Abstract
In this paper, we report on a general approach for the detection of a specific tumoural biomarker directly in serum. Such detection is made possible using a protein-binding peptide selected through an improved phage display technique and then conjugated to engineered microparticles (MPs). Protein biomarkers represent an unlimited source of information for non-invasive diagnostic and prognostic tests; MP-based assays are becoming largely used in manipulation of soluble biomarkers, but their direct use in serum is hampered by the complex biomolecular environment. Our technique overcomes the current limitations as it produces a selective MP—engineered with an antifouling layer—that ‘captures’ the relevant protein staying impervious to the background. Our system succeeds in fishing-out the human tumour necrosis factor alpha directly in serum with a high selectivity degree. Our method could have great impact in soluble protein manipulation and detection for a wide variety of diagnostic applications.
Keywords: microparticles for diagnosis, phage display selection, soluble cancer biomarker
1. Introduction
Diagnostic biomarkers are a key element in research, diagnostics and targeted therapeutics [1,2]. Recently, great emphasis has being placed on identifying circulating biomarkers, especially present in blood (plasma and serum), able to provide information on the body's response to cancer, as well as on the relationship between a tumour cell development and its environment [3–7]. Among them, recombinant human tumour necrosis factor alpha (rhTNFα) is one of the most investigated biomarkers because several studies demonstrated its potential application in diagnostic and therapeutic applications [8–10]. Indeed, rhTNFα is an inflammatory cytokine mainly secreted in response to a diverse range of stresses [8,9], overexpressed in the microenvironment of many tumours and responsible, in most cases, for their progression [11,12].
Many microparticle (MP)-based platforms with a broad range of biotechnological and medical applications have been proposed for protein fishing [13], biomarker detection [14], DNA sequencing and cell analysis [15]. Such bio-analytical methods for soluble biomarker detection offer many advantages: a larger surface for target capture, faster detection, equal sensitivity to surface functionalized microchips, easy bead handling and collecting procedure, multiplexing, a small volume of sample (less than a nanolitre), assay miniaturization and high-throughput screening [16–18]. MP-based platforms always need a moiety, which binds the target with high specificity and affinity. In order to detect protein biomarkers, one of the commonly used molecules is represented by a short peptide. However, the choice of an efficient amino acid sequence is not trivial and requires computational predictions and screening experiments. The combinatorial peptide libraries, especially random peptide libraries displayed on filamentous phages (phage display selection) is one of the mostly used method for screening [19,20]. Nevertheless, conventional procedures suffer from several drawbacks, such as the need for a large quantity of targets, poor control of stringency during washings [21–24] and high level of non-specific binders. Indeed selected peptides are often able to interact more with the solid supports rather than with the target [25–27]. To date, the literature only offers examples of phage display techniques for the screening of targeting moieties against soluble biomarkers adsorbed on solid support [28–30]. However, such techniques do not guarantee an optimal exposure, nor preserve the native structure of the soluble biomarker, thus impairing an efficient screening outcome [31,32].
Recently, phage display screening in microfluidic chips has demonstrated precise control over washing stringency on living adherent cells [33], against streptavidin-conjugated beads [34] or immobilizing a streptavidin target onto a substrate [35]. Beyond the choice of the targeting molecule, MPs materials also play a crucial role in bioassay, especially for biomarkers when in biological fluids.
Magnetic MP-based bioassays have the further advantage of quick, easy and gentle separation of biological compounds, because the particles can be manipulated using external magnetic fields for mixing [36], display [37], separation [38], encoding [39] and immunoassays [40,41] in microfluidics. However, many materials, including magnetic beads, lack antifouling properties especially for detection applications, where the background can strongly interfere with the detection. Polyethyleneglycol (PEG) is a well-known polymer for crating antifouling interfaces [42]. Furthermore, additional issues arise from the bio-conjugation of peptide to MPs. Indeed, the ideal bioassay platform requires a flexible, specific and controlled conjugation on antifouling interfaces of selected peptide. Several examples show how peptide can be conjugated to MP. However, none of them report about the use of antifouling surfaces such as PEG decorated with targeting peptide for diagnostic applications [43,44].
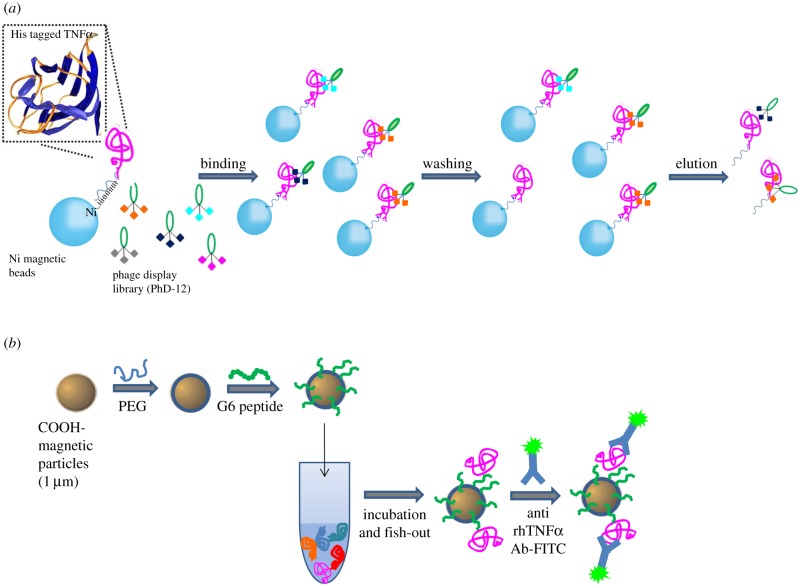
Here, we propose an integrated system capable of selectively binding circulating biomarkers in complex biological mediums. The proposed approach consists of two steps (figure 1): the identification of the binding peptide through an improved phage display technology (figure 1a), and its bio-conjugation to magnetic MPs after an antifouling treatment (figure 1b). In particular, G6 peptide was screened and its binding to rhTNFα was investigated by isothermal titration calorimetry (ITC) experiments and molecular docking (MD) simulations. After pegylation, G6 peptide was conjugated on magnetic MPs and the system was assessed in the detection of rhTNFα in human serum.
Figure 1.
Graphical scheme for the design and development of MP-based bioassay for fishing-out of soluble biomarkers. (a) Experimental scheme for selection of phage-displayed peptides with high affinity and specificity for rhTNFα performed on magnetic nickel-coated beads. (b) Graphical representation of integrated system for detection or fishing-out of any soluble biomarker in complex biological medium. This MP-based bioassay consists of selective capturing of target protein due to the presence of a specific binding peptide (previously selected by modified phage display procedure). The binding event is then detected by immunofluorescence measurements.
2. Material and methods
2.1. Chemicals and materials
TBS, bovine serum albumin (BSA), Tween 20, disodium hydrogen phosphate (Na2HPO4), sodium hydrogen phosphate (NaHPO4) salt, sodium chloride (NaCl), polyethylene glycol 8000 (PEG8000), p-nitrophenyl phosphate tablets, IPTG, X-gal, imidazole, fluorescein isothiocyanate (FITC), disodium carbonate (Na2CO3), sodium carbonate monobasic (NaHCO3), amino ethyl methacrylate hydrochloride (AEMA), MES, N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide (EDC), polyethylene glycol dimethacrylate (PEGMA), methylene bisacrylamide (MBA), potassium persulfate (KPS), tetramethylethylenediamine (TEMED) were purchased from Sigma-Aldrich (St Louis, MO, USA); the Ph.D.-12TM phage display peptide library kit was purchased from New England Biolabs (Beverly, MA, USA); hexahistidine-tagged recombinant human TNFα (6His-rhTNFα) was supplied by Prospec (East Brunswick, NJ, USA); nickel-coated magnetic beads (NiMB: ø 10 µm, ρ 1.1 g dm−3) were purchased from Kisker (Steinfurt, Germany); nickel-coated 96 multiwell plates were purchased from Thermo Scientific (Waltham, MA, USA); poly-dimethylsiloxane (PDMS) from Emanuele Mascherpa S.p.A (Milan, Italy), Dynabeads MyOne carboxylic (MP) from Invitrogen Dynal AS (Oslo, Norway). Reagents for peptide synthesis (Fmoc-protected amino acids, resins, activation and deprotection reagents) were from Novabiochem (Laufelfingen, Switzerland) and InBios (Naples, Italy). Solvents for peptide synthesis and HPLC analyses were from Romil (Dublin, Ireland); reversed phase columns for peptide analysis and the LC-MS system were supplied from Thermo Fisher (Milan, Italy). Solid-phase peptide synthesis were performed on a fully automated multichannel peptide synthesizer Syro I (Multisynthech, Germany). Pooled human serum from healthy donors was supplied by Lonza (Life Technology Ltd, Paisley, UK).
2.2. Selection of peptides binding to 6His-rhTNFα by phage display
The selection procedure provided with the Ph.D.-12 peptide library kit was modified in order to perform the phage screening on magnetic beads. For each selection cycle, first 500 ng of 6His-rhTNFα and 1 × 1011 plaque-forming units (pfu) of phages were mixed in 1 ml of binding buffer (50 mM sodium phosphate, 300 mM sodium chloride, pH 8) overnight at 4°C for the first step and 1 h at RT for the followings. Next the solution containing the proteins and phages mixture was incubated with 1 mg of NiMB for 1 h at RT. The NiMB were collected and washed several times with 1 ml of washing buffer (50 mM sodium phosphate, 300 mM sodium chloride, 0.05% Tween 20, pH 8) first and then with binding buffer to eliminate the residual Tween 20. The number of washing cycles was increased by three at each biopanning as well as the ratio of Tween containing washings. The bead pool was finally eluted in 500 µl of elution buffer (50 mM sodium phosphate, 300 mM sodium chloride, 300 mM imidazole, pH 8), titrated and amplified for next panning. After four rounds of biopanning, the positive phage clones were selected and sequenced.
2.3. ELISA
The specific binding of the positive phage clones to the 6His-rhTNFα was tested by performing ELISA. Briefly, 96-well nickel-coated plates were used for coating 6His-rhTNFα (1.5 µg per well in TBS) overnight at 4°C. The wells were blocked with 3% BSA in TBS at 37°C for 2 h. Anti-M13 polyclonal antibody (1 : 1000) (Sigma-Aldrich) was added and incubated at 37°C for 1 h, washed and incubated with secondary IgG AP conjugated (1 : 800). At the end, the bound phages were detected using p-nitrophenyl phosphate as the substrate, and the colour intensity was determined spectrophotometrically at 405 nm.
2.4. Phage clone fluorochrome labelling
Phage fluorochrome labelling was performed according to Jaye et al. protocol [45].
2.5. Microfluidic chip for phage selection
The microfluidic device consisted of a bottom glass layer and a top polymer layer, 4–5 mm thick. Channel dimensions were: 4 cm × 1 mm × 100 µm (L × W × H) for the washing chamber, and 1 cm × 400 µm × 100 µm for the separation chamber. It was fabricated by using an SU-8 master mould with features depth of 100 µm, obtained using a laser two-dimensional writer (DWL 66FS, Heidelberg Instruments, Heidelberg, Germany). From this master, a complementary PDMS layer was prepared by replica moulding (10 : 1 PDMS : cross-linker mixture at 90°C for 45 min). The device inlet and outlet ports were punched with a sharpened 30 gauge needle. The PDMS was then irreversibly bonded to a standard glass slide by treating the PMDS and glass with an oxygen plasma (50 mW, 20% oxygen, 1 min) in a plasma chamber (Plasma cleaner Femto UHP, Diener Electronic, Ebhausen, Germany) and immediately placing them in contact. The slots for the magnets were cut by hand with a scalpel blade. NdBFe magnets of 5 mm on each side (W-05-G, Supermagnete, Gottmadingen, Germany) were pressed into the slots at a position of approximately 1 mm away from the channel wall. The experiments into the microdevice used an Olympus inverted microscope (IX71, Olympus, Palo Alto, CA, USA) at 20× magnification, equipped with a programmable CCD camera (IGV-B0620, IMPERX, Boca Raton, FL, USA). The on-chip washing was performed using washing buffer. The buffer solutions were injected into the microfluidic device using 5-ml plastic syringes placed in programmable syringe pumps (neMESYS, Cetoni, Korbussen, Germany) and connected to the associated inlet ports via PTFE tubing with ID 0.8 mm. Similarly, outlet ports were connected to glass vials to recover waste and washed material. The syringe pumps delivered the buffers into the washing chamber and the separation chamber at flow rates of 10 and 35 µl min−1, respectively. To eliminate non-specifically or weakly bound phages from magnetic beads, 1 ml of sample containing 1 mg of nickel magnetic beads carrying the 6His-rhTNFα bounded to ΦG6 or Φwt was loaded into a vial and continuously injected into the microfluidic chip at a flow rate of 36 µl min−1 for 1 h, using a peristaltic pump (Peristaltic Pump HP, Medorex, Nörten-Hardenberg, Germany) connected to the inlet port via PTFE tubing with ID 0.8 mm.
2.6. Molecular recognition energy model in peptide docking
2.6.1. Dataset
We considered the 3L9J ID from the PDB data bank as a reference structure of the human TNFα protein [46,47]. The 3L9J structure includes the protein in a monomeric form and the antagonist with a good crystallographic resolution. From this structure, we removed the crystallization water molecules and the antagonist and added hydrogen atoms.
From the G6 and H4 peptides provided by phage display experiments, we extracted the common sequence hG6 (SSYYPQ) and performed a minimum energy structure optimization in a β-sheet conformation at the Amber/CPCM level of theory [48,49].
2.6.2. Molecular docking procedure
A computational protocol was adopted by using the AutoDock suite of programs [50,51]. The grid map was made of 84 × 62 × 126 points with spacing of 0.608 Å. Several docking runs were performed by activating different torsional degrees of freedom of the peptide structure and by freezing the others. Each docking search produced a set of 200 peptide structures for a total amount of 2000 guess structures representing our interacting statistical ensemble. Analysing these structures, we localized two predominant regions of interaction between the peptide and the TNFα. Therefore, we reduced the grid maps to these areas and repeated docking simulations. We built a new statistical ensemble made of 2000 peptide structures for each region. From all these structures, we extracted those showing a significant interaction energy with the TNFα.
2.6.3. Energy analysis
We extracted 22 structures belonging to the lower region as the starting point for a more accurate energetic analysis. On the selected structures we performed minimum energy optimization by the AMBER force field by allowing the peptide to relax on the frozen protein structure. On the peptide–protein optimized structures, we calculated the interaction energy Eint by the following equation:
where ETNFα–peptide, ETNFα and Epeptide are the energy of the protein–peptide complex, the isolated protein and the isolated peptide, respectively.
2.7. Peptide synthesis
Preparative RP-HPLC was carried out on a Shimadzu LC-8A, equipped with a SPD-M10 AV detector and with a Phenomenex C18 Jupiter column (50 × 22 mm ID; 10 µm). LC-MS analyses were carried out on a LCQ DECA XP Ion Trap mass spectrometer equipped with an OPTON ESI source, operating at 4.2 kV needle voltage and 320°C with a complete Surveyor HPLC system. Narrow bore 50 × 2 mm C18 BioBasic LC-MS columns were used for these analyses. G6 peptide and an unrelated sequence (DTC(Acm)RQTFRSH) were synthesized in the amine/amidate version, employing the solid-phase method and following standard Fmoc strategies. Activation of amino acids was achieved using HBTU/HOBt/DIEA (1 : 1 : 2). Peptides were removed from the resin by treatment with a TFA/TIS/H2O (90 : 5 : 5, v/v/v) mixture for 90 min at room temperature. Products were purified by RP-HPLC applying a linear gradient of 0.1% TFA CH3CN in 0.1% TFA water from 5 to 65% over 12 min. Peptides purity (95%) and identity were confirmed by LC-MS.
2.8. Isothermal titration calorimetry studies
ITC experiments were carried out with an iTC200 calorimeter (Microcal/GE Healthcare). G6 peptide (1 mM) was titrated into a solution of rhTNFα protein (25 µM). Data were fitted to a single-binding-site model with Origin software (GE Healthcare). Similar ITC studies were conducted with a solution of an unrelated (UNR) peptide (1 mM) and TNFα protein (25 µM). Protein was dialysed against 10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% Tween-20 (pH 7.7) overnight, and all further dilutions of protein and peptide for ITC were made using the leftover external dialysate.
2.9. Preparation of core-shell magnetic particles and G6 peptide conjugation
One microgram of carboxylated magnetic MPs was conditioned in 250 µl of MES, 100 mM pH 4.8, at 4°C for 1 h with occasional vortexing. To this suspension, EDC 500 mM and an excess of AEMA were added in a final volume of 500 µl and the reaction mixture shaken in the dark at 4°C overnight. The functionalized beads were precipitated down by a magnet, the supernatant removed and the precipitate washed in water three times.
To synthetize the PEG coating, 500 µg of MP were purged with nitrogen and then a mixture of PEGDMA (28.0 mg), MBA (1.0 mg), KPS (1.84 mg), acrylic acid (5.56 mg) and finally 10 µl of TEMED was added. Polymerization was carried out for 1 h at RT under sonication. Afterwards, the particles were washed in PBS buffer.
One hundred micrograms of PEG-MP were first washed twice in 50 mM MES, pH 5.5; then EDC/NHS solution 100 mM dissolved in coupling buffer was added and left in shaker for 30 min at RT. After incubation, the supernatants were removed and 5 nmol of G6 and an UNR peptide were added to the PEG-MP solutions. The coupling efficiency was evaluated by RP-HPLC analysis by comparing the peak area of peptide solution before and after the reaction. RP-HPLC (Waters 2795) is equipped with a Photodiode Array detector (Waters 2996), using a narrow bore 50 × 2 mm C18 Biobasic column, 300 Å, 3 mm (ThermoElectron), and applying a gradient of CH3CN, 0.1% TFA (Solvent B) from 5 to 70%, with respect to solvent A (H2O, 0.1% TFA) over a period of 20 min.
2.10. Transmission electron microscopy
Transmission electron microscopy (TEM) observation was performed on a FEI Tecnai G2 microscope operating at an acceleration voltage of 200 kV. The specimen was prepared as follows: one drop of diluted MPs and PEG-MPs were cast on a copper EM grid covered with a thin holey carbon film and dried at RT.
2.11. rhTNFα fishing-out
The fishing experiment was performed by incubating 50 µg of G6-MP and UNR-MP (as negative control) with 7.5 µg of rhTNFα (peptide/protein ratio 2/1) in PBS (pH 7.4) buffer or human serum from healthy donors (final volume 200 µl) at RT for 30 min.
After incubation, the particles were collected by magnetic separation and the fraction of unbound rhTNFα protein in the first supernatant was measured by RP-HPLC (Waters 2795).
Simultaneously, the MPs were washed three times in PBS buffer and then incubated with FITC-conjugated monoclonal anti-human TNFα (dilution 1 : 100) in a final volume of 200 µl at 37°C for 1 h. The same amount of MPs, not incubated with protein, was incubated with antibody under the same conditions as the negative control. The MPs were washed as described above, re-suspended in 200 µl PBS and 30 µl of each sample was loaded in ibidi channels and illuminated at confocal laser scanning microscopy (Leica SP5) using an argon laser, 488 nm, and fluorescence images of MPs were collected. Objective: HCX IRAPO L 63 × 1.4 OIL, section thickness 3 µm, scan speed 800 Hz, excitation laser argon 488 nm, λem range 500–600 nm, image size 1024 × 1024 µm2.
For fluorescence emission analysis, 60 PEG-MP were selected for each sample (G6-PEG-MP, UNR-PEG-MP) to be analysed and their fluorescence quantified.
All captured images were analysed with a public domain image-processing program, ImageJ (v. 1, 43i, NIH, Bethesda, MD, USA). The images were briefly thresholded by the Otsu algorithm and then processed with the ImageJ Analyze Particles function to computationally determine the number of single fluorescent particles in the range of 1.3 µm.
The fluorescence mean and standard deviation of each sample were calculated and Student's t-test was used to compare them (p-value < 0.001). The experimental uncertainty represents the standard error of the mean of three replicate assays.
2.12. CLSM fluorescence measurements
Thirty microlitres of magnetic beads after target-phage contact (T0) and after washing cycle in batch or device were loaded onto µ-slide channels (Ibidi, Martinsried, Germany), illuminated at confocal laser scanning microscopy (Leica SP5) using an argon laser, 488 nm, and fluorescence images of MPs were collected. Objective: HCX IRAPO L 25.0 × 0.95 WATER, section thickness 440 µm, scan speed 400 Hz, excitation laser argon 488 nm, λem range 500–600 nm, image size 620 × 620 µm2.
For fluorescence emission analysis, 60 magnetic beads were selected for each sample (T0, batch washing and device washing) to be analysed and their fluorescence quantified.
All captured images were analysed with a public domain image-processing program, ImageJ (v. 1,43i, NIH, Bethesda, MD, USA). The images were briefly thresholded by the Otsu algorithm and then processed with the ImageJ Analyze Particles function to computationally determine the number of single fluorescent particles in the range of 10 µm.
The fluorescence mean and standard deviation of each sample were calculated and Student's t-test was used to compare them (p-value < 0.001). The experimental uncertainty represents the standard error of the mean of three replicate assay.
3. Results
3.1. Identification of phage-displayed peptides binding to rhTNFα
Phage clones able to bind to rhTNFα were selected by incubation of a 12-mer linear random peptide library (PhD-12 mer library) with the protein. In order to minimize overall target surface exposition during the screening, a new procedure was applied to immobilize the target: NiMB were used to capture the recombinant 6His-rhTNFα to provide a bead-based support on which to perform the phage display screening (figure 1a). Four rounds of affinity selection were performed, as described in Material and methods. The elution titration curve showed that the maximum enrichment level was reached after four rounds of panning against rhTNFα-immobilized beads. This demonstrated that phage clones binding to rhTNFα had been enriched from the library after affinity selection (data not shown). The resulting phage pool was screened to identify the strongest binding phages and their relative peptide sequences. ELISA experiments were performed by binding 6His-rhTNFα on nickel-coated multiwell plates in order to prevent passive adsorption on the surface of the wells. The amount of coated protein was brought to an optimal surface concentration without multilayer adsorption (see the electronic supplementary material, figure S1). Six positive clones showing the strongest binding (figure 2a) were selected and sequenced. The derived sequences display significant consensus and a high number of conserved residues, as outlined in red in figure 2b. The binding fold for each phage clone is reported as the ratio of its specific signal and the one generated by Φwt. ΦG6 and ΦH4 show the strongest binding (4.27 and 3.95, respectively) and their related peptide sequence reveals an eight-residue identity out of 12. As the selected phage ΦG6 showed the highest specificity against the target, ΦG6 and its corresponding peptide were chosen for further investigation. Indeed, the specificity of ΦG6 was investigated by dose–response experiments. Different amounts of ΦG6 (1010–107 pfu) were added to constant concentrations of coated 6His-rhTNFα. A linear increase in binding signals is observed for ΦG6, but not for wt signals, suggesting a specific recognition of phage ΦG6 towards the target (see the electronic supplementary material, figure S2).
Figure 2.
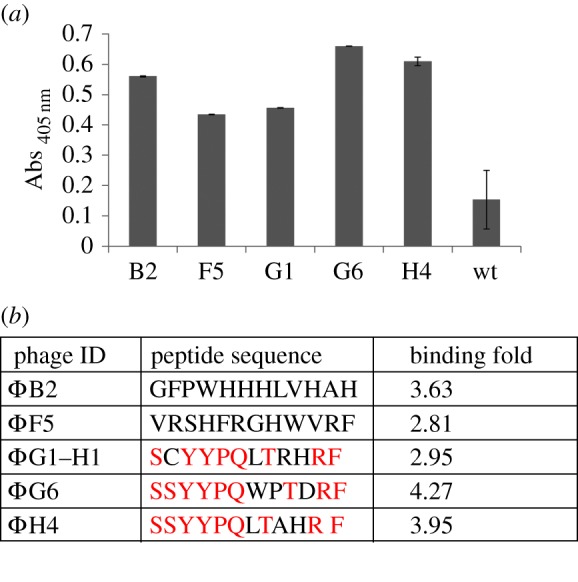
Results from de novo selection from a random PhD12 peptide library using NiMB complexed to hexahistidine-tagged recombinant human TNFα (6His-rhTNFα). (a) Binding specificity versus 6His-rhTNFα of the six best phage selected after three rounds of selection panning using the NiMB::6His-rhTNFα. (b) List of the resulting peptide sequences with the strongest specificity against rhTNFα target shows a consensus sequence in at least four clones. Conserved amino acids are reported in red.
3.2. Microfluidic phage selection: chip design
We performed a comparison between the washing efficiency of the fixed-volume washing approach used in the conventional batch process and the continuous process in the microfluidic chip. The microfluidic chip consisted of different elements: three inlets, a washing chamber containing an array of pillars, a separation chamber and two outlets (figure 3a). Through the inlets, the sample (inlet 1) and the buffer solutions (inlets 2 and 3) were continuously injected into the washing chamber. Target protein-conjugated magnetic beads were transferred to the upper wall of the washing chamber by means of four external magnets placed along the channel. Therefore, they passed from the sample solution stream into the washing buffer stream. Then, magnetic beads were guided into the separation chamber and deflected from their direction of flow by means of another external magnet, passing from the washing buffer stream into another. One outlet (4) led the dirty solution to the waste collector and the other one (5) guided the washed material to the off-chip elution step and analyses.
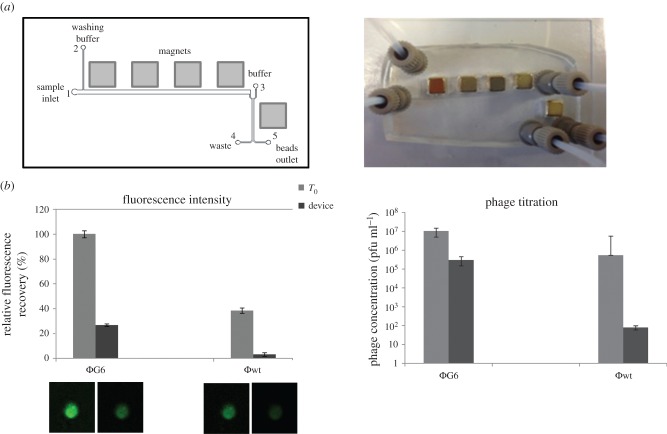
Figure 3.
Design, construction and performances of microfluidic device used for PhD selection. (a) Layout and picture of the microfluidic chip (a glass/PDMS device) used for phage selection, showing the design of the channels, external magnets and flow path. The continuous flow separation of phages is based on the transfer of target protein-conjugated magnetic beads in a continuous washing buffer by pumping through a 100 μm deep PDMS channel with a strong magnetic field perpendicularly applied. (b) Evaluation of microfluidic device washing efficiency by measuring the specific (ΦG6) and non-specific (ΦWT) residual bound phage on NiMB after one round of washing. The percentage of bound phages before and after washing step in the microfluidic device is reported as fluorescence recovery evaluated by CLSM on single particles and pfu by standard titration. (Online version in colour.)
Through the magnetic field, the magnetic beads were manipulated and transferred to the upper wall of the washing chamber, forming flagella-like chains. Beads were continuously shaken during the 1 h injection time to avoid bead settling and ensure a uniform suspension in the solution. Before starting the experiment, the syringe pumps containing the wash fluid syringe were run at a high flow rate to purge air from the device. When collecting the washed material, the magnetic beads were removed manually directly at the chip outlet.
The washing efficiency was then evaluated by comparing the residual amount of specific (ΦG6) and unspecific (Φwt) phages before and after washing by fluorescence measurement and conventional titration (figure 3b). NiMB after binding with equal amounts of fluorescein-labelled ΦG6 and Φwt were washed in batch and in chip after the manual binding procedure. Confocal microscopy images show that the phage display selection in the microfluidic device enables precise control over washing stringency during the phage selection process. ΦG6-beads produce a higher fluorescence intensity than Φwt-beads, indicating a stronger binding interaction against rhTNFα target. Moreover, after the washing cycle, the residual amount of specific ΦG6 bound phages is smaller, whereas bound Φwt-beads disappear (27% versus 3%). These results are consistent with the phage titration experiments. ΦG6 and Φwt phage titres were determined upon binding before and after the washing cycle inside the device. The titration after incubation (T0) shows that the amount of ΦG6 bound to the target is significantly higher than amount of Φwt in agreement with the specificity of ΦG6. Indeed, after washing, the wt amount—which represents the unspecific fraction—is almost zero, whereas the ΦG6 retained a titre value of 1.4 × 105 pfu ml−1 (figure 3b). Recovered beads were quantified by direct count before and after the washing process, by using a precise and handheld particle counter (Scepter 2.0 Cell Counter, Millipore, Billerica, MA, USA). Under experimental conditions, we found that 85% of the beads that entered the device were successfully recovered after the continuous washing in the microfluidic chip; whereas, 60% of the beads were recovered from the batch washing (see the electronic supplementary material, table S1). This is probably due to the loss of beads during the repeated washes during the batch processes.
3.3. G6 and H4 peptide binding prediction by molecular docking
In order to verify the specificity of the interaction between the peptide and the TNFα, we adopted a computational protocol based on docking techniques. We considered the 3L9J ID from the PDB data bank as a reference structure of the TNFα protein [46,47]. We extracted the common sequence SSYYPQ (hereafter hG6) from G6 and H4 peptides. As a first search, we performed several docking runs over the volume containing the whole protein. This search generated 2000 protein–peptide complexes, all of them involving two well-localized regions of the protein surface. Importantly, one of them (hereafter region I) covers the 132–135 sequence of the protein, which is close to the 138–141 sequence interacting with the antagonist developed by Byla et al. [47], the C-type lectin-like domain. A second search regarded a reduced volume including the two active regions. The resulting 4000 guesses of the protein–peptide complex were further analysed on the basis of the interaction energy according to the procedure detailed in the electronic supplementary material. As a refinement, 22 final structures showing the highest interaction energy were selected and further optimized by molecular mechanics. Ten protein–peptide complexes showing the highest values of the interaction energy are concentrated in region I. The complex corresponding to the most favourable interaction is reported in figure 4.
Figure 4.
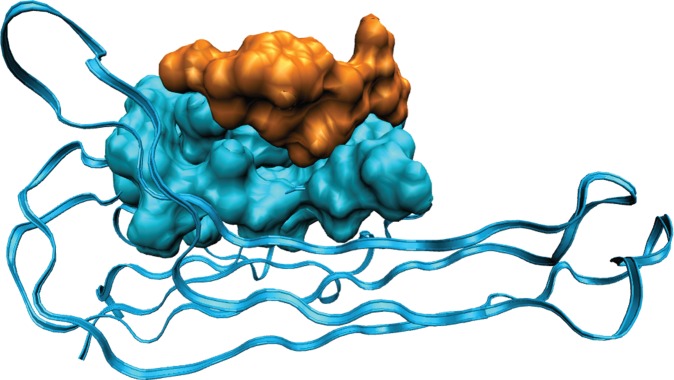
Characterization of TNFα G6 peptide binding. TNFα–hG6 peptide complex with greatest interaction energy among those individuated by MD and AMBER optimization. Most of the protein (3L9J as PDB ID) is represented as cartoon. Protein residues belonging to region I and interacting with the peptide, and the peptide itself are represented by the accessible surface in blue and orange, respectively.
3.4. Isothermal titration calorimetric studies
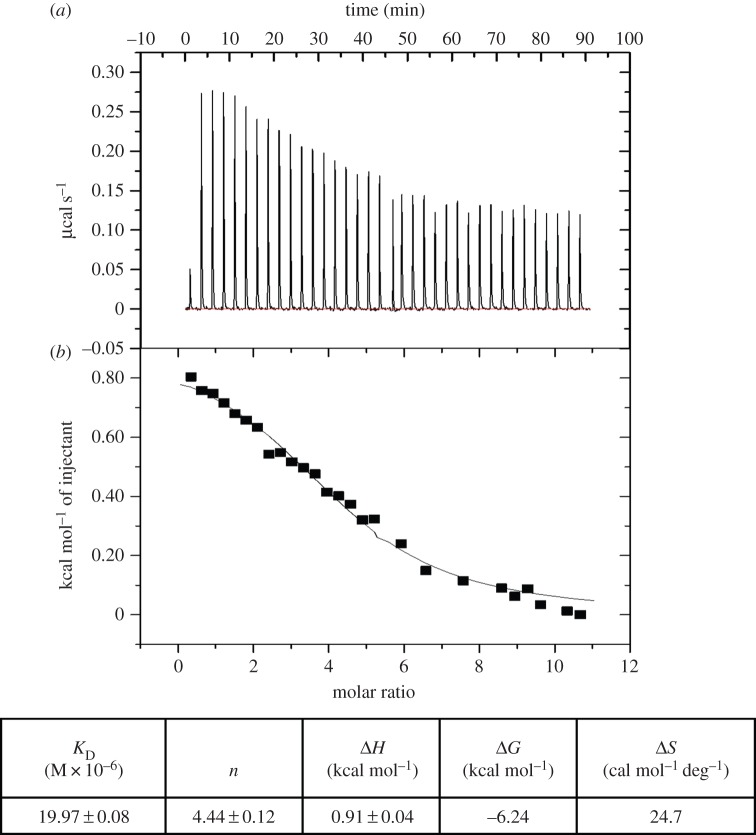
In order to quantify the rhTNFα-G6 peptide binding affinity, we performed ITC experiments. By titrating aliquots of peptide into rhTNFα solution, the upward ITC titration peaks demonstrate that the association between peptides and rhTNFα is an endothermic reaction (figure 5a). These data could be best fitted by a nonlinear ‘least-squares’ approach to the ‘one set of sites’ binding model (figure 5b), which provided the thermodynamic parameters (table in figure 5). In order to count out both non-specific interactions and artefacts resulting from ligand dilution into the protein buffer, we performed ITC experiments titrating an UNR peptide into rhTNFα solution (data not shown). The dissociation constant (KD) is approximately 10−6 M, in agreement with the affinities found for other screened TNFα-binding peptides of similar sizes [28,52].
Figure 5.
ITC studies. Calorimetric curve for rhTNFα protein (25 μM) titration with G6 peptide (1 mM) on the top. Raw and integrated data are shown in the upper and lower panels, respectively. In the lower section, data fitting was achieved with a single-binding-site model. (Online version in colour.)
3.5. rhTNFα fishing-out
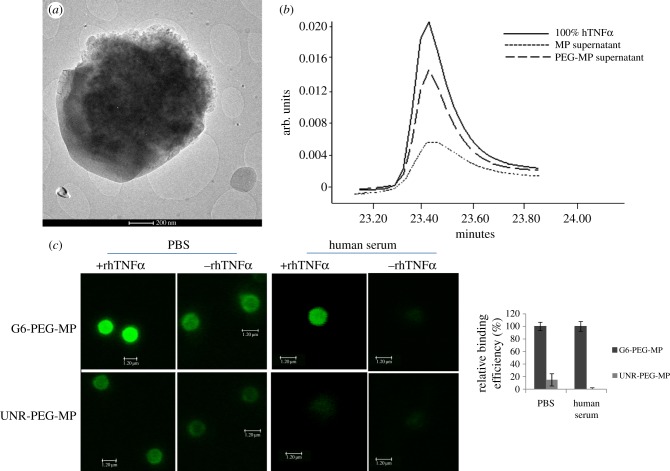
The selected peptide was conjugated to polymer decorated magnetic MPs to set up a bead-based bioassay. For the MP modification, we used a seeded polymerization for PEG shell growth on magnetic beads. The simultaneous addition of acrylic acid was necessary to provide carboxylic groups for the further peptide conjugation on the surface of the particle in order to obtain PEG surface decorated magnetic microparticles (PEG-MPs). The presence of a 160 nm shell was confirmed by TEM (figure 6a). (See the electronic supplementary material, figure S3.) The PEG contribution in conferring antifouling properties was evaluated by comparing the amount of rhTNFα not specifically adsorbed onto PEG-MPs and on plain MPs. After 30 min of incubation, the supernatants were analysed by HPLC, demonstrating that the PEG-MPs retain only 30% of rhTNFα while the plain MPs show a unspecific adsorption of 70% (figure 6b). G6 or UNR peptide was then bio-conjugated on PEG-MPs with a conjugation efficiency of about 50% (2.5 nmol) in both cases (see the electronic supplementary material, figure S4).
Figure 6.
Characterization of PEG-MP and evaluation of their capability in rhTNFα fish out. (a) TEM image of PEG-MP. (b) Chromatographic traces of rhTNFα produced by the UV detection of collected supernatant from PEG-MP and MP after incubation of the protein. (c) Evaluation of G6-PEG-MP capture efficiency. The binding event is detected by measuring the fluorescence intensity of FITC conjugated anti-human TNFα (monoclonal 1 : 100) directly on particles (image panel). Particles conjugated with UNR peptide were used as a control of unspecific signal. The experiment was performed in PBS buffer and in human serum.
In order to evaluate the ability of G6-PEG-MPs to selectively capture rhTNFα, we analysed a solution containing the protein in the ratio 1/2 with respect to immobilized peptides. The G6-PEG-MPs and UNR-PEG-MPs were isolated and the recovered protein was quantified directly by fluorescence imaging on the MPs (figure 6c) and indirectly by RP-HPLC (electronic supplementary material, figure S5). Indeed, as shown in the histogram in figure 6c, the fluorescence intensity quantification demonstrates that UNR-PEG-MPs absorb only 15% of the protein bound on G6-PEG-MPs. This is in agreement with the background level measured on the PEG-MPs. These results are consistent with HPLC analysis on collected supernatants by comparing UNR-PEG-MP and G6-PEG-MP. Quantitative analysis shows that rhTNFα efficiently binds to the G6-PEG-MP, with only 1% remaining in the supernatant. On the other hand, rhTNFα is not retained by the UNR-PEG-MPs because more than 60% of protein is in the supernatant (electronic supplementary material, figure S5).
Finally, we tested the ability of G6-PEG-MP to fish out rhTNFα even in a complex medium, like human serum, reported in figure 6c. Interestingly, the fluorescence intensity demonstrates that the G6-PEG-MPs retain the ability to selectively recognize rhTNFα even in human serum and that the complexity of the medium does not affect the background signal. Indeed, G6-PEG-MPs show high rhTNFα binding, whereas for UNR-PEG-MPs it is unappreciable.
4. Discussion
The major challenge of this study is the development of a bioassay system for the detection of soluble protein biomarkers directly in human serum. Such a system consists of engineered MP decorated with a peptide selected by phage display.
Several studies have already selected specific binding peptides against rhTNFα by phage display technology [28–30]. However, the above-referenced selections used rhTNFα targets immobilized on the surface by passive adsorption. It is well known that such adsorption may (i) mask some binding sites otherwise available in our procedure and (ii) cause conformational changes leading to selected peptides specific for the denaturated target [31,32]. Such conditions strongly affect the peptide selection output and could lead to peptides with non-specific binding. In fact, it is not common to find perfect matching between peptide sequences obtained by different selection procedures against the same target as well as between experimentally selected and computationally designed peptides.
In our case, even though the G6 sequence shows no homology with other selected sequences, it is similar to an AP ‘de novo’ peptide, which was derived from a rational design based on the binding of Z12 antibody to TNFα [53].
The binding specificity of our G6 peptide is confirmed by a prediction of binding site performed using the MD approach. MD runs, combined with the energetic analysis performed at molecular mechanics level (Amber force field) [48] as discussed in Material and methods, led to 10 protein–peptide complexes with an interaction energy values ranging from about 130 to 210 kcal mol−1. Notably, for all these cases, both the binding region and the interaction energy are in agreement with those showed by the peptide designed by Qin et al. [53]. As a matter of fact, these most stable protein–peptide complexes are all accommodated in a depressed area of the protein, interfacing a flat surface of about 800 Å. Moreover, our G6 peptide also presents important analogies with the antagonist of Byla et al. [47], the C-type lectin-like domain. On inspection of the most stable complex (figure 4), we can observe that, as it happens for the antagonist [40] serine and proline residues are responsible for a particular conformation adopted to maximize the protein–peptide interaction and to avoid steric hindrances. Furthermore, the complex shows multiple hydrogen bonds and favourable van der Waals interactions involving the serine and the tyrosine residues of the peptide with Asn46, Gln25 and Leu26.
ITC data show that the peptide binds to the protein with a dissociation constant in the low micromolar range (KD = 19.97 ± 0.08 µM) (figure 5). If compared with dissociation constants reported for other screened peptides, this KD value is lower by a factor of 10 at least [28,52].
This evidence confirms that our procedure improves the binding sites presentation of the protein during screening. It represents a novel strategy suitable for a phage display selection in which the target is linked to NiMB through a histidine tag fused at the N-terminus of the target protein (figure 1a). As a result, this screening strategy presents several advantages over conventional ones: (i) the immobilized protein retains its native conformation and is completely available for specific binding; (ii) the linkage procedure provides a universal platform to create a protein support; (iii) the elution occurs by breaking the complexation between nickel and histidine tag fused to the protein, so that the eluted phages are only those bound to the protein target. Furthermore, the use of magnetic beads opens the possibility to implement the screening in a miniaturized chip, thus accelerating the procedure, reducing the volume, increasing the repeatability and tuning the washing stringency. To this regard, we have demonstrated that the phage display selection on magnetic beads integrated in a microfluidic device allows for enhanced and controlled washing stringency leading to a minimized contribution of non-specific phage binding. Prospectively, this approach could be completely integrated in device microfluidic device with better performance and with a faster workflow.
In order to provide an integrated system capable of selectively detecting soluble rhTNFα directly in serum, the G6 peptide was coupled to magnetic MPs. Along this line, a nanometric polymer shell was coated on magnetic MPs to provide active functional sites and, at same time, antifouling properties. Here, MPs surfaces were easily modified by one pot seeded polymerization in aqueous mild conditions. In particular, we demonstrated that PEG decorated MPs showed antifouling properties against the rhTNFα compared with plain magnetic MPs. Indeed, after pegylation, the non-specific interactions of rhTNFα on PEG-MP is only 20% when compared with plain MPs, demonstrating the repellent ability of polymer decoration. Indeed, PEG is a well-known material in providing protein adsorption resistance [42]. This strategy is mostly used for drug delivery, while no reports are present in the literature with respect to bioassays [43,44]. Afterwards, the G6 peptide was conjugated on PEG-MP obtaining a surface conjugation density of 12.5 × 1012 peptide cm−2 corresponding to an average distance between each peptide of approximately 30 nm. Taking into account the TNFα monomer structure (55 Å in length along the strand direction and 25 Å across the sheet), this distance is enough to allocate the protein in trimeric conformation [54]. Therefore, we can exclude any inhibitor effect in peptide–protein binding caused by overextended peptide conjugation. On the contrary, it is possible that the peptide position can act as tridentate binders, enhancing the protein capture [55]. After G6 peptide conjugation, the ability of magnetic particles to detect rhTNFα in PBS buffer as well as in human serum was demonstrated. As a result, the complexity of human serum does not affect the capture of the rhTNFα target. Indeed, as observed in saline buffer (PBS), they showed a high selectivity of targeting also in human serum.
As result, we tailored the surface of magnetic MPs through the grafting of an antifouling polymer and a functional moiety that allows for the selective binding and magnetic removal of protein from a complex solution combining three features: (i) hydrophilic and biocompatible polymer interface, (ii) minimization of non-specific binding and of hydrophobic interaction and (iii) different chemical functionalization for the bio-conjugation [13,18,56].
Such a performance demonstrates that the described approach can play a central role in protein detection for diagnostic applications in different body fluids and open new perspectives towards its implementation in miniaturized detection devices.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Funding statement
This research was supported by Italian Ministry of University and Research (“Infrastructure for development of bio-MEMS Technologies in Advanced Sensing to apply in Environmental Monitoring, Agribusiness and Diagnostics”; PON a3_0007).
References
- 1.Barh D, Agte V, Dhawan D, Agte V, Padh H. 2012. Cancer biomarkers for diagnosis, prognosis and therapy. In Molecular and cellular therapeutics (eds D Whitehouse, R Rapley), pp. 18–68. Chichester, UK: John Wiley & Sons, Ltd. [Google Scholar]
- 2.Karley D, Gupta D, Tiwari A. 2011. Biomarker for cancer: a great promise for future. World J. Oncol. 2, 151.–. ( 10.4021/wjon352w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liotta LA, Ferrari M, Petricoin E. 2003. Clinical proteomics: written in blood. Nature 425, 905 ( 10.1038/425905a) [DOI] [PubMed] [Google Scholar]
- 4.Polanski M, Anderson NL. 2006. A list of candidate cancer biomarkers for targeted proteomics. Biomark Insight. 2, 1–48. [PMC free article] [PubMed] [Google Scholar]
- 5.Xue H, Lu B, Lai M. 2008. The cancer secretome: a reservoir of biomarkers. J. Transl. Med. 6, 52 ( 10.1186/1479-5876-6-52) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christein JD, Vickers SM, Langmead CJ, Landsittel DP, Whitcomb DC, Grizzle WE, Lokshin AE. 2011. Serum biomarker panels for the detection of pancreatic cancer. Clin. Cancer Res. 17, 805–816. ( 10.1158/1078-0432.CCR-10-0248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. 2013. Liquid biopsy: monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 10, 472–484. ( 10.1038/nrclinonc.2013.110) [DOI] [PubMed] [Google Scholar]
- 8.Balkwill F. 2006. TNF-α in promotion and progression of cancer. Cancer Metastasis Rev. 25, 409–416. ( 10.1007/s10555-006-9005-3) [DOI] [PubMed] [Google Scholar]
- 9.Balkwill F. 2009. TNF-α and cancer. Nat. Rev. Cancer 9, 361–371. ( 10.1038/nrc2628) [DOI] [PubMed] [Google Scholar]
- 10.Mocellin S, Rossi CR, Pilati P, Nitti D. 2005. Tumor necrosis factor, cancer and anticancer therapy. Cytokine Growth Factor Rev. 16, 35–53. ( 10.1016/j.cytogfr.2004.11.001) [DOI] [PubMed] [Google Scholar]
- 11.Hsiao SH, Lee MS, Lin HY, Su YC, Ho HC, Hwang JH, Lee C-C, Hung S-K. 2009. Clinical significance of measuring levels of tumor necrosis factor-alpha and soluble interleukin-2 receptor in nasopharyngeal carcinoma. Acta Otolaryngol. 129, 1519–1523. ( 10.3109/00016480902849427) [DOI] [PubMed] [Google Scholar]
- 12.Tse BWC, Scott KF, Russell PJ. 2012. Paradoxical roles of tumour necrosis factor-alpha in prostate cancer biology. Prostate Cancer 2012, 128965 ( 10.1155/2012/128965) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bong KW, Chapin SC, Doyle PS. 2010. Magnetic barcoded hydrogel microparticles for multiplexed detection. Langmuir 26, 8008–8014. ( 10.1021/la904903g) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morozov VN, Morozova TY. 2006. Active bead-linked immunoassay on protein microarrays. Anal. Chim. Acta 564, 40–52. ( 10.1016/j.aca.2005.09.068) [DOI] [PubMed] [Google Scholar]
- 15.Jiang Z, Llandro J, Mitrelias T, Bland JAC. 2006. An integrated microfluidic cell for detection, manipulation, and sorting of single micron-sized magnetic beads. J. Appl. Phys. 99, 08S105 ( 10.1063/1.2176238) [DOI] [Google Scholar]
- 16.Jun BH, et al. 2012. Quantum dots: ultrasensitive, biocompatible, quantum-dot-embedded silica nanoparticles for bioimaging. Adv. Funct. Mater. 22, 1843–1849. ( 10.1002/adfm.201102930) [DOI] [Google Scholar]
- 17.Jun BH, Kang H, Lee YS, Jeong DH. 2012. Fluorescence-based multiplex protein detection using optically encoded microbeads. Molecules 17, 2474–2490. ( 10.3390/molecules17032474) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suh SK, Chapin SC, Hatton TA, Doyle PS. 2012. Synthesis of magnetic hydrogel microparticles for bioassays and tweezer manipulation in microwells. Microfluid Nanofluid 13, 665–674. ( 10.1007/s10404-012-0977-8) [DOI] [Google Scholar]
- 19.Smith GP, Petrenko VA. 1997. Phage display. Chem. Rev. 97, 391–410. ( 10.1021/cr960065d) [DOI] [PubMed] [Google Scholar]
- 20.Causa F, et al. 2013. Evolutionary screening and adsorption behavior of engineered M13 bacteriophage and derived dodecapeptide for selective decoration of gold interfaces. J. Colloid Interface Sci. 389, 220–229. ( 10.1016/j.jcis.2012.08.046) [DOI] [PubMed] [Google Scholar]
- 21.Barbas CF, Burton DR, Scott JK, Silverman GJ. 2001. Phage display: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- 22.Nord K, Nord O, Uhlen M, Kelley B, Ljungqvist C, Nygren PA. 2001. Recombinant human factor VIII-specific affinity ligands selected from phage-displayed combinatorial libraries of protein A. Eur. J. Biochem. 268, 4269–4277. ( 10.1046/j.1432-1327.2001.02344.x) [DOI] [PubMed] [Google Scholar]
- 23.Biroccio A, Hamm J, Incitti I, De Francesco R, Tomei L. 2002. Selection of RNA aptamers that are specific and high-affinity ligands of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76, 3688–3696. ( 10.1128/JVI.76.8.3688-3696.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Legendre D, Soumillion P, Fastrez J. 1999. Engineering a regulatable enzyme for homogeneous immunoassays. Nat. Biotechnol. 17, 67–72. ( 10.1038/5243) [DOI] [PubMed] [Google Scholar]
- 25.Lu D, Shen JQ, Vil MD, Zhang HF, Jimenez X, Bohlen P, Witte L, Zhu ZP. 2003. Tailoring in vitro selection for a picomolar affinity human antibody directed against vascular endothelial growth factor receptor 2 for enhanced neutralizing activity. J. Biol. Chem. 278, 43 496–43 507. ( 10.1074/jbc.M307742200) [DOI] [PubMed] [Google Scholar]
- 26.Menendez A, Scott JK. 2005. The nature of target-unrelated peptides recovered in the screening of phage-displayed random peptide libraries with antibodies. Anal. Biochem. 336, 145–157. ( 10.1016/j.ab.2004.09.048) [DOI] [PubMed] [Google Scholar]
- 27.Zhuang GQ, Katakura Y, Furuta T, Omasa T, Kishimoto M, Suga K. 2001. A kinetic model for a biopanning process considering antigen desorption and effective antigen concentration on a solid phase. J. Biosci. Bioeng. 91, 474–481. ( 10.1016/S1389-1723) [DOI] [PubMed] [Google Scholar]
- 28.Chirinos-Rojas CL, Steward MW, Partidos CD. 1998. A peptidomimetic antagonist of TNFα-mediated cytotoxicity identified from a phage-displayed random peptide library. J. Immuol. 161, 5621–5626. [PubMed] [Google Scholar]
- 29.Zhang J, et al. 2003. Identification of anti-TNF-α peptides with consensus sequence. Biochem. Biophys. Res. Commun. 310, 1181–1187. ( 10.1016/j.bbrc.2003.09.141) [DOI] [PubMed] [Google Scholar]
- 30.Chirinos-Rojas CL, Steward MW, Partidos CD. 1999. A phage displayed mimotope inhibits tumor necrosis factor-induced cytotoxicity more effectively than the free mimotope. Immunology 96, 109–113. ( 10.1046/j.1365-2567.1999.00660.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayens CA, Willem N. 1995. Structures and stabilities of adsorbed proteins. J. Colliod Interface Sci. 169, 313–328. ( 10.1006/jcis.1995.1039) [DOI] [Google Scholar]
- 32.Roach P, Farrar D, Perry CC. 2005. Interpretation of protein adsorption: surface-induced conformational changes. Am. Chem. Soc. 127, 8168–8173. ( 10.1021/ja042898o) [DOI] [PubMed] [Google Scholar]
- 33.Wang J, et al. 2011. Selection of phage-displayed peptides on live adherent cells in microfluidic channels. Proc. Natl Acad. Sci. USA 108, 6909– 6914 ( 10.1073/pnas.1014753108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Adams JD, Turner K, Cochran FV, Gambhir SS, Soh HT. 2009. Controlling the selection stringency of phage display using a microfluidic device. Lab Chip 9, 1033–1036. ( 10.1039/b820985e) [DOI] [PubMed] [Google Scholar]
- 35.Cung K, Slater RL, Cui Y, Jones SE, Ahmad H, Naik RR. 2012. Rapid, multiplexed microfluidic phage display. Lab Chip 12, 562–565. ( 10.1039/c21c21129g) [DOI] [PubMed] [Google Scholar]
- 36.Jung JH, Kim G-Y, Seo TS. 2011. An integrated passive micromixer magnetic separation-capillary electrophoresis microdevice for rapid and multiplex pathogen detection at the single-cell level. Lab Chip 11, 3465–3470. ( 10.1039/c1lc20350a) [DOI] [PubMed] [Google Scholar]
- 37.Yin SN, Wang CF, Yu ZY, Wang J, Liu SS, Chen S. 2011. Versatile bifunctional magnetic-fluorescent responsive Janus supraballs towards the flexible bead display. Adv. Mater. 23, 2915–2919. ( 10.1002/adma.201100203) [DOI] [PubMed] [Google Scholar]
- 38.Pamme N, Manz A. 2004. On-chip free-flow magnetophoresis: continuous flow separation of magnetic particles and agglomerates. Anal. Chem. 76, 7250–7256. ( 10.1021/ac049183o) [DOI] [PubMed] [Google Scholar]
- 39.Lee H, Kim J, Kim H, Kwon S. 2010. Colour-barcoded magnetic microparticles for multiplexed bioassays. Nat. Mater. 9, 745–749. ( 10.1038/nmat2815) [DOI] [PubMed] [Google Scholar]
- 40.Choi JW, Oh KW, Thomas JH, Heineman WR, Halsall HB, Nevin JH, Helmicki AJ, Henderson HT, Ahn CH. 2001. An integrated microfluidic biochemical detection system for protein analysis with magnetic bead-based sampling capabilities. Lab Chip 2, 27–30. ( 10.1039/B107540N) [DOI] [PubMed] [Google Scholar]
- 41.Peyman SA, Iles A, Pamme N. 2009. Mobile magnetic particles as solid-supports for rapid surface-based bioanalysis in continuous flow. Lab Chip 9, 3110–3117. ( 10.1039/b904724g) [DOI] [PubMed] [Google Scholar]
- 42.Salgueiriño-Maceira V, Correa-Duarte MA. 2007. Increasing the complexity of magnetic core/shell nanocomposites for biological applications. Adv. Mater. 19, 4131–4144. ( 10.1002/adma.200700418) [DOI] [Google Scholar]
- 43.Herve K, et al. 2008. The development of stable aqueous suspensions of PEGylated SPIONs for biomedical applications. Nanotechnology 19, 465608 ( 10.1088/0957-4484/19/46/465608) [DOI] [PubMed] [Google Scholar]
- 44.Park JY, Daksha P, Lee GH, Woo S, Chang YM. 2008. Highly water-dispersible PEG surface modified ultra small superparamagnetic iron oxide nanoparticles useful for target-specific biomedical applications. Nanotechnology 19, 365603 ( 10.1088/0957-4484/19/36/365603) [DOI] [PubMed] [Google Scholar]
- 45.Jaye DL, Geigerman CM, Fuller RE, Akyildiz A, Parkos CA. 2004. Direct fluorochrome labeling of phage display library clones for studying binding specificities: applications in flow cytometry and fluorescence microscopy. J. Immunol. Methods 295, 119–127. ( 10.1016/j.jim.2004.09.011) [DOI] [PubMed] [Google Scholar]
- 46.Sussman JL, Lin D, Jiang J, Manning NO, Prilusky J, Ritter O, Abola EE. 1998. Protein data bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallogr. D Biol. Crystallogr. 54, 1078–1084. ( 10.1107/S0907444998009378) [DOI] [PubMed] [Google Scholar]
- 47.Byla P, Andersen MH, Holtet TL, Jacobsen H, Munch M, Gad HH, Thogersen HC, Hartmann R. 2010. Selection of a novel and highly specific tumor necrosis factor α (TNFα) antagonist: insight from the crystal structure of the antagonist-TNFα complex. J. Biol. Chem. 285, 12 096–12 100. ( 10.1074/jbc.M109.063305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiner SJ, Kollman PA, Case DA, Singh UC, Ghio C, Alagona G, Profeta S, Weiner P. 1984. A new force-field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 106, 765–784. ( 10.1021/ja00315a051) [DOI] [Google Scholar]
- 49.Cossi M, Rega N, Scalmani G, Barone V. 2003. Energies, structures, and electronic properties of molecules in solution with the C-PCM solvation model. J. Comput. Chem. 24, 5607–5612. ( 10.1002/jcc.10189) [DOI] [PubMed] [Google Scholar]
- 50.Goodsell DS, Morris GM, Olson AJ. 1996. Automated docking of flexible ligands: applications of AutoDock. J. Mol. Recogn. 9, 1–5. ( 10.1002/(SICI)1099-1352(199601)9:1) [DOI] [PubMed] [Google Scholar]
- 51.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 16, 2785–2791. ( 10.1002/jcc.21256). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sclavons C, Burtea C, Boutry S, Laurent S, Vander Elst L, Muller RN. 2013. Phage display screening for tumor necrosis factor-α-binding peptides: detection of inflammation in a mouse model of hepatitis. Int. J. Pept. 2013, 1–9. ( 10.1155/2013/348409) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qin W, Feng J, Li Y, Lin Z, Shen B. 2006. De novo design TNF-α antagonistic peptide based on the complex structure of TNF-α with its neutralizing monoclonal antibody Z12. J. Biotechnol. 125, 57–63. ( 10.1016/j.jbiotec.2006.01.036) [DOI] [PubMed] [Google Scholar]
- 54.Eck MJ, Sprang SR. 1989. The structure of tumor necrosis factor-α at 2.6 A resolution. J. Biol. Chem. 264, 17 595–17 605. [DOI] [PubMed] [Google Scholar]
- 55.Naffin JL, Han Y, Olivos HJ, Reddy MM, Sun T, Kodadek T. 2003. Immobilized peptides as high-affinity capture agents for self-associating proteins. Chem. Biol. 10, 251–259. ( 10.1016/S1074-5521(03)00049-8) [DOI] [PubMed] [Google Scholar]
- 56.Appleyard DC, Chapin SC, Doyle PS. 2011. Multiplexed protein quantification with barcoded hydrogel microparticles. Anal. Chem. 83, 193–199. ( 10.1021/ac1022343) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.