Abstract
In Arabidopsis thaliana expression of the B‐class MADS‐box genes APETALA3 (AP3) and PISTILLATA (PI) is confined to petals and stamens but in other plant species these genes are also transcribed in non‐flower tissues; in Solanum tuberosum they are transcribed specifically in vascular bundles leading to petals and stamens. Transcription analysis of B‐class genes in Eranthis hyemalis using reverse transcribed in situ PCR revealed that both AP3 and PI are expressed in developing vascular bundles in the tuberous rhizome, flowering stem and floral primordia. In addition, AP3 and PI transcripts are also found in stems and leaves. These results suggest a more complex role of B‐class genes in Eranthis and possible involvement in the development of vascular tissue.
Key words: Eranthis, MADS‐box genes, AP3, PI, transcription analysis, vascular bundles, RT‐ISPCR
INTRODUCTION
The APETALA3 (AP3) and PISTILLATA (PI) genes comprise the B‐class floral identity genes in higher dicots, e.g. Arabidopsis thaliana Schur and Antirrhinum majus L. (Coenet al., 1991; Meyerowitzet al., 1991; Krameret al., 1998). Both belong to the MADS‐box gene family that encodes DNA‐binding proteins. The proteins of AP3 and PI genes form heterodimers in Arabidopsis and Antirrhinum and both are essential for petal and stamen identity (Jacket al., 1992; Schwarz‐Sommeret al., 1992; Tröbneret al., 1992; Goto and Meyerowitz, 1994). Like other MIKC‐type MADS‐box genes, so far only found in plants (Alvarez‐Buyllaet al., 2000), AP3 and PI proteins contain highly conserved protein domains. The MADS‐ and K‐box are both involved in protein dimerization (Riechmannet al., 1996), and are linked together by a lesser conserved part, the I‐region and ‘finished’ by a variable C‐terminal. Transcription of AP3 and PI in Arabidopsis is observed throughout development of petals and stamens, but they are not transcribed in non‐flower tissues (Sommeret al., 1990; Jacket al., 1992; Tröbneret al., 1992; Goto and Meyerowitz, 1994; Serrano‐Cartagenaet al., 2000). The expression pattern of AP3 and PI genes in flowers of lower dicots differs to that of higher dicots, indicating that the function of the genes may be different in this group of angiosperms (Krameret al., 1998; Kramer and Irish, 1999, 2000). This finding has been connected with the theory that petals have evolved several times within the angiosperms (Takhtajan, 1991). In other higher dicots AP3 or PI genes are also transcribed in seedlings (Munsteret al., 2001), leaves (Southertonet al., 1998;Yuet al., 1999; Munsteret al., 2001) and roots (Yuet al., 1999; Munsteret al., 2001), and StDEF4 (Solanum tuberosum Walp.) is specifically expressed in vascular bundles leading to the petals and stamens (Garcia‐Marotoet al., 1993), suggesting a more complex gene function.
This paper reports on the expression pattern of an AP3/DEF and a PI/GLO gene (named EhAP3 and EhPI) in Eranthis hyemalis (L.) Salisb. (Ranunculaceae). The flower perianth of Eranthis consists of five to eight yellow tepals, also described as yellow sepals, and five to ten tubular‐shaped nectar containing honey‐leaves, also described as petals (Hiepko, 1995). The stamens are numerous and there are between two and ten carpels. Basal leaves and flowers initiate from a tuberous rhizome and three green cauline leaves arranged in a whorl form an involucre around the developing flower (Hiepko, 1995). In Ranunculaceae, nectar‐producing organs are supposed to be derived from stamens (Hiepko, 1995), and are named honey‐leaves (‘Honigblätter’; Prantl, 1887). Transcription studies are performed using reverse transcribed in situ PCR (RT–ISPCR) to obtain information about the expression of B‐class genes at the single cell level.
MATERIALS AND METHODS
Isolation of AP3 and PI cDNAs
Total RNA was extracted from flowers, leaves, stems and roots using FastRNA Kit‐Green (BIO 101, Carlsbad, USA) and genomic DNA was extracted from leaves using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). First strand cDNA synthesis was carried out in a 50 µl reaction with MuLV reverse transcriptase, RNase inhibitor, buffer II, MgCl2, dNTPs and a poly(T)16 primer, following the MuLV Reverse Transcriptase protocol (Applied Biosystems, Foster City, USA). To amplify AP3 and PI genes, the following primer sequences (according to IUPAC rules) were used in the following PCR reaction: AP3/PI MADS‐box sequence: 5′‐atmcagathaagagratmgagaa‐3′/5′‐athgagatmaaragrathgagaa‐3′ and AP3/PI K‐box sequence: 5′‐cagattttgctcaagaccgcgca‐3′/5′‐agagcatgttcaatggggataagc‐3′. The primer sequences amplify a fragment of approx. 400 bp and were designed from existing AP3 and PI sequences from six ranunculid species: Ranunculus ficaria, R. bulbosus, Delphinium ajacis, Papaver nudicaule, P. californicum and Dicentra eximia.
PCR was performed in 100 µl reactions containing buffer II, MgCl2, dNTPs, primers and Amplitaq Gold, according to the Amplitaq Gold protocol (Applied Biosystems). Amplification began with an activation step of 10 min at 95 °C, followed by 40 cycles of 40 s denaturing at 95 °C, 40 s annealing at 45 °C, and 1 min extension at 72 °C. Amplified products were analysed on a 2 % nusieve agarose gel. Bands of approx. 400 bp were cloned using the cloning PCR‐Script kit (Stratagene, La Jolla, USA) and clones were analysed based on fragment length and sequenced. Sequencing reactions (1/2 reactions in 20 µl volumes) were carried out using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit with Amplitaq DNA Polymerase, FS (Applied Biosystems) and 5 × sequencing buffer for dilution of BigDye Ready Reaction Premix (Applied Biosystems). Before gel separation the sequence reaction products were cleaned using the DyeEx Spin Kit (Qiagen). Fragments were separated on a 5 % Long Ranger gel (FMC Bioproducts, Rockland, USA) on an ABI 377 Prism DNA Sequencer (Applied Biosystems). ABI Prism 377 Collection software version 2·1 was used to evaluate the sequences.
Phylogenetic analysis
Alignment of published AP3 and PI cDNA sequences was done by hand using GeneDoc (Nicholas and Nicholas, 1997). Previously published B‐class genes were retrieved from the GenBank database (see Table 1). To avoid many missing characters in the matrix only the sequence covering the MADS‐box, I‐region and part of the K‐box was used in the analysis (alignment available at: www.bot.ku.dk/staff/boj/martin/ap3pi.txt). The Gentum gnemon gene GgM13 was used as outgroup because it is the closest relative of the B‐class genes (Beckeret al., 2000). The most parsimonious trees were generated using the PAUP* 4·0b2a package (Swofford, 1999). Characters were treated as missing when nucleotide data were not available. Phylogenetic trees were found using the heuristic search algorithm, 1000 replicate run, and random stepwise addition of sequences under the TBR (tree–bisection–reconnection) algorithm for branch swapping. Jackknife values greater than 50, generated from a 10 000 replicate run using Parsimony Jackknifer version 4·22 (Farriset al., 1996), were assigned to nodes.
Table 1.
Previously published B‐class genes retrieved from the GenBank database
| Genes | |
| Previously published species | |
| Antirrhinum majus L | AmDEF (16017), AmGLO (16023) |
| Arabidopsis thaliana Schur | AtPI (42128), AtAP3 (166607) |
| Delphinium ajacis Ledeb. | DaPI (3170477) |
| Dicentra eximia Torr. | DePI (3170467), DeAP3 (3170503) |
| Gerbera × hybrida | GhGGLO1 (4218172), GhGDEF2 (4218170) |
| Medicago sativa L. | MsNMH7 (1870205), MsNGL9 (13095570) |
| Papaver nudicaule L. | PnPI‐1 (3170463), PnAP3‐1 (3170499) |
| Ranunculus ficaria L. | RfPI‐1 (3170469), RfAP3‐1 (3170461) |
| Ranunculus bulbosus Costa | RbPI‐1 (3170471), RbAP3‐1 (3170505) |
| Sanguinaria canadensis L. | ScPI (4883905), ScAP3 (4883899) |
| Silene latifolia Britten & Rendle | SlM2 (602901), SlM3 (602903) |
| Gnetum gnemon L. | GgM13 (5019463) |
| From this work | |
| Eranthis hyemalis (L.) Salisb. | EhPI (16304399), EhAP3 (16304397) |
Genbank GI numbers in parentheses.
Tissue preparation and fixation
Plant material of Eranthis hyemalis was taken from the Botanical Garden, University of Copenhagen, Denmark. Flowers were fixed in FAA [5·5 % acetic acid, 66 % EtOH (96 %), 20 % H2O, 8·5 % paraformaldehyde (24·5 %)], placed in vacuum for 30 min and left at 5 °C overnight followed by a 20 min dehydration in 70 % EtOH. All material was embedded in ParaPlast, sectioned at 10 µm and placed on silane‐coated in situ PCR slides (Applied Biosystems). Shortly before RT–ISPCR, sections were deparaffinized in Histoclear 2 × 30 min, followed by 10 min 1 : 1 Histoclear/100 % EtOH and 2 × 10 min in 100 % EtOH. Slides were kept in 100 % EtOH until reverse transcription (usually for less than 1 h).
RT–PCR optimization
Prior to RT‐ISPCR, amplification of both EhAP3 and EhPI transcripts was optimized with specific primers (see below) in solution. Optimization was done with total RNA templates of different flower developmental stages and also on leaf, stem and root templates. RNA from flowers also covers tissue from the tuberous rhizome, because flowers initiate from this organ. The EhAP3 (264 bp) and EhPI (289 bp) gene transcripts were sequenced on an ABI 377 Prism DNA Sequencer (Applied Biosystems). The EhAP3 and EhPI primer pairs are designed and tested so they do not amplify genomic DNA, as the reverse primers span exons 3 (A–B) and 4 (B–C) in the K‐box of the B‐class genes (Sundströmet al., 1999). However, the forward primer does amplify one copy of single stranded genomic DNA during each thermal cycling giving a maximum background signal from 30 copies in each nucleus.
Reverse transcribed in situ PCR
RT–ISPCR was performed according to Johansen (1997) with the following modifications: the 50 µl RT‐mix contained 2000 U ml–1 MuLV reverse transcriptase (Applied Biosystems) and 200 U ml–1 RNase inhibitor (Applied Biosystems) in 50 mm KCL, 10 mm Tris–HCl, 5 mm MgCl2, 1 mm of each dNTP and 5 µm poly(T)16 (Applied Biosystems). As a negative control, one section on each slide was not reverse transcribed. Reactions were reverse transcribed for 10 min at room temperature followed by 20 min at 42 °C in the GeneAmp in situ apparatus. The slides were kept on ice for 5 min and then disassembled before the 40 µl PCR mix was added: 200 U ml–1 AmpliTaq DNA polymerase in 40 mm KCL, 8 mm Tris–HCl, 2·5 mm MgCl2, 0·02 mm of each dNTP (with 10 % DIG‐11‐dUTP) and 0·8 µm of the two EhAP3 or EhPI specific primers: EhAP3: 5′‐cgaagaaccggaattgtgaa‐3′, 3′‐tacgacaaagaatcggagaagg‐5′, and EhPI: 5′‐caaacaggcaggttacctactcc‐3′, 3′‐ctaaggcacctgaagggtga‐5′. PCR amplification began with a 3 min denaturing step at 94 °C followed by a 30 cycle two‐step PCR of 1 min denaturing at 94 °C and 1 min at 60 °C.
Detection of products
Primary antibodies conjugated to alkaline phosphatase were used to detect the EhAP3 and EhPI RT‐PCR products. Blocking was done in 1 % BSA‐C (Arion, Wageningen, The Netherlands) in PBS for 1 h after which 100 µl of a BSA‐C/ PBS mix containing 800 µl ultra pure water, 100 µl BSA‐C, 100 µl PBS and 10 µl of the alkaline phosphatase labelled anti‐DIG antibody (Roche, Mannheim, Germany) was applied to each section. After 1 h slides were placed under running water for 30 min and detection of alkaline phosphatase was carried out for 8–10 min using one NTB/BCIP ready‐to‐use tablet (Roche). Mounting was done in Mowiol (Sigma‐Aldrich, Steinheim, Germany).
RESULTS AND DISCUSSION
Isolation and phylogenetic analysis of Eranthis B‐class genes
The AP3‐ and PI‐specific primers amplify a fragment of approx. 400 bp. Although the primer sequences were designed from existing AP3 and PI sequences from six species of Ranunculiflorae (see Materials and Methods) they also worked in amplification of B‐class genes in several monocot species (unpubl. res.). Analysis of the Eranthis clones (17 AP3 clones and 14 PI clones) showed only amplification of B‐class genes suggesting that these primers are highly AP3‐ and PI‐gene specific.
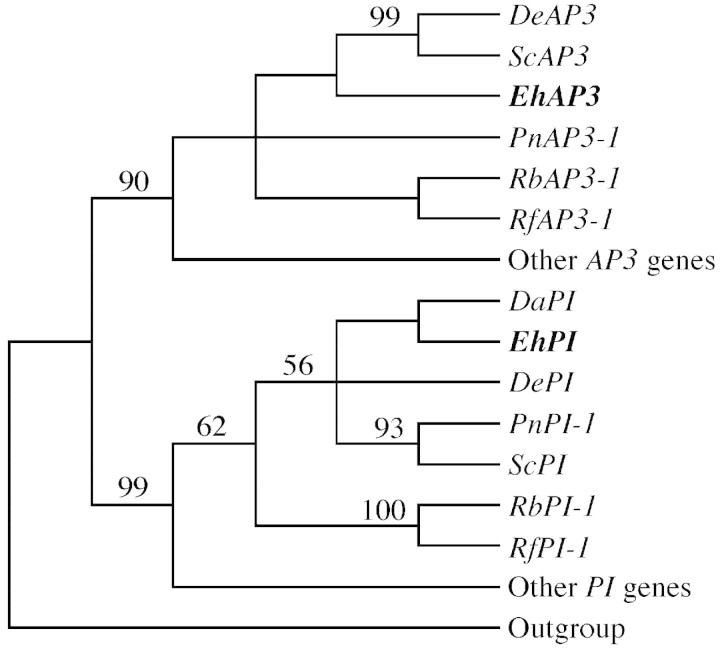
The Eranthis AP3 and PI gene sequences contain the K‐box conserved motifs (AP3 amino acids = H/QYExM and PI amino acids = KHExL), characteristic of the B‐class genes (Krameret al., 1998). A simplified phylogenetic analysis of B‐class genes shows that as expected, the Eranthis genes (named EhPI and EhAP3) are more closely related to PI and AP3 genes from other ranunculid species than to B‐class genes from more advanced dicot species (Fig. 1).
Fig. 1. Phylogenetic analysis of B‐class genes from different dicot species. The Eranthis genes (bold) group with other ranunculid AP3 and PI genes. Numbers above branches indicate jackknife values > 50. The cladogram is based on seven trees of equal length containing 295 parsimony informative characters (length 1622; consistency index = 0·42, retention index = 0·50). Other AP3 genes: AtAP3; AmDEF; GhGDEF2; MsNMH7; and SlM3. Other PI genes: AtPI; AmGLO; GhGGLO1; MsNGL9; and SlM2. Outgroup: GgM13 (see Materials and Methods).
RT–PCR and RT‐ISPCR expression analysis
Analysis of the EhAP3 and EhPI transcript in different plant organs of Eranthis was done with simple solution RT–PCR experiments on total RNA extractions. RT–PCR products of the EhAP3 and EhPI transcripts isolated from Eranthis flowers are present in both leaves and stems but not in roots (Fig. 2).
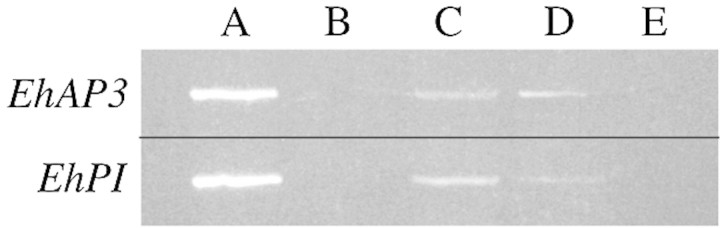
Fig. 2. Amplification of EhAP3 and EhPI on total RNA from different tissues of Eranthis using solution RT‐PCR analysis. A, Flower and tuberous rhizome; B, roots; C, leaves; D, stems; E, DNA template.
In situ transcription analysis of EhAP3 and EhPI from Eranthis is shown in Fig. 3. The expression pattern of EhAP3 and EhPI in the flower of Eranthis is not only confined to tepals, honey‐leaves and stamens but the genes are also transcribed in the cauline leaves surrounding the developing flower (Fig. 3A, C, E, G). EhAP3 and EhPI are both transcribed in developing vascular tissue, which is seen in the tuberous rhizome, through the young flower stem and in the bundles leading to the young floral organ primordia (Fig. 3C, D, G, H). In sections of more developed flowers, EhAP3 and EhPI transcripts are concentrated in the cauline leaves, tepals, honey‐leaves and in stamens restricted to the sporogeneuos tissue, but transcription in the vascular bundles is not detectable (not shown). It appears as if EhAP3 is transcribed more than EhPI, but this may simply reflect differences in efficiency of the two IS–PCR reactions. Overall, the transcription pattern of EhAP3 and EhPI in Eranthis is similar, which suggests that the protein products also form heterodimers as in flowers of Arabidopsis (AP3 and PI) and Antirrhinum (DEF and GLO) (Davieset al., 1996; Riechmannet al., 1996). However, as EhAP3 and EhPI transcripts are not confined only to tepals, honey‐leaves and stamens, as might be expected if the B‐class gene transcription pattern applies to the ABC‐model, it is unlikely that these genes only act as tepal, honey‐leaves and stamen identity genes in Eranthis.
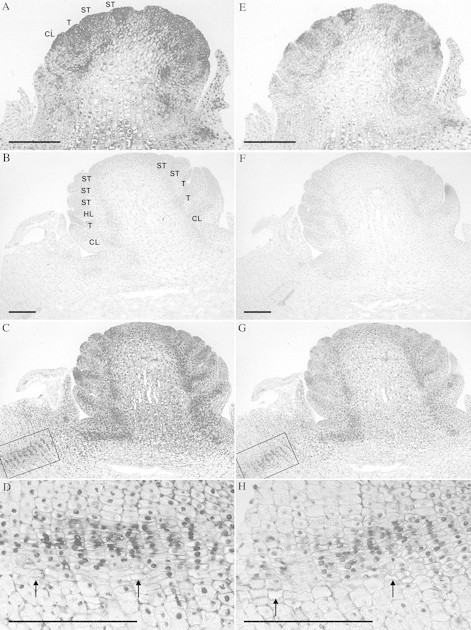
Fig. 3. Localization of EhAP3 (A–D) and EhPI (E–H) transcripts in Eranthis using reverse transcribed in situ PCR. Cauline leaves and floral primordia are starting to appear in A and E. Honey‐leaves and stamens are distinguished in C and G. Negative control in B and F (see Materials and Methods). Magnification of the vascular bundles in tissue of tuberous rhizome indicated by boxes in C and G are shown in D and H. Labels on A apply also to E and those on B to C, F and G. CL, Cauline leaf; T, tepal; HL, honey leaf; ST, stamen. Arrows in D and H indicate vessel members. Bar = 0·1 mm.
Northern blot analysis has shown that transcripts of the PI genes ZMM18/29 (Zea mays L.), EgrEGM2 (Eucalyptus grandis Maiden) and the AP3 gene GhGDEF2 (Gerbera hybrida) were also detectable in leaves (Southertonet al., 1998; Yuet al., 1999; Munsteret al., 2001). It is interesting that B‐class genes from these species and also EhAP3 and EhPI from Eranthis are transcribed in leaves because transcription of floral identity genes in leaves of Arabidopsis is inhibited by other regulatory genes (Serrano‐Cartagenaet al., 2000). This suggests that inhibition of floral identity gene transcription in leaves is not preserved throughout angiosperms and it is therefore likely that B‐class genes may have other functions besides acting as petal and stamen identity genes as in Arabidopsis (Jacket al., 1992; Goto and Meyerowitz, 1994).
Transcription of MADS‐box genes in developing vascular tissue has been observed in several plant species (Decroocqet al., 1999; Sundströmet al., 1999; Alvarez‐Buyllaet al., 2000). However, specific expression of B‐class genes in vascular bundles is only reported from the AP3 gene StDEF4 (Solanum tuberosum), where the transcript was detected in vascular bundles leading to petals and stamens (Garcia‐Marotoet al., 1993). Kramer and Irish (1999) studied B‐class gene transcription and antibody localization in flowers of two ranunculid species but did not mention B‐class gene expression in vascular bundles apart from anti‐PI antibody staining in the vascular tissue at the base of petals in the flower of Dicentra eximia Torr. However, it does appear from in situ hybridization studies on flowers of Papaver nudicaule L. (Fig. 2B, C, J, I, P and T in Kramer and Irish, 1999); that both PnAP3 and PnPI‐1 are transcribed in what is probably developing vascular strands leading to petals and stamens. This is similar to the present observations in Eranthis, except that expression in developing vascular bundles is also observed in the young flower stem and tuberous rhizome. This may suggest that B‐class genes play a role in development of vascular tissue. If B‐class genes are involved in development of vascular bundles, expression should also be detectable in roots of Eranthis. Although several solution RT–PCR experiments were performed on RNA from Eranthis roots, neither EhAP3 nor EhPI transcripts were detected. Whether this could be due to inhibition of transcripts as seen in leaves of Arabidopsis (Serrano‐Cartagenaet al., 2000) requires further investigation.
Although transcription of B‐class genes is generally restricted to petals and stamens in angiosperms, future research into B‐class genes should aim to elucidate their function in tissues other than floral tissue. One way to explore this would be to perform transformation studies by ectopic expression or co-supression of B‐class genes in species where these genes are transcribed in other organs and apparently have additional functions besides determination of petal and stamen identity.
ACKNOWLEDGEMENTS
This research was supported by grants from Familien Hede Nielsen Fond and the Botanical Institute, University of Copenhagen and the Department of Evolutionary Botany, Botanical Institute, University of Copenhagen. I thank Drs Signe Frederiksen and Bo Johansen. I also thank Kim B. Pedersen, Charlotte Hansen and Kate Jensen for technical assistance. Dr M. Gustafsson kindly commented on the manuscript. This work could not have been done without continuous support from ‘Nedløbsrøret’.
Supplementary Material
Received: 14 June 2001; Returned for revision: 15 August 2001; Accepted: 19 September 2001.
References
- Alvarez‐Buylla ER, Liljegren SJ, Pelaz S, Gold SE, Burgeff C, Ditta GS, Vergara‐Silva F, Yanofsky MF.2000. MADS‐box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant Journal 24: 457–466. [DOI] [PubMed] [Google Scholar]
- Becker A, Winter KU, Meyer B, Saedler H, Theißen, G.2000. MADS‐box gene diversity in seed plants 300 million years ago. Molecular Biology and Evolution 17: 1425–1434. [DOI] [PubMed] [Google Scholar]
- Coen ES, Doyle S, Romero JM, Elliott R, Magrath R, Carpenter R.1991. Homeotic genes controlling flower development in Antirrhinum Development (Suppl 1): 149–156.1842353 [Google Scholar]
- Davies B, Egea CM, De Andrade SE, Saedler H, Sommer H.1996. Multiple interactions amongst floral homeotic MADS box proteins. EMBO Journal 15: 4330–4343. [PMC free article] [PubMed] [Google Scholar]
- Decroocq V, Zhu X, Kauffman M, Kyozuka J, Peacock WJ, Dennis ES, Llewellyn DJ.1999. A TM3‐like MADS‐box gene from Eucalyptus expressed in both vegetative and reproductive tissues. Gene 228: 155–160. [DOI] [PubMed] [Google Scholar]
- Farris JS, Albert VA, Kaellersjoe M, Libscomb D, Kluge AG.1996. Parsimony jackknifing outperforms neighbor‐joining. Cladistics 12: 99–124. [DOI] [PubMed] [Google Scholar]
- Garcia‐Maroto F, Salamini F, Rohde W.1993. Molecular cloning and expression patterns of three alleles of the Deficiens‐homologous gene St‐Deficiens from Solanum tuberosum Plant Journal 4: 771–780. [DOI] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM.1994. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA Genes & Development 8: 1548–1560. [DOI] [PubMed] [Google Scholar]
- Hiepko P 1995. General composition of the flower. In: Engler A, Prantl K. Die Natürlichen Pflanzenfamilien. 2nd edn. Bd. 17 a. 4. Angiospermae. Ordnung Ranunculales. Fam. Ranunculaceae. Berlin: Duncker & Humblot. [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM.1992. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68: 683–697. [DOI] [PubMed] [Google Scholar]
- Johansen B 1997. In situ PCR on plant material with sub‐cellular resolution. Annals of Botany 80: 697–700. [Google Scholar]
- Kramer EM, Irish VF.1999. Evolution of genetic mechanisms controlling petal development. Nature 399: 144–148. [DOI] [PubMed] [Google Scholar]
- Kramer EM, Irish VF.2000. Evolution of the petal and stamen developmental programs: evidence from comparative studies of the lower eudicots and basal angiosperms. International Journal of Plant Sciences 161(Suppl): 29–40. [Google Scholar]
- Kramer EM, Dorit RL, Irish VF.1998. Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS‐box gene lineages. Genetics 149: 765–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz EM, Bowman JL, Brockman LL, Drews GN, Jack T, Sieburth LE, Weigel D.1991. A genetic and molecular model for flower development in Arabidopsis thaliana Development (Suppl1): 157–167. [PubMed] [Google Scholar]
- Munster T, Ursula WL, Faigl W, Werth S, Saedler H, TheiBen G.2001. Characterization of three GLOBOSA‐like MADS‐box genes from maize: evidence for ancient paralogy in one class of floral homeotic B‐function genes of grasses. Gene 262: 1–13. [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB Jr1997. GeneDoc: analysis and visualization of genetic variation. http://www.cris.com/∼Ketchup/genedoc.shtml [Google Scholar]
- Prantl K 1887. Beiträge zur Morphologie und Systematik der Ranunculaceen. Botanische Jahrbücher für Systematik 9: 225–273. [Google Scholar]
- Riechmann JL, Wang M, Meyerowitz EM.1996. DNA‐binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS Nucleic Acids Research 24: 3134–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz‐Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lonnig WE, Saedler H, Sommer H.1992. Characterization of the Antirrhinum floral homeotic MADS‐box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO Journal 11: 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano‐Cartagena J, Candela H, Robles P, Ponce MR, Perez‐Perez JM, Piqueras P, Micol JL.2000. Genetic analysis of incurvata mutants reveals three independent genetic operations at work in Arabidopsis leaf morphogenesis. Genetics 156: 1363–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer H, Beltran JP, Huijser P, Pape H, Lonnig WE, Saedler H, Schwarz SZ.1990. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO Journal 9: 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southerton SG, Marshall H, Mouradov A, Teasdale RD.1998. Eucalypt MADS‐box genes expressed in developing flowers. Plant Physiology 118: 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström J, Carlsbecker A, Svensson ME, Svenson M, Johanson U, Theissen G, Engstrom P.1999. MADS‐box genes active in developing pollen cones of Norway spruce (Picea abies) are homologous to the B‐class floral homeotic genes in angiosperms. Developmental Genetics 25: 253–266. [DOI] [PubMed] [Google Scholar]
- Swofford DL 1999. PAUP*. Phylogentic analysis using parsimony (* and other methods), Version 4·0b2a. Sunderland, Mass: Sinauer. [Google Scholar]
- Takhtajan A 1991. Evolutionary trends in flowering plants. New York: Colombia University Press. [Google Scholar]
- Tröbner W, Ramirez L, Motte P, Hue I, Huijser P, Lonnig WE, Saedler H, Sommer H, Schwarz‐Sommer Z.1992. GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO Journal 11: 4693–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Kotilainen M, Pollanen E, Mehto M, Elomaa P, Helariutta Y, Albert VA, Teeri TH.1999. Organ identity genes and modified patterns of flower development in Gerbera hybrida (Asteraceae). Plant Journal 17: 51–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.