Abstract
The genome size of coffee trees (Coffea sp.) was assessed using flow cytometry. Nuclear DNA was stained with two dyes [4′,6‐diamino‐2‐phenylindole dihydrochloride hydrate (DAPI) and propidium iodide (PI)]. Fluorescence in coffee tree nuclei (C‐PI or C‐DAPI) was compared with that of the standard, petunia (P‐PI or P‐DAPI). If there is no stoichiometric error, then the ratio between fluorescence of the target nuclei and that of the standard nuclei (R‐PI or R‐DAPI) is expected to be proportional to the genome size. Between‐tree differences in target : standard fluorescence ratios were noted in Coffea liberica var. dewevrei using propidium iodide and DAPI. For both dyes, between‐tree differences were due to a lack of proportionality when comparing locations of the coffee peak and the petunia peak. Intraspecific genome size variations clearly cannot explain variations in the target : standard fluorescence ratio. The origin of the lack of proportionality between target and standard fluorescences differed for the two dyes. With propidium iodide, there was a regression line convergence point, and no between‐tree differences were noted in this respect, whereas there was no such convergence with DAPI. An accurate estimate of genome size can thus be obtained with PI. Implications with respect to accessibility and binding mode are discussed.
Key words: Coffea liberica var. dewevrei, Petunia hybrida, propidium iodide, DAPI, dye accessibility, intraspecific variations, genome size
INTRODUCTION
The nuclear DNA content of a plant can be expressed by its DNA 1C‐value (haploid genome) or DNA 2C‐value (diploid genome). According to Bennett and Leitch (1995), DNA 1C‐values in angiosperms range from less than 0·2 pg (Arabidopsis thaliana) to 127·4 pg (Fritillaria assyriaca). The concept of DNA constancy within a species, given a constant basic chromosome number and type, is supported by the results of Bennett and Smith (1976) and Furuta et al. (1978). Observed intraspecific variations in nuclear DNA content are due to: (1) methodological problems (Greilhuber and Ebert, 1994); (2) heterochromatin modifications, as in Zea mays spp. mays (Rayburn et al., 1985); and (3) differences in dye accessibility to DNA (Darzynkiewicz et al., 1984; Evenson et al., 1986; Darzynkiewicz, 1990; Biradar and Rayburn, 1994).
The genus Coffea sub‐genus Coffea (Rubiaceae) originates from tropical regions of Africa and Madagascar. It includes more than 80 wild coffee species, all of which are diploids (2n = 22) except C. arabica (2n = 44) (Louarn, 1992). There are substantial variations in nuclear DNA content within African diploid species with a constant chromosome number (2n = 22): 2C = 0·98 to 1·78 pg (Cros et al., 1995). Within‐species between‐tree differences have also been recorded in Petunia hybrida using internal standardization (Barre et al., 1996).
Recent investigations have shown that nucleus–cytosol interactions are a source of stoichiometric error in flow cytometric assessment of nuclear DNA content in plants (Noirot et al., 2000; Price et al., 2000). For instance, cytosolic compounds present in yam leaves modify petunia nucleus fluorescence, biasing estimates by as much as 20 %. Cytosolic compounds in Coffea leaves do not have such marked effects, but differences in intraspecific nuclear DNA content could arise because of between‐tree differences in the content of cytosolic compounds.
The aim of this study was to check whether within‐species variations in Coffea nuclear DNA content represent true nuclear DNA content differences or simply differences in dye accessibility to DNA. Differences between propidium iodide (PI) and 4′,6‐diamino‐2‐phenylindole dihydrochloride hydrate (DAPI) responses were also assessed.
MATERIALS AND METHODS
Plant material
Coffealiberica spp. dewevrei and the internal standard Petunia hybrida (2C = 2·85 pg; Marie and Brown, 1993) were used in the experiments. All plant material was grown in a glasshouse under tropical climatic conditions (24 °C day/18 °C night, 70 % relative humidity).
Sample preparation
Nuclei were extracted by chopping leaves (Galbraith et al., 1983) in the lysis buffer of Dolezel et al. (1989), modified slightly (0·5 % Triton X‐100, pH = 8), and prepared just before use. The leaf weight per unit volume of buffer was approx. 400 mg ml–1 for Coffea and 250 mg ml–1 for petunia (400 000 to 2 000 000 nuclei ml–1). These leaf tissue quantities had been determined previously (Barre et al., 1996).
Petunia and Coffea leaves were chopped simultaneously in a Petri dish with lysis buffer. The solution was filtered through nylon cloth (50 µm mesh). RNase A (5 units ml–1 of nuclear suspension, boiled Boehringer Mannheim 109 169 DNase‐free; Boehringer Mannheim, Meylan, France) was added to the filtrate before storage for at least 2 h.
Each filtrate was divided for estimation of nuclear DNA content using PI and DAPI, respectively.
Staining of nuclei
Nuclei were stained with PI [95–98 % (TLC) Sigma #P 4170] or DAPI (Sigma, St Quentin, France). The optimal concentration of PI for staining was 333 µg ml–1 (curve determination in Barre et al., 1996). For DAPI, the optimal staining concentration was 10 µg ml–1 (unpubl. res.). The staining time was approx. 3 min for both dyes.
Cytometric measurements
For PI‐stained samples, a FACScan cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) was used with an argon laser (15 mW) at 488 nm with an emission pulse area of 585 nm ± 22 nm. The high voltage was set at 557 V. For DAPI‐stained samples, a FACSVantage cytometer (Becton Dickinson) was used with an argon laser (Coherent, Innova 90–6, Palo Alto, CA, USA) tuned to UV excitation at 345 nm and emission at 455 nm. The high voltage was set at 350 V.
Histograms were collected over 1024 channels. Each histogram contained 3000–5000 nuclei depending on the width of the histogram. The zero offset of the analogue‐to‐digital converter was checked with nuclei from Petunia hybrida (see Barre et al., 1996). The gain of the amplifier system was not changed. Peak locations of petunia nuclei (P‐PI and P‐DAPI for PI and DAPI, respectively) and Coffea nuclei (C‐PI and C‐DAPI) were recorded. This allowed us to compute the ratios C‐PI : P‐PI (R‐PI) and C‐DAPI : P‐DAPI (R‐DAPI).
Experimental design and statistical methods
DNA 2C‐value estimates were based on the proportionality (straight line through origin) between the locations of the standard (petunia) and sample (Coffea) peaks. The aim of the experiment was to check whether differences between coffee trees (between‐trees) were related to straight line slope differences. Ten trees of Coffea liberica spp. dewevrei were selected at random. For PI estimations, each tree was represented by two leaves, and each leaf by two extracts. For DAPI estimations, each tree was represented by three leaves with only one extract per leaf.
In a first analytical step, statistical analyses were carried out separately for each dye. For PI, a two‐way nested ANOVA model was used to test between‐tree and between‐leaf differences (with ‘leaf’ nested within ‘tree’). For DAPI, one‐way ANOVA was used to test between‐tree differences only. In addition, a one‐way ANCOVA was used to test: (1) the presence of within‐tree linear regressions between C‐PI and P‐PI (or between C‐DAPI and P‐DAPI); (2) their parallelism; and (3) their superposition.
RESULTS
Using PI, between‐tree differences (representing 28·9 % of the variance) were recorded for the target : standard ratio (R‐PI) (Table 1), whereas there were no within‐tree differences between leaves. Note the influence of Coffea on the location of the petunia peak (P‐PI) (Table 1), i.e. on the fluorescence intensity of petunia nuclei.
Table 1.
Observed means for ten C. liberica var dewevrei genotypes
| Trees | P‐PI† | C‐PI† | R‐PI | P‐DAPI† | C‐DAPI† | R‐DAPI |
| EB62 | 822·3 | 415·4 | 0.505 | 769·9 | 427·1 | 0.555 |
| EB56 | 834·5 | 413·2 | 0.495 | 771·2 | 423·5 | 0.549 |
| EB51 | 752·9 | 375·5 | 0.499 | 797·8 | 447·4 | 0.561 |
| EB58 | 872·2 | 439·1 | 0.503 | 805·0 | 444·5 | 0.552 |
| EB57 | 774·2 | 392·9 | 0.508 | 759·4 | 432·8 | 0.570 |
| EB69 | 834·3 | 419·6 | 0.503 | 760·1 | 425·4 | 0.560 |
| EB67 | 884·6 | 446·4 | 0.504 | 786·0 | 439·9 | 0.560 |
| EB65 | 885·3 | 437·1 | 0.494 | 784·9 | 422·9 | 0.539 |
| EB64 | 859·3 | 434·0 | 0.505 | 775·6 | 435·7 | 0.562 |
| EB55 | 896·3 | 450·2 | 0.502 | 774·4 | 430·1 | 0.555 |
| Tree | 3·97 * | 3·63 * | 3·38 * | 1·71 N.S. | 1·14 N.S. | 3·44* |
| Leaf | 2·21 N.S. | 1·63 N.S. | 0·48 N.S. |
(* P < 0·05; N.S., non‐significant).
ANOVA results are listed in the bottom two rows. For C‐PI, P‐PI and R‐PI, a nested ANOVA was used with two random factors: genotype; and leaf nested within genotype. Degrees of freedom were d.f.1 = 9 and d.f.2 = 10 for the genotype factor and d.f.1 = 10 and d.f.2 = 20 for the leaf factor. For P‐DAPI, C‐DAPI and R‐DAPI, a one‐way ANOVA was used with genotype as a random factor; degrees of freedom were d.f.1 = 9 and d.f.2 = 10.
† In arbitrary channel units.
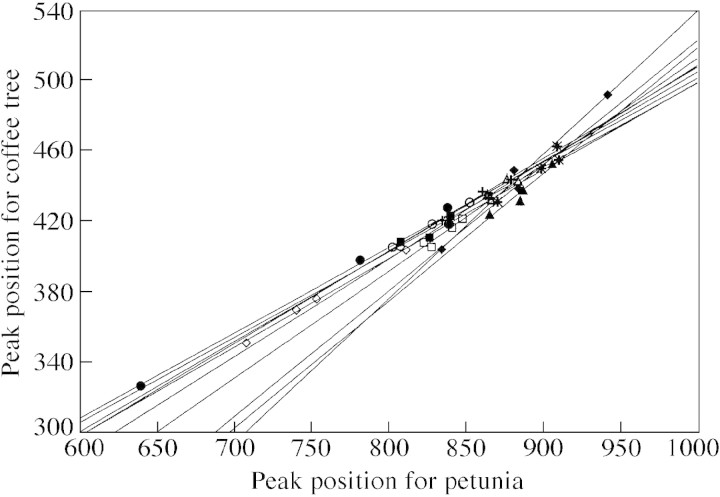
The DNA 2C‐value estimation was based on the assumption that there is proportionality (straight line through origin) between the locations of the Coffea peak (C‐PI) and the petunia peak (P‐PI) (Fig. 1). A within‐tree linear regression was noted (F1, 20 = 430·4; P < 0·001), but the straight lines differed in terms of their slopes (F9, 20 = 4·68; P < 0·01) and did not go through the origin (Table 2). Moreover, a P‐PI value existed for which C‐PI values would be identical, i.e. for which there would be no R‐PI differences. This is evidence that between‐tree differences in R‐PI do not represent genome size differences. The fact that a straight line did not intersect the origin clearly showed that the Coffea liberica spp. dewevrei cytosol effect on nuclei fluorescence differed in C. liberica spp. dewevrei and in petunia. In addition, the slope differences highlighted differential effects of Coffea cytosol on both nuclei according to the particular Coffea tree.
Fig. 1. Within‐tree relationships comparing the standard peak location (P‐PI) and the coffee peak location (C‐PI). Each symbol represents one tree.
Table 2.
Within‐species variations in linear regression parameters comparing the locations of the standard peak and the coffee peak
| Coffee tree | Slope | Intersect |
| EB62 | 0·528 | –19·0 |
| EB56 | 0·611 | –96·6 |
| EB51 | 0·506 | –5·5 |
| EB58 | 0·556 | –46·3 |
| EB57 | 0·487 | 15·7 |
| EB69 | 0·486 | 13·7 |
| EB67 | 0·824 | –282·5 |
| EB65 | 0·726 | –205·6 |
| EB64 | 0·515 | –8·7 |
| EB55 | 0·718 | –193·3 |
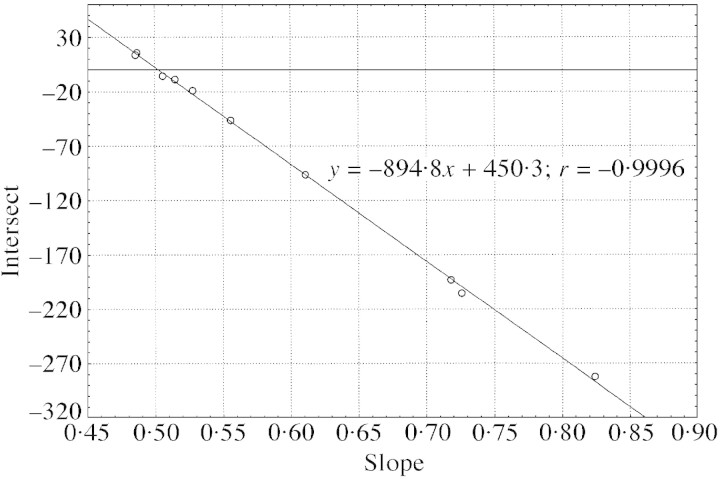
These data are plotted in Fig. 2.
If linear regressions intersect at a point where R‐PI differences disappear, a negative and linear relationship is expected between intercepts and slopes (Fig. 2). Using the fitted equation y = –894·8x + 450·3, we can estimate its coordinates (C‐PI = 450·3; P‐PI = 894·8) and then predict the R‐PI ratio without stoichiometric error (R‐PI = 0·5032). Consequently, the nuclear DNA content of Coffea liberica spp. dewevrei was estimated as 2C = 1·434 pg using PI.
Fig. 2. Relationship between slopes and intersects for within‐tree linear regressions intersecting at one point (450·3; 894·8).
The absence of within‐tree variations between leaves (noted above) using PI was taken into account for the DAPI experiments. Consequently, one‐way ANOVA was used instead of the former nested ANOVA model. Between‐tree differences, representing 44·6 % of the variance, were also noted for the R‐DAPI target : standard ratio (Table 1). In contrast to PI, there was no effect of the Coffea tree cytosol on the location of the petunia peak (P‐DAPI).
Linear regressions were calculated between C‐DAPI and P‐DAPI (F1, 10 = 95·4; P < 0·001). In contrast to the PI results, the straight lines could be considered parallel (F9, 10 = 0·30, P > 0·05), but not identical (F9, 19 = 3·86; P < 0·01). Consequently, there were no petunia peaks in areas where there were no between‐tree differences in R‐DAPI.
DISCUSSION
Stoichiometric error has been noted when estimating nuclear DNA content using both Feulgen cytophotometric (Greilhuber, 1988) and flow cytometric methods (Noirot et al., 2000). The two main findings in the present study on Coffea liberica var dewevrei were that: (1) intraspecific variation in genome size is an artefact; and (2) the nature of the artefact differs according to the dye used (DAPI and PI). Our discussion will thus focus on two points: (1) the pseudo‐intraspecific genome size variation in relation to dye accessibility; and (2) the difference between dyes in inducing pseudo‐intraspecific variation in relation to the binding mode.
Pseudo‐intraspecific variations in genome size and dye accessibility
Peak locations should be constant for a given sample when physical cytometer settings (voltage, gain) remain unchanged. Moreover, the target : standard ratio should be constant for a given DNA content so long as the proportionality between target and standard fluorescence is respected. There are no theoretical reasons for a modification of the ratio when the dye concentration varies. Nevertheless, such changes have been recorded when using mithramycine (Galbraith et al., 1983) or PI (Barre et al., 1996). A saturating concentration was used to stabilize the ratio: with 50 µg ml–1 PI, the nuclear DNA content estimates of Brassica campestris ranged from 0·95 to 1·27 pg, whereas this range decreased markedly (1·03 pg ± 0·02) when the PI concentration was doubled (Arumuganathan and Earle, 1991). The use of a saturating dye concentration to stabilize DNA estimates implies differences in DNA dye affinity between the sample and standard. This represents the first type of accessibility difference.
In a previous study on Coffea species using PI, a bias was observed in nuclear DNA content estimation, even when a saturating concentration of dye was used (Barre et al., 1996): the target peak location was not strictly proportional to the standard, and consequently estimates of DNA content varied. This represents a second type of accessibility difference. Such stoichiometric error due to the presence of cytosolic compounds acting on nuclei fluorescence has also been observed using a yam leaf extract (without nuclei and boiled), which modified the location of the petunia peak, leading to a 20 % bias in genome size (Noirot et al., 2000).
Between‐dye differences in pseudo‐intraspecific variations and binding mode
Observed between‐dye differences should be interpreted according to known differences in their DNA binding affinities: binding‐site number and/or binding mode.
Godelle et al. (1993) proposed that binding‐site number differences (BSND) were the reason why ethidium bromide estimates were not equal to the sum of Hoechst (AT‐base‐specific) and mithramycine (GC‐base‐specific) estimates. Nevertheless, BSND depend on the genome base composition and cannot explain the parallelism or convergence of straight lines obtained for target and standard nuclear fluorescence.
The binding site mode differentiates between dyes that bind preferentially to AT‐rich (DAPI, Hoechst) or GC‐rich regions (mithramycine) as compared with intercalating dyes such as PI or ethidium bromide. The binding site mode has two main consequences: (1) with AT‐ or GC‐binding, estimates for genomes with different AT : GC ratios in their nuclear DNA cannot be compared (Dolezel et al., 1998); and (2) dye accessibility to DNA differs markedly depending on heterochromatin condensation. Concerning DAPI and PI, several studies have demonstrated clearly that PI and DAPI estimations of genome size are correlated with measurements obtained by Feulgen densitometry (Michaelson et al., 1991; Dolezel et al., 1998). This relationship is weaker for DAPI (R2 = 0·59) than for PI (R2 = 0·998) (Dolezel et al., 1998). Our comparison included trees of the same species and consequently between‐dye variations could not be attributed to AT : GC ratio changes. Moreover, differences in binding site modes have to be taken into account when explaining the absence of convergence for within‐tree regressions obtained when comparing coffee and petunia fluorescence with DAPI.
In contrast, the convergence of within‐tree regressions obtained when comparing coffee and petunia fluorescence with PI could be used to correct the bias and obtain an accurate estimate of genome size. This is another reason why PI is preferred for estimating genome size in plants.
Prospects
In nuclear DNA content estimation, between‐tree differences noted in the cytosolic stochiometric error suggest that there is within‐species genetic variation in the biochemical compounds that are present in the cytosol. Further studies should thus focus on identifying these compounds. Two pathways could be explored: (1) comparing Coffea trees in terms of the biochemical composition of their leaves, and then establishing a multiple regression between the slope of the within‐tree regression and the contents of the various compounds; and (2) using an interspecific cross between C. liberica dewevrei and C. pseudozanguebariae, for which a genetic map has been obtained (Ky et al., 2000), and locating QTLs for stochiometric error. Co‐location with other QTLs involving biochemical compounds could help to identify the causal agent.
Supplementary Material
Received: 1 November 2001; Accepted: 27 November 2001.
References
- ArumuganathanK, Earle ED.1991. Estimation of nuclear DNA content of plants by flow cytometry. Plant Molecular Biology Report 9: 229–233. [Google Scholar]
- BarreP, Noirot M, Louarn J, Duperray C, Hamon S.1996. Reliable flow cytometric estimation of nuclear DNA content in coffee trees. Cytometry 24: 32–38. [DOI] [PubMed] [Google Scholar]
- BennettMD, Smith JB.1976. Nuclear DNA amounts in angiosperms. Philosophical Transactions of the Royal Society of London B 274: 227–274. [DOI] [PubMed] [Google Scholar]
- BennettMD, Leitch IJ.1995. Nuclear DNA amounts in angiosperms. Annals of Botany 76: 113–176. [Google Scholar]
- BiradarDP, Rayburn AL.1994. Flow cytometric probing of chromatin condensation in maize diploid nuclei. New Phytologist 126: 31–35. [Google Scholar]
- CrosJ, Combes MC, Chabrillange N, Duperray C, Monnot des Angles A, Hamon S.1995. Nuclear DNA content in the subgenus Coffea (Rubiaceae): inter‐ and intra‐specific variation in African species. Canadian Journal of Botany 73: 14–20. [Google Scholar]
- DarzynkiewiczZ.1990. Acid‐induced denaturation of DNA in situ as a probe of chromatin structure. In: Darzynkiewicz Z, Crissman HA, eds. Methods in cell biology Vol. 33 New York: Academic Press, 337–352. [DOI] [PubMed] [Google Scholar]
- DarzynkiewiczZ, Traganos F, Kapuscinski J, Staiano‐Coico L, Melamed MR.1984. Accessibility of DNA in situ to various fluorochromes: Relationship to chromatin changes during erythroid differentiation of Friend leukemia cells. Cytometry 5: 355–363. [DOI] [PubMed] [Google Scholar]
- DolezelJ, Binarova P, Lucretti S.1989. Analysis of nuclear DNA content in plant cells by flow cytometry. Biologia Plantarum 31: 113–120. [Google Scholar]
- DolezelJ, Greilhuber J, Lucretti S, Meister A, Lysak MA, Nardi L, Obermayer R.1998. Plant genome size estimation by flow cytometry: inter‐laboratory comparison. Annals of Botany 82: 17–26. [Google Scholar]
- EvensonD, Darzynkiewicz Z, Jost L, Janca F, Ballachey B.1986. Changes in accessibility to DNA to various fluorochromes during spermatogenesis. Cytometry 7: 45–53. [DOI] [PubMed] [Google Scholar]
- FurutaY, Nishikawa K, Haji T.1978. Uniformity of nuclear DNA content in Triticum monococcum Japanese Journal of Genetics 53: 361–366. [Google Scholar]
- GalbraithDW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E.1983. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051. [DOI] [PubMed] [Google Scholar]
- GodelleB, Cartier D, Marie D, Brown SC, Siljak‐Yakovlev S.1993. Heterochromatin study demonstrating the non‐linearity of fluorometry useful for calculating genomic base composition. Cytometry 14: 618–626. [DOI] [PubMed] [Google Scholar]
- GreilhuberJ.1988. ‘Self‐tanning’—a new and important source of stoichiometric error in cytophotometric determination of nuclear DNA content in plants. Plant Systematics and Evolution 158: 87–96. [Google Scholar]
- GreilhuberJ, Ebert I.1994. Genome size variation in Pisum sativum Genome 37: 646–655. [DOI] [PubMed] [Google Scholar]
- KyCL, Barre P, Lorieux M, Trouslot P, Akaffou S, Louarn J, Charrier A, Hamon S, Noirot M.2000. Interspecific genetic linkage map, segregation distortion and genetic conversion in coffee (Coffea sp.). Theoretical and Applied Genetics 101: 669–676. [Google Scholar]
- LouarnJ.1992. La fertilité des hybrides interspécifiques et les relations génomiques entre caféiers diploïdes d’origine africaine (genre Coffea L. sous‐genre coffea). Thesis, ORSTOM, Paris. [Google Scholar]
- MarieD, Brown SC.1993. A cytometric exercise in plant DNA histograms, with 2C values for 70 species. Biology of the Cell 78: 41–51. [DOI] [PubMed] [Google Scholar]
- MichaelsonMJ, Price HJ, Ellison JR, Johnston JS.1991. Comparison of plant DNA contents determined by Feulgen microspectrophotometry and laser flow cytometry. American Journal of Botany 78: 183–188. [Google Scholar]
- NoirotM, Barre P, Louarn J, Duperray C, Hamon S.2000. Nucleus–cytosol interactions—a source of stoichiometric error in flow cytometric estimation of nuclear DNA content in plants. Annals of Botany 86: 309–316. [Google Scholar]
- PriceH, Hodnett G, Johnston JS.2000. Sunflower (Helianthus annuus) leaves contain compounds that reduce nuclear propidium iodide fluorescence. Annals of Botany 86: 929–934. [Google Scholar]
- RayburnAL, Price HJ, Smith JD, Gold JR.1985. C‐band heterochromatin and DNA content in Zea mays American Journal of Botany 72: 1610–1617. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.