Abstract
Fractionation of complex samples at the cellular, subcellular, protein or peptide level is an indispensable strategy to improve the sensitivity in mass spectrometry-based proteomic profiling. This study revisits, evaluates, and compares the most common gel-based protein separation techniques i.e., 1-D SDS PAGE, preparative 1-D SDS PAGE, isoelectric focusing in immobilized pH gradients (IEF-IPG), and 2-D PAGE in their performance as fractionation approaches in nanoLC-ESI-MS/MS analysis of a mixture of protein standards and mitochondrial extracts isolated from rat liver. This work demonstrates that all the above techniques provide complementary protein identification results, but 1-D SDS PAGE and IEF-IPG had the highest number of identifications. The IEF-IPG technique resulted in the highest average number of detected peptides per protein. The 2-D PAGE was evaluated as a protein fractionation approach. This work shows that the recovery of proteins and resulting proteolytic digests is highly dependent on the total volume of the gel matrix. The performed comparison of the fractionation techniques demonstrates the potential of a combination of orthogonal 1-D SDS PAGE and IEF-IPG for the improved sensitivity of profiling without significant decrease in throughput.
Keywords: isoelectric focusing, liquid chromatography tandem mass spectrometry, polyacrylamide gel electrophoresis, proteomic profiling, sample fractionation
Introduction
Proteomics, a rapidly emerging field aimed towards profiling the total protein constituency of cells, tissues and biological fluids, relies heavily on two major pillars: fractionation techniques and mass spectrometry analyses. Due to the high complexity of diverse biological samples, various liquid phase and gel-based separation and fractionation techniques were developed for proteins and resulting proteolytic digests in order to make mass spectrometry (MS) based proteomic profiling more efficient and sensitive [1–2]. Wide dynamic ranges of analyte concentrations in biological samples, limited peak capacity of conventional nanoflow rate reversed-phase (RP) liquid chromatography, that is commonly used as an upfront separation technique, co-elution of several analytes competing for charges in the ionization process, and the insufficient (though progressively improving) throughput in tandem MS data acquisition rate make the development and evaluation of effective fractionation strategies a rather “hot” topic in analytical research [3]. Substantial efforts were made in improving fractionation techniques at the peptide, protein, subcellular and cellular levels to enhance the analytical power of bottom-up and top-down MS-based proteomics [4–5]. The bottom-up proteomic technique is currently more common and mature despite the fact that the sample complexity issue is artificially increased by the very principle of the approach, where proteins are digested into multiple peptides using proteolytic enzymes. Substantial progress has been made in the development of multidimensional separation techniques at the peptide level, where some chromatographic techniques such as strong cation exchange (SCX) [6–7], strong anion exchange (SAX) [8–9], reversed-phase (RP) [8, 10–11], hydrophilic interaction liquid chromatography (HILIC) [7] and electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) [6–7, 12], or capillary isoelectric focusing (CIEF) [13–14] techniques were used as the first dimension of fractionation prior to RP coupled to shotgun MS analysis. Fractionation at the peptide level is easier to automate, however several groups have previously demonstrated that fractionation at the protein level can be more effective and can yield higher profiling sensitivity [15–17]. Despite the attractiveness of fractionation strategies for reducing the complexity of the sample to improve sensitivity, the dilemma is that fractionation can be deleterious for the analysis of samples of limited availability due to sample loss at each fractionation stage [18–20].
This study revisits the most common techniques used for fractionation of proteins in proteomic applications. Most widespread protein fractionation techniques can be divided in two main categories: gel-based and gel-free. For example, all types of one- and two-dimensional electrophoresis, such as SDS-PAGE, non-denaturing or native PAGE, and isoelectric focusing using immobilized pH gradient gel strips, are gel-based [2, 21–22]. The principle of protein fractionation in gels could be based on a protein’s molecular weight (MW), isoelectric point (pI) or both. Gel Liquid Chromatography Tandem Mass Spectrometry (GeLC- MS/MS) [23–24] is an example of one of the most common and rather efficient gel-based fractionation technique that can be used in any biomedical laboratory. In GeLC-MS/MS, the sample is first separated using 1-D SDS-PAGE, then the gel is sliced into an appropriate number of gel fractions (bands), each band is subjected to in-gel enzymatic digestion, and finally, the resulting digests are analyzed by nanoLC MS/MS [2, 25].
Gel-based techniques are inexpensive, powerful, simple and easy to use in fractionation of complex protein mixtures, removal of interfering contaminants, assessment of sample amount and complexity using in-gel total protein staining [18, 26]. The resolving power of the gel-based techniques may be insufficient to separate individual proteins of similar MWs or pIs in a complex sample. However, these techniques can be effectively used for isolation of substantially simplified protein mixtures of proteins with a similar physical and structural properties (MWs or pIs) [27]. Common challenges in using gel-based approaches are poor recovery of proteins and resulting digests from the gel matrix, significant manual involvement in running and processing the gels, poor gel-to-gel reproducibility, limitations in MW and pI ranges, poor separation performance for proteins of extreme MW, pI, and hydrophobicity[28–30].
Gel-free fractionation commonly used in proteomics utilize different liquid phase chromatographic methods such as reversed-phase, size-exclusion, ion exchange, affinity chromatography, chromatofocusing, isoelectrofocusing [31–34] and combinations of those (Multidimensional Protein Identification Technology (MudPIT) [32, 35], Combined FRActional Diagonal Chromatography (COFRADIC) [36–37]). Protein or peptide fractionation in gel-free methods uses different physicochemical principles of separation (e.g. size, charge, structure, hydrophobicity, chemostructural moieties). Compared to gel-based methods, gel-free fractionation methods demonstrate improved sample recovery, reproducibility, throughput, compatibility with automation and easiness of coupling with mass spectrometry. However, gel-free techniques typically require specialized equipment and high level of operator expertise [25, 35, 38]. The liquid-phase separation techniques are traditionally used for fractionation of protein digests in bottom-up proteomics. Nevertheless, the recent technological improvement of separation tools for intact proteins makes the top-down proteomic approach increasingly more appealing, especially for characterization of post-translational modifications [5, 39–40].
There are several recently developed techniques that combine both in-gel and in-solution separation fractionation of proteins and peptides (e.g., Agilent OFFGEL fractionation, Protein Discovery GelFree System, and continuous flow electrophoresis) [41–44]. For instance, the OFFGEL technique exploits the high resolution and separation power of the pH gradient (IPG) immobilized in a gel strip to develop a pI gradient in solution for protein or peptide separations [45–46]. The GelFree technique, similar to the continuous flow electrophoresis approach, allows one to fractionate proteins of the sample by size in a gel column and elute the fractions into solution [47–48].
In this study, proteomic profiling sensitivity and dynamic range has been compared for several combinations of gel-based fractionation methods (i.e., 1-D PAGE, 1-D Preparative PAGE, IEF-IPG, and 2-D PAGE) followed by LC-ESI-MS/MS analysis. While 1-D PAGE and IEF-IPG yielded the highest profiling sensitivity and dynamic range, all techniques provided complimentary protein identification results. The IEF-IPG technique demonstrated the highest ratio of detected peptides per protein that can be beneficial for quantitative and structural characterization of proteins in various large-scale biomedical applications.
Materials and methods
Samples
Four series of protein standards collectively representing 42 different proteins were used for modeling purposes: (i) unstained broad range protein standards (212, 158.2, 116.4, 97.2, 66.4, 55.6, 42.7, 36.5, 26.6, 20, 14.3, 6.5, and 3.4 kDa) and (ii) prestained broad range protein standards (175, 80, 58, 46, 30, 25, 17, and 7 kDa) from JULE (Milford, CT, USA), (iii) prestained recombinant proteins (250, 150, 100, 75, 50, 37, 25, 20, 15, and 10 kDa) from BioRad (Hercules, CA, USA) and (iv) unstained recombinant proteins (200, 150, 120, 100, 70, 50, 40, 30, 20, 15, and 10 kDa) from Lonza (Rockland, ME, USA). The markers were mixed as 1:5:25:100 to cover approximately two orders of magnitude. Four sample loads are 8, 16, 33, 131 µg of total protein. The sample conductivity for all samples was adjusted by several steps of centrifugal ultrafiltration with IEF buffer using 10 kDa MWCO filters. (Millipore Corporation, Danvers, MA, USA) at 9,000 RCF to ≤ 300µS/cm prior to analysis [49–50]. The enriched mitochondrial extracts were isolated from rat livers according to the published protocol were kindly provided by Drs. I.G. Stavrovskaya and B.S. Kristal (Brigham and Women's Hospital, Boston) [51].
The samples were homogenized and lysed in ion-exchanged IEF buffer (7M urea, 2M thiourea, 4% CHAPS). The supernatants from the mitochondria homogenates and mixtures of protein standards were reduced and alkylated with 5 mM TBP and 10 mM acrylamide in 25 mM ammonium bicarbonate, pH 8.0 at 37°C for 90 minutes. The alkylation reaction was quenched by the addition of 50 mM DTT. The resulting mitochondrial protein extracts were cleanedup and concentrated using 10 kDa MWCO filters as described above. The protein concentration of mitochondrial protein extracts was 7.2 mg/mL. The total protein amount used per each fractionation technique was approximately 144 µg.
Gel-Based Fractionation
For 1-D PAGE and 1-D Preparative PAGE, the samples were diluted in sample buffer (63 mM Tris HCl, 10% glycerol, 2% SDS, 0.0025% bromophenol blue, pH 6.8) that was supplemented with 50mM DTT and loaded onto Criterion 8–16% Tris-HCl gels using Tris-Glycine-SDS as running buffer. For IEF, the samples were diluted in ion-exchanged 7M urea, 2M thiourea, 4% CHAPS supplemented with 50mM DTT. Duplicate IPGs were each hydrated overnight with 200 µL of sample. IEF-IPG was performed for 100,000 total Volt hours followed by equilibration of the IPG strips for 2× 10 minutes in 375mM Tris-HCl pH 8.8 containing 3M urea, 3% SDS, 50mM DTT, and 0.005% phenol red. Afterwards, one of IPG strips for each sample was transferred onto 8–16% PAGE using 50 mM MOPS, 50 mM Tris, 0.1% SDS, 1 mM EDTA, pH 7.7. PAGE was performed for 90min at 100 V constant voltage. Gels were fixed in 25% ethanol, 10% acetic acid for 1 hour, and stained overnight with KUMASI stabilized colloidal Coomassie stain (Focus Proteomics, Cleveland, OH, USA), and destained in 10% methanol, 2% acetic acid [52].
Gel Slicing and Digestion
To analyze the entire gel, it was cut in square or rectangular sections. Twenty rectangular slices resulted from each 1-D PAGE (15×85×1 mm cut into 15×4.5×1 mm slices), 1-D Preparative PAGE (135×85×1 mm cut into 135×4.5×1 mm slices), or IEF-IPG gel (120 × 3 × 0.5 mm cut into 6.5×3×0.5 mm slices). Every 2-D PAGE gel was sliced in 100 square slices using a sterile scalpel and a printed grid with 135×85 mm squares into 6.5×4.5×1 mm slices (Figure 1). Gel slices were minced, washed with deionized water, destained with 25mM ammonium bicarbonate (ABC), 50% ACN, and dried with ACN before the reduction step. Reduction and alkylation was performed respectively with 5mM TCEP in 25mM ABC pH 8.6 for 45 min and 10mM acrylamide for 45min at 37°C. A 15min wash with 25mM ABC 50% ACN removed excess acrylamide. The gel slices were washed with ACN for 2min and dried. An aliquot of 0.5 µg/µL of sequencing-grade trypsin (Promega, Madison, WI) was added to each gel slice to an approximate enzyme: substrate mass ratio of 1:25. The samples were incubated at 40°C in a shaker for 19 h. The digests were collected and combined with the extracts resulting from the following washes of the gel plugs with 50 mL of 25mM ABC, pH8.6;50 mL of 2% formic acid in 50% ACN; and 2% formic acid in aqueous 10% isopropanol 85% ACN. The digests were concentrated to the volume of approximately 5 mL and brought to the volume of 45 mL with aqueous 2% ACN 0.5% formic acid 0.1% trifluoroacetic acid (TFA).
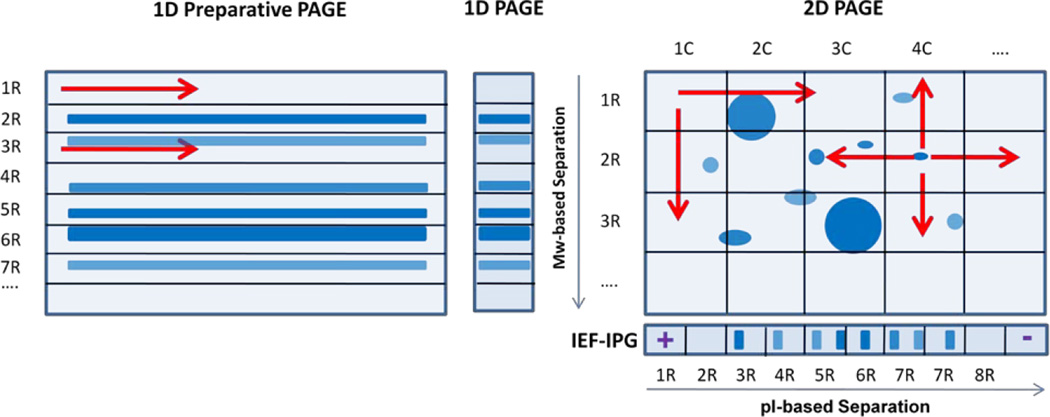
Figure 1.
Schematic representation of the four sample fractionation techniques used in the study: 1-D Preparative PAGE, 1-D PAGE, 2-D PAGE, and IEF-IPG on immobilized pH gradient strips.
To compare the above fractionation techniques, equal number of fractions per gel was used. As described above, we used twenty fractions per gel for the main comparison. The 2-D PAGE gel was sliced into ten rectangular horizontal slices and ten vertical slices that resulted in 100 gel squares (Figure 1). Each gel fraction was digested in separate Eppendorf tubes and the resulting digest was divided 50/50 and placed into two tubes (100 in total). The digests resulted from the same horizontal slice were pooled together into a single tube and the same principle was used for pooling digests derived from vertical slices (Figure 1). The total number of the pooled fractions was equal to twenty in this particular experiment. To fractionate a gel in higher or lower numbers of fractions, the same principle was used for pooling while the total number of vertical and horizontal cuts was adjusted accordingly. This technique allowed for grouping proteins with similar pIs or similar MWs in the same fractions.
Other materials and methods including LC MS protein identification, GO term enrichment analysis and statistical validation are provided in Supplemental Materials.
Results and Discussion
Fractionation on a protein level has been shown to be an effective way to improve the comprehensiveness of proteomic profiling [15, 24]. In this study, we evaluated several gel-based fractionation techniques using both well-characterized protein standards and complex “real life” samples. Also, we evaluated the 2-D GeLC-MS/MS technique as a fractionation tool for proteomic profiling.
Two-dimensional gel electrophoresis is an effective, yet laborious, method for characterization of proteomes and up-front separation prior to in-gel digestion of individually excised gel plugs and MS-based profiling. This technique was successfully applied to different tasks of quantitative proteomics [53]. Extensive proteomic catalogues and repositories were developed for numerous tissue specimens and organisms using 2-D PAGEs [54–55]. Automated 2-D gel robotic stations for spot excision, in-gel digestion, and sample cleanup are commercially available; however, the throughput of processing and MS-based analysis of individual protein spots is relatively low for becoming competitive with shotgun proteomic approaches. Mass spectrometry instrumental techniques are advancing rapidly in their detection sensitivity and quantitative accuracy [56–58]. Simple but efficient protein fractionation methods are becoming increasingly important for both bottom-up and top-down exhaustive proteomic profiling in combination with advanced MS instrumental capabilities [39–40, 59]. The 2-D PAGE technique combines the separation power of isoelectric focusing and size fractionation. Considering the efficiency of the conventional GeLC-MS profiling technique, we assessed the performance of 2-D GeLC-MS technique and compared its pre-fractionation efficiency to that of 1-D GeLC-MS without expanding the number of fractions ultimately impacting the throughput of profiling [60–61]. We have evaluated an approach for pooling the horizontal and vertical fractions of 2-D gel slices to make the number of fractions equal between the compared fractionation techniques (Figure 1).
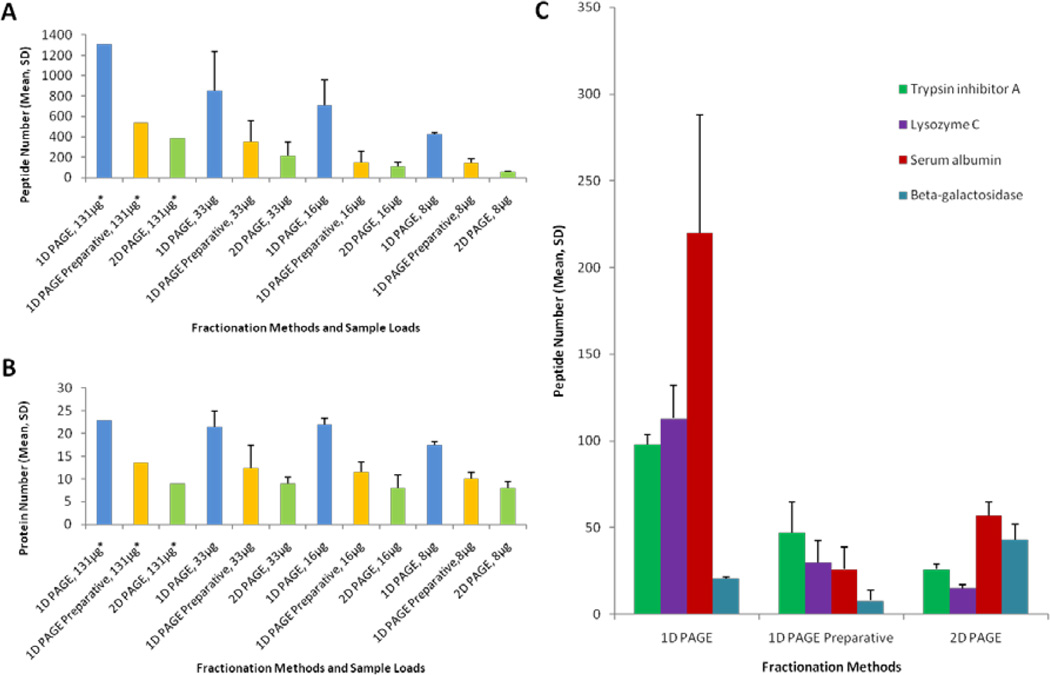
Initially, we analyzed protein standards with molecular masses (Mr) from 6.5 to 220 kDa mixed at different concentrations to cover four orders of magnitude in the dynamic range using 1-D PAGE, 1-D Preparative PAGE, and 2-D PAGE for different protein loads spanning over one order of magnitude. The whole set of experiments examined the total dynamic range of protein amounts of over three orders of magnitude (0.06–100 µg). As expected, the majority of protein standards were unambiguously detected in the in-gel digested samples using all applied fractionation techniques and sample loads. However, the experiment revealed quite noticeable differences in recoveries of peptides and proteins between the fractionation techniques that were especially pronounced for lower-abundance proteins. Typically, we found that the numbers of identified proteins and peptides in 1-D PAGE samples were higher than 1-D Preparative PAGE and 2-D PAGE fractions. In case of 1-D PAGE, higher numbers of identified peptides were found for proteins that were present at higher concentrations in all four tested protein loads (Fig. 2A, 2B). On the contrary, 2-D PAGE and 1-D preparative PAGE techniques gave higher number of identified peptides for two (out of 28) moderate- and lower-abundance proteins: beta-galactosidase and hemoglobin subunit alpha (Fig. 2). Also four proteins, including serum albumin and ovalbumin, resulted in higher peptide numbers and sequence coverage values in 2-D PAGE than 1-D Preparative PAGE (Fig. 2C). While this experiment with protein standards did not provide statistical grounds for making firm conclusions, we have noticed that the 2-D-gel based technique outperformed 1-D-gel based ones in the cases of where proteins were either well-focused into medium size spots or where proteins had multiple isoforms that were spanned across large portions of the gel in both Mr and pI directions.
Figure 2.
A set of 28 protein standards at four different protein loads was fractionated using three techniques: 1-D PAGE, 1-D Preparative PAGE and 2-D PAGE. Mean number of peptide or protein identifications and standard deviation are shown for all experiments. Fractionation techniques are compared in their profiling depth in detection of peptides (A) and proteins (B). The rest of the plotted values satisfy the above probability threshold. Illustrative examples of protein recovery for the applied fractionation approaches (C). A single technical replicate was performed for experiments marked with an asterisk (*). The 1-D PAGE experiments were performed using three gel lanes for separation of the sample at the level of 131 µg and a single lane at all other loads.
We further compared the protein fractionation techniques using mitochondria extract from rat liver as a complex biological sample (Suppl. Figure 1). Equivalent aliquots of the resulting mitochondrial lysate were fractionated into the same number of fractions using 1-D PAGE, 1-D Preparative 1-D PAGE, 2-D PAGE, and IEF-IPG, as shown in Fig. 1 and described above in Materials and Methods section. We expected some complementarities of the IEF-based and size-based PAGE protein separation techniques in their effect on proteomic profiling due to their orthogonality. The 2-D PAGE technique includes isoelectrofocusing on immobilized pH gradient strips but some sample losses are expected at the stage of protein transfer from the strip to the gel and due to irreversible entrapment of some proteins and digests in the gel matrix.
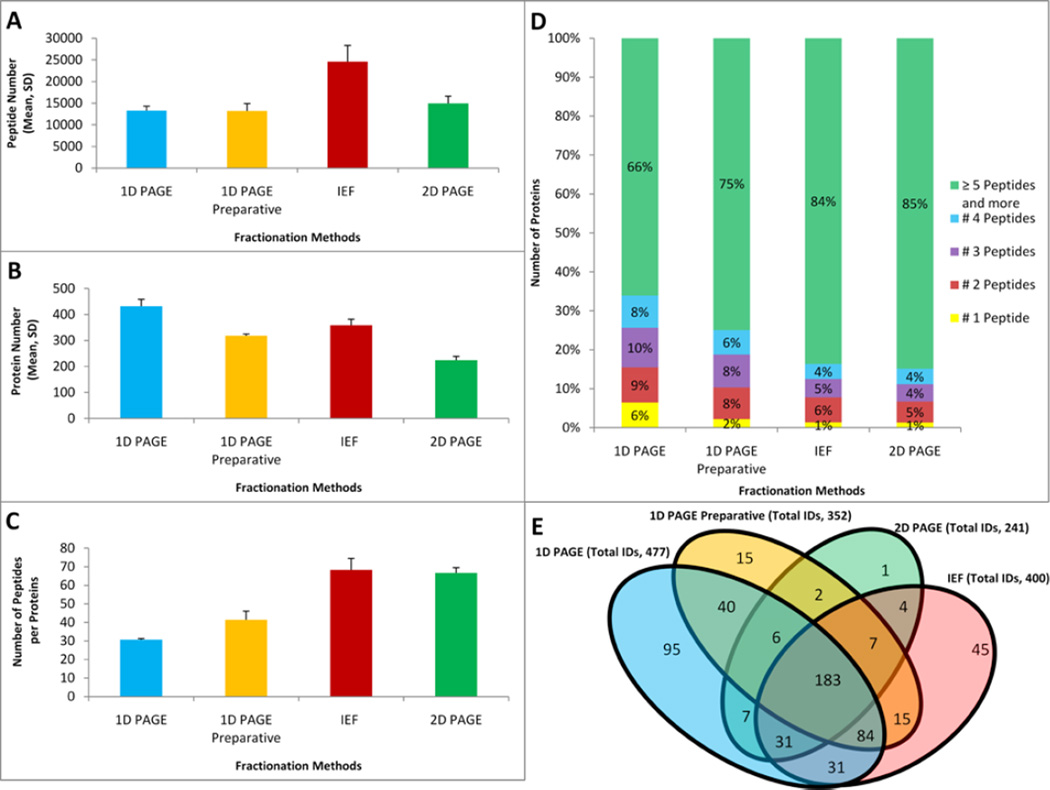
The LC-MS based profiling of protein digests resulting from the fractions obtained by the alternative techniques identified different “winners” in analytical depth at peptide and protein identification levels. While the IEF-IPG technique resulted in the highest number of detected peptides (24,600) followed by the 1-D PAGE method with approximately 46% fewer peptide IDs (13,290), the 1-D PAGE resulted in ~16% more protein IDs (477) than the IEF-IPG method (400) for the same amount of the starting material and the same number of fractions (Figure 3). The 2-D PAGE and 1-D PAGE Preparative fractionation technique resulted in lower but similar profiling sensitivity on the peptide level (14,945 and 13,182, correspondingly). The 2-D PAGE-based fractionation led to the lowest profiling sensitivity at the protein level (241). We attribute the lower profiling sensitivity of the latter fractionation technique mainly to higher losses of proteins and tryptic peptides in the gel matrix due to the substantially higher ratio of the gel volume to the sample amount. Additional sample loss is caused by an incomplete sample transfer from the first to the second dimensional SDS-gel. The increased porosity of IPG strips was recently suggested to tackle the latter source of sample losses [62–63]. Interestingly, the IEF-IPG technique resulted in slightly (~16%) fewer detected proteins than the 1-D PAGE approach for the same number of fractions and identical amount of the sample. Nevertheless, using IEF-IPG provided a twice higher number of positively identified peptides per protein. The 2-D PAGE-based technique also provided relatively high average peptide per protein ratio even though the number of identified proteins was twice lower in comparison to 1-D PAGE (Figures 3A, C). Improved recovery of peptides per protein is particularly important in quantitative label-free or stable isotope labeling applications, biomarker discovery, reliability of identification for low abundance proteins, and comprehensive characterization of protein structural features (e.g. post-translational modifications, domains, motifs, isoforms). The 1-D PAGE fractionation technique led to detection of 153 protein identifications (~32% of all proteins) based on 4 or fewer identified peptides while the IEF-IPG method yielded only 64 (~16%) proteins detected by the same peptide number. Similarly, the 2-D PAGE approach allowed for identified of 33 (~14%) proteins based on identification of four or fewer peptides (Figure 3D). Different fractionation techniques demonstrated a significant overlap of detected proteins 183 (32%). The IEF-IPG and 1-D PAGE-based methods showed the largest overlapping sets. However, the same IEF-IPG and 1-D PAGE-based techniques allowed for detection of 45 and 95 unique proteins, correspondingly. Despite being the most effective, the 1-D-gel fractionation missed approximately 90 proteins detected by other fractionation methods.
Figure 3.
Comparisons of the fractionation techniques in the resulting depth of LC-MS/MS proteomic profiling of mitochondria fractions extracted from rat liver. Profiling results are demonstrated using three different metrics: mean number of total identified peptides (A), identified proteins (B), and peptide per protein ratio (C) for each fractionation technique. All fractionation experiments were performed using 144 µg of total protein and each LC-MS/MS replicate consumed a 25% aliquot of a sample. Error bars illustrate standard deviations between LC-MS/MS replicate runs. (D) Fractions of proteomes corresponding to the mean numbers of peptides per protein identification. (E) Venn diagrams demonstrating unique and overlapping proteins of proteomes recovered by each fractionation technique.
The hierarchical clustering analysis of overlapping and non-overlapping proteins recovered by alternative techniques revealed fifteen patterns (Supplementary Table 1 and 2). More proteins could be identified by 1-D PAGE in comparison to the other methods, but there are a remarkable number of proteins (~90) that were identified by the methods other than this one (Figure 3E). Also, it should be noted that IEF-IPG and 1-D PAGE showed the most similarity (the largest overlap in detected proteins) among the other methods (even with 1-D preparative PAGE). As expected, the sum of the methods yielded the most of detected proteins (Supplementary Table 1&2). All techniques, including 2-D PAGE that resulted in detection of 14 and 16 unique proteins not detected by 1-D PAGE and IEF-IPG, respectively, proved to provide complementary results.
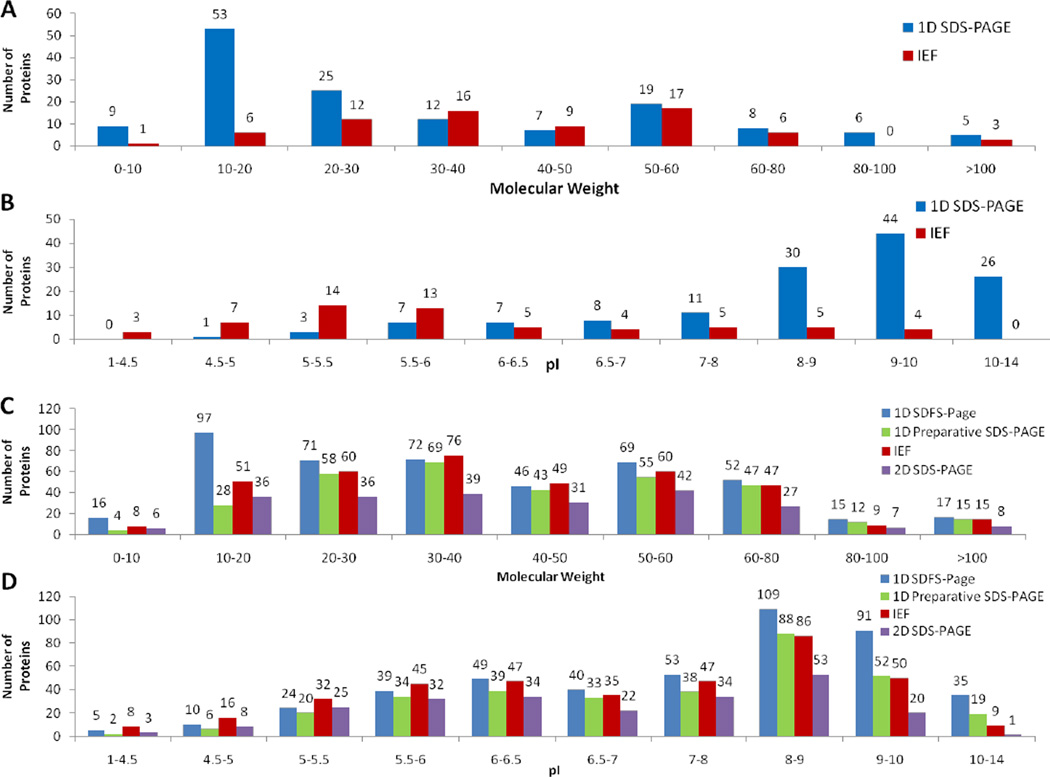
We applied the independent two-tailed Student’s t-test to compare pI and MW distributions of proteins uniquely recovered by each of the tested fractionation techniques pair-wise. We observed that there are significant differences in pI values of proteins detected using IEF-IPG and 1-D PAGE (p-value < 0.05) that also can be seen in the histograms shown in Figure 4B. As expected, the IEF-IPG technique showed significantly lower recovery of proteins with pIs surpassing the ranges of the IPG strip, and especially of alkaline proteins that are typically troublesome in IEF [2, 25]. The IEF-IPG technique performed better for proteins with mid-range pIs. The 2-D PAGE fractionation technique demonstrated a similar trend for pI distribution of recovered proteins. The performed comparative analysis of MW distribution of recovered proteins demonstrated significant loss of low Mr proteins from IPG strips (Figure 4).
Figure 4.
Molecular mass (A) and pI (B) distributions for unique (A, B) and total proteins (C, D) recovered by alternative fractionation techniques in protein extracts isolated from rat liver mitochondria.
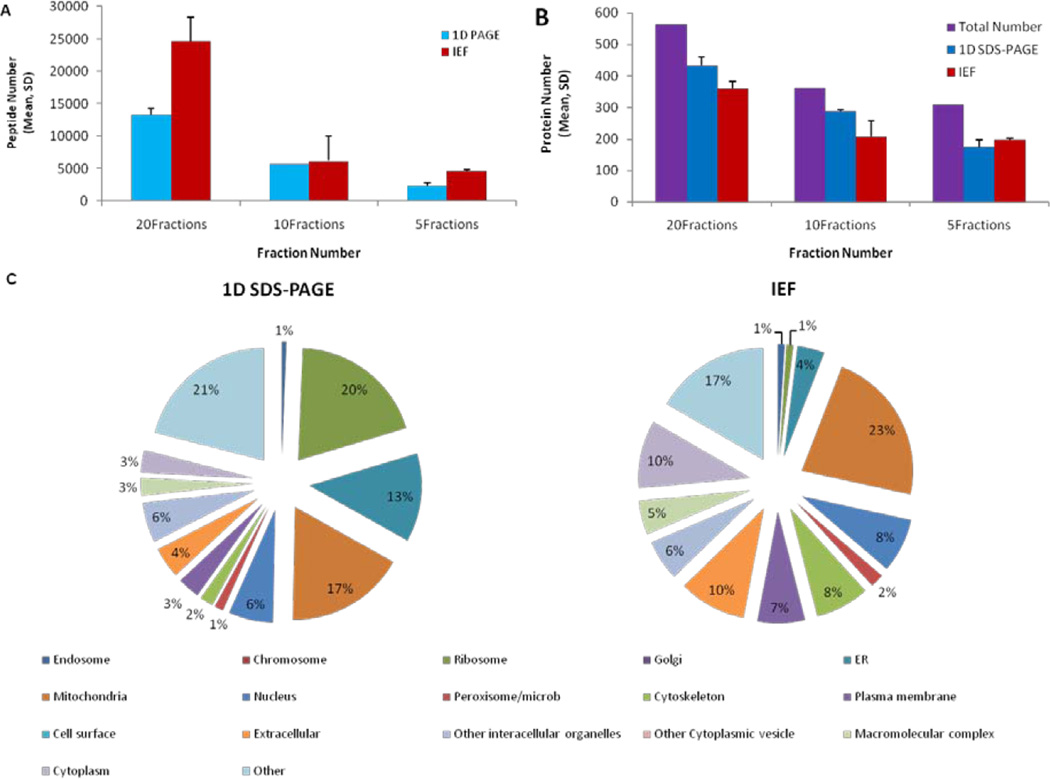
Next, we examined the effect of fraction numbers on the sensitivity of mass spectrometry-based proteomic profiling. The 1-D PAGE and IEF-IPG techniques as the two most effectual fractionation techniques from the ones we examined were used for this purpose. We compared the numbers of detected peptides and proteins for digests resulting from 5, 10, and 20 IEF-IPG or 1-D PAGE fractions. As expected, the depth of proteomic profiling decreased when the number of fractions was decreased (Figure 5). The 1-D PAGE method was slightly (~10%) more advantageous for protein identification than the IEF-IPG one when the sample was separated in 20 or 10 fractions. When the sample was divided in only five fractions, the differences in protein and peptide recovery became insignificant mainly because the MS instrument was overwhelmed with the sample complexity and the depth of analysis suffered from the ion suppression effect and insufficient speed of the MS/MS data acquisition duty cycle (Figure 4).
Figure 5.
Analytical depth of proteomic profiling of the rat mitochondria samples for various numbers of fractions (A, B). Graphical representation of the results for gene ontology cellular component term enrichment analysis for proteins uniquely detected by 1-D PAGE (left) and IEF-IPG (right) fractionation techniques in mitochondrial fractions isolated from rat liver. While the total sets of proteins recovered by each fractionation technique do not reveal any major differences in overall protein distribution (Supplementary Figure 1), the unique identifications that were obtained using IEF-IPG and 1-D PAGE highlight the most drastic differences (C).
We performed gene ontology (GO) term enrichment analysis for protein sets detected using both IEF-IPG and 1-D PAGE fractionation techniques (20 fractions). As expected, the majority of proteins detected in the mitochondria enriched fraction isolated from rat liver corresponded to mitochondria-related GO terms. Since the isolation of homogenous mitochondria is virtually impossible, the presence of other organelles reflected in identification of corresponding functional and localization GO terms was also expected. The total sets of recovered proteomes did not demonstrate any substantial differences in relative distribution of proteins related to certain cellular components or biological functions. However, the proteins uniquely recovered by using a specific technique led to determination of more noticeable changes. For instance, the IEF-IPG technique visibly enriched for cytoplasmic and extracellular proteins in comparison to the 1-D PAGE-based fractionation. At the same time the 1-D PAGE technique resulted in moderate enrichment of ribosomal, nuclear and ER proteins (Figure 5).
Concluding Remarks
We compared the efficiency of four protein separation techniques commonly used in prefractionation followed by LC-MS/MS-based profiling: 1-D PAGE, 1-D Preparative PAGE, 2-D PAGE, and IEF-IPG. The 1-DPAGE and IEF-IPG methods proved to be the most effectual and yielded the highest numbers of identified peptides and proteins at identical protein loads, numbers of fractions, and conditions of downstream nanoLC-MS/MS analyses. Each fractionation technique allowed for detection of unique proteins that can be attributed to differences in separation mechanisms and, in some extent, to the stochastic manner of data acquisition in tandem MS. The least efficient fractionation techniques 2-D PAGE and 1-D Preparative PAGE that required the highest gel volume to sample amount ratios suffered mainly due to low sample recovery from the gel matrix. A parsimonious approach of pooling fractions in the 2-D PAGE technique has been also evaluated. Its performance as an analytical technique suffered from a substantial sample dilution, especially for the low fraction numbers tested in this work.
The efficiency of 2-D gels for resolving individual proteins, protein isoforms, and identification of proteins in individual spots points out the potential of this technique for fractionation of complex samples when a higher number of fractions are collected and analyzed. The 1-D Preparative PAGE fractionation technique provided significantly lower profiling sensitivity than the common (analytical) 1-D PAGE utilizing one or two gel lanes that resulted in vastly lower volumes of the gel matrix per identical sample amount. Nevertheless, the 1-D Preparative PAGE can be an effective fractionation strategy in proteomic profiling experiments requiring higher amounts of total protein, where scaling up the analytical gel experiments becomes unfeasible due to the limited loading and separation capacities of a single gel lane.
The IEF-IPG proved to be a powerful fractionation tool that in our hands provided a slightly lower depth of proteomic profiling than the 1-D PAGE technique but a relatively large set of uniquely identified proteins undetected by 1-D PAGE. The IEF-IPG technique demonstrated its ability to recover approximately two-fold more peptides detected per each protein on average that the 1-D PAGE technique, which is especially attractive for quantitative and structural proteomic applications. As it was expected for the most commonly used pH 3–10 IPG strips, the IEF-IPG method revealed substantial losses of both highly acidic and highly basic proteins. The IEF-IPG method also demonstrated higher loss of proteins with low MW that were most likely depleted from IPG strips due to low retention in the gel matrix and subsequent losses during the equilibration and washing steps.
The comparison of results obtained for different numbers of fractions used to separate proteins from mitochondrial lysate using IEF-IPG and 1-D PAGE techniques support the above finding on the similarities in the resulting sensitivity of protein profiling. Differences in separation mechanisms, amounts of gel matrices, and limitations of techniques led to detection of complementary sets of proteins representing different biological functions and cell localization. These differences in protein recovery and complementarity of the 1-D PAGE and IEF-IPG techniques suggest the effectiveness of their combined utilization in proteomic profiling experiments.
Supplementary Material
Acknowledgements
We are grateful to Drs. Stavrovskaya I.G. and Kristal B.S. (Brigham and Women's Hospital, Boston) for providing mitochondrial extracts. We are thankful to the Department of Genetics and Complex Diseases at the Harvard University School of Public Health and the National Elite Foundation of Iran for funding support. This work was conducted with support from Harvard Catalyst | The Harvard Clinical and Translational Science Center (NIH Award #UL1 RR 025758 and financial contributions from Harvard University and its affiliated academic health care centers). The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic health care centers, the National Center for Research Resources, or the National Institutes of Health.
Abbreviations
- 1-D Preparative PAGE
one-dimensional preparative SDS polyacrylamide gel electrophoresis
- IEF-IPG
isoelectric focusing in immobilized pH gradient gel
References
- 1.Lambert JP, Ethier M, Smith JC, Figeys D. Anal Chem. 2005;77:3771–3787. doi: 10.1021/ac050586d. [DOI] [PubMed] [Google Scholar]
- 2.Baggerman G, Vierstraete E, De Loof A, Schoofs L. Comb Chem High Throughput Screen. 2005;8:669–677. doi: 10.2174/138620705774962490. [DOI] [PubMed] [Google Scholar]
- 3.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 4.Phizicky E, Bastiaens PIH, Zhu H, Snyder M, Fields S. Nature. 2003;422:208–215. doi: 10.1038/nature01512. [DOI] [PubMed] [Google Scholar]
- 5.Kelleher NL, Lin HY, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. J Am Chem Soc. 1999;121:806–812. [Google Scholar]
- 6.Hao P, Qian J, Ren Y, Sze SK. J Proteome Res. 2011;10:5568–5574. doi: 10.1021/pr2007686. [DOI] [PubMed] [Google Scholar]
- 7.Zarei M, Sprenger A, Metzger F, Gretzmeier C, Dengjel J. J Proteome Res. 2011;10:3474–3483. doi: 10.1021/pr200092z. [DOI] [PubMed] [Google Scholar]
- 8.Zhou F, Sikorski TW, Ficarro SB, Webber JT, Marto JA. Anal Chem. 2011;83:6996–7005. doi: 10.1021/ac200639v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong M, Wu M, Wang F, Qin H, Han G, Dong J, Wu Ra, Ye M, Liu Z, Zou H. Anal Chem. 2011;82:2907–2915. doi: 10.1021/ac902907w. [DOI] [PubMed] [Google Scholar]
- 10.Kong RPW, Siu SO, Lee SSM, Lo C, Chu IK. J Chromatogr A. 2011;1218:3681–3688. doi: 10.1016/j.chroma.2011.04.022. [DOI] [PubMed] [Google Scholar]
- 11.Phillips HL, Williamson JC, van Elburg KA, Snijders APL, Wright PC, Dickman MJ. Proteomics. 2009;10:2950–2960. doi: 10.1002/pmic.200900669. [DOI] [PubMed] [Google Scholar]
- 12.Chien KY, Liu HC, Goshe MB. J Proteome Res. 2011;10:4041–4053. doi: 10.1021/pr2002403. [DOI] [PubMed] [Google Scholar]
- 13.Mokaddem M, Gareil P, Varenne A. Electrophoresis. 2009;30:4040–4048. doi: 10.1002/elps.200900091. [DOI] [PubMed] [Google Scholar]
- 14.Cheng C, Lu JJ, Wang X, Roberts J, Liu S. Electrophoresis. 2010;31:2614–2621. doi: 10.1002/elps.201000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt MW, Houseman A, Ivanov AR, Wolf DA. Mol Syst Biol. 2007;3:79. doi: 10.1038/msb4100117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paulo JA, Urrutia R, Banks PA, Conwell DL, Steen H. J Proteomics. 2011;75:708–717. doi: 10.1016/j.jprot.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thakur SS, Geiger T, Chatterjee B, Bandilla P, Fröhlich F, Cox J, Mann M. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thakur D, Rejtar T, Wang D, Bones J, Cha S, Clodfelder-Miller B, Richardson E, Binns S, Dahiya S, Sgroi D, Karger BL. J Chromatogr A. 2011;1218:8168–8174. doi: 10.1016/j.chroma.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cox B, Emili A. Nat. Protocols. 2006;1:1872–1878. doi: 10.1038/nprot.2006.273. [DOI] [PubMed] [Google Scholar]
- 20.Shen Y, Zhang R, Moore RJ, Kim J, Metz TO, Hixson KK, Zhao R, Livesay EA, Udseth HR, Smith RD. Anal Chem. 2005;77:3090–3100. doi: 10.1021/ac0483062. [DOI] [PubMed] [Google Scholar]
- 21.Shapiro AL, VE aMJJ. Biochem Biophys Res Commun. 1967;28:815–820. doi: 10.1016/0006-291x(67)90391-9. [DOI] [PubMed] [Google Scholar]
- 22.Righetti PG. Isoelectric Focusing Theory, Methodology and Applications. Amsterdam: Elsevier; 1983. [Google Scholar]
- 23.Ivanov AR, Lazarev AV. Sample Preparation in Biological Mass Spectrometry. New York: Springer; 2011. [Google Scholar]
- 24.Schirle M, Heurtier M, Kuster B. Mol Cell Proteomics. 2003;2:1297–1305. doi: 10.1074/mcp.M300087-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Simpson RJ. Purifying Proteins for Proteomics A Laboratory Manual. New York: Cold Spring Harbor; 2004. [Google Scholar]
- 26.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. Nat. Protocols. 2007;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 27.Görg A, Obermaier C, Boguth G, Harder A, Scheibe B, Wildgruber R, Weiss W. Electrophoresis. 2000;21:1037–1053. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1037::AID-ELPS1037>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 28.Hellman U. Exs. 2000;88:43–54. doi: 10.1007/978-3-0348-8458-7_3. [DOI] [PubMed] [Google Scholar]
- 29.Granvogl B, Gruber P, Eichacker LA. Proteomics. 2007;7:642–654. doi: 10.1002/pmic.200600607. [DOI] [PubMed] [Google Scholar]
- 30.Albright JC, Dassenko DJ, Mohamed EA, Beussman DJ. Biochem Mol Biol Educ. 2009;37:49–55. doi: 10.1002/bmb.20259. [DOI] [PubMed] [Google Scholar]
- 31.Romijn EP, Krijgsveld J, Heck AJR. J Chromatogr A. 2003;1000:589–608. doi: 10.1016/s0021-9673(03)00178-x. [DOI] [PubMed] [Google Scholar]
- 32.Peng JM, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 33.Lu B, McClatchy DB, Kim JY, Yates JR. Proteomics. 2008;8:3947–3955. doi: 10.1002/pmic.200800120. [DOI] [PubMed] [Google Scholar]
- 34.America AHP, Cordewener JHG. Proteomics. 2008;8:731–749. doi: 10.1002/pmic.200700694. [DOI] [PubMed] [Google Scholar]
- 35.Motoyama A, Yates JR. Anal Chem. 2008;80:7187–7193. doi: 10.1021/ac8013669. [DOI] [PubMed] [Google Scholar]
- 36.Gevaert K, Van Damme P, Ghesquiere B, Impens F, Martens L, Helsens K, Vandekerckhove J. Proteomics. 2007;7:2698–2718. doi: 10.1002/pmic.200700114. [DOI] [PubMed] [Google Scholar]
- 37.Gevaert K, Van Damme P, Martens L, Vandekerckhove J. Anal Biochem. 2005;345:18–29. doi: 10.1016/j.ab.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 38.Chen G, Pramanik BN. Drug Discov Today. 2009;14:465–471. doi: 10.1016/j.drudis.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 39.Durbin KR, Tran JC, Zamdborg L, Sweet SMM, Catherman AD, Lee JE, Li M, Kellie JF, Kelleher NL. Proteomics. 2010;10:3589–3597. doi: 10.1002/pmic.201000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tran JC, Zamdborg L, Ahlf DR, Lee JE, Catherman AD, Durbin KR, Tipton JD, Vellaichamy A, Kellie JF, Li M, Wu C, Sweet SMM, Early BP, Siuti N, LeDuc RD, Compton PD, Thomas PM, Kelleher NL. Nature. 2011;480:254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arrey TN, Rietschel B, Papasotiriou DG, Bornemann S, Baeumlisberger D, Karas M, Meyer B. Anal Chem. 2011;82:2145–2149. doi: 10.1021/ac902776h. [DOI] [PubMed] [Google Scholar]
- 42.Michel PE, Reymond F, Arnaud IL, Josserand J, Girault HH, Rossier JS. Electrophoresis. 2003;24:3–11. doi: 10.1002/elps.200390030. [DOI] [PubMed] [Google Scholar]
- 43.Michel PE, Crettaz D, Morier P, Heller M, Gallot D, Tissot JD, Reymond F, Rossier JS. Electrophoresis. 2006;27:1169–1181. doi: 10.1002/elps.200500680. [DOI] [PubMed] [Google Scholar]
- 44.Waller LN, Shores K, Knapp DR. J Proteome Res. 2008;7:4577–4584. doi: 10.1021/pr8001518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geiser L, Dayon L, Vaezzadeh AR, Hochstrasser DF. In: Protein Chromatography: Methods and Protocols. Walls DLST, editor. 2011. pp. 459–472. [DOI] [PubMed] [Google Scholar]
- 46.Heller M, Michel PE, Morier P, Crettaz D, Wenz C, Tissot JD, Reymond F, Rossier JS. Electrophoresis. 2005;26:1174–1188. doi: 10.1002/elps.200410106. [DOI] [PubMed] [Google Scholar]
- 47.Hoffmann P, Ji H, Moritz RL, Connolly LM, Frecklington DF, Layton MJ, Eddes JS, Simpson RJ. Proteomics. 2001;1:807–818. doi: 10.1002/1615-9861(200107)1:7<807::AID-PROT807>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 48.Valcarcel M, Arce L, Rıos A. J Chromatogr A. 2001;924:3–30. doi: 10.1016/s0021-9673(01)00898-6. [DOI] [PubMed] [Google Scholar]
- 49.Smejkal GB, Bauer DJ. Am Biotechnol Lab. 2010;28:24–27. [Google Scholar]
- 50.Smejkal GB, Li C, Robinson MH, Lazarev AV, Lawrence NP, Chernokalskaya E. Journal of Proteome Research. 2006;5:983–987. doi: 10.1021/pr050439w. [DOI] [PubMed] [Google Scholar]
- 51.Stavrovskaya IG, Baranov SV, Guo X, Davies SS, Roberts Ii LJ, Kristal BS. Free Radic Biol Med. 2010;49:567–579. doi: 10.1016/j.freeradbiomed.2010.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wijte D, de Jong AL, Mol MAE, van Baar BLM, Heck AJR. J Proteome Res. 2006;5:2033–2038. doi: 10.1021/pr060076f. [DOI] [PubMed] [Google Scholar]
- 53.Friedman DB, Lilley KS. Clinical Proteomics: Methods and Protocols. 2008:93–124. [Google Scholar]
- 54.Hoogland C, Mostaguir K, Sanchez JC, Hochstrasser DF, Appel RD. Proteomics. 2004;4:2352–2356. doi: 10.1002/pmic.200300830. [DOI] [PubMed] [Google Scholar]
- 55.Hoogland C, Sanchez JC, Tonella L, Bairoch A, Hochstrasser DF, Appel RD. Nucleic Acids Res. 1998;26:332–333. doi: 10.1093/nar/26.1.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McLafferty FW. Annu Rev Anal Chem (Palo Alto Calif) 2011;4:1–22. doi: 10.1146/annurev-anchem-061010-114018. [DOI] [PubMed] [Google Scholar]
- 57.Hu Q, Noll RJ, Li H, Makarov A, Hardman M, Graham Cooks R. J Mass Spectrom. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 58.Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Mol Cell Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 59.Nilsson T, Mann M, Aebersold R, Yates JR, Bairoch A, Bergeron JJM. Nat Meth. 2010;7:681–685. doi: 10.1038/nmeth0910-681. [DOI] [PubMed] [Google Scholar]
- 60.Paulo JA, Lee LS, Wu B, Repas K, Mortele KJ, Banks PA, Steen H, Conwell DL. Pancreas. 2010;39:889–896. doi: 10.1097/MPA.0b013e3181cf16f4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kristjansdottir K, Wolfgeher D, Lucius N, Angulo DS, Kron SJ. J Proteome Res. 2008;7:2812–2824. doi: 10.1021/pr700816k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Candiano G, Musante L, Bruschi M, Ghiggeri GM, Herbert B, Antonucci F, Righetti PG. Electrophoresis. 2002;23:292–297. doi: 10.1002/1522-2683(200202)23:2<292::AID-ELPS292>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 63.Bruschi M, Musante L, Candiano G, Ghiggeri GM, Herbert B, Antonucci F, Righetti PG. Proteomics. 2003;3:821–825. doi: 10.1002/pmic.200300361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.