Abstract
Objective
Using gene expression microarrays and reverse transcription with quantitative polymerase chain reaction (RT-qPCR), we have recently identified several novel genes that are differentially expressed in the neonatal male versus female mouse cortex/hippocampus (Armoskus et al.). Since perinatal testosterone (T) secreted by the developing testes masculinizes cortical and hippocampal structures and the behaviors regulated by these brain regions, we hypothesized that sexually dimorphic expression of specific selected genes in these areas might be regulated by T during early development.
Methods
To test our hypothesis, we treated timed pregnant female mice daily with vehicle or testosterone propionate (TP) starting on embryonic day 16 until the day of birth. The cortex/hippocampus was collected from vehicle- and TP-treated, male and female neonatal pups. Total RNA was extracted from these brain tissues, followed by RT-qPCR to measure relative mRNA levels of seven sex chromosome genes and three autosomal genes that have previously showed sex differences.
Results
The effect of prenatal TP was confirmed as it stimulated Dhcr24 expression in the neonatal mouse cortex/hippocampus and increased the anogenital distance in females. We found a significant effect of sex, but not TP, on expression of three Y-linked (Ddx3y, Eif2s3y, and Kdm5d), four X-linked (Eif2s3x, Kdm6a, Mid1, and Xist), and one autosomal (Klk8) genes in the neonatal mouse cortex/hippocampus.
Conclusion
Although most of the selected genes are not directly regulated by prenatal T, their sexually dimorphic expression might play an important role in the control of sexually differentiated cognitive and social behaviors as well as in the etiology of sex-biased neurological disorders and mental illnesses.
Keywords: Androgenization, Cerebral cortex, Hippocampus, Sex-biased gene, Sex chromosome, Sexual differentiation, Testosterone
Introduction
In many species, including mice and humans, sexual differentiation is essential for the development of distinct brain structure and function between the sexes [1-4]. Besides physiological functions, sex differences are also noted in the prevalence and symptomatology of many neurological diseases and mental illnesses, such as depression, schizophrenia, and autism [5-8]. Therefore, determination of the mechanisms controlling sexual dimorphism in brain structure and function will enhance our understanding of the underlying causes of the sex bias in neurological disorders and assist development of new treatments for these diseases.
Similar to humans, sexual differentiation in mice is determined by the two sex chromosomes: embryos receiving two X chromosomes become females and those with one X and one Y chromosome develop as males. During early embryogenesis, the Sry gene in the sex determining region of the Y chromosome encodes a transcription factor expressed in male undifferentiated gonads to induce testicular development [9]. The developing testes secrete testosterone (T) with two perinatal peaks; one occurring around 16-17 days after conception (embryonic days 16 and 17, E16 and E17) and the other peaking a couple of hours after birth [10-12]. The rises in circulating T during the late gestation and immediately after birth are essential for masculinization and/or defeminization of certain behaviors and their underlying neural circuits (organizational effect) [10,13].
To masculinize brain structures and behaviors, T acts directly on androgen receptor (AR) and/or indirectly on estrogen receptors (ERs) via local aromatization of T to estradiol (E2) [3,14,15]. While the effects of estrogens on the developing brain are one of the primary controls of masculinization in rodents, they are less important in human masculinization [16]. ARs and ERs are abundant in the rodent brain, including the cortex and hippocampus [17-20], and functional steroid receptors are important for sex differences in many of the cognitive and social behaviors controlled by these brain regions, such as learning and memory [1,2,21].
Structural differences in male and female cortex and hippocampus are associated with distinct behavioral phenotypes between the sexes. In adult mice, males possess a thicker cortex than females, and this difference depends on circulating hormones, not on sex chromosomes [22]. The rat hippocampus shows increased neurogenesis during the first week of life, which is associated with T exposure [23]. However, the molecular mechanisms underlying the actions of sex steroids on sex differences in expression of downstream target genes remain unclear [24,25].
We have previously observed sex differences in histone modifications in the mouse cortex/hippocampus with more H3 acetylation on K9/14 (H3K9/14Ac) in males around embryonic day 18 (E18) and the day of birth (PN0) and more H3 trimethylation on K9 (H3K9Me3) in males around PN0 and 6 days after birth (PN6) [26]. Sex differences in H3K9/14Ac were regulated by T as a significant increase in H3K9/14Ac, but not H3K9Me3, was found in prenatally T-treated female neonates. Since transcriptional regulation of brain-specific genes by histone modifications is associated with changes in neural function and behavior in mice and rats [27-29], our findings suggest that T might alter gene expression in the mouse brain around the time of birth.
Using gene expression microarrays with reverse transcription quantitative polymerase chain reaction, we have recently identified a group of sex-biased genes expressed in the neonatal mouse cortex/ hippocampus [30]. Based on the organizational effect of T on brain sexual differentiation, we hypothesized that some of these sexually dimorphic genes might be regulated by perinatal T, leading to the development of distinct neural functions and behaviors between the sexes. To test our hypothesis, we determined mRNA levels of three Y-linked (Ddx3y, Eif2s3y, and Kdm5d), four X-linked (Eif2s3x, Kdm6a, Mid1, and Xist), and four autosomal genes (Dab2, Dhcr24, Klk8, and Prkdc) in the cortex/hippocampus of newborn mice prenatally treated with testosterone propionate (TP).
Sex-biased expression of most sex chromosome genes studied here has been previously observed in the whole brain of embryonic, neonatal, and adult mice, and sex differences in their expression might be dependent on the sex chromosomal complement, rather than on gonadal hormones [31-34]. However, no experiment has been conducted in the neonatal cortex/hippocampus to investigate the direct effect of T on their expression, particularly in prenatally androgenized mice. Kumamoto and Oshio (2013) showed that fetal exposure of female mice to bisphenol A (BPA) disrupted X-chromosome inactivation by reducing Xist expression in the cerebrum [35]. Based on the weak estrogenic transcriptional activities of BPA, it may be hypothesized that prenatal T, locally converted to E2 by aromatase, alters Xist expression to regulate other X-linked gene expression in the neonatal mouse cortex/hippocampus. Therefore, to investigate a possible role of T in the regulation of sexual dimorphism in gene expression, we included seven sex chromosome genes along with four autosomal genes.
Materials and Methods
Animals
Adult male and female C57BL/6J mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and housed in the Animal Care Facility at California State University, Long Beach (CSULB). Mice were kept on a 12:12 light-dark cycle (lights on at 0600). Harlan Teklad LM-485 Mouse/Rat Sterilizable Diet (#7012; Madison, WI) and water were provided ad libitum. Neonatal pups used in these studies were generated by breeding the adult mice. After pairing with a male, the females were examined daily, and the day on which a mating plug was observed was designated as embryonic day 0 (E0). Males were removed during late gestation (E14-E17). All experimental procedures were approved by CSULB Institutional Animal Care and Use Committee in accordance with Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) guidelines.
Experimental design
Beginning on E16, pregnant female mice were treated daily with subcutaneous injections of vehicle (0.05 ml sesame oil) or TP (100 μg dissolved in 0.05 ml sesame oil; Sigma-Aldrich, St. Louis, MO). On the day of birth (PN0), neonatal pups (n=11 per sex per treatment) were sacrificed by decapitation. All pups were randomly selected from nine litters produced by seven dams (2-6 pups per litter). Vehicle-treated male and female neonates came from four (two male-dominant and two female-dominant) and five (three female-dominant and two male-dominant) litters, respectively. TP-treated males were collected from four litters (two female-dominant and two male-dominant) while TP-treated females were from three litters (two female-dominant and one male-dominant). The brains were immediately removed and coronally blocked between 2.55 mm and 4.95 mm from the most rostral end based on the Atlas of the Developing Mouse Brain at E17.5, P0, and, P6 [36]. The top half of the block, including the cerebral cortex and hippocampus, was dissected out, frozen on dry ice, and stored at −80°C until processing for RNA extraction. Expression of selected sexually dimorphic genes was quantified by RT-qPCR. The sex of the pups was determined first by examining the presence of pigmentation in the anogenital regions; males showed a pigment spot over the scrotum whereas females lacked pigmentation [37]. Testes were then dissected out from the males to further confirm their sex. In addition, the anogenital distance (AGD) was measured from these neonates to determine masculinization effects of TP on peripheral tissue. Using digital calipers, two measurements were taken under magnification: one from the top of the anus to the bottom of the genitalia (inner distance), and a second from the bottom of anus to the top of the genitalia (outer distance). Measurements were averaged and subjected to statistical analysis.
RNA extraction and cDNA synthesis
Total RNA was extracted from the mouse cortex/hippocampus using the RNeasy® Lipid Tissue Mini kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Using a Bio-Rad SmartSpec™ Plus spectrophotometer (Hercules, CA), the RNA concentration of each sample (1:50 dilution in 10 mM Tris, pH = 7.5) was determined by measuring the absorbance at 260 nm (A260), and its purity was determined by the ratio of the absorbance measured at 260 nm (A260) and 280 nm (A280). The ratios of A260/A280 between 1.9 and 2.1 were considered acceptable [38]. The cDNA templates were synthesized from 1 μg extracted RNA in 20-μl reactions using the Bio-Rad iScript™ cDNA synthesis kit with a mix of oligo(dT) and random hexamer primers. The reverse transcription was performed at 25°C for 5 min, 42°C for 30 min, and 85°C for 5 min in a Bio-Rad MyCycler™ thermocycler.
PCR primer design and testing
We first looked through the literature and Harvard PrimerBank (http://pga.mgh.harvard.edu/primerbank/) to find PCR primers for selected genes. In the absence of found primers, new primers were designed and checked for their suitability using the Primer3 program (http://primer3.wi.mit.edu/). Primers were checked for specificity using NCBI's Primer-BLAST program (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) against RefSeq RNA to ensure no non-specific matches. When designing primers that could match multiple transcript variants, their sequences were aligned in ClustalW (http://www.genome.jp/tools/clustalw/), and only the primers that amplified a consensus region were used.
All PCR primers were purchased from Eurofins MWG Operon (Huntsville, AL) and listed in Table 1 with the annealing temperatures and sizes of the resulting PCR products in base pairs (bp).
Table 1.
List of the PCR primer sequences.
| Gene Symbol | RefSeq Accession | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Product Size (bp) |
Annealing Temperature (°C) |
|---|---|---|---|---|---|
| Actb* | NM_007393.3 | CCAGATCATGTTTGAGACCTTCAA | CCAGAGGCATACAGGGACAGC | 78 | 59 |
| Dab2 | NM_001008702.2 | TCATTCTCCCTATGGCTTGG | GCTGAGCAGAATCCAGTTCC | 83 | 60 |
| Ddx3y | NM_012008.2 | GGGTCTGTGATAAGGACAGTTCA | CACGACCACCAATACCATCATAG | 177 | 60 |
| Dhcr24 | NM_053272.2 | CTCTGGGTGCGAGTGAAGG | TTCCCGGACCTGTTTCTGGAT | 195 | 60 |
| Eif2s3x | NM_012010.3 | GGTGAGGGTGGAGTGACTCT | TTCCCATGAGCTACGTGACCA | 152 | 60 |
| Eif2s3y | NM_012011.1 | GGTGCTGTTGGAGCATTACC | TCAACTCGTCGGCTTAGAGC | 269 | 58 |
| Kdm5d | NM_011419.3 | CCAGGATCTGACGACTTTCTACC | TTCTCCGCAATGGGTCTGATT | 113 | 58 |
| Kdm6a | NM_009483.1 | CGGGCGGACAAAAGAAGAAC | CATAGACTTGCATCAGATCCTCC | 203 | 60 |
| Klk8 | NM_008940.2 | GATCCTGGAAGGTCGAGAGTG | CTGCTCCGGCTGATCTCTG | 193 | 58 |
| Midi | NM_183151.1 | CTGTGACGGCACCTGTCTC | AAACGGCTGACTGTTGGTCTT | 205 | 58 |
| Prkdc | NM_011159.2 | CTGTGACGGCACCTGTCTC | TCTCAATCTGAGGACGAATTGC | 203 | 58 |
| Rpl13a* | NM_009438.5 | ATGAGGTCGGGTGGAAGTACC | CAGGAGTCCGTTGGTCTTGAG | 179 | 62 |
| Xist | NR_001570.2 | GCTGGTTCGTCTATCTTGTGGG | GGATCCTGCACTGGATGAGT | 225 | 58 |
indicates a reference/housekeeping gene.
All primers were tested to ensure suitability for PCR amplification using GoTaq® Hot Start Green Master Mix (Promega, Madison, WI) according to the manufacturers’ protocol for 40 cycles. Initial tests used an annealing temperature of 2°C below the lower melting temperature of the two the primers. PCR reactions were conducted on two or three independent samples, with at least one male and one female sample, along with a negative, no template control (NTC). PCR products were run on a 1.5 % agarose gel, visualized by ethidium bromide staining, and compared to a 100bp DNA ladder (Fisher Scientific, Pittsburgh, PA). Primers were rejected as unsuitable for quantification if they featured multiple products, no products, visible product in the NTC, or a product that was not of the size predicted based on RefSeq transcripts.
Four dilution stocks were prepared, two from PN0 male cortex/ hippocampus cDNA and two from PN0 females, with 8 serial dilutions ranging from 1:8 to 1:1024, dilution being measured from the output of cDNA synthesis. Reactions include 4 μl of sample at a specific dilution, 1 μl each of forward and reverse primers at 840 nM, and 6 μl of ABsolute™ QPCR SYBR® Green Mix (Waltham, MA). Reactions were run on an Agilent Stratagene® MX3000P™ qPCR system with MxPro QPCR software (Santa Clara, CA) used for determining cycle of threshold (Ct)-values. Reactions were run with an initial 15 min denaturation/polymerase-activation step at 95°C, followed by 40 cycles of 15 s at 95°C, 1 min at the annealing temperature, and 30 s at 72°C; qPCR amplification was followed by a dissociation step with 1 min at 95°C, 30 s at 60°C, and 30 s at 95°C. Results were analyzed by linear regression with log base 2 of the dilution as the predictor and Ct of the reaction as the response. Primers were rejected as unsuitable for quantification if the average R2 of the regression was below 0.95, Ct values at the 1:16 dilution were above 30, or the dissociation curve showed multiple peaks.
RT-qPCR
RT-qPCR was conducted to measure relative mRNA levels under the same conditions as described in the dilution tests, with the sample in the reaction being 4 μl of 1:16 diluted cDNA along with a no-template control which used water in place of cDNA sample. Two replicate reactions for each sample were run and their Ct values averaged before further analysis. For a given sample unnormalized relative quantification (RQ) values for gene expression were calculated using Pfaffl's method against a baseline of vehicle-treated females for all cases except Y chromosome genes; for Y chromosome genes a baseline of vehicle-treated males was used [39,40]. RQ values of the target gene were divided by the geometric mean of the RQ values of the two stable reference genes (Actb and Rpl13a) to yield normalized relative quantification (NRQ) values [41]. NRQ values more than two standard deviations from their group mean were trimmed to two standard deviations from the group mean in the same direction. NRQ values were used as measurements of gene expression and subjected to statistical testing.
Statistical analyses
The data from individual groups were first tested for normality of the residuals by the Anderson-Darling test and next tested for equality of variances using Levene's test; if significant deviations were detected (p<0.05), a log transformation was applied to the data. After showing normality and homogeneity of variances, the data were then analyzed by two-way ANOVA to evaluate the effects of sex, TP treatment, and their interaction. When two-way ANOVA showed significant interactions, Tukey's post-hoc test was used to determine which groups were significantly different. If the variances were not homogeneous after log transformation, Bonferroni corrected t-tests were performed for multiple comparisons. For Y-chromosome genes, due to deviations from normality in female samples, Kruskal-Wallis was run to compare all four groups, followed by a two-tailed, two-sample t-test to compare vehicle-treated and TP-treated males. Analyses were run in Minitab. Data are shown as mean±SEM.
Results
Mouse body weight, tissue weight, and RNA yield
No significant sex differences in mean body weight, tissue weight, or RNA yield were observed among vehicle- and TP-treated, male and female neonatal mice (Table 2). Interestingly, a significant effect of TP treatment was found on brain tissue weight, with greater cortex/ hippocampus weight in TP-treated mice than vehicle-treated controls (p = 0.022). However, the tissue weight difference was reversed in RNA yields, as total RNA amounts extracted from TP-treated samples tended to be lower than vehicle-treated controls (p = 0.067).
Table 2.
Mouse weights, brain tissue weights, and RNA yields in prenatally vehicle- and testosterone propionate (TP)-treated mice. Data are shown as mean±SEM.
| Group | N | Mouse Body Weight (g) | Cortex/Hippocampus Weight (mg) | RNA Yield (Mg) |
|---|---|---|---|---|
| Vehicle | ||||
| Females | 11 | 1.30 ± 0.04 | 18.68 ± 1.42 | 49.73 ± 3.89 |
| Males | 11 | 1.31 ± 0.03 | 18.38 ± 1.97 | 39.77 ± 2.23 |
| TP | ||||
| Females | 11 | 1.24 ± 0.02 | 24.09 ± 1.29 | 36.26 ± 2.50 |
| Males | 11 | 1.25 ± 0.02 | 22.27 ± 1.70 | 39.92 ± 5.19 |
Effect of prenatal TP on AGD
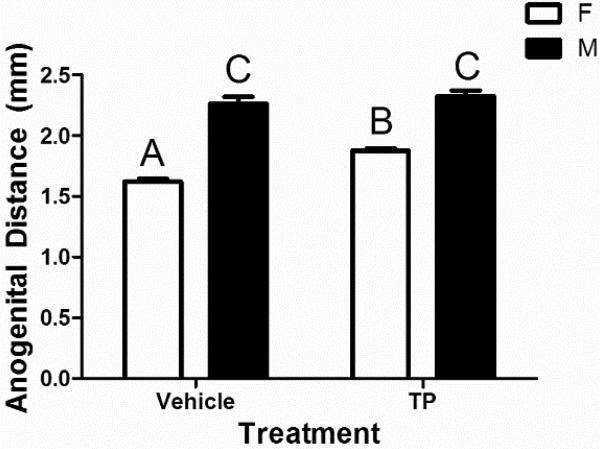
Two-way ANOVA of mean AGD of the neonatal mice receiving prenatal treatment of vehicle or TP revealed significant effects of sex (p<0.001), treatment (p = 0.005), and their interaction (p = 0.021) (Figure 1). TP had no effect on AGD in males, but significantly increased AGD (16%) in females, from 1.62 ± 0.02 mm (vehicle, n = 7) to 1.88 ± 0.02 mm (TP, n = 10) (p<0.001). Although the AGD of TP-treated females remained significantly shorter than vehicle-treated males (p<0.001), prenatal exposure of female mice to TP during late gestation resulted in partial masculinization of their external genitalia.
Figure 1.
Anogenital distance measurements of neonatal female (F) and male (M) mice prenatally treated with either vehicle or testosterone propionate (TP). Different letters indicate significant differences between two groups based on post-hoc Tukey's test (p<0.05). Bars are mean±SEM.
Effect of prenatal TP on Y chromosome gene expression in the neonatal cortex/hippocampus
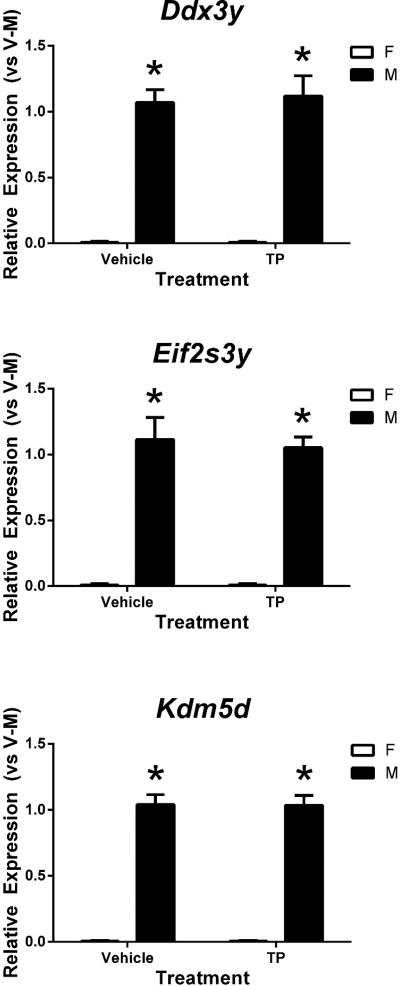
TP administration had no effect on expression of the three Y chromosome genes, Ddx3y, Eif2s3y, and Kdm5d in the cortex/hippocampus of neonatal mice. Regardless of TP treatment, the three Y-linked genes were exclusively expressed in the cortex/hippocampus of males, not females (p<0.001) (Figure 2). In addition, relative mRNA levels of these genes were similar between TP- and vehicle-treated male neonates, indicating that prenatal exposure of male embryos to extra T did not alter Y-linked gene transcription.
Figure 2.
Effects of prenatal testosterone propionate (TP) treatment on relative mRNA levels of three Y chromosome genes in the neonatal female (F) and male (M) mouse cortex/hippocampus measured by RT-qPCR. Gene expression was quantified relative to vehicle-treated male (V-M). * indicates significant difference between females and males (p<0.05). Bars are mean±SEM.
Effect of prenatal TP on X-linked gene expression in the neonatal mouse cortex/hippocampus
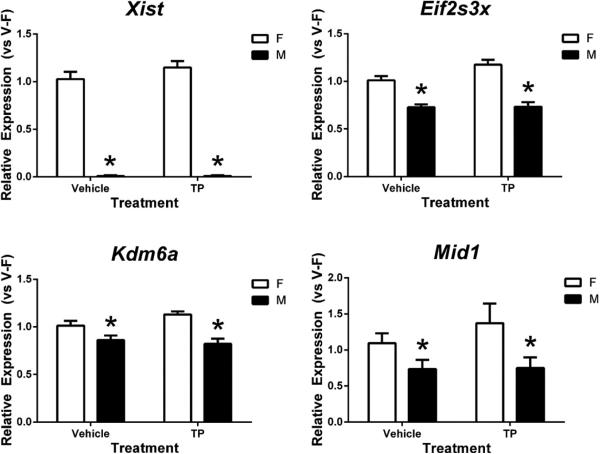
The expression of four X-linked genes (Xist, Eif2s3x, Kdm6a, and Mid1) linked or associated with X-chromosome inactivation and with escape from inactivation was greater in the neonatal cortex/ hippocampus of females than in males. The greatest difference between the sexes was in expression of Xist, which was approximately 2,500-fold higher in female pups regardless of TP treatment (p<0.001) (Figure 3, top left panel).
Figure 3.
Effects of prenatal testosterone propionate (TP) on expression of four X-linked genes in the neonatal cortex/ hippocampus of female (F) and male (M) mice. Gene expression quantified relative to vehicle-treated female (V-F). * indicates significant difference between females and males (p<0.05). Bars are mean ± SEM.
In contrast, the other three X-linked genes, Eif2s3x, Kdm6a, and Mid1, were expressed in both sexes, and showed a 1.39-, 1.12- and 1.45-fold higher expression, respectively, in the female cortex/ hippocampus compared to that of the male (Figure 3). No significant effect of TP treatment or its interaction with sex on these four X chromosome genes was observed, but interestingly, there was a trend of TP effect on Eif2s3x expression (p = 0.067) and a 1.25-fold change in Mid1 mRNA levels of TP-treated females as compared to vehicle-treated controls (1.37 ± 0.29 vs. 1.09 ± 0.14).
Effect of TP on autosomal gene expression in the neonatal mouse cortex/hippocampus
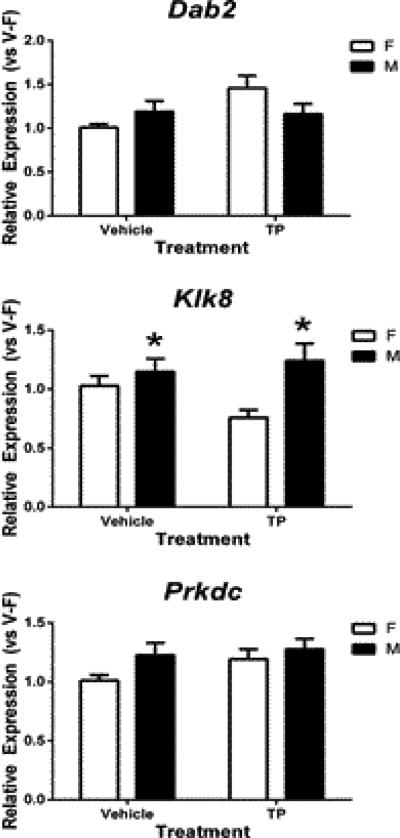
In the neonatal mouse cortex/hippocampus, Dab2 expression showed no significant effect of sex or TP, but there was a strong interaction (p = 0.052) (Figure 4, top panel). In vehicle-treated male mice, Dab2 mRNA levels (1.19 ± 0.13) increased to 119% of female controls (1.01 ± 0.04); however, this trend was reversed in TP-treated animals with 1.25 times greater expression in females (1.46 ± 0.15) as compared to males (1.16 ± 0.12).
Figure 4.
Effects of prenatal testosterone propionate (TP) treatment on expression of three autosomal genes in the neonatal female (F) and male (M) mouse cortex/hippocampus. Gene expression was normalized relative to the average of vehicled-treated female (V-F). *indicates significant difference between females and males (p<0.05). Bars are mean ± SEM.
In contrast, Klk8 expression in the neonatal mouse cortex/ hippocampus was sex dependent, and was higher in males than females regardless of treatment (p = 0.003) (Figure 4, middle panel). Interestingly, TP treatment seems to enhance the difference in Klk8 expression between the sexes; TP-treated male pups showed a 1.64-fold increase in Klk8 expression over TP-treated females while there was only a 1.12-fold change between vehicle-treated male and female mice.
There were no differences in Prkdc expression in the neonatal cortex/hippocampus between the sexes (p = 0.079), or in response to TP (p = 0.132), nor was there an interaction (p = 0.446) (Figure 4, bottom panel). Vehicle-treated male mice (1.21 ± 0.10) showed a 1.21-fold increase in Prkdc expression as compared to vehicle-treated females (1.01 ± 0.05), and this masculine trend seemed to be dependent on testosterone because increased Prkdc mRNA levels were observed in TP-treated females (1.18 ± 0.08).
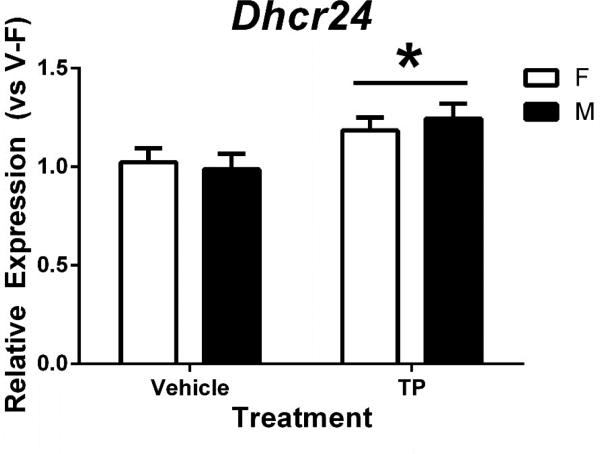
Unlike the other three autosomal genes shown above, Dhcr24 showed no hint of sex difference; however, TP treatment significantly increased Dhcr24 gene expression in the cortex/hippocampus of both male and female pups, with 1.21-fold higher expression in TP-treated mice than in vehicle-treated controls (p = 0.047) (Figure 5).
Figure 5.
Effects of prenatal testosterone propionate (TP) treatment on relative mRNA levels of Dhcr24 in the neonatal female (F) and male (M) mouse cortex/hippocampus. Gene expression quantified by RT-qPCR relative to vehicle-treated female (V-F). *indicates significant difference between vehicle and TP treatment (p<0.05). Bars are mean ± SEM.
Discussion
AGD is a sexually dimorphic trait that has been used as an anatomical marker of prenatal androgen exposure in both rodents and humans [42-44]. In mice, the AGD is positively correlated with T exposure in utero as it increases and becomes more male-like in female pups when their uterine neighbors are male [45]. In addition, female rats exposed to TP on gestation days 14-18 display increased AGD after birth [46,47]. Further, in utero treatment with flutamide, an androgen receptor antagonist that blocks the action of androgen, decreases the AGD in males on the day of birth [48]. In the current study, the AGD in female pups treated prenatally with TP was intermediate between that of vehicle-treated females and male controls (Figure 1), which is consistent with results obtained in rats [46]. Several factors might contribute to the inability of prenatal TP treatment to fully masculinize the female external genitalia. First, the dose of TP used might increase circulating T to levels intermediate between the low levels characteristic of female fetuses and the higher physiological male levels. Second, TP treatment was initiated 2 days after the testicular development had begun; thus TP-treated female embryos were exposed to T later than males. Last, development of the external genitalia is controlled by a combination of androgen-dependent and -independent mechanisms [49]. Nevertheless, the foregoing results confirm that TP treatment of pregnant dams results in partial masculinization of peripheral genital tissues of their female pups.
The mouse Y chromosome consists of a small pseudoautosomal region (PAR), and the remaining non-recombining region (NRY), which contains 27 protein-coding genes, including Ddx3y, Eif2s3y, and Kdm5d [50]. Ddx3y protein is an ATP-dependent RNA helicase with a conserved DEAD (Asp-Glu-Ala-Asp) box motif, and many of the DEAD box proteins are implicated in RNA metabolism, splicing, and translation [51]. Eif2s3y encodes subunit 3 of the translation elongation and initiation factor 2 (eIF2), which determines the rate of protein translation [52,53]. Kdm5d protein demethylates di- and trimethylated lysine 4 of histone H3 (H3K4), and di- and tri-methylation of H3K4 are often observed on the active gene promoter associated with increased transcription [54,55]. Since females do not have a Y chromosome, the NRY genes are exclusively expressed in male mouse brains [31-34]. Expression of six NRY genes, including Ddx3y, Eif2s3y, and Kdm5d, was reported to occur exclusively in male embryonic and neonatal brains [31]. This study used the four core genotypes (FCG) mouse model, XX gonadal males (XXSry) and females (XX) as well as XY gonadal males (XY-Sry) and females (XY-), thereby demonstrating that neonatal XY− females and XY−Sry males both displayed similar mRNA levels of these Y chromosome genes regardless of their gonadal sex. These results indicated that expression of Y-linked genes, such as Ddx3y, Eif2s3y, and Kdm5d, might not be regulated by testicular androgens. The results presented herein not only confirm male-specific expression of Ddx3y, Eif2s3y, and Kdm5d in the neonatal mouse cortex/hippocampus (Figure 2), but also provide direct evidence to support androgen-independent regulation of these Y-linked genes in the male mouse brain.
In contrast to males, that have two distinct sex chromosomes, X and Y, female mammals have two X chromosomes, one of which is transcriptionally silenced by X-inactivation, which equalizes the dosage of X-linked genes between the sexes. Using RNA sequencing in a female diploid Mus musculus×Mus spretus cell line, 13 X-linked genes, including Eif2s3x, Kdm6a, Mid1, and Xist, are found to be transcribed from the inactive X chromosome while most genes on the inactive X are silenced [56]. Eif2s3x, Kdm6a, and Xist have also been confirmed as X escape genes in the neonatal mouse brain [57]. The Xist gene encodes a large untranslated RNA responsible for mediating X inactivation [58]. Because Xist is only transcribed from the inactive X chromosome, it is expressed at much higher levels in female tissues [32,59]. Eif2s3x and Kdm6a have functional homologues, Eif2s3y and Uty, on the Y chromosome, respectively, so female expression of these two X escape genes is believed to balance their gene products between the sexes [31,60,61]. Eif2s3x encodes subunit three of eIF2 and is nearly identical to its homologous Y-linked copy, Eif2s3y [62]. Kdm6a encodes a histone lysine demethylase that removes methylation at lysine 27 of histone H3 (H3K27), an epigenetic mark linked to gene repression [63,64]. In humans, KDM6A gene mutations are associated with Kabuki syndrome, a rare genetic disease with developmental delay, mild to moderate intellectual disability, and postnatal growth retardation [65]. The Mid1 gene was shown to span the PAR boundary of the mouse X and Y chromosomes, with its first three exons in the X-unique region and the remaining 7 exons in the PAR [66]. Since the three most 5’ exons are required for its transcription, Mid1 is only expressed from the X, not the Y, chromosome, yielding female-biased expression of Mid1. In mice, lack of Mid1 disrupts the specification of the midbrain/cerebellar boundary, resulting in an abnormal development of the most anterior cerebellar lobes [67]. In humans, MID1 mutation has been identified as responsible for the X-linked form of Opitz G/BBB syndrome, characterized by facial anomalies, laryngotracheoesophageal defects, and genitourinary abnormalities [68,69].
The current results confirm female-biased expression of Xist, Eif2s3x, Kdm6a, and Mid1, consistent with our previous study [30]. In addition, they demonstrate that their expression is not dependent on prenatal TP levels, indicating that in the neonatal mouse cortex/ hippocampus, these four X-linked genes are mainly regulated by X-chromosomal gene dosage, rather than sex steroids (Figure 3). Our results are supported by a recent study using the Y* mouse model with four different genotypes, normal XX females (XX), 1X females, (XY*X), 2X males (XXY*), and 1X males (XY*) [33]. Expression of Eif2s3x, Kdm6a, and Xist in the whole brains of 2X mouse embryos (XX and XXY*) was greater than that in 1X embryos (XY*X and XY*) and was independent of their gonads. In the same study, there was no sexual dimorphism in Mid1 expression in embryonic mouse brains. Mid1 is located in the PAR of the mouse X and Y chromosomes, and the chromosomal rearrangement of this region in the Y* mouse might disrupt its differential expression between the sexes.
In contrast to the study by Kumamoto and Oshio (2013), prenatal TP treatment had no effect on Xist mRNA levels in females, suggesting that AR or ERs may not involve regulation of Xist expression and that suppression of Xist expression by BPA might be mediated by other receptor systems and molecular targets. In addition, Eif2s3x, Kdm6a, and Mid1 are transcribed from both the active and inactive X chromosomes, and based on the <2-fold difference in expression between the sexes, we speculate that there are unequal transcription rates from the two alleles of these X escape genes in the neonatal female cortex/hippocampus. Interestingly, among these three genes, a trend of increased Eif2s3x mRNA levels was elicited by prenatal TP (Figure 3), implying that X escape gene expression might be modulated by sex steroids and their receptors during early development. The potential hormonal regulation of Eif2s3x expression by sex steroids is in part supported by the presence of a potential estrogen responsive element (ERE) located within a 1 kb region downstream of its transcription start site (unpublished observation).
The autosomal gene, Dab2, encodes a novel phosphoprotein and in the developing CNS, its expression is primarily restricted to secretory cells, immature neuroepithelial cells, endothelial cells, and in populations of mononuclear phagocytes [70]. Dab2 is up-regulated in degenerated rat ventral prostate by androgen deprivation [71], suggesting that its expression in brain might be dependent on T. The Klk8 (kallikrein related-peptidase 8) gene, located on mouse chromosome 7, encodes a tryptic serine protease that processes extracellular matrix [72,73]. Klk8 is highly expressed in the mouse brain and concentrated in the hippocampus, anterior olfactory nucleus, septal nuclei, diagonal band of Broca, amygdala, and limbic cortex [74]. Klk8 knockout mice show aberrant synapse formation in the hippocampus, suppression of long-term potentiation, and impaired spatial memory [75-77] as well as increased anxiety [78]. Prkdc is found on mouse chromosome 16 and encodes the catalytic subunit of the DNA-dependent protein kinase, which is involved in DNA double strand break repair and cell death induced by DNA damage [79,80]. Prkdc knockout mice show increased germ cell apoptosis that leads to testicular degeneration [81]. In addition, double knockout of Prkdc together with Atm (ataxia telangiectasia mutated homolog) leads to lethality tied to greatly increased apoptosis of postmitotic neurons, suggesting an important role in neurodevelopment [82].
Among the four autosomal genes tested in the present study, we confirmed male-biased expression of Klk8 in the neonatal mouse cortex/hippocampus (Figure 4).
Although sex differences in Dab2 and Prkdc mRNA levels were insignificant, vehicle-treated male mice displayed an approximate 20% increase in their expression as compared to females (18% for Dab2 and 21% for Prkdc), showing a trend toward male bias with the fold changes comparable to those we observed previously [30]. This variability between studies could be due to the sample size as we used fewer animals per group (n=10-11) in the current study than previously (n=16-18) to accommodate the 96-well qPCR platform, which might render our results underpowered to detect the sex difference. In addition, although prenatal TP treatment did not change mRNA levels of these three genes in the neonatal mouse cortex/ hippocampus, but the interactions between sex and TP treatment for expression of both Dab2 (p = 0.052) and Klk8 (p = 0.051) approached significance, strongly suggesting that these genes might be partially modulated or influenced by prenatal TP. One possible explanation for this observation is the heterogeneity of cell types in the cortex and hippocampus. Immunoreactivity for intracellular AR is abundant in the adult rat cerebral cortex, but limited to the pyramidal neurons within layers II/III and V/VI [83]. This finding suggests that the action of T on gene expression might occur specifically within sub-populations of cortical or hippocampal cells, rather than globally throughout the entire brain, which may mask the significance of T on expression of the autosomal genes.
To determine whether prenatal TP treatment exerts an effect on brain Dhcr24, we assessed mRNA levels of Dhcr24 in the cortex/ hippocampus of vehicle- and TP-treated neonates. Dhcr24 encodes 3-beta-hydroxysterol delta-24-reductase, which converts desmosterol to cholesterol [84]. Dhcr24 also mediates anti-apoptotic and anti-oxidative activities by inhibiting the activation of caspase 3, one of the key modulators of apoptosis, and binding to p53 to displace Mdm2, resulting in p53, a tumor suppressor, accumulation [85]. Dhcr24 gene is activated by ER in neurons and glia cells [86] and by AR in prostate cells [87] and rat glioma C6 cells [88]. The androgen-responsiveness of Dhcr24 may be mediated by the direct action of AR because an androgen responsive element (ARE) is present in the human DHCR24 promoter (-4384 to -2892 bp) [87]. In the present study, we found no sex difference in Dhcr24 mRNA levels of the neonatal mouse cortex/ hippocampus, but its expression was increased by prenatal TP treatment in both sexes (Figure 5), confirming that Dhcr24 is an androgen-regulated gene. The lack of a sex difference in Dhcr24 mRNA levels might be due to insufficient secretion of endogenous T by the developing testes.
In summary, we have confirmed sex differentiated expression of seven sex chromosome genes (Ddx3y, Eif2s3y, Eif2s3x, Kdm5d, Kdm6a, Mid1, and Xist) and one autosomal gene (Klk8) in the neonatal mouse cortex/hippocampus, and demonstrated that their expression is not regulated by T during late gestation. Further, our results suggest that expression of these genes is elicited mainly by the actions of the sex chromosomes which thereby genetically program the cortex and hippocampus, ultimately resulting in sexual dimorphism in social and cognitive behaviors as well as in the underlying neural circuits. In addition, based on trends of altered expression in some of the above genes and the presence of steroid receptors in subpopulations of cortical and hippocampal cells, future studies will focus on the roles of these genes in cells expressing AR and/or ERs.
Acknowledgements
We thank Courtney Donovan, Michelle Thai, and Kathy Trang for their technical assistance. The authors are also grateful to Dr. Sandra Legan for her valuable comments and suggestions that help improve this manuscript. This work was supported by California State University Program for Education and Research in Biotechnology (CSUPERB) Faculty-Student Collaborative Research Seed Grant, and National Institutes of Health Grants SC3GM102051 and R25GM0500089.
Footnotes
Citation: Armoskus C, Mota T, Moreira D, Tsai HW (2014) Effects of Prenatal Testosterone Exposure on Sexually Dimorphic Gene Expression in the Neonatal Mouse Cortex and Hippocampus. J Steroids Hormon Sci 5: 139. doi:10.4172/2157-7536.1000139
References
- 1.Frick KM, Gresack JE. Sex differences in the behavioral response to spatial and object novelty in adult C57BL/6 mice. Behav Neurosci. 2003;117:1283–1291. doi: 10.1037/0735-7044.117.6.1283. [DOI] [PubMed] [Google Scholar]
- 2.Sutcliffe JS, Marshall KM, Neill JC. Influence of gender on working and spatial memory in the novel object recognition task in the rat. Behav Brain Res. 2007;177:117–125. doi: 10.1016/j.bbr.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 3.Hines M. Prenatal testosterone and gender-related behaviour. Eur J Endocrinol. 2006;155(Suppl 1):S115–121. doi: 10.1530/eje.1.02236. [DOI] [PubMed] [Google Scholar]
- 4.Bonthuis PJ, Cox KH, Searcy BT, Kumar P, Tobet S, et al. Of mice and rats: key species variations in the sexual differentiation of brain and behavior. Front Neuroendocrinol. 2010;31:341–358. doi: 10.1016/j.yfrne.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baron-Cohen S, Knickmeyer RC, Belmonte MK. Sex differences in the brain: implications for explaining autism. Science. 2005;310:819–823. doi: 10.1126/science.1115455. [DOI] [PubMed] [Google Scholar]
- 6.Deng Z, Sobell JL, Knowles JA. Epigenetic alterations in schizophrenia. 2010;8:358–365. [Google Scholar]
- 7.Amenson CS, Lewinsohn PM. An investigation into the observed sex difference in prevalence of unipolar depression. J Abnorm Psychol. 1981;90:1–13. doi: 10.1037//0021-843x.90.1.1. [DOI] [PubMed] [Google Scholar]
- 8.Conley CS, Rudolph KD. The emerging sex difference in adolescent depression: interacting contributions of puberty and peer stress. Dev Psychopathol. 2009;21:593–620. doi: 10.1017/S0954579409000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kopsida E, Stergiakouli E, Lynn PM, Wilkinson LS, Davies W. The Role of the Y Chromosome in Brain Function. Open Neuroendocrinol J. 2009;2:20–30. doi: 10.2174/1876528900902010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Motelica-Heino I, Castanier M, Corbier P, Edwards DA, Roffi J. Testosterone levels in plasma and testes of neonatal mice. J Steroid Biochem. 1988;31:283–286. doi: 10.1016/0022-4731(88)90351-2. [DOI] [PubMed] [Google Scholar]
- 11.Ward IL, Ward OB, Affuso JD, Long WD, 3rd, French JA, et al. Fetal testosterone surge: specific modulations induced in male rats by maternal stress and/or alcohol consumption. Horm Behav. 2003;43:531–539. doi: 10.1016/s0018-506x(03)00061-8. [DOI] [PubMed] [Google Scholar]
- 12.Vom Saal FS, Bronson FH. Sexual characteristics of adult female mice are correlated with their blood testosterone levels during prenatal development. Science. 1980;208:597–599. doi: 10.1126/science.7367881. [DOI] [PubMed] [Google Scholar]
- 13.Phoenix CH, Goy RW, Gerall AA, Young WC. Organizing action of prenatally administered testosterone propionate on the tissues mediating mating behavior in the female guinea pig. Endocrinology. 1959;65:369–382. doi: 10.1210/endo-65-3-369. [DOI] [PubMed] [Google Scholar]
- 14.Davis EC, Popper P, Gorski RA. The role of apoptosis in sexual differentiation of the rat sexually dimorphic nucleus of the preoptic area. Brain Res. 1996;734:10–18. [PubMed] [Google Scholar]
- 15.Forger NG. Control of cell number in the sexually dimorphic brain and spinal cord. J Neuroendocrinol. 2009;21:393–399. doi: 10.1111/j.1365-2826.2009.01825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris JA, Jordan CL, Breedlove SM. Sexual differentiation of the vertebrate nervous system. Nat Neurosci. 2004;7:1034–1039. doi: 10.1038/nn1325. [DOI] [PubMed] [Google Scholar]
- 17.Shughrue PJ, Merchenthaler I. Distribution of estrogen receptor beta immunoreactivity in the rat central nervous system. J Comp Neurol. 2001;436:64–81. [PubMed] [Google Scholar]
- 18.Lu SF, McKenna SE, Cologer-Clifford A, Nau EA, Simon NG. Androgen receptor in mouse brain: sex differences and similarities in autoregulation. Endocrinology. 1998;139:1594–1601. doi: 10.1210/endo.139.4.5863. [DOI] [PubMed] [Google Scholar]
- 19.Shah NM, Pisapia DJ, Maniatis S, Mendelsohn MM, Nemes A, et al. Visualizing sexual dimorphism in the brain. Neuron. 2004;43:313–319. doi: 10.1016/j.neuron.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 20.DonCarlos LL, Sarkey S, Lorenz B, Azcoitia I, Garcia-Ovejero D, et al. Novel cellular phenotypes and subcellular sites for androgen action in the forebrain. Neuroscience. 2006;138:801–807. doi: 10.1016/j.neuroscience.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 21.Rizk A, Robertson J, Raber J. Behavioral performance of tfm mice supports the beneficial role of androgen receptors in spatial learning and memory. Brain Res. 2005;1034:132–138. doi: 10.1016/j.brainres.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Markham JA, Jurgens HA, Auger CJ, De Vries GJ, Arnold AP, et al. Sex differences in mouse cortical thickness are independent of the complement of sex chromosomes. Neuroscience. 2003;116:71–75. doi: 10.1016/s0306-4522(02)00554-7. [DOI] [PubMed] [Google Scholar]
- 23.Zhang JM, Konkle AT, Zup SL, McCarthy MM. Impact of sex and hormones on new cells in the developing rat hippocampus: a novel source of sex dimorphism? Eur J Neurosci. 2008;27:791–800. doi: 10.1111/j.1460-9568.2008.06073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerr JE, Allore RJ, Beck SG, Handa RJ. Distribution and hormonal regulation of androgen receptor (AR) and AR messenger ribonucleic acid in the rat hippocampus. Endocrinology. 1995;136:3213–3221. doi: 10.1210/endo.136.8.7628354. [DOI] [PubMed] [Google Scholar]
- 25.Ivanova T, Beyer C. Ontogenetic expression and sex differences of aromatase and estrogen receptor-alpha/beta mRNA in the mouse hippocampus. Cell Tissue Res. 2000;300:231–237. doi: 10.1007/s004410000199. [DOI] [PubMed] [Google Scholar]
- 26.Tsai HW, Grant PA, Rissman EF. Sex differences in histone modifications in the neonatal mouse brain. Epigenetics. 2009;4:47–53. doi: 10.4161/epi.4.1.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forger NG, Rosen GJ, Waters EM, Jacob D, Simerly RB, et al. Deletion of Bax eliminates sex differences in the mouse forebrain. Proc Natl Acad Sci U S A. 2004;101:13666–13671. doi: 10.1073/pnas.0404644101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy MM, Auger AP, Bale TL, De Vries GJ, Dunn GA, et al. The epigenetics of sex differences in the brain. J Neurosci. 2009;29:12815–12823. doi: 10.1523/JNEUROSCI.3331-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray EK, Hien A, de Vries GJ, Forger NG. Epigenetic control of sexual differentiation of the bed nucleus of the stria terminalis. Endocrinology. 2009;150:4241–4247. doi: 10.1210/en.2009-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Armoskus C, Moreira D, Bollinger K, Jimenez O, Taniguchi S, et al. Identification of sexually dimorphic genes in the neonatal mouse cortex and hippocampus. Brain Res. 2014;1562:23–38. doi: 10.1016/j.brainres.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J, Burgoyne PS, Arnold AP. Sex differences in sex chromosome gene expression in mouse brain. Hum Mol Genet. 2002;11:1409–1419. doi: 10.1093/hmg/11.12.1409. [DOI] [PubMed] [Google Scholar]
- 32.Dewing P, Shi T, Horvath S, Vilain E. Sexually dimorphic gene expression in mouse brain precedes gonadal differentiation. Brain Res Mol Brain Res. 2003;118:82–90. doi: 10.1016/s0169-328x(03)00339-5. [DOI] [PubMed] [Google Scholar]
- 33.Wolstenholme JT, Rissman EF, Bekiranov S. Sexual differentiation in the developing mouse brain: contributions of sex chromosome genes. Genes Brain Behav. 2013;12:166–180. doi: 10.1111/gbb.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan HX, Yang W, Zhang R, Chen L, Tang L, et al. Protein-tyrosine phosphatase PCP-2 inhibits beta-catenin signaling and increases E-cadherin-dependent cell adhesion. J Biol Chem. 2006;281:15423–15433. doi: 10.1074/jbc.M602607200. [DOI] [PubMed] [Google Scholar]
- 35.Kumamoto T, Oshio S. Effect of fetal exposure to bisphenol A on brain mediated by X-chromosome inactivation. J Toxicol Sci. 2013;38:485–494. doi: 10.2131/jts.38.485. [DOI] [PubMed] [Google Scholar]
- 36.Paxinos G, Halliday G, Watson C, Koutcherov Y, Wang H. Atlas of the Developing Mouse Brain at E17.5, P0, and P6. Academic Press; Burlington, MA 01803, USA: 2007. [Google Scholar]
- 37.Wolterink-Donselaar IG, Meerding JM, Fernandes C. A method for gender determination in newborn dark pigmented mice. Lab Anim (NY) 2009;38:35–38. doi: 10.1038/laban0109-35. [DOI] [PubMed] [Google Scholar]
- 38.White P. Evaluating Concentration and Purity of RNA. University of Pennsylvania - Functional Genomics Core; 2004. pp. 1–2. [Google Scholar]
- 39.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faber K, Hughes C. Anogenital distance at birth as a predictor of volume of the sexually dimorphic nucleus of the preoptic area of the hypothalamus and pituitary responsiveness in castrated adult rats. Biol Reprod. 1992;46:101–104. doi: 10.1095/biolreprod46.1.101. [DOI] [PubMed] [Google Scholar]
- 43.Graham S, Gandelman R. The expression of ano-genital distance data in the mouse. Physiol Behav. 1986;36:103–104. doi: 10.1016/0031-9384(86)90081-8. [DOI] [PubMed] [Google Scholar]
- 44.Dean A, Sharpe RM. Clinical review: Anogenital distance or digit length ratio as measures of fetal androgen exposure: relationship to male reproductive development and its disorders. J Clin Endocrinol Metab. 2013;98:2230–2238. doi: 10.1210/jc.2012-4057. [DOI] [PubMed] [Google Scholar]
- 45.Hurd PL, Bailey AA, Gongal PA, Yan RH, Greer JJ, et al. Intrauterine position effects on anogenital distance and digit ratio in male and female mice. Arch Sex Behav. 2008;37:9–18. doi: 10.1007/s10508-007-9259-z. [DOI] [PubMed] [Google Scholar]
- 46.Hotchkiss AK, Lambright CS, Ostby JS, Parks-Saldutti L, Vandenbergh JG, et al. Prenatal Testosterone Exposure Permanently Masculinizes Anogenital Distance, Nipple Development, and Reproductive Tract Morphology in Female Sprague-Dawley Rats. Toxicological Sciences. 2007;96:335–345. doi: 10.1093/toxsci/kfm002. [DOI] [PubMed] [Google Scholar]
- 47.Wolf CJ, Hotchkiss A, Ostby JS, LeBlanc GA, Gray LE., Jr Effects of prenatal testosterone propionate on the sexual development of male and female rats: a dose-response study. Toxicol Sci. 2002;65:71–86. doi: 10.1093/toxsci/65.1.71. [DOI] [PubMed] [Google Scholar]
- 48.McIntyre BS, Barlow NJ, Foster PM. Androgen-mediated development in male rat offspring exposed to flutamide in utero: permanence and correlation of early postnatal changes in anogenital distance and nipple retention with malformations in androgen-dependent tissues. Toxicol Sci. 2001;62:236–249. doi: 10.1093/toxsci/62.2.236. [DOI] [PubMed] [Google Scholar]
- 49.Blaschko SD, Cunha GR, Baskin LS. Molecular mechanisms of external genitalia development. Differentiation. 2012;84:261–268. doi: 10.1016/j.diff.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 51.Abdelhaleem M. RNA helicases: regulators of differentiation. Clin Biochem. 2005;38:499–503. doi: 10.1016/j.clinbiochem.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 52.Proud CG. eIF2 and the control of cell physiology. Semin Cell Dev Biol. 2005;16:3–12. doi: 10.1016/j.semcdb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 53.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci U S A. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 56.Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20:614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X, Soloway PD, Clark AG. Paternally biased X inactivation in mouse neonatal brain. Genome Biol. 2010;11:R79. doi: 10.1186/gb-2010-11-7-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Senner CE, Brockdorff N. Xist gene regulation at the onset of X inactivation. Curr Opin Genet Dev. 2009;19:122–126. doi: 10.1016/j.gde.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 59.Yang X, Schadt EE, Wang S, Wang H, Arnold AP, et al. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu J, Deng X, Disteche CM. Sex-specific expression of the X-linked histone demethylase gene Jarid1c in brain. PLoS One. 2008;3:e2553. doi: 10.1371/journal.pone.0002553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu J, Watkins R, Arnold AP. Sexually dimorphic expression of the X-linked gene Eif2s3x mRNA but not protein in mouse brain. Gene Expr Patterns. 2006;6:146–155. doi: 10.1016/j.modgep.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 62.Ehrmann IE, Ellis PS, Mazeyrat S, Duthie S, Brockdorff N, et al. Characterization of genes encoding translation initiation factor eIF-2gamma in mouse and human: sex chromosome localization, escape from X-inactivation and evolution. Hum Mol Genet. 1998;7:1725–1737. doi: 10.1093/hmg/7.11.1725. [DOI] [PubMed] [Google Scholar]
- 63.Xu J, Andreassi M. Reversible histone methylation regulates brain gene expression and behavior. Horm Behav. 2011;59:383–392. doi: 10.1016/j.yhbeh.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 65.Lederer D, Grisart B, Digilio MC, Benoit V, Crespin M, et al. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am J Hum Genet. 2012;90:119–124. doi: 10.1016/j.ajhg.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Palmer S, Perry J, Kipling D, Ashworth A. A gene spans the pseudoautosomal boundary in mice. Proc Natl Acad Sci U S A. 1997;94:12030–12035. doi: 10.1073/pnas.94.22.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lancioni A, Pizzo M, Fontanella B, Ferrentino R, Napolitano LM, et al. Lack of Mid1, the mouse ortholog of the Opitz syndrome gene, causes abnormal development of the anterior cerebellar vermis. J Neurosci. 2010;30:2880–2887. doi: 10.1523/JNEUROSCI.4196-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dal Zotto L, Quaderi NA, Elliott R, Lingerfelter PA, Carrel L, et al. The mouse Mid1 gene: implications for the pathogenesis of Opitz syndrome and the evolution of the mammalian pseudoautosomal region. Hum Mol Genet. 1998;7:489–499. doi: 10.1093/hmg/7.3.489. [DOI] [PubMed] [Google Scholar]
- 69.Quaderi NA, Schweiger S, Gaudenz K, Franco B, Rugarli EI, et al. Opitz G/BBB syndrome, a defect of midline development, is due to mutations in a new RING finger gene on Xp22. Nat Genet. 1997;17:285–291. doi: 10.1038/ng1197-285. [DOI] [PubMed] [Google Scholar]
- 70.Cheung KK, Mok SC, Rezaie P, Chan WY. Dynamic expression of Dab2 in the mouse embryonic central nervous system. BMC Dev Biol. 2008;8:76. doi: 10.1186/1471-213X-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhoul J, Hernandez G, Tu SW, Huang CL, Tseng CP, Hsieh JT. The role of DOC-2/DAB2 in modulating androgen receptor-mediated cell growth via the nongenomic c-Src-mediated pathway in normal prostatic epithelium and cancer. Cancer Res. 2005;65:9906–9913. doi: 10.1158/0008-5472.CAN-05-1481. [DOI] [PubMed] [Google Scholar]
- 72.Oka T, Akisada M, Okabe A, Sakurai K, Shiosaka S, et al. Extracellular serine protease neuropsin (KLK8) modulates neurite outgrowth and fasciculation of mouse hippocampal neurons in culture. Neurosci Lett. 2002;321:141–144. doi: 10.1016/s0304-3940(01)02470-3. [DOI] [PubMed] [Google Scholar]
- 73.Harada A, Ishikawa Y, Shiosaka S. Regulation of hippocampal neuropsin (KLK8) mRNA by steroid hormones. 2004;43:552. [Google Scholar]
- 74.Suzuki J, Yoshida S, Chen ZL, Momota Y, Kato K, et al. Ontogeny of neuropsin mRNA expression in the mouse brain. Neurosci Res. 1995;23:345–351. doi: 10.1016/0168-0102(95)00960-2. [DOI] [PubMed] [Google Scholar]
- 75.Hirata A, Yoshida S, Inoue N, Matsumoto-Miyai K, Ninomiya A, et al. Abnormalities of synapses and neurons in the hippocampus of neuropsin-deficient mice. Mol Cell Neurosci. 2001;17:600–610. doi: 10.1006/mcne.2000.0945. [DOI] [PubMed] [Google Scholar]
- 76.Terayama R, Bando Y, Murakami K, Kato K, Kishibe M, et al. Neuropsin promotes oligodendrocyte death, demyelination and axonal degeneration after spinal cord injury. Neuroscience. 2007;148:175–187. doi: 10.1016/j.neuroscience.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 77.Yoshida S. Klk8, a multifunctional protease in the brain and skin: analysis of knockout mice. Biol Chem. 2010;391:375–380. doi: 10.1515/BC.2010.034. [DOI] [PubMed] [Google Scholar]
- 78.Horii Y, Yamasaki N, Miyakawa T, Shiosaka S. Increased anxiety-like behavior in neuropsin (kallikrein-related peptidase 8) gene-deficient mice. Behav Neurosci. 2008;122:498–504. doi: 10.1037/0735-7044.122.3.498. [DOI] [PubMed] [Google Scholar]
- 79.Burma S, Chen DJ. Role of DNA-PK in the cellular response to DNA double-strand breaks. DNA Repair (Amst) 2004;3:909–918. doi: 10.1016/j.dnarep.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 80.Norbury CJ, Zhivotovsky B. DNA damage-induced apoptosis. Oncogene. 2004;23:2797–2808. doi: 10.1038/sj.onc.1207532. [DOI] [PubMed] [Google Scholar]
- 81.Maser RS, Wong K, Sahin E, Xia H, Naylor M, et al. DNA-Dependent Protein Kinase Catalytic Subunit Is Not Required for Dysfunctional Telomere Fusion and Checkpoint Response in the Telomerase-Deficient Mouse. Molecular and Cellular Biology. 2007;27:2253–2265. doi: 10.1128/MCB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gladdy RA, Nutter LMJ, Kunath T, Danska JS, Guidos CJ. p53-Independent Apoptosis Disrupts Early Organogenesis in Embryos Lacking Both Ataxia-Telangiectasia Mutated and Prkdc. Molecular Cancer Research. 2006;4:311–318. doi: 10.1158/1541-7786.MCR-05-0258. [DOI] [PubMed] [Google Scholar]
- 83.Kritzer M. The distribution of immunoreactivity for intracellular androgen receptors in the cerebral cortex of hormonally intact adult male and female rats: localization in pyramidal neurons making corticocortical connections. Cereb Cortex. 2004;14:268–280. doi: 10.1093/cercor/bhg127. [DOI] [PubMed] [Google Scholar]
- 84.Waterham HR1, Koster J, Romeijn GJ, Hennekam RC, Vreken P, et al. Mutations in the 3beta-hydroxysterol Delta24-reductase gene cause desmosterolosis, an autosomal recessive disorder of cholesterol biosynthesis. Am J Hum Genet. 2001;69:685–694. doi: 10.1086/323473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu C, Miloslavskaya I, Demontis S, Maestro R, Galaktionov K. Regulation of cellular response to oncogenic and oxidative stress by Seladin-1. Nature. 2004;432:640–645. doi: 10.1038/nature03173. [DOI] [PubMed] [Google Scholar]
- 86.Peri A, Danza G, Serio M. Seladin-1 as a target of estrogen receptor activation in the brain: a new gene for a rather old story? J Endocrinol Invest. 2005;28:285–293. doi: 10.1007/BF03345387. [DOI] [PubMed] [Google Scholar]
- 87.Bonaccorsi L, Luciani P, Nesi G, Mannucci E, Deledda C, et al. Androgen receptor regulation of the seladin-1/DHCR24 gene: altered expression in prostate cancer. Lab Invest. 2008;88:1049–1056. doi: 10.1038/labinvest.2008.80. [DOI] [PubMed] [Google Scholar]
- 88.Zu H, Wu J, Zhang J, Yu M, Hong Z. Testosterone up-regulates seladin-1 expression by iAR and PI3-K/Akt signaling pathway in C6 cells. Neurosci Lett. 2012;514:122–126. doi: 10.1016/j.neulet.2012.02.072. [DOI] [PubMed] [Google Scholar]