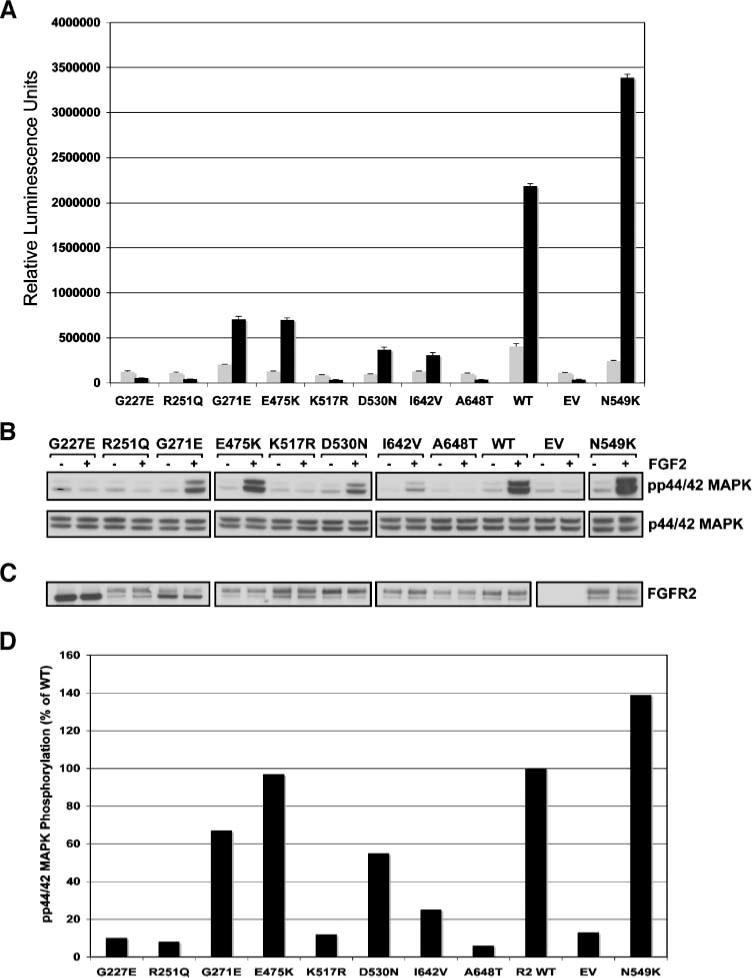
Cell proliferation in response to FGF2 in BaF3 cell lines stably transduced with wild-type and mutant FGFR2_I RES_Neo expression constructs. A. All novel mutations identified in melanoma result in a decrease in BaF3 proliferation when compared with wild-type FGFR2. Gray columns, basal proliferation in the absence of ligand; black columns, proliferation in response to 1 nmol/L FGF2. Columns, average of at least two independent experiments assayed in triplicate. B. Analysis of phospho-p44/42 MAPK on stimulation with exogenous rFGF2 shows that, with the exception of the N549K positive control, mutations impair the ability of the receptor to activate this pro-proliferation pathway compared with wild-type receptor. C. Western blot showing equal expression of FGFR2 by all mutants except empty vector in the absence (−) and presence (+) of FGF2. D. Graphical representation of p44/42 MAPK phosphorylation expressed as a percentage of phosphorylation (on ligand stimulation) of that of wild-type receptor.
As a further assessment of receptor impairment due to mutation, we examined the ability of the receptor to activate the pro-proliferative p44/42 mitogen-activated protein kinase (MAPK) signaling pathway in response to exogenous rFGF2. With the exception of the N549K known activating mutant, all mutations resulted in a reduction to varying levels in the ability of the receptor to induce phosphorylation of p44/42 MAPK (Fig. 6B–D).
Reintroduction of FGFR2 into Melanoma Cells
We next sought to determine whether reintroduction of FGFR2 could suppress proliferation of melanoma cells. Reverse transcription-PCR with isoform-specific primers and subsequent sequencing revealed normal human melanocytes and the majority of melanoma cell lines expressed the mesenchymal FGFR2c isoform (data not shown). For this reason, our reintroduction studies used the FGFR2c isoform. We overexpressed FGFR2c in melanoma cell lines that lacked FGFR2 expression by reverse transcription-PCR (SBcl2) and in melanoma cell lines that expressed mutant FGFR2 (AO4, D22, UACC2534) and assessed proliferation. Surprisingly, reintroduction of FGFR2 failed to suppress the proliferation of these melanoma cell lines (Fig. 7A). Expression of FGFR2 following lentiviral transduction was confirmed by Western blot analysis for each cell line (Fig. 7B). Similar results were also achieved in both the D22 and UACC2534 melanoma cell lines (data not shown).
FIGURE 7.
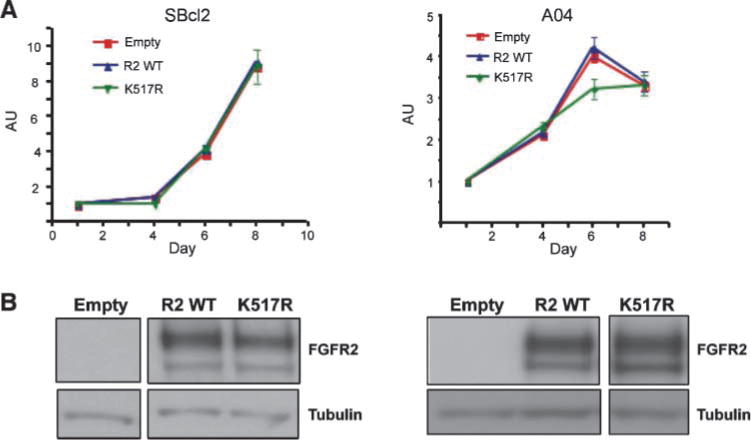
Reintroduction of FGFR2 failed to suppress proliferation of SBcl2 or AO4 melanoma cells. A. Melanoma cell lines SBcl2 and AO4 were stably transduced with empty vector, FGFR2, or FGFR2 K517R lentivirus, and proliferation was assessed using the sulforhodamine B assay. Overexpression of FGFR2 or kinase-dead FGFR2 K517R had no effect on the proliferation of these melanoma cell lines. B. FGFR2 expression was confirmed by Western blot analysis.
Discussion
In this study, we report novel FGFR2 mutations in 15 of 116 melanoma cell lines and 8 of 100 uncultured metastases and primary tumors. To determine the ratio of nonsynonymous to synonymous mutations in our data set, we went back and examined the synonymous variants we identified. Because we did not have constitutional DNA for these samples, we excluded them as probable somatic mutations if they had been previously identified in dbSNP. Several additional synonymous variants occurred alongside each other in multiple individuals, consistent with the presence of a rare germ-line haplotype block. Following the exclusion of these putative SNPs, four synonymous variants remained; therefore, the conservative ratio of nonsynonymous to synonymous mutations in our data set was 23:4, which exceeds the 2:1 ratio one would expect if these were random passenger mutations, and supporting a role for these mutations in melanoma pathogenesis. Furthermore, the mutation spectrum is characteristic of those induced by UV radiation (i.e., 70% of the mutations were C:G>T:A substitutions with one CC:GG>TT:AA tandem mutation). The presence of characteristic UV mutations in FGFR2 suggests that mutation of this gene is possibly an early event in transformation, and we anticipate that further sequencing analysis in a greater number of primary tumors and nevi will shed light on when during melanoma development these mutations arise. It should be noted that the mutation frequency reported in this study may be an underestimate. This is based on the fact that 40 cell lines were screened by denaturing high-performance liquid chromatography rather than sequencing, exons 2 to 5 were not screened in this latter panel of cell lines, and not all primary tumors provided high-quality sequence for all exons.
Insights gained from mapping these mutations onto known FGFR2 crystal structure, combined with in vitro and in vivo functional studies, show that these mutations lead to loss of receptor activity through different mechanisms, including loss of ligand binding (R251Q), incomplete processing of the receptor and retention in the ER (V248D, G227E, G271E), and complete or partial loss of tyrosine kinase activity (A648T, D530N, I642V). Future studies confirming the structural predictions described herein may also reveal that FGFR2 loss of function can occur through impaired FGF- and heparan sulfate–induced receptor dimerization and reduced binding to PLCγ.
The loss-of-function nature of our identified melanoma mutations is also consistent with published literature. In support of D530N resulting in FGFR2 loss of function, a germ-line D771N mutation at the equivalent codon in RET has been associated with Hirschsprung’s disease, commonly due to loss-of-function mutations in RET (23). In addition, while these studies were under way, the A648T FGFR2 mutation was identified in the germ line of affected patients in two independent families with autosomal dominant lacrimo-ariculo-dento-digital (LADD) syndrome (OMIM 149730; ref. 24). Consistent with our in vitro and in vivo data, a recent study showed that the A648T mutation abrogated the tyrosine kinase activity of FGFR2 (25). As discussed earlier, we propose that this mutation creates a steric barrier for the movement of the kinase N-lobe toward the C-lobe, which is predicted to take place when the kinase transits from the basal “low” activity state to the “active” state. However, precise definition of the mechanism by which A648T mutation impairs FGFR2 kinase activity awaits the resolution of crystal structure of the mutant kinase domain as was recently carried out for an A628T LADD mutation. In contrast to the A648T mutation, the A628T mutation maps onto the catalytic loop of the kinase domain, and the crystal structure of the mutant kinase shows that this mutation acts by introducing steric conflicts that alter the productive arrangement of the neighboring key catalytic residues (26).
A comparison of the mutations identified in melanoma to those our lab previously identified in endometrial cancer highlights the differences in the mutation spectra (Fig. 8). The majority of the endometrial mutations (17 of 20) are identical to FGFR2 and FGFR3 mutations previously reported in the germ line, a finding similar to that observed for FGFR3 where a comparison of FGFR3 mutations reported in cancer with germ-line mutations associated with skeletal disorders shows a remarkable concordance (reviewed in refs. 27, 28). In contrast, the more than 20 FGFR2 mutations we identified in melanoma were all novel, supporting our findings that the mutations are distinct and different from the many gain-of-function mutations previously identified in the FGFRs.
FIGURE 8.
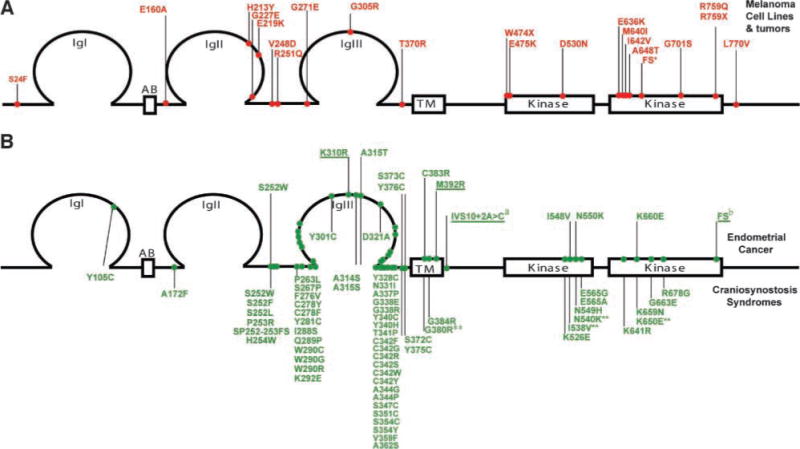
Schematic representation of FGFR2 mutations highlighting the spectrum difference of somatic mutations in melanoma compared with endometrial cancer, as well as germ-line mutations in craniosynostosis syndromes. A. Novel somatic mutations in FGFR2 identified in melanoma cell lines and uncultured tumors are presented in red above the schematic representation of the protein and numbered relative to FGFR2c (NP_000132.1). B. Somatic mutations in FGFR2 identified in primary endometrial cancers and cell lines are presented in green (top) above the schematic representation of the protein and are numbered relative to FGFR2b (NP_075259.2; ref. 12). Bottom, germ-line mutations associated with a variety of craniosynostosis syndromes and numbered relative to FGFR2c (NP_000132.1; http://www.hgmd.cf.ac.uk/ac/index.php). Novel mutations are underlined. Four somatic FGFR2 endometrial mutations, although not previously reported in the germ line, have an identical missense change reported in the paralogous position in FGFR3c in a skeletal chondrodysplasia (indicated with **; http://www.hgmd.cf.ac.uk/ac/index.php). Note that the majority of somatic mutations in endometrial cancer parallel those identified in the germ line, whereas every melanoma mutation was novel.
This discovery of loss-of-function mutations in a receptor tyrosine kinase is unexpected, given the conventional wisdom that activation of receptor tyrosine kinases (e.g., KIT, FLT3, and FGFR3) drives tumorigenesis and our previous report documenting the presence of activating mutations in FGFR2 in endometrial cancer (12). However, it should not be surprising given that a variety of receptor tyrosine kinase signaling pathways can induce either proliferation or differentiation depending on the cell type. Indeed, Cre-Lox transgenic mice, with FGFR2b having been deleted in cells expressing keratin V, develop spontaneous papillomas. Moreover, following 7,12-dimethylbenz(a)anthracene and 12-O-tetradecanoyl-phorbol-13-acetate treatment, these transgenic mice developed an increased number of papillomas and carcinomas compared with wild-type mice, providing support that FGFR2 can act as a tumor suppressor gene in some cell types (29). Furthermore, loss of FGFR2 expression has been associated with several cancers including prostate and bladder (30–32). Reintroduction of FGFR2 resulted in decreased growth in vitro and reduced tumorigenicity in vivo of bladder carcinoma cells (33), prostate tumor cells (34–36), and salivary adenocarcinoma cells (37).
Based on these data and our structural, biochemical, and biological data supporting the characterization of the FGFR2 mutations identified in melanoma as loss of function, it was indeed surprising that reintroduction of FGFR2 failed to suppress proliferation of melanoma cell lines. There are several possible explanations for these apparent disparate results. One difference between the previously published reintroduction results and the current melanoma study is that they involve different FGFR2 splice forms. The previous studies were all carried out in cells of epithelial origin with the FGFR2 IIIb isoform, whereas our studies evaluated proliferation following reintroduction of FGFR2 IIIc, the isoform of FGFR2 expressed in mesenchymal cells and in the melanocytes, which are derived from the neural crest. The lack of suppression of proliferation of melanoma cells following overexpression of FGFR2 may therefore reflect a cell lineage–specific function of FGFR2. Another possible explanation is that loss or abrogation of FGFR2 is an early event and those melanoma cell lines have acquired additional genetic aberrations that prevent suppression of proliferation following FGFR2 reintroduction. Alternatively, abrogation of FGFR2 may not contribute to melanoma pathogenesis by removing a growth inhibitory signal. Indeed, although tumor suppressors classically function to regulate cell cycle, accumulating evidence suggests that additional subsets of genes exist, which function to suppress tumorigenesis not by inhibiting proliferation but by altering other aspects of the malignant phenotype, including loss of contact inhibition, angiogenesis, cell migration, and metastasis. Therefore, the pathogenic consequence of loss of FGFR2 function may not directly involve cell cycle regulation but instead affect another aspect of tumorigenesis. The caveat exists that loss of FGFR2 might not be a driver of cell transformation but rather these mutations occur as a consequence of the oncogenic state; however, their selection in 10% of melanoma samples would argue that they must provide a selective advantage to the cells in which they arose. This is consistent with a relatively high frequency and high nonsynonymous to synonymous ratio arguing against their presence as passenger mutations.
Elucidation of the mechanism by which abrogation of FGFR2 contributes to melanoma is also complicated by our current lack of understanding about the differing cellular functions of the multiple splice forms of these receptors, particularly in the melanocytic system. There are additional splice variants affecting the ligand binding domain (2 Ig, 3 Ig), cytoplasmic juxtamembrane domain (+VT, −VT), and COOH-terminal domain of FGFR2 (C1, C3), and the role of these splice variants in normal melanocyte function and melanoma pathogenesis is currently unknown. Our lab is currently investigating the relative expression levels of these various isoforms and their functional significance in melanocytes and melanoma. FGFR2 may also have cellular functions distinct from typical receptor tyrosine kinase signaling. There have been reports that FGFR1 binds CBPand RSK in the nucleus (38), and we and others have observed that FGFR2 localized to the nucleus in various cell types (39).8 Lastly, we and others have shown that FGFR2 can heterodimerize with FGFR1 at least in vitro (40)8 and FGFRs have also been shown to bind to other molecules at the cell surface, including neural cell adhesion molecule (41). We postulate that a more complete understanding of FGFR biology will allow us to identify the mechanism by which loss of FGFR2 contributes to melanoma initiation and/or progression.
In conclusion, we provide strong genetic, biochemical, in vitro and in vivo evidence that loss-of-function FGFR2 mutations occur in a subset of melanomas. Although we have yet to uncover the biological consequence of loss of FGFR2 function in melanoma, we propose that FGFR2 should join the list of those genes that play context-dependent opposing roles in tumorigenesis. Although many genes have been shown to have context-dependent opposing roles in cancer (e.g., Notch and TGFβ; ref. 42), FGFR2 may be the first gene in which both loss-of-function and gain-of-function mutations have been reported in different tissue types. Ongoing work in our laboratory is targeted toward understanding the molecular mechanisms by which loss of FGFR2 contributes to melanoma pathogenesis.
Materials and Methods
See Supplementary Methods for details on mutation detection.
Expression, Refolding, and Purification of Ectodomain of FGFR2
DNA fragment encoding residues 147 to 366 of FGFR2c was amplified by PCR and subcloned into pET-28a bacterial expression vector using Nco I and Hin dIII cloning sites. Point mutations (G227E, V248D, R251Q, S252W, and G271E) were introduced using QuikChange site-directed mutagenesis kit (Stratagene). BL21(DE3) competent cells were transformed with wild-type or mutant expression constructs and protein expression was induced with 1 mmol/L isopropyl-L-thio-B-D-galactopyranoside for 5 h at 37 °C. Cells were lysed using French press and centrifuged; the inclusion bodies containing wild-type or mutant proteins were dissolved in 6 mol/L guanidinium hydrochloride plus 10 mmol/L DTT in 100 mmol/L Tris-HCl buffer (pH 8.0). The solubilized FGFR2 proteins were subjected to an in vitro refolding by slow dialysis against 25 mmol/L HEPES buffer (pH 7.5) containing 150 mmol/L NaCl, 10% glycerol, and L-cysteine. The correctly refolded wild-type and mutant proteins were then sequentially purified by heparin affinity and size exclusion chromatography. Surface plasmon resonance analysis was done as described previously (19).
Expression and Purification of FGFR2 Kinase Domain
The DNA fragment encoding residues 458 to 768 of human FGFR2 was subcloned into pET bacterial expression vectors with an NH2-terminal 6×His-tag to aid in protein purification. Point mutations (E475K, D530N, I642V, and A648T) were introduced using QuikChange site-directed mutagenesis kit (Stratagene). The bacterial strain BL21(DE3) cells were transformed with wild-type or mutant FGFR2 kinase expression constructs, and protein expression was induced with 1 mmol/L isopropyl-L-thio-B-D-galactopyranoside for 4 h at ambient temperature. The cells were then lysed using a French press, and the soluble kinase proteins were purified by using sequential Ni2+-chelating, anion exchange and size exclusion chromatography. The purity of the proteins was estimated to be >98% based on SDS-PAGE analysis. Purified wild-type and mutant kinase proteins were concentrated to at least 10 mg/mL using a Centricon-10. The autophosphorylation activity of the wild-type and mutant kinases was compared using a continuous spectrophotometric kinase assay according to the published protocol (43).
Site-Directed Mutagenesis of Full-length FGFR2
All mutations were introduced to the pcDNA3_FGFR2 wild-type plasmid (NM_000141) using the QuikChange II XL site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions. Additionally, mutagenesis primers were designed to introduce a novel but silent (noncoding change) restriction enzyme recognition site to allow for fast, preliminary screening of clones. The full list of primer sequences can be found in Supplementary Table S1. After restriction enzyme screening, the entire coding sequence of FGFR2 was sequenced for each clone to confirm the presence of the intended mutation and to ensure that no other mutations were introduced during the mutagenesis process. Plasmid DNA was then isolated using Qiagen EndoFree Plasmid DNA Isolation kits, and DNA with A260/A280 > 1.8 was used in all subsequent experiments.
Cell Lines and Transfections
All cell lines described in this series of experiments were maintained under standard culturing conditions as described by the American Type Culture Collection or Deutsche Sammlung von Mikroorganismen und Zellkulturen (BaF3 cell line). Transient transfections of 1 μg total DNA (consisting of 250 μg wild-type or mutant FGFR2 + 750 μg empty plasmid) were achieved with FuGene 6 (Roche) at a 3:1 ratio of reagent/DNA according to the manufacturer’s instructions.
Cell Lysis, Endoglycosidase Digestions, and Western Blotting
At 36 to 48 h posttransfection, cells were washed with ice-cold PBS and lysed in ice-cold lysis buffer [20 mmol/L HEPES (pH 7.4), 1% Triton X-100, 2 mmol/L EGTA, 10% glycerol, Complete Protease Inhibitor Cocktail (Roche), 1 mmol/L NaF, 1 mmol/L Na3VO4, 100 μmol/L phenylmethylsulfonyl fluoride]. For PNGase and EndoHf digestions, 20 μg of lysate were subjected to digestion according to the manufacturer’s recommendations. Protein was resolved on 3% to 8% Tris-acetate gels and transferred onto polyvinylidene difluoride membranes (Invitrogen). After blocking the membranes with 5% skim milk/TBST, membranes were probed with 1:2,000 Bek C-17 (Santa Cruz Biotechnology).
Immunofluorescence
Cells were transfected as described and plated onto eight-well chamber slides. After 36 to 48 h, cells were fixed with 4% paraformaldehyde, permeabilized in 0.2% Triton, and incubated with 1:300 Bek C-17 and 1:1,000 anti–protein disulfide isomerase (Invitrogen) and then detected with goat ant-rabbit Cy3 (1:200; Jackson ImmunoResearch) and goat anti-mouse Alexa Fluor 488 (1:1,000; Invitrogen), respectively. Slides were mounted with Pro Long Gold anti-fade reagent and imaged under a Zeiss LSM 5 Pascal confocal microscope.
Lentivirus Production
293FT cells on poly-D-lysine–coated plates were transfected with lentiviral packaging vectors and pTYF.FGFR2.IRES.Neo (or mutant FGFR2) using Superfect (Qiagen) at a 4:1 Superfect/DNA ratio as per manufacturer’s instructions. Media containing the virus were collected at 24 and 40 h, pooled, filtered through a 0.45-um low protein binding filter, and stored at −80°C until use. Viral stocks were titered in HEK293 cells following serial dilution in a six-well plate, selection for 14 d in 800 μg/mL geneticin, staining with crystal violet, and colony counting.
BaF3 Proliferation Assays
BaF3 cells (5 × 105) were infected with a multiplicity of infection of 1 (5 × 105 viral transducing units) for 23 h, then placed under selective pressure (1,200 μg/mL geneticin) for 14 d in the presence of 5 ng/mL IL-3. Transduced, stably selected BaF3 cells containing wild-type and mutant FGFR2 were maintained under selection in RPMI/10% fetal bovine serum containing 50 nmol/L β-mercaptoethanol, 100 units/mL penicillin/100 μg/mL streptomycin sulfate, 1,000 μg/mL geneticin, and supplemented with 5 ng/mL murine IL-3 (R&D Sytems). Before the proliferation assay, cells were counted and washed in the above media containing no IL-3. Cells were plated at 1 × 104 per well in triplicate in a 96-well plate in media containing 16.7 ng/mL (1 nmol/L) FGF2 and 10 μg/mL heparin. Half the volume of media was removed at day 3 and replaced with an equal volume containing fresh ligand at 2× concentration and no IL-3. Proliferation was assessed using the ViaLight Plus Cell Proliferation/Cytotoxity Kit (Lonza Rockland, Inc.) according to the manufacturer’s instructions.
Melanoma Proliferation Assays
SbCl2 melanoma cells were cultured in 2% Tumor Medium containing a 4:1 mixture of MCDB 153 medium with 1.5 g/L sodium bicarbonate and Leibovitz’s L-15 medium with 2 mmol/L L-glutamine supplemented with 0.005 mg/mL bovine insulin, 1.68 mmol/L CaCl2, and 2% fetal bovine serum. AO4, D22, and UACC2534 cells were grown in RPMI supplemented with 10% fetal bovine serum. SBcl2, AO4, D22, and UACC2534 melanoma cells were transduced with empty vector, FGFR2, or kinase-dead FGFR2 (K517R) at a multiplicity of infection of 2 in the presence of 6 μg/mL polybrene. Cells were selected for at least 14 d in 500 to 800 μg/mL G418. Cells (2,000–4,000) were plated per well in a 96-well plate in full growth media. The next day, cells were washed in PBS and switched to low-serum media overnight (0.5% fetal bovine serum for SbCl2 and 1% for AO4, D22, and UACC2534). Cells were then stimulated with 10 ng/mL FGF2 and 10 μg/mL heparin in low-serum media and proliferation was assessed using the sulforhodamine B assay. Fresh FGF2 and heparin were added on day 4.
p44/42 Phosphorylation
To examine p44/42 MAPK phosphorylation, cells were washed in PBS and resuspended in BaF media containing no fetal bovine serum and no IL-3 (BaF starve media) for 2 h. Cells were pelleted and resuspended in either BaF starve media with 10 μg/mL heparin only (unstimulated or “−”) or BaF starve media with heparin and 1nmol/L rFGF2 (stimulated or “+”) for 10 min at 37°C. After 10 min, the cells were pelleted by brief centrifugation and immediately lysed in ice-cold lysis buffer. Twenty micrograms of total protein were resolved on a 4% to 12% Bis Tris gel and probed for phosho-p44/42 MAPK. Blots were then stripped and reprobed for total p44/42 MAPK (Cell Signaling Technologies).
Phosphorylation was quantitated using Image J9 and presented as the ratio of phosphorylated over total p44/42 MAPK for each mutant divided by the same ratio for the FGF2-stimulated wild-type receptor.
Supplementary Material
Acknowledgments
We thank Nick Hayward, Peter Parsons, Chris Schmidt, and Kathy Brown (Queensland Institute of Medical Research, Brisbane, Australia) for supplying melanoma cell lines; Dr. Daniel Donoghue (Dept. of Chemistry and Biochemistry, UCSD, San Diego, CA) for providing FGFR2 expression constructs; and Michael Berens for helpful discussions and critical reading of the manuscript.
Grant support: Harry J. Lloyd Charitable Trust, Melanoma Research Foundation, and Leslie Ann Ballard Foundation (P.M. Pollock); NIH grant 2R01 DE 13686-08 (M. Mohammadi); and American Cancer Society Postdoctoral Fellowship grant PF-07-215-01-TBE (S.A. Byron).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Unpublished results.
References
- 1.Halaban R. Growth factors and melanomas. Semin Oncol. 1996;23:673–81. [PubMed] [Google Scholar]
- 2.Lazar-Molnar E, Hegyesi H, Toth S, Falus A. Autocrine and paracrine regulation by cytokines and growth factors in melanoma. Cytokine. 2000;12:547–54. doi: 10.1006/cyto.1999.0614. [DOI] [PubMed] [Google Scholar]
- 3.Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;(2):REVIEWS3005. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nesbit M, Nesbit HK, Bennett J, et al. Basic fibroblast growth factor induces a transformed phenotype in normal human melanocytes. Oncogene. 1999;18:6469–76. doi: 10.1038/sj.onc.1203066. [DOI] [PubMed] [Google Scholar]
- 5.Meier F, Caroli U, Satyamoorthy K, et al. Fibroblast growth factor-2 but not Mel-CAM and/or β3 integrin promotes progression of melanocytes to melanoma. Exp Dermatol. 2003;12:296–306. doi: 10.1034/j.1600-0625.2003.120310.x. [DOI] [PubMed] [Google Scholar]
- 6.Halaban R, Kwon BS, Ghosh S, Delli Bovi P, Baird A. bFGF as an autocrine growth factor for human melanomas. Oncogene Res. 1988;3:177–86. [PubMed] [Google Scholar]
- 7.Becker D, Meier CB, Herlyn M. Proliferation of human malignant melanomas is inhibited by antisense oligodeoxynucleotides targeted against basic fibroblast growth factor. EMBO J. 1989;8:3685–91. doi: 10.1002/j.1460-2075.1989.tb08543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker D, Lee PL, Rodeck U, Herlyn M. Inhibition of the fibroblast growth factor receptor 1 (FGFR-1) gene in human melanocytes and malignant melanomas leads to inhibition of proliferation and signs indicative of differentiation. Oncogene. 1992;7:2303–13. [PubMed] [Google Scholar]
- 9.Xerri L, Battyani Z, Grob JJ, et al. Expression of FGF1 and FGFR1 in human melanoma tissues. Melanoma Res. 1996;6:223–30. doi: 10.1097/00008390-199606000-00005. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Becker D. Antisense targeting of basic fibroblast growth factor and fibroblast growth factor receptor-1 in human melanomas blocks intratumoral angiogenesis and tumor growth. Nat Med. 1997;3:887–93. doi: 10.1038/nm0897-887. [DOI] [PubMed] [Google Scholar]
- 11.Valesky M, Spang AJ, Fisher GW, Farkas DL, Becker D. Noninvasive dynamic fluorescence imaging of human melanomas reveals that targeted inhibition of bFGF or FGFR-1 in melanoma cells blocks tumor growth by apoptosis. Mol Med. 2002;8:103–12. [PMC free article] [PubMed] [Google Scholar]
- 12.Pollock PM, Gartside MG, Dejeza LC, et al. Frequent activating FGFR2 mutations in endometrial carcinomas parallel germline mutations associated with craniosynostosis and skeletal dysplasia syndromes. Oncogene. 2007;26:7158–62. doi: 10.1038/sj.onc.1210529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephens P, Edkins S, Davies H, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–2. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 14.Schlessinger J, Plotnikov AN, Ibrahimi OA, et al. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6:743–50. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 15.Chen H, Ma J, Li W, et al. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol Cell. 2007;27:717–30. doi: 10.1016/j.molcel.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Xu CF, Ma J, et al. A crystallographic snapshot of tyrosine trans-phosphorylation in action. Proc Natl Acad Sci U S A. 2008;105:19660–5. doi: 10.1073/pnas.0807752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ibrahimi OA, Eliseenkova AV, Plotnikov AN, Yu K, Ornitz DM, Mohammadi M. Structural basis for fibroblast growth factor receptor 2 activation in Apert syndrome. Proc Natl Acad Sci U S A. 2001;98:7182–7. doi: 10.1073/pnas.121183798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu K, Herr AB, Waksman G, Ornitz DM. Loss of fibroblast growth factor receptor 2 ligand-binding specificity in Apert syndrome. Proc Natl Acad Sci U S A. 2000;97:14536–41. doi: 10.1073/pnas.97.26.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ibrahimi OA, Zhang F, Eliseenkova AV, Itoh N, Linhardt RJ, Mohammadi M. Biochemical analysis of pathogenic ligand-dependent FGFR2 mutations suggests distinct pathophysiological mechanisms for craniofacial and limb abnormalities. Hum Mol Genet. 2004;13:2313–24. doi: 10.1093/hmg/ddh235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatch NE, Hudson M, Seto ML, Cunningham ML, Bothwell M. Intracellular retention, degradation, and signaling of glycosylation-deficient FGFR2 and craniosynostosis syndrome-associated FGFR2C278F. J Biol Chem. 2006;281:27292–305. doi: 10.1074/jbc.M600448200. [DOI] [PubMed] [Google Scholar]
- 21.Meyer AN, Gastwirt RF, Schlaepfer DD, Donoghue DJ. The cytoplasmic tyrosine kinase Pyk2 as a novel effector of fibroblast growth factor receptor 3 activation. J Biol Chem. 2004;279:28450–7. doi: 10.1074/jbc.M403335200. [DOI] [PubMed] [Google Scholar]
- 22.Naski MC, Wang Q, Xu J, Ornitz DM. Graded activation of fibroblast growth factor receptor 3 by mutations causing achondroplasia and thanatophoric dysplasia. Nat Genet. 1996;13:233–7. doi: 10.1038/ng0696-233. [DOI] [PubMed] [Google Scholar]
- 23.Julies MG, Moore SW, Kotze MJ, du Plessis L. Novel RET mutations in Hirschsprung’s disease patients from the diverse South African population. Eur J Hum Genet. 2001;9:419–23. doi: 10.1038/sj.ejhg.5200650. [DOI] [PubMed] [Google Scholar]
- 24.Rohmann E, Brunner HG, Kayserili H, et al. Mutations in different components of FGF signaling in LADD syndrome. Nat Genet. 2006;38:414–7. doi: 10.1038/ng1757. [DOI] [PubMed] [Google Scholar]
- 25.Shams I, Rohmann E, Eswarakumar VP, et al. Lacrimo-auriculo-dento-digital syndrome is caused by reduced activity of the fibroblast growth factor 10 (FGF10)-FGF receptor 2 signaling pathway. Mol Cell Biol. 2007;27:6903–12. doi: 10.1128/MCB.00544-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lew ED, Bae JH, Rohmann E, Wollnik B, Schlessinger J. Structural basis for reduced FGFR2 activity in LADD syndrome: implications for FGFR autoinhibition and activation. Proc Natl Acad Sci U S A. 2007;104:19802–7. doi: 10.1073/pnas.0709905104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.L’Hote CG, Knowles MA. Cell responses to FGFR3 signalling: growth, differentiation and apoptosis. Exp Cell Res. 2005;304:417–31. doi: 10.1016/j.yexcr.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Logie A, Dunois-Larde C, Rosty C, et al. Activating mutations of the tyrosine kinase receptor FGFR3 are associated with benign skin tumors in mice and humans. Hum Mol Genet. 2005;14:1153–60. doi: 10.1093/hmg/ddi127. [DOI] [PubMed] [Google Scholar]
- 29.Grose R, Fantl V, Werner S, et al. The role of fibroblast growth factor receptor 2b in skin homeostasis and cancer development. EMBO J. 2007;26:1268–78. doi: 10.1038/sj.emboj.7601583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamaguchi F, Saya H, Bruner JM, Morrison RS. Differential expression of two fibroblast growth factor-receptor genes is associated with malignant progression in human astrocytomas. Proc Natl Acad Sci U S A. 1994;91:484–8. doi: 10.1073/pnas.91.2.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naimi B, Latil A, Fournier G, Mangin P, Cussenot O, Berthon P. Down-regulation of (IIIb) and (IIIc) isoforms of fibroblast growth factor receptor 2 (FGFR2) is associated with malignant progression in human prostate. Prostate. 2002;52:245–52. doi: 10.1002/pros.10104. [DOI] [PubMed] [Google Scholar]
- 32.Diez de Medina SG, Chopin D, El Marjou A, et al. Decreased expression of keratinocyte growth factor receptor in a subset of human transitional cell bladder carcinomas. Oncogene. 1997;14:323–30. doi: 10.1038/sj.onc.1200830. [DOI] [PubMed] [Google Scholar]
- 33.Ricol D, Cappellen D, El Marjou A, et al. Tumour suppressive properties of fibroblast growth factor receptor 2-IIIb in human bladder cancer. Oncogene. 1999;18:7234–43. doi: 10.1038/sj.onc.1203186. [DOI] [PubMed] [Google Scholar]
- 34.Feng S, Wang F, Matsubara A, Kan M, McKeehan WL. Fibroblast growth factor receptor 2 limits and receptor 1 accelerates tumorigenicity of prostate epithelial cells. Cancer Res. 1997;57:5369–78. [PubMed] [Google Scholar]
- 35.Matsubara A, Kan M, Feng S, McKeehan WL. Inhibition of growth of malignant rat prostate tumor cells by restoration of fibroblast growth factor receptor 2. Cancer Res. 1998;58:1509–14. [PubMed] [Google Scholar]
- 36.Yasumoto H, Matsubara A, Mutaguchi K, Usui T, McKeehan WL. Restoration of fibroblast growth factor receptor2 suppresses growth and tumorigenicity of malignant human prostate carcinoma PC-3 cells. Prostate. 2004;61:236–42. doi: 10.1002/pros.20093. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Wang H, Toratani S, et al. Growth inhibition by keratinocyte growth factor receptor of human salivary adenocarcinoma cells through induction of differentiation and apoptosis. Proc Natl Acad Sci U S A. 2001;98:11336–40. doi: 10.1073/pnas.191377098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang X, Stachowiak EK, Dunham-Ems SM, Klejbor I, Stachowiak MK. Control of CREB-binding protein signaling by nuclear fibroblast growth factor receptor-1: a novel mechanism of gene regulation. J Biol Chem. 2005;280:28451–62. doi: 10.1074/jbc.M504400200. [DOI] [PubMed] [Google Scholar]
- 39.Kim Y, Bingham N, Sekido R, Parker KL, Lovell-Badge R, Capel B. Fibroblast growth factor receptor 2 regulates proliferation and Sertoli differentiation during male sex determination. Proc Natl Acad Sci U S A. 2007;104:16558–63. doi: 10.1073/pnas.0702581104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ueno H, Gunn M, Dell K, Tseng A, Jr, Williams L. A truncated form of fibroblast growth factor receptor 1 inhibits signal transduction by multiple types of fibroblast growth factor receptor. J Biol Chem. 1992;267:1470–6. [PubMed] [Google Scholar]
- 41.Kiselyov VV, Skladchikova G, Hinsby AM, et al. Structural basis for a direct interaction between FGFR1 and NCAM and evidence for a regulatory role of ATP. Structure. 2003;11:691–701. doi: 10.1016/s0969-2126(03)00096-0. [DOI] [PubMed] [Google Scholar]
- 42.Rowland BD, Peeper DS. KLF4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer. 2006;6:11–23. doi: 10.1038/nrc1780. [DOI] [PubMed] [Google Scholar]
- 43.Barker SC, Kassel DB, Weigl D, Huang X, Luther MA, Knight WB. Characterization of pp60c-src tyrosine kinase activities using a continuous assay: autoactivation of the enzyme is an intermolecular autophosphorylation process. Biochemistry. 1995;34:14843–51. doi: 10.1021/bi00045a027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


