SUMMARY
Small RNAs impact several cellular processes through gene regulation. Argonaute proteins bind small RNAs to form effector complexes that control transcriptional and post-transcriptional gene expression. PIWI proteins belong to the Argonaute protein family, and bind PIWI-interacting RNAs (piRNAs). They are highly abundant in the germline, but are also expressed in some somatic tissues. The PIWI/piRNA pathway has a role in transposon repression in Drosophila, which occurs both by epigenetic regulation and post-transcriptional degradation of transposon mRNAs. These functions are conserved, but clear differences in the extent and mechanism of transposon repression exist between species. Mutations in piwi genes lead to the upregulation of transposon mRNAs. It is hypothesized that this increased transposon mobilization leads to genomic instability and thus sterility, although no causal link has been established between transposon upregulation and genome instability. An alternative scenario could be that piwi mutations directly affect genomic instability, and thus lead to increased transposon expression. We propose that the PIWI/piRNA pathway controls genome stability in several ways: suppression of transposons, direct regulation of chromatin architecture and regulation of genes that control important biological processes related to genome stability. The PIWI/piRNA pathway also regulates at least some, if not many, protein-coding genes, which further lends support to the idea that piwi genes may have broader functions beyond transposon repression. An intriguing possibility is that the PIWI/piRNA pathway is using transposon sequences to coordinate the expression of large groups of genes to regulate cellular function.
INTRODUCTION
Small RNA pathways have diverse roles in regulating gene expression in eukaryotic organisms. Post-transcriptional gene silencing via translational repression and mRNA degradation, is ubiquitous in animals, plants, and fungi (Ghildiyal and Zamore, 2009). In addition, small RNAs are able to direct heterochromatin formation in both fission yeast and plants, thereby silencing gene transcription (Martienssen et al., 2008). The profound impact of small RNA pathways on gene regulation is obvious from the significant roles they play in a variety of biological processes including stem cell self-renewal and differentiation (Gangaraju and Lin, 2009; Subramanyam and Blelloch, 2011), various aspects of animal development (Stefani and Slack, 2008), germline development (Saxe and Lin, 2011), and human diseases including cancer (Esteller, 2011). It is increasingly clear that small RNA pathways exert significant control over the expression of large numbers of genes, and therefore can exert significant influence over gene networks.
Three major classes of small RNAs have been identified in animals: microRNAs (miRNAs), small interfering RNAs (siRNAs), and PIWI-interacting RNAs (piRNAs), and each class operates in a distinct pathway (reviewed in Ghildiyal and Zamore, 2009). Mature small RNAs associate with Argonaute proteins and guide them to their sites of action—for example, to cleave target RNAs or direct epigenetic changes on chromatin (Hammond et al., 2001; Liu et al., 2004a). Phylogenetic analysis clearly distinguishes two subfamilies of Argonaute proteins: the AGO and PIWI subfamilies (Mochizuki et al., 2002). AGO proteins are ubiquitously expressed in animal tissues and bind both miRNAs and siRNAs, whereas PIWI subfamily proteins bind piRNAs and exhibit more restricted expression patterns that include germline and adult stem cells (reviewed in Juliano et al., 2011). The founding member of the PIWI family was identified as an essential gene for the maintenance of fertility in Drosophila. Subsequent work demonstrated evolutionary conservation in germline expression and the requirement for fertility in Caenorhabditis elegans, zebrafish, and mice (Lin and Spradling, 1997; Cox et al., 1998; Deng and Lin, 2002; Houwing et al., 2007). Although originally identified in the germline, PIWI proteins are also expressed in somatic tissues, including different kinds of adult stem cells that reside within these tissues (see Table 1 for comprehensive classification of expression patterns). piRNAs are approximately 23–31 nucleotides long and are highly expressed in the Drosophila germline and mouse testes (Aravin et al., 2006; Girard et al., 2006; Grivna et al., 2006a; Saito et al., 2006; Vagin et al., 2006; Brennecke et al., 2007). piRNA populations exhibit stark differences when compared to miRNA populations. miRNAs are often conserved between species and exhibit limited diversity. For example, the human genome is predicted to encode between 1,000 and 10,000 miRNA precursor sequences (Bentwich et al., 2005; Miranda et al., 2006). By contrast, one species has hundreds of thousands unique piRNA sequences, and these sequences are not conserved between species. This sequence complexity of piRNA populations makes functional deductions challenging. In Drosophila, the majority of piRNAs share sequences with repetitive elements such as transposons. This observation lends support to the widely accepted hypothesis that the PIWI/piRNA pathway represses transposon expression in the germline (Brennecke et al., 2007; Gunawardane et al., 2007). Indeed, when PIWI proteins are depleted in Drosophila, transposon levels increase (Reiss et al., 2004; Sarot et al., 2004; Savitsky et al., 2006).
TABLE 1.
Survey of Piwi Proteins Across Animal Phylogeny: Somatic Expression Versus Germline Expression and Nuclear Expression Versus Cytoplasmic Expression
| Organism | Homologues | Localization | Expression | |
|---|---|---|---|---|
| Germline* | Soma* | |||
| Fly | Piwi | Cytoplasm, Nucleus | PGCs (Megosh et al, 2006) |
Embryonic somatic cells (Megosh et al, 2006) |
| Nucleus | Ovary-GSCs, 16 cell cysts. Oocyte, Nurse cells (Cox et al, 1998) |
Ovary - Terminal filament, Epithelial sheath cells, Follicle cells (Cox et al, 1998) |
||
| Testis - GSCs, Gonialblasts (Cox et al, 2000) |
Testis - Hub cells, Somatic stem cells, Cyst progenitor cells (Cox et al, 2000) |
|||
| Salivary Gland (Brower-Toland et al, 2007) |
||||
| Aub | Cytoplasm | PGCs (Harris and Macdonald, 2001) | Embryonic soma (Harris and Macdonald, 2001) | |
| Ovary - GSCs, all cyst cells, Oocyte, Nurse cells (Brennecke et al, 2007) |
||||
| Testis - GSCs, Gonialblasts, Spermatogonia, Spermatocytes (Nishida et al, 2007) |
||||
| Ago3 | Cytoplasm | Ovary - Germline stem cells, All cyst cells, Oocyte, Nurse cells (Gunawardane et al, 2007) |
Ovary- Faint in follicle cells (Gunawardane et al, 2007) Cap cells (Brennecke et al, 2007) |
|
| Testis - GSCs, Gonialblasts, Spermatogoni a (Nagao et al, 2010) |
||||
| Mouse | MIWI | Cytoplasm, Nucleus (Dense body) (Beyret and Lin, 2011) |
Testis -Meiotic spermatocytes, Elongating spermatids (Deng and Lin, 2002) |
|
| Unknown | Pancreas (Yan et al, 2011) Brain, Heart, Liver, Lung, Kidney (Lee et al, 2011) |
|||
| MILI | Cytoplasm, Nucleus (Dense body) (Beyret and Lin, 2011) |
Testis - GSCs, Gonocytes, Spermatogonia, Meiotic spermatocytes, Spermatids (Kuramochi-Miyagawa et al. 2004, Unhavaithaya et al. 2009, Wang et al. 2009) |
||
| Unknown | Mesenchymal stem cells (Wu et al, 2010) |
|||
| MIWI2 | N ucleus, Cytoplasm | Testis - GSCs (Aravin et al, 2008) |
Testis - Sertoli cells (Carmell et al, 2007) |
|
| Zebrafish | Ziwi | Cytoplasm | PGCs (Houwing et al, 2007) |
|
| Ovary - Oogonia, Stage 1 oocytes (Houwing et al, 2007) | ||||
| Testis - Spermatogonia, Spermatocytes (Houwing et al, 2007) | ||||
| Zili | Nucleus | PGCs (Houwing et al, 2008) |
Embryonic soma (Sun et al, 2010) |
|
| Cytoplasm, Nucleus | Ovary - Oogonia, Stage l-IV Oocytes (Houwing et al, 2008) |
|||
| Cytoplasm | Testis - Spermatogonia, Spermatocytes, Spermatids (Houwing et al, 2008) |
|||
| Frog | Xiwi | Cytoplasm, Nucleus(Mitotic/Meiotic Spindles) |
Embryos Stage 1–20. Germline/Somatic separation unknown(Wilczynska et al, 2009) | |
| Ovary - Stage I - IV occytes, Mature oocytes (Lau et al, 2009) |
||||
| Unknown | Testis (Lau et al, 2009) | |||
| Xili | Unknown | Embryos Stage 1–42. Germline/Somatic separation unknown (Wilczynska et al, 2009) |
||
| Cytoplasm | Ovary - Stage I - IV occytes, Mature oocytes (Wilczynska et al, 2009) |
|||
| Unknown | Testis (Wilczynska et al, 2009) | |||
| Nematode | PRG1 | Cytoplasm | Gonad- Germline stem cells, Mitotic/meiotic germ cells, Mature oocytes (Batista et al, 2008) |
|
| PRG2 | Unknown | Unknown | Unknown | |
| Human | hiwi (piwil1) | Cytoplasm(Qiao et al, 2002) Nucleus, HEK 293T cells (Sugimoto et al, 2007) |
mRNA Testis - Spermatocytes, Round spermatids (Qiao et al, 2002) |
|
| Unknown |
mRNA Hematopoietic stem cells (Sharma et al, 2001) |
|||
|
mRNA Various tissues inc. Kidney, Heart, Brain, Liver, Muscle, Pancreas etc. (Sharma et al, 2001) |
||||
| hili (piwiL2) | Nucleus, HEK293T cells (Sugimoto et al, 2007) |
cDNA Testis (Sasaki et al, 2003) |
||
| piwiL3 | Nucleus, HEK293T cells Sugimoto et al, 2007) |
cDNA Testis (Sasaki et al, 2003) |
||
| hiwi2 (piwiL4) | Nucleus, HEK293T cells (Sugimoto et al, 2007) |
cDNA Testis (Sasaki et al, 2003) |
cDNABone marrow, Leukocytes, Pancreas (Sasaki et al, 2003) mRNA Various tissues inc. Spleen, Lung, Liver, Brain, Heart, Kidney, Ovary etc. (Sugimoto et al, 2007) |
|
| Sea Slug(Aplysia) | Piwi | Unknown | Ovotestis (Rajasethupathy et al, 2012) | |
| Nucleus | Brain (Rajasethupathy et al, 2012) | |||
| Sea Urchin | Seawi | Cytoplasm, Nucleus(Mitotic Spindle) |
Embryonic soma (Rodriguez et al, 2005) |
|
| Ovary - Oocytes (Rodriguez et al, 2005) |
||||
| Planarian | smedwi-1 | Unknown | mRNA Neoblasts (Reddien et al, 2005) | |
| smedwi- 2 | Unknown | mRNA Neoblasts (Reddien et al, 2005) | ||
| Polychaete Annelid (Capitella teleta) |
ct-piwi1 | Unknown |
mRNA PGCs (Giani et al, 2011) |
mRNA Somatic cells of embryo (Giani et al, 2011) |
|
mRNA Larvae - Brain, foregut, mesoderm (Giani et al, 2011) | ||||
|
mRNA Immature oocytes (Giani et al, 2011) |
mRNA Genital ducts (Giani et al, 2011) |
|||
|
mRNA Posterior growth zone (Giani et al, 2011) |
||||
| ct-piwi2 | Unknown |
mRNA Primordial germ cells (Giani et al, 2011) |
mRNA Embryonic soma (Giani et al, 2011) | |
| mRNA Larvae - Brain, foregut, mesoderm (Giani et al, 2011) | ||||
|
mRNA Immature oocytes (Giani et al, 2011) |
mRNA Genital ducts (Giani et al, 2011) |
|||
|
mRNA Posterior growth zone (Giani et al, 2011) |
||||
| Colonial Ascidian |
Piwi | Nucleus, Cytoplasm | Gonadal primordia of ovaries and testes (Brown et al, 2009) |
|
| Hemocytes (Brown et al, 2009) |
||||
| Endostyle-Epithelial cells, (Brown et al, 2009) | ||||
| EfPiwiA | Unknown |
mRNA Pluripotent stem cells - Archaeocytes (Funayama et al, 2010) |
||
|
mRNA Choanocytes (Funayama et al, 2010) |
||||
| EfPiwiB | Unknown |
mRNA Pluripotent stem cells - Archaeocytes (Funayama et al, 2010) |
||
|
mRNA Choanocytes (Funayama et al, 2010) |
||||
| Ctenophore | PpiPiwi1 | Unknown |
mRNA Female gonad - Oocytes and nurse cells (Alie et al, 2011) |
|
|
mRNA Male gonad - Developing spermatocytes (Alie et al, 2011) |
||||
|
mRNA Somatic stem cells of tentacle root, comb rows and aboral sensory complex (Alie et al, 2011) |
||||
| PpiPiwi2 | Unknown |
mRNA Female gonad - Oocytes and nurse cells (Alie et al, 2011) |
||
|
mRNA Male gonad - Developing spermatocytes (Alie et al, 2011) |
||||
|
mRNA Unidentified cells of somatic origin (Alie et al, 2011) |
||||
Analogous expression in somatic, germline tissues are placed side by side. Blank boxes indicate no experimental evidence for PIWI expression.
Transposable elements are mobile genetic fragments that are able to self-propagate, thereby achieving high abundance in eukaryotic genomes (Table 2). Transposons are split into two classes based on their mode of replication. Class 1 elements, or retrotransposons, utilize reverse transcriptase to replicate via an RNA intermediate. Representatives include autonomous elements that encode their own reverse transcriptase, such as the long terminal repeat elements (LTRs) and the long interspersed nuclear elements (LINEs), which do not contain LTRs. Non-autonomous retrotransposons, such as short interspersed nuclear elements (SINEs), which do not encode their own reverse transcriptase, also exist and usually depend on autonomous elements for their transposition. Class-2 elements are DNA transposons and can also be autonomous or non-autonomous. The transposase encoded by a DNA transposon can directly cut and paste transposon sequences or can be copied by rolling-circle DNA replication (Wicker et al., 2007; Rebollo et al., 2012). Uncontrolled transposition is a threat to genomic integrity and may be especially important to control in the animal germline, where genetic information is stored and passed on to future generations.
TABLE 2.
Survey of piRNA Sequencing and Mapping Data Across Animal Phylogeny
| Organism and Tissue |
% Repetetive Sequence in Genome |
Sequenced piRNAs |
% piRNAs mapping to transposons* |
% mapping to coding genes |
Ping pong signature |
References |
|---|---|---|---|---|---|---|
| Fly Ovary | 10% Sela et al., 2010 |
Piwi | 77% | ~5% | Yes | Brennecke et al., 2007 |
| Aubergine | 68% | ~5% | ||||
| Argonaute 3 | 78% | ~1% | ||||
| Fly Testis | Aubergine | 7% | 0.5% | Nagao et al., 2010 | ||
| Argonaute 3 | 54% | 4% | ||||
|
Fly Cultured Somatic Ovarian Cells |
Piwi | 54% | 17% | No | Saito et al., 2009 | |
|
Mouse, Adult Testis |
37.5% Waterston et al., 2002 |
MIWI | 17% | ~1–2% | No |
Girard et al., 2006; Aravin et al., 2006; Grivna et al., 2006a |
| MIWI | 15% | |||||
|
Mouse, 10 dpp Testis |
MILI | 35% | 29% | Yes | Aravin et al., 2007b | |
|
Mouse, 16.5 dpc Fetal Testis |
MIWI2 | 76% | ~1–2% | Yes | Aravin et al., 2008 | |
| MILI | 46% | 5% | ||||
|
Zebrafish Ovary |
26% Sela et al., 2010 |
Ziwi | 48% | 16% | Yes | Houwing et al., 2008 |
| Zili | 36% | 17% | ||||
|
Zebrafish Testis |
Ziwi | 32% | 22% | |||
| Zili | 23% | 19% | ||||
| Frog Oocyte | ~33% Hellsten et al., 2010 |
Xiwi | 23% | 19% | Yes (Total small RNA sequencing) | Lau et al., 2009 |
| Nematode | 9% Sela et al., 2010 |
PRG-1 | 21U RNAs map to two large genomic clusters, not tranpsoson-enriched |
No |
Batista et al., 2008; Das et al., 2008 |
|
| Rat Testis | ~40% Gibbs et al., 2004 |
RIWI | 20% | ~1% | Unknown | Lau et al., 2006 |
| Planarians | 31% Friedlander et al., 2009 |
Putative | 32% | Unknown | Yes | Friedlander et al., 2009 |
| Sea Anemone | 25% Putnam et al., 2007 |
Putative | Unkno | wn | Yes | Grimson et al., 2008 |
| Sponge | 20–30% Erpenbeck et al., 2012 |
Putative | Unkno | wn | Yes | Grimson et al., 2008 |
Mapping of piRNAs to transposon sequences is more difficult in animals with poorly-annotated genomes, and the reader should check references to determine how these data were obtained.
Klattenhoff et al. (2007) observed that mutations in the PIWI/piRNA pathway lead to increased DNA double-stranded breaks in Drosophila ovarian germ cells. It was thus proposed that the increase in DNA double-stranded breaks could be due to the upregulation of transposons and their mobilization, although it was astutely pointed out by these authors that an alternative explanation is equally possible: mutations in the PIWI/piRNA pathway could lead to DNA damage, which then triggers the upregulation of transposons. Furthermore, evidence of transposon up-regulation is largely measured at the RNA level. Therefore, it is not yet understood if increased levels of transposon RNA correlates with increased transposon mobilization in PIWI/piRNA-pathway mutants. The hypothesis that the PIWI/piRNA pathway functions primarily to silence transposons in the germline has gained enormous traction in the past few years, and we will discuss the evidence supporting this idea. We will also discuss evidence that disruptions in genome stability cause transposon upregulation, consistent with the alternative possibility that the PIWI/piRNA pathway directly regulates processes that affect genome stability. PIWI mutant animals appear to have pleiotropic phenotypes, which likely indicates that the PIWI/piRNA pathway is regulating several biological processes, the misregulation of which could cause genomic instability. A consequence of this instability could be transposon upregulation.
It is likely that these two alternatives are not mutually exclusive and feed off of each other; PIWI proteins most probably regulate genome stability via several different methods, aided by the vast number and diversity of associated piRNAs. The loss of PIWI proteins and piRNAs thus results in a complex scenario of widespread loss of genomic integrity that need not be solely due to transposon upregulation. We will additionally discuss increasing evidence for PIWI protein-mediated regulation of non-transposon gene expression both transcriptionally and post-transcriptionally, which further suggests a much broader role for the PIWI/piRNA pathway in controlling biological processes.
piRNAs AND TRANSPOSON REPRESSION IN DROSOPHILA
Evidence accumulated from many studies in Drosophila suggests that the PIWI/piRNA pathway functions to repress transposons in the germline (Malone et al., 2009; Lau, 2010; Saito and Siomi, 2010; Senti and Brennecke, 2010; Siomi et al., 2010b, 2011). This is thought to occur at both the transcriptional and post-transcriptional levels. Important clues about PIWIs, piRNAs, and their association with transposon repression were gained from pioneering work in Drosophila ovaries. piRNAs were identified in the Drosophila ovary by sequencing the small RNAs specifically associated with PIWI proteins. About 80% of the piRNAs identified from the Drosophila ovary (both germ and somatic cells) map to repeat sequences, and the vast majority of these are transposons or transposon remnants (Brennecke et al., 2007). This is a significant enrichment for transposon sequences, as only 10% of the Drosophila genome is composed of repetitive elements (Table 2; Sela et al., 2010). Drosophila has three PIWI proteins: Piwi (the founding member of the Argonaute family), Aubergine (Aub), and Argonaute 3 (Ago3), and each binds a distinct population of piRNAs (Lin and Spradling, 1997; Brennecke et al., 2007).1 Piwi protein is nuclear and found in both germ and somatic cells of the ovary, whereas Aub and Ago3 are cytoplasmic, enriched in the perinuclear nuage, and are primarily restricted to germ cells (Cox et al., 2000; Harris and Macdonald, 2001; Brennecke et al., 2007).
Post-transcriptional repression of transposons can occur concurrently with piRNA biogenesis. As it is currently understood, this process can be described in three steps in the Drosophila female ovary: (1) Transcription of long, single-stranded precursors from piRNA cluster loci (Figs. 1 and 2); (2) Processing of precursor transcripts into piRNAs by primary biogenesis (while the details of primary piRNA biogenesis have been worked out in ovarian somatic cells, it is also thought to occur in germ cells; Fig. 1); and (3) The production of secondary piRNAs by ping-pong biogenesis mediated by Aub and Ago3. The last step is concomitant with post-transcriptional silencing of functional transposon mRNAs in germ cells (Fig. 2). Primary and secondary processing likely occurs in specialized cytoplasmic compartments. While there are obvious correlative links between biogenesis and post-transcriptional repression, many open questions remain. In addition, post-transcriptional degradation of transposons also likely occurs in processing bodies via mRNA deadenylation and exonuclease degradation (Lim et al., 2009). Finally, transposon repression also occurs at the transcriptional level and is directed by Piwi/piRNA complexes in the nucleus.
Figure 1.
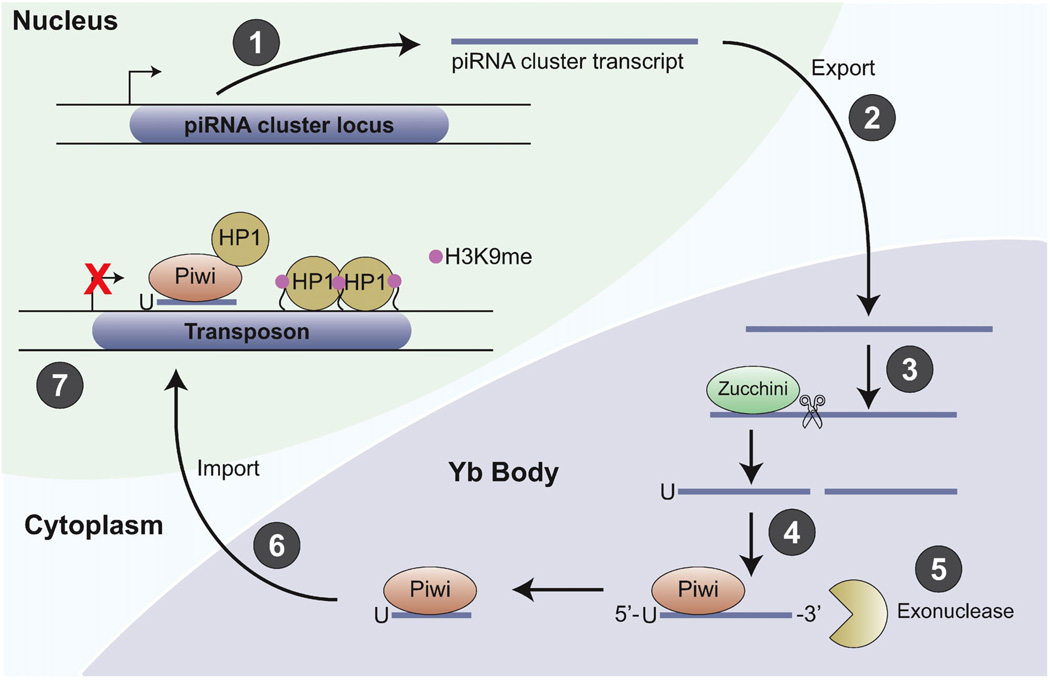
Primary piRNA processing in Drosophila ovarian somatic cells. The mechanism of primary piRNA processing is best understood in the somatic cells of the Drosophila ovary, but appears to be a conserved process that occurs in the Drosophila female germ cells and in other animals. Processing occurs as follows: (1) Primary processing starts with transcription of long, single-strand precursors from piRNA clusters in the genome. These loci often consist largely of dead transposon sequences. (2) Primary transcripts are exportedto the nucleus byan unknown mechanism, and primary processing occurs in cytoplasmic Yb Bodies. (3) Primary transcripts are cleaved into intermediate-sized RNAs by the endonuclease Zucchini. (4) Processed RNAs with a 5′-uridine are selected and bound by Piwi; the remaining RNA intermediates are likely unstable. (5) An unidentified exonuclease trims the 3′-end to create the mature piRNA. (6) The Piwi/piRNA complex is imported into the nucleus by an unknown mechanism; where (7) it is required for H3K9 methylation and transcriptional silencing of transposons. Piwi directly binds Heterochromatin Protein 1 (HP1) and thus may recruit HP1 to the chromatin.
Figure 2.
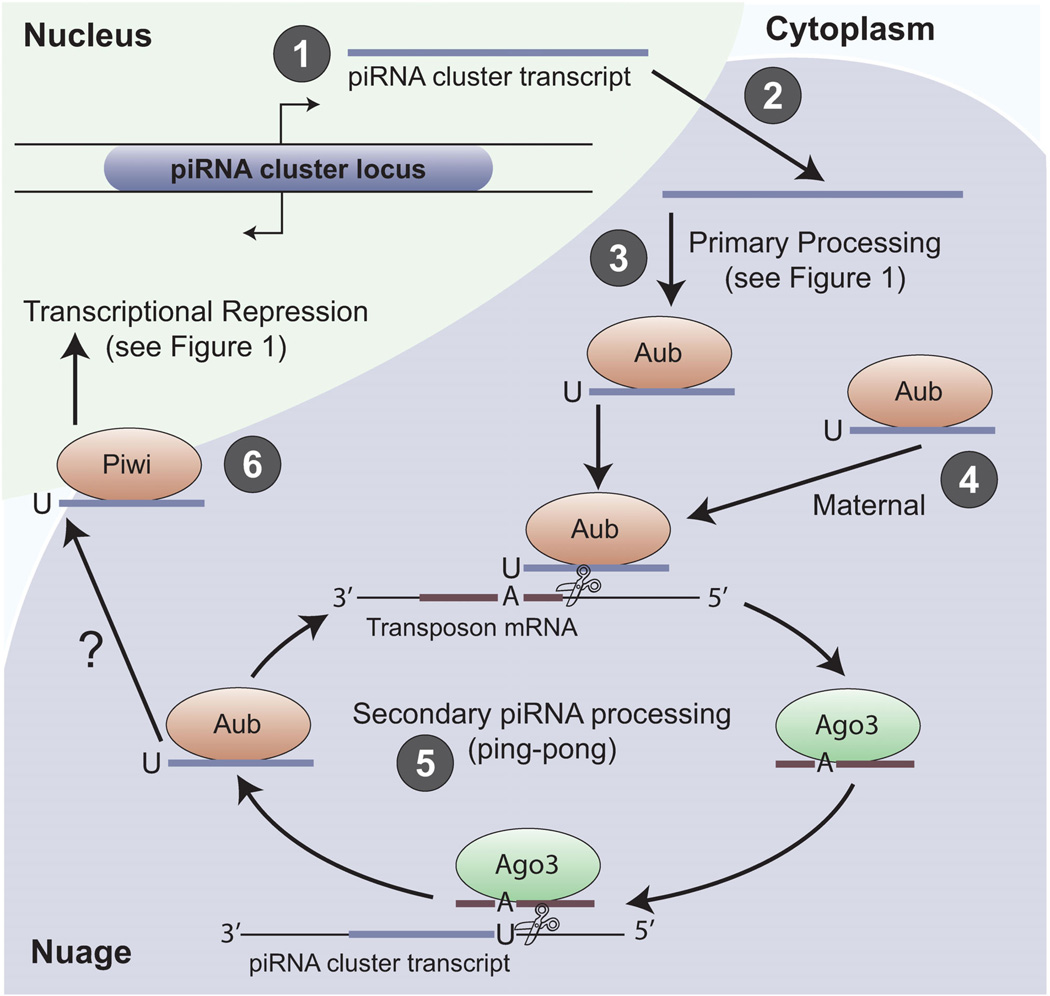
Secondary piRNA processing in Drosophila ovarian germ cells. The mechanism of secondary piRNA biogenesis is best understood in Drosophila germ cells, but the signatures of this mechanism have been observed in animals from sponges to mice (see Table 2). Processing occurs as follows: (1) Primary transcripts are synthesized from piRNA cluster loci, which have bidirectional promoters in Drosophila female germ cells. Primary transcripts consist largely of dead transposon sequences oriented in both the sense and antisense directions. (2) Transcripts are exported and (3) processed into primary piRNAs that are antisense to active transposons (see Fig. 1). In Drosophila, both Piwi and Aub are capable of binding primary piRNAs. Secondary piRNA processing (also called ping-pong processing) requires the input of mature Aub/piRNA complexes from either primary processing or from (4) maternal contribution. The relative importance of these two sources of PIWI/piRNA complexes is not understood, and may depend on the type of transposon being silenced. (5) Aub/piRNA complexes bind to active transposon mRNAs, resulting in cleavage to form the 5′-end of a new piRNA; this slicing activity of PIWI proteins has only been shown in vitro. It is unknown how the 3′-end of the piRNA is formed, but it likely occurs by 3′-end trimming similar to primary piRNA processing. The newly formed piRNA is sense to active transposon mRNAs and bound by Ago3. This complex can direct the formation of new piRNAs from piRNA cluster loci, thus reinforcing the antisense nature of piRNAs bound to Aub. (6) Primary piRNAs bound to Piwi are transported into the nucleus to transcriptionally silence transposons, similar to Figure 1, but it is unclear if these piRNAs are made from ping-pong processing or solely from primary piRNA biogenesis.
piRNA Biogenesis May Be Coupled to Post-Transcriptional Repression of Transposons
Step1:Transcription of piRNA precursor transcripts
The first step of piRNA biogenesis is the transcription of long, single-stranded RNA precursors from regions in the genome called piRNA cluster loci (Figs. 1 and 2). Approximately 92% of piRNAs in the Drosophila ovary are derived from piRNA cluster loci: these regions comprise 3.5% of the genome and are enriched in non-functional transposon remnants (Brennecke et al., 2007). The piRNA clusters that are active in germ cells are largely dual-stranded, which means that piRNAs originate from both genomic strands, while somatic piRNA clusters are largely derived from only one strand (Brennecke et al., 2007; Malone et al., 2009). The presence of long precursor transcripts has been clearly demonstrated, but very little is currently understood about the transcriptional regulation of these transcripts (Brennecke et al., 2007; Klattenhoff et al., 2009). A recent study suggests that RNA Polymerase II is used to transcribe piRNA cluster transcripts in mice (Gu et al., 2012), but a comprehensive understanding of the identity of the promoters and the transcriptional machinery required is still a wide-open question. Of the 142 clusters identified in the Drosophila genome, only seven are found in euchromatic regions (Brennecke et al., 2007). This begs the question, how are the precursor transcripts transcribed if they are embedded in highly heterochromatic regions? Interestingly, an HP1 (heterochromatin protein 1) homolog, Rhino, binds to dual-strand clusters and is required for their transcription (Klattenhoff et al., 2009). Mutations in rhino result in the collapse of germline piRNA biogenesis and upregulation of transposon RNAs (Klattenhoff et al., 2009). Rhino complexes with Cutoff, a novel homolog of the yeast transcription termination factor Rai1; mutations in cutoff display the same phenotypes as rhino mutants (Pane et al., 2011). The histone methyl transferase SETDB1, which normally promotes heterochromatin formation, is also required for transcription of piRNA precursor transcripts (Rangan et al., 2011). Piwi protein itself functions as an epigenetic modulator and is required for the transcription of the piRNA 3R-TAS1 from the telomeric region of chromosome 3 (Yin and Lin, 2007). Finally, both piwi and ago2 (which generally binds siRNAs) are required for the transcription of a transgene integrated in a piRNA cluster locus (Moshkovich and Lei, 2010). Taken together, these data suggest a piRNA cluster-specific chromatin state that allows for their transcription in highly heterochromatic regions.
Step 2: Primary piRNA biogenesis in somatic cells and the germline
Primary transcripts from piRNA clusters are processed into piRNAs by a mechanism termed primary piRNA biogenesis (Fig. 1). The details are largely being worked out in Drosophila ovarian somatic cells, where only one PIWI protein (Piwi) is expressed and piRNAs are made solely by this mechanism. A similar primary piRNA biogenesis pathway seems to occur in germ cells, but there are notable differences, which are discussed in detail below (see Cellular compartmentalization of piRNA biogenesis section). The majority of piRNAs produced via primary piRNA biogenesis in somatic cells are made from a single-stranded piRNA locus called flamenco (Li et al., 2009; Malone et al., 2009). Transposon remnants in this locus are overwhelmingly oriented in the antisense orientation, thus resulting in the production of antisense Piwi-bound piRNAs with a strong bias for uridine at the 5′ position (Malone et al., 2009). The piRNA pathway in the somatic cells of the ovary represses the expression of several retrotransposons, including many from the gypsy family (Prud’homme et al., 1995; Mevel-Ninio et al., 2007; Desset et al., 2008). This repression is required to maintain the integrity of the germline because these retrotransposons are capable of forming viral particles and invading the neighboring oocyte (Pelisson et al., 1994; Chalvet et al., 1999; Leblanc et al., 2000; Brasset et al., 2006).
Primary piRNA biogenesis is generally thought to involve cleavage of primary transcripts into smaller pieces, binding to PIWI proteins, and then trimming them to the final piRNA size. It is well established that primary processing requires the endonuclease Zucchini (zuc), which appears to non-selectively cut piRNA primary transcripts into smaller and perhaps variably sized pieces (Pane et al., 2007; Olivieri et al., 2010; Nishimasu et al., 2012). Drosophila zuc mutants have an increased abundance of piRNA primary transcripts (Haase et al., 2010). It is likely that initial processing occurs in the cytoplasm where Zuc protein is localized, although nothing is understood about the export of primary piRNA transcripts from the nucleus prior to their processing. It appears that the RNA fragments initially cut by Zuc are subsequently bound by Piwi. Evidence that the initial fragments bound to Piwi are longer than the mature piRNAs comes from mouse testes, where it was demonstrated that MIWI and MILI (mouse PIWI homologs, see Table 1) bind to piRNA precursors that are identical to mature piRNAs at the 5′-end, but are extended at the 3′-end (Vourekas et al., 2012). In addition, incubating Siwi (silkworm Piwi) with artificial piRNA precursors in vitro demonstrates that Siwi preferentially binds RNA fragments with a 5′-uridine (Kawaoka et al., 2011b). This suggests a model by which Zuc randomly cuts piRNA precursor transcripts and Piwi selects those that have a 5′-uridine; the remaining are likely unstable and therefore degraded. This could be similar to the preference of human Ago2 for miRNAs with either an adenine or uridine at the 5′-end, which bind to a specific pocket in Ago2 (Frank et al., 2010). piRNA precursors bound to Piwi are trimmed to the appropriate size at the 3′-end by an unidentified exonuclease, followed by 2′-O-methylation of the 3′-end by Hen1 (Saito et al., 2007; Kawaoka et al., 2011b). It is possible that the size of the mature piRNA is dictated simply by the footprint of the bound Piwi protein, which would explain why different Piwi protein homologs preferentially associate with piRNAs of different sizes (Brennecke et al., 2007).
Step 3: Secondary piRNA biogenesis (ping-pong) in the germline may be coupled with post-transcriptional transposon repression
In Drosophila female germ cells, primary piRNAs are predominantly antisense to transposon coding regions and are thought to trigger secondary piRNA biogenesis, which occurs as a back-and-forth mechanism between Aub and Ago3 (Fig. 2). Aub-bound primary piRNAs are predominantly antisense (83%), whereas Ago3-bound secondary piRNAs are pre-dominantly sense to transposons (75%). Aub in complex with piRNAs may target transposon RNAs for degradation by processing them into secondary piRNAs (Brennecke et al., 2007). These secondary piRNAs, in complex with Ago3, can direct the production of more antisense piRNAs from piRNA cluster transcripts. This process is thus termed ping-pong piRNA biogenesis to signify the back-and-forth nature of the mechanism (Brennecke et al., 2007). These new Aub-bound piRNAs are called primary piRNAs, but the definition here is murky because it is impossible to tell if these piRNAs were produced by primary or secondary piRNA biogenesis. The ping-pong mechanism produces a recognizable signature in piRNA sequences. So-called primary piRNAs have a uridine bias at the 5′-end and secondary piRNAs have an adenine bias at the 10th position. Furthermore, a 10-base complementary overlap is observed between primary and secondary RNAs, which is predicted to occur because Argonaute proteins are known to slice their target RNAs 5′ to the base paired with the 10th nucleotide of the small RNA guide (Brennecke et al., 2007; Gunawardane et al., 2007). This complementary overlap is particularly strong when comparing Ago3-bound and Aub-bound piRNAs (48%; Brennecke et al., 2007). Piwi function in flies is completely independent of its slicing activity, which strongly suggests that Piwi does not participate in secondary piRNA biogenesis (Darricarrere et al., 2013). Unfortunately, slicer mutants in Aub and Ago3 have not yet been analyzed to definitively test the model that these proteins are responsible for slicing RNA substrates during secondary piRNA biogenesis.
An intriguing aspect of the ping-pong model is that piRNA biogenesis is linked to the post-transcriptional repression of transposons, such that the production of sense piRNAs from a specific transposon will seed the production of yet more antisense piRNAs from precursor transcripts that will then target that particular transposon for degradation. piRNA cluster loci that are expressed in the germline consist of transposon remnants oriented randomly in the sense or antisense direction (Brennecke et al., 2007; Malone et al., 2009), although antisense piRNAs predominate in both the Aub- and Piwi-bound populations and the total piRNA population (Brennecke et al., 2007). Ago3-bound sense piRNAs are derived from active transposon mRNAs and may act to amplify the production of antisense piRNAs from piRNA clusters, thus giving rise to the antisense bias. In support of this, antisense piRNA populations collapse in ago3 mutant ovaries and transposon mRNA levels are highly upregulated (Li et al., 2009). Furthermore, piRNAs bound to Aub showed no antisense bias in the absence of Ago3 (Li et al., 2009). An intriguing study supports this model by demonstrating that the transcription of functional copies of the I-element retrotransposon are required for the production of sufficient I-element piRNAs and silencing of the functional I-element RNA (Chambeyron et al., 2008). I (Inducer) Drosophila strains contain 10 functional I-element copies in euchromatin, whereas R (Reactive) strains have no functional copies (both strains have non-functional copies in the heterochromatin). The I strains produce sufficient numbers of antisense piRNAs against the I-element to promote silencing; R strains do not (Brennecke et al., 2008; Chambeyron et al., 2008). This demonstrates that for the I-element, functional copies of a retrotransposon are required for transposon silencing.
It is important to note that post-transcriptional degradation of transposon RNAs likely does not occur exclusively by piRNA biogenesis. In the Drosophila ovary, PIWI proteins, piRNAs, and retrotransposon RNAs co-localize with proteins involved in mRNA degradation. Mutations in these genes leads to the accumulation of retrotransposon transcripts, which strongly suggests that retrotransposon RNAs can also be degraded by the same mechanisms as mRNAs (Lim et al., 2009).
Cellular compartmentalization of piRNA biogenesis
The bulk of primary piRNA biogenesis in ovarian somatic cells likely occurs in cytoplasmic granules called Yb bodies, so named because they were first identified by the accumulation of the TUDOR-domain-containing protein Yb (Szakmary et al., 2009; Olivieri et al., 2010; Qi et al., 2010; Saito et al., 2010; Fig. 1). Both the putative RNA helicase Armitage (Armi) and the TUDOR-domain-containing protein Vreteno (Vret) are required for primary piRNA biogenesis and also localize to the Yb bodies (Olivieri et al., 2010; Saito et al., 2010; Handler et al., 2011; Zamparini et al., 2011). It is not clear how Armi and Vret function in the primary piRNA pathway, although Armi, Vret, Piwi, and Yb have been demonstrated by immunoprecipation experiments to be in a common complex (Haase et al., 2010; Olivieri et al., 2010; Saito et al., 2010; Handler et al., 2011). Interestingly, in zuc mutants, Piwi protein is lost from the nucleus and accumulates both diffusely in the cytoplasm and in perinuclear spots coincident with Yb bodies (Olivieri et al., 2010). Furthermore, Armi, Vret, and Yb accumulate in significantly more massive Yb bodies in zuc mutants (Olivieri et al., 2010; Saito et al., 2010; Handler et al., 2011). This suggests that the loss of Zuc disrupts the dynamics of the downstream proteins in the pathway, perhaps causing these proteins to accumulate at sites where they normally transit through temporarily. In zuc, armi, and vret mutants, Piwi is not loaded with piRNAs and accumulates in the cytoplasm, strongly suggesting that piRNA loading onto Piwi takes place in the cytoplasm and is required for Piwi transport into the nucleus (Olivieri et al., 2010; Saito et al., 2010; Handler et al., 2011; Zamparini et al., 2011). Indeed, constituitively cytoplasmic Piwi mutants that are missing their nuclear localization signal are still loaded with mature piRNAs, and a Piwi mutant that cannot load piRNAs does not localize to the nucleus (Saito et al., 2009, 2010). In further support of this, two other nuclear PIWI homologs in distantly related animals require piRNA loading for nuclear localization: (1) In mouse male germ cells, MIWI2 loses nuclear localization in mili mutants, where MIWI2 no longer associates with piRNAs (Aravin et al., 2008; Zheng et al., 2010); and (2) In Tetrahymena, association with mature small RNAs is required for the nuclear localization of the Piwi homolog Twi1p (Noto et al., 2010). Thus, piRNA loading of PIWI is a conserved requirement for nuclear localization.
Several of the same genes are required in the germline for primary piRNA biogenesis, including zuc, armi, and vret (Olivieri et al., 2010; Handler et al., 2011; Zamparini et al., 2011). By contrast, Yb is specific for the somatic cells and Yb bodies do not exist in germ cells. Instead, primary piRNA biogenesis may occur in the nuage where Armi and Vret accumulate (Fig. 2; Lim and Kai, 2007; Pane et al., 2007; Handler et al., 2011). The function of Yb may be replaced in the germline by two closely related proteins called brother and sister of Yb (Handler et al., 2011). In zuc mutants, Piwi is delocalized from germline nuclei into clouds around the nucleus that also contain Armi (Olivieri et al., 2010). This is reminiscent of piRNA pathway protein mislocalization in somatic zuc mutants, and suggests that primary piRNA biogenesis in the Drosophila female germline may be similar to the mechanism observed in the somatic cells of the ovary. Secondary piRNA biogenesis likely also occurs in the nuage where both Aub and Ago3 are found (Brennecke et al., 2007).
Open questions about the relationship between piRNA biogenesis and transposon repression
The details of the ping-pong model are still under investigation, and many open questions remain, including the following examples:
How is the ping-pong cycle initiated? It was initially proposed that Aub is loaded with piRNAs that are made by the primary piRNA pathway, and that these Aub/piRNA complexes can then initiate secondary piRNA biogenesis (Brennecke et al., 2007; Gunawardane et al., 2007). In support of this model, ectopically expressed Aub in an ovarian somatic cell line is loaded with almost the identical piRNA population as Piwi (Olivieri et al., 2012). Considering that secondary piRNA biogenesis cannot occur in this cell line due to the lack of Ago3 expression, this strongly supports the hypothesis that Aub can be loaded with piRNAs that are made by primary biogenesis. Furthermore, when Ago3 is ectopically expressed in these cells, it is not loaded with piRNAs, which suggests that Ago3 can only load piRNAs made by secondary piRNA biogenesis (Olivieri et al., 2012). Yet, maternally inherited piRNAs can also initiate secondary piRNA biogenesis, which offers an alternative source of piRNAs to start the ping-pong cycle (Brennecke et al., 2008; Kawaoka et al., 2011a). The relative importance of primary piRNA biogenesis versus maternally inherited piRNAs in initiating the ping-pong cycle remains to be determined, although a recent study suggests that this may be different for different transposons (Olivieri et al., 2012). Olivieri and coworkers found that germline transposons could be split into two classes: (A) Transposons that can be processed by ping-pong in the absence of primary biogenesis (i.e., mutations in armi or zuc) and (B) Transposons that require primary piRNA biogenesis factors to maintain the ping-pong cycle (Olivieri et al., 2012). The authors speculate that class A transposons could rely on maternally loaded piRNAs to support ping-pong biogenesis.
As discussed above, Piwi is involved in primary piRNA biogenesis, but is this role specific for the somatic cells of the gonad where it is the only PIWI protein expressed? In the germline, both Aub and Piwi bind primary piRNAs (i.e., piRNAs with a 5′-uridine bias), and thus it is not completely clear how the functions of Aub and Piwi are delineated in the germ cells. Phylogenetic analysis demonstrates that Piwi and Aub are the products of a recent gene-duplication event, and could share similar functions (Juliano et al., 2011). In the silkworm, where it was demonstrated that Siwi can selectively bind 5′-uridine RNAs, there are only two Piwi proteins: Siwi (related to both Aub and Piwi in fly) and BmAgo3 (related to fly Ago3; Kawaoka et al., 2008, 2011b). Therefore, it could be Aub that is selecting the 5′-uridine products of primary transcript processing either in addition to, or instead of, PIWI in the female germ cells of Drosophila.
The ping-pong model is predicated on Aub and Ago3 being able to slice their target RNAs, but is this activity required for piRNA biogenesis? Slicing activity has been demonstrated for all three Drosophila PIWI proteins in vitro, but an in vivo requirement for the catalytic residues of Aub or Ago3 have not been demonstrated (Saito et al., 2006; Gunawardane et al., 2007). Further, Piwi slicing activity is not required for piRNA biogenesis, thus it likely does not significantly participate in ping-pong piRNA biogenesis (Darricarrere et al., 2013). It has, however, been established that the catalytic activity of MILI in mice is required for piRNA biogenesis (details discussed below; De Fazio et al., 2011).
How does the total piRNA population of the Drosophila ovary remain biased for the antisense orientation (i.e., Aub and Piwi-bound piRNAs are much more abundant than Ago3-bound piRNAs)? If functional transposon RNAs are readily processed into sense piRNAs, it is unclear what becomes of these piRNAs; perhaps they are selectively degraded and/or piRNAs are less stable when not associated with PIWI proteins.
Currently it is thought that the repression of transposons in the Drosophila female germline occurs both transcriptionally (see below) and post-transcriptionally, but what is the relative importance of these two mechanisms? It is possible, for example, that post-transcriptional repression of transposons is not significant, and instead the ping-pong cycle is required to fuel the production of piRNAs that will be used in transcriptional silencing.
Several genes are required for piRNA production in Drosophila female germ cells. Obtaining a better understanding of the molecular functions of these genes in piRNA biogenesis will help answer some of these outstanding questions. These genes include RNA helicases (spindle-E and vasa) and a host of TUDOR-domain containing genes (krimper, tejas, qin, tudor, and kumo) (Lim and Kai, 2007; Malone et al., 2009; Nishida et al., 2009; Patil and Kai, 2010; Zhang et al., 2011; Anand and Kai, 2012). A recent study combined epistatic analysis and comparisons between the piRNA populations of different germline knockdowns to group these factors in discrete steps (Olivieri et al., 2012). First, as described above, zuc and armi are required only for primary piRNA biogenesis. By contrast, spindle-E, vasa, and krimper were found to be required only for secondary piRNA biogenesis. A recent study suggests an additional function for Vasa in the transport of cluster piRNA transcripts from the nucleus to the nuage, which occurs prior to primary piRNA biogenesis (Zhang et al., 2012). Finally, some genes are required for both primary and secondary biogenesis: vret, brother and sister of Yb, and Shutdown (shu). [Although it should be noted that two previous studies demonstrate that vret is dispensable for ping-pong biogenesis (Handler et al., 2011; Zamparini et al., 2011).] This last category of genes may be required to complete down stream steps that are common to both primary and secondary piRNA processing, such as piRNA loading and the maturation of a PIWI/piRNA complex (Olivieri et al., 2012). For example, shu binds the chaperone protein Hsp90 and is required for both primary and secondary piRNA biogenesis (Olivieri et al., 2012; Preall et al., 2012). In shu-mutant Drosophila ovaries, all transposable element-derived piRNA populations completely collapse (Allan and Ratajczak, 2011; Olivieri et al., 2012; Preall et al., 2012). Additionally, mutations in shu that abrogate Hsp90 binding cannot restore piRNA levels in the shu mutant, and epistatic analysis places shu downstream of other piRNA biogenesis factors (Olivieri et al., 2012). These data support a model by which Hsp90 and its co-chaperone shu are required to load piRNAs onto Argonaute proteins, regardless of how (i.e., by which pathway) those piRNAs are produced. This hypothesis is supported by the fact that Hsp90 is required to load siRNA duplexes onto Argonaute proteins in plants and Drosophila, and the Hsp90 co-chaperone Cyclophilin40 is required for siRNA loading in plants (Iki et al., 2010, 2012; Iwasaki et al., 2010; Miyoshi et al., 2010).
Transcriptional Silencing of Transposons by PIWI/piRNA Complexes in the Nucleus
In addition to functioning in piRNA biogenesis, the nuclear protein Piwi, in association with mature piRNAs, may be an effector of transposon silencing by epigenetic mechanisms. In support of this, transposons are derepressed when Piwi nuclear localization is disrupted (Klenov et al., 2011). Piwi protein bound to a transposon-derived piRNA produced by aub/ago3 ping-pong biogenesis is thought to translocate to the nucleus and to silence transposons epigenetically (Figs. 1 and 2). This is supported by three observations: (1) When Piwi is knocked down specifically in germ cells, transposon expression increases, piRNA levels go up (presumably due to increased levels of functional transposon mRNAs being funneled into secondary piRNA biogenesis), and Aub remains localized to the nuage (Wang and Elgin, 2011). (2) In aub and ago3 mutants, piRNA levels are reduced and Piwi protein is no longer localized to the nucleus (Li et al., 2009; Wang and Elgin, 2011). (3) In both piwi and aub mutants, there is a loss of repressive chromatin marks at transposon loci (Klenov et al., 2007, 2011; Wang and Elgin, 2011; Sienski et al., 2012). Piwi’s association with HP1a offers an attractive mechanistic model for chromatin regulation via recruitment of HP1a to transposon loci (Brower-Toland et al., 2007). In support of this model, a new study demonstrates that binding of Piwi/piRNA complexes to ectopic euchromatic sites recruits HP1, leads to increases in repressive chromatin marks, and a loss of RNA Polymerase II binding (Huang et al., 2013). Thus, piRNAs may act as sequence-specific guides to recruit epigenetic machinery to particular chromatin sites.
A recent study further supports the model that Piwi/piRNA complexes in the nucleus can direct epigenetic silencing of transposons in cultured ovarian somatic cells (Sienski et al., 2012). In this case, piRNAs are made by primary processing rather than by Ago3/Aub, which are not expressed in ovarian somatic cells. In piwi knockdown ovarian somatic cells, both total levels, as measured by RNA-seq, and levels of nascent transcript, as measured by global run-on sequencing (GRO-seq), of transposon RNA increase (Sienski et al., 2012). Furthermore, RNA Polymerase II occupancy is increased and H3K9me3 levels are decreased (Sienski et al., 2012). These data strongly support a model in which piwi is required for the epigenetic silencing of transposable elements in ovarian somatic cells, and are consistent with the in vivo data described above.
Is Transposon Repression Conserved in Animal Germlines?
The function of the PIWI/piRNA pathway to repress transposons is well established in the Drosophila ovary, yet evidence from the male germline suggests that the pathway could function more broadly. In the fly testes, less than 10% of the piRNAs bound to Aub are transposon-derived and 54% of Ago3-bound piRNAs are transposon-derived (Table 2; Nagao et al., 2010). Although there is evidence that these piRNAs are produced by the ping-pong mechanism, transposon levels do not significantly increase in aub and ago3 mutant testes by RT-PCR, contrary to what is observed in Drosophila ovaries (Brennecke et al., 2007; Nagao et al., 2010). By contrast, repression of specific transposons in the male Drosophila germline is dependent on piwi (Kalmykova et al., 2005). Thus, while the piRNA pathway does play some role in Drosophila testis in repressing transposons, this may be independent of the ping-pong cycle. Whole genome transposon depression needs to be performed in Drosophila testis, however, before definitive conclusions can be made. In the mouse testis, there is evidence that the pathway is required for the repression of LINE1 transposons (Reuter et al., 2011). But given that piRNAs derived from transposons in the adult mouse testis are lower than would be expected by chance, there are very likely other functions as well (Table 2). Furthermore, there is no enrichment for transposon sequences in the piRNA populations of the zebrafish or rat testis (Table 2). Therefore, there may be conserved spermatogenesis-specific functions for the PIWI/piRNA pathway that go beyond transposon control. In addition, significant numbers of piRNAs map to protein-coding genes (Table 2), which implies that the PIWI/piRNA pathway could be directly regulating these genes; documented cases of this are discussed in detail below.
Transposon upregulation after PIWI mutation has been reported in the fly, mouse, zebrafish, and C. elegans (Kalmykova et al., 2005; Aravin et al., 2007b; Brennecke et al., 2007; Carmell et al., 2007; Batista et al., 2008; Das et al., 2008; Houwing et al., 2008; Kuramochi-Miyagawa et al., 2008). Yet, as discussed further below, there is variability in the extent of upregulation, the number of transposon families affected, and in the mechanism of repression. piRNAs are remarkably enriched for transposon sequences in the Drosophila ovary; the percentage of piRNAs that map to transposons is approximately seven-times higher than the percentage of transposon sequences in the genome (Table 2). A survey of piRNA mapping data currently available indicates that this trend may not be true for animals generally. The highest transposon enrichment seen in other animals is twofold, and in many cases, there is a depletion of transposon sequences (Table 2). This does not mean that transposon repression is not a vital function of the PIWI/piRNA pathway in these animals, but it certainly suggests that additional functions are likely.
OTHER MECHANISMS OF FOREIGN DNA REPRESSION BY THE PIWI/piRNA PATHWAY
The PIWI/piRNA pathway may have a conserved role in recognizing and silencing foreign DNA, such as transposon sequences, although the actual mechanisms of this process appear to vary significantly between organisms. Repression of transposons by the path way in the mouse testes does share some significant similarities with the Drosophila ovary, such as ping pong-mediated post-transcriptional repression and epigenetic silencing. On the other hand, there are also clear mechanistic differences between PIWI/piRNA function in the fly and mouse. In C. elegans and the ciliate Tetrahymena, the pathway also recognizes and represses foreign DNA, but the mechanisms are strikingly different. Thus, although the function of the path way to repress foreign DNA may be widely conserved, the method by which this is achieved is divergent.
Transposon Repression by the PIWI/piRNA Pathway in Mouse Testes
All three PIWI homologues in mice, miwi, mili, and miwi2, are required for fertility in males (Deng and Lin, 2002; Kuramochi-Miyagawa et al., 2004; Carmell et al., 2007). piRNAs are abundantly expressed in the testes of mice that are at least 14 days old (14 days post-partum), where meiosis has progressed to the pachytene stage and mili and miwi are expressed (Aravin et al., 2006; Girard et al., 2006; Grivna et al., 2006a). Sequencing of these pachytene piRNAs from adult mouse testes reveals that they have no observable ping-pong signature, and they are depleted for repeat sequences (Aravin et al., 2006, 2007a; Girard et al., 2006; Grivna et al., 2006b). Nevertheless, when the catalytic domain of MIWI is mutated, piRNA biogenesis is unaffected but expression of the LINE1 retro-transposon class increases in mouse testes (Reuter et al., 2011). These data suggest that MIWI represses LINE1 retrotransposons in the adult testes by cleaving the RNA in a ping-pong-independent manner.
In the pre-natal mouse testes (16.5 days post-coitum), where miwi2 and mili are expressed, piRNA populations are biased for transposon sequences and exhibit the ping-pong signature (5′-uridine piRNAs and 10th position adenosine piRNAs) for LINE1 and IAP (Intracisternal A-particle) retro-transposons (Aravin et al., 2008). Furthermore, both mili and miwi2 mutants exhibit increased RNA levels of LINE1 and IAP retrotransposons (Aravin et al., 2007b; Carmell et al., 2007; Kuramochi-Miyagawa et al., 2008). MIWI2 and MILI localize to cytoplasmic granules in male fetal germ cells, where they may function in the ping-pong biogenesis pathway to post-transcriptionally repress retrotransposons. In contrast to flies, where primary piRNAs are derived from cluster transcripts, murine primary piRNAs (5′-uridine bias) bound to MILI are sense (likely derived from the RNAs of functional transposons) and the secondary piRNAs (10th position adenosine bias) bound to MIWI2 are antisense (likely produced from piRNA cluster transcripts; Aravin et al., 2008). This model may not be correct, however, because a catalytic mutation in MILI, but not MIWI2, leads to decreased piRNA populations and increased retrotransposon expression (De Fazio et al., 2011). Therefore, an intra-MILI ping-pong cycle may exist to produce piRNAs in the pre-natal mouse testes (De Fazio et al., 2011). Indeed, when pre-pachytene piRNAs were sequenced from 10-days post-partum testes where only mili is expressed, evidence for ping-pong amplification was found, which supports the existence of an intra-MILI ping-pong cycle (Aravin et al., 2007b). This is similar to an intra-Aub ping-pong cycle that is detected in Drosophila ovaries in addition to the typical Aub-Ago3 ping-pong cycle (Li et al., 2009; Zhang et al., 2011).
Both mili and miwi2 mutants exhibit a loss of methylation at retrotransposon promoters in mouse testes, suggesting epigenetic repression by the PIWI/piRNA pathway (Kuramochi-Miyagawa et al., 2008). MIWI2 is a nuclear protein and therefore may have a role in directing the methylation of transposon sequences during the period of de novo DNA methylation that occurs in the male just before birth (Hajkova et al., 2002; Kato et al., 2007; Aravin et al., 2008). piRNAs derived from ping-pong biogenesis could be loaded onto MIWI2, then MIWI2/piRNA complex may enter the nucleus to direct transcriptional silencing of transposons (Aravin et al., 2008; De Fazio et al., 2011). A direct role for MIWI2 in DNA methylation has not been definitively demonstrated, however.
While the details remain to be worked out, it is already clear that the mechanisms of piRNA biogenesis and transposon repression are complex and there are significant differences between species, despite the similarities in the genes required for the function of the PIWI/piRNA pathway between Drosophila and mice. For example, the mouse zuc homolog is also required for primary piRNA biogenesis by cleaving of single-stranded piRNA precursor transcripts (Watanabe et al., 2011a; Ipsaro et al., 2012). The mouse putative DEAD box helicase MOV10L1 is related to the Drosophila armi, and may also be required for piRNA biogenesis because there is a lack of all mature piRNAs in mov10l1 mutant testes (Frost et al., 2010; Zheng et al., 2010).The mouse homolog of the DEAD box helicase Vasa, Mvh, has been implicated in piRNA biogenesis in fetal male germ cells, which is consistent with its function in the Drosophila ovary (Kuramochi-Miyagawa et al., 2010). Finally, several TUDOR-domain containing proteins are also required in both mouse and Drosophila for proper PIWI/piRNA pathway function, which has been previously reviewed (Siomi et al., 2010a).
The PIWI/piRNA Pathway Functions to Stably Silence Foreign DNA Over Many Generations in C. elegans
C. elegans has two Piwi homologs, prg-1 and prg-2, which are 91% identical and are a product of a recent gene duplication as other Caenorhabditis species have only one prg gene (Das et al., 2008). Mutations in prg-1 lead to reduced fertility and temperature-sensitive sterility, where-as prg-2 mutants show no obvious defects (Batista et al., 2008; Wang and Reinke, 2008). PRG-1 binds to a class of RNAs (21U-RNAs) that, like piRNAs have a 5′-uridine bias, but are only 21 nucleotides long (Fig. 3). The presence of 21U-RNAs is dependent upon prg-1, and both PRG-1 and 21U-RNAs are restricted to the germline (Batista et al., 2008; Das et al., 2008; Wang and Reinke, 2008). When mapped back to the genome, 21U-RNAs are largely found in two genomic clusters on chromosome IV, concentrated in between protein coding genes and in introns (Ruby et al., 2006). In contrast to the long precursor transcripts required for piRNA production in mice and flies, 21U-RNAs are transcribed as individual transcription units that are approximately 25–26 nucleotides long, and are subsequently shortened to 21 nucleotides (Ruby et al., 2006; Cecere et al., 2012; Gu et al., 2012; Fig. 3). While the 21U-RNAs are not enriched for transposon sequences, two studies found that the DNA transposon family Tc3 is upregulated approximately fourfold in prg-1 mutants (Batista et al., 2008; Das et al., 2008). prg-1 acts upstream of the C. elegans endogenous siRNA pathway (or 22G–RNAs) to regulate Tc3 expression; this pathway was previously shown to silence transposons in the C. elegans germline (Sijen and Plasterk, 2003; Das et al., 2008). Transposon repression could thus be happening through the endogenous siRNA pathway and not directly through the piRNA pathway. Several recent studies have now demonstrated that PRG-1 and associated 21U-RNAs act to recognize and silence foreign DNA, such as an introduced transgene, in the germline (Ashe et al., 2012; Lee et al., 2012; Shirayama et al., 2012; Fig. 3). Targeting of PRG-1 to the mRNA of a single-copy transgene triggers the production of 22G–RNAs from surrounding regions via RNA-dependent RNA polymerase (Ashe et al., 2012; Bagijn et al., 2012; Lee et al., 2012; Shirayama et al., 2012). Subsequently, these 22G–RNAs are loaded into germline Argonaute proteins that translocate to the nucleus and direct epigenetic silencing of the region. The silencing of the transgene is then stable over many generations (Ashe et al., 2012; Shirayama et al., 2012). It is not yet clear how the PIWI pathway is able to recognize self from non-self in C. elegans, although it was noted that there are endogenous 21U-RNAs that could imperfectly recognize the GFP transgene (Shirayama et al., 2012). This is clearly an exciting discovery that may shed light on a conserved function for the PIWI/piRNA pathway in repressing the expression of foreign DNA.
Figure 3.
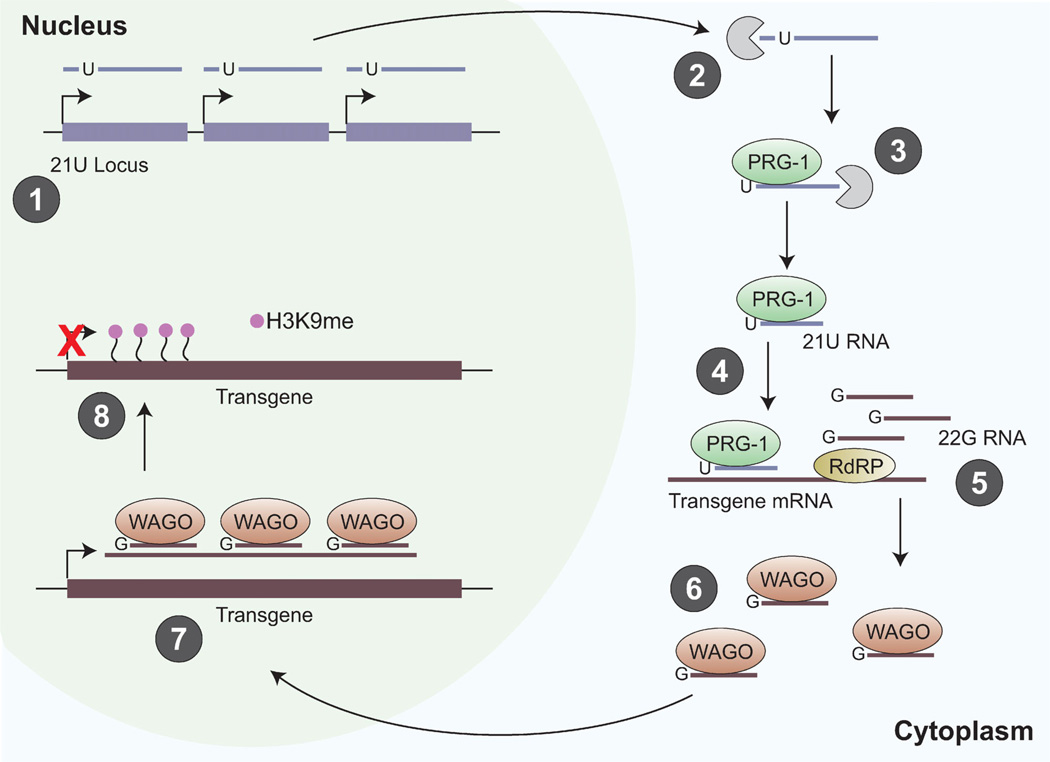
The PIWI/piRNA pathway in C. elegans silences foreign DNA in the germline. C. elegans 2 1U–RNAs are 21-nucleotides long and have a 5’-uridine. 21U-RNAs are considered the worm piRNAs because they bind to PRG-1, a PIWI protein homolog. (1) 21U–RNA precursors are transcribed from individual transcription units within each 21U locus. The 21U–RNA precursors are capped, small RNAs approximately 25–26 nucleotides long, with a uridine at the third position. (2) Precursor transcripts are truncated by two bases at the 5′-end, leaving a uridine at the 5′-end. (3) 5’-uridine RNA precursors are bound to PRG-1 and trimmed at the 3′-end to form the mature 21U–RNA. (4) The PRG-1/21U–RNA complex recognizes the mRNA of foreign DNA, for example a single-copy transgene. The basis of this recognition is not understood, but may be by imperfect base-pairing between the 21U–RNA and the target. (5) PRG-1 recruits RNA-dependent RNA Polymerase (RdRP) to produce 22G–RNAs from surrounding regions on the mRNA. (6) The 22G–RNAs are bound by germline-specific worm Argonaute proteins (WAGOs) and transported into the nucleus. (7) WAGO/22G–RNA complexes bind to nascent transgene transcripts, which results in H3K9 methylation and transgene silencing that lasts several generations.
The PIWI/piRNA Pathway Is Required for DNA Elimination in the Somatic Macronucleus of Ciliates
The ciliates are a group of protozoans typified by cilia structures and nuclear dimorphism. The diploid micronucleus (germline) contains a transcriptionally silent, complete copy of the genome that will be passed on to the next generation. By contrast, the polyploid macronucleus (somatic) has undergone DNA elimination and serves as the template for gene transcription (Prescott, 1994). During sexual reproduction, the micronucleus undergoes meiosis and exchanges haploid nuclei with a mating partner. The old macronucleus is lost and the newly formed zygotic nucleus divides to form a new micronucleus and macronucleus. The macronucleus subsequently undergoes DNA elimination, a process by which repetitive DNA, transposons, and unidentified AT-rich regions, collectively known as internal eliminated sequences (IES), are removed from the macronuclues, leaving behind the genes that will be transcribed (reviewed in Chalker and Yao, 2011). The mechanisms of DNA elimination have been best worked out in Tetrahymena, where the PIWI homolog TWI1 is required for this process (Mochizuki et al., 2002). scanRNAs (27–30 nt long) are made in the micronucleus in a Dicer-dependent fashion (which differs from metazoan piRNAs), and are loaded onto Twi1p (Mochizuki et al., 2002; Mochizuki and Gorovsky, 2005) (Fig. 4). The Twi1p/scanRNA complex directs H3K9 methylation and HP1 binding, thus marking DNA for elimination in the macronucleus (Liu et al., 2004b, 2007) (Fig. 4). The process shares significant similarities with the transcriptional transposon silencing described above for Drosophila. In Tetrahymena, however, the DNA is eliminated rather than transcriptionally silenced. A recent study in a distantly related ciliate, Oxytricha, demonstrated that a similar pathway is used. Instead of marking sequences for elimination, the scanRNAs (or piRNAs) are produced from the maternal macronuclear genome and thus mark sequences for protection in the new macronucleus (Fang et al., 2012). Despite the large evolutionary distance and differences in life strategy, the role for the PIWI/piRNA pathway in elimination of transposon and other repeat sequences from the ciliate somatic genome points to an ancient relationship between Piwi, piRNAs, and foreign DNA regulation.
Figure 4.
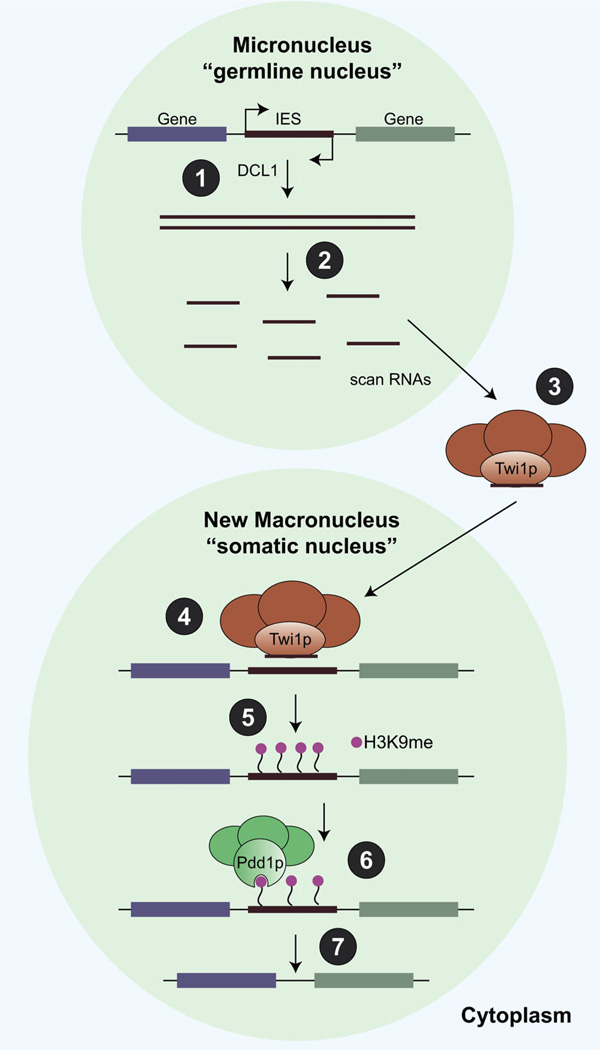
The PIWI/piRNA pathway marks repeat sequences for elimination in the somatic macronucleus of Tetrahymena. (1) Bidirectional transcription in the micronucleus creates double-stranded RNAs, which are a substrate for Dicer (DCL2). (2) Long-stranded RNAs are processed into scanRNAs by a Dicer-dependent mechanism. Scan RNAs are made from all sequences in the micronucleus, but only those made against internal eliminated sequences (IES) are shown above. (3) Scan RNAs are exported from the micronucleus (germline nucleus) and bind to a protein complex that contains the PIWI homolog Twi1p. Before entering the new macronucleus (somatic nucleus), Twi1p/scanRNA complexes enter the old macronucleus and scanRNAs that have homology against the DNA are destroyed (not shown in figure). (4) Twi1p/scanRNA complexes that remain are homologous only to IES regions, and are imported into the new macronucleus. (5) The Twi1p/scanRNA complexes mark IES regions for elimination by directing H3K9 methylation. (6) The Pdd1 protein contains a chromoshadow domain, which recognizes the H3K9 methylation on IES regions and recruits the machinery required for IES excision (7) leaving behind DNA enriched only for transcribed genes.
THE PIWI/piRNA PATHWAY AND THE MAINTENANCE OF GENOMIC INTEGRITY
Mutations in the PIWI/piRNA pathway lead to a dramatic upregulation of phosphorylated H2Ax, which is generally thought to mark unrepaired DNA double-stranded breaks. Furthermore, axis-determination defects observed in some Drosophila PIWI/piRNA-pathway mutants are partially rescued by inactivation of DNA damage signaling (Khurana et al., 2010). The currently accepted hypothesis suggests that uncontrolled transposition is the cause of DNA damage in PIWI/piRNA-pathway mutants. Support for this hypothesis arises from two general observations: (1) Several piRNAs map to transposons and (2) transposon mRNA levels are highly upregulated in piRNA pathway-deficient Drosophila ovaries and mouse testes (see above for details). Transcripts of functional transposons even accumulate within the oocyte nucleus in mutant backgrounds (Chambeyron et al., 2008), and integration of one particular transposon into the genome occurs in male Drosophila piwi mutants (Kalmykova et al., 2005). Yet, several piRNAs do not map to transposons (Table 2), evidence for transposon integration is scarce, and a comprehensive connection between transposon mobilization and DNA damage has not been elucidated. Causality thus remains undetermined, especially in light of the complex relationship between transposon mobilization and genome instability that we will discuss in this section.
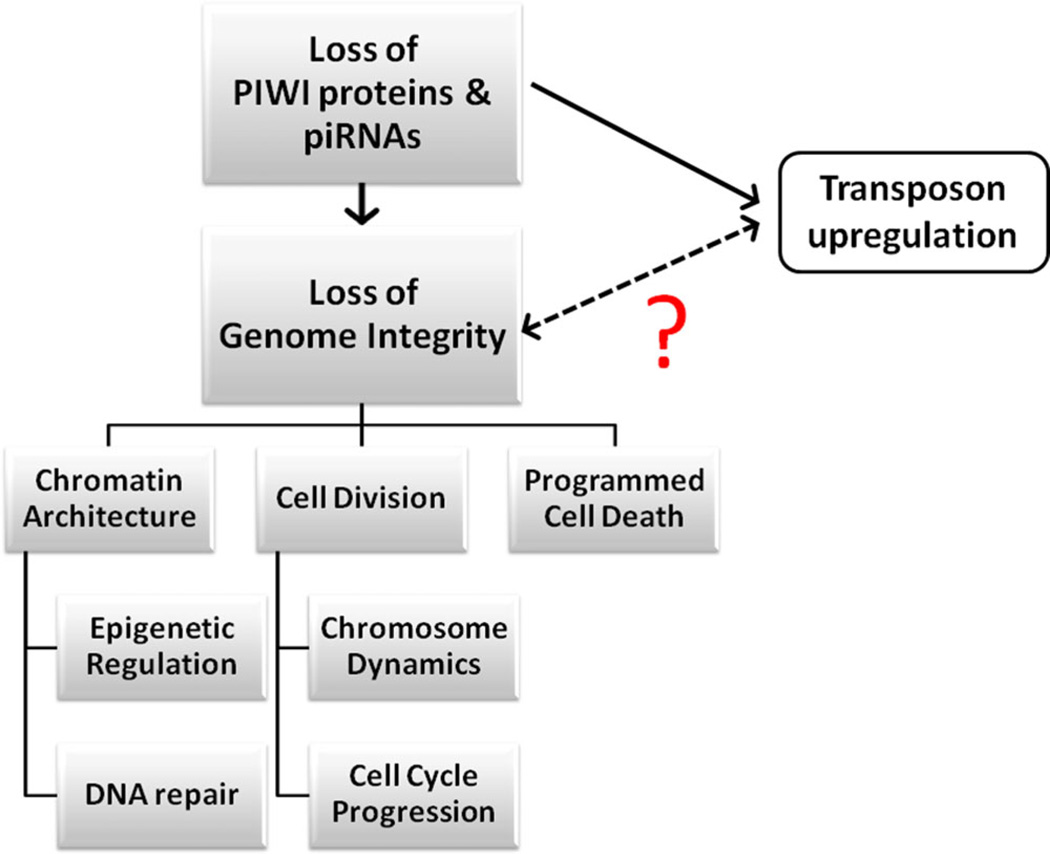
Barbara McClintock’s genomic stress hypothesis, which proposes transposon mobilization as an innate defense against stress, highlights the equally probable alternative that transposon upregulation is a consequence of genome instability rather than a cause (McClintock, 1984). Defects in PIWI mutants are immediate, and effects are seen within one generation, which may not be expected for transposon insertion-inflicted damage. In addition, it is not entirely clear that the upregulation of transposons alone could have the long-lasting, deleterious effects observed in PIWI/piRNA-pathway mutants. Finally, there is increasing evidence supporting the alternative that the pathway could directly regulate genome stability via the regulation of chromosome architecture, cell division, and apoptosis (Fig. 5). All of these possibilities will be discussed below in detail, and are important to keep in mind as we move forward in untangling the complex relationship between the PIWI/piRNA pathway and transposon regulation.
Figure 5.
The relationship between PIWI/piRNA pathway depletion, transposon upregulation, and the loss of genomic integrity. One currently accepted hypothesis suggests that the loss of the PIWI/piRNA pathway leads to uncontrolled transposon mobilization, which causes a loss of genomic integrity. An equally probable alternative is that the pathway could directly impact genome stability, which could then lead to the transposon upregulation seen in mutants. In this scenario, transposon upregulation is a symptom of the loss of genomic integrity rather than the cause. The double-headed arrow indicates the uncer-tain relationship between transposon upregulation and the loss of genome integrity in PIWI mutants. Emerging roles in the maintenance of chromosome architecture via epigenetic regulation and DNA repair; cell division via the regulation of chromosome dynamics and progression through the cell cycle; and programmed cell death offer experimental evidence for this alternate possibility.
The Link Between Transposons and Genome Stability
DNA damage can cause increases in transposition
A growing body of evidence suggests that DNA damage induced by both exogenous and endogenous sources can cause the mobilization of both DNA and RNA transposons in a variety of organisms. For example, the Ty element in yeast, an LTR retrotransposon, is mobilized in response to DNA damage-inducing agents such as UV light and 4-nitroquinoline-1-oxide (4NQO; Bradshaw and McEntee, 1989). The mobility of Ty1 is regulated by factors involved in the overall maintenance of genome integrity, such as telomere maintenance, DNA repair, suppression of DNA recombination, and DNA-damage response path-ways (Scholes et al., 2001). The loss of any of these regulators contributes to the hypermobility of Ty1, indicating that changes in genome integrity can modulate transposition. In telomerase mutants that underwent telomere erosion, Ty1 retrotransposition increased in parallel to the shortening of telomere DNA (Scholes et al., 2003). Activation of a DNA-damage signaling pathway was essential to this process, suggesting that an increase in retrotransposon mobility could be part of the cellular response to DNA damage. A later study found that activation of S-phase checkpoint pathways in yeast, via the replication stress pathway and/or the DNA damage pathway, is also able to increase Ty1 mobility, giving further support to this idea (Curcio et al., 2007).
Several results in Drosophila indicate that transposon mobilization in response to DNA damage is conserved in multicellular animals. First, injecting healthy male Drosophila with Mitomycin D to induce DNA damage results in a high mutation frequency in offspring; this is a result of genomic rearrangements due to excision of the gypsy transposon (Georgiev et al., 1990). Second, heat shock induces the mobility of an LTR transposon of the copia family (Ratner et al., 1992). Third, the effects of hybrid dysgenesis, which is thought to be due to transposon upregulation, are increased when parental females are treated with gamma rays and inhibitors of DNA replication (Bregliano et al., 1995). Furthermore, sub-lethal doses of gamma irradiation led to increases in excision of the P-element, a well studied DNA transposon (Handler and Gomez, 1997).
Several cell culture experiments demonstrate that transposon mobilization in response to DNA damage also occurs in vertebrates. Exposure of apoptosis-resistant murine and human cells to DNA-damaging agents increased SINE RNA levels and endogenous reverse transcriptase activity, thus indicating increased mobilization (Rudin and Thompson, 2001). SINE elements do not have their own reverse transcriptase activity, and are thought to utilize LINE family proteins for transposition. Indeed, it was later shown that LINE-1 retrotransposition increases upon gamma irradiation of cultured cells (Farkash et al., 2006). Oxidative stress, a common source of endogenous DNA damage, was also found to increase LINE-1 activity in human neuronal precursor cells as observed by anincrease in LINE-1 RNA levels as well as transposition events (Giorgi et al., 2011). This was also observed in yeast, where an increase in levels of reactive oxygen species resulted in increased mobility of Ty1 (Stoycheva et al., 2010). Thus, a broad survey of the literature points to a conserved phenomenon: transposon mobility in response to genotoxic stress. It is therefore important to keep this in mind when observing the effects of PIWI/piRNA pathway mutations on genomic integrity. The possibility that transposon upregulation is at least in part a response to increased DNA damage triggered by mutations in the PIWI/piRNA pathway remains a distinct possibility.
Transposon mobilization and the induction of immediate and lasting damage
It is generally assumed that rampant transposition is an obvious source of genome instability, but it is not clear if these effects are in fact immediately catastrophic to the organism. It is important to understand if damage induced by mobilization of transposons sufficiently explains the various defects observed in PIWI/piRNA-pathway mutants. For example, in male mice mutant for PIWI homologues, germ cells exhibit increased DNA damage, increased apoptosis, a block in meiosis, and ultimately a complete lack of fertility (Deng and Lin, 2002; Kuramochi-Miyagawa et al., 2004; Carmell et al., 2007). Similarly, PIWI/piRNA pathway-depleted Drosophila show a wide range of developmental defects, and any embryos laid are ultimately unable to develop (Cox et al., 1998; Harris and Macdonald, 2001; Li et al., 2009). Can the DNA damage induced by upregulation of transposons explain the results of mutations in the PIWI/piRNA pathway?
Drosophila hybrid dysgenesis models offer a good source of comparison since transposons are highly over-active while the PIWI/piRNA pathway is presumably intact. Hybrid dysgenesis is a syndrome resulting from an intraspecies cross. Paternal transposons not present on the maternal side are introduced into a zygote and triggers genetic instability due to a lack of protection on the part of the zygote against the newly introduced transposon(s) (Bregliano et al., 1980). The most obvious and common consequence of hybrid dysgenesis is sterility resulting from gonadal atrophy, very similar to flies lacking PIWI proteins and piRNAs. Additionally, eggs laid by dysgenic flies do not hatch. As dysgenic flies age, however, fertility is restored; a phenomenon that has puzzled researchers ever since it was first described (Bucheton, 1979). A recent paper connected the piRNA pathway to this syndrome by showing that new transposons introduced to dysgenic progeny are gradually silenced through the production of de novo piRNAs as flies age. These new piRNAs are produced both from paternally inherited piRNA clusters and resident element transposition into piRNA clusters, where they template new piRNA production (Khurana et al., 2011).
A major difference does however exist between PIWI/piRNA pathway mutant flies and hybrid dysgenesis models. Hybrid dysgenesis results in the misregulation of just one transposon during oogenesis whereas a large number of transposable elements appear de-repressed in PIWI-mutant flies (Bingham et al., 1982; Bucheton et al., 1984; Brennecke et al., 2007). This would suggest that any phenotype seen in dysgenic flies should be greatly exacer-bated in PIWI mutants. Yet, embryos laid by young dysgenic females undergo catastrophic meiosis, which causes death within the first embryonic cell division, unlike embryos depleted of PIWI proteins and piRNAs, which are able to proceed further in development (Lavige, 1986; Khurana et al., 2010). Overall this suggests that while overactive transposons could certainly cause infertility, the upregulation of transposons in hybrid dysgenesis models does not exactly phenocopy PIWI/piRNA-pathway mutants. Thus, transposon upregulation may not fully explain the myriad of phenotypes observed in PIWI/piRNA-pathway mutants.
Few studies show that overactive transposons can directly induce genome instability due to the sole presence of unrepaired DNA double-stranded breaks. In the most well-cited example, increases in DNA double-stranded breaks due to over-activity of LINE-1 were observed in mammalian cell culture, but these were repaired 48 hr after the insult and no consequent increase in apoptosis was reported (Gasior et al., 2006). This is significantly different from PIWI/piRNA mutants, where unrepaired DNA double-stranded breaks persist, and are often accompanied by rampant apoptosis (Kuramochi-Miyagawa et al., 2004; Carmell et al., 2007; Houwing et al., 2007; Watanabe et al., 2011a). If the yeast genome is artificially overloaded with retroelements, no defects in growth rate or gross morphology are observed, but there is increased sensitivity to DNA-damaging agents and DNA-replication errors, which then lead to loss of genomic integrity. In combination with the results obtained in mammalian cells, this suggests that increases in retrotransposon abundance need not be immediately deleterious to the genome, unless perhaps defects in the DNA-replication and error-prevention machinery exist (Scheifele et al., 2009).
Two primary sources of genome instability can arise from uncontrolled transposition. The first category involves physical rearrangements due to ectopic recombination. This occurs primarily due to the presence of homology to transposable elements interspersed through the genome. Second, the regulation of gene expression can be altered via de novo insertion near regulatory elements or into coding sequences (Hedges and Deininger, 2007). Importantly, transposon upregulation does not generally lead to increased amounts of unrepaired DNA double-stranded breaks that go unresolved, as is seen in PIWI/piRNA-pathway mutants where DNA double-stranded breaks persist (Gasior et al., 2006; Hedges and Deininger, 2007; Robert et al., 2008; Scheifele et al., 2009; Huefner et al., 2011). A careful study of the spatial and temporal regulation of transposons correlated to the type of genomic damage sustained in PIWI mutants is required to resolve the root cause of defects seen.
PIWI Proteins and Their Role in the Regulation of Chromatin Architecture
The regulation of chromatin organization has a large impact on genome function (Van Bortle and Corces, 2012). Recent work suggests that the PIWI/piRNA pathway is an epigenetic regulator. While a connection to genome stability is still nebulous, independent work ascribing a role for the pathway in DNA repair via modulation of chromatin architecture suggests that PIWI/piRNA pathway participation in the regulation of chromatin could have a direct impact on genome integrity.
Epigenetic regulation mediated by the PIWI/piRNA pathway
Mutations in piwi and aub suppress position-effect variegation (Pal-Bhadra et al., 2004), which occurs when a gene is situated near heterochromatin and is thus variably expressed from cell-to-cell. This is typified in the fly eye by a P-element mediated insertion of a tandem repeat of the white gene (required for red eye pigment), which induces heterochromatin formation and results in a mosaic red and white eye color (Dorer and Henikoff, 1994). Piwi and Heterochromatin protein-1 (HP1) physically interact, as demonstrated by immunoprecipitation from embryo extracts and co-localize on polytene chromosomes, along with the repressive chromatin mark H3K9 methylation (Pal-Bhadra et al., 2004; Brower-Toland et al., 2007). Mutations in piwi and aub lead to re-distribution of HP1 along the chromosome, and a decrease in H3K9 methylation (Pal-Bhadra et al., 2004). These results taken together suggest that the PIWI/piRNA pathway could be directing heterochromatin formation by the recruitment of HP1a.
This hypothesis is supported by the demonstration that inserting transposon 1360 into a normally euchromatic site induced heterochromatin formation, and that the potential piRNA binding sites in transposon 1360 are required for this phenomenon (Sentmanat and Elgin, 2012). The authors of this study suggest that heterochromatin domains are set up in the embryo by the PIWI/piRNA pathway, and are subse-quently propagated in the adult tissue by chromatin marks. It is important to note that the euchromatic sites tested in this study are adjacent to heterochromatin sites, which could indicate that Piwi is only capable of spreading heterochromatin rather than initiating de novo heterochromatin formation. A recent study demonstrates that ectopic recruitment of Piwi/piRNA complexes to euchromatic sites does promote de novo heterochromatin formation, however, and loss of Piwi perturbs the epigenetic state of the genome, suggesting that Piwi/piRNAs complexes are a global regulator of chromatin (Huang et al., 2013). piwi is also required for the transgenerational suppression of cryptic phenotypes in the fly, and epigenetic silencing initiated by Piwi in the worm is very stable and can be inherited over several generations (Gangaraju et al., 2011; Shirayama et al., 2012). These studies additionally point to a vital role for the pathway in initiating stable chromatin states.
A potentially conflicting study shows that P-element insertions into piRNA clusters require the PIWI/piRNA pathway to be expressed, and there is an increase of HP1 at cluster loci in Piwi/pRNA pathway mutants. These data are seemingly contrary to previous reports of the PIWI/piRNA pathway promoting heterochromatin formation (Moshkovich and Lei, 2010), but may be related to the piRNA cluster loci examined, which are transcribed despite being embedded in heterochromatin. Piwi was also shown to promote euchromatin marks at the telomere region of chromosome 3, thus promoting the production of piRNA 3R-TAS1 (Yin and Lin, 2007). Therefore, PIWI/piRNA path-way function may depend on chromatin context. Further-more, Piwi localization on polytene chromosomes is differentially sensitive to different RNase enzymes, suggesting both RNA/RNA and DNA/RNA hybrids are used at different locations for targeting PIWI/piRNA complexes to the chromatin (Brower-Toland et al., 2007).
DNA repair mediated by the PIWI/piRNA pathway
Recent studies point to the possibility of a direct role for the PIWI/piRNA pathway in repairing DNA damage. Both Mili and Hili, respectively murine and human PIWI proteins, have a role in maintaining an open chromatin state that aids DNA repair. In cells damaged with cisplatin, chromatin relaxation usually occurs via histone acetylation to facilitate repair. Acetylase activity was decreased in mili mutants, thus impeding chromatin relaxation and repair after cisplatin insult. Furthermore, enhanced PIWI protein expression was described as a key factor in the cisplatin resistance of human ovarian cancer cells (Wang et al., 2011b). In a separate study, mili knock-out cells were deficient in DNA repair after irradiation and cisplatin insult. A variety of DNA double-stranded break repair defects and increased susceptibility to apoptosis were observed, and these were again attributed to chromatin relaxation defects due to impaired histone acetylation (Yin et al., 2011). An interesting study examining proteins associated with Alu retrotransposon-derived piRNAs also indicates a role for the pathway in the maintenance of genome stability. Lentiviral knockdown of these piRNAs were found to reinstate self-renewal of senescent human adipocytes stem cells (Wang et al., 2011a). Analysis of nuclear interactors of Alu-derived piRNAs revealed proteins associated with DNA repair and chromatin remodeling (Blackwell et al., 2012), suggesting that the cytotoxicity associated with the upregulation of Alu-associated piRNAs is due to a loss of efficient DNA repair and chromatin regulation, which leads to genome instability. Stable suppression of Alu transcription thus probably reverses senescence by reinstating appropriate DNA repair and maintaining chromosome architecture.
Small RNAs may have a conserved role in repairing DNA double-stranded breaks. In both Arabidopsis and human cells, small RNAs (double-stranded break-induced small RNAs or diRNAs) were produced from regions in the immediate vicinity of the targeted break sites, and these small RNAs were required for efficient repair. It was proposed that the small RNAs function as guide molecules for histone modifications around the DNA double-stranded breaks, which could then facilitate recruitment of repair components (Wei et al., 2012). It would be interesting to test if piRNA production increases specifically at DNA double-stranded break sites, or if the PIWI/piRNA pathway only functions more widely in chromatin relaxation to aid DNA repair. Modulation of chromatin is a key factor in most DNA repair pathways (Cann and Dellaire, 2011), and PIWI’s involvement in this facet of genome stability is intriguing especially given its role as an epigenetic regulator (Lin and Yin, 2008).
PIWI Proteins and Their Role in Cell Division
Normal cell division hinges on coordination between the regulation of chromosome dynamics and regulation of progression through the cell cycle. Recent studies highlight an emerging role for PIWIs/piRNAs in both aspects of cell division, and fully understanding these functions will require careful scrutiny.
Regulation of chromosome dynamics
Centromeres and telomeres, major structural components of the chromosomes, are comprised of heterochromatin. Therefore, the organization of heterochromatin is essential for accurate chromosome dynamics during both mitosis and meiosis (Dernburg et al., 1996; Bernard et al., 2001). As discussed above, Drosophila Piwi is enriched at constitutive heterochromatin domains and associates with HP1. Piwi depletion impacts not only HP1 localization, but also key histone modifications traditionally associated with gene silencing, which suggests an involvement in chromatin organization (Pal-Bhadra et al., 2004). The loss of HP1 and inappropriate histone modification are increasingly being associated with defects during mitotic and meiotic progression due to kinetochore depletion, centromere abnormalities leading to segregation defects, and telomere shortening, all of which could compromise chromosome stability leading to abnormal cell division (Dialynas et al., 2008; Heit et al., 2009; Xu et al., 2009).
Telomeres were first described as structures capping and protecting the ends of chromosomes to prevent chromosome fusions. A second, important end-elongation function was later discovered, which is required to maintain the fidelity of DNA replication. Thus telomeres have an important role in the cell cycle. Drosophila is unique in that telomeres are maintained by the mobilization of three telomere-localized retrotransposons, HeT-A, TART, and TAHRE, instead of the telomerase enzyme (Pardue and DeBaryshe, 2011). Sub-telomeric sequences produce piRNAs that map to these transposons, and transcripts of all three retrotransposons are upregulated with the loss of PIWI proteins, suggesting a role in telomere dynamics (Brennecke et al., 2007; Shpiz et al., 2007, 2009). Heterozygous mutations in aub and spindle E increase retrotrans-position events to broken chromosome ends, as was shown in an elegant assay utilizing de novo telomere formation at induced, broken chromosome ends to study the frequency of telomere mobilization events (Savitsky et al., 2006). The PIWI/piRNA pathway is thus likely capable of regulating telomere length by regulating retrotransposons. Functional conclusions are hard to make based on this study, however, since flies with only one copy of aub or spindle E did not display an appreciable change in HeT-A or TART retro-transposon activity without the use of this assay. While this could merely be because Drosophila telomeres are extremely long, and any phenotype would require generations before being noticed, PIWI/piRNA involvement in telomere elongation could also be occurring only in a broken chromosome situation.
More recently, a role for piRNA pathway proteins was shown in facilitating chromosome-end protection via recruitment of the telomere-capping complex (Khurana et al., 2010). Telomere fusions in Drosophila embryos depleted of aub and armi indicate a requirement for the PIWI/piRNA pathway in telomere resolution. Indeed, telomeric recruitment of HP1 and HOAP (HP1/origin of replication-associated protein), components of the telomere-protection complex (TPC), did not occur in mutants. Thus, a sub-population of telomere-specific piRNAs may be required to recruit the TPC to chromosome ends, which are mediated by Aub and Armi and are unique among piRNA pathway proteins for this function. Depleting components required for non-homologous end joining (NHEJ) in a mutant background rescued the telomere fusion defect in aub and armi mutants, suggesting NHEJ as a mechanism for telomere ligation. Yet, studies of telomere fusions in TPC mutants in Drosophila do not suggest the involvement of NHEJ in this process (Rong, 2008). The contribution of NHEJ in this situation is thus puzzling, and might suggest mis-regulation of the pathway in a PIWI-mutant background. The proposed small RNA involvement in this process also suggests sequence-specificity requirements conferred by the piRNAs. In Drosophila, however, capping does not require sequence specificity, that is, any sequence at the end of a broken chromosome can be capped (Rong, 2008). Dissection of the functional requirement of this subpopulation of piRNAs and examination of the exact mechanism underlying telomere fusions seen will therefore prove to be very interesting.
PIWI proteins and associated piRNAs can also directly impact chromosome condensation and segregation. During mitosis, condensin loading begins at the peri-centromeric region and spreads to cover the chromosome arms, reaching a maximum at anaphase to facilitate chromosome separation (Oliveira et al., 2007). In Drosophila germ cells, mitotic bodies composed of piRNA pathway proteins bind to peri-centromeric, piRNA-producing loci. Mutants display aberrant condensin loading, leading to a delay in chromosome condensation and segregation defects (Pek and Kai, 2011). This finding suggests a model in which recruitment of the condensin complex is piRNA-guided and implicates the PIWI/piRNA pathway in another facet of chromosome structural organization.
Regulation of cell cycle progression
While PIWI/ piRNA requirement in stem cell self-renewal has garnered a lot of attention, important roles that have been emerging in parallel are in the regulation of both mitosis and meiosis. The mitotic proliferation of primordial germ cells during development and maintenance of adult stem cells via mitosis are universal themes (Kimble, 2011). While a direct involvement in the regulation of mitosis has not been adequately explored, the PIWI family’s role in sustaining an adequate population of germ cells appears to be conserved. In adult flies, the self-renewing divisions of germ line stem cells are governed by somatically expressed Piwi. In contrast, Piwi within germline stem cells acts cell-autonomously as a mitotic promoter; loss of Piwi within germline stem cells reduces the rate of division while overexpression increases the rate of stem cell divisions (Cox et al., 1998, 2000). It is interesting to note that this function of Piwi is genetically separable from transposon repression; Piwi mutants that cannot localize to the nucleus are able to rescue germline stem cell defects, but not transposon repression (Klenov et al., 2011). Similarly in C. elegans, depletion of prg1 and 2 drastically reduces the mitotic proliferation zone at the distal end of each gonadal arm. Within this shortened zone, the number of cells that do undergo mitosis have a highly reduced mitotic index, suggesting that Piwi in C. elegans is capable not only of modulating the number of cells undergoing mitosis but also the rate at which they go through mitosis (Cox et al., 1998). Zebrafish and mouse orthologues of Piwi are additionally implicated in the maintenance of germ cells. The loss of Ziwi and Zili triggers massive apoptosis, resulting in the complete loss of germ cells within 40 days of development, while miwi2 mutants display similar phenotypes with regards to male germ cells (Carmell et al., 2007; Houwing et al., 2007, 2008).
A requirement for PIWI proteins during meiotic progression was first found in mice, where MILI and MIWI2 depletion results in male germ-cell arrest at prophase of meiosis I. In contrast, miwi, which is expressed during later steps of spermatogenesis, is required at post-meiotic stages of spermatogenesis (discussed further below; Deng and Lin, 2002; Kuramochi-Miyagawa et al., 2004; Carmell et al., 2007; Houwing et al., 2007, 2008). A detailed examination of meiotic defects in Miwr−/−; Milir−/− mice, which results in a loss of all PIWI/piRNA function in the adult testes, suggests that the mechanics of meiotic prophase, including homolog pairing and synapsis formation, occur normally. Beyret and Lin speculate that the modification of epigenetic status or perturbation of recombination dynamics could explain the arrest phenotypes seen. Interestingly, the same study found that piRNAs are significantly upregulated during the first wave of meiosis in mice, concurrent with an upregulation of their protein partners, providing additional evidence for a potential function in the regulation of entry or progression through meisois (Beyret and Lin, 2011).
This putative function in facilitating meiotic progression appears to be conserved. Two meiotic checkpoints have been described in Drosophila: a DNA-damage checkpoint and a pachytene checkpoint (Joyce and McKim, 2011; Lake and Hawley, 2012). While activation of the DNA-damage checkpoint has been demonstrated in PIWI/piRNA mutant oogenesis, this may not indicate a specific meiotic involvement, but a general requirement for genome stability in the accurate completion of meiosis (Ghabrial et al., 1998; Klattenhoff et al., 2007). Yet a recent paper described the involvement of Maelstrom, a PIWI/piRNA pathway component, in repression of the pachytene checkpoint, which is commonly triggered by a failure in chromosome synapsis, and merits exploration of possible defects in the mechanics of meiosis in Drosophila PIWI/piRNA-pathway mutants (Pek et al., 2012).
A meiotic-progression function has also been ascribed to zili in zebrafish. Houwing and colleagues show that oocytes laid by heterozygotic females depleted of one copy of zili arrest at meiosis I despite being normally fertilized. It is important to note that this defect is seen without an increase in transposon transcripts. Along with the observation that a sizeable percentage of piRNAs in zebrafish map to genic instead of transposon regions (Table 2), this suggests that the regulation of meiosis by PIWIs and associated piRNAs could be separable from transposon regulation (Houwing et al., 2008).
A role for the PIWI/piRNA pathway in the regulation of cell division outside the germline is also emerging through studies in the sea urchin embryo, fly embryo, and mouse. As described above, vasa is a key player in the PIWI/piRNA pathway (Arkov and Ramos, 2010; Kuramochi-Miyagawa et al., 2010). A recent study in the sea urchin embryo shows an interesting function for Vasa in mitotic progression via the regulation of mitotic cyclins. Vasa localizes to spindles and separating chromatids during mitosis, and is dependent on regulation by cyclin-dependent kinases (Cdks) for this localization. Depletion of Vasa in early embryos perturbs cyclin B translation and arrests the cell cycle at M-phase, suggesting an essential involvement in cell cycle progression (Yajima and Wessel, 2011). A recently identified function of Piwi proteins in regulation of maternal transcript destruction during the maternal-to-zygotic transition in early embryos also suggests the potential for mitotic regulation (Rouget et al., 2010). While the study focused on the misregulation of nanos translation, there are other targets identified in the study. While they do not look at cyclinB, this gene is also a target of mRNA decay at the maternal-to-zygotic transition and may be also targeted by the PIWI/piRNA pathway (Benoit et al., 2009). Considering the variety of targets aberrantly stabilized by loss of some piRNA pathway components, it is not a stretch to consider inappropriate regulation of cyclin B mRNAs, levels of which are essential to the normal progression of mitosis during Drosophila embryogenesis (McCleland et al., 2009).
In adult mouse mesenchymal stem cells, MILI and associated piRNA expression are found in the cytoplasm selectively in mitotic cells. An analysis of potential MILI target genes revealed a non-random enrichment of cell cycle and microtubule regulation associated genes suggesting an involvement in the regulation of proliferation. The authors go on to show that the depletion of MILI from these cells pushes the cells into mitosis and increases proliferation. This is in contrast to piwi mutations in the germline, where depletion results in decreased proliferation. This indicates a role for the PIWI/piRNA pathway in the somatic cell cycle that may or may not be analogous to its functions in the regulation of germ cell divisions (Wu et al., 2010).
These studies suggest the PIWI/piRNA pathway exerts either a direct or indirect role in ensuring the fidelity of the cell cycle. Work implicating human and mouse homologues of PIWI proteins in the development of cancer highlights the importance of exploring this model further (Suzuki et al., 2012).
PIWI Proteins and Their Role in Programmed Cell Death
PIWI function appears to be intertwined with the induction of apoptosis, as seen by drastic increases in apoptosis in the germ cells of both mice and zebrafish lacking PIWI homologues. This phenotype was first detected in miwi-mutant mice, which show survival deficiencies of spermatognia, spermatocytes, and spermatids. These defects are seen only in the second wave of spermatogenesis, and thus are attributed to indirect effects of an earlier spermiogenic arrest (Deng and Lin, 2002). In both miwi2- and mili-mutant mice, however, apoptosis appears to begin concurrently with spermatogenesis arrest at meiosis I of prophase, suggesting that the two defects are linked (Kuramochi-Miyagawa et al., 2004; Carmell et al., 2007). Similar results are seen in other murine piRNA pathway mutants, including Tdrd1, Tdrd5, Tdrd9, Mov10L1, Mael, and MitoPLD (Chuma et al., 2006; Soper et al., 2008; Shoji et al., 2009; Frost et al., 2010; Watanabe et al., 2011a; Yabuta et al., 2011). This correlation between the onset of germ-cell defects and apoptosis induction is also seen in zebrafish, where loss of Ziwi results in depletion of germ cells and an increase in apoptosis. A partial loss of Ziwi, to a degree that allows for the survival of germ cells, unexpectedly causes increases in apoptosis that become evident in adulthood. This indicates that ziwi mutants have a defect that leads to apoptosis independent of Ziwi’s roles in germ cell development (Houwing et al., 2007). Although Zili-depleted zebrafish also lose germ cells, no evidence for an increase in apoptosis is seen, suggesting distinct mechanisms for each zebrafish PIWI homolog in maintaining germ cells (Houwing et al., 2008).
While apoptosis could be an indirect consequence of PIWI’s other biological functions going awry, especially when an association with DNA damage signaling is taken into consideration (Klattenhoff and Theurkauf, 2008), the possibility of a more direct involvement in apoptotic signaling is coming to light based on studies in cancer cell lines. The anti-apoptotic gene BCL-XL is a potential downstream target of Piwil2 in cancer cells; STAT3, a positive regulator of BCL-XL was additionally found to be upregulated upon PIWI expression. In addition, the removal of Piwil2 increased apoptosis whereas overexpression decreased apoptosis. Findings in NIH-3T3 cells were confirmed in mouse and human cancer cell lines as well as in breast cancer stem cells (Lee et al., 2006, 2010). A more mechanistic understanding of PIWI’s role in modulating this path-way was recently described in HeLa cells, where its anti-apoptotic function may derive primarily from the repression of p53 via formation of a tripartite, nuclear complex of PIWIL2, STAT3, and SRC. In the nucleus, the complex induces silencing of the p53 promoter via histone modification, suggesting that PIWIL2 control of p53 expression is transcriptional (Lu et al., 2012).
An alternative mechanism for PIWI proteins in modulating apoptosis involves the activation of pro-survival factors. On examination of PIWI expression in pre-cancer stem cell lines, a large number of previously unknown variants of PIWI were discovered. These variants were detected in cancer stem cell lines, mouse and human tumor cell lines, as well as in primary and metastatic tissue samples. The expression of one variant in particular, PL2L60, resulted in the nuclear localization of NF-κB and subsequent upregulation of BCL2, a pro survival factor. Indeed breast cancer cells overexpressing PL2L60 form tumorous nodules when transplanted into mice (Ye et al., 2010).
It is important keep in mind that most studies exploring a direct function for PIWI in apoptosis have been limited to cancer cells, which is a specialized model system. While increased apoptosis is also observed in the germline after depletion of PIWI/piRNA pathway function, the genomic instability induced by these mutations could explain the increased cell death observed. Testing for a direct role of the PIWI/piRNA pathway in apoptosis using physiologically normal cells is required to draw any definitive conclusions.
What Is the Relationship Between the PIWI/ piRNA Pathway, Transposons, and Genome Stability?
PIWI/piRNA pathway mutations lead to transposon up-regulation, increased DNA double-stranded breaks, and increased rates of apoptosis. A proposed explanation of these observations is that the PIWI/piRNA pathway functions primarily in transposon repression, and that the unbridled expression of transposons in a PIWI/piRNA pathway mutant leads to genomic instability. In light of the data reviewed in this section, an alternative interpretation for the phenotypes observed in PIWI/piRNA mutants can be put forth. If the PIWI/piRNA path way is itself required to regulate or maintain genome stability, then mutations in the pathway may lead to such phenotypes as unresolved double-strand DNA breaks. Furthermore, the persistent DNA damage in these mutants could ultimately lead to transposon upregulation. This scenario does not exclude the role of the Piwi/piRNA pathway in repressing transposon transcription and translation, which has been well documented and is likely required for the long-term fitness of the organism. Still, it has not yet been definitively shown that transposon upregulation directly leads to the immediate and catastrophic myriad of phenotypes observed in PIWI/piRNA-pathway mutants. Rather, the PIWI/piRNA pathway appears to control a multitude of processes, and thus mutant phenotypes are likely pleiotropic. The upregulation of transposon sequences in PIWI/piRNA-pathway mutants is likely due in part to a direct role for the pathway in repressing transposon transcription and degrading transposon transcripts, but could further be exacerbated by a loss of genome stability. It is clear that the relationship between transposons, DNA damage, and genome stability is fraught with complexity. Every possibility needs to be considered when trying to delineate PIWI function in the context of transposon upregulation.
REGULATION OF NON-TRANSPOSON GENES BY THE PIWI/piRNA PATHWAY
A growing number of studies now supports the conclusion that the PIWI/piRNA pathway functions not only to repress transposons, but also to regulate protein-coding genes. Throughout animal phylogeny, PIWI proteins are found in both the nucleus and the cytoplasm (summarized in Table 1). Additionally, gene regulation can occur at both the transcriptional and post-transcriptional levels.
Transcriptional/Epigenetic Regulation of Protein Coding Genes
Two studies demonstrate that the PIWI/piRNA pathway represses the expression of a gene by the methylation of its promoter: (1) Rasgrf1 in the mouse and (2) CREB2 in the sea slug (Watanabe et al., 2011b; Rajasethupathy et al., 2012). In the mouse, imprinted genes have monoallelic expression due to the methylation of either the maternal or paternal allele. Differential methylation is erased in primordial germ cells and is subsequently established de novo in male prospermatogonia (Sasaki and Matsui, 2008). The PIWI/piRNA pathway is required for re-establishing methylation for imprinting at the Rasgrf1 gene locus (Watanabe et al., 2011b). Hundreds of piRNAs that are derived from a piRNA cluster and resemble LTR transposons map antisense to a specific region in the Rasgrf1 locus. A non-coding RNA is also transcribed from this region, so it was proposed that the MIWI2/piRNA effector complex is targeting the nascent transcript of this non-coding RNA, which subsequently recruits methylation machinery (Watanabe et al., 2011b).This model resembles transcriptional silencing in Schizosaccharomyces pombe and Arabidopsis, where siRNA/Argonaute complexes bind nascent RNAs and recruit epigenetic modifiers (Buhler et al., 2006; Wierzbicki et al., 2009). The PIWI/piRNA pathway is also required for methylation of the CREB2 promoter in the Aplysia (sea slug) central nervous system (Rajasethupathy et al., 2012). CREB2 is a transcription factor that inhibits long-term memory, thus silencing of the gene by the PIWI/piRNA pathway is required to establish long-term memories (Bartsch et al., 1995; Rajasethupathy et al., 2012). PIWI/piRNA complexes are expressed in the nucleus of Aplysia nerve cells, and a specific piRNA (aca-pIR-F) is required for the methylation of CREB2. Interestingly, a putative sequence near the CREB2 transcription start site allows for PIWI/piRNA (aca-piR-F) complex binding and recruitment of methylation machinery (Rajasethupathy et al., 2012).
As Drosophila do not have DNA methylation, the PIWI/ piRNA pathway directs the epigenetic silencing of transposons in the Drosophila ovary by H3K9 methylation. It is not yet clear if this mechanism is also used to regulate protein-coding genes in Drosophila. Intriguingly, one study suggests that piRNAs produced from the 3′-untranslated region (UTR) of traffic jam mRNA in Drosophila ovarian somatic cells may be required to repress the transcription of the FasIII gene (Saito et al., 2009). In addition, it was recently demonstrated that insertion of a transposon into a protein-coding gene in ovarian somatic cells leads to Piwi-dependent silencing of that gene (Sienski et al., 2012). Thus, it is possible that the PIWI/piRNA pathway functions to repress protein-coding genes at the transcriptional level in Drosophila, similar to the mechanisms observed in mouse and sea slug.
Post-Transcriptional Regulation of Protein Coding Genes
Several PIWI protein homologs are cytoplasmic (Table 1), and are thus poised to regulate gene expression at the post-transcriptional level. It is well established that Argonaute proteins, in complex with miRNAs, target mRNAs for translational repression and ultimately degradation (Bazzini et al., 2012). PIWI/piRNA effector complexes could work in a similar manner, but data supporting such a mechanism are still scarce. Work in the Drosophila ovary demonstrated that components of the PIWI/piRNA pathway co-localize with a subset of processing bodies, which were thus named pi-bodies (Lim et al., 2009). Processing bodies are a site of mRNA degradation and contain enzymes responsible for the de-capping and degradation of RNA (reviewed in Eulalio et al., 2007). PIWI proteins, piRNAs, processing body enzymes, and retrotransposon transcripts co-localize in pi-bodies. Furthermore, mutations in the PIWI/piRNA pathway or processing body genes lead to the stabilization of HeT-A retrotransposon mRNA (Lim et al., 2009). This suggests that the PIWI/piRNA pathway can direct degradation of transposon RNAs via post-transcriptional mechanisms similar to those used to degrade mRNAs in processing bodies. This may be a conserved function of the pathway, as similar co-localization was observed in the mouse fetal testis between PIWI/piRNA pathway proteins and processing body enzymes; these bodies were termed piP-bodies (Aravin et al., 2009).
Data identifying non-transposon mRNA targets that are degraded by the PIWI/piRNA pathway are still rare. In one well-documented example, the pathway targets maternally loaded mRNAs for deadenylation and subsequent degradation at the maternal-to-zygotic transition in Drosophila embryos (Rouget et al., 2010). Using nanos mRNA as a model, it was demonstrated that transposable element-like sequences in the 3′-UTR are targeted by Aub and/or Ago3 in complex with piRNAs derived from transposon sequences. Transposable element-like sequences were also found in the 3′-UTRs of several other mRNAs that are turned over at the maternal-to-zygotic transition, suggesting a widespread mechanism for clearing mRNAs (Rouget et al., 2010). There are piRNAs complementary to the vasa transcript in the Drosophila testes, and Vasa protein levels are increased in aub and ago3 mutants (Nishida et al., 2007; Li et al., 2009). Furthermore, Aub/piRNA complexes immunoprecipitated from the testes can slice vasa transcripts in vitro, suggesting a potential mechanism for repressing vasa in vivo (Nishida et al., 2007). Although there are relatively few reported examples of the PIWI/ piRNA pathway targeting mRNAs for transcriptional silencing, a large number of piRNAs are derived directly from the 3′-UTRs of select genes (Robine et al., 2009). This suggests that the pathway may repress gene expression by turning mRNAs into piRNAs. An alternative, but not mutually exclusive, possibility is that piRNAs derived from 3′-UTRs are subsequently used to regulate the expression of other genes.
In contrast to known roles for Argonaute proteins in directing mRNA silencing, MIWI is required for protecting mRNAs that function in spermiogenesis, the final stage of spermatogenesis in which mature sperm are formed from spermatids (Deng and Lin, 2002; Vourekas et al., 2012). In the first description of the miwi-mutant mouse, it was noted that the phenotype was very similar to the CREM-mutant phenotype; CREM is a transcription factor and master regulator of spermiogenesis (Fimia et al., 2001; Deng and Lin, 2002). In miwi-mutant testes, the transcriptional targets of CREM are lost and in wild-type testes, MIWI directly interacts with the CREM target mRNAs (Deng and Lin, 2002). The authors speculated that MIWI binding could function to stabilize the CREM target mRNAs, which was demonstrated to be the case 10 years later (Vourekas et al., 2012). Using high-throughput sequencing of RNA isolated by cross-linking immunoprecipitation (HITS-CLIP), it was found that MIWI coats several hundred mRNAs, and this population is enriched for genes required in the post-meiotic stages of spermatogenesis (Vourekas et al., 2012). The genes required for spermiogenesis are transcribed earlier in spermatogenesis before chromatin compaction occurs, and the mRNAs are stored in messenger ribonucleoprotein particles(mRNPs; Bagarova et al., 2010). MIWI proteins bound to repressed mRNAs accumulate in these mRNPs, but these mRNAs are lost in miwi-mutant testis (Vourekas et al., 2012). The repressed mRNPs were isolated, and it was found that piRNAs are absent from the mRNPs, thus suggesting that MIWI can directly bind to mRNA without a piRNA guide. Partially consistent with this, an earlier study found that MIWI associates with polysomes; these isolated complexes also included the repressed mRNPs (Grivna et al., 2006b; Bagarova et al., 2010). Yet, piRNAs were also found in association with polysomes, suggesting that PIWI/piRNA complexes are required for post-transcriptional control, perhaps in addition to stabilizing mRNAs for spermiogenesis. MILI is also associated with polysomes, and overall protein synthesis is significantly reduced in mili-mutant testes (Unhavaithaya et al., 2009). Finally, both MIWI and MILI associate with core components of the translational machinery and thus may function broadly in regulating translation (Grivna et al., 2006b; Unhavaithaya et al., 2009). The function of PIWI proteins in stabilizing mRNAs required during spermiogenesis may be conserved. In C. elegans, over 500 mRNAs are down-regulated in prg-1 mutants; these mRNAs are enriched for spermatogenesis genes, especially those required in later stages (Wang and Reinke, 2008). prg-1 mutants are temperature-sensitive sterile, perhaps indicating that mRNA protection is para-mount at increased temperatures (Batista et al., 2008; Wang and Reinke, 2008).
CAN THE PIWI/piRNA PATHWAY HARNESS THE REGULATORY POWER OF TRANSPOSONS?
In the more than 60 years since the initial discovery of transposons by Barbara McClintock (McClintock, 1951), the impact of transposable elements on genomes has been hotly debated. On the one hand, they are considered genomic parasites that exist because of their selfish nature and thus have deleterious effects on the fitness of their host genomes (Doolittle and Sapienza, 1980; Orgel and Crick, 1980). On the other hand, they may prove useful to their host genomes by providing a mechanism for inventing novel regulatory networks (McClintock, 1951; Britten and Davidson, 1969). Transposable element insertion into the genome can directly affect gene expression in several ways. For example, insertions into open reading frames are generally deleterious and selected against (Medstrand et al., 2002). By contrast, insertions into regulatory regions are common: 18.1% of mice transcription start sites and 31.4% of human transcription start sites have transposable element insertions (Faulkner et al., 2009). Ultimately the propagation of transposable elements requires the survival of their host. Thus, transposable elements regulate their own mobilization such that they can co-exist with the host genome (Rebollo et al., 2012). Transposable elements have evolved to interact with host regulatory genes and thus transposable element regulatory regions can be co-opted by the host genome for gene regulation. There-fore, the relationship between host genomes and transposable elements is not strictly antagonistic; instead, there is a complex interplay between the two. It is important to keep this in mind when considering the relationship between transposon regulation and the PIWI/piRNA pathway.
In an adult organism, maintaining homeostasis requires coordinately regulating large groups of genes both at the transcriptional and post-transcriptional levels. For example, in stem-cell driven processes such as gametogenesis or hematopoiesis, individual cells transit through many inter-mediate states before completing differentiation. This requires synchronized transitions in gene expression levels, which need to be regulated with precision. Transposon sequences inserted into regulatory regions could potentially impact transcription, mRNA stability, or translation. Furthermore, transposon mobilization allows for the insertion of common regulatory sequences throughout the genome. We propose that the PIWI/piRNA pathway is well-positioned to regulate large groups of genes with common transposon-derived sequences due to its intimate relation-ship with transposons. Therefore the PIWI/piRNA pathway could be coordinately regulating large numbers of genes required for normal function, either maintaining homeostasis or transitioning through cellular states.
In addition to regulating gene expression, specific targeting of piRNAs to complementary sites on chromatin could impact chromosome structure through the recruitment of epigenetic regulators, DNA repair machinery, and molecules involved in the coordinated movement of chromatin during the cell cycle. Disruption of these processes along with abnormal alterations of gene expression levels in PIWI/piRNA-pathway mutants could lead to the variety of phenotypes observed. It is probable that the regulatory control of the PIWI/piRNA pathway reaches far beyond our current understanding. Research has largely focused on its function as a transposon repressor in the germline. Yet, PIWI proteins are also expressed in the soma, can regulate gene expression, including protein-coding genes, on several different levels, and can modulate chromatin architecture. A deeper exploration of these additional functions is warranted and should prove very illuminating.
ACKNOWLEDGMENTS
We would like to thank Gary Wessel, Stefan Materna, and Toshiaki Watanabe for valuable comments and helpful discussion. We also thank Haifan Lin for his support. Finally, we are extremely grateful to an anonymous reviewer for insightful feedback that greatly improved the manuscript. Research by S.R.M. and C.E.J. is supported by an NIH Pioneer Award (DP1CA174418) and a Mathers award to Haifan Lin, and by an NIH F32 (GM9037222) award to C.E.J.
Grant sponsor: NIH Pioneer;
Grant number: DP1CA174418;
Grant sponsor: Mathers;
Grant sponsor: NIH F32;
Grant number: GM9037222
Glossary
Abbreviations used
- AGO
Argonaute sub-family of Argonaute proteins
- Armi
Armitage
- Aub
Aubergine
- HP1
heterochromatin protein 1
- LINE
long interspersed nuclear element
- LTR
long terminal repeat elements
- miRNAs
microRNAs
- piRNAs
PIWI-interacting RNAs
- PIWI
Piwi sub-family of Argo-naute proteins
- SINE
short interspersed nuclear element
- siRNAs
small-interfering RNAs
- UTR
untranslated region
- Vret
Vreteno
- Zuc
Zucchini
Footnotes
We will use “PIWI” throughout this review to mean the PIWI protein family and “Piwi” to mean the specific protein in Drosophila.
NOTE ADDED IN PROOF
The mouse homolog of shutdown, Fkbp6, binds to Hsp90 and is required for repression of LINE1 in the testis and is involved in piRNA biogenesis. This demonstrates the conserved nature of the Hsp90 machinery for piRNA biogenesis (Xiol et al., 2012).
REFERENCES
- Alie A, Leclere L, Jager M, Dayraud C, Chang P, Le Guyader H, Queinnec E, Manuel M. Somatic stem cells express Piwi and Vasa genes in an adult ctenophore: Ancient association of germline genes with stemness. Dev Biol. 2011;350:183–197. doi: 10.1016/j.ydbio.2010.10.019. [DOI] [PubMed] [Google Scholar]
- Allan RK, Ratajczak T. Versatile TPR domains accommo-date different modes of target protein recognition and function. Cell Stress Chaperones. 2011;16:353–367. doi: 10.1007/s12192-010-0248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A, Kai T. The tudor domain protein kumo is required to assemble the nuage and to generate germline piRNAs in Drosophila. EMBO J. 2012;31:870–882. doi: 10.1038/emboj.2011.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007a;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007b;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 2009;5:e1000764. doi: 10.1371/journal.pgen.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkov AL, Ramos A. Building RNA-protein granules: Insight from the germline. Trends Cell Biol. 2010;20:482–490. doi: 10.1016/j.tcb.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, Pintacuda G, Sakaguchi A, Sarkies P, Ahmed S, Miska EA. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans . Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagarova J, Chowdhury TA, Kimura M, Kleene KC. Identification of elements in the Smcp 5′ and 3′ UTR that repress translation and promote the formation of heavy inactive mRNPs in spermatids by analysis of mutations in transgenic mice. Reproduction. 2010;140:853–864. doi: 10.1530/REP-10-0323. [DOI] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science. 2012;337:574–578. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch D, Ghirardi M, Skehel PA, Karl KA, Herder SP, Chen M, Bailey CH, Kandel ER. Aplysia CREB2 represses long-term facilitation: Relief of repression converts transient facilitation into long-term functional and structural change. Cell. 1995;83:979–992. doi: 10.1016/0092-8674(95)90213-9. [DOI] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, Conte D, Jr, Luo S, Schroth GP, Carrington JC, Bartel DP, Mello CC. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans . Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit B, He CH, Zhang F, Votruba SM, Tadros W, Westwood JT, Smibert CA, Lipshitz HD, Theurkauf WE. An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development. 2009;136:923–932. doi: 10.1242/dev.031815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Beyret E, Lin H. Pinpointing the expression of piRNAs and function of the PIWI protein subfamily during spermatogenesis in the mouse. Dev Biol. 2011;355:215–226. doi: 10.1016/j.ydbio.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham PM, Kidwell MG, Rubin GM. The molecular basis of P-M hybrid dysgenesis: The role of the P element, a P-strain-specific transposon family. Cell. 1982;29:995–1004. doi: 10.1016/0092-8674(82)90463-9. [DOI] [PubMed] [Google Scholar]
- Blackwell BJ, Lopez MF, Wang J, Krastins B, Sarracino D, Tollervey JR, Dobke M, Jordan IK, Lunyak VV. Protein interactions with piALU RNA indicates putative participation of retroRNA in the cell cycle, DNA repair and chromatin assembly. Mob Genet Elem. 2012;2:26–35. doi: 10.4161/mge.19032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw VA, McEntee K. DNA damage activates transcription and transposition of yeast Ty retrotransposons. Mol Gen Genet. 1989;218:465–474. doi: 10.1007/BF00332411. [DOI] [PubMed] [Google Scholar]
- Brasset E, Taddei AR, Arnaud F, Faye B, Fausto AM, Mazzini M, Giorgi F, Vaury C. Viral particles of the endogenous retrovirus ZAM from Drosophila melanogaster use a pre-existing endosome/exosome pathway for transfer to the oocyte. Retrovirology. 2006;3:25. doi: 10.1186/1742-4690-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregliano JC, Picard G, Bucheton A, Pelisson A, Lavige JM, L’Heritier P. Hybrid dysgenesis in Drosophila melanogaster. Science. 1980;207:606–611. doi: 10.1126/science.6766221. [DOI] [PubMed] [Google Scholar]
- Bregliano JC, Laurencon A, Degroote F. Evidence for an inducible repair-recombination system in the female germ line of Drosophila melanogaster. I. Induction by inhibitors of nucleotide synthesis and by gamma rays. Genetics. 1995;141:571–578. doi: 10.1093/genetics/141.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. Gene regulation for higher cells: A theory. Science. 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown FD, Tiozzo S, Roux MM, Ishizuka K, Swalla BJ, De Tomaso AW. Early lineage specification of long-lived germline precursors in the colonial ascidian Botryllus schlosseri. Development. 2009;136:3485–3494. doi: 10.1242/dev.037754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheton A. Non-Mendelian female sterility in Drosophila melanogaster: Influence of aging and thermic treatments. III. Cumulative effects induced by these factors. Genetics. 1979;93:131–142. doi: 10.1093/genetics/93.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheton A, Paro R, Sang HM, Pelisson A, Finnegan DJ. The molecular basis of I-R hybrid dysgenesis in Drosophila melanogaster: Identification, cloning, and properties of the I factor. Cell. 1984;38:153–163. doi: 10.1016/0092-8674(84)90536-1. [DOI] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi-and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Cann KL, Dellaire G. Heterochromatin and the DNA damage response: The need to relax. Biochem Cell Biol. 2011;89:45–60. doi: 10.1139/O10-113. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Cecere G, Zheng GX, Mansisidor AR, Klymko KE, Grishok A. Promoters recognized by forkhead proteins exist for individual 21U-RNAs. Mol Cell. 2012;47:734–745. doi: 10.1016/j.molcel.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Yao MC. DNA elimination in ciliates: Transposon domestication and genome surveillance. Annu Rev Genet. 2011;45:227–246. doi: 10.1146/annurev-genet-110410-132432. [DOI] [PubMed] [Google Scholar]
- Chalvet F, Teysset L, Terzian C, Prud’homme N, Santamaria P, Bucheton A, Pelisson A. Proviral amplification of the Gypsy endogenous retrovirus of Drosophila melanogaster involves env-independent invasion of the female germline. EMBO J. 1999;18:2659–2669. doi: 10.1093/emboj/18.9.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Popkova A, Payen-Groschene G, Brun C, Laouini D, Pelisson A, Bucheton A. piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. Proc Natl Acad Sci USA. 2008;105:14964–14969. doi: 10.1073/pnas.0805943105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Kitamura K, Kasai S, Fujioka M, Hiyoshi M, Takamune K, Noce T, Nakatsuji N. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci USA. 2006;103:15894–15899. doi: 10.1073/pnas.0601878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Curcio MJ, Kenny AE, Moore S, Garfinkel DJ, Weintraub M, Gamache ER, Scholes DT. S-phase checkpoint pathways stimulate the mobility of the retrovirus-like transposon Ty1. Mol Cell Biol. 2007;27:8874–8885. doi: 10.1128/MCB.01095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darricarrere N, Liu N, Watanabe T, Lin H. Function of Piwi, a nuclear Piwi/Argonaute protein, is independent of its slicer activity. Proc Natl Acad Sci USA. 2013;110:1297–1302. doi: 10.1073/pnas.1213283110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, Matthews N, Berezikov E, Ketting RF, Tavare S, Miska EA. Piwi and piRNAs act upstream of an endogenous siRNA path-way to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O’Carroll D. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature. 2011;480:259–263. doi: 10.1038/nature10547. [DOI] [PubMed] [Google Scholar]
- Deng W, Lin H. Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, Sedat JW, Hawley RS. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell. 1996;86:135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]
- Desset S, Buchon N, Meignin C, Coiffet M, Vaury C. In Drosophila melanogaster the COM locus directs the somatic silencing of two retrotransposons through both Piwi-dependent and -independent pathways. PLoS ONE. 2008;3:e1526. doi: 10.1371/journal.pone.0001526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dialynas GK, Vitalini MW, Wallrath LL. Linking heterochromatin protein 1 (HP1) to cancer progression. Mutat Res. 2008;647:13–20. doi: 10.1016/j.mrfmmm.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- Dorer DR, Henikoff S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 1994;77:993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- Erpenbeck D, Schmitz J, Churakov G, Huchon D, Worheide G, Degnan BM. First evidence of miniature transposable elements in sponges (Porifera) Hydrobiologia. 2012;687:43–47. [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: At the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Fang W, Wang X, Bracht JR, Nowacki M, Landweber LF. Piwi-interacting RNAs protect DNA against loss during oxytricha genome rearrangement. Cell. 2012;151:1243–1255. doi: 10.1016/j.cell.2012.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkash EA, Kao GD, Horman SR, Prak ET. Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic Acids Res. 2006;34:1196–1204. doi: 10.1093/nar/gkj522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, Waki K, Hornig N, Arakawa T, Takahashi H, Kawai J, Forrest AR, Suzuki H, Hayashizaki Y, Hume DA, Orlando V, Grimmond SM, Carninci P. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- Fimia GM, Morlon A, Macho B, De Cesare D, Sassone-Corsi P. Transcriptional cascades during spermatogenesis: Pivotal role of CREM and ACT. Mol Cell Endocrinol. 2001;179:17–23. doi: 10.1016/s0303-7207(01)00463-4. [DOI] [PubMed] [Google Scholar]
- Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- Friedlander MR, Adamidi C, Han T, Lebedeva S, Isenbarger TA, Hirst M, Marra M, Nusbaum C, Lee WL, Jenkin JC, Sanchez Alvarado A, Kim JK, Rajewsky N. High-resolution profiling and discovery of planarian small RNAs. Proc Natl Acad Sci U S A. 2009;106:11546–11551. doi: 10.1073/pnas.0905222106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost RJ, Hamra FK, Richardson JA, Qi X, Bassel-Duby R, Olson EN. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci USA. 2010;107:11847–11852. doi: 10.1073/pnas.1007158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funayama N, Nakatsukasa M, Mohri K, Masuda Y, Agata K. Piwi expression in archeocytes and choanocytes in demosponges: insights into the stem cell system in demosponges. Evol Dev. 2010;12:275–287. doi: 10.1111/j.1525-142X.2010.00413.x. [DOI] [PubMed] [Google Scholar]
- Gangaraju VK, Lin H. MicroRNAs: Key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaraju VK, Yin H, Weiner MM, Wang J, Huang XA, Lin H. Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nat Genet. 2011;43:153–158. doi: 10.1038/ng.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev PG, Korochkina SE, Georgieva SG, Gerasimova TI. Mitomycin C induces genomic rearrangements involving transposable elements in Drosophila melanogaster. Mol Gen Genet. 1990;220:229–233. doi: 10.1007/BF00260486. [DOI] [PubMed] [Google Scholar]
- Ghabrial A, Ray RP, Schupbach T. Okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes Dev. 1998;12:2711–2723. doi: 10.1101/gad.12.17.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giani VC, Jr, Yamaguchi E, Boyle MJ, Seaver EC. Somatic and germline expression of piwi during development and regeneration in the marine polychaete annelid Capitella teleta. EvoDevo. 2011;2:10. doi: 10.1186/2041-9139-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Giorgi G, Marcantonio P, Del Re B. LINE-1 retrotransposition in human neuroblastoma cells is affected by oxidative stress. Cell Tissue Res. 2011;346:383–391. doi: 10.1007/s00441-011-1289-0. [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006a;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci USA. 2006b;103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Lee HC, Chaves D, Youngman EM, Pazour GJ, Conte D, Jr, Mello CC. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell. 2012;151:1488–1500. doi: 10.1016/j.cell.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, Pappin DJ, Chen C, Gordon A, Hannon GJ. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev. 2010;24:2499–2504. doi: 10.1101/gad.1968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Handler AM, Gomez SP. Pelement excision in Drosophila is stimulated by gamma-irradiation in transient embryonic assays. Genet Res. 1997;70:75–78. doi: 10.1017/s0016672397002759. [DOI] [PubMed] [Google Scholar]
- Handler D, Olivieri D, Novatchkova M, Gruber FS, Meixner K, Mechtler K, Stark A, Sachidanandam R, Brennecke J. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J. 2011;30:3977–3993. doi: 10.1038/emboj.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Hedges DJ, Deininger PL. Inviting instability: Transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat Res. 2007;616:46–59. doi: 10.1016/j.mrfmmm.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heit R, Rattner JB, Chan GK, Hendzel MJ. G2 histone methylation is required for the proper segregation of chromosomes. J Cell Sci. 2009;122:2957–2968. doi: 10.1242/jcs.045351. [DOI] [PubMed] [Google Scholar]
- Hellsten U, et al. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RH, Hannon GJ, Draper BW, Ketting RF. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Houwing S, Berezikov E, Ketting RF. Zili is required for germ cell differentiation and meiosis in zebrafish. Embo J. 2008;27:2702–2711. doi: 10.1038/emboj.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XA, Yin H, Sweeney S, Raha D, Snyder M, Lin H. A major epigenetic programming mechanism guided by piRNAs. Dev Cell. 2013;24:502–516. doi: 10.1016/j.devcel.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huefner ND, Mizuno Y, Weil CF, Korf I, Britt AB. Breadth by depth: Expanding our understanding of the repair of transposon-induced DNA double strand breaks via deep-sequencing. DNA Repair. 2011;10:1023–1033. doi: 10.1016/j.dnarep.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Iki T, Yoshikawa M, Nishikiori M, Jaudal MC, Matsumoto-Yokoyama E, Mitsuhara I, Meshi T, Ishikawa M. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol Cell. 2010;39:282–291. doi: 10.1016/j.molcel.2010.05.014. [DOI] [PubMed] [Google Scholar]
- Iki T, Yoshikawa M, Meshi T, Ishikawa M. Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J. 2012;31:267–278. doi: 10.1038/emboj.2011.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature. 2012;491:279–283. doi: 10.1038/nature11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell. 2010;39:292–299. doi: 10.1016/j.molcel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- Joyce EF, McKim KS. Meiotic checkpoints and the interchro-mosomal effect on crossing over in Drosophila females. Fly. 2011;5:134–140. doi: 10.4161/fly.5.2.14767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano C, Wang J, Lin H. Uniting germline and stem cells: The function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet. 2011;45:447–469. doi: 10.1146/annurev-genet-110410-132541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmykova AI, Klenov MS, Gvozdev VA. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 2005;33:2052–2059. doi: 10.1093/nar/gki323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet. 2007;16:2272–2280. doi: 10.1093/hmg/ddm179. [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Minami K, Katsuma S, Mita K, Shimada T. Developmentally synchronized expression of two Bombyx mori Piwi subfamily genes, SIWI and BmAGO3 in germ-line cells. Biochem Biophys Res Commun. 2008;367:755–760. doi: 10.1016/j.bbrc.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Arai Y, Kadota K, Suzuki Y, Hara K, Sugano S, Shimizu K, Tomari Y, Shimada T, Katsuma S. Zygotic amplification of secondary piRNAs during silkworm embryogenesis. RNA. 2011a;17:1401–1407. doi: 10.1261/rna.2709411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka S, Izumi N, Katsuma S, Tomari Y. 3′ end formation of PIWI-interacting RNAs in vitro. Mol Cell. 2011b;43:1015–1022. doi: 10.1016/j.molcel.2011.07.029. [DOI] [PubMed] [Google Scholar]
- Khurana JS, Xu J, Weng Z, Theurkauf WE. Distinct functions for the Drosophila piRNA pathway in genome maintenance and telomere protection. PLoS Genet. 2010;6:e1001246. doi: 10.1371/journal.pgen.1001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana JS, Wang J, Xu J, Koppetsch BS, Thomson TC, Nowosielska A, Li C, Zamore PD, Weng Z, Theurkauf WE. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell. 2011;147:1551–1563. doi: 10.1016/j.cell.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J. Molecular regulation of the mitosis/meiosis decision in multicellular organisms. Cold Spring Harb Perspect Biol. 2011;3:a002683. doi: 10.1101/cshperspect.a002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, Seitz H, Zamore PD, Weng Z, Theurkauf WE. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Sokolova OA, Yakushev EY, Stolyarenko AD, Mikhaleva EA, Lavrov SA, Gvozdev VA. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc Natl Acad Sci USA. 2011;108:18760–18765. doi: 10.1073/pnas.1106676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, Totoki Y, Shibata T, Kimura T, Nakatsuji N, Noce T, Sasaki H, Nakano T. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 2010;24:887–892. doi: 10.1101/gad.1902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake CM, Hawley RS. The molecular control of meiotic chromosomal behavior: Events in early meiotic prophase in Drosophila oocytes. Annu Rev Physiol. 2012;74:425–451. doi: 10.1146/annurev-physiol-020911-153342. [DOI] [PubMed] [Google Scholar]
- Lau NC, Ohsumi T, Borowsky M, Kingston RE, Blower MD. Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi. EMBO J. 2009;28:2945–2958. doi: 10.1038/emboj.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC. Small RNAs in the animal gonad: Guarding genomes and guiding development. Int J Biochem Cell Biol. 2010;42:1334–1347. doi: 10.1016/j.biocel.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavige JM. I-R system of hybrid dysgenesis in Drosophila melanogaster: Further data on the arrest of development of the embryos from SF females. Biol Cell. 1986;56:207–216. [Google Scholar]
- Leblanc P, Desset S, Giorgi F, Taddei AR, Fausto AM, Mazzini M, Dastugue B, Vaury C. Life cycle of an endogenous retrovirus, ZAM, in Drosophila melanogaster. J Virol. 2000;74:10658–10669. doi: 10.1128/jvi.74.22.10658-10669.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Banerjee S, Zhou H, Jammalamadaka A, Arcila M, Manjunath BS, Kosik KS. Identification of piRNAs in the central nervous system. RNA. 2011;17:1090–1099. doi: 10.1261/rna.2565011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Schutte D, Wulf G, Fuzesi L, Radzun HJ, Schweyer S, Engel W, Nayernia K. Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Hum Mol Genet. 2006;15:201–211. doi: 10.1093/hmg/ddi430. [DOI] [PubMed] [Google Scholar]
- Lee JH, Jung C, Javadian-Elyaderani P, Schweyer S, Schutte D, Shoukier M, Karimi-Busheri F, Weinfeld M, Rasouli-Nia A, Hengstler JG, Mantilla A, Soleimanpour-Lichaei HR, Engel W, Robson CN, Nayernia K. Pathways of proliferation and antiapoptosis driven in breast cancer stem cells by stem cell protein piwil2. Cancer Res. 2010;70:4569–4579. doi: 10.1158/0008-5472.CAN-09-2670. [DOI] [PubMed] [Google Scholar]
- Lee HC, Gu W, Shirayama M, Youngman E, Conte D, Jr, Mello CC. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell. 2012;150:78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, Kittler EL, Zapp ML, Klattenhoff C, Schulz N, Theurkauf WE, Weng Z, Zamore PD. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci USA. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Tao L, Kai T. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J Cell Biol. 2009;186:333–342. doi: 10.1083/jcb.200904063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Lin H, Yin H. A novel epigenetic mechanism in Drosophila somatic cells mediated by Piwi and piRNAs. Cold Spring Harb Symp Quant Biol. 2008;73:273–281. doi: 10.1101/sqb.2008.73.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004a;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Liu Y, Mochizuki K, Gorovsky MA. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc Natl Acad Sci USA. 2004b;101:1679–1684. doi: 10.1073/pnas.0305421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 2007;21:1530–1545. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Zhang K, Li C, Yao Y, Tao D, Liu Y, Zhang S, Ma Y. Piwil2 suppresses p53 by inducing phosphorylation of signal transducer and activator of transcription 3 in tumor cells. PLoS ONE. 2012;7:e30999. doi: 10.1371/journal.pone.0030999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachi-danandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen RA, Kloc A, Slotkin RK, Tanurdzic M. Epigenetic inheritance and reprogramming in plants and fission yeast. Cold Spring Harb Symp Quant Biol. 2008;73:265–271. doi: 10.1101/sqb.2008.73.062. [DOI] [PubMed] [Google Scholar]
- McCleland ML, Farrell JA, O’Farrell PH. Influence of cyclin type and dose on mitotic entry and progression in the early Drosophila embryo. J Cell Biol. 2009;184:639–646. doi: 10.1083/jcb.200810012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. Chromosome organization and genic expression. Cold Spring Harb Symp Quant Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- Medstrand P, van de Lagemaat LN, Mager DL. Retroelement distributions in the human genome: Variations associated with age and proximity to genes. Genome Res. 2002;12:1483–1495. doi: 10.1101/gr.388902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megosh HB, Cox DN, Campbell C, Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Mevel-Ninio M, Pelisson A, Kinder J, Campos AR, Bucheton A. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics. 2007;175:1615–1624. doi: 10.1534/genetics.106.068106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Takeuchi A, Siomi H, Siomi MC. A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat Struct Mol Biol. 2010;17:1024–1026. doi: 10.1038/nsmb.1875. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- Moshkovich N, Lei EP. HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet. 2010;6:e1000880. doi: 10.1371/journal.pgen.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao A, Mituyama T, Huang H, Chen D, Siomi MC, Siomi H. Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA. 2010;16:2503–2515. doi: 10.1261/rna.2270710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, Siomi H, Siomi MC. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA. 2007;13:1911–1922. doi: 10.1261/rna.744307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Okada TN, Kawamura T, Mituyama T, Kawamura Y, Inagaki S, Huang H, Chen D, Kodama T, Siomi H, Siomi MC. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 2009;28:3820–3831. doi: 10.1038/emboj.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, Ishitani R, Siomi H, Siomi MC, Nureki O. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491:284–287. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- Noto T, Kurth HM, Kataoka K, Aronica L, DeSouza LV, Siu KW, Pearlman RE, Gorovsky MA, Mochizuki K. The Tetrahymena argonaute-binding protein Giw1p directs a mature argonaute-siRNA complex to the nucleus. Cell. 2010;140:692–703. doi: 10.1016/j.cell.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira RA, Heidmann S, Sunkel CE. Condensin I binds chromatin early in prophase and displays a highly dynamic association with Drosophila mitotic chromosomes. Chromo-soma. 2007;116:259–274. doi: 10.1007/s00412-007-0097-5. [DOI] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010;29:3301–3317. doi: 10.1038/emboj.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Senti KA, Subramanian S, Sachidanandam R, Brennecke J. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in drosophila. Mol Cell. 2012;47:954–969. doi: 10.1016/j.molcel.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel LE, Crick FH. Selfish DNA: The ultimate parasite. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schupbach T. Zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Jiang P, Zhao DY, Singh M, Schupbach T. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. EMBO J. 2011;30:4601–4615. doi: 10.1038/emboj.2011.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue ML, DeBaryshe PG. Retrotransposons that maintain chromosome ends. Proc Natl Acad Sci USA. 2011;108:20317–20324. doi: 10.1073/pnas.1100278108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil VS, Kai T. Repression of retroelements in drosophila germline via piRNA pathway by the tudor domain protein tejas. Curr Biol. 2010;20:724–730. doi: 10.1016/j.cub.2010.02.046. [DOI] [PubMed] [Google Scholar]
- Pek JW, Kai T. DEAD-box RNA helicase Belle/DDX3 and the RNA interference pathway promote mitotic chromosome segregation. Proc Natl Acad Sci USA. 2011;108:12007–12012. doi: 10.1073/pnas.1106245108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pek JW, Ng BF, Kai T. Polo-mediated phosphorylation of Maelstrom regulates oocyte determination during oogenesis in Drosophila. Development. 2012;139:4505–4513. doi: 10.1242/dev.082867. [DOI] [PubMed] [Google Scholar]
- Pelisson A, Song SU, Prud’homme N, Smith PA, Bucheton A, Corces VG. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 1994;13:4401–4411. doi: 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preall JB, Czech B, Guzzardo PM, Muerdter F, Hannon GJ. Shutdown is a component of the Drosophila piRNA biogenesis machinery. RNA. 2012;18:1446–1457. doi: 10.1261/rna.034405.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme N, Gans M, Masson M, Terzian C, Bucheton A. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics. 1995;139:697–711. doi: 10.1093/genetics/139.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, Jurka J, Genikhovich G, Grigoriev IV, Lucas SM, Steele RE, Finnerty JR, Technau U, Martindale MQ, Rokhsar DS. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Qi H, Watanabe T, Ku HY, Liu N, Zhong M, Lin H. The Yb Body, a major site for piwi-associated RNA biogenesis and a gateway for piwi expression and transport to the nucleus in somatic cells. J Biol Chem. 2010;286:3789–3797. doi: 10.1074/jbc.M110.193888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao D, Zeeman AM, Deng W, Looijenga LH, Lin H. Molecular characterization of hiwi, a human member of the piwi gene family whose overexpression is correlated to seminomas. Oncogene. 2002;21:3988–3999. doi: 10.1038/sj.onc.1205505. [DOI] [PubMed] [Google Scholar]
- Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, Kandel ER. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149:693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P, Malone CD, Navarro C, Newbold SP, Hayes PS, Sachidanandam R, Hannon GJ, Lehmann R. piRNA production requires heterochromatin formation in Drosophila. Curr Biol. 2011;21:1373–1379. doi: 10.1016/j.cub.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner VA, Zabanov SA, Kolesnikova OV, Vasilyeva LA. Induction of the mobile genetic element Dm-412 transpositions in the Drosophila genome by heat shock treatment. Proc Natl Acad Sci USA. 1992;89:5650–5654. doi: 10.1073/pnas.89.12.5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL. Transposable elements: An abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 2012;46:21–42. doi: 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sanchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Reiss D, Josse T, Anxolabehere D, Ronsseray S. aubergine mutations in Drosophila melanogaster impair P cytotype determination by telomeric P elements inserted in heterochromatin. Mol Genet Genomics. 2004;272:336–343. doi: 10.1007/s00438-004-1061-1. [DOI] [PubMed] [Google Scholar]
- Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011;480:264–267. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- Robert VJ, Davis MW, Jorgensen EM, Bessereau JL. Gene conversion and end-joining-repair double-strand breaks in the Caenorhabditis elegans germline. Genetics. 2008;180:673–679. doi: 10.1534/genetics.108.089698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC. A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr Biol. 2009;19:2066–2076. doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AJ, Seipel SA, Hamill DR, Romancino DPMDIC, Suprenant KA, Bonder EM. Seawi–a sea urchin piwi/argonaute family member is a component of MT-RNP complexes. RNA. 2005;11:646–656. doi: 10.1261/rna.7198205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong YS. Telomere capping in Drosophila: Dealing with chromosome ends that most resemble DNA breaks. Chromosoma. 2008;117:235–242. doi: 10.1007/s00412-007-0144-2. [DOI] [PubMed] [Google Scholar]
- Rouget C, Papin C, Boureux A, Meunier AC, Franco B, Robine N, Lai EC, Pelisson A, Simonelig M. Maternal mRNA dead-enylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467:1128–1132. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans . Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Rudin CM, Thompson CB. Transcriptional activation of short interspersed elements by DNA-damaging agents. Genes Chromosomes Cancer. 2001;30:64–71. [PubMed] [Google Scholar]
- Saito K, Siomi MC. Small RNA-mediated quiescence of transposable elements in animals. Dev Cell. 2010;19:687–697. doi: 10.1016/j.devcel.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura YOno Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010;24:2493–2498. doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot E, Payen-Groschene G, Bucheton A, Pelisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y. Epigenetic events in mammalian germcell development: Reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Shiohama A, Minoshima S, Shimizu N. Identification of eight members of the Argonaute family in the human genome small star, filled. Genomics. 2003;82:323–330. doi: 10.1016/s0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxe JP, Lin H. Small noncoding RNAs in the germline. Cold Spring Harb Perspect Biol. 2011;3:a002717. doi: 10.1101/cshperspect.a002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheifele LZ, Cost GJ, Zupancic ML, Caputo EM, Boeke JD. Retrotransposon overdose and genome integrity. Proc Natl Acad Sci USA. 2009;106:13927–13932. doi: 10.1073/pnas.0906552106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes DT, Banerjee M, Bowen B, Curcio MJ. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics. 2001;159:1449–1465. doi: 10.1093/genetics/159.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes DT, Kenny AE, Gamache ER, Mou Z, Curcio MJ. Activation of a LTR-retrotransposon by telomere erosion. Proc Natl Acad Sci USA. 2003;100:15736–15741. doi: 10.1073/pnas.2136609100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela N, Kim E, Ast G. The role of transposable elements in the evolution of non-mammalian vertebrates and invertebrates. Genome Biol. 2010;11:R59. doi: 10.1186/gb-2010-11-6-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti KA, Brennecke J. The piRNA pathway: A fly’s per-spective on the guardian of the genome. Trends Genet. 2010;26:499–509. doi: 10.1016/j.tig.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentmanat MF, Elgin SC. Ectopic assembly of heterochromatin in Drosophila melanogaster triggered by transposable elements. Proc Natl Acad Sci USA. 2012;109:14104–14109. doi: 10.1073/pnas.1207036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma AK, Nelson MC, Brandt JE, Wessman M, Mahmud N, Weller KP, Hoffman R. Human CD34(+) stem cells express the hiwi gene, a human homologue of the Drosophila gene piwi. Blood. 2001;97:426–434. doi: 10.1182/blood.v97.2.426. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Jr, Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, Hata K, Martin SL, Noce T, Kuramochi-Miyagawa S, Nakano T, Sasaki H, Pillai RS, Nakatsuji N, Chuma S. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17:775–787. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Shpiz S, Kwon D, Uneva A, Kim M, Klenov M, Rozovsky Y, Georgiev P, Savitsky M, Kalmykova A. Characterization of Drosophila telomeric retroelement TAHRE: Transcription, transpositions, and RNAi-based regulation of expression. Mol Biol Evol. 2007;24:2535–2545. doi: 10.1093/molbev/msm205. [DOI] [PubMed] [Google Scholar]
- Shpiz S, Kwon D, Rozovsky Y, Kalmykova A. rasiRNA pathway controls antisense expression of Drosophila telomeric retrotransposons in the nucleus. Nucleic Acids Res. 2009;37:268–278. doi: 10.1093/nar/gkn960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Donertas D, Brennecke J. Transcriptional silencing of transposons by piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Plasterk RH. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–314. doi: 10.1038/nature02107. [DOI] [PubMed] [Google Scholar]
- Siomi MC, Mannen T, Siomi H. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev. 2010a;24:636–646. doi: 10.1101/gad.1899210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Miyoshi T, Siomi H. piRNA-mediated silencing in Drosophila germlines. Semin Cell Dev Biol. 2010b;21:754–759. doi: 10.1016/j.semcdb.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Soper SFC, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15:285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Stoycheva T, Pesheva M, Venkov P. The role of reactive oxygen species in the induction of Ty1 retrotransposition in Saccharomyces cerevisiae. Yeast. 2010;27:259–267. doi: 10.1002/yea.1749. [DOI] [PubMed] [Google Scholar]
- Subramanyam D, Blelloch R. From microRNAs to targets: Pathway discovery in cell fate transitions. Curr Opin Genet Dev. 2011;21:498–503. doi: 10.1016/j.gde.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Kage H, Aki N, Sano A, Kitagawa H, Nagase T, Yatomi Y, Ohishi N, Takai D. The induction of H3K9 methylation by PIWIL4 at the p16Ink4a locus. Biochem Biophys Res Commun. 2007;359:497–502. doi: 10.1016/j.bbrc.2007.05.136. [DOI] [PubMed] [Google Scholar]
- Sun H, Li D, Chen S, Liu Y, Liao X, Deng W, Li N, Zeng M, Tao D, Ma Y. Zili inhibits transforming growth factor-beta signaling by interacting with Smad4. J Biol Chem. 2010;285:4243–4250. doi: 10.1074/jbc.M109.079533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Honda S, Kirino Y. PIWI expression and function in cancer. Front Genet. 2012;3:204. doi: 10.3389/fgene.2012.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakmary A, Reedy M, Qi H, Lin H. The Yb protein defines a novel organelle and regulates male germline stem cell self-renewal in Drosophila melanogaster. J Cell Biol. 2009;185:613–627. doi: 10.1083/jcb.200903034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, Nakano T, Lin H. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J Biol Chem. 2009;284:6507–6519. doi: 10.1074/jbc.M809104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Van Bortle K, Corces VG. Nuclear organization and genome function. Annu Rev Cell Dev Biol. 2012;28:163–187. doi: 10.1146/annurev-cellbio-101011-155824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory BD, Mourelatos Z. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol. 2012;19:773–781. doi: 10.1038/nsmb.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SH, Elgin SC. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc Natl Acad Sci USA. 2011;108:21164–21169. doi: 10.1073/pnas.1107892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Reinke V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol. 2008;18:861–867. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Saxe J, Tanaka T, Chuma S, Lin H. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr Biol. 2009;19:640–644. doi: 10.1016/j.cub.2009.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Geesman GJ, Hostikka SL, Atallah M, Blackwell B, Lee E, Cook PJ, Pasaniuc B, Shariat G, Halperin E, Dobke M, Rosenfeld MG, Jordan IK, Lunyak VV. Inhibition of activated pericentromeric SINE/Alu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell Cycle. 2011a;10:3016–3030. doi: 10.4161/cc.10.17.17543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QE, Han C, Milum K, Wani AA. Stem cell protein Piwil2 modulates chromatin modifications upon cisplatin treatment. Mutat Res. 2011b;708:59–68. doi: 10.1016/j.mrfmmm.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, Noce T, Nakano T, Nakatsuji N, Lin H, Sasaki H. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell. 2011a;20:364–375. doi: 10.1016/j.devcel.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, Gotoh K, Hiura H, Arima T, Fujiyama A, Sado T, Shibata T, Nakano T, Lin H, Ichiyanagi K, Soloway PD, Sasaki H. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011b;332:848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang YG, Qi Y. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynska A, Minshall N, Armisen J, Miska EA, Standart N. Two Piwi proteins, Xiwi and Xili, are expressed in the Xenopus female germline. RNA. 2009;15:337–345. doi: 10.1261/rna.1422509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Ma Q, Shehadeh LA, Wilson A, Xia L, Yu H, Webster KA. Expression of the Argonaute protein PiwiL2 and piRNAs in adult mouse mesenchymal stem cells. Biochem Biophys Res Commun. 2010;396:915–920. doi: 10.1016/j.bbrc.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiol J, Cora E, Koglgruber R, Chuma S, Subramanian S, Hosokawa M, Reuter M, Yang Z, Berninger P, Palencia A, Benes V, Penninger J, Sachidanandam R, Pillai RS. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol Cell. 2012;47:970–979. doi: 10.1016/j.molcel.2012.07.019. [DOI] [PubMed] [Google Scholar]
- Xu D, Bai J, Duan Q, Costa M, Dai W. Covalent modifications of histones during mitosis and meiosis. Cell Cycle. 2009;8:3688–3694. doi: 10.4161/cc.8.22.9908. [DOI] [PubMed] [Google Scholar]
- Yabuta Y, Ohta H, Abe T, Kurimoto K, Chuma S, Saitou M. TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J Cell Biol. 2011;192:781–795. doi: 10.1083/jcb.201009043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M, Wessel GM. The DEAD-box RNA helicase Vasa functions in embryonic mitotic progression in the sea urchin. Development. 2011;138:2217–2222. doi: 10.1242/dev.065052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Hu HY, Jiang X, Maierhofer V, Neb E, He LP, Hu YP, Hu HP, Li N, Chen W, Khaitovich P. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 2011;39:6596–6607. doi: 10.1093/nar/gkr298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Yin DT, Chen L, Zhou Q, Shen R, He G, Yan Q, Tong Z, Issekutz AC, Shapiro CL, Barsky SH, Lin H, Li JJ, Gao JX. Identification of Piwil2-like (PL2L) proteins that promote tumorigenesis. PLoS ONE. 2010;5:e13406. doi: 10.1371/journal.pone.0013406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- Yin DT, Wang Q, Chen L, Liu MY, Han C, Yan Q, Shen R, He G, Duan W, Li JJ, Wani A, Gao JX. Germline stem cell gene PIWIL2 mediates DNA repair through relaxation of chromatin. PLoS ONE. 2011;6:e27154. doi: 10.1371/journal.pone.0027154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamparini AL, Davis MY, Malone CD, Vieira E, Zavadil J, Sachidanandam R, Hannon GJ, Lehmann R. Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila. Development. 2011;138:4039–4050. doi: 10.1242/dev.069187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xu J, Koppetsch BS, Wang J, Tipping C, Ma S, Weng Z, Theurkauf WE, Zamore PD. Heterotypic piRNA Ping-Pong requires qin, a protein with both E3 ligase and Tudor domains. Mol Cell. 2011;44:572–584. doi: 10.1016/j.molcel.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, Weng Z, Theurkauf WE. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 2012;151:871–884. doi: 10.1016/j.cell.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, McLaughlin KJ, Stark A, Sachidanandam R, Pillai RS, Wang PJ. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci USA. 2010;107:11841–11846. doi: 10.1073/pnas.1003953107. [DOI] [PMC free article] [PubMed] [Google Scholar]