Abstract
Since their identification in 1994, cancer stem cells (CSCs) have been objects of intensive study. Their properties and mechanisms of formation have together become a major focus of current cancer research, in part because of their enhanced ability to initiate and fuel tumor growth and their intrinsic resistance to conventional therapeutics. The discovery that activation in carcinoma cells of the epithelial-to-mesenchymal transition (EMT) program can give rise to cells with stem-like properties has provided one possible mechanism explaining how CSCs arise and presented a possible avenue for their therapeutic manipulation. This review addresses the more recent developments in CSC research, focusing on carcinomas that are able to undergo an EMT. We discuss the signaling pathways that create these cells, cell-intrinsic mechanisms that could be exploited for their selective elimination or induction of their differentiation, and the role of the tumor microenvironment in sustaining them. Finally, we propose ways to exploit our current knowledge of their complex biology to design novel therapies to eliminate them.
Keywords: Cancer Stem Cell, CSC, Epithelial-Mesenchymal Transition, EMT, Targeting
The Cancer Stem Cell paradigm
A semantic dispute?
The cancer stem cell (CSC) hypothesis posits the existence of subpopulations of neoplastic cells within a tumor that exhibit an elevated ability to seed new tumors upon experimental implantation in appropriate animal hosts. Implicit in this power is the ability of such cells to divide asymmetrically, yielding daughters that remain as CSCs (the trait of self-renewal) as well as daughters that differentiate into the neoplastic cells forming the bulk of the tumor. The reference to this notion as a hypothesis stems from the early days of CSC research, when their existence was the object of skepticism and intense debate. However, in light of a large body of supporting research reported in recent years, the existence of multiple subpopulations within a tumor with distinct tumor-initiating powers is no longer a matter of speculation and hypothesis. Accordingly, the use of the term “cancer stem cell paradigm” now seems to be more appropriate (see Box 1 for a brief history of CSC research).
BOX 1. A brief history of modern CSC research.
The field of CSC research was first launched by the experimental demonstration that certain minority subpopulations of primary human acute myeloid leukemias (AML) could propagate disease in immunodeficient mouse hosts at higher frequencies than the bulk populations of leukemic cells forming these neoplasms151. These leukemia-initiating cells were studied when grown as xenografts in severe combined immunodeficient (SCID) mouse hosts, and as such, were termed SCID leukemia-initiating cells. They were found to exhibit cell-surface antigen marker phenotypes similar to those of normal SCID-repopulating cells, implying that the cell-of-origin for this disease was closely allied to a hematopoietic stem cell240. These observations led, over the ensuing decade, to hunts for corresponding CSCs in solid tumors, resulting in the discovery of such populations in breast20, 28, brain11, prostate14, ovarian241, colon 155, 176, 177, liver13, 158, lung12 and pancreatic tumors13. In each case, these CSC subpopulations, which usually exist as minority subpopulations within tumors, have been defined operationally by their elevated tumor-initiating ability relative to that of corresponding majority populations of neoplastic cells in various tumors (Fig 1).
Despite repeated successes in identifying such tumor-initiating subpopulations of cells within individual tumors, there has been substantial skepticism and controversy arising from the fact that these repopulation studies were carried out in immunocompromised mice, which lack an adaptive immune system and whose tissue microenvironments may therefore differ substantially from those present in humans242. Additionally, studies in several cancers, such as AML and melanoma, have shown that CSCs need not be rare subpopulations and could be as frequent as 1 in 4 cells 119, 243. It is now well accepted that certain types of cancer, such as melanoma, do not closely follow the CSC paradigm developed from studying carcinomas, whereas many others, such as breast cancer, do indeed do so. Moreover, studies in genetically engineered mouse models (GEMMs) have also demonstrated the presence of CSCs in certain leukemias and breast cancer, providing direct support for the CSC model in syngeneic models 244, 245.
More recently, three lineage-tracing studies have provided unequivocal evidence of the existence of CSCs in syngeneic models of authochthonously arising tumors. One group used a GEMM that expresses ΔTK-IRES-GFP transgene driven by the promoter of the Nestin gene that is normally expressed in neural stem cells; this transgene ensured expression of both the thymidine kinase (TK) and green fluorescent proteins in stem cells. Upon crossing these Nes-ΔTK-GFP mice with a GEMM strain engineered to develop glioblastomas at high rates (Mut7 mice), the authors observed the presence of a minor quiescent GFP+ subpopulation. Moreover, upon treatment with a chemotherapeutic agent, temozolomide (TMZ), the bulk of the highly proliferative GFP- compartment in each tumor was eliminated, resulting in the preferential survival of the GFP+ CSCs, which expanded thereafter to give rise to a relapsed tumor 37. This ability of the tumors to regrow following TMZ treatment was lost upon treatment with ganciclovir, which eradicated those cells expressing the TK gene, i.e., the Nestin-expressing tumor-initiating population.
A different strategy was followed by a second group, which employed a GEMM that expresses yellow-fluorescent protein (YFP) in the keratin-14-expressing cells of the basal layer of the skin epidermis, doing so conditionally in response to Cre-mediated recombination. This labeling strategy was used to trace carcinogen-induced squamous skin papillomas, revealing that the bulk of such tumors had limited proliferative capacity. However, ~20% of cells possessed the capacity for long-term propagation and were capable of giving rise to progeny that constituted the majority of the tumor. This indicated that a subset of the tumor cells, the CSCs, were capable of generating the bulk of the tumor, ostensibly by undergoing asymmetric division. During the transition from a benign papilloma to a more malignant squamous cell carcinoma, an increase in the numbers of these long-term replicating CSCs was accompanied by a concurrent decrease in the number of differentiated cells. The increase in proportion of CSCs correlates with the increase in aggressiveness of the tumor upon transition to a full-blown carcinoma246.
A small population of Lgr5+ cells residing in the intestinal crypt has been found to have the potential to divide both symmetrically and asymmetrically and has previously been identified as the stem cell in this organ247. Quite recently, a third group employed Lgr5-specific Apc mutant mice; in these mice, the Apc mutation leads to aberrant activation of the Wnt pathway specifically in Lgr5-expressing intestinal stem cells. These mice were crossed with multicolor Cre reporter mice in which activation of Cre recombinase, by administration of tamoxifen, enables single Lgr5+ stem cells to randomly adopt one of four alternative fluorescent labels. This led to the formation of single-colored tumors that consisted of several cell types, indicative of the presence of individual Lgr5+ CSCs, each of which could give rise to a tumor containing several distinct cell types. Additionally, when a second low dose of tamoxifen was administered, a few of the Lgr5+ CSCs changed to a different color following a pulse of Cre activation. This gave rise to a stream of cells in the newly displayed color, showing that these CSCs were consistently a source that could replenish the bulk of cells in each of the observed adenomas 248.
These studies have verified the existence of CSCs in three different tumor models, eliminating major doubts about the existence of such populations within the syngeneic tumor microenvironments of autochthonously arising tumors. Moreover, these studies provided compelling evidence that such CSCs adhere to the stem-cell model by self-renewing and at the same time generating progenitors that have lost their stemness and proceed to form the bulk of a tumor.
Beyond debates about the existence of CSCs are yet others surrounding the terms used to describe these cells. Participants of The 2011 Working Conference on CSCs have outlined guidelines on how to define these cells depending on the biological system in which they are being studied1. Initially used by Edmund Beecher Wilson in 18962, the term “stem cell” has been associated with normal development for almost a century before its use in the context of cancer in the late 1980s3,4. The century-long use of the term “stem cell” in the context of normal embryonic and adult development precluded, in the minds of some, its use in other contexts, notably those associated with neoplasia. While normal stem cells (SCs) often exhibit an ability to differentiate into multiple distinct cell types, to date most CSCs are not known to differentiate into more than a single cell type – the cells composing the bulk of the tumor. However, evidence for multilineage differentiation potential of CSCs has been reported in colon carcinomas and leukemias5,6, providing further basis for their residence at the apex of a hierarchy and possessing core traits of self-renewal and differentiation, as do normal SCs.
While the phenotypes of normal stem cells seem to be fixed and therefore easier to identify, the phenotypes of CSCs are complex, variable from one tumor to another, and often affected by the abnormalities resulting from the process of neoplastic transformation; hence CSCs are often difficult to rigorously define by associating them with traits beyond their shared functional trait of tumor-initiating ability. Moreover, the existence of CSCs within tumors implies that cancer cells sharing a common genetic make-up can nevertheless exist in at least two alternative phenotypic states – CSCs and non-CSCs.
Intra-tumoral heterogeneity and CSCs – two sides to the same coin?
The existence of several forms of intra-tumoral heterogeneity has been discussed in detail elsewhere 7. Taking breast cancer as an example, exome and whole-genome sequencing efforts have shown that majority of these tumors have more than one driver mutation, with a large proportion of abnormalities affecting so-called passenger genes 8. The presence of such a large number of genetic abnormalities could lead to the occurrence of different subpopulations within the tumor, each of these possessing different combinations of genetically derived predispositions for growth, survival and dominance in the tumor microenvironment. Moreover, single-cell sequencing has enabled the identification of distinct subpopulations within human breast cancer samples, identifying one of these as the dominant, metastasis-seeding population 9. In addition to the contributions to intratumoral heterogeneity of genetic alterations, cancer cells also carry heritable epigenetic alterations, which may serve equally well to generate phenotypically distinct subpopulations within tumors.
The existence of CSCs represents an entirely distinct dimension of intratumoral heterogeneity. Thus, each of the above-described subpopulations of carcinoma cells within a tumor may carry its own group of CSCs. By definition, the CSCs and non-CSCs within such a subpopulation will share the heritable genetic and epigenetic alterations that define that subpopulation. With the advent of new technologies, this form of heterogeneity is now more accessible to scientific inquiry, but the causes and effects of non-genetic heterogeneity are still poorly understood. A more comprehensive review of intratumoral heterogeneity and cancer stem cells has been published recently 10.
Properties of CSCs
Markers used for CSC isolation
CSCs have been identified in a variety of carcinomas in various frequencies using combinations of cell-surface antigens. Indeed, the ability to use cell-surface markers to physically isolate distinct subpopulations of neoplastic cells with differing biological properties represents the most compelling demonstration that cancer cells can reside in multiple, alternative phenotypic states within a given tumor. Of note, several cell-surface markers used for the isolation of CSCs represent cell-surface antigens expressed by normal adult stem cells e.g., the CD133 antigen has been used as a CSC marker in various cancers, including those of the brain11, lung12, pancreatic13 and prostate14 among others, while also being a reliable marker of stem/progenitor cells in normal adult tissues, such as the brain15, kidney16, liver17 and prostate18. Thus, earlier studies of normal tissues uncovered a series of these antigens that subsequently proved useful for segregating CSCs from non-CSCs within tumors.
In addition to such cell-surface markers, certain intracellular proteins have also been useful for studying normal and cancer SCs. Aldehyde Dehydrogenase I (ALDH1) is a protein that has been used to mark CSCs in several cancers including leukemias19, carcinomas of the breast20, colon21, liver22, lung23 and pancreas24, among others. The “side population” (SP) that is capable of efflux of the Hoechst 33342 dye represents another non-cell-surface marker that has been used for the isolation of normal and cancer SCs25–27. The most commonly used markers and marker combinations are summarized in Table 1.
Table 1.
List of commonly used CSC markers, their expression and function in normal tissue
| Marker | Expression in normal tissue |
Normal Function | Reported to be CSC marker in |
|---|---|---|---|
| CD34 | Hematopoietic stem and progenitors147,148, endothelial cells149 |
Regulator of cell adhesion150 |
Hematological malignancies in combination with CD38151 |
| CD38 | Hematopoietic cells, skelet al and heart muscle, proximal convoluted tubules of kidney, normal adult prostate152 |
Ecto-enzyme involved in signal transduction, calcium signaling and cell adhesion152 |
Hematological malignancies in combination with CD34151 |
| CD44 | Leukocytes, epithelial cells, Endothelial cells, mesenchymal cells153 |
Varied functions including cell adhesion and migration, cell-cell interaction, cell signaling, leukocyte attachment and rolling154 |
Breast in combination with CD2428, Colon in combination with EpCAM155Gastric156Head and Neck157Liver in combination with CD90158Ovarian in combination with c-Kit158Pancreatic in combination with EpCAM and CD24159Prostate in combination with integrin α2β1 and CD13314 |
| CD24 | B cells, follicular dendritic cells, granulocytes, epithelial cells160 |
B-cell proliferation and maturation160. Function in other tissue is poorly understood. |
Breast28, Gastric in combination with CD44161Pancreatic in combination with EpCAM and CD44159 |
| CD90 | Fetal liver cells and thymocytes162, hematopoietic stem and progenitor cells163 mesenchymal stromal cells164, activated endothelial cells165, neuronal cells166 |
Regulation of cell adhesion167, signal transduction in T cells168 |
Brain169Liver170 and Lung tumors171 |
| CD133 | Hematopoietic stem and progenitor cells172 endothelial progenitor cells173, fetal neural stem cells174, renal stem cells16 prostate stem cells18 |
Poorly understood |
Brain175Colon176,177 Endometrial178Liver179Lung with ABCG2 or CXCR4180Ovarian181 Pancreatic in combination with CXCR413Prostate in combination with integrin α2β1 and CD4414 |
| ALDH | Epithelial cells of the esophagus, stomach, intestine, colon, liver, mammary gland and pancreas. Endocrine cells of the adrenal, thyroid and salivary glands, haematopoietic cells182. |
Conversion of aldehydes generated by metabolic processes into carboxylic acids183. Function in ester hydrolysis and as an antioxidant184. Involved in retinoic acid signaling184. |
Breast20Colon185Head and Neck186Liver in combination with CD13322Melanoma187 and Pancreatic tumors |
| Hoechst33342 exclusion (Side population) |
Hematopoietic stem cells25 mammary stem/progenitor cells188,189, pulmonary stem cells190, cardiac |
Not an endogenous marker/protein, but based on the ability of cells to efflux Hoechst33342 involving the |
Astrocytomas195Gastrointestinal tumors196Gliomas197 Hepatocellular carcinomas198Lung cancer199Thyroid cancer200 |
| progenitors191, hepatocyte progenitors192, neural stem and progenitors193 and keratinocytes progenitors194. |
ABC transporter trans- membrane proteins25. |
The similarity of antigen display by normal tissue SCs and the CSCs from corresponding neoplastic tissues would seem to lend further support to the notion that CSCs are, at least in certain respects, bona fide SCs. In the eyes of some, this would seem to indicate that CSCs derive directly from the SCs in the normal tissue-of-origin of a tumor. However, as discussed in Box 2, alternative hypotheses describing the origins of CSCs must also to be entertained.
BOX 2. How do CSCs arise?
Carcinomas usually arise as a consequence of a succession of genetic and epigenetic alterations occurring over many years if not decades; the accumulation of these changes leads ultimately to expression of the multiple neoplastic traits associated with highly malignant cells and thus aggressive tumors. This raises the question of whether CSCs are invariably present within incipient tumors at these various stages of tumor progression. One insight – nothing more than a speculation at present – comes from the observed similarities between normal tissue SCs and CSCs cited above: If the cells in fully normal epithelial tissues existing prior to tumor initiation contain subpopulations of SCs, and if progressed, highly malignant populations of carcinoma cells arising at the last step of tumor progression contain SCs (i.e., CSCs), then it seems plausible that each of the cell populations formed at intermediate stages of tumor progression also contains its own subpopulation of SCs. Stated in a more general way, all populations of epithelial cells, independent of their stage of tumor progression, may contain subpopulations of SCs. Indeed, this may even hold true for populations of epithelial cells propagated in vitro.
If SCs of various genetic configurations were to be found to be present at all of the distinct stages of multi-step tumor progression, this would seem to suggest, in turn, that each SC subpopulation derives directly from a SC subpopulation existing in the previous stage of tumor progression, i.e., one SC subpopulation evolves directly into the SCs present in the successor subpopulation driving growth of the next, incrementally more aggressive stage of tumor progression. Although attractive for its simplicity, this model carries with it several biological problems, including the fact that SCs, including CSCs represent relatively small target populations that generally are not highly active mitotically. Given this, a more plausible model is that transit-amplifying/progenitor cells are the targets of mutation during multi-step tumorigenesis, and that they introduce their acquired mutations into corresponding SC pools via a process of spontaneous dedifferentiation73, 74.
Despite the growing list of CSC markers, several of these have been reported not to be uniformly useful in identifying CSCs. For example, although the CD44+CD24− profile was used in early studies of breast CSCs, the authors report that not all breast cancer cell populations could be stratified using this set of markers28. In fact, several of these markers, including CD44, CD90 and CD34, play roles in cell adhesion and attachment and have been thought to favor the survival of cells in the relatively harsh environment of immunodeficient mice, arguing that the procedure of xenotransplantation may inadvertently select for the outgrowth of cells expressing these cell-surface proteins29. This might suggest that, in the long run, it may be more profitable to employ markers that function physiologically to support the CSC phenotype, thereby ensuring close linkage between marker display and residence in the CSC state. For instance, components of the signaling pathways that play an essential role in the biology of colorectal CSCs, such as that driven by canonical Wnt signalling, may eventually yield highly specific and thus highly useful CSC markers30. A more detailed discussion of CSC markers has been recently published31.
Epithelial-to-mesenchymal transition and the stem-cell state
The cell-biological program termed the epithelial-mesenchymal transition (EMT) was initially studied because of the critical roles that it plays in many of the cell-type interconversions underlying organogenesis during normal development. During passage through an EMT, epithelial cells lose their differentiated characteristics of cell-cell adhesion and lack of motility and acquire instead the traits of mesenchymal cells that confer on them migratory and invasive powers along with elevated resistance to apoptosis32. In the context of carcinoma pathogenesis, the EMT has been increasingly linked to the ability of carcinoma cells to invade locally and disseminate to distant anatomical sites, where they may then initiate metastases33.
In general, activation of an EMT program in both normal and neoplastic cells appears to require heterotypic signaling between these cells and neighboring stromal cells. Thus, stromal signals, largely in the form of secreted factors, are released by various stromal cell types and impinge on nearby epithelial cells, resulting in the induction of intracellular signaling cascades in the latter. These pathways lead to the expression of transcription factors (TFs) that orchestrate the EMT program (EMT-TFs) and regulate various target genes whose expression ultimately results in acquisition of mesenchymal cell traits.
Evidence that the EMT process contributes to the progression of a variety of carcinoma types is rapidly accumulating from diverse laboratories34–38. The EMT program has also been shown to result in the generation of epithelial cells that have stem-like properties 39, 40. This appears to be true for both normal and neoplastic mammary epithelial stem cells, the latter representing cells that exhibit CSC-like properties39,40. Indeed, currently available evidence is compatible with the notion that epithelial cells in the normal mammary gland employ components of the EMT program as the main route for entering into the SC state41. It remains to be seen whether other epithelial tissues rely similarly on versions of the EMT program to generate their own normal and, by extension, neoplastic SCs. It appears that, depending on the EMT-TFs involved, epithelial cells may enter into the SC state, the mesenchymal state, or both. Moreover, while not yet reported, accumulating evidence suggests that normal and neoplastic SCs arising in epithelial tissues generally exhibit a mixture of epithelial and mesenchymal traits, indicating that they have advanced only partially through an EMT program, as discussed in more detail below41.
Signaling pathways characteristic of EMT-induced CSCs
Since the EMT is a key program generating CSCs, it has become important to elucidate the signaling pathways responsible for activation of this program and for maintenance of cells in the resulting mesenchymal (or quasi-mesenchymal) state. As mentioned, in the case of carcinomas, the EMT is often and perhaps invariably induced through the convergence of various signals deriving from the tumor stroma, including extracellular matrix components such as collagen, as well as secreted factors, such as TGF-β, canonical and non-canonical Wnts42. Such signalling cascades induce expression of the EMT-TFs mentioned above, which include members of the Snail, Twist and Zeb family of proteins, among others. These proteins, acting as transcription factors, are responsible for orchestrating the gene expression programs that activate effectors of EMT phenotype, doing so through the repression of epithelial genes and the activation of mesenchymal genes.
The TGF-β pathway is the first and best-studied signalling cascade operating to induce the EMT program in various epithelial tissue types43. Binding of TGF-β ligand induces dimerization of the types I and II TGF-β receptors, leading in turn to the phosphorylation of Smad2 and Smad3, which form a complex with Smad4; once formed, the resulting transcription factor complex migrates to the nucleus, where it can induce, among other responses, a transcriptional program that mediates the acquisition of mesenchymal properties and suppression of epithelial traits43. In the context of cancer, this canonical TGF-β pathway is also known to collaborate with several other pathways including ERK44, p38 MAP Kinase45, Wnt-β-catenin46 and PI3 kinase47 in promoting mesenchymal and migratory properties of cancer cells. Similarly, both canonical and non-canonical types of Wnt signaling are responsible for induction of an EMT and stem-like properties in various tissue types48–50.
Recent studies have unveiled a novel collaboration between the aforementioned pathways in the induction of an EMT; this signalling operates through paracrine signals originating ostensibly in the tumor stroma as well as autocrine signals derived from the carcinoma cells themselves50. Thus, the paracrine signals trigger expression of an EMT program in carcinoma cells, which is subsequently maintained by autocrine signals generated by these neoplastic cells. Such autocrine signals, at least in one case examined, involve the same signalling factors that previously triggered initiation of the EMT program. Stated differently, these autocrine signals serve to maintain the resulting mesenchymal state in a self-perpetuating fashion in the absence of further extrinsic signals coming from the stroma; such persistent signalling would seem to assist individual tumor cells to migrate and invade through foreign tissue while maintaining, in a cell-autonomous fashion their EMT-associated traits; in the longer term, these wandering cells may disseminate to distant sites in the body, i.e., metastasize. Consistent with this notion, recent studies of the circulating tumor cells (CTCs) released by primary carcinomas have shown expression of the WNT2 gene whose product increases propensity for metastasis37.
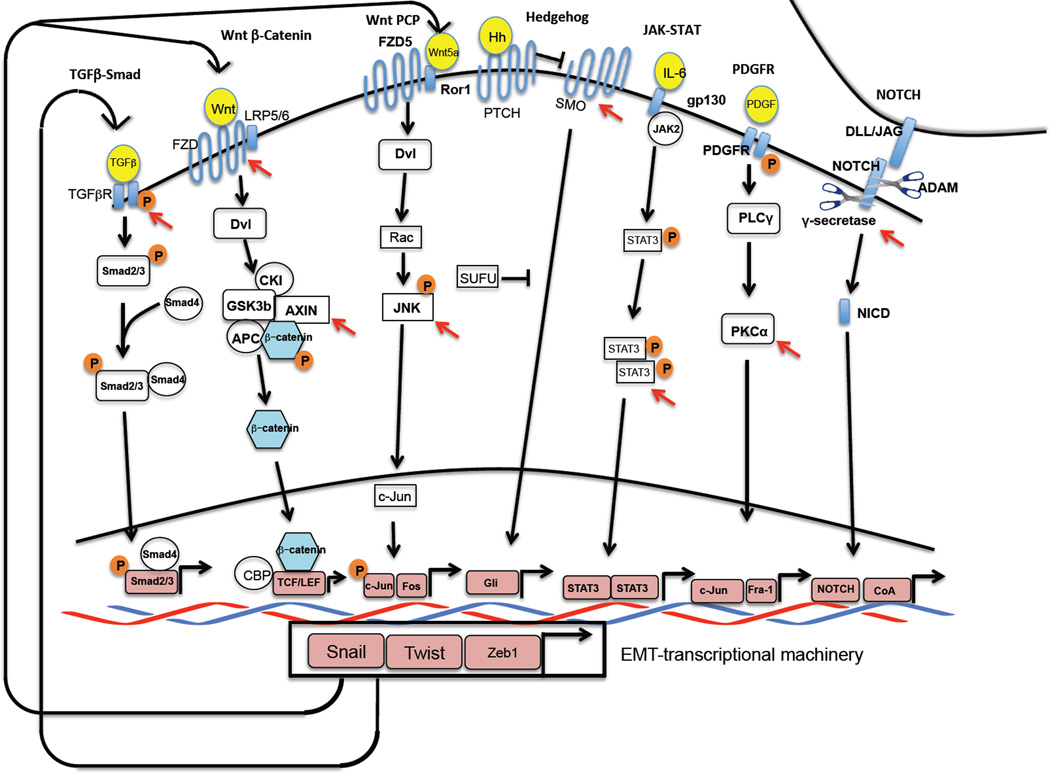
Autocrine signaling loops may also serve an alternative function: to perpetuate residence of cells in the epithelial state. For example, the autocrine production by epithelial cells of bone morphogenetic proteins (BMPs), Gremlin, DKK1 and SFRP serve as inhibitors of autocrine TGF-β and Wnt signalling respectively, protecting the epithelial cells from inadvertent activation of signalling that would lead to activation of the EMT program; conversely, the shutdown of these secreted inhibitors in mesenchymal cells opens the door to autocrine TGF-β and Wnt signaling, permitting activation of mesenchymal gene expression programs. Given the apparent close connection between the EMT program and the CSC state, these same dynamics would seem to apply as well to entrance into and out of this phenotypic state. Other signalling pathways that are implicated in the induction and maintenance of CSC traits include Prostaglandin E251, Hedgehog52, NOTCH53 and PDGFR54; a summary of these pathways is presented on Figure 2.
Figure 2.
Signaling pathways employed by CSCs: Distinct combinations of paracrine and juxtracrine signals induce the EMT in different contexts. A representation of signaling pathways that CSCs have been reported to depend on is shown here. The transcription factors that are downstream of these signaling pathways act as co-factors and cooperate with the EMT-inducing transcription factors such as Snail to induce and maintain the mesenchymal/CSC state. The EMT program also activate several positive, self-reinforcing feedback loops in order to maintain cells in a mesenchymal/CSC state; shown here are three involving canonical and non-canonical Wnts as well as TGF-β. Nodes of the pathway that could be open to therapeutic targeting are indicated by red arrowheads.
The interplay between such autocrine signals produced by carcinoma cells and paracrine signals arising in the tumor stroma presumably creates a complex array of cues that dictate the extent of epithelial/non-CSC and mesenchymal/CSC characteristics displayed by carcinoma cells. The possibility that these signals can function in an essentially unlimited number of combinations implies the existence of multiple distinct phenotypic states between the fully differentiated, strictly epithelial state and the fully mesenchymal state, these two states representing the extremes of the EMT program.
In truth, the extent of epithelial vs. mesenchymal polarization that carcinoma cells undergo within actual human tumors is poorly resolved at present. It seems increasingly likely that carcinoma cells that have activated an EMT program usually enter into a state in which certain epithelial markers are retained while mesenchymal are newly acquired, resulting in what is often termed a “partial EMT”. Accordingly, cells that have passed entirely through an EMT program and have thus undergone a “complete EMT” resemble transdifferentiated cells of the mesenchymal mesodermal lineage and lose the epithelial vs. mesenchymal plasticity that is required for expression of tumor-initiating properties and is created by a partial EMT55,56. Moreover, bona fide epithelial SCs – both normal and neoplastic – would seem to arise from cells that have such mixed epithelial/mesenchymal properties.
Resistance to conventional drugs
Chemotherapy and radiotherapy have been the treatments of choice for the past half-century, often affording remarkable reductions in tumor burden. As mentioned in passing earlier, induction of an EMT leads to the acquisition of resistance to both forms of therapy, a phenomenon that has been documented in the greatest detail in breast and ovarian cancers57, 58. Chemoresistance has also been shown to be higher in tumors that harbor a gene signature indicative of desmoplastic or reactive stroma, which is consistent with the notion that signals secreted by a reactive stroma play a major role in the induction of an EMT59. In light of the complex regulation of the CSC state and its maintenance, how might one utilize our current knowledge of signal transduction biochemistry to specifically target this treatment-resistant subpopulation?
At present, we possess only an incomplete understanding of the actual biochemical and cell-physiologic mechanisms underlying the intrinsic chemo- and radioresistance of tumor cells that have passed, even partially, through an EMT. Moreover, resistance to cytotoxic treatments may also be attributable to the lower proliferative rate that results from the acquisition of mesenchymal properties60,61. Indeed, CSCs from a variety of tumors have been shown to be slow cycling and to exhibit an increased level of quiescence compared to the majority populations of cancer cells within certain tumors62,63. Additionally, the resistance to chemotherapy in normal stem cells has been attributed to high-level expression of anti-apoptotic proteins64 and to ABC transporters that are capable of efflux of the Hoechst 33342 dye, creating the “side population (SP)” observed upon fluorescence-activated cell sorting (FACS) fractionation of tumor cell populations25,65,66; these mechanisms could also operate to confer similar properties on CSCs.
A recent study using a genetically engineered mouse model (GEMM) of glioblastoma development has shown that a quiescent, population of tumor cells survives treatment with temozolomide and regenerates the tumor by differentiating into populations of highly proliferative cells37. This finding demonstrates directly that the CSCs in this tumor exhibit elevated resistance to chemotherapy, and that purely cytotoxic treatment regimens that target cycling cells are bound to fail unless accompanied by a targeted therapy that specifically targets these small, phenotypically distinct subpopulations.
Therapeutic Targeting of CSCs – what has been done so far?
Understanding of CSC-dependent signaling pathways
The identification and characterization of CSCs has revealed the need for specific molecular therapies that target the key signalling pathways supporting these cells and their residence in the CSC state. As described above, CSCs and normal SCs share a number of properties. This explains why signalling pathways, such as those activated by Wnt, TGF-β, NOTCH and Hedgehog – all known to be essential for the self-renewal properties of normal adult stem cells 52, 53, 67, 68 – are emerging as attractive targets whose inactivation may allow elimination of CSCs,
Studies of CSCs and the EMT program have led to a preliminary understanding of the signaling pathways that these cells preferentially employ, examples of which are illustrated in Fig 2. From these studies it has become evident that these pathways are highly context-dependent and several of them may actively collaborate to maintain residence in the CSC state, one example being the aforementioned activation of both TGF-β and Wnt signalling pathways in the maintenance of mammary CSCs50. These extracellular signalling channels may offer opportunities for interdicting these pathways in the extracellular space through, for example, neutralizing antibodies.
Screens to identify novel targeted therapeutics
Pharmacology has been profoundly changed by the ability to screen large, complex chemical compound libraries in order to identify chemical species that target specific proteins within cells. In the case of CSCs, chemical screening for agents that specifically target these cells has been a challenge due to their rarity and the inability to propagate in culture CSC populations isolated by flow cytometry. One strategy to circumvent this hurdle has involved the recent screening of mammary epithelial cells that have been forced experimentally to undergo an EMT and thus have acquired certain CSC characteristics, including increased tumor-initiating ability in vivo39, 40.
One group has carried out a 16,000-compound library screen in order to identify compounds that could preferentially kill EMT-induced CSCs; these CSCs were derived through knockdown of E-cadherin, an alteration known to favour activation of the EMT program57. Through this screen, this group showed that salinomycin pretreatment of CSCs resulted in a ~100-fold decrease in tumor-seeding ability relative to the conventional agent, paclitaxel. Similar screens have subsequently been carried out by others to identify compounds that preferentially target glioblastoma CSCs69, ovarian CSCs70, breast CSCs71 and AML LSCs72.
In principle, such screens can allow the identification of novel modulators of cell phenotype and do so in an unbiased way. However, these studies also bring to light the still-incomplete understanding that we possess of CSCs and the pathways that these cells depend on. Like other similar strategies of drug development, such screens should be used as starting points for further functional studies that reveal at a mechanistic level precisely how these agents actually work. Additionally, screens like these are carried out in two-dimensional cultures in the absence of components that would ordinarily be present in the tumor microenvironment e.g., the extracellular matrix, stromal cells including fibroblasts, myofibroblasts and immune cells, as well as endothelial cells forming microvessels. Such deficiencies must be taken into account when attempting to extrapolate the results of these screens to the behaviour of CSCs in vivo.
Ideally a future anti-CSC therapy, using agents such as those cited above, should eliminate the pool of cancer cells that are intrinsically resistant to conventional therapies, while a concomitantly administered conventional agent would eliminate the non-CSC cells, which are known to be susceptible to existing cytotoxic therapies. Importantly, elimination of the CSCs on its own may not suffice to induce an acceptable, durable clinical response, since new CSCs may be generated in CSC-depleted tumors via the spontaneous dedifferentiation of non-CSCs – a consequence of cellular plasticity that enables the emergence of de novo CSCs from differentiated cells73, 74.
Therapeutic targeting of CSCs – other strategies and the road ahead
Targeting the tumor microenvironment
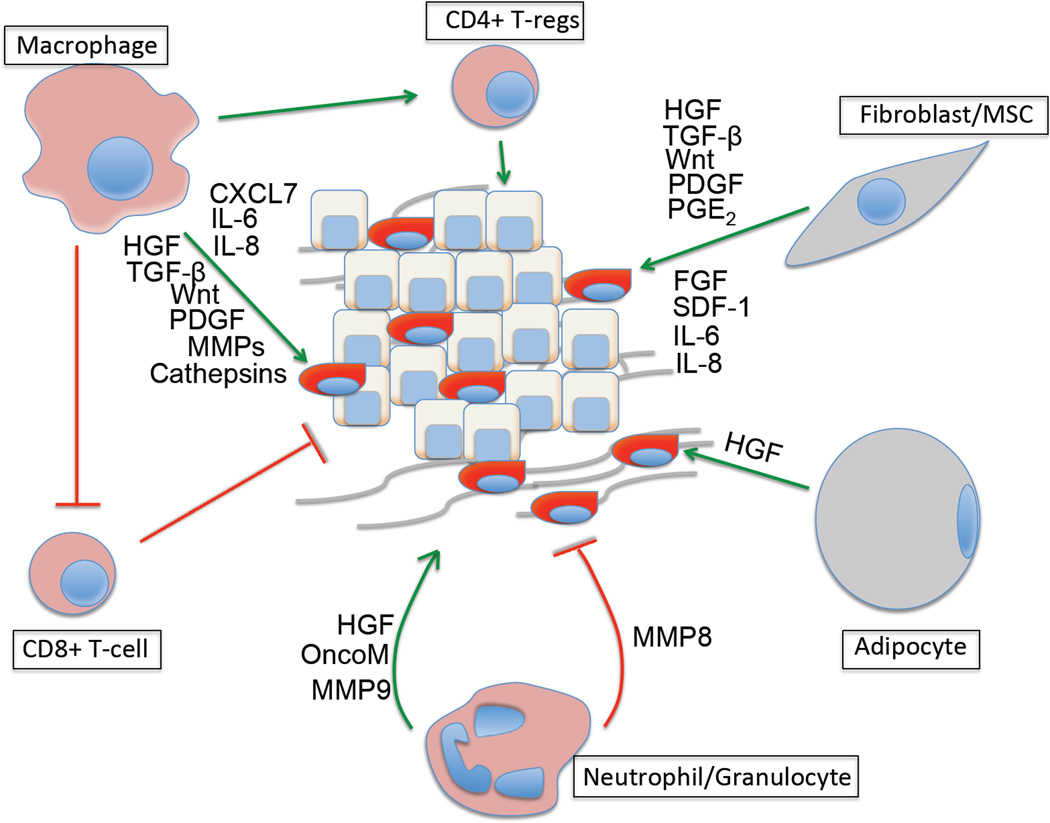
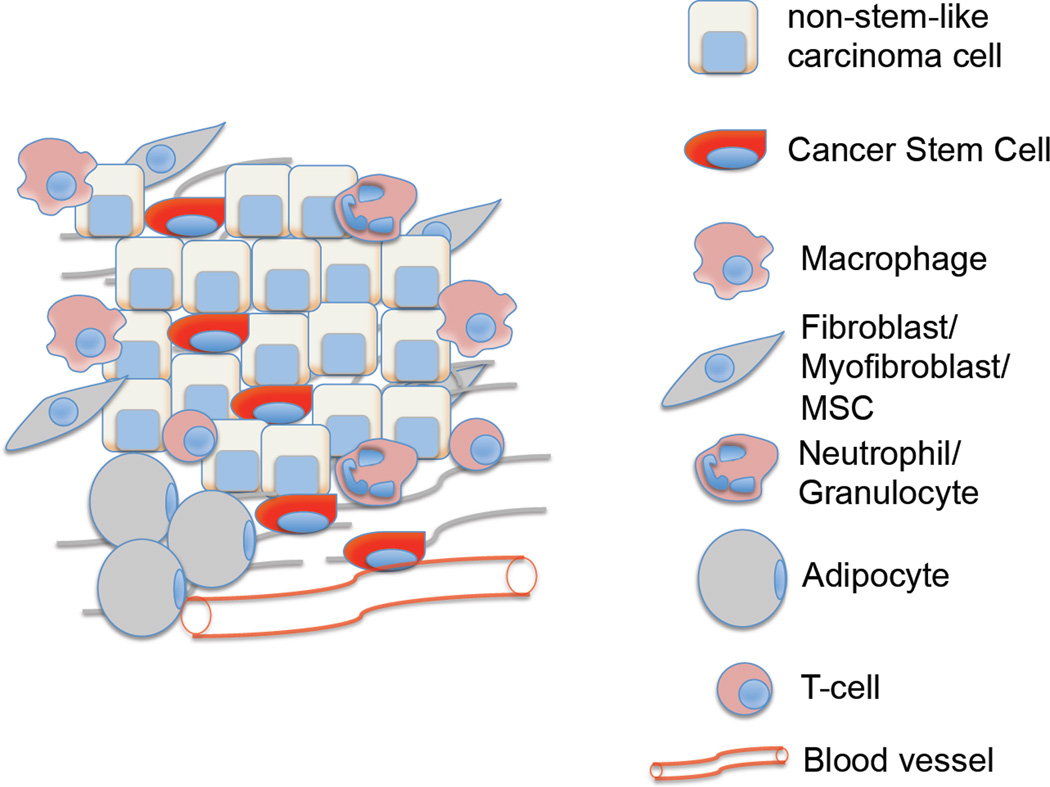
Direct targeting of CSCs represents one major strategy for eliminating these cells and thus the tumors that they support. However, alternative strategies have been suggested by the rapidly growing information on the tumor microenvironment and its role in triggering activation of an EMT program in carcinoma cells and possible entrance of these cells into the CSC state. As mentioned above, heterotypic signals arising in the tumor-associated stroma are often responsible for activating this program in nearby carcinoma cells. Prominent among the signal-emitting cells of the stroma are fibroblasts, myofibroblasts, adipocytes and mesenchymal stem cells (MSCs), infiltrating immune cells such as macrophages and neutrophils, as well as endothelial cells that make up the walls of blood vessels that extend through the tumor (See Fig 3; Box 3). Detailed reviews of these stromal components have been published previously75, 76. In addition, the extracellular matrix (ECM) assembled by these cells also has strong effects on invading carcinoma cells 77.
Figure 3.
Impact of stromal cells and secreted factors on CSCs – Each stromal cell type depicted here influences the tumor by secreting factors that stimulate the formation of CSCs and help maintain the residence of already-formed CSCs in the SC state. Summarized here are some of the major factors that secreted by each cell type that are known to impact CSCs. Green arrow – Tumor promotion, Red crossbar – Tumor inhibition.
BOX 3. What constitutes the tumor microenvironment?
The tumor microenvironment, in addition to harboring carcinoma cells, consist of various components that play a major role in influencing the outcome of the malignancy. These can be widely classified into three main groups – cells of hematopoietic origin, cells of mesenchymal origin and non-cellular components. Tumors of different origins and different stages of progression will inevitably contain components in various proportions. Nonetheless, these three classes of stromal components represent the most abundant elements present in the microenvironment of solid tumors.
Cells of hematopoietic origin
This compartment consists of cells that arise in the bone marrow and can be subdivided into cells of the lymphoid lineage consisting of T-cells, B-cells and NK-cells, and those of the myeloid lineage that include macrophages, neutrophils and myeloid-derived suppressor cells (MDSC). The roles of different subsets of T-cells in tumor promotion249 and tumor elimination250 have been well studied and are the subject of recent advances in the development of novel immunotherapies126. The alternate activation of macrophages and their ability to promote tumor development and progression have also been well documented251. Similarly, each of the other constituent cell types has either a positive or negative effect on the outcome of the tumor. Importantly, interactions between different cell types within the stroma can also play a major role on tumor progression, as has been shown for CD4+ T cells and macrophages252, for example. Other cell types that play a major role in tumorigenesis but are not abundantly present in the tumor microenvironment per se include platelets and dendritic cells.
Cells of mesenchymal origin
These comprise of cells derived from the mesenchyme and include fibroblasts, myofibroblasts, mesenchymal stem cells (MSCs), adipocytes and endothelial cells. Myofibroblasts and MSCs derived from the bone marrow have been shown to directly support CSCs by creating a favorable niche and facilitating tumor progression78, 253. Adipocytes, until recently were thought of only as energy storage houses; however, recent studies reveal the importance of factors that they secrete (e.g., hepatocyte growth factor; HGF) in tumor progression254. Endothelial cells and pericytes that constitute the walls of blood vessels play a major role in vascular functionality, angiogenesis as well as regulating cancer cell dissemination.255
Non-cellular components
The major non-cellular component of the tumor microenvironment is the extracellular matrix (ECM), which consists of many distinct components including proteins, glycoproteins and proteoglycans that enable its functions both structurally and functionally77. The ECM can be subdivided into the more compact basement membrane, which is a specialized ECM rich in type IV collagen, laminin and fibronectin, and the interstitial matrix, which consists of fibrillar collagens, proteoglycans and glycoproteins that contribute to the tensile strength of the tissue77.The ECM is involved in the formation of a SC niche and while it acts to maintain tissue architecture and prevent cancer cell invasion, abnormal ECM has been shown to promote tumor progression and tumor angiogenesis77.
Together these components make up the tumor microenvironment, which represents a complex habitat involving myriad interactions between cell types and ECM, each of them playing a role in influencing tumor outcome.
In the case of colon carcinomas, the interactions between the carcinoma cells and stromal cells, specifically myofibroblasts, have been shown to be important in inducing and maintaining a more stem-like state in the former30, showing directly that the stroma can play a major role in the generation of CSCs. Moreover, interactions between certain classes of carcinoma cells and MSCs induce the latter to secrete Prostaglandin E2 (PGE2), which is then responsible for the activation of the β-catenin signaling in the carcinoma cells; once activated, this signaling promote their acquisition of a CSC-phenotype78. Similar reciprocal interactions also exist in breast cancers, where MSCs recruited from the bone marrow interact with carcinoma cells via paracrine cytokine signalling involving CXCL7 and IL-6, which are responsible for stimulating the self-renewal of the neoplastic cells79. Hence, MSCs secrete cytokines and growth factors that together create a suitable niche enabling carcinoma cells to acquire and maintain stemness. Similar roles have also been reported for tumor-associated macrophages (TAMs), which secrete factors such as IL-6 that activate the JAK-STAT pathway within the tumor cells, enhancing their tumorigenicity and resistance to chemotherapy by imparting CSC properties to them80–82.
In principle, the rapidly accumulating insights into the paracrine signaling pathways activating and sustaining the CSC program should provide insights for targeting CSCs; such a focused approach would represent an alternative to the untargeted use of high throughput screening described above. For example, one means of blocking pathways activated by stroma-derived signals could employ antagonists of the EP4 PGE2 receptor, such as the small molecule RQ-1598683, thereby reversing the tumor-promoting effects that MSCs have on carcinoma cells. Similarly, inhibitors of STAT3 DNA-binding, such as the small molecule NSC 7485984, may be effective in blocking immune cell-activated IL-6/STAT3 signaling that supports CSC properties in certain carcinomas.
Of course, such approaches may also result in inhibition of signaling cascades in normal cells in which these signals play critical roles. For example, Wnt signaling, operating via intracellular β-catenin signaling, is known to be essential for the regulation and homeostasis of intestinal stem cells85, 86, and the use of Wnt inhibitors might therefore lead to a depletion of the normal resident stem cell population that is responsible for continuous regeneration of the intestinal epithelium. However, the dependency of colorectal CSCs on β-catenin signaling might exceed that of the normal intestinal stem cells, yielding a favorable therapeutic index, such that a low dose treatment might still be able to deplete CSC activity without significantly affecting normal organ homeostasis. Indeed, several clinical trials using inhibitors of Wnt signaling are currently underway, such as a currently ongoing phase I study of an oral inhibitor of Porcupine (LGK974), an enzyme involved in the post-translational maturation of Wnt proteins, for treating patients with advanced breast and pancreatic cancer (http://clinicaltrials.gov/ct2/show/study/NCT01351103).
An alternative approach to disrupting stromal signals might involve preventing the cellular sources of these signals from being recruited into the tumor stroma in the first place. Thus, MSCs are often recruited into the tumor-associated stroma by carcinoma cell-derived IL-887. In such cases, inhibition of this homing signal might be effective in preventing MSC localizing to the tumor stroma and the resulting development of a supportive CSC niche. Similar tropic signals have also been reported for macrophages, which home to the primary tumor in response to factors such as CSF-1 and several chemokines, including CCL2, CCL5 and CXCL1288–90. Antibodies that block the CSF-1 receptor (CSF1R) have been developed and shown to be effective at reducing the numbers of tumor-associated macrophages homing to tumors in syngeneic mouse tumor models91. Similarly, inhibitors of the tyrosine kinase activity of CSF1R have also been developed and shown to inhibit the tumor-promoting effects of macrophages, including the acquired resistance of tumor cells to chemotherapy92. A CSF1R kinase inhibitor, the small molecule ARRY-382, has recently gone through phase I clinical trials in patients with metastatic cancers (http://clinicaltrials.gov/ct2/show/NCT01316822) and may be the forerunner of a large repertoire of agents that are used either singly or in combination with other drugs to prevent macrophage homing.
Inhibition of CSC-dependent pathways
One strategy to block the initiation of the EMT program as well as entrance into and maintenance of the CSC state has already been suggested by findings cited earlier. Thus, epithelial non-CSCs cells synthesize and secrete high levels of physiologic inhibitors of Wnt signaling, such as SFRP and DKK proteins, which act at the cell surface to block ligand binding-mediated activation of Frizzled receptor signalling 50. As argued above, these secreted inhibitors ostensibly serve to reinforce residence in the epithelial state. Similarly, secreted inhibitors of the TGF-β pathway, including Gremlin and BMPs 4, 6, 7, 9, 10, are expressed and secreted at high levels in the non-CSCs, where they appear to block another pathway (involving TGF-β) that is critical for maintenance of the non-CSC/epithelial state50. Derivatives of these secreted inhibitory molecules, acting in the extracellular space, could become potential therapeutics that prevent stochastic inter-conversion between the CSCs and non-CSC states, tilting the balance in favour of the non-CSC state. We note that therapeutic proteins like these are difficult and expensive to produce in large quantities and often difficult to deliver into the interstices of complex tissues including tumors.
Given the importance of Wnt signaling for the induction of the CSC state, several low molecular weight inhibitors of this pathway have been developed. One of the first inhibitors to target the Wnt pathway was ICG-001, a small molecule that blocks the ability of CREB-binding protein (CBP), a co-factor for a large number of transcription factors93, to act as a co-activator of the β-catenin-TCF complex, the transcription factor complex activated by canonical Wnt signalling 94. An inhibitor of the previously cited Porcupine enzyme (the small molecule IWP295 or LGK97496), a membrane-bound O-acyltransferase responsible for the post-translational maturation of Wnts via palmitoylation, has also been developed for the inhibition of the pathway. These inhibitors may be effective in preventing secretion of active Wnt molecules by the stromal cells, thereby blocking the paracrine signalling that triggers formation of new CSCs; in addition, they may block the autocrine signalling that is used by existing CSCs to maintain their residence in the SC state. Recent studies have also identified a role for yet another β-catenin co-factor, YAP1, which forms a complex β-catenin and is essential for the formation of β-catenin-driven cancers97. Reviews detailing the targeting of various aspects of the Wnt98 and TGF-β99 pathways have been published elsewhere.
Several molecules targeting various nodes of the Hedgehog and NOTCH pathways are also coming into prominence for their ability to target CSCs. For example, IPI926, a derivative of the natural product SMO antagonist cyclopamine100, is undergoing clinical trials for malignancies, such as basal cell carcinomas101 and metastatic pancreatic cancer, in combination with chemotherapeutic drugs such as gemcitabine (http://clinicaltrials.gov/show/NCT01130142). Similar trials have also been initiated for the SMO competitive small molecule antagonist GDC-0449 against various tumors 102, 103, with FDA approval being granted to treat adults with advanced basal cell carcinoma. MK-0752, a small molecule gamma secretase inhibitor, is under clinical trials for the treatment of advanced solid tumors104, whereas antibodies targeting Delta-like ligand 4 of NOTCH105 are also promising candidates to reduce CSC frequency of breast tumor xenografts and other solid tumors106. A list of inhibitors that target pathways that are preferentially utilized by CSCs and may possess therapeutic utility is summarized in Table 2. Unanswered by these studies are the therapeutic indices that these agents will exhibit, that is, will they in fact target neoplastic CSCs far more effectively than the SCs residing in corresponding normal tissues?
Table 2.
List of pathways that may be beneficial for the specific targeting of CSCs and potential candidates in clinical development
| Pathway | Targets | Compounds | Potential Indications | Clinical development |
|---|---|---|---|---|
| Wnt | Porcupine | IWP95 or LGK97496 |
Breast cancer – Maintenance of breast CSCs through recruitment of Wnt ligands201 |
Phase I trial for malignancies dependent on Wnt ligands including melanoma, breast and pancreatic malignancies http://clinicaltrials.gov/show/NCT01351103 |
| Pancreatic cancer – Circulating tumor cells show high levels of WNT2 expression202 | ||||
| β-catenin: - Interaction of β- catenin with TCF, CBP - Tankyrase inhibition for stabilization of AXIN2 |
ICG-00194 or iCRT203 or OXT-328204 or XAV-939205 or IWR95 |
Colon cancer – Required for maintenance of Colon CSCs through signals from stroma30 |
CWP232291206 which induces β-catenin degradation is under Phase I trials for AML and CML http://clinicaltrials.gov/ct2/show/NCT01398462 Phase I trials for advanced solid tumors http://clinicaltrials.gov/ct2/show/NCT01302405 |
|
| AML and CML – Could prove effective in MLL-fusion driven AML CSCs and Imatinib resistant CSCs in CML207,208 | ||||
| Breast cancer – Degradation of β-catenin blocks the generation of breast CSCs204 | ||||
| Squamous cell carcinoma – β- catenin required for maintenance of CSCs and tumor progression209 | ||||
| TGF-β | TGF-βRI | LY2157299210 | Breast cancer – Increased TGF- β signaling in CSCs of chemotherapy-resistant breast cancer122 |
LY2157299 in Phase I trials for recurrent malignant glioma http://clinicaltrials.gov/ct2/show/NCT01682187 |
| Glioma – Inhibition of TGF-β decreases glioma-initiating cell population211 | ||||
| Pancreatic cancer - TGF-β induces an EMT in CSCs promoting metastasis212 | ||||
| TGF-β | GC1008213 a.k.a. Fresolimumab or AP12009214 |
As above | Phase I trial of GC1008 in Glioma http://clinicaltrials.gov/ct2/show/NCT01472731, Renal cell carcinoma and Melanoma http://clinicaltrials.gov/ct2/show/NCT00356460 and metastatic breast cancer http://clinicaltrials.gov/ct2/show/NCT01401062 Phase I trial of AP12009 in Melanoma, Pancreatic and Colorectal neoplasms http://clinicaltrials.gov/ct2/show/NCT00844064 |
|
| NOTCH | Gamma-secretase | MK-0752215 or PF03084014216 or R04929097217 |
Breast cancer – NOTCH signaling regulates breast CSCs218 |
Phase I trial of MK-0752 in advanced breast cancer http://clinicaltrials.gov/ct2/show/NCT00106145 Phase I trial of PF-03084014 in metastatic breast cancer http://clinicaltrials.gov/ct2/show/NCT01876251 and glioma Phase I trial of R04929097 in advanced solid tumors217 |
| Ovarian cancer – NOTCH signaling is critical for the regulation of CSCs and platinum resistance219 | ||||
| Glioblastoma – CSCs receive NOTCH signals from endothelial cells in stroma220 | ||||
| Pancreatic cancer – NOTCH inhibition induces apoptosis in pancreatic CSCs221 | ||||
| DLL4 | Anti-DLL4 Ab106, 222 |
Colon cancer – Blocking DLL4 reduces tumor-initiating frequency106 |
Phase I trial of MEDI0639 in advanced solid tumors http://clinicaltrials.gov/ct2/show/NCT01577745 |
|
| Hedgehog | SMO | Cyclopamine22 3 or GDC-0449224 or LDE225225 |
Glioblastoma – Self-renewal of CSCs requires Hh-Gli signaling226,227 |
Phase I trial of LDE225 in advanced solid tumors http://clinicaltrials.gov/ct2/show/NCT01576666 Phase I trial of a combination of NOTCH and SMO inhibition in metastatic breast cancer http://clinicaltrials.gov/ct2/show/NCT01071564 |
| Breast cancer – Hh signaling regulates self-renewal of breast CSCs52 | ||||
| Pancreatic cancer – Hh inhibition reverses the EMT and tumor metastasis228 | ||||
| CML – Expansion of LSCs is dependent on activation of Hh229 | ||||
| Multiple myeloma – Hh signaling important for maintenance of CSCs230 | ||||
| JAK- STAT |
IL-6 | CNT0328231 or Tocilizumab232 AZD1480233 Compound1234,235 |
Glioblastoma – STAT3 required for multipotency of CSCs236 |
Phase II trial of CTN0328 in advanced prostate cancer http://clinicaltrials.gov/ct2/show/results/NCT00433446 Phase I trial of OPB-31121 in advanced solid tumors http://clinicaltrials.gov/ct2/show/study/NCT00955812 Phase I trial of WP1066 in glioblastoma and other solid tumors http://clinicaltrials.gov/ct2/show/NCT01904123 |
| Colon cancer – STAT3 required for survival and proliferation of CSCs237 | ||||
| JAK1/2 | Prostate cancer – Blockade of JAK-STAT pathway inhibits CSC function238 |
|||
| STAT3 | Breast cancer – JAK-STAT pathway required for breast CSC signaling82 |
|||
| PDGFR | PDGFR/PKCα | Bisindolylmal eimide I239 |
Breast cancer – PDGFR signaling required for survival of breast CSCs54 |
Phase II trial of ISIS 3521 in metastatic breast cancer http://clinicaltrials.gov/ct2/show/NCT00003236 |
Differentiation therapy to curb stem-like properties of CSCs
Despite being intensively pursued for almost four decades, the search for ideal targeted anti-cancer drugs has yielded relatively few successes. One of the first successes of targeted therapy for any type of cancer involved the use all-trans retinoic acid (ATRA), which was given to patients suffering from acute promyelocytic leukemia107. Following treatment with ATRA, leukemic promyelocytes are relieved of their block of differentiation and differentiate into mature granulocytes. The success of this therapy has led to the concept that differentiation therapy may be effectively used to treat other forms of cancer. In the case of CSCs, notably those in carcinomas, the notion would be to induce their exit from the CSC state into the more differentiated epithelial state of non-CSCs.
Other agents that have similarly been shown to be beneficial in differentiation therapy include (i) phorbol myristate acetate (PMA), which is capable of inducing leukemic cell differentiation by, among other pathways, protein kinase C (PKC)-mediated induction of transcription factors such as PU.1 and AP1108; (ii) hexamethylamine bisacetamide (HMBA), which induces differentiation of human leukemia cells through several distinct mechanisms109; (iii) dimethylsulfoxide (DMSO), which induces differentiation of human promyelocytic leukemias110; and (iv) vitamin D3 which induces maturation of several leukemic cell lines111.
Yet other compounds such as suberoylanilide hydroxamic acid (SAHA), a histone deacetylase inhibitor (HDACi), have been shown to induce differentiation of human breast cancer cells112 and endometrial carcinomas113. The inhibition of HDACs is thought to release the repression of gene promoters that play major roles in the processes of differentiation and cell cycle arrest, thereby permitting their expression. Another functional class of compounds that induce cell differentiation is represented by 5-azacytidine, a chemical analogue of cytidine, which acts by inactivating the DNA methyltransferases that catalyze methylation of CpG islands and associated transcriptional repression at gene promoters114.
The current explosion in the field of epigenetics research (defined here as the study of various types of chromatin modification) is uncovering a large number of histone methyltransferases and histone demethylases115. Modulation of these enzymes might also serve to increase the transcription of genes that are important for CSC differentiation or silence those that are required for cell migration and invasion. For example, the promoter of the CDH1 gene, which encodes E-cadherin, the keystone of the epithelial state, is a target of silencing in many cancers through the actions of the polycomb repressive complex (PRC2), which imposes the K27me3 repressive mark on histone H3116. Hence, inhibitors of histone-modifying enzymes, such as the methyltransferase EZH2117, could act as differentiation-inducing agents of CSCs that have undergone an EMT, acting by inducing the re-expression of E-cadherin and thereby restoring epithelial properties. In fact, the small molecule EZH2 inhibitor E7438 is currently under phase I/II clinical trials for advanced solid tumors and B-cell lymphomas (http://clinicaltrials.gov/ct2/show/NCT01897571). A more detailed review of epigenetic regulation of EMT and CSCs has recently been published118.
All differentiation-inducing agents act on the premise that a block of terminal differentiation is one of the major characteristics of the neoplastic state. The realization of this phenomenon came from studying leukemias and analyzing the phenotypes of cancer cells in relation to the normal differentiation hierarchy of the hematopoietic system. Thus, acute myeloid leukemia caused by the MLL-AF9 chromosomal translocation leads to the expansion of leukemic stem cell populations that exhibit a cell-surface antigen profile similar to that of granulocyte-monocyte progenitors119. In the case of MLL-AF9 leukemias, it is known that cells are maintained in the less-differentiated progenitor stage by the fusion protein, which activates target genes such as HOXA9/MEIS1 that are responsible for coordinating a downstream CSC program through aberrant methylation of H3K79 by the histone methyltransferase DOT1L120. Hence, inhibitors of DOT1L may specifically act to disrupt the MLL-AF9-induced transcriptional program that fuels these cancers121.
Similar to an argument made earlier, a clinically effective differentiation therapy would need to be combined with chemotherapy in order to eradicate both CSC- and non-CSC populations within a tumor. In fact, more recent studies have focused on such combinations that involve inhibition of TGF-β, which results in differentiation of triple-negative breast cancer CSCs, in combination with paclitaxel, which should eradicate the rapidly cycling, non-CSCs forming the bulk of tumors under treatment122. Targeted therapy would presumably act by depleting CSC potential, giving rise to non-CSC counterparts that are more susceptible to chemotherapy. In fact, the use of agents that specifically target CSCs in combination with conventional chemotherapy has already been evaluated in clinical trials designed to gauge their efficacy in combination with radiotherapy for metastatic breast cancers (http://clinicaltrials.gov/ct2/show/NCT01401062).
One proviso must be cited here, suggested by observations that disseminated carcinoma cells must be able to undergo a mesenchymal-to-epithelial transition (MET) at their site of dissemination in order to generate the mixed CSC/non-CSC populations that appear to be critical to the robust growth of a tumor. 123, 124. Hence inducing CSC differentiation by inducing an MET, while still inhibiting CSC formation and maintenance in the primary tumor, may inadvertently support the process metastatic colonization at distant sites. The complex dynamics of these reversible processes operating during the invasion-metastasis cascade remain to be elucidated.
Directed immunotherapy against CSCs
The last decade has seen resurgence of the idea of directing the cancer patient’s immune system against tumor-specific and tumor-associated antigens. Strategies that exploit our understanding of the immune system for cancer therapy include the use of specific peptides that are derived from tumor antigens, which could be used for cancer vaccination as immunotherapy, the use of dendritic-cells as vaccines to activate adaptive immune cells against specific antigens, and the blockade of immune checkpoints that inhibit anti-tumor immune responses125. T-cell responses to a particular antigen are dictated by the nature of their interaction with an antigen-presenting cell, and depend on co-stimulatory and inhibitors signals that are delivered upon the binding of specific ligands from antigen-presenting cells to complementary receptors on T-cells. The modulation of these so-called immune checkpoints through the use of antagonists of inhibitory signals has been recognized as a major avenue for the development of novel immune-based therapies 125. In fact, drugs such as Ipilimumab, a CTLA4-blocking antibody, have been found to confer clear benefit in treating melanoma 126, 127, with clinical trials underway for several other malignancies including prostate cancer (http://clinicaltrials.gov/ct2/show/NCT01194271) and ovarian cancer (http://clinicaltrials.gov/ct2/show/NCT01611558). Accordingly, the exploitation of the immune system may yield another dimension of anti-CSC therapy by directing immunocytes to recognize CSCs through the unique collection of cell-surface antigens that these cells display. We note, however, another process that may deflect immune attacks on tumors: the autocrine secretion of TGF-β (a known immunosuppressant) by CSCs may create zones around these cells that are protected from attack by various cellular components of the immune system50,128. In addition, the CSC-secreted TGF-β may cause the formation of regulatory T cells (Tregs) that have additional immunosuppressive effects on other subclasses of T lymphocytes129.
As described earlier, the identification of CSC cell-surface markers (see Table 1) has enabled the isolation of these cells from primary tumors. Such markers could be used to survey tumor antigens that are preferentially expressed on the CSCs compared to the bulk of tumor cells and reveal novel vulnerabilities that could be exploited to design stem cell-specific immunotherapies. Additionally, the expression levels of certain potentially inhibitory receptors that could attenuate an immune response could be higher in CSCs compared to non-CSCs; when these inhibitory receptors are blocked, a more uniform immune response against populations of both CSCs and non-CSCs may possibly be achieved, resulting in long-term responses.
Exploiting metabolic differences to target CSCs
The field of cancer metabolism has undergone a revival over the past decade with renewed interest in the Warburg effect, which proposes that cancer cells generate ATP through glycolysis rather than oxidative phosphorylation, doing so even under non-hypoxic conditions130. Through several key breakthrough studies, the role of cell metabolism has evolved into an active area of cancer research with the development of novel therapeutics based on our understanding of the signaling regulating metabolic pathways 131, including temsirolimus and everolimus, inhibitors of mTORC1, which are approved to treat several cancers132–139.
Hypoxia within a tumor can lead to alterations in the metabolism of cancer cells through the inhibition of mTOR signaling140. The hypoxic conditions that are apparent in many if not most tumors have also been shown to lead to the induction of an EMT and the generation of CSCs141, 142. This leads to the speculation that hypoxia-induced metabolic changes could contribute to the mesenchymal/CSC phenotype. Indeed, a recent study reported a metabolic switch to glycolysis that occurs upon activation of the EMT-inducing transcription factor Snail and that blocking of this switch through ectopic expression of fructose-1,6-biphosphatase (FBP1) abrogates the ability of Snail to impart mesenchymal/stem-like properties to breast cancer cells143. Other studies also show the components of the mevalonate metabolic pathway to be important in the generation of breast CSCs, with inhibition of the pathway using hydroxy-3-methylglutaryl CoA reductase blockers, resulting in reduction of CSC properties144. These studies suggest that alteration of the metabolic phenotype could be an essential step required for entrance into the CSC state, and that study of the differences in metabolism between CSCs and non-CSCs may reveal novel ways by which we can specifically target these cells.
Conclusions
The avenues discussed above represent only a portion of the possible ways by which CSCs could be targeted therapeutically. For example, another strategy that has not been discussed here involves the delivery of short-hairpin or small-interfering RNAs that can induce a CSC-specific knock-down of critical genes identified through large-scale screening. Synthetic lethality studies might also uncover novel dependencies of CSCs that could be candidates for future targeting studies.
There are, however, multiple hurdles that have to be surmounted before we can effectively eliminate these cells. First, it is apparent that CSCs exists in a specific niche formed by multiple cell types and distinct signaling molecules, including growth factors, cytokines and ECM components. Hence, studying the biological properties of CSCs in isolation or performing screens with them in the absence of a surrounding relevant niche is hardly guaranteed to yield results that can be translated to responses occurring in vivo. Second, our lack of appropriate experimental systems to accurately model human tumors means that, at least at present, we are forced to study human tumors in immunodeficient mice in the absence of an adaptive immune system or study mouse tumors in a syngeneic setting. Neither of these experimental settings accurately recapitulates the biological complexity of the tumors encountered in the oncology clinic.
The current practice of modeling human tumors by the alteration of one or two genetic loci in the cells of mice cannot provide a fair representation of the intricate nature of the disease and may not lead to the development of therapies that translate directly humans. Moreover, mouse and human cells have fundamental differences in their intracellular wiring, which lead to differences in the processes underlying tumorigenesis in these two species145 . Hence, we need to devise novel and appropriate ways to study primary human tumor xenografts in ways that preserves their CSC subpopulations in physiologic states similar to those that existed prior to the isolation from patients. The use of mice that harbor a human immune system (humanized mice146) may partly compensate for the lack of an adaptive immune system; for example, the macrophages that home to tumors growing as xenografts in such mice would be of human origin, secreting factors that may serve to model a CSC niche more similar to the one found in the human body. Ultimately, using better models, we should be able to develop a more detailed, nuanced understanding of the essential nature of CSCs and the mechanisms supporting them within human tumors.
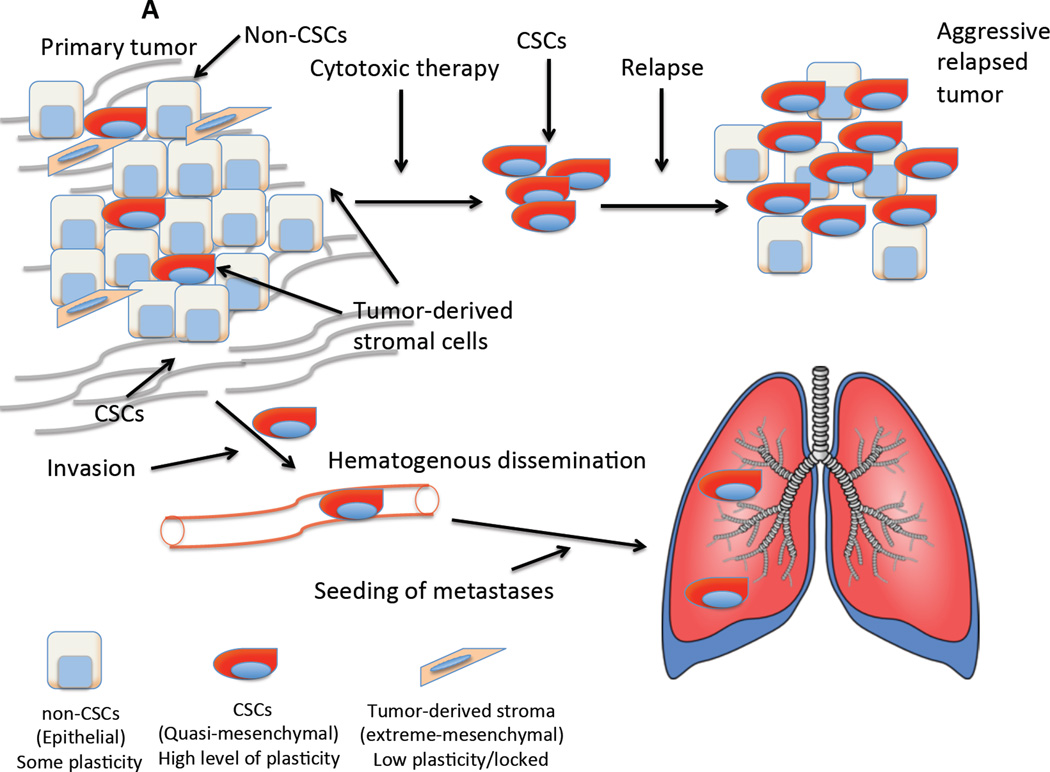
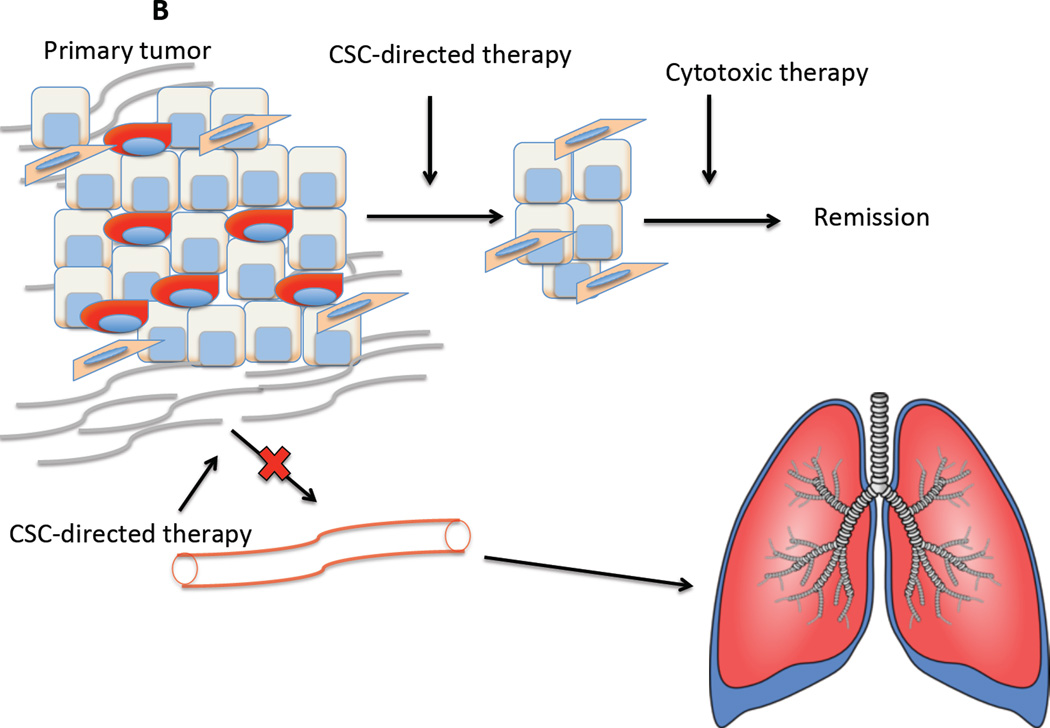
Figure 1.
Why specific targeting of CSCs is important. (A) Why specific targeting of CSCs is important: Two major issues confound our ability to treat cancers – their ability to resist cytotoxic therapy and their propensity to form metastases. Tumors are a heterogeneous mixtures of epithelial cells that consist of CSCs that have mixed epithelial-mesenchymal phenotypes and non-stem cells which are epithelial. Carcinoma cells may also be able to undergo a complete EMT, generating cells that resemble stromal-like fibroblasts and mesenchymal stem cells that may create a permissive niche. (B) The use of specific inhibitors of CSCs would attenuate their ability to survive conventional cytotoxic therapies and relapse. Loss of the mesenchymal/CSC population would also retard metastatic spread of the tumor.
Figure.
Illustration of tumor microenvironment showing all major constituents mentioned above
Acknowledgements
The authors thank Dr. Brian Bierie, Dr. Jordan Krall, Dr. Wai Leong Tam, Ms. Katharina Kober and other members of the Weinberg laboratory for helpful discussions. D.R.P was supported by a C.J. Martin Overseas Biomedical Fellowship from the National Health and Medical Research Council of Australia. The Weinberg laboratory is supported by grants from the Breast Cancer Research Foundation (BCRF), the National Institute of Health (NIH; U54-CA163109), the Samuel Waxman Cancer Research Foundation, Ludwig Center for Molecular Oncology at MIT and the Department of Defense US Army (Grant 1210095). R.A.W. is an American Cancer Society Research Professor and a Daniel K. Ludwig Cancer Research Professor
References
- 1.Valent P, et al. Cancer stem cell definitions and terminology: the devil is in the details. Nat Rev Cancer. 2012;12:767–775. doi: 10.1038/nrc3368. [DOI] [PubMed] [Google Scholar]
- 2.Wilson EB. In: The cell in development and inheritance. Osborn HF, editor. New York: The Macmillan Company; 1896. [Google Scholar]
- 3.Pierce GB, Speers WC. Tumors as caricatures of the process of tissue renewal: prospects for therapy by directing differentiation. Cancer Res. 1988;48:1996–12004. [PubMed] [Google Scholar]
- 4.Carney DN, Gazdar AF, Bunn PA, Jr, Guccion JG. Demonstration of the stem cell nature of clonogenic tumor cells from lung cancer patients. Stem Cells. 1982;1:149–164. [PubMed] [Google Scholar]
- 5.Wiseman DH, Greystoke BF, Somervaille TC. The variety of leukemic stem cells in myeloid malignancy. Oncogene. 2013 doi: 10.1038/onc.2013.269. [DOI] [PubMed] [Google Scholar]
- 6.Vermeulen L, et al. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci U S A. 2008;105:13427–13432. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 8.Stephens PJ, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Singh SK, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 12.Eramo A, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504–514. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 13.Hermann PC, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 15.Lee A, et al. Isolation of neural stem cells from the postnatal cerebellum. Nat Neurosci. 2005;8:723–729. doi: 10.1038/nn1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bussolati B, et al. Isolation of renal progenitor cells from adult human kidney. Am J Pathol. 2005;166:545–555. doi: 10.1016/S0002-9440(10)62276-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.am Esch JS, 2nd, et al. Portal application of autologous CD133+bone marrow cells to the liver: a novel concept to support hepatic regeneration. Stem Cells. 2005;23:463–470. doi: 10.1634/stemcells.2004-0283. [DOI] [PubMed] [Google Scholar]
- 18.Richardson GD, et al. CD133, a novel marker for human prostatic epithelial stem cells. J Cell Sci. 2004;117:3539–3545. doi: 10.1242/jcs.01222. [DOI] [PubMed] [Google Scholar]
- 19.Cheung AM, et al. Aldehyde dehydrogenase activity in leukemic blasts defines a subgroup of acute myeloid leukemia with adverse prognosis and superior NOD/SCID engrafting potential. Leukemia. 2007;21:1423–1430. doi: 10.1038/sj.leu.2404721. [DOI] [PubMed] [Google Scholar]
- 20.Ginestier C, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carpentino JE, et al. Aldehyde dehydrogenase-expressing colon stem cells contribute to tumorigenesis in the transition from colitis to cancer. Cancer Res. 2009;69:8208–8215. doi: 10.1158/0008-5472.CAN-09-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma S, et al. Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations. Mol Cancer Res. 2008;6:1146–1153. doi: 10.1158/1541-7786.MCR-08-0035. [DOI] [PubMed] [Google Scholar]
- 23.Jiang F, et al. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res. 2009;7:330–338. doi: 10.1158/1541-7786.MCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasheed ZA, et al. Prognostic significance of tumorigenic cells with mesenchymal features in pancreatic adenocarcinoma. J Natl Cancer Inst. 2010;102:340–351. doi: 10.1093/jnci/djp535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li R, Wu X, Wei H, Tian S. Characterization of side population cells isolated from the gastric cancer cell line SGC-7901. Oncol Lett. 2013;5:877–883. doi: 10.3892/ol.2013.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li XX, et al. Emodin as an effective agent in targeting cancer stem-like side population cells of gallbladder carcinoma. Stem Cells Dev. 2013;22:554–566. doi: 10.1089/scd.2011.0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kemper K, Grandela C, Medema JP. Molecular identification and targeting of colorectal cancer stem cells. Oncotarget. 2010;1:387–395. doi: 10.18632/oncotarget.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vermeulen L, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 31.Medema JP. Cancer stem cells: the challenges ahead. Nat Cell Biol. 2013;15:338–344. doi: 10.1038/ncb2717. [DOI] [PubMed] [Google Scholar]
- 32.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 33.Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol. 2011;27:347–376. doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- 34.Puhr M, et al. Epithelial-to-mesenchymal transition leads to docetaxel resistance in prostate cancer and is mediated by reduced expression of miR-200cand miR-205. Am J Pathol. 2012;181:2188–2201. doi: 10.1016/j.ajpath.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 35.Rhim AD, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu M, et al. Circulating Breast Tumor Cells Exhibit Dynamic Changes in Epithelial and Mesenchymal Composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 39.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morel AP, et al. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo W, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 43.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 44.Ellenrieder V, et al. Transforming growth factor beta1 treatment leads to an epithelial-mesenchymal transdifferentiation of pancreatic cancer cells requiring extracellular signal-regulated kinase 2 activation. Cancer Res. 2001;61:4222–4228. [PubMed] [Google Scholar]
- 45.Bakin AV, Rinehart C, Tomlinson AK, Arteaga CL. p38 mitogen-activated protein kinase is required for TGFbeta-mediated fibroblastic transdifferentiation and cell migration. J Cell Sci. 2002;115:3193–3206. doi: 10.1242/jcs.115.15.3193. [DOI] [PubMed] [Google Scholar]
- 46.Masszi A, et al. Integrity of cell-cell contacts is a critical regulator of TGF-beta 1-induced epithelial-to-myofibroblast transition: role for beta-catenin. Am J Pathol. 2004;165:1955–1967. doi: 10.1016/s0002-9440(10)63247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bakin AV, Tomlinson AK, Bhowmick NA, Moses HL, Arteaga CL. Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J Biol Chem. 2000;275:36803–10. doi: 10.1074/jbc.M005912200. [DOI] [PubMed] [Google Scholar]
- 48.Yook JI, Li XY, Ota I, Fearon ER, Weiss SJ. Wnt-dependent regulation of the E-cadherin repressor snail. J Biol Chem. 2005;280:11740–11748. doi: 10.1074/jbc.M413878200. [DOI] [PubMed] [Google Scholar]
- 49.Yook JI, et al. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol. 2006;8:1398–1406. doi: 10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
- 50.Scheel C, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li HJ, Reinhardt F, Herschman HR, Weinberg RA. Cancer-stimulated mesenchymal stem cells create a carcinoma stem cell niche via prostaglandin E2 signaling. Cancer discovery. 2012;2:840–855. doi: 10.1158/2159-8290.CD-12-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu S, et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66:6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bouras T, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 54.Tam WL, et al. Protein Kinase C alpha Is a Central Signaling Node and Therapeutic Target for Breast Cancer Stem Cells. Cancer Cell. 2013;24:347–364. doi: 10.1016/j.ccr.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Celia-Terrassa T, et al. Epithelial-mesenchymal transition can suppress major attributes of human epithelial tumor-initiating cells. J Clin Invest. 2012;122:1849–18468. doi: 10.1172/JCI59218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Battula VL, et al. Epithelial-mesenchymal transition-derived cells exhibit multilineage differentiation potential similar to mesenchymal stem cells. Stem Cells. 2010;28:1435–1445. doi: 10.1002/stem.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gupta PB, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kurrey NK, et al. Snail and slug mediate radioresistance and chemoresistance by antagonizing p53-mediated apoptosis and acquiring a stem-like phenotype in ovarian cancer cells. Stem Cells. 2009;27:2059–2068. doi: 10.1002/stem.154. [DOI] [PubMed] [Google Scholar]
- 59.Farmer P, et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009;15:68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
- 60.Moore N, Lyle S. Quiescent, slow-cycling stem cell populations in cancer: a review of the evidence and discussion of significance. J Oncol. 2011;2011 doi: 10.1155/2011/396076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anjomshoaa A, et al. Slow proliferation as a biological feature of colorectal cancer metastasis. Br J Cancer. 2009;101:822–828. doi: 10.1038/sj.bjc.6605229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roesch A, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pece S, et al. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 64.Feuerhake F, Sigg W, Hofter EA, Dimpfl T, Welsch U. Immunohistochemical analysis of Bcl-2 and Bax expression in relation to cell turnover and epithelial differentiation markers in the non-lactating human mammary gland epithelium. Cell Tissue Res. 2000;299:47–58. doi: 10.1007/s004419900127. [DOI] [PubMed] [Google Scholar]
- 65.Kondo T, Setoguchi T, Taga T. Persistence of a small subpopulation of cancer stem-like cells in the C6 glioma cell line. Proc Natl Acad Sci U S A. 2004;101:781–786. doi: 10.1073/pnas.0307618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou S, et al. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat Med. 2001;7:1028–1034. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
- 67.Brennan KR, Brown AM. Wnt proteins in mammary development and cancer. J Mammary Gland Biol Neoplasia. 2004;9:119–131. doi: 10.1023/B:JOMG.0000037157.94207.33. [DOI] [PubMed] [Google Scholar]
- 68.Tan AR, Alexe G, Reiss M. Transforming growth factor-beta signaling: emerging stem cell target in metastatic breast cancer? Breast Cancer Res Treat. 2009;115:453–495. doi: 10.1007/s10549-008-0184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Visnyei K, et al. A molecular screening approach to identify and characterize inhibitors of glioblastoma stem cells. Molecular cancer therapeutics. 2011;10:1818–1828. doi: 10.1158/1535-7163.MCT-11-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mezencev R, Wang L, McDonald JF. Identification of inhibitors of ovarian cancer stem-like cells by high-throughput screening. Journal of ovarian research. 2012;5:30. doi: 10.1186/1757-2215-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carmody LC, et al. Phenotypic high-throughput screening elucidates target pathway in breast cancer stem cell-like cells. Journal of biomolecular screening. 2012;17:1204–1210. doi: 10.1177/1087057112458317. [DOI] [PubMed] [Google Scholar]
- 72.Sachlos E, et al. Identification of drugs including a dopamine receptor antagonist that selectively target cancer stem cells. Cell. 2012;149:1284–1297. doi: 10.1016/j.cell.2012.03.049. [DOI] [PubMed] [Google Scholar]
- 73.Chaffer CL, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gupta PB, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 75.Mueller MM, Fusenig NE. Friends or foes - bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 76.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 77.Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li HJ, Reinhardt F, Herschman HR, Weinberg RA. Cancer-stimulated mesenchymal stem cells create a carcinoma stem cell niche via prostaglandin E2 signaling. Cancer Discov. 2012;2:840–855. doi: 10.1158/2159-8290.CD-12-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu S, et al. Breast cancer stem cells are regulated by mesenchymal stem cells through cytokine networks. Cancer Res. 2011;71:614–624. doi: 10.1158/0008-5472.CAN-10-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jinushi M, et al. Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells. Proc Natl Acad Sci U S A. 2011;108:12425–12430. doi: 10.1073/pnas.1106645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang J, et al. Tumor-associated macrophages regulate murine breast cancer stem cells through a novel paracrine EGFR/Stat3/Sox-2 signaling pathway. Stem Cells. 2013;31:248–258. doi: 10.1002/stem.1281. [DOI] [PubMed] [Google Scholar]
- 82.Marotta LL, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44(+)CD24(−) stem cell-like breast cancer cells in human tumors. J Clin Invest. 2011;121:2723–2735. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ma X, et al. A prostaglandin E (PGE) receptor EP4 antagonist protects natural killer cells from PGE-mediated immunosuppression and inhibits breast cancer metastasis. Oncoimmunology. 2013;2:e22647. doi: 10.4161/onci.22647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lin L, et al. The STAT3 inhibitor NSC 74859 is effective in hepatocellular cancers with disrupted TGF-beta signaling. Oncogene. 2009;28:961–972. doi: 10.1038/onc.2008.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pinto D, Gregorieff A, Begthel H, Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Booth C, Brady G, Potten CS. Crowd control in the crypt. Nat Med. 2002;8:1360–1361. doi: 10.1038/nm1202-1360. [DOI] [PubMed] [Google Scholar]
- 87.Birnbaum T, et al. Malignant gliomas actively recruit bone marrow stromal cells by secreting angiogenic cytokines. J Neurooncol. 2007;83:241–247. doi: 10.1007/s11060-007-9332-4. [DOI] [PubMed] [Google Scholar]
- 88.Lin EY, Nguyen AV, Russell RG, Pollard JW. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med. 2001;193:727–740. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Green CE, et al. Chemoattractant signaling between tumor cells and macrophages regulates cancer cell migration, metastasis and neovascularization. PLoS One. 2009;4:e6713. doi: 10.1371/journal.pone.0006713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sica A, Allavena P, Mantovani A. Cancer related inflammation: the macrophage connection. Cancer Lett. 2008;267:204–215. doi: 10.1016/j.canlet.2008.03.028. [DOI] [PubMed] [Google Scholar]
- 91.MacDonald KP, et al. An antibody against the colony-stimulating factor 1 receptor depletes the resident subset of monocytes and tissue- and tumor-associated macrophages but does not inhibit inflammation. Blood. 2010;116:3955–3963. doi: 10.1182/blood-2010-02-266296. [DOI] [PubMed] [Google Scholar]
- 92.DeNardo DG, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011;1:54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 94.Emami KH, et al. A small molecule inhibitor of beta-catenin/CREB-binding protein transcription [corrected] Proc Natl Acad Sci U S A. 2004;101:12682–12687. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen B, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5:100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu J, et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc Natl Acad Sci U S A. 2013;110:20224–20229. doi: 10.1073/pnas.1314239110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rosenbluh J, et al. beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell. 2012;151:1457–1473. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 99.Katz LH, et al. Targeting TGF-beta signaling in cancer. Expert Opin Ther Targets. 2013 doi: 10.1517/14728222.2013.782287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tremblay MR, et al. Discovery of a potent and orally active hedgehog pathway antagonist (IPI-926) J Med Chem. 2009;52:4400–4418. doi: 10.1021/jm900305z. [DOI] [PubMed] [Google Scholar]
- 101.Jimeno A, et al. Phase I Study of the Hedgehog Pathway Inhibitor IPI-926 in Adult Patients with Solid Tumors. Clin Cancer Res. 2013;19:2766–2774. doi: 10.1158/1078-0432.CCR-12-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Graham RA, et al. Pharmacokinetics of hedgehog pathway inhibitor vismodegib (GDC-0449) in patients with locally advanced or metastatic solid tumors: the role of alpha-1-acid glycoprotein binding. Clin Cancer Res. 2011;17:2512–2520. doi: 10.1158/1078-0432.CCR-10-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.LoRusso PM, et al. Phase I trial of hedgehog pathway inhibitor vismodegib (GDC-0449) in patients with refractory, locally advanced or metastatic solid tumors. Clin Cancer Res. 2011;17:2502–2511. doi: 10.1158/1078-0432.CCR-10-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Krop I, et al. Phase I pharmacologic and pharmacodynamic study of the gamma secretase (Notch) inhibitor MK-0752 in adult patients with advanced solid tumors. J Clin Oncol. 2012;30:2307–2313. doi: 10.1200/JCO.2011.39.1540. [DOI] [PubMed] [Google Scholar]
- 105.Ridgway J, et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444:1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- 106.Hoey T, et al. DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell Stem Cell. 2009;5:168–177. doi: 10.1016/j.stem.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 107.Warrell RP, Jr, de The H, Wang ZY, Degos L. Acute promyelocytic leukemia. N Engl J Med. 1993;329:177–189. doi: 10.1056/NEJM199307153290307. [DOI] [PubMed] [Google Scholar]
- 108.Carey JO, Posekany KJ, deVente JE, Pettit GR, Ways DK. Phorbol ester-stimulated phosphorylation of PU.1: association with leukemic cell growth inhibition. Blood. 1996;87:4316–4324. [PubMed] [Google Scholar]
- 109.Wu H, et al. Reduction in lactate accumulation correlates with differentiation-induced terminal cell division of leukemia cells. Differentiation. 1991;48:51–58. doi: 10.1111/j.1432-0436.1991.tb00242.x. [DOI] [PubMed] [Google Scholar]
- 110.Arcangeli A, et al. Polar/apolar compounds induce leukemia cell differentiation by modulating cell-surface potential. Proc Natl Acad Sci U S A. 1993;90:5858–5862. doi: 10.1073/pnas.90.12.5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Olsson I, Gullberg U, Ivhed I, Nilsson K. Induction of differentiation of the human histiocytic lymphoma cell line U-937 by 1 alpha,25-dihydroxycholecalciferol. Cancer Res. 1983;43:5862–5867. [PubMed] [Google Scholar]
- 112.Munster PN, et al. The histone deacetylase inhibitor suberoylanilide hydroxamic acid induces differentiation of human breast cancer cells. Cancer Res. 2001;61:8492–8497. [PubMed] [Google Scholar]
- 113.Uchida H, Maruyama T, Nagashima T, Asada H, Yoshimura Y. Histone deacetylase inhibitors induce differentiation of human endometrial adenocarcinoma cells through up-regulation of glycodelin. Endocrinology. 2005;146:5365–5373. doi: 10.1210/en.2005-0359. [DOI] [PubMed] [Google Scholar]
- 114.Momparler RL. Epigenetic therapy of cancer with 5-aza-2'-deoxycytidine (decitabine) Semin Oncol. 2005;32:443–451. doi: 10.1053/j.seminoncol.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 115.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased inhuman cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cao Q, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27:7274–7284. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 118.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Krivtsov AV, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 120.Bernt KM, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Daigle SR, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bhola NE, et al. TGF-beta inhibition enhances chemotherapy action against triple-negative breast cancer. J Clin Invest. 2013;123:1348–1358. doi: 10.1172/JCI65416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ocana OH, et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22:709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 124.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22:725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271:1734–1736. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 128.Yoshimura A, Muto G. TGF-beta function in immune suppression. Curr Top Microbiol Immunol. 2011;350:127–147. doi: 10.1007/82_2010_87. [DOI] [PubMed] [Google Scholar]
- 129.Wan YY, Flavell RA. 'Yin-Yang' functions of transforming growth factor-beta and T regulatory cells in immune regulation. Immunol Rev. 2007;220:199–213. doi: 10.1111/j.1600-065X.2007.00565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. J Gen Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tennant DA, Duran RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 132.Elstrom RL, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 133.Ji H, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 134.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 135.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gross S, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207:339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wouters BG, Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat Rev Cancer. 2008;8:851–864. doi: 10.1038/nrc2501. [DOI] [PubMed] [Google Scholar]
- 141.Cannito S, et al. Redox mechanisms switch on hypoxia-dependent epithelial-mesenchymal transition in cancer cells. Carcinogenesis. 2008;29:2267–2278. doi: 10.1093/carcin/bgn216. [DOI] [PubMed] [Google Scholar]
- 142.Li Z, Rich JN. Hypoxia and hypoxia inducible factors in cancer stem cell maintenance. Curr Top Microbiol Immunol. 2010;345:21–30. doi: 10.1007/82_2010_75. [DOI] [PubMed] [Google Scholar]
- 143.Dong C, et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 2013;23:316–331. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ginestier C, et al. Mevalonate metabolism regulates Basal breast cancer stem cells and is a potential therapeutic target. Stem Cells. 2012;30:1327–1337. doi: 10.1002/stem.1122. [DOI] [PubMed] [Google Scholar]
- 145.Rangarajan A, Weinberg RA. Opinion: Comparative biology of mouse versus human cells: modelling human cancer in mice. Nat Rev Cancer. 2003;3:952–959. doi: 10.1038/nrc1235. [DOI] [PubMed] [Google Scholar]
- 146.Hiramatsu H, et al. Complete reconstitution of human lymphocytes from cord blood CD34+ cells using the NOD/SCID/gammacnull mice model. Blood. 2003;102:873–880. doi: 10.1182/blood-2002-09-2755. [DOI] [PubMed] [Google Scholar]
- 147.Civin CI, et al. Antigenic analysis of hematopoiesis. III A hematopoietic progenitor cell surface antigen defined by a monoclonal antibody raised against KG-1a cells. J Immunol. 1984;133:157–165. [PubMed] [Google Scholar]
- 148.Andrews RG, Singer JW, Bernstein ID. Precursors of colony-forming cells in humans can be distinguished from colony-forming cells by expression of the CD33 and CD34 antigens and light scatter properties. J Exp Med. 1989;169:1721–1731. doi: 10.1084/jem.169.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lin G, Finger E, Gutierrez-Ramos JC. Expression of CD34 in endothelial cells, hematopoietic progenitors and nervous cells in fetal and adult mouse tissues. Eur J Immunol. 1995;25:1508–1516. doi: 10.1002/eji.1830250606. [DOI] [PubMed] [Google Scholar]
- 150.Healy L, et al. The stem cell antigen CD34 functions as a regulator of hemopoietic cell adhesion. Proc Natl Acad Sci U S A. 1995;92:12240–12244. doi: 10.1073/pnas.92.26.12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lapidot T, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 152.Ferrero E, Malavasi F. The metamorphosis of a molecule: from soluble enzyme to the leukocyte receptor CD38. J Leukoc Biol. 1999;65:151–161. doi: 10.1002/jlb.65.2.151. [DOI] [PubMed] [Google Scholar]
- 153.Borland G, Ross JA, Guy K. Forms and functions of CD44. Immunology. 1998;93:139–148. doi: 10.1046/j.1365-2567.1998.00431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 155.Dalerba P, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl AcadSci U S A. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Takaishi S, et al. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells. 2009;27:1006–1020. doi: 10.1002/stem.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Prince ME, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007;104:973–8. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yang ZF, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 159.Li C, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 160.Kristiansen G, Sammar M, Altevogt P. Tumour biological aspects of CD24, a mucin-like adhesion molecule. J Mol Histol. 2004;35:255–262. doi: 10.1023/b:hijo.0000032357.16261.c5. [DOI] [PubMed] [Google Scholar]
- 161.Zhang C, Li C, He F, Cai Y, Yang H. Identification of CD44+CD24+ gastric cancer stem cells. J Cancer Res Clin Oncol. 2011;137:1679–1686. doi: 10.1007/s00432-011-1038-5. [DOI] [PubMed] [Google Scholar]
- 162.McKenzie JL, Fabre JW. Human thy-1: unusual localization and possible functional significance in lymphoid tissues. J Immunol. 1981;126:843–850. [PubMed] [Google Scholar]
- 163.Mayani H, Lansdorp PM. Thy-1 expression is linked to functional properties of primitive hematopoietic progenitor cells from human umbilical cord blood. Blood. 1994;83:2410–2417. [PubMed] [Google Scholar]
- 164.Dominici M, et al. Minimal criteria for defining multipotent mesenchymal stromal cells The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 165.Wetzel A, et al. Human Thy-1 (CD90) on activated endothelial cells is a counterreceptor for the leukocyte integrin Mac-1 (CD11b/CD18) J Immunol. 2004;172:3850–3859. doi: 10.4049/jimmunol.172.6.3850. [DOI] [PubMed] [Google Scholar]
- 166.Avalos AM, Labra CV, Quest AF, Leyton L. Signaling triggered by Thy-1 interaction with beta 3 integrin on astrocytes is an essential step towards unraveling neuronal Thy-1 function. Biol Res. 2002;35:231–238. doi: 10.4067/s0716-97602002000200015. [DOI] [PubMed] [Google Scholar]
- 167.Rege TA, Hagood JS. Thy-1, a versatile modulator of signaling affecting cellular adhesion, proliferation, survival, and cytokine/growth factor responses. Biochim Biophys Acta. 2006;1763:991–999. doi: 10.1016/j.bbamcr.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kroczek RA, Gunter KC, Germain RN, Shevach EM. Thy-1 functions as a signal transduction molecule in T lymphocytes and transfected B lymphocytes. Nature. 1986;322:181–184. doi: 10.1038/322181a0. [DOI] [PubMed] [Google Scholar]
- 169.He J, et al. CD90 is identified as a candidate marker for cancer stem cells in primary high-grade gliomas using tissue microarrays. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.010744. M111 010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yang ZF, et al. Identification of local and circulating cancer stem cells in human liver cancer. Hepatology. 2008;47:919–928. doi: 10.1002/hep.22082. [DOI] [PubMed] [Google Scholar]
- 171.Donnenberg VS, Landreneau RJ, Donnenberg AD. Tumorigenic stem and progenitor cells: implications for the therapeutic index of anti-cancer agents. J Control Release. 2007;122:385–91. doi: 10.1016/j.jconrel.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Buhring HJ, et al. Expression of novel surface antigens on early hematopoietic cells. Ann N Y Acad Sci. 1999;872:25–38. doi: 10.1111/j.1749-6632.1999.tb08450.x. discussion 38–9. [DOI] [PubMed] [Google Scholar]
- 173.Gehling UM, et al. In vitro differentiation of endothelial cells from AC133-positive progenitor cells. Blood. 2000;95:3106–3112. [PubMed] [Google Scholar]
- 174.Uchida N, et al. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci U S A. 2000;97:14720–14725. doi: 10.1073/pnas.97.26.14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 176.Ricci-Vitiani L, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 177.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 178.Rutella S, et al. Cells with characteristics of cancer stem/progenitor cells express the CD133 antigen in human endometrial tumors. Clin Cancer Res. 2009;15:4299–4311. doi: 10.1158/1078-0432.CCR-08-1883. [DOI] [PubMed] [Google Scholar]
- 179.Ma S, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–2556. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 180.Bertolini G, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009;106:16281–16286. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ferrandina G, et al. Expression of CD133–1 and CD133–2 in ovarian cancer. Int J Gynecol Cancer. 2008;18:506–514. doi: 10.1111/j.1525-1438.2007.01056.x. [DOI] [PubMed] [Google Scholar]
- 182.Deng S, et al. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS One. 2010;5:e10277. doi: 10.1371/journal.pone.0010277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4:697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Black W, Vasiliou V. The aldehyde dehydrogenase gene superfamily resource center. Hum Genomics. 2009;4:136–142. doi: 10.1186/1479-7364-4-2-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Huang EH, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69:3382–3389. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chen YC, et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem Biophys Res Commun. 2009;385:307–313. doi: 10.1016/j.bbrc.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 187.Boonyaratanakornkit JB, et al. Selection of tumorigenic melanoma cells using ALDH. J Invest Dermatol. 2010;130:2799–2808. doi: 10.1038/jid.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Alvi AJ, et al. Functional and molecular characterisation of mammary side population cells. Breast Cancer Res. 2003;5:R1–R8. doi: 10.1186/bcr547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Majka SM, et al. Identification of novel resident pulmonary stem cells: form and function of the lung side population. Stem Cells. 2005;23:1073–1081. doi: 10.1634/stemcells.2005-0039. [DOI] [PubMed] [Google Scholar]
- 191.Martin CM, et al. Persistent expression of the ATP-binding cassette transporter, Abcg2, identifies cardiac SP cells in the developing and adult heart. Dev Biol. 2004;265:262–275. doi: 10.1016/j.ydbio.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 192.Hussain SZ, et al. Side population cells derived from adult human liver generate hepatocyte-like cells in vitro. Dig DisSci. 2005;50:1755–1763. doi: 10.1007/s10620-005-2933-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kim M, Morshead CM. Distinct populations of forebrain neural stem and progenitor cells can be isolated using side-population analysis. J Neurosci. 2003;23:10703–10709. doi: 10.1523/JNEUROSCI.23-33-10703.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Larderet G, et al. Human side population keratinocytes exhibit long-term proliferative potential and a specific gene expression profile and can form a pluristratified epidermis. Stem Cells. 2006;24:965–974. doi: 10.1634/stemcells.2005-0196. [DOI] [PubMed] [Google Scholar]
- 195.Chua C, et al. Characterization of a side population of astrocytoma cells in response to temozolomide. J Neurosurg. 2008;109:856–866. doi: 10.3171/JNS/2008/109/11/0856. [DOI] [PubMed] [Google Scholar]
- 196.Haraguchi N, et al. Characterization of a side population of cancer cells from human gastrointestinal system. Stem Cells. 2006;24:506–513. doi: 10.1634/stemcells.2005-0282. [DOI] [PubMed] [Google Scholar]
- 197.Bleau AM, et al. PTEN/PI3K/Akt pathway regulates the side population phenotype and ABCG2 activity in glioma tumor stem-like cells. Cell Stem Cell. 2009;4:226–235. doi: 10.1016/j.stem.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chiba T, et al. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. Hepatology. 2006;44:240–251. doi: 10.1002/hep.21227. [DOI] [PubMed] [Google Scholar]
- 199.Ho MM, Ng AV, Lam S, Hung JY. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007;67:4827–4833. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- 200.Mitsutake N, et al. Characterization of side population in thyroid cancer cell lines: cancer stem-like cells are enriched partly but not exclusively. Endocrinology. 2007;148:1797–1803. doi: 10.1210/en.2006-1553. [DOI] [PubMed] [Google Scholar]
- 201.Malanchi I, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2012;481:85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- 202.Yu M, et al. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 2012;487:510–513. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gonsalves FC, et al. An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. Proc Natl Acad Sci U S A. 2011;108:5954–5963. doi: 10.1073/pnas.1017496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhu C, et al. Phosphosulindac (OXT-328) selectively targets breast cancer stem cells in vitro and in human breast cancer xenografts. Stem Cells. 2012;30:2065–2075. doi: 10.1002/stem.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Huang SM, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 206.Garber K. Drugging the Wnt pathway: problems and progress. J Natl Cancer Inst. 2009;101:548–550. doi: 10.1093/jnci/djp084. [DOI] [PubMed] [Google Scholar]
- 207.Heidel FH, et al. Genetic and pharmacologic inhibition of beta-catenin targets imatinib-resistant leukemia stem cells in CML. Cell Stem Cell. 2012;10:412–424. doi: 10.1016/j.stem.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Yeung J, et al. beta-Catenin mediates the establishment and drug resistance of MLL leukemic stem cells. Cancer Cell. 2010;18:606–618. doi: 10.1016/j.ccr.2010.10.032. [DOI] [PubMed] [Google Scholar]
- 209.Malanchi I, et al. Cutaneous cancer stem cell maintenance is dependent on beta-catenin signalling. Nature. 2008;452:650–653. doi: 10.1038/nature06835. [DOI] [PubMed] [Google Scholar]
- 210.Bueno L, et al. Semi-mechanistic modelling of the tumour growth inhibitory effects of LY2157299, a new type I receptor TGF-beta kinase antagonist, in mice. Eur J Cancer. 2008;44:142–150. doi: 10.1016/j.ejca.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 211.Anido J, et al. TGF-beta Receptor Inhibitors Target the CD44(high)/Id1(high) Glioma-Initiating Cell Population in Human Glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 212.Kabashima A, et al. Side population of pancreatic cancer cells predominates in TGF-beta-mediated epithelial to mesenchymal transition and invasion. Int J Cancer. 2009;124:2771–2779. doi: 10.1002/ijc.24349. [DOI] [PubMed] [Google Scholar]
- 213.Trachtman H, et al. A phase, psingle-dose study of fresolimumab, an anti-TGF-beta antibody, in treatment-resistant primary focal segmental glomerulosclerosis. Kidney Int. 2011;79:1236–1243. doi: 10.1038/ki.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Schlingensiepen KH, et al. Targeted tumor therapy with the TGF-beta 2 antisense compound AP 12009. Cytokine Growth Factor Rev. 2006;17:129–139. doi: 10.1016/j.cytogfr.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 215.Olson RE, Albright CF. Recent progress in the medicinal chemistry of gamma-secretase inhibitors. Curr Top Med Chem. 2008;8:17–33. doi: 10.2174/156802608783334088. [DOI] [PubMed] [Google Scholar]
- 216.Lanz TA, et al. Pharmacodynamics and pharmacokinetics of the gamma-secretase inhibitor PF-3084014. J Pharmacol Exp Ther. 2010;334:269–277. doi: 10.1124/jpet.110.167379. [DOI] [PubMed] [Google Scholar]
- 217.Richter S, et al. A phase I study of the oral gamma secretase inhibitor R04929097 in combination with gemcitabine in patients with advanced solid tumors (PHL-078/CTEP 8575) Invest New Drugs. 2013 doi: 10.1007/s10637-013-9965-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Harrison H, et al. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 2010;70:709–718. doi: 10.1158/0008-5472.CAN-09-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.McAuliffe SM, et al. Targeting Notch, a key pathway for ovarian cancer stem cells, sensitizes tumors to platinum therapy. Proc Natl Acad Sci U S A. 2012;109:E2939–E2948. doi: 10.1073/pnas.1206400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zhu TS, et al. Endothelial cells create a stem cell niche in glioblastoma by providing NOTCH ligands that nurture self-renewal of cancer stem-like cells. Cancer Res. 2011;71:6061–6072. doi: 10.1158/0008-5472.CAN-10-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Palagani V, et al. Epithelial mesenchymal transition and pancreatic tumor initiating CD44+/EpCAM+ cells are inhibited by gamma-secretase inhibitor IX. PLoS One. 2012;7:e46514. doi: 10.1371/journal.pone.0046514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Jenkins DW, et al. MEDI0639: a novel therapeutic antibody targeting Dll4 modulates endothelial cell function and angiogenesis in vivo. Mol Cancer Ther. 2012;11:1650–1660. doi: 10.1158/1535-7163.MCT-11-1027. [DOI] [PubMed] [Google Scholar]
- 223.Cooper MK, Porter JA, Young KE, Beachy PA. Teratogen-mediated inhibition of target tissue response to Shh signaling. Science. 1998;280:1603–1607. doi: 10.1126/science.280.5369.1603. [DOI] [PubMed] [Google Scholar]
- 224.Robarge KD, et al. GDC-0449-a potent inhibitor of the hedgehog pathway. Bioorg Med Chem Lett. 2009;19:5576–5581. doi: 10.1016/j.bmcl.2009.08.049. [DOI] [PubMed] [Google Scholar]
- 225.Buonamici S, et al. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci Transl Med. 2010;2:51ra70. doi: 10.1126/scitranslmed.3001599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Bar EE, et al. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells. 2007;25:2524–2533. doi: 10.1634/stemcells.2007-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Feldmann G, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–2196. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Dierks C, et al. Expansion of Bcr-Abl-positive leukemic stem cells is dependent on Hedgehog pathway activation. Cancer Cell. 2008;14:238–249. doi: 10.1016/j.ccr.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 230.Peacock CD, et al. Hedgehog signaling maintains a tumor stem cell compartment in multiple myeloma. Proc Natl Acad Sci U S A. 2007;104:4048–4053. doi: 10.1073/pnas.0611682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Wallner L, et al. Inhibition of interleukin-6 with CNTO328, an anti-interleukin-6 monoclonal antibody, inhibits conversion of androgen-dependent prostate cancer to an androgen-independentphenotype in orchiectomized mice. Cancer Res. 2006;66:3087–3095. doi: 10.1158/0008-5472.CAN-05-3447. [DOI] [PubMed] [Google Scholar]
- 232.Nakashima Y, et al. Clinical evaluation of tocilizumab for patients with active rheumatoid arthritis refractory to anti-TNF biologics: tocilizumab in combination with methotrexate. Mod Rheumatol. 2010;20:343–352. doi: 10.1007/s10165-010-0290-x. [DOI] [PubMed] [Google Scholar]
- 233.Hedvat M, et al. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell. 2009;16:487–497. doi: 10.1016/j.ccr.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Song H, Wang R, Wang S, Lin J. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc Natl Acad Sci U S A. 2005;102:4700–4705. doi: 10.1073/pnas.0409894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Debnath B, Xu S, Neamati N. Small molecule inhibitors of signal transducer and activator of transcription 3 (Stat3) protein. J Med Chem. 2012;55:6645–6668. doi: 10.1021/jm300207s. [DOI] [PubMed] [Google Scholar]
- 236.Sherry MM, Reeves A, Wu JK, Cochran BH. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27:2383–2392. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Lin L, et al. STAT3 is necessary for proliferation and survival in colon cancer-initiating cells. Cancer Res. 2011;71:7226–7237. doi: 10.1158/0008-5472.CAN-10-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Kroon P, et al. JAK-STAT blockade inhibits tumor initiation and clonogenic recovery of prostate cancer stem-like cells. Cancer Res. 2013;73:5288–5298. doi: 10.1158/0008-5472.CAN-13-0874. [DOI] [PubMed] [Google Scholar]
- 239.Toullec D, et al. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J Biol Chem. 1991;266:15771–15781. [PubMed] [Google Scholar]
- 240.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 241.Bapat SA, Mali AM, Koppikar CB, Kurrey NK. Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res. 2005;65:3025–3029. doi: 10.1158/0008-5472.CAN-04-3931. [DOI] [PubMed] [Google Scholar]
- 242.Kelly PN, Dakic A, Adams JM, Nutt SL, Strasser A. Tumor growth need not be driven by rare cancer stem cells. Science. 2007;317:337. doi: 10.1126/science.1142596. [DOI] [PubMed] [Google Scholar]
- 243.Quintana E, et al. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Deshpande AJ, et al. Acute myeloid leukemia is propagated by a leukemic stem cell with lymphoid characteristics in a mouse model of CALM/AF10-positive leukemia. Cancer Cell. 2006;10:363–374. doi: 10.1016/j.ccr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 245.Cho RW, et al. Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1 murine breast tumors. Stem Cells. 2008;26:364–371. doi: 10.1634/stemcells.2007-0440. [DOI] [PubMed] [Google Scholar]
- 246.Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488:527–530. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 248.Schepers AG, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 249.Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol. 2006;6:295–307. doi: 10.1038/nri1806. [DOI] [PubMed] [Google Scholar]
- 250.Restifo NP, Dudley ME, Rosenberg SA. Adoptive immunotherapy for cancer: harnessing the T cell response. Nat Rev Immunol. 2012;12:269–281. doi: 10.1038/nri3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4:71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 252.DeNardo DG, et al. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Quante M, et al. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell. 2011;19:257–272. doi: 10.1016/j.ccr.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Dirat B, et al. Cancer-associated adipocytes exhibit an activated phenotype and contribute to breast cancer invasion. Cancer Res. 2011;71:2455–2465. doi: 10.1158/0008-5472.CAN-10-3323. [DOI] [PubMed] [Google Scholar]
- 255.Butler JM, Kobayashi H, Rafii S. Instructive role of the vascular niche in promoting tumour growth and tissue repair by angiocrine factors. Nat Rev Cancer. 2010;10:138–146. doi: 10.1038/nrc2791. [DOI] [PMC free article] [PubMed] [Google Scholar]