Abstract
Tsetse flies (Glossina sp.) that transmit trypanosomes causing human (and animal) African trypanosomiasis (HAT and AAT, respectively) harbor symbiotic microorganisms, including the obligate primary symbiont Wigglesworthia glossinidia. A relationship between Wigglesworthia and tsetse fly infection by trypanosomes has been suggested, as removal of the symbiont results in a higher susceptibility to midgut infection in adult flies. To investigate this relationship and to decipher the role of W. glossinidia in the fly's susceptibility to trypanosome infection, we challenged flies with trypanosomes and subsequently analyzed and compared the transcriptomes of W. glossinidia from susceptible and refractory tsetse flies at three time points (3, 10, and 20 days). More than 200 W. glossinidia genes were found to be differentially expressed between susceptible and refractory flies. The high specificity of these differentially expressed genes makes it possible to distinguish Wigglesworthia inhabiting these two distinct groups of flies. Furthermore, gene expression patterns were observed to evolve during the infection time course, such that very few differentially expressed genes were found in common in Wigglesworthia from the 3-, 10- and 20-day post-feeding fly samples. The overall results clearly demonstrate that the taking up of trypanosomes by flies, regardless of whether flies proceed with the developmental program of Trypanosoma brucei gambiense, strongly alters gene expression in Wigglesworthia. These results therefore provide a novel framework for studies that aim to decrease or even abolish tsetse fly vector competence.
Keywords: Wigglesworthia, tsetse fly, trypanosomes, tripartite interactions, transcriptome
Introduction
Human African trypanosomiasis (HAT), or sleeping sickness, is a neglected vector-borne parasitic disease caused by protozoa belonging to the Trypanosoma genus. Two of the subspecies, Trypanosoma brucei gambiense and T. b. rhodesiense, are responsible for the chronic form of HAT in western and central Africa, and the acute form of the disease in eastern Africa, respectively (Kennedy, 2008).
Although there are drugs to treat this disease, they have limited effectiveness and produce harmful side effects (Cattand et al., 2001; Barrett, 2006). The repeated use of some drugs has led to the emergence of resistant forms of different trypanosome species (De Koning, 2001; Matovu et al., 2001). Consequently, the exploration for novel strategies must continue, including alternative vector-based strategies (Rio et al., 2004).
To be transmitted, the parasite must first establish itself within the insect midgut following an infectious blood meal, and then mature in the salivary glands or mouthparts (depending on the trypanosome species) (Vickerman et al., 1988; Van den Abbeele et al., 1999). Tsetse flies are normally refractory to trypanosome infections, and typically exhibit an infection rate of less than 50%, even under ideal laboratory conditions (Ravel et al., 2003), while field infection rates rarely exceed 10% of the fly population (Moloo et al., 1986; Frézil and Cuisance, 1994; Maudlin and Welburn, 1994). Furthermore, many infected flies fail to produce mature parasites and therefore never become infective (Moloo et al., 1986; Dukes et al., 1989; Frézil and Cuisance, 1994; Maudlin and Welburn, 1994; Jamonneau et al., 2004). This ability to acquire the parasite, facilitate its maturation, and transmit it to a mammalian host is known as vector competence.
Tsetse flies harbor 3 different symbiotic microorganisms (Aksoy, 2000), including Sodalis glossinidius (Cheng and Aksoy, 1999; Dale and Maudlin, 1999) and the obligate primary symbiont Wigglesworthia glossinidia (Aksoy, 1995a; Aksoy et al., 1995). Both are members of the Enterobacteriaceae family, inhabit the fly's gut, and are vertically transmitted to the intrauterine-developing larvae (Cheng and Aksoy, 1999). Wigglesworthia is localized intracellularly in specialized host cells of the anterior midgut and extracellularly in the female milk glands (Balmand et al., 2013). Sodalis has a broad tissular tropism and can also be found in the hemolymph, salivary glands, etc. (Cheng and Aksoy, 1999). The third symbiont, Wolbachia (O'Neill et al., 1993), belongs to the Rickettsiaceae family and causes cytoplasmic incompatibility in tsetse flies (Alam et al., 2011).
S. glossinidius has been suspected of being involved in the trypanosome establishment process (Maudlin and Ellis, 1985; Welburn et al., 1993). Epidemiological investigations conducted in several HAT foci in Cameroon have demonstrated an association between the presence of this symbiont and the establishment of trypanosome species and subspecies in the fly midgut (Farikou et al., 2010).
Wigglesworthia has coevolved with Glossina species, and develops in the fly's bacteriocytes where it encodes a plethora of vitamins and biosynthetic products that may promote host reproduction as well as fly nutrition throughout its development (Akman et al., 2002; Rio et al., 2012). In the absence of Wigglesworthia, vitamin supplementation of the blood meal can partially restore host fertility (Nogge, 1976, 1982). The presence of Wigglesworthia is necessary during the larval stages for proper immune development (Weiss et al., 2011, 2012). The presence of larval microbiota also contributes to the development of the adult peritrophic matrix (a structure that separates epithelial cells from the lumen content), which regulates immune induction following the trypanosome challenge to the fly (Weiss et al., 2013).
The role of Wigglesworthia in tsetse fly susceptibility to trypanosome infection remains largely unknown. A relationship between Wigglesworthia and trypanosome infection has been suggested, since removal of the symbiont results in higher susceptibility to midgut infection in older, non-teneral flies (Pais et al., 2008; Wang et al., 2009; Snyder and Rio, 2013). It was also previously reported that G. morsitans morsitans (Gmm) demonstrates much higher vector competence than G. brevipalpis (Gb) (Harley, 1971; Moloo and Kutuza, 1988; Moloo et al., 1994). Recently, comparison of the Wigglesworthia spp. genomes from Gmm and Gb revealed metabolomic differences between the primary symbiont strains harbored by these two fly species (Rio et al., 2012; Snyder and Rio, 2013). One such differences involves the chorismate, phenylalanine, and folate biosynthetic pathways, which are present in Wigglesworthia from Gmm but not Gb. Interestingly, African trypanosomes are auxotrophic for phenylalanine and folate; their genome, however, encodes the chorismate and folate transporters, allowing the parasite to absorb these molecules from their environment (Berriman et al., 2005; Jackson et al., 2014). This may explain why Gmm, which harbors a Wigglesworthia strain capable of providing the trypanosomes with the different metabolites they are unable to synthesize, is more susceptible to parasite infection than Gb (Rio et al., 2012; Snyder and Rio, 2013).
The mechanisms controlling tsetse fly infection by trypanosomes thus appear to be very complex. Nevertheless, sequencing and annotation of the Wigglesworthia genome from the tsetse fly (Akman et al., 2002) has enabled analysis of the gene expression profiles of these bacteria, allowing the identification of genes or pathways involved in specific traits. In this context, we have challenged a group of tsetse flies with trypanosomes and subsequently analyzed the W. glossinidia transcriptome from susceptible and refractory tsetse flies at three time points, in order to identify the differentially expressed genes between bacteria from these different tsetse fly groups. In addition, we were willing to perform the experiment under strictly the same conditions as those used to identify the differentially expressed genes of Sodalis (Hamidou Soumana et al., 2014). The investigations on Wigglesworthia transcriptome were, thus, performed on the samples that were previously used to analyse the Sodalis transcriptome.
Materials and methods
Ethics statement
The experimental protocols involving animals were approved by the Ethics Committee and the Veterinary Department of the Centre International de Recherche Agronomique pour le Développement (CIRAD), Montpellier, France (specific approval numbers 12TRYP03, 12TRYP04, and 12TRYP06). The experiments were conducted according to internationally recognized guidelines (French regulation).
T. b. gambiense strain
The S7/2/2 T. b. gambiense strain used in this study was isolated in 2002 from HAT patients in the Bonon sleeping sickness focus, located in Ivory Coast (Ravel et al., 2006).
Experimental and sampling procedures
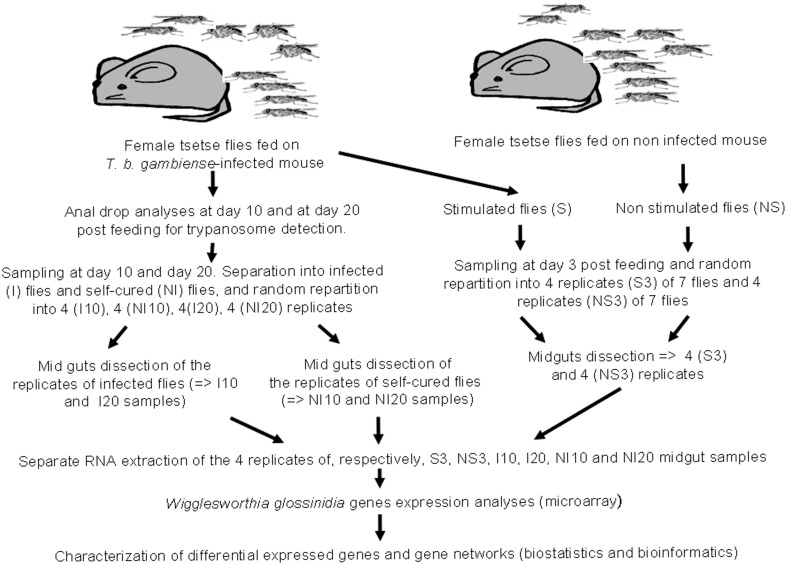
Wigglesworthia transcriptome experiments were performed on the same samples as those previously used to identify the differential expressed genes of Sodalis (Hamidou Soumana et al., 2014). The main experimental steps are summarized in Figure 1, and described below. All the flies under experiment were fed on infected or non-infected balb/cj mice.
Figure 1.
Design depicting experimental infection, sampling procedures, and microarray data analysis.
A set of 100 teneral (less than 32 h old) female G. p. gambiensis flies from the laboratory colony were fed on non-infected mice, and a second set of 900 teneral (less than 32 h old) female flies from the same colony were fed on trypanosome-infected mice. Subsequently, non-gorged flies were removed from the experiment. After their blood meal, both gorged flies and control flies, were maintained by feeding on uninfected rabbit, 3 days per week, until the end of each experimental condition (e.g., 3, 10, and 20 days post-feeding). Sampling times were chosen according to a previously determined time course of susceptible fly infection by trypanosomes (Ravel et al., 2003), so as to compare the expression profile of Wigglesworthia at the different stages of the infection process. The 3-day and 10-day sampling times were selected to target differentially expressed genes involved in early events associated with trypanosome entry into the midgut and with the establishment of infection, respectively. The 20-day sampling time point was chosen to target the expression of genes involved in events occurring relatively late in trypanosome infection time course. The first set of flies (uninfected flies) was composed of four groups (four biological replicates), each of seven flies randomly chosen. Three days after their blood meal on uninfected mice, all flies from each group were dissected and the corresponding 7 midguts were pooled, yielding 4 pools of 7 midguts. These 4 biological replicates were then analyzed separately, by RNA extraction, for their transcriptome. These biological replicates are referred to as “NS 3 day samples” (NS for “non-stimulated” flies or flies not exposed to trypanosome infections).
A stabilate of T. b. gambiense S7/2/2 was thawed at room temperature and 0.2 ml was injected intraperitoneally into balb/cj mice, as described in Hamidou Soumana et al. (2014), in order to feed the second set of flies an infected blood meal. To monitor parasitemia, tail blood was examined using a phase-contrast microscope at 400× magnification. Teneral (less than 32 h old) female flies were fed on T. b. gambiense-infected mice displaying parasitemia levels ranging from 16 to 64 × 106 parasites/ml. This set of flies was then separated into three groups. Three days after ingesting the infected blood meal, the first group was subdivided into 4 subgroups of 7 flies. All flies from each subgroup were dissected separately, and the 7 corresponding midguts were pooled, yielding 4 pools of 7 midguts (i.e., 4 biological replicates), referred to as “S 3 day samples” (S for “stimulated” flies). Following dissection, the presence of trypanosomes in the blood meal of these flies was confirmed by microscopy.
The second and third groups consist of flies that were dissected 10 and 20 days after feeding on the infected blood meal, respectively. Before dissection, the status of each fly (e.g., trypanosome infected or non-infected) was controlled by collecting anal drops from each fly, either at 10 (second group) or 20 days (third group) following their blood meal on infected mice. DNA was extracted from these anal drops using the Chelex method, as described by Ravel et al. (2003), and the presence of trypanosomes was investigated by PCR using the TBR1 and TBR2 primers (Moser et al., 1989). The identification of trypanosomes in anal drops indicates midgut infections, and flies were considered uninfected if the PCR assays were negative for the anal drops.
Based on these PCR results, tsetse flies from the second and third groups were subdivided by infection status into a subgroup of flies with trypanosome-infected midguts (e.g., a positive PCR result for the anal drop test), and a subgroup of flies without midgut infection (e.g., a negative PCR result for the anal drop test). The prevalence of infection in the second group was less than 5%, whereas it was greater than 10% in the third group. Four biological replicates were processed for each subgroup of the second and third groups, as described above for “S 3 day samples.” Samples collected 10 days after the blood meal are referred to as “I 10-day samples” and “NI 10-day samples,” and samples collected after 20 days are labeled “I 20-day samples” and “NI 20-day samples” (I and NI signify infected and non-infected flies, respectively) (Figure 1). Finally, the experiment included a total of 24 samples, consisting of 6 sets of 4 replicates each of 7 flies/7 pooled midguts; exceptionally, “I 10-day samples” and “NI 10-day samples” only grouped 3 flies, owing to the weak infection prevalence at the corresponding sampling time.
RNA extraction
RNA was extracted from the pooled midguts of each biological replicate using TRIzol reagent (Gibco-BRL, France), according to the manufacturer's protocol. One previous study has suggested that TRIzol extraction may degrade some bacterial RNA samples (Jahn et al., 2008). The study further indicates that reproducible transcript profiling requires an investigation of RNA integrity before use, and that RNA Integrity Number values acceptable for some biological systems may not work well in other biological systems. Thus, the experiments reported here were performed in quadruplicate, and the integrity of each RNA sample (as well as the absence of contaminating DNA) was checked on an Agilent RNA 6000 Bioanalyzer. The quantification of each RNA sample was performed using the Agilent RNA 6000 Nano kit (Agilent Technologies, France).
Oligonucleotide microarray design
A genome-wide Wigglesworthia transcriptome analysis from G. p. gambiensis was performed using the Agilent Technologies oligonucleotide microarray format (www.agilent.com). The custom-made density array (8 × 15 K format; AMADID 050087, Agilent Technologies) was designed with 60-mer oligos specific to the 673 genes of the W. glossinidia chromosome from G. morsitans morsitans (NCBI Reference Sequence: NC_016893.1) (Akman et al., 2002). For each gene, 10 different unique probes designed by Hybrigenics (Clermont-Ferrand, France) were used. The Agilent design uses the uniqueness of probe sequences as one of the criteria for probe selection, to avoid cross-hybridization with non-target genes.
The details of the array design and sample description are available at the Gene Expression Omnibus (GEO) under the accession number GPL18427. The details of the expression data are available at GEO under the accession number GSE55931.
cDNA preparation and microarray hybridization
Cy3 dCTP direct cDNA labeling was performed with 100 ng of total RNA using the Low Input Quick Amp Labeling Kit One-Color (Agilent Technologies, France). After CyDye-labeled cDNA production, samples were carefully matched and hybridized onto the custom-made whole W. glossinidia Genome Oligo Microarrays. Hybridization was performed at 65°C for 17 h at 60 rpm. Microarrays were then scanned using an Agilent Technologies scanner.
Microarray data analysis, gene ontology analysis and downstream annotation
The microarray approach has been used to analyse W. glossinidia gene expression, as its reliability had been validated by qPCR in our previous investigation on Sodalis differential gene expression (Hamidou Soumana et al., 2014). All microarray data are MIAME-compliant, and the raw and normalized data have been deposited in the MIAME-compliant GEO database (Edgar et al., 2002) (GEO accession number GSE55931). Individual microarray quality was evaluated based on the QC report, pair-wise MA plots, and box plots. The results were extracted using the Feature Extraction software 11.0.1 (Agilent Technologies). The raw data files were imported to GeneSpring GX (version 12.0, Agilent Technologies), which was used for background adjustment, quantile normalization of data (Bolstad et al., 2003; Smyth et al., 2005), log-transformation, and gene clustering for the 24 microarrays.
Gene ontology was performed only on genes with significant differential expression, using the GeneSpring database. An unpaired t-test was used for statistical analysis (Qin et al., 2012). A p-value below 0.05 was considered as statistically significant and indicates significant differences between groups (e.g., between NS 3-day samples/S 3-day samples, between I 10-day samples/NI 10-day samples, and between I 20-day samples/NI 20-day samples). The representation of each differentially expressed gene within each ontology category (including categories for molecular function, cellular component, and biological processes) was measured. A corrected p-value (Benjamini-Yekutelli correction) lower than 0.5 indicates over-representation of the differentially expressed genes within that particular category.
Wigglesworthia gene expression data were further analyzed via two-dimensional hierarchical clustering, while three-dimensional clustering of samples was analyzed by principle component analysis (PCA). Hierarchical clustering of differentially expressed genes was performed using average linkage and the Pearson-centered distance metrics (Claverie, 1999). Gene trees were created to group similar genes and to improve visualization of the data (Butte, 2002). The relationship between genes is represented by a tree in this type of clustering, where branch length reflects the degree of similarity between genes. PCA was performed within GeneSpring on infected (or stimulated) vs. non-infected (or non-stimulated) conditions at the different experimental infection times using the list of genes.
Results
Principal component and hierarchical clustering analysis of the Wigglesworthia transcription profiles
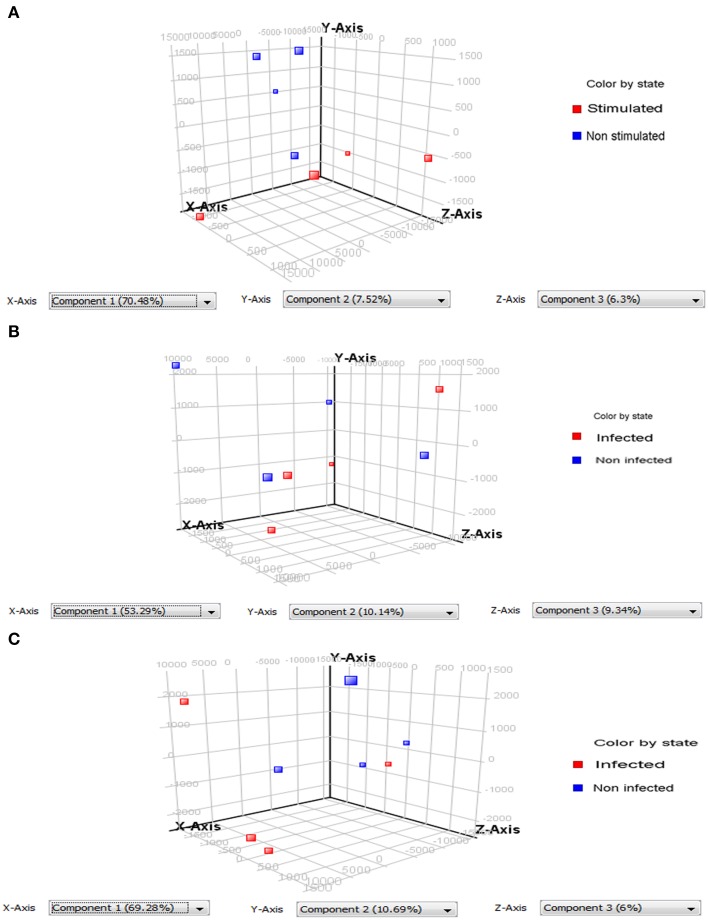
PCA of the differentially expressed Wigglesworthia genes evidenced two types of results: (a) the ability to discriminate infected (or stimulated) from non-infected (or non-stimulated) flies; and (b) the existence of variability between the replicates (Figure 2). The quality of the discrimination between infected and non-infected flies however depends on the extent of the diversity between the repeats. Our results show that PCA can correctly discriminate stimulated from non-stimulated flies (3-day sampling; Figure 2A) and infected from non-infected flies at the 20-day sampling (Figure 2C), even though in each case one repeat was slightly different from the other three. The repeats were even more dispersed at the 10-day sampling, such that the infected samples could not be perfectly discriminated from the non-infected samples (Figure 2B).
Figure 2.
Principle component analysis (PCA) performed on differentially expressed Wigglesworthia genes. Two groups containing 4 replicates each were compared: samples from infected or stimulated flies (red squares), and from non-infected or non-stimulated flies (blue squares). (A) PCA on differentially expressed Wiggleswortia genes from stimulated and non-stimulated flies, sampled at day 3 after fly feeding. Stimulated and non-stimulated flies, were fed on trypanosome-infected and non-infected mice, respectively; (B) PCA on differentially expressed Wiggleswortia genes from infected and non-infected flies sampled at day 10 after fly feeding. (C) PCA on differentially expressed Wiggleswortia genes from infected and non-infected flies sampled at day 20 after fly feeding. For (B,C), both groups of flies were fed on infected mice; the infected group comprised flies in which anal drops were trypanosome-positive, whereas the non-infected group contained flies in which anal drops were trypanosome-negative (even though they had ingested an infected blood meal).
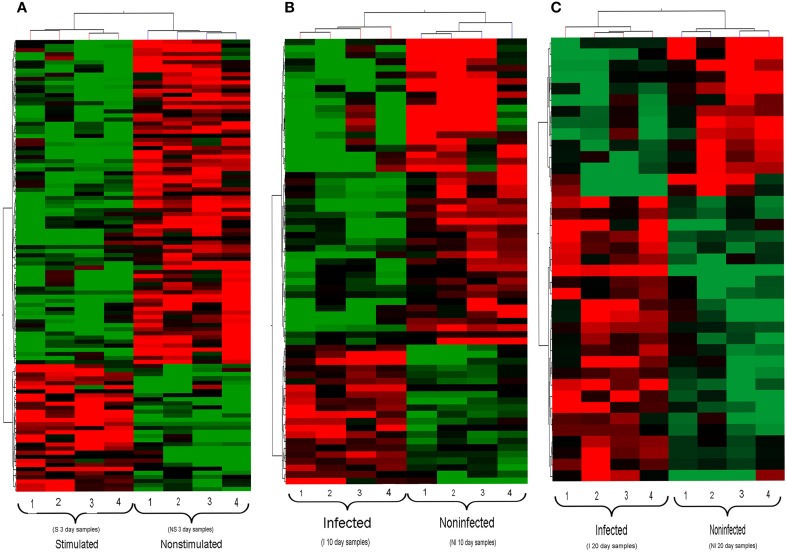
Hierarchical clustering analysis of the transcription profiles displayed by Wigglesworthia from stimulated and non-stimulated flies at 3 days (Figure 3A), and from infected and non-infected flies at 10 and 20 days (Figures 3B,C, respectively), clearly made Wigglesworthia expression profiles unique for each group. Clustering of the Wigglesworthia samples was based on the expression levels of: 103 genes with significant differential expression in stimulated vs. non-stimulated tsetse flies; 61 genes with significant differential expression in infected vs. non-infected tsetse flies, 10 days after ingesting the trypanosome-infected blood meal; and 38 genes with significant differential expression in infected vs. non-infected tsetse flies, 20 days after ingesting the trypanosome-infected blood meal.
Figure 3.
Hierarchical clustering of Wigglesworthia genes. For (A–C), each group of flies is comprised of 4 replicates. (A) Genes from stimulated and non-stimulated tsetse flies, 3 days after ingestion of a trypanosome-infected blood meal or non-infected meal, respectively. (B) Genes from infected and non-infected tsetse flies, 10 days after ingestion of a trypanosome-infected blood meal. (C) Genes from infected and non-infected tsetse flies, 20 days after ingestion of a trypanosome-infected blood meal. The set of Wigglesworthia genes was extracted from the full data set (673 genes and 5115 probes) using a t-test (p < 0.05). The two-dimensional hierarchical cluster was performed on the basis of the gene expression profiles of each sample, using the GeneSpring GX software (version 12.0). Each row represents one Wigglesworthia gene and each column represents one Wigglesworthia sample. The intensity of the red and green colors, respectively up- and down-regulated genes, indicates the corresponding expression levels.
Gene expression profiling of Wigglesworthia in tsetse flies that are refractory and susceptible to trypanosomes
Microarray analysis of the whole Wigglesworthia genome expression comparing stimulated (3 days) flies to non-stimulated flies identified a total of 103 genes with significant differential expression (p < 0.05 according to t-tests, n = 4 Table 1). Of these genes, 73 were down-regulated and 30 were up-regulated in stimulated flies and non-stimulated flies, respectively. 79 genes exhibited a fold-change greater than 1.1 and 2 genes exhibited a fold-change greater than 1.5. 61 genes with significant differential expression (p < 0.05 according to t-tests, n = 4; Table 2), were identified by comparing gene expression profiles of infected and non-infected flies, 10 days after the blood meal on infected mice. 42 genes were down-regulated and 19 genes were up-regulated in infected flies and non-infected flies, respectively. 54 genes exhibited a fold-change greater than 1.1 and 6 genes exhibited a fold-change greater than 1.5. Thirty-eight Wigglesworthia genes were significantly differentially expressed (p < 0.05 according to t-tests, n = 4; Table 3) between infected and non-infected flies, 20 days after the blood meal on infected mice. 14 genes were down-regulated and 24 genes were up-regulated in infected flies and non-infected flies, respectively. 38 genes exhibited a fold-change higher than 1.1, 8 genes exhibited a fold-change greater than 1.5, 3 genes exhibited a fold-change greater than 2.0 and 1 gene (thiamine biosynthesis ThiGH complex subunit) exhibited a fold-change higher than 3.0.
Table 1.
Wigglesworthia genes displaying significant differential expression in trypanosome stimulated vs. non-stimulated flies, 3 days post-blood meal uptake.
| Gene symbols | Expression | GenBank | Name |
|---|---|---|---|
| cspE | Down | WIGMOR_0606__cspE | DNA-binding transcriptional repressor |
| secY | Down | WIGMOR_0186__secY | Preprotein translocase membrane subunit |
| groL | Down | WIGMOR_0521__groL | Cpn60 chaperonin GroEL, large subunit of GroESL |
| Down | WIGMOR_0357 | MarR family transcriptional regulator | |
| sucC | Down | WIGMOR_0345__sucC | Succinyl-CoA synthetase, beta subunit |
| aspS | Down | WIGMOR_0121__aspS | Aspartyl-tRNA synthetase |
| ycfH | Down | WIGMOR_0102__ycfH | Putative metallodependent hydrolase |
| rpoC | Down | WIGMOR_0228__rpoC | RNA polymerase, beta prime subunit |
| motA | Down | WIGMOR_0035__motA | Proton conductor component of flagella motor |
| ybgF | Down | WIGMOR_0352__ybgF | SecB-dependent secretory protein |
| flhC | Down | WIGMOR_0034__flhC | Family flagellar transcriptional activator |
| accC | Down | WIGMOR_0138__accC | Acetyl-CoA carboxylase, biotin carboxylase subunit |
| yccA | Down | WIGMOR_0220__yccA | Inner membrane protein |
| lpxA | Down | WIGMOR_0385__lpxA | UDP-N-acetylglucosamine acyltransferase |
| grpE | Down | WIGMOR_0116__grpE | Pseudo |
| rpoZ | Down | WIGMOR_0359__rpoZ | RNA polymerase subunit omega |
| alaS | Down | WIGMOR_0548__alaS | Alanyl-tRNA synthetase |
| fusA | Down | WIGMOR_0032__fusA | GTP-binding protein chain elongation factor EF-G |
| Down | WIGMOR_0040 | Family flagellar basal-body P-ring formation protein | |
| mltC | Down | WIGMOR_0079__mltC | Membrane-bound lytic murein transglycosylase C |
| thiL | Down | WIGMOR_0293__thiL | Thiamin-monophosphate kinase |
| znuA | Down | WIGMOR_0125__znuA | Periplasmic component of a high-affinity zinc uptake System |
| mrcB | Down | WIGMOR_0148__mrcB | Penicillin-binding protein 1B (PBP1B) |
| gntY | Down | WIGMOR_0476__gntY | Putative gluconate transport associate protein |
| tmk | Down | WIGMOR_0100__tmk | Thymidylate kinase |
| secA | Down | WIGMOR_0585__secA | Preprotein translocase subunit, ATPase |
| gyrA | Down | WIGMOR_0315__gyrA | DNA gyrase subunit A |
| hemF | Down | WIGMOR_0163__hemF | Coproporphyrinogen III oxidase |
| rplP | Down | WIGMOR_0199__rplP | 50S ribosomal protein L16 |
| rpsG | Down | WIGMOR_0031__rpsG | 30S ribosomal protein S7 |
| ftsA | Down | WIGMOR_0582__ftsA | Cell division protein |
| groS | Down | WIGMOR_0520__groS | Cpn10 chaperonin GroES, small subunit of GroESL |
| def | Down | WIGMOR_0262__def | Peptide deformylase |
| Down | WIGMOR_0349 | TolA family protein | |
| putA | Down | WIGMOR_0327__putA | Transcriptional repressor/proline dehydrogenase/delta-1- pyrroline-5-carboxylate dehydrogenase |
| serS | Down | WIGMOR_0269__serS | Seryl-tRNA synthetase |
| ispG | Down | WIGMOR_0176__ispG | 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate Synthase |
| hslU | Down | WIGMOR_0502__hslU | Molecular chaperone and ATPase component of HslUV protease |
| thiE | Down | WIGMOR_0249__thiE | Thiamin phosphate synthase |
| Down | WIGMOR_0310 | ncRNA | |
| rpsR | Down | WIGMOR_0517__rpsR | 30S ribosomal protein S18 |
| sucA | Down | WIGMOR_0343__sucA | Thiamin-requiring 2-oxoglutarate decarboxylase |
| fliN | Down | WIGMOR_0056__fliN | Flagellar motor switching and energizing component |
| kdsA | Down | WIGMOR_0424__kdsA | 3-deoxy-D-manno-octulosonate 8-phosphate synthase |
| bioC | Down | WIGMOR_0304__bioC | Malonyl-CoA methyltransferase |
| fliF | Down | WIGMOR_0064__fliF | Flagellar basal-body MS-ring and collar protein |
| Down | WIGMOR_0083 | Pyrroline-5-carboxylate reductase | |
| holE | Down | WIGMOR_0570__holE | DNA polymerase III subunit theta |
| pyrI | Down | WIGMOR_0154__pyrI | Aspartate carbamoyltransferase, regulatory subunit |
| tyrS | Down | WIGMOR_0429__tyrS | Tyrosyl-tRNA synthetase |
| dapF | Down | WIGMOR_0508__dapF | Diaminopimelate epimerase |
| Down | WIGMOR_0443 | YhbN family protein | |
| dapA | Down | WIGMOR_0437__dapA | Dihydrodipicolinate synthase |
| ydjM | Down | WIGMOR_0156__ydjM | Putative inner membrane protein |
| fmt | Down | WIGMOR_0263__fmt | 10-formyltetrahydrofolate:L-methionyl- tRNA(fMet) N-formyltransferase |
| gpsA | Down | WIGMOR_0210__gpsA | Glycerol-3-phosphate dehydrogenase |
| recB | Down | WIGMOR_0512__recB | Exonuclease V subunit beta |
| pepA | Down | WIGMOR_0300__pepA | Aminopeptidase A |
| folD | Down | WIGMOR_0539__folD | Bifunctional 5,10-methylene-tetrahydrofolate dehydrogenase/5,10-methylene-tetrahydrofolate cyclohydrolase |
| rpsK | Down | WIGMOR_0183__rpsK | 30S ribosomal protein S11 |
| ileS | Down | WIGMOR_0483__ileS | Isoleucyl-tRNA synthetase |
| motB | Down | WIGMOR_0036__motB | Family proton-channel complex protein |
| ycfH | Down | WIGMOR_0102__ycfH | Putative metallodependent hydrolase |
| Down | WIGMOR_0595 | GltJ family glutamate/aspartate transport system permease protein | |
| rpsI | Down | WIGMOR_0645__rpsI | 30S ribosomal protein S9 |
| rpoH | Down | WIGMOR_0076__rpoH | RNA polymerase, sigma 32 (sigma H) factor |
| Down | WIGMOR_0325 | tRNA | |
| ribB | Down | WIGMOR_0545__ribB | 3,4-dihydroxy-2-butanone-4-phosphate synthase |
| acpP | Down | WIGMOR_0097__acpP | Acyl carrier protein (ACP) |
| nadA | Down | WIGMOR_0624__nadA | Quinolinate synthase subunit A |
| pal | Up | WIGMOR_0351__pal | Peptidoglycan-associated outer membrane lipoprotein |
| Up | WIGMOR_0666 | tRNA | |
| yebA | Up | WIGMOR_0126__yebA | Putative peptidase |
| rplX | Up | WIGMOR_0195__rplX | 50S ribosomal protein L24 |
| yggV | Up | WIGMOR_0082__yggV | dITP/XTP pyrophosphatase |
| guaB | Up | WIGMOR_0672__guaB | IMP dehydrogenase |
| Up | WIGMOR_0558 | tRNA | |
| rplB | Up | WIGMOR_0203__rplB | 50S ribosomal protein L2 |
| hflC | Up | WIGMOR_0597__hflC | Modulator for HflB protease specific for phage lambda cII repressor |
| rnhA | Up | WIGMOR_0072__rnhA | Ribonuclease HI |
| Up | WIGMOR_0662 | tRNA | |
| sucD | Up | WIGMOR_0346__sucD | Succinyl-CoA synthetase subunit alpha |
| purD | Up | WIGMOR_0245__purD | Phosphoribosylglycinamide synthetase phosphoribosylamine-glycine ligase |
| metG | Up | WIGMOR_0214__metG | Methionyl-tRNA synthetase |
| rpsC | Up | WIGMOR_0200__rpsC | 30S ribosomal protein S3 |
| flgE | Up | WIGMOR_0044__flgE | Flagellar hook protein |
| zapA | Up | WIGMOR_0328__zapA | Protein that localizes to the cytokinetic ring |
| fumC | Up | WIGMOR_0426__fumC | Fumarate hydratase |
| Up | WIGMOR_0128 | tRNA | |
| Up | WIGMOR_0135 | tRNA | |
| metK | Up | WIGMOR_0454__metK | Methionine adenosyltransferase 1 |
| infC | Up | WIGMOR_0086__infC | Protein chain initiation factor IF-3 |
| sucB | Up | WIGMOR_0344__sucB | Dihydrolipoyltranssuccinase |
| ftsH | Up | WIGMOR_0553__ftsH | Subunit of integral membrane ATP-dependent zinc metallopeptidase |
| Up | WIGMOR_0134 | 16S ribosomal RNA | |
| htpX | Up | WIGMOR_0111__htpX | Putative endopeptidase |
| Up | WIGMOR_0237 | tRNA | |
| rnhA | Up | WIGMOR_0072__rnhA | Ribonuclease HI |
| mreB | Up | WIGMOR_0141__mreB | Family actin-like cell wall component |
| ksgA | Up | WIGMOR_0022__ksgA | S-adenosylmethionine-6-N′,N′-adenosyl (rRNA) dimethyltransferase |
Wigglesworthia genes with significant differential expression, p < 0.05.
Table 2.
Wigglesworthia genes displaying significant differential expression in trypanosome infected vs. non-infected flies, 10 days post-infected blood meal uptake.
| Gene symbols | Expression | GenBank | Name |
|---|---|---|---|
| mreB | Down | WIGMOR_0141__mreB | Family actin-like cell wall component |
| lolE | Down | WIGMOR_0108__lolE | Membrane component of an ABC superfamily outer membrane-specific lipoprotein transporter |
| Down | WIGMOR_0238 | tRNA | |
| Down | WIGMOR_0236 | Translation elongation factor Tu | |
| yadG | Down | WIGMOR_0498__yadG | Putative ATP-binding component of an ABC superfamily transporter |
| Down | WIGMOR_0160 | tRNA | |
| htpX | Down | WIGMOR_0111__htpX | Putative endopeptidase |
| accD | Down | WIGMOR_0463__accD | Acetyl-CoA carboxylase carboxyl transferase, beta subunit |
| kdsA | Down | WIGMOR_0424__kdsA | 3-deoxy-D-manno-octulosonate 8-phosphate synthase |
| rplP | Down | WIGMOR_0199__rplP | 50S ribosomal protein L16 |
| clpX | Down | WIGMOR_0629__clpX | ATPase and specificity subunit of ClpX-ClpP ATP-dependent serine protease |
| secA | Down | WIGMOR_0585__secA | Preprotein translocase subunit, ATPase |
| hslV | Down | WIGMOR_0501__hslV | Peptidase component of the HslUV protease |
| Down | WIGMOR_0369 | tRNA | |
| Down | WIGMOR_0435 | tRNA | |
| rpsC | Down | WIGMOR_0200__rpsC | 30S ribosomal protein S3 |
| flgE | Down | WIGMOR_0044__flgE | Flagellar hook protein |
| gmk | Down | WIGMOR_0358__gmk | Guanylate kinase |
| nusG | Down | WIGMOR_0234__nusG | Transcription termination factor |
| yadG | Down | WIGMOR_0498__yadG | Putative ATP-binding component of an ABC superfamily transporter |
| sucB | Down | WIGMOR_0344__sucB | Dihydrolipoyltranssuccinase |
| pgk | Down | WIGMOR_0469__pgk | Phosphoglycerate kinase |
| mraW | Down | WIGMOR_0572__mraW | S-adenosyl-dependent methyltransferase |
| mreB | Down | WIGMOR_0141__mreB | Family actin-like cell wall component |
| acpP | Down | WIGMOR_0097__acpP | Acyl carrier protein (ACP) |
| mdlB | Down | WIGMOR_0567__mdlB | Putative ATP-binding component of multidrug ABC transporter |
| cysS | Down | WIGMOR_0538__cysS | Cysteinyl-tRNA synthetase |
| leuS | Down | WIGMOR_0615__leuS | Leucyl-tRNA synthetase |
| nusG | Down | WIGMOR_0234__nusG | Transcription termination factor |
| mreB | Down | WIGMOR_0141__mreB | Family actin-like cell wall component |
| atpA | Down | WIGMOR_0006__atpA | F1 sector of membrane-bound ATP synthase, alpha subunit |
| nadA | Down | WIGMOR_0624__nadA | Quinolinate synthase subunit A |
| bioA | Down | WIGMOR_0307__bioA | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase |
| Down | WIGMOR_0549 | tRNA | |
| Down | WIGMOR_0558 | tRNA | |
| ftsA | Down | WIGMOR_0582__ftsA | Cell division protein |
| fbaA | Down | WIGMOR_0468__fbaA | Fructose-bisphosphate aldolase |
| atpD | Down | WIGMOR_0008__atpD | F1 sector of membrane-bound ATP synthase, beta subunit |
| flhC | Down | WIGMOR_0034__flhC | Family flagellar transcriptional activator |
| alaS | Down | WIGMOR_0548__alaS | Alanyl-tRNA synthetase |
| Down | WIGMOR_0128 | tRNA | |
| proS | Down | WIGMOR_0145__proS | Prolyl-tRNA synthetase |
| rpsQ | Down | WIGMOR_0197__rpsQ | 30S ribosomal protein S17 |
| Down | WIGMOR_0458 | tRNA | |
| Down | WIGMOR_0134 | 16S ribosomal RNA | |
| Up | WIGMOR_0310 | ncRNA | |
| ribB | Up | WIGMOR_0545__ribB | 3,4-dihydroxy-2-butanone-4-phosphate synthase |
| Up | WIGMOR_0310 | ncRNA | |
| pnp | Up | WIGMOR_0564__pnp | Polynucleotide phosphorylase/polyadenylase |
| gyrB | Up | WIGMOR_0018__gyrB | DNA gyrase subunit B |
| rpsK | Up | WIGMOR_0183__rpsK | 30S ribosomal protein S11 |
| pepA | Up | WIGMOR_0300__pepA | Aminopeptidase A |
| flhD | Up | WIGMOR_0033__flhD | Family flagellar transcriptional activator |
| skp | Up | WIGMOR_0382__skp | Periplasmic chaperone |
| rpoH | Up | WIGMOR_0076__rpoH | RNA polymerase, sigma 32 (sigma H) factor |
| hemL | Up | WIGMOR_0077__hemL | Glutamate-1-semialdehyde aminotransferase |
| panB | Up | WIGMOR_0311__panB | 3-methyl-2-oxobutanoate hydroxymethyltransferase |
| groS | Up | WIGMOR_0520__groS | Cpn10 chaperonin GroES, small subunit of GroESL |
| groL | Up | WIGMOR_0521__groL | Cpn60 chaperonin GroEL, large subunit of GroESL |
| truA | Up | WIGMOR_0464__truA | tRNA pseudouridine synthase A |
| sufS | Up | WIGMOR_0410__sufS | PLP-dependent selenocysteine lyase |
| prlC | Up | WIGMOR_0367__prlC | Oligopeptidase A |
| rlmB | Up | WIGMOR_0605__rlmB | 23S rRNA Gm2251-methyltransferase |
| pykA | Up | WIGMOR_0648__pykA | Pyruvate kinase II |
Wigglesworthia genes with significant differential expression, p < 0.05.
Table 3.
Wigglesworthia genes displaying significant differential expression in trypanosome-infected vs. non-infected flies, 20 days post-infected blood meal uptake.
| Gene symbols | Expression | GenBank | Name |
|---|---|---|---|
| cls | Down | WIGMOR_0400__cls | Cardiolipin synthase 1 |
| rpoC | Down | WIGMOR_0228__rpoC | RNA polymerase, beta prime subunit |
| aroA | Down | WIGMOR_0272__aroA | 3-phosphoshikimate 1-Carboxyvinyltransferase |
| flgC | Down | WIGMOR_0042__flgC | Flagellar component of cell-proximal portion of basal-body rod |
| yoaE | Down | WIGMOR_0112__yoaE | Hypothetical protein |
| alaS | Down | WIGMOR_0548__alaS | Alanyl-tRNA synthetase |
| asnS | Down | WIGMOR_0515__asnS | Asparaginyl tRNA synthetase |
| purF | Down | WIGMOR_0460__purF | Amidophosphoribosyltransferase |
| thiE | Down | WIGMOR_0249__thiE | Thiamin phosphate synthase |
| yadG | Down | WIGMOR_0498__yadG | Putative ATP-binding component of an ABC superfamily transporter |
| fusA | Down | WIGMOR_0032__fusA | GTP-binding protein chain elongation factor EF-G |
| groL | Down | WIGMOR_0521__groL | Cpn60 chaperonin GroEL, large subunit of GroESL |
| metG | Down | WIGMOR_0214__metG | Methionyl-tRNA synthetase |
| gpmA | Down | WIGMOR_0568__gpmA | Phosphoglyceromutase |
| nadA | Up | WIGMOR_0624__nadA | Quinolinate synthase subunit A |
| thiH | Up | WIGMOR_0253__thiH | Thiamin biosynthesis ThiGH complex subunit |
| pgi | Up | WIGMOR_0144__pgi | Glucosephosphate isomerase |
| Up | WIGMOR_0290 | tRNA | |
| Up | WIGMOR_0147 | tRNA | |
| rpsB | Up | WIGMOR_0374__rpsB | 30S ribosomal protein S2 |
| pepA | Up | WIGMOR_0300__pepA | Aminopeptidase A |
| pheS | Up | WIGMOR_0089__pheS | Phenylalanine tRNA synthetase subunit alpha |
| hns | Up | WIGMOR_0399__hns | Global DNA-binding transcriptional dual regulator H-NS |
| sufA | Up | WIGMOR_0406__sufA | Fe-S cluster assembly protein |
| Up | WIGMOR_0092 | Ribonuclease E | |
| ahpC | Up | WIGMOR_0270__ahpC | Alkyl hydroperoxide reductase C22 protein |
| leuS | Up | WIGMOR_0615__leuS | Leucyl-tRNA synthetase |
| rnt | Up | WIGMOR_0432__rnt | Ribonuclease T (RNase T) |
| sufA | Up | WIGMOR_0406__sufA | Fe-S cluster assembly protein |
| pyrG | Up | WIGMOR_0413__pyrG | CTP synthetase |
| yidC | Up | WIGMOR_0013__yidC | Membrane insertion protein |
| rpoH | Up | WIGMOR_0076__rpoH | RNA polymerase, sigma 32 (sigma H) Factor |
| ruvC | Up | WIGMOR_0122__ruvC | Endonuclease component of RuvABC resolvasome |
| nrdF | Up | WIGMOR_0676__nrdF | Ferritin-like ribonucleoside-diphosphate reductase 2, beta subunit |
| bamA | Up | WIGMOR_0381__bamA | Outer membrane beta-barrel protein assembly factor |
| sufC | Up | WIGMOR_0408__sufC | Fe-S cluster assembly transport protein |
| skp | Up | WIGMOR_0382__skp | Periplasmic chaperone |
| gshA | Up | WIGMOR_0534__gshA | Gamma-glutamate-cysteine ligase |
| ftsY | Up | WIGMOR_0075__ftsY | Signal recognition particle protein |
Wigglesworthia genes with significantl differential expression, p < 0.05.
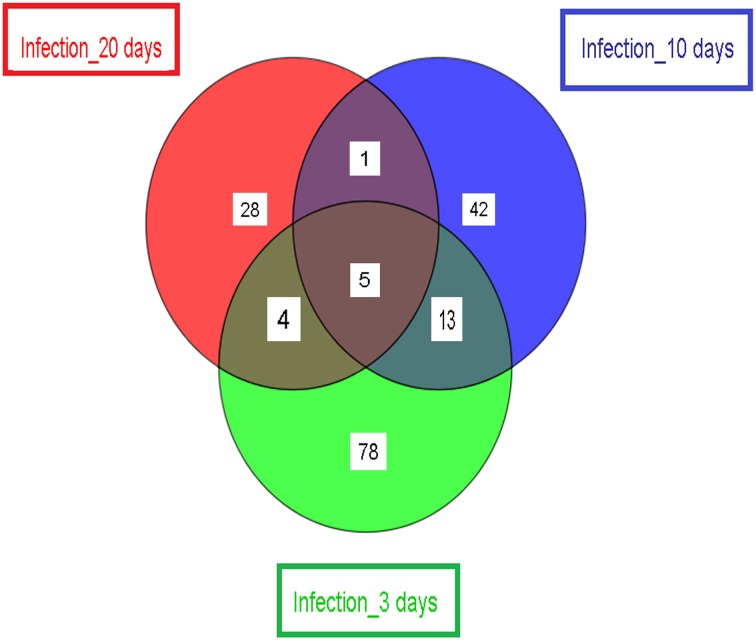
Finally, we compared the significantly differentially expressed Wigglesworthia genes identified by t-test in the infected (or stimulated) vs. non-infected (or non-stimulated) tsetse flies at the three selected time points (3, 10, and 20 days). The results, presented in Figure 4, demonstrate that only 5 genes (WIGMOR_0521__groL: Cpn60 chaperonin GroEL; WIGMOR_0076__rpoH: RNA polymerase, sigma 32 (sigma H) factor; WIGMOR_0624__nadA: quinolinate synthase subunit A; WIGMOR_0548__alaS: alanyl-tRNA synthetase and WIGMOR_0300__pepA: aminopeptidase A) were common at all three sampling times; 78, 42, and 28 genes were specific to the 3-, 10-, and 20-day samples, respectively (Supplementary Table S1); 13 genes were found in both 3- and 10-day samples; 4 genes were found in both 3- and 20-day samples; and only one gene was found in both 10- and 20-day samples (Supplementary Table S1).
Figure 4.
Schematic representation showing the number of significant differentially expressed Wigglesworthia genes. Samples are from day 3 (stimulated/non-stimulated flies) and days 10 and 20 (infected/non-infected flies). The number of differentially expressed genes (specific to the sampling time point) is displayed, as well as the number of differentially expressed genes shared by two or more samples.
GO annotation of the list of differentially expressed genes
We hypothesized that some of the differentially expressed genes would be involved in downstream molecular pathways that are important for maintaining susceptibility or refractoriness to trypanosome infection. To identify which types of global cellular processes or specific molecular functions are responsive, the list of differentially expressed genes obtained comparing the different conditions (stimulated/non-stimulated, infected/non-infected) from the three time points (3, 10, and 20 days) was annotated using the GO database (Blake and Harris, 2008). Each of the genes was assigned to molecular function, cellular component, and biological processes categories as designated by the GO database. Those GO categories significantly over-represented in the list of differentially expressed genes match a greater number of differentially expressed genes than would be expected by chance.
A significant over-representation of genes involved in “binding and metabolic processes” was identified in the Gene Ontology annotation of differentially expressed genes from 3-day samples (Table 4). This includes genes that encode proteins involved in protein synthesis as well as chaperonins (Cpn60 and Cpn10), which were all down-regulated in stimulated flies. The categories “developmental processes,” “morphogenesis,” and “cellular processes” displayed the greatest proportion of differentially expressed genes in the 10-day samples (Table 5), among which both chaperonins (Cpn60 and Cpn10) were up-regulated in infected flies. Surprisingly, no significant enrichment was obtained from differentially expressed Wigglesworthia genes in the 20-day fly samples.
Table 4.
Enriched Gene Ontology terms of differentially expressed genesa in trypanosome-stimulated vs. non-stimulated flies, 3 days post-blood meal uptake.
| GO accession | GO Termb | p-Value | Corrected p-valuec | Wigglesworthia genes |
|---|---|---|---|---|
| GO:0044267 | Cellular protein metabolic process | 6.505E-4 | 0.2539 | rpsK, rplP, rpsC, rplB, fmt, rpsG, fusA, rpsR, groS, groL, rpsI, infC |
| GO:0030554 | Adenyl nucleotide binding | 7.432E-4 | 0.2539 | groS, groL, ftsH, secA |
| GO:0032559 | Adenyl ribonucleotide binding | 7.432E-4 | 0.2539 | groS, groL, ftsH, secA |
| GO:0043170 | Macromolecule metabolic process | 2.837E-4 | 0.2539 | rpsK, rplP, rpsC, rplB, ksgA, rpoC, fmt, rpsG, fusA, rpoZ, recB, groS, groL, ftsH, rpsI, rnhA, rpoH, infC, rpsR |
| GO:004328 | ||||
| GO:0005524 | ATP binding | 7.432E-4 | 0.2539 | groS, groL, ftsH, secA |
| GO:0044238 | Primary metabolic process | 6.480E-4 | 0.25 | pyrI, rpsK, rplP, rpsC, rplB, ksgA, rpoC, fmt, gyrA, rpsG, fusA, rpoZ, recB, rpsR, groS, groL, ftsH, nadA, rpsI, rnhA, rpoH, infC, tmk |
| GO:0005515 | Protein binding | 3.805E-4 | 0.25 | groL, secA |
| |GO:0045308 | ||||
| GO:0019538 | Protein metabolic process | 6.859E-4 | 0.25 | rpsK, rplP, rpsC, rplB, fmt, rpsG, fusA, rpsR, groS, groL, ftsH, rpsI, infC |
| |GO:0006411 | ||||
| GO:0071704 | Organic substance metabolic process | 8.857E-4 | 0.2689 | pyrI, rpsK, rplP, rpsC, rplB, ksgA, rpoC, fmt, gyrA, rpsG, fusA, rpoZ, recB, rpsR, groS, groL, ribB, ftsH, nadA, rpsI, rnhA, rpoH, infC, tmk |
| GO:0044260 | Cellular macromolecule metabolic | 0.0012 | 0.3325 | rpsK, rplP, rpsC, rplB, ksgA, rpoC, fmt, rpsG, fusA, rpoZ, rpsR, groS, groL, rpsI, rpoH, infC |
| |GO:0034960 | Process | |||
| GO:0001883 | Purine nucleoside binding | 0.0030 | 0.4902 | fusA, groS, groL, ftsH, secA |
This set of genes was extracted from the full data set (673 genes and 5115 probes) using a t-test (p < 0.05).
GO terms that were overrepresented in the set of differentially expressed genes.
P-value after application of the Benjamini-Yekutelli correction (P < 0.5).
Table 5.
Enriched Gene Ontology terms of differentially expressed genesa in trypanosome-infected vs. non-infected flies, 10 days post-infected blood meal uptake.
| GO accession | GO Termb | p-Value | Corrected p-valuec | Wigglesworthia genes |
|---|---|---|---|---|
| GO:0048856 | Anatomical structure development | 1.508E-4 | 0.0555 | mreB |
| GO:0048869 | Cellular developmental process | 1.508E-4 | 0.0555 | mreB |
| GO:0032989 | Cellular component morphogenesis | 1.508E-4 | 0.0555 | mreB |
| GO:0009653 | Anatomical structure morphogenesis | 1.508E-4 | 0.0555 | mreB |
| GO:0044767 | Single-organism developmental process | 1.508E-4 | 0.0555 | mreB |
| GO:0000902 | Cell morphogenesis | 1.508E-4 | 0.0555 | mreB |
| |GO:0007148 | ||||
| |GO:0045790 | ||||
| |GO:0045791 | ||||
| GO:0009987 | Cellular process | 0.001 | 0.3449 | atpA, atpD, mreB, gyrB, rpsK, rpsQ, rplP, rpsC, gmk, flgE, truA, pgk, hslV, groS, groL, ribB, ftsA, nadA, clpX, pykA, rpoH |
| |GO:0008151 | ||||
| |GO:0050875 | ||||
| GO:0032502 | Developmental process | 0.001 | 0.3656 | mreB |
This set of genes was extracted from the full data set (673 genes and 5115 probes) using a t-test (p < 0.05).
GO terms that were overrepresented in the set of differentially expressed genes.
P-value after application of the Benjamini-Yekutelli correction (P < 0.5).
Discussion
W. glossinidia, the obligate symbiont of the tsetse fly, is involved in a large portion of physiological events in tsetse, including fly susceptibility or refractoriness to trypanosome infection. Several mechanisms have previously been reported, such as the modification of tsetse fly immunity or the supply of different nutrients (Pais et al., 2008; Wang et al., 2009; Rio et al., 2012; Snyder and Rio, 2013).
Presently, very few is known on the involvement of Wigglesworthia in tsetse flies vector competence, thus the reason of our study, a global transcriptomic approach which is the first one on the W. glossinidia gene differential expression. The analyses were performed at 3, 10, and 20 days post-feeding, in order to target differentially expressed genes involved in early events associated with trypanosome entry into the midgut (3 days sampling), with the establishment of infection (10 days sampling), and with events occurring relatively late in trypanosome infection time course, respectively.
Among the 673 Wigglesworthia genes, we identified (over the three sampling points) approximately 200 genes with significant expression changes in stimulated vs. non-stimulated flies (3-day sampling) and in infected vs. refractory flies (10- and 20-day samplings). The differences in the levels of gene expression were usually relatively weak (1.1–1.7-fold over- or under-expression), they nevertheless were statistically significant, suggestive of a biologically meaningful variation.
Only five genes were found in common when the list of genes showing modified expression in Wigglesworthia from 3-day samples was compared to those from 10- and 20-day samples. This result suggests the occurrence of highly complex and specific interactions that evolve during the time spent following the fly's challenge with trypanosomes.
PCA of the differentially expressed Wigglesworthia genes appears as a rather poor powerful tool for distinguishing stimulated from non-stimulated flies, as well as infected from non-infected flies. In contrast, clustering analysis of the gene expression data from the samples resulted in a hierarchical tree that discriminates stimulated from non-stimulated flies (3 days post-infected blood meal), and infected from non-infected flies (10 and 20 days post-infected blood meal). This clearly indicates that the expression of the Wigglesworthia genes not only evolves with time spent after the fly's infected blood meal (e.g., 3, 10, or 20 days), but that it also depends on the fly's status (stimulated or non-stimulated; infected or self-cured). These results are in line with those of Hamidou Soumana et al. (2014), who noted that trypanosome-responsive Sodalis glossinidius genes interact in well-defined patterns during the infection time course, making it possible to distinguish flies that are susceptible from those that are refractory to parasite infection.
Many of the Wigglesworthia genes that are down-regulated in trypanosome-stimulated and infected flies are involved in protein synthesis, such as the genes encoding ribosomal proteins, polymerases, elongation factors, etc. This was a somewhat surprising finding, as some of these proteins may be involved in pathogen survival (Nandan et al., 2002). Strikingly, we also observed that the expression of Cpn60 chaperonin GroEL, Cpn10 chaperonin GroES, and ncRNA (as well as genes implicated in the transport of bacterial toxins and in the synthesis of thiamine) were down-regulated in Wigglesworthia from stimulated flies (3 days post-infected blood meal), whereas the expression of the same three genes was up-regulated in Wigglesworthia from infected flies (10 days post-infected blood meal).
Interestingly, it has been reported that acute infection of mammalian cells with several types of viruses often results in the induction of heat-shock protein expression (Santoro, 1994), as observed for the up-regulation of chaperone proteins 24 h after infection with DENV (Chen et al., 2011). These studies support our findings for infected tsetse flies (10 days), in which two up-regulated genes (Cpn60 and Cpn10) from Wigglesworthia encode proteins involved in protein folding. Along these lines, the expression of a 60 kDa chaperone in the fly midgut was first described by Aksoy (1995b) and was reported once again by other researchers in 2002 (Haines et al., 2002). Several hypotheses have been made by Haines et al. (2002) which could explain the overexpression of genes encoding these chaperones:
Chaperones may be required for bacterial survival in the hostile environment of the tsetse midgut.
In other obligate endosymbionts, non-chaperonin activities have been reported for chaperones among which prevention of disassembly of invading microbes (Filichkin et al., 1997), protection from proteolytic degradation (Evans et al., 1992). In this context, Wigglesworthia may secrete chaperones that could bind proteins produced by the tsetse fly, thus protecting the trypanosome from the fly immune system.
Enterobacter aerogenes, for example, produces a chaperone that functions as an insect toxin, which contributes to paralyzing the ant-lion's insect prey (Yoshida et al., 2001; Haines et al., 2002). This chaperone contains four key residues (Val 100, Asn 101, Asp 338, and Ala 471) that are crucial for toxicity. The Wigglesworthia chaperone possesses three (Val 100, Asn 101, Ala 471) out of four of these crucial residues. This observation suggests that the W. glossinidia chaperones (Haines et al., 2002) (which have 86% homology with the E. aerogenes chaperone) could also function as toxins involved in the attrition process of trypanosomes during the early steps of their developmental cycle (occuring after the first 3 days following the infected blood meal).
Down-regulation of chaperone genes 3 days post-infected blood meal could allow further development of infection. This demonstrates the intricacy of regulations by chaperones, which depends on the time spent since the infected blood meal.
The development of trypanosome infection is complex, and several different molecules could be involved. For example, we have found that Δ(1)-pyrroline-5-carboxylate (P5C) dehydrogenase was down-expressed in tsetse flies stimulated by trypanosomes (3 days after an infected blood meal). The two-step oxidation of proline is catalyzed by proline oxidase and P5C dehydrogenase, to produce P5C and glutamate. When exogenous proline is supplied by the activities of proline oxidase and P5C reductase (conversion of P5C to proline), then impairment of P5C dehydrogenase activity can causes the P5C-proline cyclization. This proline is oxidized by the proline oxidase-FAD complex that delivers electrons to the electron transport chain and to O2, leading to mitochondrial reactive oxygen species (ROS) over-production. Coupled activity of proline oxidase and P5C dehydrogenase is therefore important for maintaining ROS homeostasis. In Trypanosoma cruzi, proline is involved in a variety of biological processes that are essential for pathogenesis (Contreras et al., 1985; Homsy et al., 1989; Martins et al., 2009).
In the case of tsetse flies that were fed on an infected blood meal, down-regulation of their P5C dehydrogenase could be due to a restriction in proline production at day 3 upon trypanosome ingestion. Furthermore, this may decrease the production of proline-rich proteins like tsetse EP (Haines et al., 2010) (involved in immunity).
The TolA (Gaspar et al., 2000) and UDP-N-Acetyl-glucosamine Acyltransferase are enzymes involved in the biosynthetic pathway of the lipopolysaccharide membrane, which acts as a barrier to the entry of antibacterial compounds (Nikaido, 1989). The Glossina scavenger peptidoglycan receptor PGRP-LB, plays a role in the detection and elimination of midgut trypanosomes (Wang et al., 2009). Furthermore, the virulence of some bacteria was shown to depend on secreted and cell wall peptidoglycan-associated virulence factors (Zawadzka-Skomial et al., 2006). At 3 days post-infected blood meal, the TolA and UDP-N-Acetyl-glucosamine Acyltransferase genes were down-regulated in Wigglesworthia from trypanosome-stimulated flies. This could favor a decrease in the stimulation of the tsetse immune response and, in turn, promote further establishment of trypanosomes.
Other proteins have been shown to be down-regulated in flies fed on infected blood meal, such as glycolytic enzymes. A number of these proteins, including glyceraldehyde-3-phosphate dehydrogenases (GAPDH), have been found to exhibit non-glycolytic functions which contribute to the ability of several bacterial pathogens to invade tissues (Boyle and Lottenberg, 1997; Tunio et al., 2010). In fact, viral infection usually causes the stimulation of glycolytic enzyme production in the midguts of Aedes aegypti infected with chikungunya and dengue-2 viruses (Tchankouo-Nguetcheu et al., 2010). The glycolytic enzyme GAPDH is also involved in the energetic metabolism of bloodstream trypanosomes, and can be considered as a virulence factor (Cronín et al., 1989). However, all of these processes do not likely occur in trypanosome-stimulated flies, since the glycolytic enzymes were shown to be down-regulated 3 days after flies were challenged with trypanosomes.
Therefore, our results show that trypanosome ingestion strongly alters gene expression of Wigglesworthia, even though the flies do not proceed with the developmental program of T. b. gambiense.
Ten days following an infected blood meal, the expression of several genes, such as quinolinate synthase, were down-regulated in Wigglesworthia from infected flies (as compared to non-infected flies). This enzyme allows the synthesis of quinolinic acid, a toxic molecule that has been detected in the central nervous system of patients displaying AIDS and meningitis (Eads et al., 1997).
In previous investigations, Weiss et al. (2011) showed that Wigglesworthia must be present during the immature larval stages for the tsetse fly immune system to develop and function properly at the adult stage. The artificial elimination of Wigglesworthia from larvae will compromise the fly immune system development throughout the development of the flies and the adult tsetse flies will display an immature immune system. In turn susceptibility to gut trypanosome infection in adult flies will be increased; in contrast, adult flies carrying Wigglesworthia will be highly resistant (Wang et al., 2009).
In addition, it has been suggested that differences in the susceptibility between Gmm and Gb are in turn caused by differences in the shikimate biosynthetic capabilities (phenylanine, folate and chorismate) of the Wigglesworthia strain they harbor (Rio et al., 2012). Our analyses failed to reveal such a role of the corresponding pathway in the vector competence of G. p. gambiensis for T. b. gambiense. Indeed, genes involved in this pathway were not found to be differentially expressed in stimulated flies at 3 days or in infected flies at 10 days, whereas some genes were found to be down-expressed (such as aro A) in infected flies at 20 days post-feeding.
Our study showed that, in trypanosomes stimulated flies (3 days post-infected bloodmeal), the expression of some Wigglesworthia genes are downregulated. This observation means that, despite its crucial role in fly resistance to trypanosome (as demonstrated by Wang et al., 2009; Weiss et al., 2011), the resistance level can be modulated by external factors which could favor further establishment of trypanosome. In the case of infected flies one of these external factors could be the trypanosome itself, against which the symbiont was unable to protect its host fly.
So the involvement of Wigglesworthia symbionts and fly immune system in tsetse fly infection by trypanosomes seems to be very complex; it is necessary but not sufficient to ensure fly resistance as demonstrated by the success of fly (carrying Wigglesworthia) infections performed out by number of investigators.
At the 3-day sampling time, we compared the gene expression of Wigglesworthia harbored within flies that ingested a non-infected blood meal to that of the symbiont of flies that ingested an infected blood meal. The blood of mice infected by trypanosomes may be quite different from non-infected mouse blood, as it transports a high concentration of trypanosomes but also may contain a range of other molecules (trypanosome-secreted proteins and other metabolites, mice reactive or degradation products, etc.). Thus, the question: besides the trypanosome itself, do these molecules, if any, that are not present in the non-infected blood meal ingested by control flies, contribute to alter the gene expression of the symbiont from flies having got the infected meal? In which extend? There is no response to this question. However, and whatever it may be, the altered gene expression of the symbiont from flies fed upon an infected meal is directly caused by the trypanosome presence, and possibly via molecules that it produced (or induced) during its development within the mammalian host. This question does not raise regarding the 10- and 20-day samples, as both groups of flies under comparison ingested the same first infected blood meal. Despite this similarity, the gene expressions differed once the flies of one group became trypanosome-infected (susceptible flies) and that of the second group self-cured the ingested trypanosomes (refractory flies).
Surprisingly, even genes encoding ribosomal proteins were differentially expressed; more surprisingly was that some of these genes were up-regulated whereas others were down-regulated. For example, in stimulated flies (S3) vs. non-stimulated flies (NS3), 50S ribosomal protein L2 and L24, and 30S ribosomal protein S3 were over-expressed whereas 50S ribosomal protein L16, and 30S ribosomal protein S7, S9, S11, and S18 were down regulated. Similar results were recorded especially in infected flies vs. refractory flies in day 10 samples (I10 vs. NI10). Similar results have been reported by Wang et al. (2013) on roots of Arabidopsis either phosphate- or iron-deficient vs. control roots. The authors considered the resulting alteration in ribosome composition as “a mechanism by which plants adapt to changing environmental conditions.” In our “model,” the stress is caused by the fly ingested trypanosomes, and results, among others, in differences in expression of genes encoding ribosomal proteins. Are these modifications involved in W. glosssinidia adaptation to changing environment caused by trypanosome invasion?
Finally, our results demonstrate that some genes considered as house-keeping genes are differentially expressed although their expression usually showed little variation. Fly infection cannot be compared with a classical physiological progression which could possibly account for the induction of unusual variations, even in the expression of such genes. Some of these genes also encode proteins displaying alternative functions (GAPDH, for example); alternative gene expression regulation may therefore occur regarding these genes.
The final objective of the global investigation we have undertaken is to decipher the molecular cross-talk between the tsetse fly, its symbionts, Sodalis and Wigglesworthia, and the trypanosome, in order to identify genes controlling crucial steps of fly infection by the parasite. The present study is the second step of this global investigation we have begun with the differential expression of Sodalis genes performed on the same biological samples. The levels of Wiggleworthia differentially expressed genes were lower than those recorded for Sodalis differentially expressed genes. Possibly the “flexibility” of gene expression is lesser in Wigglesworthia then in other bacteria; as an obligate symbiont, its fundamental physiological characteristics cannot be drastically modified otherwise it would no more fulfill its crucial role allowing fly survival. Nevertheless, differential expression of Wigglesworthia genes has been recorded, the corresponding genes identified and GO annotation performed. The results clearly demonstrate fly infection by trypanosomes to affect the symbiont transcriptomic machinery. Further investigations are necessary to assess the involvement of some of these differentially expressed genes in the parasite development process, and to evaluate their significance in the frame of an anti-vector competence strategy for combatting sleeping sickness.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the “Région Languedoc-Roussillon—Appel d'Offre Chercheur d'Avenir 2011,” the “Service de Coopération et d'Action Culturelle de l'Ambassade de France au Niger,” and the “Institut de Recherche pour le Développement” for their financial support. I. Hamidou Soumana is a PhD student supported by the Niger French Embassy, Service de Coopération et d'Action Culturelle (SCAC).
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00620/abstract
References
- Akman L., Yamashita A., Watanabe H., Oshima K., Shiba T., Hattori M., et al. (2002). Genome sequence of the endocellular obligate symbiont of tsetse, Wigglesworthia glossinidia. Nat. Genet. 32, 402–407. 10.1038/ng986 [DOI] [PubMed] [Google Scholar]
- Aksoy S. (1995a). Wigglesworthia gen. nov. and Wigglesworthia glossinidia sp. nov., taxa consisting of the mycetocyte-associated, primary endosymbionts of tsetse flies. Int. J. Syst. Bacteriol. 45, 848–851. 10.1099/00207713-45-4-848 [DOI] [PubMed] [Google Scholar]
- Aksoy S. (1995b). Molecular analysis of the endosymbionts of tsetse flies: 16S rDNA locus and over-expression of a chaperonin. Insect Mol. Biol. 4, 23–29. 10.1111/j.1365-2583.1995.tb00004.x [DOI] [PubMed] [Google Scholar]
- Aksoy S. (2000). Tsetse-a haven for microorganisms. Parasitol. Today 16, 114–118. 10.1016/S0169-4758(99)01606-3 [DOI] [PubMed] [Google Scholar]
- Aksoy S., Pourhosseini A. A., Chow A. (1995). Mycetome endosymbionts of tsetse flies constitute a distinct lineage related to Enterobacteriaceae. Insect Mol. Biol. 4, 15–22. 10.1111/j.1365-2583.1995.tb00003.x [DOI] [PubMed] [Google Scholar]
- Alam U., Medlock J., Brelsfoard C., Pais R., Lohs C., Balmand S., et al. (2011). Wolbachia symbiont infections induce strong cytoplasmic incompatibility in the tsetse fly Glossina morsitans. PLoS Pathog. 7:e1002415. 10.1371/journal.ppat.1002415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmand S., Lohs C., Aksoy S., Heddi A. (2013). Tissue distribution and transmission routes for the tsetse fly endosymbionts. J. Invertebr. Pathol. 112(Suppl.), S116–S122. 10.1016/j.jip.2012.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett M. P. (2006). The rise and fall of sleeping sickness. Lancet 367, 1377–1378. 10.1016/S0140-6736(06)68591-7 [DOI] [PubMed] [Google Scholar]
- Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D. C., et al. (2005). The genome of the African trypanosome Trypanosoma brucei. Science 309, 416–422. 10.1126/science.1112642 [DOI] [PubMed] [Google Scholar]
- Blake J. A., Harris M. A. (2008). The Gene Ontology (GO) project: structured vocabularies for molecular biology and their application to genome and expression analysis. Curr. Protoc. Bioinformatics 7:7.2 10.1002/0471250953.bi0702s23 [DOI] [PubMed] [Google Scholar]
- Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193. 10.1093/bioinformatics/19.2.185 [DOI] [PubMed] [Google Scholar]
- Boyle M. D., Lottenberg R. (1997). Plasminogen activation by invasive human pathogens. Thromb. Haemost. 77, 1–10. [PubMed] [Google Scholar]
- Butte A. (2002). The use and analysis of microarray data. Nat. Rev. Drug Discov. 1, 951–960. 10.1038/nrd961 [DOI] [PubMed] [Google Scholar]
- Cattand P., Jannin J., Lucas P. (2001). Sleeping sickness surveillance: an essential step towards elimination. Trop. Med. Int. Health 6, 348–361. 10.1046/j.1365-3156.2001.00669.x [DOI] [PubMed] [Google Scholar]
- Chen T. H., Tang P., Yang C. F., Kao L. H., Lo Y. P., Chuang C. K., et al. (2011). Antioxidant defense is one of the mechanisms by which mosquito cells survive dengue 2 viral infection. Virology 410, 410–417. 10.1016/j.virol.2010.12.013 [DOI] [PubMed] [Google Scholar]
- Cheng Q., Aksoy S. (1999). Tissue tropism, transmission and expression of foreign genes in vivo in midgut symbionts of tsetse flies. Insect Mol. Biol. 8, 125–132. 10.1046/j.1365-2583.1999.810125.x [DOI] [PubMed] [Google Scholar]
- Claverie J. M. (1999). Computational methods for the identification of differential and coordinated gene expression. Hum. Mol. Genet. 8, 1821–1832. 10.1093/hmg/8.10.1821 [DOI] [PubMed] [Google Scholar]
- Contreras V. T., Morel C. M., Goldenberg S. (1985). Stage specific gene expression precedes morphological changes during Trypanosoma cruzi metacyclogenesis. Mol. Biochem. Parasitol. 4, 83–96 10.1016/0166-6851(85)90108-2 [DOI] [PubMed] [Google Scholar]
- Cronín C. N., Nolan D. P., Voorheis H. P. (1989). The enzymes of the classical pentose phosphate pathway display differential activities in procyclic and bloodstream forms of Trypanosoma brucei. FEBS Lett. 244, 26–30. 10.1016/0014-5793(89)81154-8 [DOI] [PubMed] [Google Scholar]
- Dale C., Maudlin I. (1999). Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int. J. Syst. Bacteriol. 49, 267–275. 10.1099/00207713-49-1-267 [DOI] [PubMed] [Google Scholar]
- De Koning H. P. (2001). Transporters in African trypanosomes: role in drug action and resistance. Int. J. Parasitol. 31, 512–522. 10.1016/S0020-7519(01)00167-9 [DOI] [PubMed] [Google Scholar]
- Dukes P., Kaukas A., Hudson K. M., Asonganyi T., Gashumba J. K. (1989). A new method for isolating Trypanosoma brucei gambiense from sleeping sickness patients. Trans. R. Soc. Trop. Med. Hyg. 83, 636–639. 10.1016/0035-9203(89)90379-9 [DOI] [PubMed] [Google Scholar]
- Eads J. C., Ozturk D., Wexler T. B., Grubmeyer C., Sacchettini J. C. (1997). A new function for a common fold: the crystal structure of quinolinic acid phosphoribosyltransferase. Structure 5, 47–58. 10.1016/S0969-2126(97)00165-2 [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A. E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D. J., Jr., Evans D. G., Engstrand L., Graham D. Y. (1992). Urease-associated heat shock protein of Helicobacter pylori. Infect. Immun. 60, 2125–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farikou O., Njiokou F., Mbida Mbida J. A., Njitchouang G. R., Nana Djeunga H., Asonganyi T., et al. (2010). Tripartite interactions between tsetse flies, Sodalis glossinidius and trypanosomes-an epidemiological approach in two historical human African trypanosomiasis foci in Cameroon. Infect. Genet. Evol. 10, 115–121. 10.1016/j.meegid.2009.10.008 [DOI] [PubMed] [Google Scholar]
- Filichkin S. A., Brumfield S., Filichkin T. P., Young M. J. (1997). In vitro interactions of the aphid endosymbiotic SymL chaperonin with barley yellow dwarf virus. J. Virol. 71, 569–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frézil J. L., Cuisance D. (1994). Trypanosomiasis, diseases with future: prospects and uncertainty. Bull. Soc. Pathol. Exot. 87, 391–393. [PubMed] [Google Scholar]
- Gaspar J. A., Thomas J. A., Marolda C. L., Valvano M. A. (2000). Surface expression of O-specific lipopolysaccharide in Escherichia coli requires the function of the TolA protein. Mol. Microbiol. 38, 262–275. 10.1046/j.1365-2958.2000.02094.x [DOI] [PubMed] [Google Scholar]
- Haines L. R., Haddow J. D., Aksoy S., Gooding R. H., Pearson T. W. (2002). The major protein in the midgut of teneral Glossina morsitans morsitans is a molecular chaperone from the endosymbiotic bacterium Wigglesworthia glossinidia. Insect Biochem. Mol. Biol. 32, 1429–1438. 10.1016/S0965-1748(02)00063-2 [DOI] [PubMed] [Google Scholar]
- Haines L. R., Lehane S. M., Pearson T. W., Lehane M. J. (2010). Tsetse EP protein protects the fly midgut from trypanosome establishment. PLoS Pathog. 6:e1000793. 10.1371/journal.ppat.1000793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidou Soumana I., Loriod B., Ravel S., Tchicaya B., Simo G., Rihet P., et al. (2014). The transcriptional signatures of Sodalis glossinidius in the Glossina palpalis gambiensis flies negative for Trypanosoma brucei gambiense contrast with those of this symbiont in tsetse flies positive for the parasite: possible involvement of a Sodalis-hosted prophage in fly Trypanosoma refractoriness? Infect. Genet. Evol. 24, 41–56. 10.1016/j.meegid.2014.03.005 [DOI] [PubMed] [Google Scholar]
- Harley J. M. (1971). Comparison of the susceptibility of infection with Trypanosoma rhodesiense of Glossina pallidipes, G. morsitans, G. fuscipes and G. brevipalpis. Ann. Trop. Med. Parasitol. 65, 185–189. [DOI] [PubMed] [Google Scholar]
- Homsy J. J., Granger B., Krassner S. M. (1989). Some factors inducing formation of metacyclic stages of Trypanosoma cruzi. J. Protozool. 36, 150–153. 10.1111/j.1550-7408.1989.tb01063.x [DOI] [PubMed] [Google Scholar]
- Jackson A. P., Sanders M., Berry A., McQuillan J., Aslett M. A., Quail M. A., et al. (2014). The genome sequence of Trypanosoma brucei gambiense, causative agent of chronic human african trypanosomiasis. PLoS Negl. Trop. Dis. 4:e658. 10.1371/journal.pntd.0000658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn C. E., Charkowski A. O., Willis D. K. (2008). Evaluation of isolation methods and RNA integrity for bacterial RNA quantitation. J. Microbiol. Methods. 75, 318–324. 10.1016/j.mimet.2008.07.004 [DOI] [PubMed] [Google Scholar]
- Jamonneau V., Ravel S., Koffi M., Kaba D., Zeze D. G., Ndri L., et al. (2004). Mixed infections of trypanosomes in tsetse and pigs and their epidemiological significance in a sleeping sickness focus of Cote d'Ivoire. Parasitology 129, 693–702. 10.1017/S0031182004005876 [DOI] [PubMed] [Google Scholar]
- Kennedy P. G. (2008). The continuing problem of human African trypanosomiasis (sleeping sickness). Ann. Neurol. 64, 116–126. 10.1002/ana.21429 [DOI] [PubMed] [Google Scholar]
- Martins R. M., Covarrubias C., Rojas R. G., Silber A. M., Yoshida N. (2009). Use of L- proline and ATP production by Trypanosoma cruzi metacyclic forms as requirements for host cell invasion. Infect. Immun. 77, 3023–3032. 10.1128/IAI.00138-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matovu E., Seebeck T., Enyaru J. C. K., Kaminsky R. (2001). Drug resistance in Trypanosoma brucei spp., the causative agents of sleeping sickness in man and nagana in cattle. Microbes Infect. 3, 763–770. 10.1016/S1286-4579(01)01432-0 [DOI] [PubMed] [Google Scholar]
- Maudlin I., Ellis D. S. (1985). Association between intracellular Rickettsia-like infections of midgut cells and susceptibility to trypanosome infection in Glossina spp. Z. Parasitenkd. 71, 683–687. 10.1007/BF00925601 [DOI] [PubMed] [Google Scholar]
- Maudlin I., Welburn S. C. (1994). Maturation of trypanosome infections in tsetse. Exp. Parasitol. 79, 202–205. 10.1006/expr.1994.1081 [DOI] [PubMed] [Google Scholar]
- Moloo S. K., Asonganyi T., Jenni L. (1986). Cyclical development of Trypanosoma brucei gambiense from cattle and goats in Glossina. Acta Trop. 43, 407–408. [PubMed] [Google Scholar]
- Moloo S. K., Kabata J. M., Sabwa C. L. (1994). A study on the maturation of procyclic Trypanosoma brucei brucei in Glossina morsitans centralis and G. brevipalpis. Med. Vet. Entomol. 8, 369–374. 10.1111/j.1365-2915.1994.tb00100.x [DOI] [PubMed] [Google Scholar]
- Moloo S. K., Kutuza S. B. (1988). Comparative study on the susceptibility of different Glossina species to Trypanosoma brucei brucei infection. Trop. Med. Parasitol. 39, 211–213. [PubMed] [Google Scholar]
- Moser D. R., Cook G. A., Ochs D. E., Bailey C. P., McKane M. R., Donelson J. E. (1989). Detection of Trypanosoma congolense and Trypanosoma brucei subspecies by DNA amplification using the polymerase chain reaction. Parasitology 99, 57–66. 10.1017/S0031182000061023 [DOI] [PubMed] [Google Scholar]
- Nandan D., Yi T., Lopez M., Lai C., Reiner N. E. (2002). Leishmania EF-1alpha activates the Src homology 2 domain containing tyrosine phosphatase SHP-1 leading to macrophage deactivation. J. Biol. Chem. 277, 50190–50197. 10.1074/jbc.M209210200 [DOI] [PubMed] [Google Scholar]
- Nikaido H. (1989). Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob. Agents Chemother. 33, 1831–1836. 10.1128/AAC.33.11.1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogge G. (1976). Sterility in tsetse flies (Glossina morsitans Westwood) caused by loss of symbionts. Experentia 32, 995–996. 10.1007/BF01933932 [DOI] [PubMed] [Google Scholar]
- Nogge G. (1982). Significance of symbionts for the maintenance of an optional nutritional state for successful reproduction in hematophagous arthropods. Parasitology 82, 299–304. [Google Scholar]
- O'Neill S. L., Gooding R. H., Aksoy S. (1993). Phylogenetically distant symbiotic microorganisms reside in Glossina midgut and ovary tissues. Med. Vet. Entomol. 7, 377–383. [DOI] [PubMed] [Google Scholar]
- Pais R., Lohs C., Wu Y. N., Wang J. W., Aksoy S. (2008). The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl. Environ. Microbiol. 74, 5965–5974. 10.1128/AEM.00741-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X. Y., Kojima Y., Mizuno K., Ueoka K., Muroya K., Miyado M., et al. (2012). Identification of novel low-dose bisphenol a targets in human foreskin fibroblast cells derived from hypospadias patients. PLoS ONE 7:e36711. 10.1371/journal.pone.0036711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel S., Grébaut P., Cuisance D., Cuny G. (2003). Monitoring the developmental status of Trypanosoma brucei gambiense in the tsetse fly by means of PCR analysis of anal and saliva drops. Acta Trop. 88, 161–165. 10.1016/S0001-706X(03)00191-8 [DOI] [PubMed] [Google Scholar]
- Ravel S., Patrel D., Koffi M., Jamonneau V., Cuny G. (2006). Cyclical transmission of Trypanosoma brucei gambiense in Glossina palpalis gambiensis displays great differences among field isolates. Acta Trop. 100, 151–155. 10.1016/j.actatropica.2006.09.011 [DOI] [PubMed] [Google Scholar]
- Rio R. V., Hu Y., Aksoy S. (2004). Strategies of the home-team: symbioses exploited for vector-borne disease control. Trends Microbiol. 12, 325–336. 10.1016/j.tim.2004.05.001 [DOI] [PubMed] [Google Scholar]
- Rio R. V. M., Symula R. E., Wang J., Lohs C., Wu Y.-N., Snyder A. K., et al. (2012). Insight into the transmission biology and species-specific functional capabilities of tsetse (Diptera: Glossinidae) obligate symbiont Wigglesworthia. MBio 3, e00240–e00211. 10.1128/mBio.00240-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro M. G. (1994). Heat shock proteins and virus replication: hsp70s as mediators of the antiviral effects of prostaglandins. Experientia 50, 1039–1047. 10.1007/BF01923459 [DOI] [PubMed] [Google Scholar]
- Smyth G. K., Michaud J., Scott H. S. (2005). Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21, 2067–2075. 10.1093/bioinformatics/bti270 [DOI] [PubMed] [Google Scholar]
- Snyder A. K., Rio R. V. (2013). Interwoven biology of the tsetse holobiont. J. Bacteriol. 195, 4322–4330. 10.1128/JB.00487-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchankouo-Nguetcheu S., Khun H., Pincet L., Roux P., Bahut M., Huerre M., et al. (2010). Differential protein modulation in midguts of Aedes aegypti infected with chikungunya and dengue 2 viruses. PLoS ONE 5:10. 10.1371/journal.pone.0013149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunio S. A., Oldfield N. J., Berry A., Ala'Aldeen D. A., Wooldridge K. G., Turner D. P. (2010). The moonlighting protein fructose-1, 6-bisphosphate aldolase of Neisseria meningitidis: surface localization and role in host cell adhesion. Mol. Microbiol. 76, 605–615. 10.1111/j.1365-2958.2010.07098.x [DOI] [PubMed] [Google Scholar]
- Van den Abbeele J., Claes Y., Bockstaele D., Ray D., Coosemans M. (1999). Trypanosoma brucei spp. development in the tsetse fly: characterization of the post-mesocyclic stages in the foregut and proboscis. Parasitology 118, 469–478. 10.1017/S0031182099004217 [DOI] [PubMed] [Google Scholar]
- Vickerman K., Tetley L., Hendry A., Turner C. M. (1988). Biology of African trypanosomes in the tsetse fly. Biol. Cell 64, 109–119. 10.1016/0248-4900(88)90070-6 [DOI] [PubMed] [Google Scholar]
- Wang J., Lan P., Gao H., Zheng L., Li W., Schmidt W. (2013). Expression changes of ribosomal proteins in phosphate- and iron-deficient Arabidopsis roots predict stress-specific alterations in ribosome composition. BMC Genomics. 14:783. 10.1186/1471-2164-14-783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wu Y., Yang G., Aksoy S. (2009). Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc. Natl. Acad. Sci. U.S.A. 106, 12133–12138. 10.1073/pnas.0901226106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B. L., Maltz M., Aksoy S. (2012). Obligate symbionts activate immune system development in the tsetse fly. J. Immunol. 188, 3395–3403. 10.4049/jimmunol.1103691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B. L., Wang J., Maltz M. A., Wu Y., Aksoy S. (2013). Trypanosome infection establishment in the tsetse fly gut is influenced by microbiome-regulated host immune barriers. PLoS Pathog. 9:e1003318. 10.1371/journal.ppat.1003318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B. L., Wang J. W., Aksoy S. (2011). Tsetse immune system maturation requires the presence of obligate symbionts in larvae. PLoS Biol. 9:e1000619. 10.1371/journal.pbio.1000619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn S. C., Arnold K., Maudlin I., Gooday G. W. (1993). Rickettsia-like organisms and chitinase production in relation to transmission of trypanosomes by tsetse flies. Parasitology 107, 141–145. 10.1017/S003118200006724X [DOI] [PubMed] [Google Scholar]
- Yoshida N., Oeda K., Watanabe E., Mikami T., Fukita Y., Nishimura K., et al. (2001). Chaperonin turned insect toxin. Nature 411:44. 10.1038/35075148 [DOI] [PubMed] [Google Scholar]
- Zawadzka-Skomial J., Markiewicz Z., Nguyen-Distèche M., Devreese B., Frère J. M., Terrak M. (2006). Characterization of the bifunctional glycosyltransferase/acyltransferase penicillin-binding protein 4 of Listeria monocytogenes. J. Bacteriol. 188, 1875–1881. 10.1128/JB.188.5.1875-1881.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.