Abstract
Recent reports suggest that autoantibodies directed to aberrantly glycosylated mucins, in particular MUC1 and MUC4, are found in patients with colorectal cancer. There is, however, limited information on the autoantibody levels before clinical diagnosis, and their utility in cancer screening in the general population. In our study, we have generated O-glycosylated synthetic MUC1 and MUC4 peptides in vitro, to mimic cancer-associated glycoforms, and displayed these on microarrays. The assay’s performance was tested through an initial screening of serum samples taken from patients at the time of colorectal cancer diagnosis and healthy controls. Subsequently, the selected biomarkers were evaluated in a blinded nested case–control study using stored serum samples from among the 50,640 women randomized to the multimodal arm of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS), where women gave annual blood samples for several years. Cases were 97 postmenopausal women who developed colorectal cancer after recruitment and were age-matched to 97 women without any history of cancer. MUC1-STn and MUC1-Core3 IgG autoantibodies identified cases with 8.2 and 13.4% sensitivity, respectively, at 95% specificity. IgA to MUC4 glycoforms were unable to discriminate between cases and controls in the UKCTOCS sera. Additional analysis was undertaken by combining the data of MUC1-STn and MUC1-Core3 with previously generated data on autoantibodies to p53 peptides, which increased the sensitivity to 32.0% at 95% specificity. These findings suggest that a combination of antibody signatures may have a role as part of a biomarker panel for the early detection of colorectal cancer.
Keywords: MUC1, MUC4, glycosylation, early diagnosis, microarray, seromic profiling, biomarkers, cancer, autoantibodies, rectum, colon
What’s new? —
Serum antibodies against tumour-associated antigens (TAAs) have shown promise as biomarkers for early cancer detection. In this study, the authors asked whether autoantibodies to specific glycopeptides correlated with colorectal cancer (CRC). They found that an assay for autoantibodies to aberrant glycosylated MUC1 predicted CRC with 95% specificity—but with low sensitivity. However, when the assay combined MUC1 with p53, the sensitivity increased to 32%. These finding suggest that a combination of antibody signatures may eventually enable a biomarker panel for the early detection of CRC.
Autoantibodies against tumor-associated antigens (TAAs) are emerging as potent biomarkers for early cancer detection.1,2 However, autoantibodies to single targets do not possess sufficient diagnostic sensitivity and specificity to be used as screening tests. It is generally believed that multiplexed assays with several autoantibody targets will be required to achieve the performance characteristics necessary for cancer screening.2 Many informative cancer-associated antigens have been identified among intracellular proteins, including GPR78,3 p53,4 NY-ESO-15 and CDC25.6 In addition, some cell membrane glycoproteins have been identified as potential biomarkers, such as MUC1,7 HER28 and mesothelin.9 The combination of these and other markers has provided encouraging results, but higher sensitivity and specificity are required for screening of the general population.
A key feature of neoplastic cells is alterations in the post-translational modification of proteins and other molecules.10–12 Leading among them are alterations in O-linked glycosylation, which involves incomplete elongation of O-glycans on surface proteins creating immunogenic epitopes, such as Tn (GalNAc), STn (NeuAcα2,6GalNAc) and T (Galβ3GalNAc) antigens.13,14 Highly glycosylated mucin proteins such as MUC1 and MUC415 are overexpressed and aberrantly glycosylated in many carcinomas.16,17 The autoantibody response to such MUC1-associated O-glycopeptide epitopes has been demonstrated in patients with breast, ovarian, prostate and colorectal cancer.7,18,19 Both IgG autoantibodies to MUC1 glycoforms and IgA autoantibodies to MUC4 have been found in colorectal cancer patients.18 Using inhibition studies, epitope mapping and purification of autoantibodies with recombinant proteins, IgA and IgG responses have been shown to be highly specific to a combined epitope of an O-glycan and protein backbone.18,20 In contrast, IgM antibodies are unsuitable markers of disease as they recognize all glycoforms of the glycopeptides.19
We now report on the autoantibody signatures to MUC1 and MUC4 glycopeptides in colorectal cancer in both a clinical case–control set and a blinded nested case–control study with prospective collection of samples before outcome ascertainment.21 The latter sample set from UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS) includes serial samples taken before clinical diagnosis of colorectal cancer together with serial samples from age-matched controls. The design gave us a unique opportunity to assess development of an immunological response to aberrantly glycosylated MUC1 and MUC4 over time, and compare it to our previously reported p53 autoantibody profile in the same sample set.22 In addition, as the humoral immune response may have a prognostic impact, we analyzed the association of autoantibody response before diagnosis and mortality.
Material and Methods
Sample description
Human sera were obtained from two different sources: Asterand and the UKCTOCS.23
“Time of diagnosis” clinical sample set from Asterand
The “time of diagnosis” sample set consisted of 157 colorectal cancer patients (86 males and 71 females; mean age 59.3 ± 8.0 years) and 40 controls (20 males and 20 females; mean age 38.7 ± 7.7 years). The sample set included 69 Stage I, 69 Stage II and 19 Stage III patients. Controls were healthy volunteers from the same source. All cancer sera were taken at the time of diagnosis before cancer treatment, including surgery (further clinical information is provided in Supporting Information Table 1).
Sample set from the UKCTOCS
Serum samples from individuals participating in the multimodal arm of UKCTOCS trial23 were included. In this trial, 50,640 postmenopausal women were randomized to the multimodal group between 2001 and 2005, and donated samples annually until 2011. A rigid protocol was followed at all 13 trial centers with regard to sample collection.22 All women were flagged by the Health and Social Care Information Centre (HSCIC) for cancers and deaths. For our study, up-to-date cancer registry data were obtained in December 2009, yielding a median follow of 6.8 (interquartile range, IQR 5.9–8.4) years. Colorectal cancer cases were defined by cancer registration of malignant neoplasm of the colon, rectosigmoid junction or rectum (ICD-10 codes C18, C19 and C20) and a death certificate where the above ICD-10 codes were primary or contributory causes of death. Colorectal cancer notification was received in 101 women with serial samples, 97 of whom had given consent for secondary studies. All cancer sera were collected after randomization to the trial, but before diagnosis. Controls were women from the same trial center who had no history of any cancer at last follow-up and who had donated serial serum samples during the same period. Controls were matched to the cancers by age (within 5 years) at sample before diagnosis in a 1:1 ratio. Three controls were subsequently excluded from the analysis owing to notification of cancer diagnosis at final follow-up (February 2011). The final cohort consisted of 97 cases (297 samples) and 94 controls (286 serial samples). Baseline characteristics of the women have been previously published.22,24 The median age at randomization of women was 65.0 (25th–75th centiles, 61.3–70.4) for cases and 65.0 (25th–75th centiles, 60.2–69.7) for controls. For our study, the samples were retrieved from the off-site cryorepository and allowed to thaw briefly before being aliquoted into 2D barcoded tubes (0.5-ml Tracker Tubes in Loborack, MP52325, Micronics, High Wycombe, UK). The tubes were kept at −80°C until they were shipped to the University of Copenhagen where they were assayed blinded to the case–control status.
Synthetic peptides and recombinant proteins
Synthetic peptides were synthesized at Schaefer-N, Copenhagen, Denmark. Synthesis of recombinant mucin glycoprotein fragments in Escherichia coli was as follows: N- or C-terminally 6× His- and T7-tagged recombinant fragment of MUC2 and two MUC4 fragments [short (s) and long (L)] were produced in E. coli as previously described.18 Gene sequences were inserted into the bacterial expression vectors pET22 (Novagen), pET28 (Merck Millipore, Darmstadt, Germany) or pET28 (minus), a modified pET 28 vector without N-terminal tags and expressed in Rosetta2 (Novagen), nickel purified using NiNTA agarose (Qiagen, Venlo, NL) followed by HPLC purification before and after in vitro O-glycosylation. Products were analyzed by SDS-PAGE and MALDI-TOF mass spectrometry on a Voyager-DE™ PRO workstation (Applied Biosystems, Life Technologies, Carlsbad, CA) using 2,5-dihydroxybenzoic acid (Sigma, St. Louis, Missouri) as matrix.
Microarray
MUC1, MUC2 and MUC4 glycopeptides and glycoproteins were printed on Schott Nexterion® Slide H MPX 48 (Schott AG, Mainz, Germany) as previously described.18 In brief, triplicates of all peptides, in 150 mM sodium phosphate pH 8.5 with 0.005% CHAPS, were printed on a BioRobotics MicroGrid II spotter (Genomics Solution) with a 0.21-mm pitch using Stealth 3B Micro Spotting Pins (Telechem International ArrayIt Division). After printing, slides were incubated for 1 hr in a humidified hybridization chamber with 75% relative humidity and stored until use at −20°C. Before use unspotted slide areas were blocked for 1 hr with 50 mM ethanolamine in 50 mM sodium borate, pH 8.5. Human sera were diluted 1:4 and were incubated in a closed container with gentle agitation for 1 hr, washed thrice in PBS-Tween, followed by 1-hr incubation with appropriate secondary antibodies. Human IgG antibodies were detected with Cy3-conjugated goat anti-human IgG (Fc specific) (Sigma) diluted 1:4,000 in PBS-T. Human IgA antibodies were detected with Cy3-conjugated goat anti-human IgA (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA,). After incubation with secondary antibodies the slides were washed thrice in PBS-T, and after the final wash, slides were rinsed briefly in H2O, dried by centrifugation (200g) and scanned in a ProScanArray HT Microarray Scanner (PerkinElmer) followed by image analysis with ProScanArray Express 4.0 software (PerkinElmer). Each spot was done in three replicates and the mean value of relative fluorescence units (RFUs) was used. For comparison, slides were scanned with identical scanning parameters. The intra-assay coefficient of variation (CV) of spot triplicates was less than 10%. Interassay CVs were less than 30% for peptides with RFU values in the dynamic range.
Statistics
For each peptide, sensitivity was determined at 95% specificity for differential diagnosis in the “time of diagnosis” and for screening in the UKCTOCS sample set. Reactivity was defined as elevated if above the cutoff defined by the 95th in the control samples. To investigate if combining the autoantibody information to several key peptides improves predictive performance, a multivariate classifier approach was evaluated. Four different discriminant analysis (DA) methods were assessed (linear, quadratic, logistic and kth nearest neighbor DA) for their performance characteristics using leave-one-out cross-validation, where the discriminant function was determined with each sample left out in turn, and then used to classify that sample. This helped limit the upward bias in sensitivity resulting from using a classification rule on the same sample it was derived from. Subsequently, the predictive ability to identify colorectal cancer cases was further explored by combining the identified MUC1 autoantibody data with previously generated autoantibody data to p53 peptide in the same sample set. The p53 set was restricted to the seven autoantibodies to p53 peptide epitopes (-9, -10, -25, -34, -43, -44 and -58) that detected autoantibodies in 10% or more of the cancer patients at a specificity of 95% in both the “time of diagnosis” and UKCTOCS samples sets in our previous study.22 A maximum of two peptide autoantibodies from each study was allowed in the combined classifier to prevent overfitting and keep any biomarker combination clinically feasible. As quadratic DA had proven to be the best approach, only this method was used here, and again leave-one-out cross-validation was used.
To investigate whether autoantibodies to MUC1 or MUC4 peptides were prognostic markers related to risk of death from colorectal cancer, a “competing risks” regression model was used where the event of interest was death with colorectal cancer as primary or contributory cause, and a death from any other cause was considered a competing risk. All models were adjusted for age at sample. Data were analyzed and plotted using Stata 12 and GraphPad Prism software.
Results
Differential diagnosis using “time of diagnosis” set
IgG autoantibodies to MUC1 glycopeptides
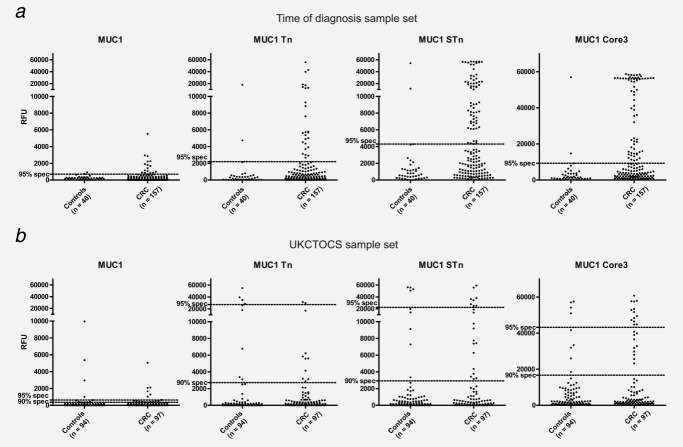
Sensitivity for detection of colorectal cancer was 16.6% (26/157), 42.0% (66/157) and 42.0% (66/157) for MUC1-Tn, MUC1-STn and MUC1-Core3, respectively, at 95% specificity (Table 1). Combining MUC1-STn and MUC1-Core3 resulted in a sensitivity of 44.6% (70/157) at 95% specificity. In keeping with our previous results, the presence of autoantibodies to MUC1-STn or MUC1-Core3 was not related to stage (Supporting Information Fig. 1).
Table 1.
IgG autoantibodies to MUC1 glycopeptides and p53 peptides
| Time of diagnosis CRC Stage I–III (n = 157) | UKCTOCS cases (n = 97) | |||
|---|---|---|---|---|
| Sensitivity (%) | Sensitivity (%) | |||
| Peptide | 95% specificity | 90% specificity | 95% specificity | 90% specificity |
| MUC1 | 10.2 (16/157) | 19.7 (31/157) | 7.2 (7/97) | 17.5 (17/97) |
| MUC1-Tn | 16.6 (26/157) | 35.7 (56/157) | 2.1 (2/97) | 11.4 (11/97) |
| MUC1-STn | 42.0 (66/157) | 50.3 (79/157) | 8.2 (8/97) | 21.6 (21/97) |
| MUC1-Core3 | 42.0 (66/157) | 48.4 (76/157) | 13.4 (13/97) | 23.7 (24/97) |
| MUC1-Core3 + MU1-STn | 44.6 (70/157) | 59.9 (94/157) | 14.4 (14/97) | 21.6 (21/97) |
| MUC1-STn + p53-10 + p53-25 | 20.4 (32/157) | 34.4 (54/157) | 24.7 (24/97) | 32.0 (31/97) |
| MUC1-Core3 + p53-10 + p53-25 | 35.0 (55/157) | 44.0 (69/157) | 26.8 (26/97) | 41.2 (40/97) |
| MUC1-Core3 + MUC1-STn + p53-10 + p53-25 | 47.8 (75/157) | 52.2 (82/157) | 32.0 (31/97) | 35.1 (34/97) |
| MUC1-Core3 + MUC1-STn + p53-58 | 47.8 (75/157) | 59.9 (94/157) | 19.6 (19/97) | 25.8 (25/97) |
| MUC1-Core3 + MUC1-STn + p53-43 | 54.8 (86/157) | 61.1 (96/157) | 23.7 (23/97) | 32.0 (31/97) |
Combination of MUC1 and p53
The combination of autoantibodies to p53 peptides, p53-43 and p53-58 increased the predictive ability to MUC1-STn and MUC1-Core3 reactivity. Autoantibodies to p53-43, MUC1-STn and MUC1-Core3 resulted in the highest sensitivity of 54.8% (86/157) at 95% specificity (Table 1).
IgA autoantibodies to MUC4 glycopeptides
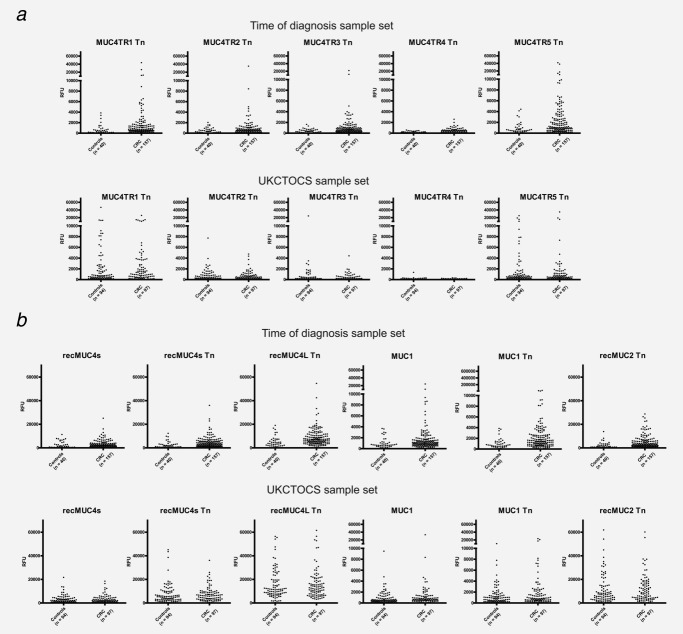
Similar to our previous findings,18 we found increased IgA reactivity to the five short GalNAc-glycosylated MUC4 tandem repeats (MUC4TR1–5) as well as the two recombinant fragments, which cover several MUC4 tandem repeats. The sensitivity varied between 9.6 (15/157) and 22.9% (36/157) at 95% specificity (Table 2 and Fig. 1). Combination of IgG MUC1 autoantibodies with IgA MUC4 antibodies made little difference to the performance of the MUC1 autoantibodies alone in the “time of diagnosis” sample set. Only MUC4TR5Tn appeared to add to the discriminant power of the MUC1 autoantibodies, increasing the AUC for the respective ROC curves by 0.046 (p-value for test of equal ROC curves = 0.031).
Table 2.
IgA autoantibodies to Tn-MUC1, 2 and 4
| Time of diagnosis CRC Stage I–III (n = 157) | UKCTOCS (n = 97) | |
|---|---|---|
| Peptide | Sensitivity (%)1 | Sensitivity (%)1 |
| MUC4TR1 Tn | 10.8 (17/157) | 7.2 (7/97) |
| MUC4TR2 Tn | 9.6 (15/157) | 6.4 (6/97) |
| MUC4TR3 Tn | 17.8 (28/157) | 3.1 (3/97) |
| MUC4TR4 Tn | 22.9 (36/157) | 2.0 (2/97) |
| MUC4TR5 Tn | 19.7 (31/157) | 3.1 (3/97) |
| recMUC4s Tn | 9.6 (15/157) | 8.2 (8/97) |
| recMUC4L Tn | 12.1 (19/157) | 6.2 (6/97) |
| MUC1 Tn | 19.1 (30/157) | 10.3 (10/97) |
| recMUC2 Tn | 17.8 (28/157) | 5.2 (5/97) |
At 95% specificity.
Figure 1.
IgA autoantibodies to MUC4 tandem repeat glycopeptides and mucins proteins in “time of diagnosis” and UKCTOCS set. (a) DOTPLOT of serum IgA autoantibodies to MUC4 tandem repeat glycopeptides measured by peptide-array assay and expressed as relative fluorescence units (RFUs) (y-axis). (b) DOTPLOT of serum IgA autoantibodies binding to recombinant mucin glycoproteins measured by peptide-array assay and expressed as RFUs (y-axis). For UKCTOCS set, serum from last sample before diagnosis in colorectal cancer cases (n = 97) and last sample taken for controls (n = 94) were used.
Screening using the case–control set from UKCTOCS
In the sample closest to diagnosis (median years before diagnosis 0.57 years, IQR 0.25–0.91), the sensitivity for colorectal cancer at 95% specificity was 8.2% (8/97) for MUC1-STn and 13.4% (13/97) for MUC1-Core3 (Fig. 2). Combining MUC1-STn and MUC1-Core3 reactivity resulted in a sensitivity of 14.4% (14/97) at 95% specificity (Table 1). No difference in autoantibody sensitivity for colorectal cancer was detected in hormone replacement therapy (HRT) versus non-HRT users. No difference was found for IgA autoantibodies to nonglycosylated and glycosylated MUC4 peptides between cases and controls (Table 2 and Fig. 1).
Figure 2.
IgG autoantibodies to MUC1 glycopeptides in “time of diagnosis” and UKCTOCS set. DOTPLOT of serum IgG autoantibodies binding to MUC1 glycopeptides measured by peptide-array assay and expressed as relative fluorescence units (RFUs) (y-axis). (a) Serum from healthy individuals (n = 40) and colorectal cancer patients (n = 157) in “time of diagnosis” set. Dotted lines represent 95% specificity. (b) Serum from last sample before diagnosis in colorectal cancer cases (n = 97) and last sample taken for controls (n = 94) from UKCTOCS set. Dotted lines indicate 90 or 95% specificity.
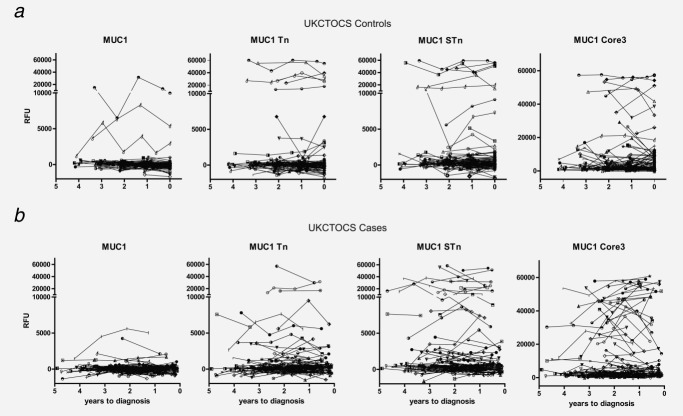
The appearance of autoantibodies to MUC1-STn and MUC1-Core3 was assessed over time in the serially collected samples before diagnosis in cases and matched serial samples in controls. In all MUC1-STn- or MUC1-Core3-positive cancer patients, detectable autoantibodies were detected in all serial samples taken before diagnosis (Fig. 3) with a median lead time of 2.23 (IQR 1.69–2.82) and 2.41 years (IQR 1.75–2.72), respectively. Of seven 15-mer p53 peptides from the whole p53 sequence previously reported,22 autoantibodies to p53-10 and p53-25 showed the highest added value to the MUC1 glycopeptide responses. On combining MUC1-STn and MUC1-Core3 autoantibodies with autoantibodies to these two p53 peptides improved sensitivity from 14.4 (14/97) to 32.0% (31/97) at 95% specificity (Table 1).
Figure 3.
Autoantibodies in prediagnostic serial samples from colorectal cancer patients (n = 97) and serial samples from controls (n = 94) to MUC1 glycopeptides. Each graph represents the autoantibody reactivity to MUC1 glycopeptides. Each symbol represents a single control or case individual. (a) Number of years to final serum sample taken is indicated for the controls on the x-axis, and (B) number of years before diagnosis for the cases is indicated on the x-axis. y-Axis represents relative fluorescence units (RFUs).
Prognosis
Of the 97 women with colorectal cancer, 35 died before censorship on June 13, 2013 (except for two subjects from Northern Ireland, censored on August 24, 2012). After correction for multiple testing, only MUC4TR5 of all the MUC1 or MUC4 autoantibodies was significantly (p = 0.000011) related to risk of death in patients with colorectal cancer. An increase in levels of 1 standard deviation was related to a 12.7% (95% CI: 6.86%, 18.9%) increase in the subhazard ratio (SHR) for death from colorectal cancer. In comparison, three (p53-4, p53-14 and p53-25) of the 18 p53 peptides analyzed were significantly (p < 0.05) related to risk of death, with increasing values associated with increasing risk of CR death (SHR > 1).
Discussion
To our knowledge, this is the first study that has investigated autoantibody responses to aberrantly O-glycosylated MUC1 and MUC4 in prospectively collected sample sets obtained before clinical diagnosis of colorectal cancer. MUC1-STn and MUC1-Core3 IgG autoantibodies identified cases with 8.2 and 13.4% sensitivity, respectively, at 95% specificity, whereas IgA to MUC4 glycoforms were unable to discriminate between cases and controls. Combining MUC1-STn and MUC1-Core3 reactivity and previously generated p53 peptide reactivity increased sensitivity to 32.0% at 95% specificity. These findings suggest that a combination of antibody signatures may have a role in a panel of early detection markers for colorectal cancer.
In the context of differential diagnosis, the sensitivity (at 95% specificity) of the autoantibody response to MUC1-STn and MUC1-Core3 in the clinical (time of diagnosis) set was 42% for each peptide. Although lower, this was comparable to our previous findings (MUC1-STn 56.9%; MUC1-Core3 44.8%) in other clinical sets.18 However, when we assessed sensitivity (at 95% specificity) in the context of screening using the preclinical sample closest to diagnosis in the population-based UKCTOCS set, it fell to 8.2% for MUC1-STn and 13.4% for MUC1-Core3 reactivity. Furthermore, although we found a difference in MUC4 IgA autoantibody reactivity between patients and controls in the clinical “time of diagnosis samples” set, there was no difference in the preclinical samples from women from UKCTOCS who developed colorectal cancer and age-matched controls. The lower sensitivity in the preclinical sample set is in keeping with findings from other studies using the less biased PRoBE (prospective specimen collection before outcome ascertainment and retrospective-blinded evaluation) study design21 for evaluating cancer biomarkers for screening.25 It reflects both the fact that the UKCTOCS set is an unbiased population set of colorectal cancer patients, while all patients who donate sera for research at time of diagnosis of their cancer are subgroups of the colorectal patient population and such cohorts are subject to a host of biases related to their mode of recruitment.
An increased number of controls in UKCTOCS had MUC1-STn and MUC1-Core3 autoantibodies compared to the low anti-MUC1 reactivity observed in the “time of diagnosis” controls. One explanation for the difference between the two sample sets is the lack of appropriate matching in the “time of diagnosis” sample set. Importantly, “time of diagnosis” control sera were from healthy volunteers with an average age of 38.7 years, compared with an average age of 59.3 years of cancer patients in the same set. The “time of diagnosis” controls are therefore likely to have an underrepresentation of individuals with other medical conditions.26 In addition, the higher average age of the UKCTOCS control group could have resulted in more individuals with inflammatory and other noncancer-related medical conditions that might have contributed to higher levels of MUC1-Core3/STn reactivity. This is supported by the trend toward higher levels of autoantibodies to MUC1-Core3 with higher age (Supporting Information Fig. 2). In this context, it is noteworthy that increased levels of autoantibodies to aberrantly glycosylated MUC1 have been found in patients with gastrointestinal inflammation.18,27 Another obvious difference between the two sets is that the “time of diagnosis” set also included male individuals. However, this is unlikely to have contributed to the differences, as there was no difference in autoantibody profile between males and females in the “time of diagnosis” set. The “time of diagnosis” cases were also not matched to controls for country of residence with a large proportion of the cancer cases being European Caucasians, while controls were predominantly American Caucasians. In contrast, the UKCTOCS controls were drawn from the same populations as the cancers and were therefore very closely matched to the cancers on most baseline characteristics including ethnicity and age. There is also the possibility that some of the MUC1-Core3 and -STn reactivity in UKCTOCS controls could reflect the presence of occult undiagnosed cancers in the controls. This would be in accordance with the previously described upregulation of STn in premalignant colonic tissue.28,29 As the UKCTOCS cohort is continually being followed up though cancer registries, it should be possible in due course to address this. Such issues are common in biomarkers studies using clinical sets to assess performance characteristics.30
Although a lower percentage of the UKCTOCS cancer patients were positive for MUC1-Core3 and MUC1-STn compared to the “time of diagnosis” sample set, we did not detect notable differences between the two sample sets for p53 autoantibodies.22 This indicates that autoantibodies are stable and do not degrade during the longer intervals to sample processing that in the UKCTOCS samples. Finally, in the UKCTOCS cohort, HRT use at recruitment was higher in those who developed colorectal cancer (p < 0.03). Although HRT has been reported to have confounding effects in protein biomarkers studies,31 we found no difference in autoantibody response to aberrant glycosylated MUC1 between HRT users and non-HRT users.
We previously found that autoantibodies to p53 could detect 25% of colorectal cancer patients at a median of 1.4 years before clinical diagnosis.22 Combining autoantibodies to aberrant glycosylated MUC1 and p53 increased the proportion of colorectal cancer cases detected to 32% at 95% specificity. However, a panel with MUC1 and p53 does not possess high enough sensitivity as a first-line screening test. Previous studies have shown that a larger antigen panel can improve sensitivity in clinical sets.8,32–35 Such antigen panels could be relevant if combined with current screening modalities that include a number of validated tests with high specificities and reasonable sensitivities (e.g., sigmoidoscopy, fecal occult blood and fecal DNA). Limited compliance is a major issue in relation to screening using the current tests. Furthermore, colonoscopy as first-line screen for colorectal cancer has recently been questioned given society’s current lack of capacity for primary colonoscopy screening.36 Incorporation of a blood test for autoantibodies in combination with other biomarkers, such as methylated SEPT9 DNA,37–39 into a “risk assessment evaluation” test may help to improve compliance with screening.
Presence of autoantibodies to MUC4TR5, p53-4, p53-14 and p53-25 was significantly related to risk of death due to colorectal cancer, with increasing values associated with increasing risk. Autoantibodies to p53 have previously been shown to be associated with decreased survival of patients with esophageal, colorectal and urological cancers.40–42 In ovarian cancer, however, a large study with 227 patients showed no correlation,43 whereas other studies have shown improved survival.44,45 A potential explanation for the survival benefits described in some studies could be that IgG immunity to p53 and other tumor antigens reflect the presence of antigen-specific T cells associated with increased survival. In contrast to the findings for p53, it was a surprise that there was an association between autoantibodies to MUC4TR5 and risk of death due to colorectal cancer. In this context, however, it should be noted that MUC4TR5 did not discriminate between cases and controls and only rather low levels of reactivity were observed. Further studies are clearly needed to verify the association between risk of death and autoantibodies to MUC4TR5. Finally, we found no association between risk of death due to colorectal cancer and MUC1 autoantibodies. Autoantibodies to MUC1 have been shown to be of prognostic value for poor clinical response and reduced overall survival in ovarian cancer,46 and other studies have shown association of autoantibodies to MUC1 to be associated with improved survival in breast cancer,47 gastric cancer,48 pancreatic cancer49 and lung cancer.50
In conclusion, although in clinical samples autoantibodies to glycosylated MUC1 and MUC4 showed encouraging sensitivity for differential diagnosis of colorectal cancer, their sensitivity for colorectal cancer screening in a population-based prediagnostic sample set was lower. However, MUC1 autoantibody signatures combined with p53 autoantibody signatures did detect additional patients than either alone, suggesting that autoantibody signatures may aid early detection of colorectal cancer. Further validation of these findings is warranted.
Acknowledgments
The authors express their gratitude to Karin Uch Hansen and Susanne Johannesen for excellent technical assistance. They are particularly grateful to the women throughout the UK who are participating in the UKCTOCS trial, to the entire medical, nursing and administrative staff who work on the UKCTOCS and to the independent members of the numerous oversight committees. The UKCTOCS trial was core funded by the Medical Research Council, Cancer Research UK and the Department of Health, with additional support from the Eve Appeal, Special Trustees of Bart’s and the London and Special Trustees of University College London Hospital (UCLH). A large portion of this work was done at UCLH/UCL within the “women’s health theme” of the National Institute for Health Research UCLH/UCL Comprehensive Biomedical Research Centre supported by the Department of Health. The researchers are independent from the funders.
Supporting Information
Additional Supporting Information may be found in the online version of this article
References
- Lu H, Goodell V, Disis ML. Humoral immunity directed against tumor-associated antigens as potential biomarkers for the early diagnosis of cancer. J Proteome Res. 2008;7:1388–94. doi: 10.1021/pr700818f. [DOI] [PubMed] [Google Scholar]
- Tainsky MA. Genomic and proteomic biomarkers for cancer: a multitude of opportunities. Biochim Biophys Acta. 2009;1796:176–93. doi: 10.1016/j.bbcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz PJ, Kim J, Do KA, et al. Fingerprinting the circulating repertoire of antibodies from cancer patients. Nat Biotechnol. 2003;21:57–63. doi: 10.1038/nbt774. [DOI] [PubMed] [Google Scholar]
- Lubin R, Schlichtholz B, Bengoufa D, et al. Analysis of p53 antibodies in patients with various cancers define B-cell epitopes of human p53: distribution on primary structure and exposure on protein surface. Cancer Res. 1993;53:5872–6. [PubMed] [Google Scholar]
- Kawabata R, Wada H, Isobe M, et al. Antibody response against NY-ESO-1 in CHP-NY-ESO-1 vaccinated patients. Int J Cancer. 2007;120:2178–84. doi: 10.1002/ijc.22583. [DOI] [PubMed] [Google Scholar]
- Liu WL, Zhang G, Wang JY, et al. Proteomics-based identification of autoantibody against CDC25B as a novel serum marker in esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2008;375:440–5. doi: 10.1016/j.bbrc.2008.08.039. [DOI] [PubMed] [Google Scholar]
- Snijdewint FG, von Mensdorff-Pouilly S, Karuntu-Wanamarta AH, et al. Cellular and humoral immune responses to MUC1 mucin and tandem-repeat peptides in ovarian cancer patients and controls. Cancer Immunol Immunother. 1999;48:47–55. doi: 10.1007/s002620050547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman C, Murray A, Chakrabarti J, et al. Autoantibodies in breast cancer: their use as an aid to early diagnosis. Ann Oncol. 2007;18:868–73. doi: 10.1093/annonc/mdm007. [DOI] [PubMed] [Google Scholar]
- Hellstrom I, Friedman E, Verch T, et al. Anti-mesothelin antibodies and circulating mesothelin relate to the clinical state in ovarian cancer patients. Cancer Epidemiol Biomarkers Prev. 2008;17:1520–6. doi: 10.1158/1055-9965.EPI-08-0039. [DOI] [PubMed] [Google Scholar]
- Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- Hakomori S. Glycosylation defining cancer malignancy: new wine in an old bottle. Proc Natl Acad Sci USA. 2002;99:10231–3. doi: 10.1073/pnas.172380699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldova R, Wormald MR, Dwek RA, et al. Glycosylation changes on serum glycoproteins in ovarian cancer may contribute to disease pathogenesis. Dis Markers. 2008;25:219–32. doi: 10.1155/2008/601583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer GF. T and Tn, general carcinoma autoantigens. Science. 1984;224:1198–206. doi: 10.1126/science.6729450. [DOI] [PubMed] [Google Scholar]
- Tarp MA, Clausen H. Mucin-type O-glycosylation and its potential use in drug and vaccine development. Biochim Biophys Acta. 2008;1780:546–63. doi: 10.1016/j.bbagen.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- Hattrup CL, Gendler SJ. Structure and function of the cell surface (tethered) mucins. Annu Rev Physiol. 2008;70:431–57. doi: 10.1146/annurev.physiol.70.113006.100659. [DOI] [PubMed] [Google Scholar]
- Taylor-Papadimitriou J, Burchell J, Miles DW, et al. MUC1 and cancer. Biochim Biophys Acta. 1999;1455:301–13. doi: 10.1016/s0925-4439(99)00055-1. [DOI] [PubMed] [Google Scholar]
- Pedersen JW, Blixt O, Bennett EP, et al. Seromic profiling of colorectal cancer patients with novel glycopeptide microarray. Int J Cancer. 2011;128:1860–71. doi: 10.1002/ijc.25778. [DOI] [PubMed] [Google Scholar]
- Wandall HH, Blixt O, Tarp MA, et al. Cancer biomarkers defined by autoantibody signatures to aberrant O-glycopeptide epitopes. Cancer Res. 2010;70:1306–13. doi: 10.1158/0008-5472.CAN-09-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt O, Clo E, Nudelman AS, et al. A high-throughput O-glycopeptide discovery platform for seromic profiling. J Proteome Res. 2010;9:5250–61. doi: 10.1021/pr1005229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe MS, Feng Z, Janes H, et al. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst. 2008;100:1432–8. doi: 10.1093/jnci/djn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen JW, Gentry-Maharaj A, Fourkala EO, et al. Early detection of cancer in the general population: a blinded case-control study of p53 autoantibodies in colorectal cancer. Br J Cancer. 2013;108:107–14. doi: 10.1038/bjc.2012.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon U, Gentry-Maharaj A, Ryan A, et al. Recruitment to multicentre trials—lessons from UKCTOCS: descriptive study. BMJ. 2008;337:a2079. doi: 10.1136/bmj.a2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon U, Gentry-Maharaj A, Hallett R, et al. Sensitivity and specificity of multimodal and ultrasound screening for ovarian cancer, and stage distribution of detected cancers: results of the prevalence screen of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS) Lancet Oncol. 2009;10:327–40. doi: 10.1016/S1470-2045(09)70026-9. [DOI] [PubMed] [Google Scholar]
- Cramer DW, Bast RC, Jr, Berg CD, et al. Ovarian cancer biomarker performance in prostate, lung, colorectal, and ovarian cancer screening trial specimens. Cancer Prev Res (Phila) 2011;4:365–74. doi: 10.1158/1940-6207.CAPR-10-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chechlinska M, Kowalewska M, Nowak R. Systemic inflammation as a confounding factor in cancer biomarker discovery and validation. Nat Rev Cancer. 2010;10:2–3. doi: 10.1038/nrc2782. [DOI] [PubMed] [Google Scholar]
- Furr AE, Ranganathan S, Finn OJ. Aberrant expression of MUC1 mucin in pediatric inflammatory bowel disease. Pediatr Dev Pathol. 2010;13:24–31. doi: 10.2350/08-06-0479.1. [DOI] [PubMed] [Google Scholar]
- Itzkowitz SH, Bloom EJ, Lau TS, et al. Mucin associated Tn and sialosyl-Tn antigen expression in colorectal polyps. Gut. 1992;33:518–23. doi: 10.1136/gut.33.4.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzkowitz SH, Young E, Dubois D, et al. Sialosyl-Tn antigen is prevalent and precedes dysplasia in ulcerative colitis: a retrospective case-control study. Gastroenterology. 1996;110:694–704. doi: 10.1053/gast.1996.v110.pm8608878. [DOI] [PubMed] [Google Scholar]
- Diamandis EP. Cancer biomarkers: can we turn recent failures into success? J Natl Cancer Inst. 2010;102:1462–7. doi: 10.1093/jnci/djq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitteri SJ, Hanash SM. Confounding effects of hormone replacement therapy in protein biomarker studies. Cancer Epidemiol Biomarkers Prev. 2011;20:134–9. doi: 10.1158/1055-9965.EPI-10-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman CJ, Thorpe AJ, Murray A, et al. Immunobiomarkers in small cell lung cancer: potential early cancer signals. Clin Cancer Res. 2011;17:1474–80. doi: 10.1158/1078-0432.CCR-10-1363. [DOI] [PubMed] [Google Scholar]
- Boyle P, Chapman CJ, Holdenrieder S, et al. Clinical validation of an autoantibody test for lung cancer. Ann Oncol. 2011;22:383–9. doi: 10.1093/annonc/mdq361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnjatic S, Ritter E, Buchler MW, et al. Seromic profiling of ovarian and pancreatic cancer. Proc Natl Acad Sci USA. 2010;107:5088–93. doi: 10.1073/pnas.0914213107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Choi G, Li L, et al. Occurrence of autoantibodies to annexin I, 14-3-3 theta and LAMR1 in prediagnostic lung cancer sera. J Clin Oncol. 2008;26:5060–6. doi: 10.1200/JCO.2008.16.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilschut JA, Habbema JD, van Leerdam ME, et al. Fecal occult blood testing when colonoscopy capacity is limited. J Natl Cancer Inst. 2011;103:1741–51. doi: 10.1093/jnci/djr385. [DOI] [PubMed] [Google Scholar]
- Church TR, Wandell M, Lofton-Day C, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2013 doi: 10.1136/gutjnl-2012-304149. doi: 10.1136/gutjnl-2012-304149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deVos T, Tetzner R, Model F, et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem. 2009;55:1337–46. doi: 10.1373/clinchem.2008.115808. [DOI] [PubMed] [Google Scholar]
- Tanzer M, Balluff B, Distler J, et al. Performance of epigenetic markers SEPT9 and ALX4 in plasma for detection of colorectal precancerous lesions. PLoS One. 2010;5:e9061. doi: 10.1371/journal.pone.0009061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang C, Unteregger G, Kartarius S, et al. p53 autoantibodies in patients with urological tumours. Br J Urol. 1998;82:721–6. doi: 10.1046/j.1464-410x.1998.00814.x. [DOI] [PubMed] [Google Scholar]
- Bergqvist AS, Bergqvist M, Brattstrom D, et al. Serum p53 autoantibodies as prognostic marker in patients with oesophageal carcinoma. Anticancer Res. 2001;21:4141–5. [PubMed] [Google Scholar]
- Tang R, Ko MC, Wang JY, et al. Humoral response to p53 in human colorectal tumors: a prospective study of 1,209 patients. Int J Cancer. 2001;94:859–63. doi: 10.1002/ijc.1541. [DOI] [PubMed] [Google Scholar]
- Hogdall EV, Hogdall CK, Blaakaer J, et al. P53 autoantibodies in sera from Danish ovarian cancer patients and their correlation with clinical data and prognosis. APMIS. 2002;110:545–53. doi: 10.1034/j.1600-0463.2002.11007805.x. [DOI] [PubMed] [Google Scholar]
- Goodell V, Salazar LG, Urban N, et al. Antibody immunity to the p53 oncogenic protein is a prognostic indicator in ovarian cancer. J Clin Oncol. 2006;24:762–8. doi: 10.1200/JCO.2005.03.2813. [DOI] [PubMed] [Google Scholar]
- Anderson KS, Wong J, Vitonis A, et al. p53 autoantibodies as potential detection and prognostic biomarkers in serous ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:859–68. doi: 10.1158/1055-9965.EPI-09-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budiu RA, Mantia-Smaldone G, Elishaev E, et al. Soluble MUC1 and serum MUC1-specific antibodies are potential prognostic biomarkers for platinum-resistant ovarian cancer. Cancer Immunol Immunother. 2011;60:975–84. doi: 10.1007/s00262-011-1010-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mensdorff-Pouilly S, Verstraeten AA, Kenemans P, et al. Survival in early breast cancer patients is favorably influenced by a natural humoral immune response to polymorphic epithelial mucin. J Clin Oncol. 2000;18:574–83. doi: 10.1200/JCO.2000.18.3.574. [DOI] [PubMed] [Google Scholar]
- Kurtenkov O, Klaamas K, Mensdorff-Pouilly S, et al. Humoral immune response to MUC1 and to the Thomsen-Friedenreich (TF) glycotope in patients with gastric cancer: relation to survival. Acta Oncol. 2007;46:316–23. doi: 10.1080/02841860601055441. [DOI] [PubMed] [Google Scholar]
- Hamanaka Y, Suehiro Y, Fukui M, et al. Circulating anti-MUC1 IgG antibodies as a favorable prognostic factor for pancreatic cancer. Int J Cancer. 2003;103:97–100. doi: 10.1002/ijc.10801. [DOI] [PubMed] [Google Scholar]
- Hirasawa Y, Kohno N, Yokoyama A, et al. Natural autoantibody to MUC1 is a prognostic indicator for non-small cell lung cancer. Am J Respir Crit Care Med. 2000;161:589–94. doi: 10.1164/ajrccm.161.2.9905028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.