Abstract
The ‘Oxalis tuberosa alliance’ is a group of Andean Oxalis species allied to the Andean tuber crop O. tuberosa Molina (Oxalidaceae), commonly known as ‘oca’. As part of a larger project studying the origins of polyploidy and domestication of cultivated oca, flow cytometry was used to survey DNA ploidy levels among Bolivian and Peruvian accessions of alliance members. In addition, this study provided a first assessment of C‐values in the alliance by estimating nuclear DNA contents of these accessions using chicken erythrocytes as internal standard. Ten Bolivian accessions of cultivated O. tuberosa were confirmed to be octoploid, with a mean nuclear DNA content of approx. 3·6 pg/2C. Two Peruvian wild Oxalis species, O. phaeotricha and O. picchensis, were inferred to be tetraploid (both with approx. 1·67 pg/2C), the latter being one of the putative progenitors of O. tuberosa identified by chloroplast‐expressed glutamine synthetase data in prior work. The remaining accessions (from 78 populations provisionally identified as 35 species) were DNA diploid, with nuclear DNA contents varying from 0·79 to 1·34 pg/2C.
Key words: Andean crops, C‐value, DNA ploidy, flow cytometry, nuclear DNA content, oca, Oxalidaceae, Oxalistuberosa, octoploid, ploidy levels, polyploidy
INTRODUCTION
The ‘Oxalis tuberosa alliance’ was first described on cytological grounds by de Azkue and Martínez (1990) as a group of morphologically similar Oxalis species related to the Andean tuber crop oca [O. tuberosa Molina (Oxalidaceae)] that shares x = 8, a base chromosome number that is rare in Oxalis. The genus overall has base chromosome numbers varying from five to 12, with seven being most frequent (Marks, 1956; Talledo and Escobar, 1995). In addition to the dozen diploid and polyploid species studied by de Azkue and Martínez (1990), a few other Oxalis species have been reported with x = 8 (Table 1). Additional Oxalis species for which cytological information is as yet lacking also appear to belong to the group, based on the morphological similarities they share with members of the alliance (Emshwiller, 1999a, b). The members of the alliance are primarily Andean, being found in highlands from Central America and northern Venezuela to northwestern Argentina (Emshwiller, 2002). Although the species of the alliance were classified by Knuth (e.g. 1930) in several sections, the more recent treatment by Lourteig (2000) places almost all members of the alliance in two sections, Lotoideae Lourteig and Herrerae R. Knuth, with a small number in section Ortgieseae R. Knuth. De Azkue and Martínez (1990) found domesticated O. tuberosa to be the only octoploid in the alliance, with 2n = 8x = 64. This chromosome number for oca has since been confirmed by several workers, but different chromosome counts have been reported in both older and more recent reports, some of which differ markedly (see citations in Table 2).
Table 1.
Chromosome numbers reported for x = 8 Oxalis other than O. tuberosa
| Species* | 2n | Reference |
| O. herrerae R. Knuth | 16 | de Azkue and Martínez (1990) |
| Valladolid (1996) | ||
| O. medicaginea Kunth | 16 | de Azkue and Martínez (1990) |
| O. mollissima (Rusby) R. Knuth | 16 | de Azkue and Martínez (1990) |
| O. oblongiformis R. Knuth | 16 | de Azkue and Martínez (1990) |
| O. peduncularis H.B.K | 16 | Diers (1961) |
| de Azkue and Martínez (1990) | ||
| Valladolid (1996) | ||
| O. picchensis R. Knuth† | 16 | Valladolid (1996) |
| O. ptychoclada Diels | 16 | Favarger and Huynh (1965) |
| Huynh (1965) | ||
| O. subintegra R. Knuth | 16 | de Azkue and Martínez (1990) |
| O. tabaconasensis R. Knuth | 16 | de Azkue and Martínez (1990) |
| O. aff. villosula R. Knuth‡ | 16 | de Azkue and Martínez (1990) |
| O. sp.§ | 16 | de Azkue and Martínez (1990) |
| O. melilotoides Zuccarini | 16 | Valladolid (1996) |
| 32 | Brücher (1969) | |
| O. lotoides Kunth | 32 | de Azkue and Martínez (1990) |
| O. spiralis R. & P. ex G. Don | 16 | Favarger and Huynh (1965) |
| 48 | Huynh (1965) | |
| Brücher (1969)(as O. pubescens Kunth) | ||
| de Azkue and Martínez (1990) | ||
| Mathew (1958)(as O. pubescens Kunth) | ||
| O. nubigena Walpers | 48–50 | Diers (1961) |
| O. spp. (others unidentified) | 16 | Valladolid (1996) |
| 48 | Diers (1961) | |
| O. andina Britton¶ | 16 | de Azkue (2000) |
* Because of the difficulties of identifying Oxalis species, it is possible that some determinations may be incorrect.
† My examination of this plant, in CIP glasshouses, lead me to question this identification. It may be O. rigidicaulis R. Knuth, a taxon that I believe, contrary to Lourteig (2000), may be distinct from O. boliviana Britton.
‡O. villosula is probably a synonym of O. san‐miguellii or O. urubambensis, which are both endemic in southern Peru. The plant studied by de Azkue and Martínez was collected in Churuja, the same area as one of the accessions sampled here (EE807), of a species that I designate as Oxalis sp. ‘V’ (see Table 3).
§ This plant was collected in the vicinity of Machu Picchu, suggesting that it might be O. san‐miguellii R. Knuth or O. urubambensis R. Knuth, both of which are found in that area.
¶ See discussion in text about relationships of O. andina and its allies to the O. tuberosa alliance.
Table 2.
Chromosome numbers reported for Oxalis tuberosa Molina
| 2n | Reference |
| 14 | Heitz (1927) (as O. crenata Jacquin) |
| 63–64, 68–70 | Kostoff et al. (1935) |
| 66 | Cárdenas and Hawkes (1948) |
| 58–66 | Gibbs et al. (1978) |
| 64 | de Azkue and Martínez (1990) |
| 64 | Valladolid et al. (1994) |
| 64 | Medina Hinostroza (1994) |
| 14 | Talledo and Escobar (1995) |
| 64 | Valladolid Cavero (1996) |
| 24, 32, 40, 48, 64 | Guamán Calderón (1997) |
| 64 | Vinueza Vela (1997) |
| 49 | Hayano Kanashiro (1998) |
Phylogenetic analysis of DNA sequences of two nuclear loci, the internal transcribed spacer region (ITS) of nuclear ribosomal DNA and the chloroplast‐expressed isozyme of glutamine synthetase (ncpGS), has confirmed the monophyly of the O. tuberosa alliance and the inclusion in the group of octoploid oca as well as many of the morphologically similar species mentioned above that still lack cytological data. That is, sequences of all these species appear in a single clade on each of the gene trees of ITS or ncpGS data, whether analysed separately or simultaneously (Emshwiller and Doyle, 1998, 1999, 2002; Emshwiller, 2002). The O. tuberosa alliance, as recognized here, includes these other morphologically similar species that join the same clade as the original alliance species. However, although O. andina Britton has also been reported to have x = 8 (de Azkue, 2000), I exclude it from the alliance because there is no evidence from either gene that this species or its allies (a clade that forms the sister group of the alliance) were involved in the origins of oca. Data from ncpGS indicated that oca is allopolyploid, derived from hybridization of multiple species, and two wild tuber‐bearing taxa were identified as progenitor candidates, O. picchensis R. Knuth from southern Peru and an as yet unnamed taxon from Bolivia (Emshwiller and Doyle, 2002). However, several alternative hypotheses remained about oca’s origins that were all congruent with the ncpGS data (Emshwiller, 1999a), and distinguishing the most likely of these hypotheses required additional data, including information on ploidy levels of the putative progenitors. Some individuals of wild Oxalis species were heterozygous for ncpGS sequences, and ploidy information was necessary to interpret this heterozygosity as either normal allelic polymorphism or as homologous loci (Emshwiller, 1999a; Emshwiller and Doyle, 2002).
Cultivated oca has an important place in the diets and farming systems of rural communities in the Andean highlands. Knowledge of the origins of the crop, its relationships with wild allies and the ploidy levels of its close relatives is crucial for the potential use of wild relatives in future breeding programmes for oca. Information about genome size can also have practical relevance for molecular characterization by techniques such as RAPDs or AFLPs, in which the choice of primers and/or restriction enzymes is affected by the nuclear DNA content (Bennett et al., 2000). In addition, because the evolution of higher‐level polyploids is less well understood than that of tetraploids, the study of the O. tuberosa alliance and the origins of oca provides an important case study.
As part of a larger project studying the origins of cultivated oca and evolutionary relationships of members of the O. tuberosa alliance, a survey was conducted of ploidy levels among Peruvian and Bolivian populations of alliance members. The primary objectives of this survey were to use flow cytometry as an independent data source to confirm the octoploidy of cultivated oca, to determine the ploidy level of O. picchensis, one of the putative progenitors of cultivated oca identified by ncpGS data (Emshwiller, 1999a; Emshwiller and Doyle, 2002), and to survey ploidy levels in other members of the alliance as an initial assessment of frequency of polyploidy in the alliance. As a secondary objective, this flow cytometry survey provides the first exploration of the patterns of variation of DNA content within and among diploid species in the O. tuberosa alliance. Some observations of repeatable variation among species, populations or individuals are reported here to suggest avenues for further study.
Flow cytometry has frequently been used in ploidy analysis (e.g. Costich et al., 1993; Braeutigam and Braeutigam, 1996; Lysák and Doležel, 1998; Brummer et al., 1999; Gamiette et al., 1999; Dansi et al., 2001). Some of the advantages of flow cytometry for this purpose, as summarized by Doležel (1997), include its ease, rapidity, ability to detect mixoploidy and the use of a small amount of leaf tissue rather than the need for dividing cells. However, the use of flow cytometry to infer ploidy level is only appropriate when comparing accessions from the same or closely related species, where large differences in chromosome size are not expected. Although divergent members of the large genus Oxalis have chromosomes of diverse sizes (Heitz, 1927; Warburg, 1938; Marks, 1956; Brücher, 1969; Naranjo et al., 1982; de Azkue and Martínez, 1983, 1984, 1988; de Azkue, 2000), the members of the O. tuberosa alliance that have been studied are consistently reported to have small chromosomes, of roughly similar size (Brücher, 1969; Gibbs et al., 1978; de Azkue and Martínez, 1990; Valladolid et al., 1994; Valladolid, 1996). This observation parallels the low ITS sequence variation found within the alliance, contrasting with high levels of variation among divergent members of the genus overall (Tosto and Hopp, 1996; Emshwiller and Doyle, 1998). Thus, although I would be hesitant to extend this application of estimates of DNA content from flow cytometry to infer ploidy levels across the entire genus, where chromosome sizes vary greatly, the use of this technique to infer ploidy levels among members of the Oxalis tuberosa alliance is justified because of the relative uniformity of chromosome size reported among these species.
MATERIALS AND METHODS
Sampling
Most of the accessions of wild Oxalis used in this study were collected in Bolivia and Peru (Table 3), while cultivated oca accessions were kindly provided by the Programa de Investigación de la Papa (PROINPA), Cochabamba, Bolivia. Also included were a few purchased horticultural Oxalis plants that had been sampled in prior molecular studies (Emshwiller and Doyle, 1998) as well as some ‘volunteer’ seedlings that appeared in glasshouse collections. Some of the species determinations should be considered provisional (see Discussion). Although the recent treatment by Lourteig (2000) has greatly improved the taxonomy of the genus, some problems of species delimitation still remain. Pending further study, I provisionally retain the use of some names that Lourteig (2000) has reduced to synonymy, because the names reflect differences among the plants, such as molecular or ploidy differences, or morphological differences that are stable in common garden conditions. In a few cases, alternative names that have been reduced to synonymy by Lourteig (2000) are listed alongside the accepted name.
Table 3.
Accessions of Oxalis sampled for nuclear DNA content
| Accessionnumber* | Taxon | Latitude(°S) | Longitude(°W) | Altitude(m asl) | Provenance | Ploidy | DNAcontentpg/2C† | s.d. | Number ofmeasurements(number ofplants‡) |
| MHG847 | O.tuberosa Molina | 16°09′ | 69°04′ | 3850 | Bolivia, La Paz, Manco Kapac,Copacabana | 8x | 3·553 | 1(1) | |
| MHG861 | O.tuberosa Molina | 18°54′ | 66°46′ | 3750 | Bolivia, Oruro, Avaroa,Challapata | 8x | 3·693 | 1(1) | |
| MHG864 | O.tuberosa Molina | 19°19′ | 65°55′ | 3800 | Bolivia, Potosí, Tomás Frías,Yocalla | 8x | 3·664 | 0·184 | 2(1) |
| MHG865 | O.tuberosa Molina | 19°19′ | 65°55′ | 3800 | Bolivia, Potosí, Tomás Frías,Huancarani, Yocalla | 8x | 3·578 | 1(1) | |
| MHG884 | O.tuberosa Molina | 19°25′ | 65°44′ | 3800 | Bolivia, Potosí, Tomás Frías,Chiutara, Potosí | 8x | 3·543 | 0·119 | 2(1) |
| MHG891 | O.tuberosa Molina | 19°25′ | 65°44′ | 3600 | Bolivia, Potosí, Tomás Frías,Manquiri, Potosí | 8x | 3·551 | 1(1) | |
| MHG894 | O.tuberosa Molina | 19°47′ | 65°30′ | 4100 | Bolivia, Potosí,Linares, Potosí | 8x | 3·562 | 1(1) | |
| MHG897 | O.tuberosa Molina | 19°36′ | 65°24′ | 3650 | Bolivia, Potosí, Saavedra,Lagunillas, Betanzos | 8x | 3·678 | 1(1) | |
| 07 | O.tuberosa Molina | – | – | – | Bolivia | 8x | 3·623 | 1(1) | |
| Tiraque 10 | O.tuberosa Molina | – | – | – | Bolivia | 8x | 3·670 | 1(1) | |
| EE561 | O.phaeotricha Diels | 13°13′ | 72°18′ | 3300 | Peru, Cusco, Urubamba,Quebrada Tajcacabove Ollantaytambo | 4x | 1·673 | 0·022 | 3(1) |
| EE500 | O.picchensis R. Knuth | 13°34′ | 71°53′ | 3400 | Peru, Cusco, Cusco,San Jerónimo, Rdtoward Paruro | 4x | 1·676 | 0·059 | 3(1) |
| EE541 | O. spiralis R. &P. ex G. Don | 13°08′ | 72°31′ | 2400 | Peru, Cusco, Urubamba,Machu Picchu, Puente Ruinas | 2x | 1·280 | 0·115 | 2(2) |
| EE588 | O. spiralis R. &P. ex G. Don | 11°43′ | 75°04′ | Peru, Junín, Concepción,Comas | 2x | 1·216 | 0·039 | 3(1) | |
| EE602 | O. spiralis R. &P. ex G. Don | 12°00′ | 74°54′ | Peru, Junín, Huancayo,Road to Pariahuanca | 2x | 1·311 | 0·017 | 3(2) | |
| EE607 | O. spiralis R. &P. ex G. Don | 11°20′ | 75°39′ | Peru, Junín, Tarma,Acobamba—Chipián | 2x | 1·062 | 1(1) | ||
| EE618 | O. spiralis R. &P. ex G. Don | 11°15′ | 75°33′ | 2400 | Peru, Junín, Tarma,Rd to San Ramón, Carpapata | 2x | 1·106 | 1(1) | |
| EE620b | O. spiralis R. &P. ex G. Don | 11°10′ | 75°25′ | 1500 | Peru, Junín, Tarma,Rd to San Ramón, San Luis | 2x | 1·081 | 1(1) | |
| EE914 | O. spiralis R. &P. ex G. Don | 13°02′ | 72°02′ | 2800 | Peru, Cusco, Calca,Valley of Río Lares | 2x | 1·246 | 0·051 | 4(2) |
| EE927 | O. spiralis R. &P. ex G. Don | 13°08′ | 72°31′ | 2400 | Peru, Cusco, Urubamba,Aguas Calientes | 2x | 1·339 | 0·034 | 2(1) |
| Seedlings | O. spiralis R. &P. ex G. Don | – | – | – | seedlings appeared ingreenhouse collections | 2x | 1·115 | 0·022 | 6(6) |
| EE532 | O.cuzcensis R. Knuth | 13°29′ | 71°52′ | 3750 | Peru, Cusco, Calca, C.C. Picol | 2x | 1·173 | 0·026 | 4(2) |
| EE291 | O. unduavensis (Rusby)R. Knuth | 16°17′ | 67°48′ | 2940 | Bolivia, La Paz, Nor Yungas,Unduavi—Coroico Rd | 2x | 1·011 | 1(1) | |
| EE438 | O.unduavensis (Rusby)R. Knuth | 16°09′ | 68°07′ | 3050 | Bolivia, La Paz, Murillo,Zongo Valley | 2x | 1·031 | 0·052 | 3(3) |
| EE868 | O. sp. cfr. melilotoidesZuccarini | 13°05′ | 72°23′ | 2450 | Peru, Cusco, Urubamba,Lucumayu Valley, San Luis | 2x | 0·930 | 1(1) | |
| EE578 | O. petrophila R. Knuth | 11°49′ | 75°19′ | Peru, Junín, Concepción,La Esperanza, Ch’iqchi | 2x | 0·983 | 0·031 | 2(1) | |
| EE655 | O. sp. S | 14°27′ | 69°32′ | 3400 | Peru, Puno, Sandia,Cuyo Cuyo | 2x | 1·028 | 0·022 | 2(2) |
| EE706 | O. sp. S | 14°25′ | 69°29′ | 3000 | Peru, Puno, Sandia, belowCuyo Cuyo | 2x | 0·972 | 1(1) | |
| EE485 | O.lucumayensisR. Knuth ssp. lucumayensis | 13°06′ | 71°41′ | 2700 | Peru, Cusco, Paucartambo,below Challabamba | 2x | 0·929 | 1(1) | |
| EE289 | O.lucumayensis R. Knuthssp. subiens Lourteig | 16°17′ | 67°48′ | 2940 | Bolivia, La Paz, Nor Yungas,Unduavi—Coroico Rd | 2x | 0·919 | 1(1) | |
| EEx359 | O.lucumayensis R. Knuthssp. subiens Lourteig | 16°17′ | 67°50′ | 2700 | Bolivia, Cochabamba,Sud Yungas, Unduavi—Chulumani Rd | 2x | 0·986 | 1(1) | |
| EE447 | O.lucumayensis R. Knuthssp. subiens Lourteig | 16°16′ | 67°47′ | 2610 | Bolivia, La Paz, Nor Yungas,Unduavi—Coroico Rd | 2x | 0·890 | 1(1) | |
| EE294 | O.lucumayensis (hybrid?) | 16°17′ | 67°48′ | 2940 | Bolivia, La Paz, Nor Yungas,Unduavi—Coroico Rd | 2x | 0·998 | 1(1) | |
| MV s/n | O. sp. aff. lucumayensis | – | – | – | Peru, Puno, Sandia province | 2x | 0·907 | 1(1) | |
| VULC1§ | O. vulcanicola Donn. Sm. | – | – | – | ‘Central America’ purchasedfrom Merry Gardens | 2x | 0·886 | 1(1) | |
| EE604 | O.humbertii R. Knuth | 12°00′ | 74°54′ | 3300 | Peru, Junín, Huancayo,Chilifruta | 2x | 1·040 | 1(1) | |
| EE592 | O. sp. C | 11°43′ | 75°05′ | Peru, Junín, Concepción, Comas | 2x | 1·037 | 1(1) | ||
| EE777 | O.medicaginea Kunth | 06°56′ | 78°11′ | 3150 | Peru, Cajamarca, Celendín,Quilimbash | 2x | 0·836 | 1(1) | |
| EE809 | O.medicaginea Kunth | 06°14′ | 77°53′ | 2230 | Peru, Amazonas, Chachapoyas,Quebrada Molina | 2x | 0·803 | 1(1) | |
| EE810 | O.medicaginea Kunth | 06°15′ | 77°51′ | 2750 | Peru, Amazonas, Chachapoyas,Pumaurqu | 2x | 0·851 | 0·040 | 3(2) |
| EE783 | O.lotoides Kunth | 06°48′ | 77°56′ | 2720 | Peru, Amazonas, Chachapoyas,Marañón valley above Balsas | 2x | 0·881 | 1(1) | |
| EE789 | O.lotoides Kunth | 06°47′ | 77°55′ | 2930 | Peru, Amazonas, Chachapoyas,Marañón valley above Balsas | 2x | 0·864 | 0·035 | 2(2) |
| EE851 | O.lotoides Kunth | 07°24′ | 78°47′ | 3850 | Peru, Cajamarca, Contumazá,near Bosque Cachil | 2x | 0·826 | 0·014 | 2(1) |
| EE798 | O.mollis Kunth | 05°51′ | 77°58′ | 2070 | Peru, Amazonas, Bongará,Pedro Ruiz Gallo—La Florida Rd | 2x | 0·892 | 0·016 | 2(1) |
| EE356 | O.oulophora Lourteig | 16°18′ | 67°52′ | 2700 | Bolivia, La Paz, Sud Yungas,Unduavi—Chulumani Rd | 2x | 0·872 | 1(1) | |
| EE415 | O.oulophora Lourteig | 16°09′ | 68°07′ | 3160 | Bolivia, La Paz, Murillo,Valley of Río Zongo | 2x | 0·883 | 1(1) | |
| EE705 | O.marcapatensis R. Knuth | 14°25′ | 69°29′ | 3000 | Peru, Puno, Sandia,Ñakonki, below Cuyo Cuyo | 2x | 0·897 | 0·043 | 2(1) |
| EE910 | O.peduncularis Kunth | 13°12′ | 72°23′ | 2800 | Peru, Cusco, Urubamba,Misk’i Puquio, Pisqacucho | 2x | 0·985 | 0·055 | 4(2) |
| EE507 | O.peduncularis Kunth | 13°16′ | 72°11′ | 2900 | Peru, Cusco, Urubamba,Urubamba Valley, Yanahuara | 2x | 1·033 | 0·032 | 2(2) |
| EE512 | O.peduncularis Kunth | 13°14′ | 72°10′ | 3300 | Peru, Cusco, Urubamba,Quebrada Mant’anáyabove Yanahuara | 2x | 0·959 | 0·025 | 2(2) |
| EE514 | O.peduncularis Kunth | 13°14′ | 72°09′ | 3450 | Peru, Cusco, Urubamba,Quebrada Mant’anáyabove Yanahuara | 2x | 1·019 | 1(1) | |
| EE912 | O.peduncularis Kunth | 13°11′ | 72°23′ | 3490 | Peru, Cusco, Urubamba,Misk’i Puquio | 2x | 0·983 | 1(1) | |
| EE595 | O. peduncularis Kunth(O. ledigii R. Knuth) | 12°00′ | 75°09′ | Peru, Junín, Huancayo, Porbolín,km 13 from Huancayo | 2x | 0·980 | 0·024 | 2(1) | |
| EE746 | O. peduncularis Kunth(O. weberbaueri R. Knuth) | 09°00′ | 77°44′ | 3700 | Peru, Ancash, Huaylas,west side of Cordillera Blanca,Parón | 2x | 1·107 | 0·072 | 3(2) |
| EE752 | O. peduncularis Kunth(O. weberbaueri R. Knuth) | 09°00′ | 77°31′ | 3800 | Peru, Ancash, Yungáy,east side of Cordillera Blanca,Quebrada Ranicuráy | 2x | 1·055 | 1(1) | |
| EE753 | O. peduncularis Kunth(O. weberbaueri R. Knuth) | 09°05′ | 77°39′ | 3700 | Peru, Ancash, Yungáy,Llanganuco Lakes region | 2x | 1·157 | 1(1) | |
| EE850 | O. cfr. peduncularis Kunth | 07°24′ | 78°47′ | 2600 | Peru, Cajamarca, Contumazá,near Bosque Cachíl | 2x | 1·139 | 0·018 | 2(2) |
| EE853 | O. cfr. peduncularis Kunth | 07°23′ | 78°49′ | 2700 | Peru, Cajamarca, Contumazá,near Contumazá | 2x | 1·085 | 0·053 | 2(2) |
| EE778 | O.peduncularis Kunthvar. pilosa | 06°53′ | 78°07′ | 3140 | Peru, Cajamarca,Celendín, Jelig | 2x | 1·128 | 1(1) | |
| EE780 | O.peduncularis Kunthvar. pilosa | 07°06′ | 78°19′ | 3040 | Peru, Cajamarca,Cajamarca—Celedín Rd | 2x | 1·070 | 1(1) | |
| EE787 | O.peduncularis Kunthvar. pilosa | 06°50′ | 77°57′ | 1900 | Peru, Amazonas, Chachapoyas,Marañón valley above Balsas | 2x | 1·087 | 1(1) | |
| EE791 | O.peduncularis Kunthvar. pilosa | 06°48′ | 77°56′ | 2660 | Peru, Amazonas, Chachapoyas,Marañón valley above Balsas | 2x | 1·112 | 0·036 | 2(2) |
| EE792 | O.peduncularis Kunthvar. pilosa | 06°50′ | 77°57′ | 2030 | Peru, Amazonas, Chachapoyas,Marañón valley above Balsas | 2x | 1·053 | 1(1) | |
| EE804 | O.peduncularis Kunthvar. pilosa | 06°53′ | 78°07′ | 3150 | Peru, Cajamarca, Celendín, Jelig | 2x | 1·163 | 0·019 | 2(1) |
| EE807 | O. sp. V | 06°00′ | 77°57′ | 1520 | Peru, Amazonas,Bongará, Churuja | 2x | 1·043 | 0·037 | 2(2) |
| EE813 | O. sp. V | 06°13′ | 77°49′ | 2040 | Peru, Amazonas,Chachapoyas—Molinopampa Rd | 2x | 1·010 | 1(1) | |
| EE548 | O.paucartambensisR. Knuth | 13°54′ | 71°29′ | 3400 | Peru, Cusco, Quispicanchi,Paucarpata, near Cusipata | 2x | 0·894 | 0·020 | 2(2) |
| EE961 | O.paucartambensisR. Knuth | 13°24′ | 71°51′ | 3700 | Peru, Cusco, Calca,C.C. Viacha | 2x | 0·873 | 0·017 | 2(1) |
| EE962 | O.paucartambensisR. Knuth | 13°24′ | 71°51′ | 3700 | Peru, Cusco, Calca,C.C. Viacha | 2x | 0·864 | 1(1) | |
| EE865 | O.paucartambensisR. Knuth | 13°33′ | 71°52′ | 3250 | Peru, Cusco, Cusco,San Jerónimo | 2x | 0·959 | 1(1) | |
| EE553 | O.herrerae R. Knuth | 13°31′ | 71°58′ | 3400 | Peru, Cusco, Cusco,Cusco, Barrio San Blas | 2x | 0·867 | 1(1) | |
| EE560 | O.herrerae R. Knuth | 13°13′ | 72°18′ | 3300 | Peru, Cusco, Urubamba, QuebradaTajcac, above Ollantaytambo | 2x | 0·852 | 0·017 | 2(2) |
| EE911 | O.herrerae R. Knuth | 13°12′ | 72°23′ | 3400 | Peru, Cusco, Urubamba,Misk’i Puquio, Piscacucho | 2x | 0·815 | 0·033 | 2(1) |
| EE913 | O.herrerae R. Knuth | 13°03′ | 72°02′ | 3230 | Peru, Cusco, Calca, Velley ofRío Lares, Choquecancha | 2x | 0·926 | 0·027 | 2(2) |
| EE600 | O.ptychoclada Diels var.trichocarpa Lourteig | 12°00′ | 74°54′ | 3700 | Peru, Junín, Huancayo—Pariahuanca Rd, km 63,above Chilifruta | 2x | 0·950 | 0·020 | 2(1) |
| EE610 | O.ptychoclada Diels | 11°15′ | 75°35′ | 2600 | Peru, Junín, Tarma,Rd. to Huasahuasi | 2x | 0·837 | 0·008 | 2(2) |
| EE613 | O.ptychoclada Diels | 11°16′ | 75°37′ | 2400 | Peru, Junín, Tarma,Tarma—San Ramón Rd | 2x | 0·886 | 1(1) | |
| EE916 | O.ptychoclada Diels | 12°58′ | 72°04′ | 2500 | Peru, Cusco, Calca,Valley of Río Lares | 2x | 0·790 | 0·014 | 3(2) |
| EE545 | O. san‐migueliiR. Knuth | 13°08′ | 72°31′ | 2400 | Peru, Cusco, Urubamba,Machu Picchu, Puente Ruinas | 2x | 0·830 | 0·001 | 2(1) |
| EE920 | O.san‐migueliiR. Knuth | 13°08′ | 72°31′ | 2600 | Peru, Cusco, Urubamba,Machu Picchu | 2x | 0·874 | 1(1) | |
| EE926 | O. urubambensisR. Knuth | 13°08′ | 72°31′ | 2400 | Peru, Cusco, Urubamba,Aguas Calientes | 2x | 0·880 | 0·029 | 2(2) |
| EE511 | O. sp. cfr. teneriensisR. Knuth | 13°14′ | 72°10′ | 3200 | Peru, Cusco, Urubamba,Quebrada Mantanáyabove Yanahuara | 2x | 0·966 | 0·034 | 2(2) |
| EE504 | O. sp. A | 13°33′ | 72°34′ | 2100 | Peru, Cusco, Anta, near PuenteCunyac at Río Apurímac | 2x | 1·097 | 0·018 | 2(2) |
| EE786 | O.tabaconasensisR. Knuth | 06°42′ | 77°55′ | 2800 | Peru, Amazonas, Chachapoyas,above Leimebamba | 2x | 0·993 | 1(1) | |
| EE790 | O.tabaconasensisR. Knuth | 06°48′ | 77°55′ | 2700 | Peru, Amazonas, Chachapoyas,above Leimebamba | 2x | 1·051 | 0·009 | 2(1) |
| EE797 | O.tabaconasensis(O. oblongiformis)R. Knuth | 05°56′ | 77°59′ | 1460 | Peru, Amazonas, Bongará,Pedro Ruiz Gallo—La Florida Rd | 2x | 0·983 | 0·023 | 2(2) |
| EE605 | O. coralleoidesR. Knuth | 11°48′ | 75°37′ | Peru, Junín, La Oroya—Jauja Rd., Km 55 | 2x | 1·064 | 1(1) | ||
| HERR1§ | O. herreraeR. Knuth | – | – | – | ‘Peru’ purchased fromMerry Gardens | 2x | 0·794 | 1(1) | |
| PED1§ | O. peduncularis Kunth | – | – | – | ‘Peru, Ecuador’ purchased fromMerry Gardens | 2x | 0·927 | 1(1) | |
| Hybrid? | O. peduncularis ×O. herrerae? | – | – | – | Seedling that appearedin glasshouse collections whenonly purchased plants werepresent | 2x | 0·873 | 0·014 | 2(1) |
| EE921¶ | O.boliviana Britton | 13°08′ | 72°33′ | 2400 | Peru, Cusco, Urubamba,Machu Picchu | 2x | 0·957 | 1(1) | |
| EE773¶ | O.megalorrhiza Jacquin | 09°12′ | 77°42′ | 2700 | Peru, Ancash, Yungáy,below Guitarrero Cave | 2x | 0·814 | 1(1) |
Flow cytometry
Initial attempts to prepare samples using MgSO4 buffer (Arumuganathan and Earle, 1991) or Lysis Buffer LB01 (Doležel et al., 1989; Galbraith et al., 1997) did not produce satisfactory results, presumably due to the acidity of the Oxalis leaves (pH approx. 1·7), which may have exceeded the buffering capacity of the solutions. Successful sample preparation followed a two‐step protocol (Otto, 1990; Doležel and Göhde, 1995; Galbraith et al., 1997; see also Dansi et al., 2001) with simplifying modifications as posted at http://www.ueb.cas.cz/olomouc1/lcgcm/Flow/protocol/andc3.htm. Typically, a small amount of young leaves was chopped with a new razor blade in 0·5 ml of ice‐cold ‘Otto I’ solution [0·1 m citric acid monohydrate, 0·5 % (v/v) Tween 20] in a Petri dish (also over ice). In some cases about 0·5 ml of ice‐cold Otto I solution was added and mixed well with a pipette, whereas in later preparations less additional solution was added to avoid diluting the nuclei. The suspension was filtered through 50‐µm nylon mesh, and incubated at room temperature from 1 h to overnight. Centrifugation was not effective in concentrating the nuclei, so it was not continued. Two volumes of ‘Otto II’ solution (0·4 m Na2HPO4·12H2O containing 2 µl ml–1 β‐mercaptoethanol) containing propidium iodide and RNase (each at a final concentration of 50 µg ml–1) were added to the samples just before they were analysed in a cytometer (within 5 min).
Samples were run on a Becton‐Dickinson FACSCalibur flow cytometer operated by the Cornell School of Veterinary Medicine, with an argon laser exciting at 488 nm. Pulse area was detected using FL2‐A (585 mean/42 bandwidth) with a threshold at FLS 35. CellQuest software (Apple version 3·1·f 3·1·3) was used to visualize and measure histogram peaks. Initial samples used co‐chopped rice (Oryza sativa) leaves as an internal standard, but the fluorescence peak for this standard was superimposed on those of some of the diploid Oxalis, so this standard was not continued. Subsequent samples used erythrocytes from a young Babcock B300 (egg‐laying breed) chicken (CRBCs) as internal standard. Although plant standards are preferable for determination of plant nuclear DNA content in absolute units (Johnston et al., 1999; R. Price, pers. comm.; J. Doležel, pers. comm.), CRBCs are an adequate standard for ploidy determinations, and offer convenience because a single blood sample can be used for all runs. The blood was used within 2 weeks of collection, kept in a heparinized tube and diluted for use with ‘CRBC buffer II’ [140 mm NaCl, 5 % (v/v) Triton X‐100; see recipes at: http://www.ueb.cas.cz/olomouc1/lcgcm/Flow/protocol/recept.htm]. The fluorescence of CRBCs declined relative to that of the sample within 1 or 2 h (data not shown), which might indicate that the propidium iodide concentration was not optimal. Therefore all results reported here are from samples that were run as quickly as possible after addition of ‘Otto II’ solution and CRBCs. The genome size of each sample was calculated using 2·5 pg as the DNA content of CRBCs, which is an approximate mean of several reported estimates (Tiersch and Wachtel, 1991; Johnston et al., 1999; R. Price, pers. comm.; J. Doležel, pers. comm.). Although the use of animal standards for plant samples may cast some doubt upon these estimates as absolute values, the use of a consistent methodology for all samples and the repeatability of measurements of the same accessions allows for comparison of similarities and differences among species and populations.
RESULTS
Quality and reliability of the flow cytometric data
In addition to the linearity tests conducted daily by the flow cytometry facility, the instrument linearity was also monitored by comparing the proportionality of peaks of different ploidy in the same sample to the ideal ratio (see Vilhar et al., 2001). In general, the proportionality over most runs was 1–1·5 % below the expected ratio of two. Specifically, the ratio of 4C to 2C peaks over all runs averaged 1·979, and that of 8C to 4C peaks averaged 1·972. Most coefficient of variation (CV) values of the sample 2C peaks ranged between 2·3 and 7·5 (mean 5·44), with a few higher values (up to 10·8). The higher CV values were usually in samples with high background. High background noise is not unusual in the lower channel numbers (e.g. Kudo and Kimura, 2001a, b), and has been alternatively ascribed to broken cells damaged during the extraction procedure (Lagunes‐Espinoza et al., 2000) or to autofluorescence of chloroplasts in the cytosol (Galbraith et al., 1997). In addition to this background signal, anomalous additional smaller peaks were observed in profiles of several samples of older leaves of oca, but these were later confirmed to derive from contamination by glasshouse whitefly larvae (2C approx. 0·89 pg, 4C approx. 1·78 pg). All data described below, unless otherwise indicated, are based on samples that included chicken erythrocytes as internal standards.
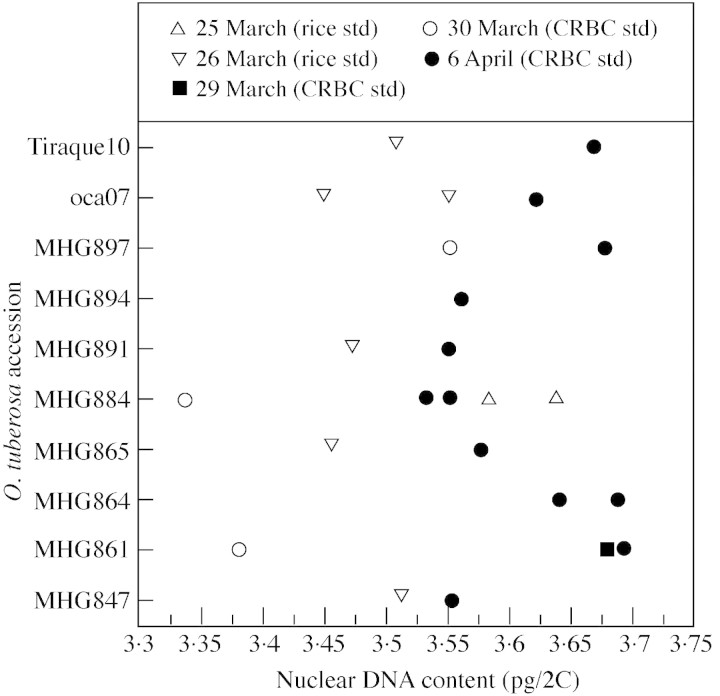
Multiple preparations from the same plant run on different days sometimes varied at least as much as the samples from different plants of the population, possibly due to instrumental fluctuations (see Kudo and Kimura, 2001a). With the exception of cultivated O. tuberosa accessions, this fluctuation was usually less than 0·1 pg. In the case of cultivated oca, most of the ten accessions were sampled on more than 1 d, using either CRBCs or rice leaves as internal standards. The day‐to‐day variation in DNA content estimates exceeded both the variation among oca accessions and that between values estimated with the two different standards (Fig. 1). Because averaging the measurements from different days into a separate mean for each accession would distort and exaggerate the differences among them, the values shown in Table 3 are those from the only day on which all oca accessions were sampled (6 Apr. 1999).
Fig. 1. Estimated values of 2C nuclear DNA content for the ten accessions of cultivated oca measured on different days, using either CRBCs or rice leaves as internal standard (0·87 pg/2C used as value for rice standard). The day‐to‐day variation (e.g. between samples run on 6 April and those run on 30 March) was greater for the cultivated O. tuberosa accessions than for wild Oxalis samples, for unknown reasons, but did not depend on the standard used. Only the values from 6 Apr. 1999 are shown in Table 3, because only on that day were all ten samples measured. The mean value of all oca accessions measured on that day is 3·610 pg/2C (s.d. = 0·061), whereas the mean for all days is 3·514 pg/2C (s.d. = 0·116) and the mean of those using rice leaves as internal standard was 3·524 pg/2C (s.d. = 0·066).
Utility of flow cytometry to infer ploidy levels of members of the O. tuberosa alliance
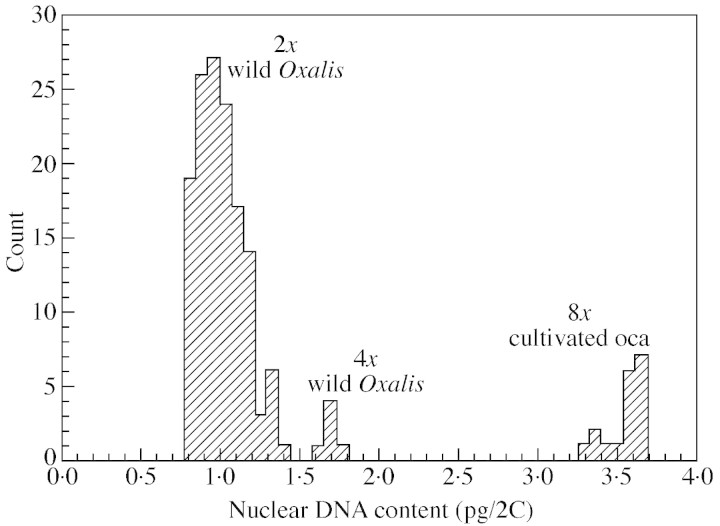
DNA ploidy levels of samples were determined by comparing their DNA contents with those of taxa of known ploidy level in various ways. The distribution of DNA content estimates for species in the alliance is discontinuous (Fig. 2), so that although there is some variation in the contents of diploid species (see below), ploidy levels can be easily inferred. The range of values observed for cultivated oca in Fig. 2 is mostly due to day‐to‐day variation, as discussed above. All estimates for cultivated O. tuberosa accessions fell in the octoploid range, with approx. four‐times the DNA content of the diploids. Specifically, cultivated O. tuberosa accessions measured on 6 Apr. 1999 varied from 3·543 to 3·693 pg (Table 3), with a mean value of 3·610 (mean for all days = 3·514, see Fig. 1). A quarter of these 2C values for oca would be 0·903 for 6 April (or 0·879 for all measurements), very close to the modal value for the diploid species (Fig. 2). Two of the wild Oxalis accessions, O. phaeotricha Diels (EE561) and O. picchensis R. Knuth (EE500), had similar 2C values of approx. 1·67 pg, considered to be in the tetraploid range. The remainder of the species in the O. tuberosa alliance had DNA contents that varied from 0·790 to 1·339 pg. These included species reported to have diploid chromosome counts (see Table 1). None of the accessions sampled appeared to be hexaploid.
Fig. 2. Distribution of all estimates of 2C values in samples of members of the Oxalis tuberosa alliance that were tested with CRBCs as internal standard in this study. The discontinuities between the ranges of diploid, tetraploid and octoploid values make them easily distinguishable. The day‐to‐day variation among the octoploid cultivated samples (see text and Fig. 1) contributes to the spread of values among those samples.
The polyploid status of O. tuberosa and O. picchensis was also confirmed by comparing the ratio of their fluorescence peaks with that of a diploid accession run together in the same tube (albeit not co‐chopped). Polyploids frequently have DNA contents that are somewhat below the ratio ‘expected’ from closely related diploids (Lysák and Doležel, 1998; Bennett et al., 2000). For instance, a tetraploid may have a DNA content somewhat less than twice that of closely related diploids. Not surprisingly, this situation seems to be the case in the O. tuberosa alliance. The ratio of the fluorescence peaks of two oca accessions, 07 and MHG847, to that of co‐run diploid O. peduncularis Kunth (accession PED1) was 3·830 and 3·941, respectively, i.e. close to four‐times the diploid, confirming their octoploid status. Tetraploidy of accession EE500 of O. picchensis was corroborated by running simultaneously with accession EE916, tentatively identified as the diploid O. ptychoclada Diels, resulting in a ratio of fluorescence peaks of 2·079. This ratio is very close to two, rather than below two, but this particular diploid species had one of the smallest genome sizes among the alliance members sampled in this study. These co‐run samples offered an additional opportunity to confirm the internal consistency of measurements by calculating the DNA contents of the polyploids based on the co‐run diploids as calibration standards (the diploids having been previously estimated using CRBC or rice internal standards). These tests resulted in estimates very similar to the direct measurements of the same polyploids (i.e. 2C estimates of 3·65 pg and 3·55 pg for two accessions of O. tuberosa based on PED1 as standard, and 1·65 pg for O. picchensis based on EE916 as standard).
In a few cases the DNA ploidy found in this study differs from ploidy levels reported previously for the same species. These cases may reflect either differences in species identifications or real variation in cytotypes. Although both are inferred in this study to be diploid, different cytotypes have been reported (see citations in Table 1) for O. lotoides Kunth (4x) and O. spiralis R. & P. ex G. Don (both 2x and 6x). Both of these are widespread species, and O. spiralis has a wide ecological amplitude as well (see below), so the finding of diploid levels in the accessions sampled here would not preclude the possibility of other cytotypes in these species. On the other hand, differing species delimitations are relevant in the case of accession EE532, identified as O. cuzcensis, which is here inferred to be DNA diploid (Table 3). I tentatively retain the name O. cuzcensis, which is reduced by Lourteig (2000) to synonymy with O. nubigena Walpers, a species that would be hexaploid according to the chromosome count of Diers (1961).
Endopolyploidy
In some of the Oxalis samples a cluster of nuclei formed a visible 8C peak. This suggests that these leaf tissues exhibit some endopolyploidy, an increase in DNA content due to endomitosis (or endoreduplication; i.e. somatic duplication of chromosomes without nuclear division). In most cases in which such a peak was discernible, the 8C peak was small relative to the 2C and 4C peaks [i.e. the number of ‘events’ (i.e. nuclei) was usually only 2–5 % of the number in the 2C peak, although in a very few samples it reached nearly 8 %). Scatter plots of FL2‐A vs. FL2‐W (pulse area vs. pulse width) of a few representative samples with relatively large 8C peaks were examined and gated (‘doublet discrimination’) to eliminate the possibility that these larger 8C peaks represented aggregated nuclei. The latter possibility is also unlikely as there was no peak in a position that would correspond to 6C (see De Rocher et al., 1990).
Endopolyploidy has been reported in many plant taxa (see Lagunes‐Espinoza et al., 2000; Kudo and Kimura, 2001a, b, and references therein), and is especially associated with large epidermal cells (Melaragno et al., 1993) and succulents with small genomes (De Rocher et al., 1990). Members of the O. tuberosa alliance all have somewhat succulent leaves with very large epidermal cells, and are often succulent throughout the plant, so the occurrence of endopolyploidy in the alliance is concordant with observations in other succulents. Endopolyploidy may explain the giant nuclei reported in the carpel mesophyll of O. ptychoclada and O. spiralis as well as in the different types of glandular hairs on the anther filaments, sepals and pedicels of the latter species (Huynh, 1965). Recent reports of endoreduplication in the pod wall of legumes (Lagunes‐Espinoza et al., 2000) may refer to a similar phenomenon to the giant nuclei observed by Huynh (1965) in carpel mesophyll in these two Oxalis species. Although O. spiralis rarely showed an 8C peak in flow cytometric profiles, this is not surprising because only vegetative tissues were sampled for this study, rather than the inflorescence organs in which glandular hairs occur in this species.
DISCUSSION
Octoploid Oxalis tuberosa
The finding of octoploid‐level DNA contents in these ten cultivated O. tuberosa accessions is consistent with the reports of 2n = 8x = 64 in well over 100 samples of cultivated oca from diverse areas of the Andes (de Azkue, 1990; Medina Hinostroza, 1994; Valladolid et al., 1994; Valladolid, 1996; Vinueza Vela, 1997). The finding of octoploid levels in cultivated oca is also consistent with previous evidence from ncpGS data, in which multiple sequence types (putative homologues) found within individual plants show fixed heterozygosity and join separate parts of the gene tree, suggesting that oca is allopolyploid and perhaps autoallopolyploid (Emshwiller, 1999a; Emshwiller and Doyle, 2002). The flow cytometric data for the ten oca accessions sampled here showed no evidence of euploid variation as reported by Guamán Calderón (1997), nor lower diploid levels consistent with the reported counts of 2n = 14 (e.g. Heitz, 1927; Talledo and Escobar, 1995). On the other hand, these flow cytometric data alone would not provide strong evidence against the report by Hayano Kanashiro (1998) that oca is 2n = 7x = 49, because heptaploid and octoploid DNA content levels might be difficult to distinguish. However, molecular, cytological and morphological data confirm oca’s close relationship with a group of species that are all based on x = 8 (de Azkue and Martínez, 1990; Tosto and Hopp, 1996; Emshwiller and Doyle, 1998, 2002; Emshwiller, 1999b). In addition, although some genotypes show a high proportion of inviable pollen (Gibbs, 1976; Trognitz et al., 2000), oca is somewhat fertile, producing copious seed in at least some circumstances (Alandia Borda, 1967; Panti Pacheco, 1972; Vallenas Ramírez, 1992; Pallares Ponce, 1998; Trognitz et al., 1998; Trognitz and Hermann, 2001), whereas odd polyploids are usually sterile (Allard, 1960). The genome size for oca reported here does not negate the possibility of aneuploidy in oca, as reflected in the slight variation in chromosome numbers in some reports (Kostoff et al., 1935; Cárdenas and Hawkes, 1948; Gibbs et al., 1978). Vegetative propagation and dispersal by humans mean that oca may not be under the same selection pressures to regain fertility by reducing meiotic abnormalities that operate in seed‐propagated species, and so some aneuploid genotypes might be preserved. However, the observation by Vinueza Vela (1997) of chromosome breakage at the nucleolus organizer region (NOR; often observed as a secondary constriction in metaphase chromosomes) might explain some of the reports of numbers above 64. Because of the small number of cultivated oca accessions sampled here, it is also possible that different accessions might have other ploidy levels. The report by Guamán Calderón (1997) of other euploid numbers is particularly interesting because the accessions she studied were from areas of Bolivia where wild tuber‐bearing Oxalis grows in close proximity to cultivated oca (pers. obs.). The existence of different ploidy levels in cultivated oca in this region would be significant because it might indicate possibilities such as crop/wild hybridization across ploidy levels, multiple domestications or the persistence of earlier domesticated forms of oca. Thus these reports should be confirmed through measurement of genome size and/or independent chromosome counts.
Tetraploid O. picchensis and O. phaeotricha
Oxalispicchensis, here inferred to be tetraploid, is one of the wild tuber‐bearing taxa identified by ncpGS data as a possible genome donor of octoploid oca (Emshwiller, 1999a; Emshwiller and Doyle, 2002). Oxalispicchensis might be autotetraploid because there was little variation in the ncpGS sequence in the two plants sampled. Oxalis phaeotricha, on the other hand, had multiple sequences of ncpGS within an individual plant (E. Emshwiller, unpubl. res.), so it might be allopolyploid. The latter species was not included in the study of oca’s origins cited above because of technical difficulties in determining its DNA sequences for ncpGS, and because enough of the sequence was available to exclude it as one of the progenitors of oca.
Information regarding the ploidy level of O. picchensis has proved useful in the study of the origins of octoploidy of O. tuberosa, by helping to eliminate some of the competing hypotheses about oca’s origins that were consistent with ncpGS data (Emshwiller, 1999a; Emshwiller and Doyle, 2002). For instance, hypotheses of the origins of polyploidy in O. tuberosa (e.g. Fig. 5·7 in Emshwiller, 1999a) included the possibility that oca arose through hybridization of a diploid O. picchensis with a hexaploid Bolivian wild tuber‐bearing taxon (i.e. 2x + 6x = 8x), a scenario that could involve polyploidization through either asexual or bilateral sexual means (sensu Mendiburu and Peloquin, 1976). This hypothesis is now eliminated by the finding that O. picchensis is tetraploid. However, no living plants were available to determine ploidy of the other putative genome donor of octoploid oca, namely Bolivian populations of wild tuber‐bearing Oxalis of a yet unnamed taxon. Thus, several alternative scenarios still remain (because various modes of polyploidization are possible, as reviewed by Mendiburu and Peloquin, 1976).
Comparison of DNA content with phylogenetic relationships
The variation in nuclear DNA content among Oxalis species sampled in this study did not form a pattern according to phylogenetic relationships, either in terms of comparisons of the alliance members with outgroup taxa or among groups within the alliance. Sampling included two Andean species, O. megalorrhiza Jacquin and O. boliviana Britton, which are not members of the O. tuberosa alliance, as indicated by their placement outside of the alliance clade in the results of phylogenetic analyses of ITS and ncpGS data (Emshwiller and Doyle, 1998, 2002). Both of these taxa were found to have DNA contents similar to those of diploid members of the O. tuberosa alliance. Chromosome numbers for Oxalis megalorrhiza (under the name O. carnosa Molina, a name frequently misapplied to O. megalorrhiza) are reported to be either 14 (Heitz, 1926, cited in Federov, 1974) or 18 (de Azkue, 2000). Oxalis boliviana has an unknown chromosome number.
There is also no pattern with respect to the subclades within the Oxalis tuberosa alliance clade that were found in phylogenetic analyses of ncpGS data (Emshwiller and Doyle, 2002). There is nearly complete overlap in the ranges of DNA contents of diploid species that joined each of the subclades within the alliance in that study. In some cases species that were sisters on the ncpGS gene tree have quite divergent DNA contents (e.g. accessions EE916 and EE504).
Intraspecific variation within diploid members of the alliance
With a few exceptions to be discussed below, the estimates for different accessions that were identified as the same species usually varied by no more than approx. 0·1 pg, i.e. no more than the day‐to‐day variation in measurements of the same plant (see Fig. 3, in which taxa are arranged in groups for purposes of the following discussion). Most cases of greater intraspecific variation are those in which there is variation in the tristylous breeding system (see below) or considerable morphological and/or ecological variation among the plants or populations. Thus, the intraspecific variation would be considered ‘orthodox’ in the sense of Greilhuber (1998), who discusses genome size differentiation among populations that are separated by geographical barriers into distinct reproductive communities. Andean environments are extremely heterogeneous in altitude, rainfall and other environmental variables over very short geographic distances (Frère et al., 1975). Gene flow among populations may be limited by the distances between appropriate habitats, or else it may have been intermittent in the past, as vegetation zones shifted during Pleistocene cycles of glaciation (Vuilleumier, 1971). Here, intraspecific sampling was not designed to test for correlation of nuclear DNA content with any environmental parameters (see review in Poggio et al., 1998), and the interaction of multiple factors might make interpretation of such patterns complex.
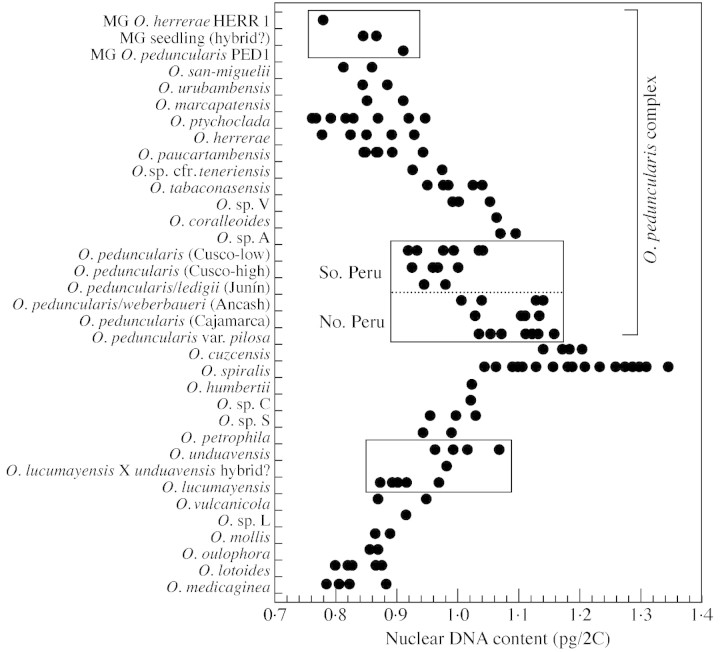
Fig. 3. Estimates of 2C values of diploid species in the O. tuberosa alliance, arranged to compare variation among and within species. The middle box illustrates one of several examples in which ‘lumping’ populations into a single species, O. peduncularis, may contribute to artificially high ‘intraspecific’ variation in genome size. The other two boxes illustrate cases of possible hybrids that have DNA contents intermediate between their putative parents. The ‘O. peduncularis complex’ indicates a group of morphologically similar taxa in which species boundaries are problematic. In prior analyses of ncpGS data (Emshwiller and Doyle, 2002), all of the accessions in this complex that were sampled for ncpGS joined the subclade designated ‘O. peduncularis clade’, which also included the sequence of O. cuzcensis. The members of the ‘O. peduncularis complex’ do not differ in their range of nuclear DNA contents from the other alliance members sampled, most of which joined the other subclade within the alliance, designated the ‘O. lotoides clade’ (other than O. petrophila, which joined neither subclade; see Emshwiller and Doyle, 2002).
Assessment of intraspecific variation in species of the O. tuberosa alliance is complicated by the aforementioned problems of species delimitation that still remain despite the clarification of Oxalis taxonomy by Lourteig (2000). The situation in some species complexes in the alliance seems to reflect the problem described by Greilhuber (1998), who asserts that intraspecific variation in genome size ‘can be a taxonomical artefact’ and provides the example of Scilla biflora L. s.l., which if ‘treated as one species, shows a two‐fold genome size variation, but if split up taxonomically, intraspecific variation diminishes to hardly more than methodological error’. The group designated here as the O. peduncularis complex is one in which these problems are especially evident. This group differs morphologically and ecologically from the rest of the alliance (Emshwiller, 1999a, b, 2002), being found in drier conditions (such as inter‐Andean valleys) than the moist forest habitats of the other alliance members. All populations in this complex join the same subclade in the ncpGS gene tree (designated ‘O. peduncularis clade’ in Emshwiller, 1999a, 2002; Emshwiller and Doyle, 2002). However, this clade also includes sequences of species that do not share the same morphological characteristics, i.e. O. picchensis and O. cuzcensis, so these species are not included here in the O. peduncularis complex.
The O. peduncularis complex corresponds roughly, but not exactly, with the taxa classified by Lourteig (2000) in section Herrerae, in which she included four species, with one subspecies and two varieties (plus autonyms), and reduced several previously published species to synonymy. However, my study of morphology of species in the complex led me to the opinion that this group includes more than four species, and that not only should some of the species names that were reduced by Lourteig be retained, but additional names may need to be proposed. Recognizing only four species in this complex would result in even higher ‘intraspecific’ variation in DNA content than that seen within the species as provisionally determined here (Fig. 3). Thus, the situation in the O. peduncularis complex/section Herrerae sensu Lourteig (2000) is similar to the case of Scilla cited by Greilhuber (1998). As an illustration, populations of O. peduncularis from different parts of Peru are separated in Fig. 3 by region of origin and variety. Populations of O. peduncularis in northern Peru have higher values (albeit overlapping) than those in southern Peru (P < 0·001), suggesting that these populations are separated by distance into at least separate gene pools, if not separate species. Genome sizes of O. paucartambensis R. Knuth and O. peduncularis var. pilosa are non‐overlapping (Fig. 3), supporting the retention of the former as a distinct species (contra Lourteig, 2000). On the other hand, morphologically different populations from high and low elevations in Cusco Department have similar DNA contents (Fig. 3). With sampling directed specifically toward these questions, estimates of nuclear genome size may be able to complement other data in helping to resolve some of the problems of species delimitation in the alliance.
The O. spiralis complex also has high intraspecific variation in genome size, which may have taxonomic implications. The plants identified here as O. spiralis not only had the largest genome sizes among the diploid species sampled, but also the widest range of apparently intraspecific variation (Fig. 3). Yet the plants sampled include little of this taxon’s broad range of morphological, geographical and ecological variation (Emshwiller, 2002), and none of the hexaploid cytotypes that have been reported for O. spiralis (see citations in Table 1). More thorough sampling in the O. spiralis complex may elucidate the relationship of DNA content to variation in environmental factors or species and/or population boundaries.
This variation in genome size in O. spiralis might be related to the tristylous breeding system. Some populations are tristylous (the norm in Oxalis), with individuals of short‐styled (S), mid‐styled (M) and long‐styled (L) forms, whereas others are semi‐homostylous and apparently self‐compatible (e.g. accession EE607 and the seedling plants that appeared in glasshouse collections). Semi‐homostyled plants also occur in other members of the O. tuberosa alliance, including cultivated oca (pers. obs., see also Carrión et al., 1995). The semi‐homostylous plants had the smaller DNA contents among the sampled O. spiralis. In some other alliance species, individuals of different style morph differed in DNA content, but results were equivocal and sampling was insufficient to test significance. Although flow‐cytometry has elucidated dimorphism in genome size in dioecious taxa and has been used in plant sex‐determination (Costich et al., 1991; Doležel and Göhde, 1995), I am not aware of any studies assessing genome size differences in heterostylous taxa.
The possibility of hybridization, suggested by the observation of plants of intermediate morphology, complicates the species delimitation problems in some parts of the alliance. Two suspected hybrid Oxalis included in this study (see boxes in Fig. 3) each had estimated 2C‐values intermediate between those of their putative parents, similarly to the interspecific hybrids in Alstroemeria L. studied by Buitendijk et al. (1997). One was a seedling that was suspected to be a hybrid between the purchased plants of O. peduncularis (PED1) and O. herrerae (HERR1), which had a DNA content intermediate between that of those accessions. The second plant, EE294, had both an intermediate morphology and an intermediate DNA content between O. lucumayensis R. Knuth ssp. subiens Lourteig and O. unduavensis R. Knuth, two species present in the area in which EE294 was collected.
ACKNOWLEDGEMENTS
The author thanks G. Ansah, K. Arumuganathan, D. Costich, E. Earle, R. Getchell, A. Mora, R. Price and especially J. Doležel for advice and/or technical training; IBTA and INRENA for permission to export plants from Bolivia and Peru; PROINPA, especially M. L. Ugarte, for the use of cultivated oca accessions; J. J. Doyle for laboratory space for sample preparation; the Cornell Greenhouse Consortium for caring for the plants; and the following people (in chronological order) for help with logistics and/or field collections in Bolivia and Peru: M. G. Asbun‐Claros, J. Miller, M. L. Ugarte, F. Terrazas, S. Guamán, T. Villarroel, J. Almanza, R. Vargas, B. Eriksen, U. Molau, A. Valladolid, G. Meza, P. Cruz, A. Castelo, A. Tupayachi, P. Núñez, R. Urrunaga, H. Flores, R. Estrada, R. Ortega, C. Arbizu, M. Ramírez, A. Andia, F. Vivanco, N. Arce, L. Torrez, M. Vallenas and I. Sánchez‐Vega. This study was made possible by the generosity of a grant from Abbott Laboratories to The Field Museum, with additional financial support from the U.S. National Science Foundation, Cornell University and a J. W. Fulbright Grant.
Supplementary Material
Received: 5 November 2001; Returned for revision: 21 January 2002; Accepted: 11 March 2002.
References
- Alandia BordaS.1967. Producción de semilla sexual en oca. Sayaña (Bolivia)2: 12–15. [Google Scholar]
- AllardRW.1960. Principles of plant breeding. New York: John Wiley & Sons. [Google Scholar]
- ArumuganathanK, Earle ED.1991. Estimation of nuclear DNA content of plants by flow cytometry. Plant Molecular Biology Reporter 9: 229–233. [Google Scholar]
- BennettMD, Bhandol P, Leitch IJ.2000. Nuclear DNA amounts in angiosperms and their modern uses: 807 new estimates. Annals of Botany 86: 859–909. [Google Scholar]
- BraeutigamS, Braeutigam E.1996. Determination of the ploidy level in the genus Hieracium subgenus Pilosella (Hill) S.F. Gray by flow cytometric DNA analysis. Folia Geobotanica & Phytotaxonomica 31: 315–321. [Google Scholar]
- BrücherH.1969. Poliploidia en especies sudamericana de Oxalis Boletín de la Sociedad Venozolana de Ciencias Naturales 28: 145–178. [Google Scholar]
- BrummerEC, Cazcarro PM, Luth D.1999. Plant genetic resources: Ploidy determination of alfalfa germplasm accessions using flow cytometry. Crop Science 39: 1202–1207. [Google Scholar]
- BuitendijkJH, Boon EJ, Ramanna MS.1997. Nuclear DNA content in twelve species of Alstroemeria L. and some of their hybrids. Annals of Botany 79: 343–353. [Google Scholar]
- CárdenasM, Hawkes JG.1948. Número de cromosomas de algunas plantas nativas cultivadas por los indios en los Andes. Revista de Agricultura Bolivia 5: 30–32. [Google Scholar]
- CarriónS, Hermann M, Trognitz B.1995. La biología reproductiva de la oca (Oxalis tuberosa Molina). Boletín de Lima 98: 48–68. [Google Scholar]
- CostichDE, Meagher TR, Yurkow EJ.1991. A rapid means of sex identification in Silene latifolia by use of flow cytometry. Plant Molecular Biology Reporter 9: 359–370. [Google Scholar]
- CostichDE, Ortiz R, Meagher TR, Bruederle LP, Vorsa N.1993. Determination of ploidy level and nuclear DNA content in blueberry by flow cytometry. Theoretical and Applied Genetics 86: 1001–1006. [DOI] [PubMed] [Google Scholar]
- DansiA, Mignouna HD, Pillay M, Zok S.2001. Ploidy variation in the cultivated yams (Dioscorea cayenensis‐Dioscorea rotundata complex) from Cameroon as determined by flow cytometry. Euphytica 119: 301–307. [Google Scholar]
- de AzkueD.2000. Chromosome diversity of South American Oxalis (Oxalidaceae). Botanical Journal of the Linnean Society 132: 143–152. [Google Scholar]
- de AzkueD, Martínez A.1983. The chromosome complements of shrubby Oxalis species from South America. Plant Systematics and Evolution 141: 187–197. [Google Scholar]
- de AzkueD, Martínez A.1984. Variación del cariotipo, volumen nuclear y contenido de ADN en siete especies de Oxalis Darwiniana 25: 267–277. [Google Scholar]
- de AzkueD, Martínez A.1988. DNA content and chromosome evolution in the shrubby Oxalis Genome 30: 52–57. [Google Scholar]
- de AzkueD, Martínez A.1990. Chromosome number of the Oxalis tuberosa alliance (Oxalidaceae). Plant Systematics and Evolution 169: 25–29. [Google Scholar]
- De RocherEJ, Harkins KR, Galbraith DW, Bohnert HJ.1990. Developmentally regulated systemic endopolyploidy in succulents with small genomes. Science 250: 99–101. [DOI] [PubMed] [Google Scholar]
- DiersL.1961. Der Anteil an Polyploiden in den Vegetationsgürteln der Westkordillere Perus. Zeitschrift für Botanik 49: 437–488. [Google Scholar]
- DoleželJ.1997. Application of flow cytometry for the study of plant genomes. Journal of Applied Genetics 38: 285–302. [Google Scholar]
- DoleželJ, Göhde W.1995. Sex determination in dioecious plants Melandrium album and M. rubrum using high‐resolution flow cytometry. Cytometry 19: 103–106. [DOI] [PubMed] [Google Scholar]
- DoleželJ, Binarova P, Lucretti S.1989. Analysis of nuclear DNA content in plant cells by flow cytometry. Biologia Plantarum 31: 113–120. [Google Scholar]
- EmshwillerE.1999a Origins of domestication and polyploidy in the Andean tuber crop Oxalis tuberosa Molina (Oxalidaceae). PhD Thesis, Cornell University, USA. [Google Scholar]
- EmshwillerE.1999b The relationships of Peruvian Oxalis species to cultivated oca. Arnaldoa 6: 117–139. [Google Scholar]
- EmshwillerE.2002. Biogeography of the ‘Oxalis tuberosa alliance’. Botanical Review 68: 128–152. [Google Scholar]
- EmshwillerE, Doyle JJ.1998. Origins of domestication and polyploidy in oca (Oxalis tuberosa: Oxalidaceae): nrDNA ITS data. American Journal of Botany 85: 975–985. [PubMed] [Google Scholar]
- EmshwillerE, Doyle JJ.1999. Chloroplast‐expressed glutamine synthetase (ncpGS): potential utility for phylogenetic studies with an example from Oxalis (Oxalidaceae). Molecular Phylogenetics and Evolution 12: 310–319. [DOI] [PubMed] [Google Scholar]
- EmshwillerE, Doyle JJ.2002. Origins of domestication and polyploidy in oca (Oxalis tuberosa: Oxalidaceae) 2: Chloroplast‐expressed glutamine synthetase (ncpGS) data. American Journal of Botany 89 (in press). [DOI] [PubMed] [Google Scholar]
- FavargerC, Huynh K.1965. Chromosome number reports IV. Taxon 14: 86–92. [Google Scholar]
- FederovAA.1974. Chromosome numbers of flowering plants Leningrad: Academy of Sciences. [Google Scholar]
- FrèreM, Rea J, Rijks JQ.1975. Estudio agroclimatológico de la zona andina. Rome: United Nations Food and Agriculture Organization (FAO), United Nations Educational, Scientific and Cultural Organization (UNESCO), Organización Meterologica Mundial (OMM) Geneva. [Google Scholar]
- GalbraithDW, Lambert GM, Macas J, Doležel J.1997. Analysis of nuclear DNA content and ploidy in higher plants. In: Robinson JP et al, eds. Current protocols in cytometry. New York: John Wiley & Sons, Inc. [DOI] [PubMed] [Google Scholar]
- GamietteF, Bakry F, Ano G.1999. Ploidy determination of some yam species (Dioscorea spp.) by flow cytometry and conventional chromosomes counting. Genetic Resources and Crop Evolution 46: 19–27. [Google Scholar]
- GibbsPE.1976. Studies on the breeding system of Oxalis tuberosa Mol. Flora 165: 129–132. [Google Scholar]
- GibbsPE, Marshall D, Brunton D.1978. Studies on the cytology of Oxalis tuberosa and Tropaeolum tuberosum Edinburgh Royal Botanical Garden – Notes 37: 215–220. [Google Scholar]
- GreilhuberJ.1998. Intraspecific variation in genome size: a critical reassessment. Annals of Botany 82: 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guamán CalderónS.1997. Conservación in situ caracterización y evaluación de la biodiversidad de oca (Oxalis tuberosa) y papalisa (Ullucus tuberosus) en Candelaria (Chapare) y Pocanche (Ayopaya). Ing. Agr. Thesis, Universidad Mayor de San Simón, Cochabamba, Bolivia. [Google Scholar]
- Hayano KanashiroAC.1998. Citogenética de Oxalis tuberosa Mol. ‘Oca’: Número Cromosómico y Analisis Cariotipico. Lic. Biol. Thesis, Universidad Ricardo Palma, Lima, Peru. [Google Scholar]
- HeitzE.1926. Der Nachweis der Chromosomen. Vergleichende Studien über ihre Zahl, Grösse und Form im Pflanzenreich. I. Zeitschrift für Botanik 18: 625–681. [Google Scholar]
- HeitzE.1927. Über multiple und aberrante Chromosomenzahlen. Abhandlungen aus dem Gebiete der Naturwissenschaften, Hamburg 21: 47–57. [Google Scholar]
- HuynhK‐L.1965. Contribution à l’étude caryologique et embryologique des Phanérogames du Pérou. Denkschriften der Schweizerischen Naturforshenden Gesellschaft – Mémoires de la Société Helvétique des Sciences Naturelles 85: 1–178. [Google Scholar]
- JohnstonJS, Bennett MD, Rayburn AL, Galbraith DW, Price HJ.1999. Reference standards for determination of DNA content of plant nuclei. American Journal of Botany 86: 609–613. [PubMed] [Google Scholar]
- KnuthR.1930. Oxalidaceae In: Engler A, ed. Das Pflanzenreich IV; Regni vegetabilis conspectus Leipzig: W. Engelmann, 1–481. [Google Scholar]
- KostoffD, Dogadkina H, Tichonowa A.1935. Chromosome number of certain Angiosperm plants (Nicotiana, Petunia, Oxalis, Secale, and Punica) Doklady Academiia Nauk URSS 3: 401. [Google Scholar]
- KudoN, Kimura Y.2001a Flow cytometric evidence for endopolyploidy in seedlings of some Brassica species. Theoretical and Applied Genetics 102: 104–110. [Google Scholar]
- KudoN, Kimura Y.2001b Patterns of endopolyploidy during seedling development in cabbage (Brassica oleracea L.). Annals of Botany 87: 275–281. [DOI] [PubMed] [Google Scholar]
- Lagunes‐EspinozaLDC, Huyghe C, Bousseau D, Barre P, Papineau J.2000. Endoreduplication occurs during pod wall development in temperate grain legumes. Annals of Botany 86: 185–190. [Google Scholar]
- LourteigA.2000. Oxalis L. subgénero Monoxalis (Small) Lourteig, Oxalis y Trifidus Lourteig. Bradea 7: 201–629. [Google Scholar]
- LysákMA, Doležel J.1998. Estimation of nuclear DNA content in Sesleria (Poaceae). Caryologia 51: 123–132. [Google Scholar]
- MarksGE.1956. Chromosome numbers in the genus Oxalis New Phytologist 55: 120–129. [Google Scholar]
- Medina HinostrozaTC.1994. Contaje cromosómico de la oca (Oxalis tuberosa Molina) conservada in vitro. Ingeniero Agrónomo Thesis, Universidad Nacional del Centro del Perú, Huancayo, Peru. [Google Scholar]
- MelaragnoJE, Mehrotra B, Coleman AW.1993. Relationship between endopolyploidy and cell size in epidermal tissues of Arabidopsis Plant Cell 5: 1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MendiburuAO, Peloquin SJ.1976. Sexual polyploidization and depolyploidization: some terminology and definitions. Theoretical and Applied Genetics 48: 137–143. [DOI] [PubMed] [Google Scholar]
- NaranjoCA, Mola LM, Poggio L, Mulgura de Romero M.1982. Estudios citotaxonómicos y evolutivos en especies herbaceas sudamericanas de Oxalis (Oxalidaceae). I. Boletín de la Sociedad Argentina de Botánica 20: 183–200. [Google Scholar]
- OttoF.1990. DAPI staining of fixed cells for high‐resolution flow cytometry of nuclear DNA. In: Crissman HA, Darzynkiewicz Z, eds. Methods in cell biology. New York: Academic Press. [DOI] [PubMed] [Google Scholar]
- Pallares PonceI.1998. Estudio de la Herencia de la Tristilía en Oca (Oxalis tuberosa Molina) (Geraniales: Oxalidaceae). Thesis, Pontificia Universidad Católica del Ecuador, Quito, Ecuador. [Google Scholar]
- Panti PachecoM.1972. Obtención de semilla botánica en Oxalis tuberosa Mol. (oca) con fines de mejoramiento. Ing. Agr. Thesis, Universidad Nacional de San Antonio Abad del Cusco, Cusco, Peru. [Google Scholar]
- PoggioL, Rosato M, Chiavarino AM, Naranjo CA.1998. Genome size and environmental correlations in maize (Zea mays ssp. mays, Poaceae). Annals of Botany 82: 107–115. [Google Scholar]
- TalledoD, Escobar C.1995. Citogenética de Oxalis tuberosa: ciclo celular y número cromosómico. Biotempo (Universidad Ricardo Palma, Lima, Peru) 2: 33–46. [Google Scholar]
- TierschTR, Wachtel SS.1991. On the evolution of genome size of birds. Journal of Heredity 82: 363–368. [DOI] [PubMed] [Google Scholar]
- TostoDS, Hopp HE.1996. Sequence analysis of the 5·8S ribosomal DNA and internal transcribed spacers (ITS1 and ITS2) from five species of the Oxalis tuberosa alliance. DNA Sequence – The Journal of Sequencing and Mapping 6: 361–364. [DOI] [PubMed] [Google Scholar]
- TrognitzBR, Hermann M.2001. Inheritance of tristyly in Oxalis tuberosa (Oxalidaceae). Heredity 86: 564–573. [DOI] [PubMed] [Google Scholar]
- TrognitzBR, Carrion S, Hermann M.2000. Expression of stylar incompatibility in the Andean clonal tuber crop oca (Oxalis tuberosa Mol. Oxalidaceae). Sexual Plant Reproduction 13: 105–111. [Google Scholar]
- TrognitzBR, Hermann M, Carrión S.1998. Germplasm conservation of oca (Oxalis tuberosa Mol.) through botanical seed. Seed formation under a system of polymorphic incompatibility. Euphytica 101: 133–141. [Google Scholar]
- ValladolidA.1996. Niveles de ploidía de la oca (Oxalis tuberosa Mol.) y sus parientes silvestres. MSc Thesis, Universidad Nacional Agraria La Molina, Lima, Peru. [Google Scholar]
- ValladolidA, Arbizu C, Talledo D.1994. Niveles de ploidía de la oca (Oxalis tuberosa Mol.) y sus parientes silvestres. Agro Sur (Universidad Austral de Chile, Facultad de Ciencias Agrarias) 22 (número especial): 11–12. [Google Scholar]
- Vallenas RamírezM.1992. Fructificación, producción y viabilidad de semilla sexual en oca (Oxalis tuberosa Mol.). In: Morales D, Vacher JJ, eds. Actas del VII Congreso Internacional Sobre Cultivos Andinos, La Paz, Bolivia. Instituto Boliviano de Tecnología Agropecuaria (IBTA); L’Institut Français de Recherche Scientifique pour le Developpement en Cooperation (ORSTOM); Centro Internacional de Investigación y Desarrollo (CIID‐Canada). [Google Scholar]
- VilharB, Greilhuber J, Dolenc Koce J, Temsh EM, Dermastia M.2001. Plant genome size measurement with DNA image cytometry. Annals of Botany 87: 719–728. [Google Scholar]
- Vinueza VelaJF.1997. Evaluación y Caracterización Citogenética de 20 Entradas de Oca (Oxalis tuberosa Molina) Recolectadas en Ecuador. Ing. Agr. Thesis, Universidad Central del Ecuador, Cutuglahua, Pichincha, Ecuador. [Google Scholar]
- VuilleumierBS.1971. Pleistocene changes in the fauna and flora of South America. Science 173: 771–780. [DOI] [PubMed] [Google Scholar]
- WarburgEF.1938. Taxonomy and relationship in the Geraniales, Part II. New Phytologist 37: 189–210. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.