Abstract
Objectives
To determine the prevalence and spectrum of mutations and genotype phenotype relationships in the largest hypertrophic cardiomyopathy (HCM) cohort to date and provide an easy, clinically applicable phenotype-derived score that provides a pretest probability for a positive HCM genetic test.
Patients and Methods
Between 1999 and 2007, 1053 unrelated patients with the clinical diagnosis of HCM (60% male, age at diagnosis 44.4 ± 19 years) had HCM genetic testing for the HCM-associated myofilament genes. Phenotyping was performed by review of electronic medical record.
Results
Overall, 359 patients (34%) were genotype positive for a putative HCM associated mutation in ≥ 1 HCM-associated gene. Univariate and multivariate analyses demonstrated echocardiographic reverse curve morphology, age at diagnosis < 45 years, MLVWT ≥ 20 mm, family history of HCM, and family history of SCD to be positive predictors of positive genetic test while hypertension was a negative predictor. A score, based on the number 6 predictors of a positive genetic test, predicted a positive genetic test ranging from 6% when only hypertension was present to 80% when all 5 positive predictor markers were present.
Conclusions
In this largest HCM cohort published to date, the overall yield of genetic testing was 34%. Although all patients were diagnosed clinically with HCM, the presence or absence of six simple clinical/echocardiographic markers predicted the likelihood of mutation-positive HCM. Phenotype-guided genetic testing with the use of the Mayo HCM Genotype Predictor score provides an easy tool for an effective genetic counseling session.
Keywords: HCM, hypertrophic cardiomyopathy, genetics, Mayo HCM genotype predictor
BACKGROUND
Diagnosed as unexplained cardiac hypertrophy in the absence of inciting factors such as uncontrolled hypertension and aortic stenosis, HCM is the most common heritable cardiovascular disease with an estimated prevalence of 1 in 500 individuals1. HCM is the most frequent identifiable cause of sudden cardiac death (SCD) among young athletes and clinically has a heterogeneous presentation with varying degrees of hypertrophy, left ventricular outflow tract obstruction, ventricular septal morphology, and symptoms, such as syncope or dyspnea2,3
This phenotypic heterogeneity is matched by the marked heterogeneity in the underlying genotype. Currently, mutations in 9 genes that encode various cardiac myofilaments have been associated in the pathogenesis of sarcomeric/myofilament-HCM with most mutations being unique, individual variants. Clinical genetic testing (either institutionally provided or commercially available) is being used increasingly in the evaluation of patients with HCM and their family members4. Although specific genotype-based treatments for HCM are not yet available, a positive genetic test result confirms the etiology of the disease, potentially guides timing for the next follow-up evaluation, and enables mutation-specific confirmatory testing of the appropriate family members in accordance with the recent HRS/EHRA and ACCF/AHA guidelines4,5. Furthermore, a genetic test result might lead to decreased frequency or even total discharge of clinical screening for genotype-negative, phenotype negative relatives when an index case has a positive genetic test resulting in significant decrease of screening associated costs for the families and their insurance providers.
Studies over the last two decades have provided valuable insights on the yield of genetic testing, genotype-phenotype relationships, and have described clinical tools to aid cardiologists and genetic counselors about whether to order genetic test. However, most of these studies have been limited by small cohort size, early genetic data, and absence of a full spectrum of variants in large populations of controls6–13. Herein, we present the prevalence of myofilament mutations and genotype-phenotype relationships in the largest cohort of unrelated patients with clinically diagnosed HCM that have been tested to date. Furthermore in this era of individualized medicine, we provide an easy to use, clinically applicable score to determine the likelihood of a positive genetic test result for patients presenting with HCM.
METHODS
Cohort
Between April 1, 1997 and February 1, 2007, 1053 unrelated patients diagnosed with HCM were referred to Mayo Clinic for clinical evaluation and were enrolled in this study approved by Mayo Clinic’s Institutional Review Board. All patients were evaluated by cardiologists who are HCM specialists and were diagnosed clinically with HCM based on presence of unexplained cardiac hypertrophy with maximum left ventricular wall thickness (MLVWT) ≥ 13 mm on echocardiography. Clinical data such as age at diagnosis, symptoms, family history of HCM and/or SCD, and HCM-related interventions were collected by review of the electronic medical record (EMR) and stored in a database blinded to the genotype. Echocardiographic septal contour was assessed as previously described and categorized as reverse curve, sigmoid, apical or neutral contour12. A common clinical finding, some patients in this cohort also had mild concomitant as a tertiahypertension or a reported history of (previously untreated) hypertension. If present in the EMR as a formal diagnosis, this was noted for these patients in our database. However, in these cases, the diagnosis of HCM was felt to be the appropriate diagnosis by experienced HCM specialists as the severity and extent of hypertrophy was out of proportion to the mild degree of concomitant hypertension.
Genetic analyses
DNA was extracted from peripheral blood lymphocytes using the Purgene DNA extraction kit (Gentra, Inc, Minneapolis, Minnesota) and stored at 4C prior to genetic analysis. All patients underwent comprehensive genetic testing for mutations in the 9 HCM-associated myofilament/sarcomeric genes (ACTC1-encoded cardiac actin (ACTC1), MYBPC3-encoded cardiac myosin binding protein C (MYBPC3), MYH7-encoded beta-myosin heavy chain (MYH7), MYL2-encoded regulatory myosin light chain (MYL2), MYL3-encoded essential myosin light chain (MYL3), TNNC1-encoded troponin C, (TNNC1), TNNI3-encoded troponin I (TNNI3), TNNT2-encoded troponin T (TNNT2) and TPM1-encoded alpha tropomyosin (TPM1). Genetic testing was performed using polymerase chain reaction (PCR), denaturing high performance liquid chromatography (DHPLC; WAVE™, Transgenomic, Omaha, NE) and direct DNA sequencing as described previously10,14. The reported mutation-detection sensitivity of DHPLC is approximately 95%15–17. In short, each translated exon of the 9 myofilament genes was amplified by PCR, after which each amplicon was subjected to DHPLC. Abnormal DHPLC profiles were further processed and subjected to direct DNA sequencing to identify the nature of the nucleotide and possible amino acid substitution. PCR and DHPLC methods and temperatures are available upon request. Genetic variants predicted to alter the protein, such as missense mutations, in-frame and frame-shift insertion/deletion mutations, canonical splice-sites (±1–4), and nonsense mutations resulting in a premature truncation were identified. The patient-identified variants were checked for their presence in i) an internal panel of reference alleles derived from 200 ostensibly healthy controls and ii) over >8000 publically available exomes from the NHLBI exome sequencing project (ESP) and 1000 genome project. Variants were considered HCM-associated if they i) were absent in all 8400 controls, or ii) present in <0.01% of controls but significantly overrepresented in HCM patients compared to the controls (p<.005). Variants were classified using standard nomenclature.
Statistical analysis
Statistical analysis was performed using JMP 9.0® statistical software (SAS Institute Inc, Cary, NC, USA). Continuous data were presented as mean ± standard deviation. Comparisons of means were analyzed by unpaired Student’s T-test for continuous variables and Fisher’s exact tests for proportions. Receiver-operator characteristic (ROC) analyses were performed to determine the cut-off for a positive genetic test for continuous variables (age diagnosis, MLVWT). Subsequent logistic regression models were used to identify predictors of positive genetic test and expressed as odds ratio (OR) and 95% confidence interval (95% CI). Values demonstrating significant differences between genotype positive and genotype negative patients by univariate analyses (age at diagnosis, MLVWT, family history of HCM, family history of SCD, reverse septal contour HCM and presence of mild concomitant hypertension) as well as female sex were entered as covariates into the logistic regression model.
Mayo HCM Genotype Predictor
Based predictors elucidated by multivariate analyses identifying predictors of a positive genetic test we created a model estimating the likelihood of a positive genetic test based on clinical parameters routinely assessed in the evaluation of patients with HCM. Herein, each independent predictor identified though the multivariate analysis was assigned equipotent weight for either a positive predictor (+1 point) or negative predictors (−1 point) of the HCM genetic test generating a score ranging from −1 to 5 for each patients. Subsequently, the yield of genetic testing was calculated for each prediction score subgroup.
RESULTS
Cohort demographics
Demographics of the study cohort are summarized in Table 1. In brief, there were more males (629, 60%) than females, mean age at diagnosis 44.4 ± 19 years; mean maximum left ventricular wall thickness (MLVWT) 21.0 ± 6mm; 47% of patients were classified as obstructive HCM based on resting LVOT gradient. Approximately one-third of patients had a familial form of disease as suggested by a positive family history of HCM and/or SCD. Although 36% of patients presented with mild, concomitant hypertension, the HCM cardiology specialists deemed the hypertension to be either controlled or the severity insufficient to account for the degree of LVH leading to a rendered clinical diagnosis of HCM anyway. Overall, the majority of patients were Caucasian (90%), while 1% of patients were African-American, 2% were other and 7% were unknown or chose not to disclose.
Table 1.
Cohort demographics
| Study Cohort |
Genotype Positive |
Genotype Negative |
P-value | |
|---|---|---|---|---|
| N (%) | 1053 | 359 (34) | 694 (66) | - |
| Sex M/F (% male) | 629/424 (60) | 210/149 (59) | 419/275 (60) | .6 |
| Age at Diagnosis (years) | 44.4 ± 19 | 36.4 ± 17 | 48.5 ± 18 | <.001 |
| Caucasian n(%) | 947 (90) | 321 (90) | 626 (90) | .7 |
| Family History of HCM n(%) | 340 (32) | 181 (51) | 159 (23) | <.001 |
| Family History of SCD n(%) | 201 (20) | 97 (28) | 104 (16) | <.001 |
| Family History of HCM and/or SCD n(%) | 394 (37) | 194 (54) | 200 (29) | <.001 |
| History of Hypertension | 396 (36) | 69 (20) | 300 (44) | <.001 |
| MLVWT (mm) | 21.0 ± 6 | 22.6 ± 6 | 20.1 ± 5 | <.001 |
| LVOT Gradient (mm Hg) | 43.9 ± 48 | 39.5 ± 42 | 46.2 ± 45 | .02 |
| Pts. with Obstructive HCM n(%) | 500 (47) | 162 (45) | 338 (49) | .3 |
| Septal Shape (%) | ||||
| Sigmoid | 425 (42.2) | 80 (24) | 345 (52) | <.001 |
| Reverse curve | 316 (31.5) | 191 (56) | 125 (19) | |
| Neutral | 194 (19.3) | 51 (15) | 143 (21) | |
| Apical | 71 (7) | 18 (15) | 53 (8) | |
| Myectomy n(%) | 467 (45) | 167 (48) | 300 (44) | .3 |
| ICD n(%) | 184 (18) | 92 (26) | 92 (14) | <.001 |
HCM, hypertrophic cardiomyopathy; ICD, implantable cardioverter defibrillator; LVOT, left ventricular outflow tract; MLVWT, maximum left ventricular wall thickness SCD, sudden cardiac death
Genotypes
After comprehensive analysis, we identified 208 rare, putatively pathogenic mutations (71 novel) in 359 (34%) patients (eTable 1). Among this subset with a positive HCM genetic test, most variants were identified in MYBPC3 (n = 96; 46%) and MYH7 (n = 74; 36%) with additional mutations found in MYL2 (n = 9), TPM1 (n = 8), TNNI3 (n = 8), TNNT2 (n = 5), TNNC1 (n = 4), ACTC1 (n = 2), and MYL3 (n = 1).
The majority of variants identified were missense variants (n = 144; 69%), however MYBPC3 showed greatest variability in type of genetic alteration. While in the remaining 8 genes, 96% of variants (108/112) were missense variants, only 38% of variants in MYPBC3 were missense; the remaining MYBPC3 variant types identified were nucleotide insertions/deletions most with a resulting frameshift (n = 36; 35%), nonsense mutations (n = 16; 17%) or intronic/splice site mutations (n = 10; 10%; eTable 1).
Most HCM-associated variants were unique to a single patient. However, a number of variants were seen more than once with some variants seen in ≥ 10 unrelated patients: MYBPC3-3330+2 T>G (17 patients), MYBPC3- Tr792Valfs*41 (15 patients), MYH7-Arg663His (14 patients), and MYBPC3- Pro955Argfs*95 (10 patients). The majority (N = 201, 97%) of variants were exclusive to the HCM cases being completely absent in > 8000 controls. However, 7 of the putative HCM-associated variants (MYBPC3-Arg502Trp, MYBPC3-Glu542Gln, MYBPC3-Trp792Arg, MYBPC-Tr792Valfs*41, MYH7-Arg663His, MYH7-Ala797Thr, and MYH7-Lys1459Asn) were seen in the public exomes, albeit at a frequency ≤ 0.01%. Compared to their ultra-rare status in the public domain, these 7 variants were overrepresented markedly among the HCM cases (eTable 1).
Yield of Genetic Testing
Overall, 359 out of 1053 patients (34%) had an HCM-associated mutation (Table 2).The majority of patients had MYBPC3-HCM (n= 182; 17% of study cohort or 51% of genotype positive patients). The second largest subgroup were patients with MYH7-HCM (n = 121) with a yield of 11% overall or 34% of the subset with genotype positive HCM (Table 2). Eighteen patients (1.7% of complete cohort (18/1053; 5% of genotype positive patients (18/359) had >1 mutation, with the majority of those patients having at least 1 variant in MYBPC3 (78%). Combined, the remaining 7 genes together (TNNI3, MYL2, TPM1, TNNC1, TNNT2, ACTC and MYL3) represented only 4% of all HCM patients and 10% of the subset with a positive genetic test (Table 2). Based on our inclusion/exclusion criteria, 57 variants, that were seen in 60 cases, were excluded as putative HCM-associated variants as they were either seen in >0.01% of controls or not overrepresented in cases versus controls (P≥.005).
Table 2.
Yield of Genetic Testing
| Accession no. | N (n = 1053) |
Yield of genetic testing among all patients (%) |
Yield among genotype positive patients (%) |
|
|---|---|---|---|---|
| Genotype positive | 359 | 34 | - | |
| MYBPC3 | NM_000256.3 | 182 | 17 | 51 |
| MYH7 | NM_000257.2 | 121 | 11 | 34 |
| Multiple | - | 18 | 1.7 | 5 |
| TNNI3 | NM_000363.4 | 13 | 1.2 | 4 |
| MYL2 | NM_000432.3 | 9 | 0.8 | 2.5 |
| TPM1 | NM_001018005.1 | 6 | 0.6 | 1.7 |
| TNNC1 | NM_003280.1 | 4 | 0.4 | 1.1 |
| TNNT2 | NM_001001430.1 | 3 | 0.3 | 0.8 |
| ACTC1 | NM_005159.4 | 2 | 0.2 | 0.6 |
| MYL3 | NM_00258.2 | 1 | 0.1 | 0.3 |
Genotype-phenotype correlations
When comparing the clinical phenotype of genotype positive to genotype negative patients, genotype positive patients exhibited a more severe phenotype than the patients who remain genetically elusive (Table 1). Overall, genotype positive patients were younger at diagnosis (36.4 ± 17 vs. 48.5 ± 18 years; P<.001), had more hypertrophy (22.6 ± 6 vs. 20.1 ± 5 mm; P<.001) and were more likely to have a family history if HCM (51% vs. 23%; P<.001) compared to genotype negative patients (Table 1). Also, genotype positive patients were more likely to have a family history of SCD (28% vs. 16%; P<.001). Patients with reverse-curve HCM had the highest yield of genetic testing with 56% of these patients having ≥ 1 HCM variant identified (P <.001). Reflecting the surgical bias of our institution, 45% of all HCM patients have had a surgical myectomy because of symptoms refractory to pharmacotherapy. However, there was no difference in rate of myectomy between genotype positive and genotype negative patients (48% vs. 44% respectively; P=.3).
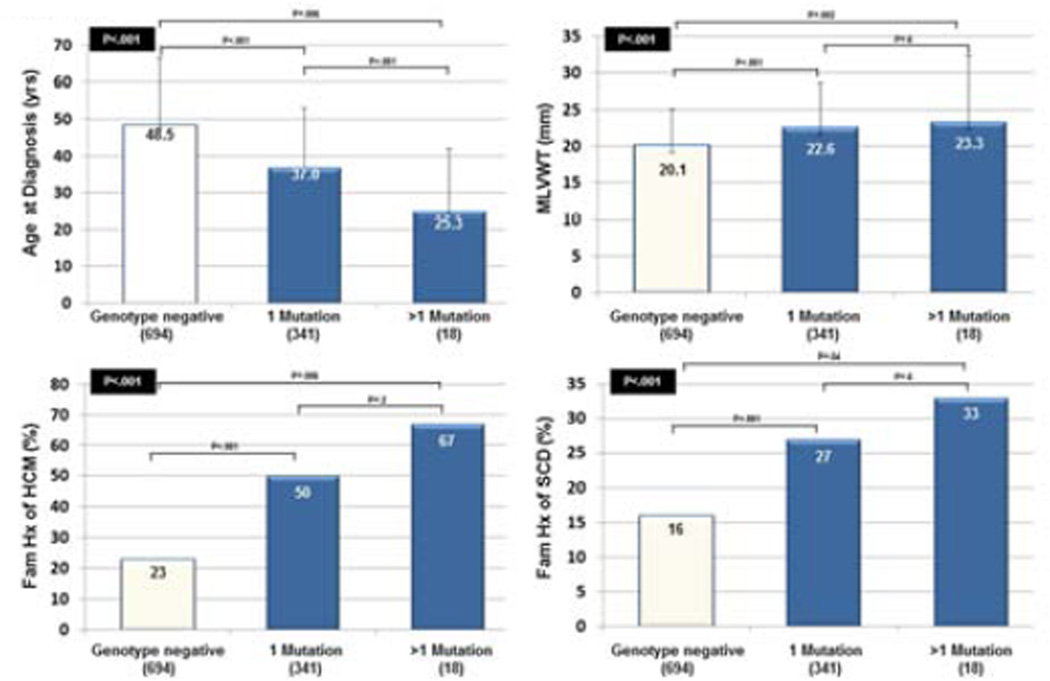
Similar to previous observations, patients with > 1 variant seemed to have worse clinical phenotype than patients with 1 variant or genotype negative patients (Figure 1). Patients with >1 mutation were significantly younger at diagnosis compared to patients with a single mutation and while the percentage of patients with a family history of HCM and family history of SCD was higher in patients with >1 mutation, due to the small number of patients (18/1053), we did not have the power to detect significant differences between these subgroups.
Figure 1. Genotype-phenotype analyses for patients with 1 or > 1 mutation.
Genotype-phenotype analyses for patients with 1 (n = 341) or > 1 mutation (n = 18) with respect to age at diagnosis (top left), maximum left ventricular wall thickness (MLVWT) (top right), family history of HCM (bottom left), and family history of SCD (bottom right). A p-value for the overall analysis is shown in the black box. P-values for analyses between each subgroup are shown above black bars.
Akin to previous observations, in this large cohort, no phenotypic differences were found between patients carrying a variant in the two most common HCM-associated genes – MYBPC3 (n = 182) and MYH7 (n = 121) making them phenotypically indistinguishable. Aside from a slight overrepresentation of MYBPC3-positive male patients (64% versus 51% men with MYH7-HCM; P=.03), they were similar with regard to age at diagnosis (37.6 ± 15 vs. 36.4 ± 19 years; P=.5), MLVWT (22.9 ± 6 vs. 22.3 ± 6 mm; P=.5), proportion of patients with reverse-curve HCM (53% vs. 61%; P=.2), family history of HCM (50% vs. 49%; P=.8) and family history of SCD (25% vs. 28%; P=.7) respectively. These and additional comparisons are summarized in eTable 2.
ROC and multivariate analyses
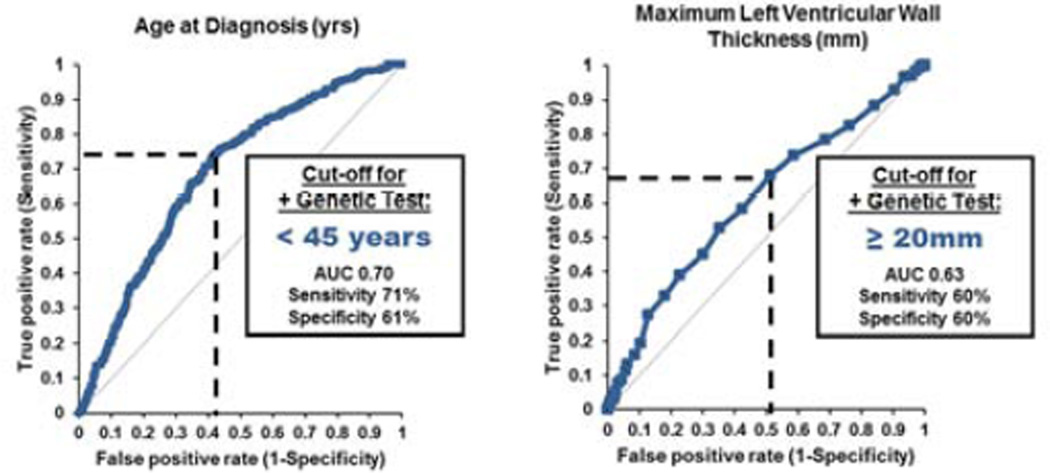
To determine the cut-off values to predict a positive genetic test for continuous variables, we performed ROC-analysis for age at diagnosis and MLVWT. This analysis revealed the cut-off to predict a positive genetic test for age at diagnosis at < 45 years (AUC 0.70; P<.001) and MLWVT at ≥ 20 mm (AUC 0.63, P<.001, Figure 2). Using these two cut-off points as well as sex, a family history of HCM, family history of SCD, reverse-curve septal morphology, and the presence of mild, concomitant hypertension, we performed Cox regression multivariate analysis to determine both positive and negative predictors of a positive genetic test.
Figure 2. ROC-analyses.
Receiver-operator characteristic (ROC) analyses for continuous variables (age at diagnosis; left, and maximum left ventricular wall thickness (right) determining the cut-off for a positive genetic test.
Akin to results derived from the univariate analyses, the multivariate model showed there were 6 parameters that were independent predictors (5 positive and 1 negative) of a positive genetic test: reverse-curve HCM, age at diagnosis, MLVWT family history of HCM, family history of SCD and presence of mild concomitant hypertension. Reverse-curve HCM was shown to be the strongest predictor of a positive genetic test (OR 2.97, 95% CI 2.20 – 4.04; P<.001; Table 3). In this dataset, both family history of HCM (OR 2.36, 95%CI 1.74–3.29; P<.001) and family history of SCD (OR1.45, 95%CI 1.01–2.01) were independent predictors of a positive genetic test. As speculated, concomitant hypertension was a negative predictor of a positive genetic test (OR 0.47, 95% CI 0.33 – 0.67; P<.001; Table 3). In our cohort, sex did not affect yield of genetic testing (OR 1.29 95% CI 0.95–1.75; P=.1; Table 3).
Table 3.
Multivariate Analysis
| OR | 95% CI | P-value | |
|---|---|---|---|
| Reverse Curve HCM | 2.97 | 2.18 – 4.04 | <.001 |
| Age at Diagnosis < 45 Years | 2.46 | 1.78 – 3.38 | <.001 |
| Family History of HCM | 2.36 | 1.74 – 3.29 | <.001 |
| Family History of SCD | 1.45 | 1.01 – 2.01 | .02 |
| MLVWT ≥ 20 mm | 1.72 | 1.26 – 2.35 | .01 |
| Female sex | 1.29 | 0.95 – 1.75 | .1 |
| Hypertension | 0.47 | 0.33 – 0.67 | <.001 |
HCM, hypertrophic cardiomyopathy; SCD, sudden cardiac death.
Mayo HCM Genotype Predictor
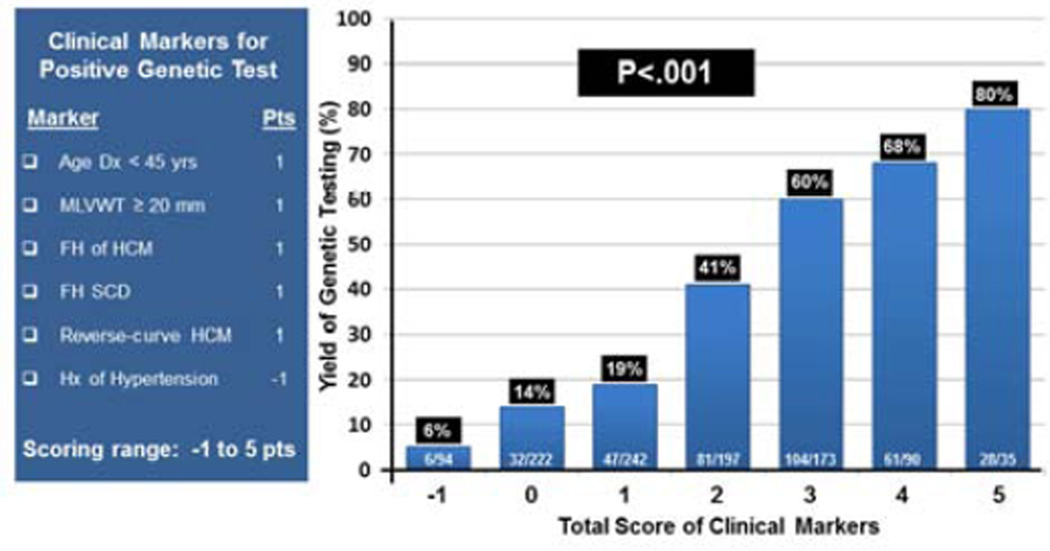
Based on these results, we determined the predictive impact of these 6 markers, that independently predicted a positive genetic test (age diagnosis <45 years, MLVWT ≥ 20mm, family history of HCM, family history of SCD, and reverse septal contour on echocardiogram) as well as the negative predictor (mild concomitant hypertension) and derived a cumulative genotype predictor score for HCM genetic testing (Figure 3). As described, each marker was given equipotent weight (+ 1 point for positive predictors, -1 point for the negative predictor of mild hypertension). Overall, the mean score was 1.6 ± 1 points and the yield of genetic testing ranged from 6% for patients with -1 point to > 80% for patients with 5 points with an incremental increase in yield between each score subgroup (P<.001, Figure 3). Thirty percent of patients (316/1053) had -1 or 0 points (yield 20%) whereas 28% of patients (298/1053) had ≥ 3 points with a yield of genetic ranging from 60% to 80%.
Figure 3. Mayo HCM Genotype Predictor Score.
Figure showing the components of the Mayo HCM Predictor Score and points attributed to each marker on the right. Bar diagram (right) showing the yield of genetic testing for each of the scored subgroups predicting a positive genetic test from 6% to over 80%.
To determine whether the scoring system could be enhanced further, we assigned weights to each of the parameters based on the odds ratios derived from the multivariate analyses. In this new score, reverse curve HCM received 3 points, age at diagnosis <45 and family history of HCM 2 points, family history of SCD and MLVWT ≥ 20 mm 1 point and hypertension -1 point. Despite a wider range of scores, this method failed to further widen the yield of genetic testing with a range of 7% for patient with -1 point to 80% for a patient with all five positive markers present (i.e. revised score = 9 points, eTable 3).
Further, as this data seems to be mostly driven by reverse septal curvature and family history of HCM, we performed analysis with a simplified score only counting these 2 parameters. While indeed the yield increased with each parameter present (0 markers gave 16% yield, 1 marker 49% and 2 markers 68%), this method failed to provide the range and associated yield of the 6 marker score (eTable 3).
DISCUSSION
Yield of genetic testing
Since the discovery of the first locus and mutation in MYH7-encoded beta-myosin heavy chain over 2 decades ago18,19, hundreds of HCM-associated mutations have been discovered. The majority of studies and most comprehensive analyses have focused on the genes encoding the various cardiac myofilaments20 with most commercial genetic testing panels for HCM including these 9 genes responsible for sarcomeric/myofilament-HCM. While the yield of genetic testing and its various genotype-phenotype relationships have been described previously, most of these studies were done in much smaller cohorts comprised of 100 to up to 500 patients with a yield of genetic testing ranging between 30 and 60%6–13.
In our current study, we present the genotype and phenotype of the largest study of unrelated patients with HCM to date comprising 1053 patients that have been evaluated clinically and comprehensively genotyped for mutations in these 9 genes. The overall yield of genetic testing on this cohort was 34% and as seen previously, the majority of mutations were found in MYBPC3 and MYH7 representing >80% of genotyped HCM (17% and 11% of all of HCM in our cohort; Table 2). Although TNNI3 and TNNT2 have been touted as relatively large contributors, these genes contributed only 4% (TNNI3) and 0.8% (TNNT2) to the subset of genotype positive HCM. Overall, only 13 (1.2%) and 5 (0.5%) patients had TNNI3-HCM or TNNT2-HCM respectively. Although multiple mutation HCM status has been reported previously in up to 5% of HCM, only 1.7% of our HCM patients had more than 1 mutation in this study.
While the yield of genetic testing in our study is consistent with the range of yields reported in the literature6–11, our seemingly low yield and low frequency of mutations in certain genes may be accounted for partially by our stringent criteria for calling a variant disease associated. While earlier studies might have allowed a variant to be called ‘disease-associated’ solely based on its absence in 50–100 reference alleles, we raised the bar and only included case-derived variants that were absent in all publically available exomes and our internal panel of 200 ostensibly healthy controls (> 8400 individuals), or seen with a frequency of <0.01% in controls but nevertheless significantly overrepresented in cases versus controls. These criteria and the rationale to include variants seen in publically available exomes sound a prudent cautionary note in calling a variant disease-causing in the era of next-generation sequencing and they are in line with recent 2012 publications by Golbus et al. as well as Bick et al. that describe the presence of known HCM-associated, likely pathogenic myofilament mutations21,22. Herein, in a subset of the >8,000 publically available exomes (3,600 patients from the Framingham Heart Study and the Jackson Heart Study), 0.3% of patients had a putative pathogenic HCM variant with 4 of these 22 patients showing clinical manifestations of HCM21. Using these criteria, only 6 variants, that were seen in both HCM patients as well as controls but at < 0.01% frequency, were included in this study as genotype positive, including a variant recently published as the first HCM mutation (MYH7-R663H) to be characterized in patient-specific, induced pluripotent stem cell-derived cardiocytes23. In their study, MYH7-Arg663His, which was seen in 14 of our unrelated HCM patients and recorded once in >8000 public domain exomes, demonstrated in vitro evidence for HCM, such as cellular enlargement, contractile arrhythmia at the single-cell level as well as calcium dysregulation. These observations further bolster the rationale to include some of these ultra-rare variants that have been documented in an apparent control subject as a bona fide HCM-associated mutation (i.e. genotype positive)23. In fact, the requirements imposed for a variant to be classified as a putative pathogenic mutation in this study may be too stringent. Further, it can be speculated that this stringent criteria might exclude possibly pathogenic variants that are overrepresented among control variants in ethnicities not matching the ethnicity of our cohort. To this end, we looked at the 55 rare, but overrepresented variants excluded and performed ethnically matched comparison (data not shown). Among these, only one variant (MYBPC3-A216T) was seen in 4 HCM cases and was overrepresented in non-Caucasian controls (4 cases) compared to Caucasian controls (1 control case). However, since only 1 of 4 HCM cases with this variant was Caucasian, this variant was not included as possibly pathogenic Lastly, compared to direct DNA Sanger sequencing-based mutation detection platforms with its estimated 99% sensitivity to detect, a few mutation-positive cases might have been missed as a result of our mutation-detection strategy that utilized DHPLC (>95% sensitivity)15–17.
As a tertiary referral center for surgical treatment of HCM, our cohort is overrepresented with patients undergoing this procedure. While 45% of our patients have undergone surgical myectomy, the estimated rate for this procedure among all patients with HCM is approximately 5–10% 24. While this certainly reflects a bias to the composition of our cohort, there was no difference in yield of genetic testing among the subset of patients with a myectomy who underwent genetic testing (yield 36%) compared to those patients who did not undergo the surgical procedure (yield 32%; P=.3).
Genotype-phenotype relationships
Consistent with previous studies, our data shows that patients with a HCM-associated mutation present with a more severe phenotype than genotype negative patients with regard to age at diagnosis, degree of hypertrophy, and family history of HCM (Table 1). Also, genotype positive patients are more likely to have a family history of SCD (28%) compared to genotype negative patients (16%; P<.001). While these data do not provide insight into the risk of SCD for the individual patient, it does suggest a genetic component to SCD risk stratification in HCM and can provide valuable information for management and follow-up of genotype positive patients with HCM and their family members with or without overt disease. Overall, genotype positive patients are significantly more likely to present with a family history of HCM and/or SCD (54%) again suggesting familial disease. Conversely, 29% of patients that remained genotype negative presented with family history of HCM and/or SCD thereby identifying the subset to focus on for novel gene discovery.
Currently, 66% of our patients remain genetically elusive. Especially among the genotype negative patients with a severe phenotype and strong family history of HCM and/or SCD (i.e. a high genotype predictor score), the door is open for discovery of new HCM-causative mutations that underlie the HCM-phenotype. On the other hand, among patients with a milder phenotype characterized by late-onset disease, sigmoidal septal shape, and possible concomitant mild hypertension (i.e. low genotype predictor score), the disease might not be primarily genetically mediated, but may instead stem from a culmination of factors such as gender, genetic modifiers, and environmental risk factors.
Mayo HCM Genotype Predictor
With (commercial) clinical genetic testing widely available, physicians and genetic counselors need tools to inform the pre-genetic test counseling session. Using the results from our multivariate models, we have developed a simple score based on the presence of clinically assessed disease parameters that can predict the likelihood of a positive genetic test. Using the Mayo HCM Genotype Predictor, the likelihood of positive genetic test ranged from 6% when only hypertension was present (−1 point) to 80% when all five positive predictors were present and concomitant hypertension was absent (5 points). Various studies have looked previously at genotype-phenotype correlations and attempted to use this to predict genotype status of an HCM patient10,13,25,26.
Most comparable to our own, Gruner et al. presented their scoring system (Toronto HCM genotype score) that is quite different and much more complex. For example, a different weight is assigned to each parameter used such as a different score for age per decade at diagnosis (-1 point for each decade > 20 (range 0 to -7 points), 4 points for female gender, 6 points for positive family history of HCM, 5 points for reverse curve and neutral HCM, and a score range for cardiac hypertrophy as expressed by ratio of maximum wall thickness (MWTH) and posterior wall thickness (PWTH) (MWTH:PWTH ratio; 0 – 4 points). Four points were subtracted for concomitant hypertension. Family history of SCD is absent in the Toronto HCM genotype score, while the authors have also chosen to include neutral contour as a positive predictor for positive genetic test. In our cohort, neutral contour has a yield of genetic testing of 15% and was not a predictor of a positive genetic test.
Most notably however, is the choice to put a significant weight on female gender (4 points) based on their multivariate analysis showing it to be a positive predictor (OR 2.0, 95% CI 1.25–3.21). However, putting this much weight on female gender might be overstated and could lead to skewed results27,28. In fact, in our large cohort, we were unable to replicate this as female sex was not a predictor of a positive genetic test (OR 1.29 (95% CI 0.95 – 1.75). Therefore, gender was excluded from our scoring system. Further attempts to enhance the Mayo HCM Genotype Predictor score by assigning different weights to each parameter were not successful. Accordingly, the most useful and easy to use scoring system seems to be our simple, equipotent assessment regarding the presence or absence of six clinical/echocardiographic parameters.
Recently published consensus statements and guidelines by HRS/EHRA and the ACCF/AHA highlight the importance of genetic counseling in the setting of genetic testing for HCM4,5. The goal of genetic counseling is to support patients in understanding and adapting to the medical, psychosocial, and familial implications of hereditary conditions, including providing thorough discussions about risks, benefits, and options available for clinical and/or genetic testing29,30. Patients make important decisions about genetic testing based on a number of determinants, including cost, insurance coverage, and perceived medical and psychosocial benefit of testing for self and family members. Other genetic counseling specialties have utilized mutation prevalence tools to aid in the genetic testing decision making process (i.e. the Myriad and Penn II tables in hereditary breast cancer counseling)31,32. Herein, the role of a cardiac genetic counselor working as part of the multi-disciplinary team treating the patient with HCM is of increasing importance in the ever evolving era of genomic and individualized medicine33.
The Mayo HCM Genotype Predictor can enhance similarly the informed decision making within the setting of HCM. Certainly, data has been available regarding elements that increase the likelihood of a positive test (family history, morphology etc.). However, without a simple clinically applicable tool that combines these factors, most patients are likely quoted a 50–60% detection rate without individualizing that estimate based upon his/her personal HCM phenotype. While the exact score or moment to pursue genetic testing is ultimately up to the provider and the HCM patient/family, the Mayo HCM Genotype Predictor score fosters and enables a well informed choice. On the one end of the spectrum, patients diagnosed with HCM (but scoring -1 point) have only a 5% chance of having a positive test which is similar to the current estimated background noise rate (estimated frequency of HCM-associated variants in healthy controls) making interpretation of the test result more complicated21,22. On the other hand, patients with scores of ≥ 3 points have an a priori yield of ≥ 60% which is higher than the overall yield of genetic testing in various cohort studies.
CONCLUSIONS
In this largest HCM cohort published to date, the overall yield of genetic testing was 34%. Although all were diagnosed clinically with HCM, the presence or absence of 6 simple clinical/echocardiographic markers predicted the likelihood of mutation-positive, sarcomeric HCM with ~5% chance when only hypertension was present to 80% chance when all five positive predictor markers were present and hypertension was absent. Although the current scoring system predicts the yield for each subgroup, the score can and should only guide the discussion as to whether or not genetic testing should be pursued. At this point in time, a minimal score should not be mandated before permitting the HCM genetic test to be ordered.
Supplementary Material
Acknowledgments
FUNDING
This publication [or project] was supported by Grant Number UL1 TR000135 from the National Center for Advancing Translational Sciences (NCATS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
ABBREVIATIONS
- HCM
Hypertrophic cardiomyopathy
- ICD
Implantable cardioverter defibrillator
- LVOT
Left ventricular outflow tract
- MLVWT
Maximum left ventricular wall thickness
- MYBPC3
Myosin binding protein C
- MYH7
Beta-myosin heavy chain
- SCD
Sudden cardiac death
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES
MJA is a consultant for Boston Scientific, Medtronic, St. Jude Medical, Inc., and Transgenomic. Intellectual property derived from MJA’s research program resulted in license agreements in 2004 between Mayo Clinic Health Solutions (formerly Mayo Medical Ventures) and PGxHealth (formerly Genaissance Pharmaceuticals, now Transgenomic) with respect to their FAMILION-LQTS and FAMILION-CPVT genetic tests but not their FAMILION-HCM genetic test. The other authors have no conflicts of interest to disclose. None of the disclosures pertain to this study and none of the companies provided financial support for this study.
REFERENCES
- 1.Maron BJ. Hypertrophic cardiomyopathy: A systematic review. JAMA. 2002;287(10):1308–1320. doi: 10.1001/jama.287.10.1308. [DOI] [PubMed] [Google Scholar]
- 2.Maron B, Epstein S, Roberts W. Causes of sudden death in competitive athletes. J Am Coll Cardiol. 1986;7(1):204–214. doi: 10.1016/s0735-1097(86)80283-2. [DOI] [PubMed] [Google Scholar]
- 3.Maron BJ, Doerer JJ, Haas TS, Tierney DM, Mueller FO. Sudden Deaths in Young Competitive Athletes: Analysis of 1866 Deaths in the United States, 1980- 2006. Circulation. 2009;119(8):1085–1092. doi: 10.1161/CIRCULATIONAHA.108.804617. [DOI] [PubMed] [Google Scholar]
- 4.Ackerman MJ, Priori SG, Willems S, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Heart Rhythm. 2011;8(8):1308–1339. doi: 10.1016/j.hrthm.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 5.Gersh BJ, Maron BJ, Bonow RO, et al. 2011 ACCF/AHA Guideline for the Diagnosis and Treatment of Hypertrophic Cardiomyopathy A Report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines Developed in Collaboration With the American Association for Thoracic Surgery, American Society of Echocardiography, American Society of Nuclear Cardiology, Heart Failure Society of America, Heart Rhythm Society, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. J Am Coll Cardiol. 2011;58(25):e212–e260. doi: 10.1016/j.jacc.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 6.Andersen PS, Havndrup O, Hougs L, et al. Diagnostic yield, interpretation, and clinical utility of mutation screening of sarcomere encoding genes in Danish hypertrophic cardiomyopathy patients and relatives. Hum Mutat. 2009;30(3):363–370. doi: 10.1002/humu.20862. [DOI] [PubMed] [Google Scholar]
- 7.Millat G, Bouvagnet P, Chevalier P, et al. Prevalence and spectrum of mutations in a cohort of 192 unrelated patients with hypertrophic cardiomyopathy. Eur J Med Genet. 2010;53(5):261–267. doi: 10.1016/j.ejmg.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Olivotto I, Girolami F, Ackerman MJ, et al. Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyopathy. Mayo Clin Proc. 2008;83(6):630–638. doi: 10.4065/83.6.630. [DOI] [PubMed] [Google Scholar]
- 9.Van Driest SL, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Yield of Genetic Testing in Hypertrophic Cardiomyopathy. Mayo Clin Proc. 2005;80(6):739–744. doi: 10.1016/S0025-6196(11)61527-9. [DOI] [PubMed] [Google Scholar]
- 10.Van Driest SL, Vasile VC, Ommen SR, et al. Myosin binding protein C mutations and compound heterozygosity in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44(9):1903–1910. doi: 10.1016/j.jacc.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 11.Erdmann J, Daehmlow S, Wischke S, et al. Mutation spectrum in a large cohort of unrelated consecutive patients with hypertrophic cardiomyopathy. Clin Genet. 2003;64(4):339–349. doi: 10.1034/j.1399-0004.2003.00151.x. [DOI] [PubMed] [Google Scholar]
- 12.Binder J, Ommen SR, Gersh BJ, et al. Echocardiography-guided genetic testing in hypertrophic cardiomyopathy: septal morphological features predict the presence of myofilament mutations. Mayo Clin Proc. 2006;81(4):459–467. doi: 10.4065/81.4.459. [DOI] [PubMed] [Google Scholar]
- 13.Van Driest SL, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Sarcomeric genotyping in hypertrophic cardiomyopathy. Mayo Clin Proc. 2005;80(4):463–469. doi: 10.1016/S0025-6196(11)63196-0. [DOI] [PubMed] [Google Scholar]
- 14.Van Driest SL, Jaeger MA, Ommen SR, et al. Comprehensive analysis of the beta-myosin heavy chain gene in 389 unrelated patients with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44(3):602–610. doi: 10.1016/j.jacc.2004.04.039. [DOI] [PubMed] [Google Scholar]
- 15.Battochio A, Mohammed S, Winthrop D, et al. Detection of c-KIT and PDGFRA gene mutations in gastrointestinal stromal tumors: comparison of DHPLC and DNA sequencing methods using a single population-based cohort. Am J Clin. Pathol. 2010;133(1):149–155. doi: 10.1309/AJCP1FNW7RGZFTYU. [DOI] [PubMed] [Google Scholar]
- 16.Gross E, Arnold N, Pfeifer K, Bandick K, Kiechle M. Identification of specific BRCA1 and BRCA2 variants by DHPLC. Hum Mutat. 2000;16(4):345–353. doi: 10.1002/1098-1004(200010)16:4<345::AID-HUMU7>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 17.Taliani MR, Roberts SC, Dukek BA, Pruthi RK, Nichols WL, Heit JA. Sensitivity and specificity of denaturing high-pressure liquid chromatography for unknown protein C gene mutations. Genet Test. 2001;5(1):39–44. doi: 10.1089/109065701750168680. [DOI] [PubMed] [Google Scholar]
- 18.Jarcho JA, McKenna W, Pare JA, et al. Mapping a gene for familial hypertrophic cardiomyopathy to chromosome 14q1. N. Engl. J. Med. 1989;321(20):1372–1378. doi: 10.1056/NEJM198911163212005. [DOI] [PubMed] [Google Scholar]
- 19.Geisterfer-Lowrance AA, Kass S, Tanigawa G, et al. A molecular basis for familial hypertrophic cardiomyopathy: A beta cardiac myosin heavy chain gene missense mutation. Cell. 1990 doi: 10.1016/0092-8674(90)90274-i. 62999-1006. [DOI] [PubMed] [Google Scholar]
- 20.Bos JM, Towbin JA, Ackerman MJ. Diagnostic, Prognostic, and Therapeutic Implications of Genetic Testing for Hypertrophic Cardiomyopathy. J Am Coll Cardiol. 2009;54(3):201–211. doi: 10.1016/j.jacc.2009.02.075. [DOI] [PubMed] [Google Scholar]
- 21.Bick AG, Flannick J, Ito K, et al. Burden of rare sarcomere gene variants in the Framingham and Jackson Heart Study cohorts. Am J Hum Genet. 2012;91(3):513–519. doi: 10.1016/j.ajhg.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golbus JR, Puckelwartz MJ, Fahrenbach JP, Dellefave-Castillo LM, Wolfgeher D, McNally EM. Population-based variation in cardiomyopathy genes. Circulation Cardiovasc Genet. 2012;5(4):391–399. doi: 10.1161/CIRCGENETICS.112.962928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan F, Lee AS, Liang P, et al. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12(1):101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maron BJ. Controversies in cardiovascular medicine. Surgical myectomy remains the primary treatment option for severely symptomatic patients with obstructive hypertrophic cardiomyopathy. Circulation. 2007;116(2):196–206. doi: 10.1161/CIRCULATIONAHA.107.691378. discussion 206. [DOI] [PubMed] [Google Scholar]
- 25.Gruner C, Ivanov J, Care M, et al. The Toronto HCM Genotype Score for Prediction of a Positive Genotype in Hypertrophic Cardiomyopathy. Circulation Cardiovasc genet. 2012 doi: 10.1161/CIRCGENETICS.112.963363. [DOI] [PubMed] [Google Scholar]
- 26.Ingles J, Sarina T, Yeates L, et al. Clinical predictors of genetic testing outcomes in hypertrophic cardiomyopathy. Genet Med. 2013 doi: 10.1038/gim.2013.44. [DOI] [PubMed] [Google Scholar]
- 27.Bos JM, Theis JL, Tajik AJ, Gersh BJ, Ommen SR, Ackerman MJ. Relationship between sex, shape, and substrate in hypertrophic cardiomyopathy. Am Heart J. 2008;155(6):1128–1134. doi: 10.1016/j.ahj.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olivotto I, Maron MS, Adabag AS, et al. Gender-related differences in the clinical presentation and outcome of hypertrophic cardiomyopathy. J Am Coll Cardiol. 2005;46(3):480–487. doi: 10.1016/j.jacc.2005.04.043. [DOI] [PubMed] [Google Scholar]
- 29.Ingles J, Zodgekar PR, Yeates L, Macciocca I, Semsarian C, Fatkin D. Guidelines for genetic testing of inherited cardiac disorders. Heart Lung Circ. 2011;20(11):681–687. doi: 10.1016/j.hlc.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 30.Ingles J, Yeates L, O'Brien L, et al. Genetic testing for inherited heart diseases: longitudinal impact on health-related quality of life. Genet Med. 2012 doi: 10.1038/gim.2012.47. [DOI] [PubMed] [Google Scholar]
- 31.Frank TS, Deffenbaugh AM, Reid JE, et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002;20(6):1480–1490. doi: 10.1200/JCO.2002.20.6.1480. [DOI] [PubMed] [Google Scholar]
- 32.Lindor NM, Johnson KJ, Harvey H, et al. Predicting BRCA1 and BRCA2 gene mutation carriers: comparison of PENN II model to previous study. Fam Cancer. 2010;9(4):495–502. doi: 10.1007/s10689-010-9348-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ingles J, Yeates L, Semsarian C. The emerging role of the cardiac genetic counselor. Heart Rhythm. 2011;8(12):1958–1962. doi: 10.1016/j.hrthm.2011.07.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.