Abstract
Smoking and obesity are each well-established risk factors for cardiovascular heart disease, which together impose earlier onset and greater severity of disease. To identify early signaling events in the response of the heart to cigarette smoke exposure within the setting of obesity, we exposed normal weight and high fat diet-induced obese (DIO) C57BL/6 mice to repeated inhaled doses of mainstream (MS) or sidestream (SS) cigarette smoke administered over a two week period, monitoring effects on both cardiac and pulmonary transcriptomes. MS smoke (250 µg wet total particulate matter (WTPM)/L, 5h/day) exposures elicited robust cellular and molecular inflammatory responses in the lung with 1466 differentially expressed pulmonary genes (p<0.01) in normal weight animals, and a muchattenuated response (463 genes) in the hearts of the same animals. In contrast, exposures to SS smoke (85 µg WTPM/L) with an equivalent CO concentration as that of MS smoke (~250 CO ppm), induced a weak pulmonary response (328 genes), but an extensive cardiac response (1590 genes). SS smoke, and to a lesser extent MS smoke preferentially elicited hypoxia- and stress-responsive genes as well as genes predicting early changes of vascular smooth muscle and endothelium, precursors of cardiovascular disease. The most sensitive smoke-induced cardiac transcriptional changes of normal weight mice were largely absent in DIO mice after smoke exposure, while genes involved in fatty acid utilization were unaffected. At the same time, smoke exposure suppressed multiple proteome maintenance genes induced in the hearts of DIO mice. Together these results underscore the sensitivity of the heart to SS smoke and reveal adaptive responses in healthy individuals that are absent in the setting of high fat diet and obesity.
Keywords: heart transcriptome, mainstream cigarette smoke, sidestream cigarette smoke, hypoxia, peroxisomal proliferator-activated receptor-α, high fat diet, obesity
Obesity and smoking each represent major risk factors for premature death from heart disease, particularly cardiovascular disease (CVD) and ischemia, with multiplicative increases in disease risk when both risk factors are present. 1–5 Exposure to second hand smoke by nonsmokers also constitutes a significant risk factor. Second hand smoke is predominantly comprised of sidestream (SS) smoke emitted from the burning tip of the cigarette (85%), with the remainder consisting of exhaled mainstream (MS) smoke. 1, 2, 5, 6 Both types of smoke are comprised qualitatively of the same complex mixture of over 4,000 chemical constituents, but many toxins (e.g., CO, nicotine, acrolein, formaldehyde, and polycyclic aromatic hydrocarbons) are increased as much as 100-fold in SS smoke relative to MS smoke, suggesting the greater toxicity of SS smoke. 7 In agreement with this suggestion, active smoking approximately doubles the risk of CVD whereas second hand smoke exposure (typically occurring at 1/100th the dose) increases this risk by 30%, i.e., a 15-fold greater disease risk from second hand smoke. 1, 2, 8, 9
Research has highlighted several specific physiological effects associated with cigarette smoking that may contribute to both acute and chronic myocardial events. These include sympathetic nerve activation, which increases both blood pressure and myocardial contractility, as well as variability in heart rate. 1, 2, 5 An immediate effect is increased cardiac afterload, which elevates the heart’s oxygen demand at the same time that the high levels of CO in cigarette smoke result in hypoxia that limits oxygen delivery to the heart. Smoking also promotes a pro-thrombotic status within the cardiovasculature through increased blood platelet activation and aggregation, endothelial dysfunction and injury, reduced fibrinolysis, and an altered profile of circulating lipids (increased levels of oxidized LDL and decreased levels of HDL); all of these characteristics may contribute to the development of atherosclerosis and the increased likelihood of myocardial infarction.
A number of these characteristics, also appear in human and animal models of obesity, but the molecular signals that initiate these changes are not well understood, nor how obesity may alter the heart’s response to cigarette smoke exposure. 10, 11 It has been proposed that obesity predisposes peripheral organs to inflammation as a result of pro-inflammatory cytokine and adipokine secretion from adipose tissue. 12–14 An additional factor is the large metabolic needs of the heart for both sufficient energy substrates and oxygen to fuel contraction; smoking limits oxygen delivery to the heart, whereas fatty acid oxidation requires high oxygen. High fat diets, common in obesity, exaggerate the normal cardiac preference for fats as fuel, where fatty acids may supply 60–90% of total ATP with the remainder supplied by glucose. Continual myocardial contraction, which requires immense amounts (kg/day in humans) of ATP, also requires substantial metabolic flexibility to accommodate changes in availability of both oxygen and oxidizable fuels.15, 16 This energetic versatility is provided by several transcription factors, primarily (i.) peroxisome proliferator activated-receptor-alpha (PPARα), a master regulator of genes for uptake and utilization of fatty acids and glucose, and (ii.) hypoxia inducible factor-1 alpha (HIF-1α), which controls genes that minimize oxygen consumption. 17, 18 A significant loss in the ability to switch between metabolisms that utilize glucose (consuming low oxygen) or fatty acids (with high oxygen consumption) is a hallmark feature of the failing heart, in which ATP levels are progressively reduced eventually resulting in energy starvation and inability to adequately perfuse the body. Deficits in metabolic flexibility of the heart have also been observed in obesity and diabetes.16 Thus, the limited oxygen supply imposed by smoking may be further compromised by the increased oxygen needs of a heart relying on a high fat diet.
To examine cardiac responses to smoking and obesity, whole genome microarrays were used to compare transcriptomes of normal weight with high fat diet-induced obese (DIO) C57BL/6 mice after administration of a regime of two weeks of repeated exposures to inhaled MS or SS cigarette smoke. Comparing cardiac transcriptional responses with those in the lungs of the same animals indicated a greater sensitivity of the heart to SS smoke as compared with the lung or with MS smoke in the heart. Moreover, the cardiac response to smoke included a large proportion of genes involved in hypoxic and stress response in non-obese mice, which was substantially suppressed in smoke-exposed DIO mice.
EXPERIMENTAL PROCEDURES
Animals
Non-obese (i.e., regular weight, RW) and diet-induced obese (DIO) male C57BL/6 mice at 13 weeks of age were purchased from Jackson Laboratories (Bar Harbor, ME). RW mice were fed throughout the study a diet consisting of pelleted PMI® 5002 Certified Rodent Diet (Richmond, IN) containing 5kcal% fat; DIO mice were fed a high calorie/high fat (60kal% fat) diet consisting of pelleted D12492 Rodent Diet (Research Diets Inc., New Brunswick, NJ) starting at 6 weeks of age for a total of nine weeks prior to the start of the 2-week exposures (i.e., a total of an 11-week high fat diet). Animals were acclimated to the AALAS-accredited animal facility as well as to the nose-only inhalation exposure restraint tubes for one week prior to the initiation of smoke exposures. Feed and water were provided ad libitum; animal rooms set to a 12 h light cycle. Health screens were performed on sentinel mice; terminal health monitoring included serological testing for antibodies to common rodent pathogens.
Experimental design
Groups of RW and DIO C57BL/6 mice were exposed to HEPA filtered air (sham controls, SC), mainstream (MS), or sidestream (SS) cigarette smoke by nose-only inhalation exposure for 5 hr/day for a total of 8 exposures over two weeks as follows: 5 consecutive days of exposure followed by 2 days with no exposure, then three days of exposure, with necropsies occurring the day following the last exposure. Target cigarette smoke exposure concentrations were 250 µg wet total particulate matter per liter (WTPM/L) of air for the MS exposures and 85 µg WTPM/L for the SS exposures, based on previous short-term MS cigarette exposure studies used to establish the tolerance of RW and DIO mice to SS smoke, which has lower particulate concentrations but higher levels of acutely toxic volatile components. Based on these results, CO concentrations were considered a limiting factor in the ability of the animals to tolerate the two-week exposure regimen without significant toxicity (i.e., over 10% loss in body weight or lethality) and thus, experimental MS and SS smoke exposures were set with equivalent CO concentrations (~250 ppm). A total of 144 mice divided into three 48-mice cohorts (8 mice per treatment group) were used for the purpose of collecting parallel biological samples for specific analyses: i.) heart and lung tissues for gene expression studies, ii.) bronchoalveolar lavage (BAL) fluid for cytology, and iii.) blood for post-exposure carboxyhemoglobin (CO-Hb) measurements. Necropsies for all animals were completed within a few hours of the light period, thereby minimizing any measured changes due to circadian rhythms.
Tissue and blood collection
On the day after the last exposure, mice from one cohort were euthanized with pentobarbital and exsanguinated prior to tissue collection. The left half of each organ refrigerated in cold RNAlater® overnight, then removed from fixative and stored at −80°C with the remainder (right heart and lung) flash-frozen and stored at −80°C for future analyses. Before analyses, RNA sample quality was evaluated for fragmentation from RNA gels as well as 28S:18S ratios. Immediately following the last exposure, mice from a separate cohort were anesthetized with isoflurane and blood collected for CO-Hb determinations using an OSM3 hemoximeter (Radiometer, Copenhagen, Denmark).
BAL fluid collection and cytology
BAL fluid was collected from isolated lungs as described previously.19–21 Cytological evaluations (viability, cell count and differentials) were performed on 50 µL of cell suspension, performing cell differentials on trypan blue-excluding (viable) cells. Three hundred nucleated white blood cells were counted to determine the number of alveolar macrophages, neutrophils, lymphocytes and eosinophils. Statistical significance was calculated by 2-factor ANOVA with a Tukey’s adjusted p-value for all pairwise comparisons.
Gene expression and pathway analysis
Whole genome microarray analysis using Affymetrix Mouse Genome 430A 2.0 chips (Affymetrix, Santa Clara, CA, USA; 22,690 probesets) was performed according to manufacturer’s instructions. Raw intensity data were quantile normalized by Robust Multi-Array Analysis (RMA) summarization,22 log2 transformed to RW-SC, and subjected to ANOVA 23 with Tukey’s multiple corrections test (p<0.01) and 5% false discovery rate calculation24 using GeneSpring v.11 (Silicon Genetics, Redwood City, CA). Functional enrichment statistics and network analysis were determined using DAVID to identify the most significant biological processes affected by smoke or obesity.25–27 Primary transcriptional regulators of genes altered by obesity or smoke exposure were deduced using MetaCore’s (GeneGo) statistical Interactome tool to measure gene interconnectedness in the experimental dataset relative to all known interactions from the Affymetrix platform. Statistical significance of over-connected interactions was calculated using a hypergeometric distribution (p<0.05, 5% FDR), where the p value represents the probability of a particular mapping arising by chance for experimental data compared with the background Affymetrix platform. Significantly over-connected transcription factors were filtered for those with connectivity ratios (Actual:Expected) > 1.5-fold.
RESULTS
Exposures and In-Life Characterization
To compare transcriptional responses of normal weight with obese mice to MS and SS smoke inhalation, we employed a dietary model of the early stages of obesity, where mice have been characterized as having impaired glucose tolerance and mild insulin resistance by 8 weeks, but do not develop overt diabetes.28–30 In the present study, half of the mice were fed a high calorie/high fat (60kcal%) diet for 9 weeks by the start of smoke exposures, attaining weights on average 21% heavier than their lean counterparts (RW; Table S1). These diets were maintained throughout the subsequent smoke exposures, which employed target doses for MS (250 µg WTPM/L) and SS (85 µg WTPM/L) smoke based upon tolerance levels previously determined to deliver the same CO levels (~250 ppm (Table S2) for both modes of smoke administration.(31, 32 Reflecting elevated CO chamber concentrations, all cigarette smoke-exposed mice exhibited comparable blood carboxyhemoglobin (CO-Hb) levels, which were elevated relative to those of sham controls exposed to filtered air (Table S1), in agreement with previous studies.19, 33
Pulmonary inflammation is induced by MS, but not SS smoke
To assess the response of the mice to each exposure condition, we monitored any pulmonary inflammation by differential cytology, identifying the abundance and type of immune cells present in the bronchoalveolar lavage (BAL) fluid (Figure S1). We find no significant increases in inflammatory cells after SS smoke exposures of either RW or DIO mice, but RW mice exposed to MS smoke showed increased infiltration of neutrophils into the BAL fluid, without significant increases in alveolar macrophages, eosinophils, or lymphocytes. On the other hand, DIO mice exposed to MS smoke exhibited a significant increase in macrophages and an almost four-fold increase in neutrophils as compared with RW mice.
Distinct transcriptional responses in the heart and lung
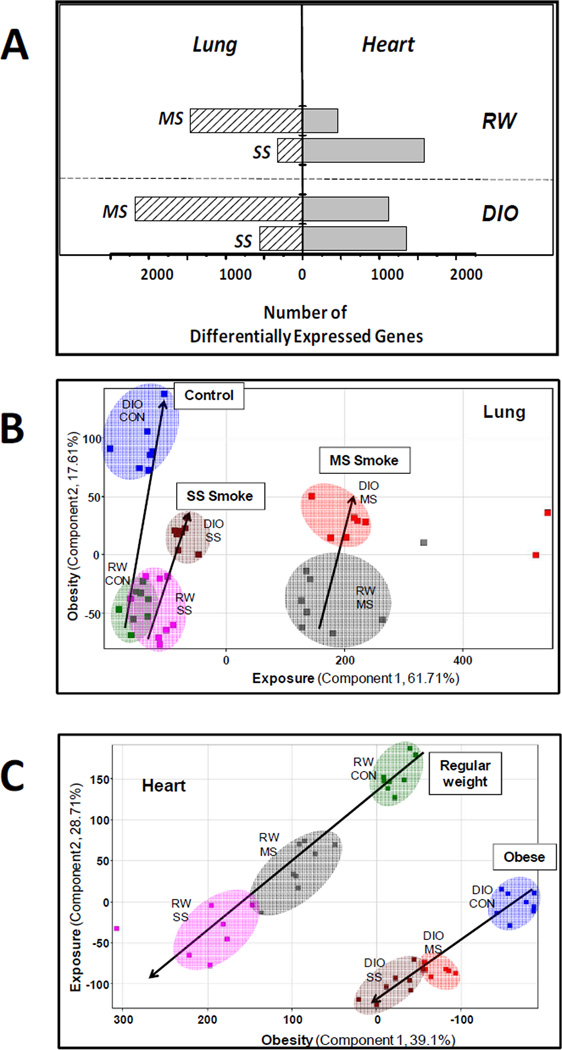
Transcriptional responses to smoke inhalation in both lungs and hearts of RW and DIO mice were evaluated using whole genome microarray analysis of total RNA extracted from lung or heart. Overall, 3012 pulmonary genes and 3252 cardiac genes were differentially expressed across the study at a significance level of p<0.01 with a 5% false discovery rate. Most of these genes are unique to either lung or heart with only ~20% (580) genes in common, suggesting a highly tissue-specific transcriptional response to smoke. Lung and heart exhibited different sensitivities to the same exposure/diet condition as evidenced by the associated numbers of significantly regulated genes (Figure 1A). For example, in the lungs of RW mice, MS smoke exposures resulted in 1466 differentially expressed genes, with a much-attenuated transcriptional response (463 genes) in the hearts of the same animals. In contrast, SS smoke exposure produced only a modest transcriptional response (328 genes) in the lung, but an almost five-fold greater number of significant cardiac gene changes (1590 genes). The response of DIO mice to smoke exposure included, on average, a larger number of regulated genes as compared with corresponding exposure groups of RW tissues, but with a similar pattern, i.e., of MS smoke exposure resulting in attenuation, and SS smoke in amplification, of the cardiac response relative to that of the lung. This preferential sensitivity of the heart to SS smoke relative to lung may indicate differential partitioning of inhaled cigarette smoke toxicants and/or different mechanisms of response. Consistent with the heart’s role as a secondary target of exposure, the average extent of cardiac gene changes was generally lower than in the lung as exemplified by the 65-fold differential expression of the most highly regulated pulmonary genes as compared with those in heart which did not exceed a 5-fold change (Table S3).
Figure 1. Differential Sensitivity of Hearts and Lung Transcriptomes to MS and SS Cigarette Smoke and High Fat Diet-Induced Obesity.
Panel A: The number of statistically significant (p<0.01) genes differentially expressed in lungs (hashed bars) or hearts (solid bars) of RW or DIO C57BL/6 mice exposed to repeated MS or SS smoke exposures as described in Experimental Procedures. Gene changes were assessed from whole genome microarray data of RNA extracted from whole tissues as described. Panels B & C: Principal Component Analysis of All 3,012 Pulmonary (B) and 3,252 Cardiac (C) Genes by treatment using normalized intensity values. Each point represents an individual animal (N=8 per group) from each treatment group using the following colored symbols: RW-SC (green), RW-MS (gray), RW-SS (magenta), DIO-SC (dark blue), DIO-MS (red), and DIO-SS (brown). Principal components analysis was performed on treatment groups with GeneSpring v.11 (Silicon Genetics) using non-transformed (ratio) normalized intensity values. Variances are described for the lung data (panel B) such that principal component 1 represents the variance on the effect of exposure and principal component 2 describes the variance due to the effect of diet phenotype (obesity). For the heart data (panel C), principal component 1 and 2 represent the variance on the effect of diet phenotype (obesity) and exposure, respectively.
Both lung and heart also exhibited significant transcriptional responses to high fat diet and obesity based on a comparison of data from sham controls of DIO with those of RW mice; the DIO lung exhibits 703 significantly regulated genes and the DIO heart exhibits 1143 significant genes (Table S3). The diet phenotype had a larger effect than exposure on the transcriptional response of the heart; in contrast, smoke exposure was the major influence on the transcriptional response of the lung, as supported by both hierarchical clustering (Figure S2) and principal component analysis (PCA; Figure 1). Hierarchical clustering, which identifies groups of genes regulated in response to the same environmental factors, was performed on the 580 common gene changes across both heart and lung and showed distinct classification of gene signatures by tissue and sub-classification by exposure condition. Notably, the lung data clustered by exposure condition (MS, SS, or SC (sham control)), whereas heart data clustered by diet (RW, DIO). Similarly PCA of the entire heart and lung datasets reflect different tissue-specific clustering patterns; i.e., exposure is the first principle component in lung, whereas diet was the first principle component in heart (Figure 1).
Cardiac Transcriptional Signatures of Exposure and Diet
Significant cardiac genes in each exposure and diet group were categorized by statistical enrichment (p<0.05) of GO biological processes with almost 40% (1059 genes) of all significant cardiac genes contributing to the resulting enriched processes (Figure S3).26, 34 Different patterns of responses are observed for each exposure/diet group based on varying levels of significance of each biological process highlighting the distinct transcriptional signatures of each treatment group. For example, MS and SS smoke each elicited different patterns of responses in the RW heart, with further difference in the DIO heart. Some of the most significant enriched processes were exhibited in the DIO heart and involved processes related to energy metabolism, e.g., lipid, ketone, carbohydrate, and phosphate metabolic processes (Figure S3). Subsequent smoke exposure suppressed these latter processes to varying degrees. All experimental groups exhibited a highly significant response in the process of cellular protein metabolism, which includes a large proportion of down-regulated genes involved in protein translation, a well-documented response to stress conditions.35–38 Many of the trends in GO processes are reiterated at the level of individual gene changes as further described.
Smoke elicits strong regulation of hypoxic and stress genes in the non-obese heart
In view of the large fraction of significant genes not falling into identified GO processes (Figure S3), we further focused on the most responsive individual genes, i.e., those exceeding 1.5 fold change in expression. As highlighted in Table 1, a significant fraction of the most smoke-responsive genes in RW hearts are related to hypoxia response, i.e., target genes of the HIF-1α transcription factor. Thus, this finding is consistent with the high levels of CO in cigarette smoke, which binds with high affinity to oxygen binding sites of hemoglobin. Despite elevated levels of circulating CO-Hb measured immediately after the last smoke exposure, the short (2–6 h) half-life of CO-Hb suggests that smoke-related hypoxia was largely resolved 24 h post exposure when heart samples were taken for microarray analysis.39 Thus, the observed sensitive regulation of multiple HIF-1α genes suggests a sustained effect of hypoxia on the cardiac transcriptome.
Table 1.
Transcriptional Responses of the RW Heart to MS- and SS-Smoke: Most Sensitive Genesa
| Fold-Change | ||||
|---|---|---|---|---|
| Gene Symbol | Gene Name | MS | SS | DIO Regb |
| Energy Metabolism | ||||
| Pdk4 | Pyruvate dehydrogenase kinase, 4 | ---- | 1.7 | Y |
| Glu1 | Glutamine synthetase | 1.4 | 1.6 | N |
| Iron homeostasis | ||||
| Slc25a37 | Mitoferrin-1 | 1.5 | 2.1 | N |
| Tfrc | Transferrin receptor | −3.4 | −4.1 | N |
| Cp | Ceruloplasmin/ferroxidase | −1.4 | −1.8 | N/Y |
| Alas1 | Aminolevulinic acid synthase 1 | −1.4 | −1.7 | N |
| Stress/Inflammation | ||||
| Ddit4 | DNA damage inducible transcript 4 (RTP301) | 1.3 | 2.1 | N |
| Mkrn1 | Makorin, ring finger protein, 1 | ---- | 1.7 | N |
| Rit1 | Ras-like, without CAAX 1, GTPase | 1.4 | 1.7 | N |
| Cebpb | CCAAT/enhancer binding protein β, transcription factor | 1.4 | 1.7 | N |
| C es3 | Carboxylesterase 3 | 1.3 | 1.5 | N |
| Nr1d1 | Rev-Erb A alpha transcription factor | −1.9 | −2.2 | N/Y |
| Nr3c1 | Glucocorticoid receptor transcription factor | ---- | −2.1 | N |
| Dbp | D site albumin promoter binding protein | −1.3 | −2.0 | N |
| Tnfsf12a | TNF receptor, member 12 (TWEAKR/Fn14) | −1.7 | −1.7 | N |
| Dct | Dopachrome tautomerase | −1.5 | −1.4 | N |
| Proliferation/cell cycle | ||||
| Bptf | Bromodomain PHD finger transcription factor | ---- | −1.9 | N |
| Ccnd1 | Cyclin D1 | −1.3 | −1.8 | N |
| Bhlhe40 | Basic helix-loop-helix family, e40 (DEC1) | −1.3 | −1.7 | N/Y |
| Sfrs3 | Splicing factor, Arg/Ser-rich 3 (SRp20) | −1.5 | −1.6 | N |
| Ion Transport | ||||
| Kcnd2 | Potassium voltage gated channel, K4.2 | ---- | −1.9 | N |
| Kcne1 | K voltage gated channel | −2.1 | −1.8 | Y |
| Slc41a3 | MgtE, Na-Mg exchanger | −1.6 | −1.5 | N |
| Vascular Tone | ||||
| Snca | a-Synuclein | ---- | 2.8 | N |
| Nampt | Nicotinamide phosphoribosyl transferase (Visfatin) | 1.4 | 1.6 | Y |
| Ptgds | Prostaglandin D2 synthase | −1.6 | −1.8 | Y |
| Nppb | Natriuretic peptide precursor B (BNP) | −1.7 | −1.8 | N/Y |
| Slmap | Sarcolemma associated protein | ---- | −1.8 | N |
| Cald1 | Caldesmon 1 | ---- | −1.8 | N |
| Npr3 | Natriuretic peptide receptor 3 | −1.4 | −1.6 | N |
Selected 30/108 significant (p<0.01) cardiac genes regulated >1.5-fold by either mainstream smoke (MS) or sidestream smoke (SS), relative to RW mice exposed to filtered air (controls). Alternate gene or gene product names are included. (The complete list of regulated cardiac genes is available in Supporting Table S3). The extent of regulation is indicated as fold-change with missing values (----) indicating no significant gene change. Highlighted entries indicate genes previously reported to be regulated by HIF-1α. 69–86
Y (yes) or N (no) indicates whether a gene was significantly regulated (p<0.01) in MS and SS smoke-exposed DIO mice; N/Y indicates that the gene was regulated in the DIO heart exposed to SS smoke, but not to MS smoke.
HIF-1α was identified as the most enriched transcription factor (p-value of 2.99×10−7) associated with genes differentially expressed in the RW-SS exposure group, based on statistical measures of the interconnectedness of genes in the experimental dataset relative to all known interactions from the Affymetrix platform. On the other hand, in the case of MS smoke-regulated genes, HIF-1α ranked as the fifth most enriched transcription factor (p value of 7.28×10−5) in the dataset. Similarly, the extent of regulation (fold-change) of multiple hypoxia-regulated genes was less or absent in mice exposed to MS as compared with SS smoke. Such differences in HIF-1α gene responses, despite identical CO doses in MS and SS smoke, highlight the complexity in hypoxia responses and possible gene interactions. For example, each of the transcription factor genes (i.e., Cebpb, Nr1d1, Nr3c1, Dbp, Bptf; Table 1) regulated by smoke exposure has multiple response elements and interactions with other transcription factors.
Sensitivity to additional stressors in cigarette smoke is reflected by the regulation of non-hypoxic genes also involved in stress and inflammatory processes, as well as inhibition of cell cycle progression and proliferation. Smoke exposure also elicits altered expression of several ion transporters that play essential roles in the cardiac action potential maintaining contractile rhythmicity, and notably, genes involved in vascular tone and endothelial dysfunction, a recognized precursor in the development of atherosclerosis. 40, 41 Among genes most sensitive to smoke exposure are several circadian genes, (Nr1d1, Dbp, Bhlhe40), consistent with previously reported disruption of circadian processes by smoking that has been linked to health disorders and myocardial infarction.42–45
Smoke exposure modifies the transcriptional response to diet-induced obesity
In view of the major influence of the diet phenotype on transcriptional responses in the heart (Figures 1 and S2), differential gene expression was assessed from a comparison of microarray data from both smokeexposed and control DIO hearts with that of RW controls, thus permitting the effects of smoke on both diet-related and non-diet-related genes to be simultaneously monitored. Cardiac genes related to diet phenotype, i.e., those differentially expressed in DIO-SC as compared with RW-SC, include 1148 significant genes. A substantial proportion of both the most highly expressed individual genes (Table 2) and enriched GO processes (Figure S3) of the DIO response involve gene changes related to metabolic adaptation to a high fat diet, i.e., multiple PPARα target genes involved in enhanced uptake and oxidation of fatty acids at the expense of glucose uptake and glycolysis.46–53 While the sensitive regulation of most DIO-related PPARα genes was virtually unmitigated by subsequent smoke exposure, several notable exceptions were observed (e.g., Serpine1, Rbp7, Slc2a1; Table 2), for which smoke exposure reversed the initial DIO regulation.
Table 2.
Transcriptional Responses of the DIO Heart to High Fat Diet and Obesity in Sham Controls (SC) and to MS or SS-Smoke Exposure: Most Sensitive Genesa
| Gene | Gene Name | Function | Fold-Change | ||
|---|---|---|---|---|---|
| PPAR-α target genes | SC | MS | SS | ||
| Pdk4 | Pyruvate dehydrogenase kinase 4 | Inhibitor of PDH | 3.6 | 3.1 | 3.9 |
| Hmgcs2 | Hydroxy-methylglutaryl-CoA synthase 2 | Ketogenesis | 3.2 | 4.3* | 4.6* |
| Acot1 | Acyl-CoA thioesterase 1, mitochondrial | Fatty Acid Uptake | 2.3 | 2.1 | 2.3 |
| Angptl4 | Angiopoietin-like 4 | Lipoprotein Lipase Inhibitor | 2.1 | 2.4 | 3.7 |
| Acot2 | Acyl-CoA thioesterase 2, soluble | Fatty Acid Uptake | 1.8 | 1.9 | 2.0 |
| Ucp3 | Uncoupling protein 3, mitochondrial | H+/OH Carrier | 1.8 | 3.7 | 2.2 |
| Fbp2 | Fructose bisphosphatase 2 | Gluconeogenesis Inhibitor | 1.7 | 1.9 | 1.9 |
| Serpine1 | Plasminogen activator inhibitor (PAI-1) | Inhibitor of Fibrinolysis | 1.7 | ---- | ---- |
| Rbp7 | Retinol binding protein 7 | Cellular retinol stabilization | 1.7 | ---- | ---- |
| Pex11a | Peroxisomal biogenesis factor 11 alpha | Peroxisome proliferation | 1.6 | 1.7 | 1.7 |
| Decr1 | 2,4-dienoyl CoA reductase1, mitochondrial | Fatty Acid Oxidation | 1.5 | 1.6* | 1.6* |
| Bdh1 | 3-Hydroxybutyrate dehydrogenase | Ketone Body Metabolism | −1.6 | −1.5 | ---- |
| Slc2a1 | Glut1 glucose transporter | Glucose Uptake | −1.7 | ---- | ---- |
| Pcx | Pyruvate carboxylase | Gluconeogenesis | −1.8 | −1.9 | −1.6 |
| Other DIO-regulated genes | |||||
| Hspa1b | Heat shock protein 1B | Protein folding | 4.9 | ---- | ---- |
| Hspa1a | Heat shock protein 1A | Protein Folding | 3.5 | ---- | ---- |
| Dnaja1 | DnaJ (Hsp40) homolog | Protein Folding | 1.8 | ---- | ---- |
| Dnajb1 | DnaJ (Hsp40) homolog | Protein Folding | 1.6 | ---- | ---- |
| Hsp90aa1 | Heat shock protein 90, alpha | Protein Folding | 1.6 | ---- | ---- |
| Mthfd2 | Methylene tetrahydrofolate dehydrogenase | Homocysteine catabolism | 2.7 | 1.7* | 2.0 |
| Chrna2 | Nicotinic cholinergenic receptor α2 | Sympathetic nerve activation | 1.8 | 1.9 | 1.7 |
| Hsd17b11 | Hydroxysteroid dehydrogenase | Estrogen, Androgen Metabolism | 1.7 | 1.6 | 1.8 |
| Sgk1 | Serum/glucocorticoid-inducible kinase 1 | Stress/Angiogenesis | 1.6 | ---- | 1.5 |
| Cth | Cystathionase | Homocysteine catabolism | −1.8 | ---- | ---- |
| Cigarette smoke-regulated genes | |||||
| Ddit4l | DNA damage inducible transcript 4-like | Negative regulator of protein synthesis | ---- | 1.5* | 2.0* |
| Txnip | Thioredoxin interacting protein | Inflammosome Activator | ---- | 1.7 | 1.9 |
| Rit1 | Ras-like, without CAAX 1 | Regulation of Stress Signaling | ---- | 1.5* | 1.6* |
| Kcne1 | Potassium voltage-gated channel, Isk | Regulation of Action Potential | ---- | −1.5 | −2.2* |
| Cp | Ceruloplasmin | Iron Homeostasis | ---- | −1.4 | −1.8* |
| Ptgds | Prostaglandin D2 synthase | Vasodilation | ---- | −1.8* | −1.7* |
| Nr1d1 | Nuclear receptor, Rev-ErbA alpha | Inflammatory Modulation | ---- | ---- | −1.7* |
| Nppb | Naturiuretic precursor peptide B | Vasodilation | ---- | ---- | −1.5 |
| Bhlhe40 | Basic helix-loop-helix family, e40 (DEC1) | Repressor of growth/differentiation | ---- | ---- | −1.5 |
| Amph | Amphiphysin | Stabilization of T-tubules | ---- | −1.8 | −1.5 |
| Lgals4 | Galectin-4 | T-cell Inhibitor | ---- | ---- | −1.5 |
| Cpxm2 | Carboxypeptidase X, member 2 | Cell Adhesion | ---- | −2.0 | −1.5 |
| Tbx20 | T-box20 transcription factor | Ventricle wall development | ---- | ---- | −1.5 |
| Nnat | Neuronatin | Inflammatory Resolution | ---- | −1.6 | −1.6 |
| Igf bp3 | Insulin growth factor binding protein 3 | IGF Carrier | ---- | −1.6 | −1.6 |
| Inhbb | Inhibin B | Repressor of growth/differentiation | ---- | ---- | −1.7 |
Significant (p<0.01) genes regulated >1.5-fold in DIO hearts following exposure to mainstream smoke (MS) or sidestream smoke (SS), or filtered air (SC) relative to RW control mice, and their associated extents of regulation (expressed as positive or negative fold-change, with missing values (----) indicating no significant differential gene expression.
Non-PPARα target genes were also sensitively regulated in the DIO heart (in the absence of smoke exposure), which included genes involved in protein folding (Hspa1a, Hspa1b, Hsp90aa1, Dnaja1, Dnajb1), and synthesis of the antioxidant, glutathione (Mthfd2, Cth); smoke exposure also resulted in loss of their DIO-related induction. Considering the entire set of 1148 significant DIO-regulated genes, 41% (557 genes) and 58% (779 genes) of these underwent a complete loss of regulation after MS and SS smoke exposure, respectively, indicating a substantial alteration by smoke of the heart’s adaptation to high fat diet and obesity.
High fat diet-induced obesity modifies the heart’s response to smoke
The DIO heart also exhibits a substantial modification in its transcriptional response to smoke relative to that of the RW heart. With respect to the number of regulated genes, the response to MS smoke is significantly enhanced (from 462 to 614 genes) in DIO hearts relative to RW hearts, whereas the response to SS smoke is dramatically reduced (from 1590 to 491 genes). At the level of the most sensitive individual genes, the smoke response of DIO hearts is characterized by the absence of much of the smoke response that occurred in healthy RW hearts (Table 1), including both hypoxic- and stress-related genes as well as the appearance of a new set of smoke-responsive genes unique to the DIO heart. Moreover, a general suppression in the extent of gene regulation is observed in the smoke response of the DIO heart, as illustrated by the low number (26) of sensitive (>1.5 fold change) SS smoke-responsive genes in DIO hearts as compared with the 106 genes sensitive to SS smoke in RW hearts (Tables 1–2).
DISCUSSION
Significance and study design
The present study is the first, to our knowledge, that addresses the in vivo transcriptional response of the heart to cigarette smoke exposure in the setting of high fat diet and obesity, and thus takes a first step towards identifying the molecular basis of adaptive responses that may lead to an increased risk of heart disease in obese smokers.54, 55 We have identified the most sensitive individual genes and collective cellular processes responsive to repeated cigarette smoke exposures in the murine heart and the substantial modification of this response in the setting of high fat diet and obesity (Tables 1–2; Figure S3), using the C57Bl/6 DIO mouse, a model that is relevant to common high calorie/ high fat dietary choices of humans. Moreover, the custom-designed smoke inhalation system that delivers highly uniform doses of both MS and SS smoke provides defined exposures not normally available for studies of human smokers. This, with lung microarray data, BAL fluid cytology, and blood CO-Hb levels has permitted a more fully informed understanding of the transcriptional response of the heart. In particular, the heart’s relatively modest transcriptional response to MS smoke can be better appreciated with the knowledge that the delivered MS smoke doses were sufficient to elicit a robust cellular inflammatory and transcriptional response in the lungs of these mice (Figures 1, S1; Table S3). More striking then, is the extensive number of cardiac genes elicited by SS smoke in the absence of any cellular pulmonary inflammation.
The DIO heart
A central finding of this study entails the substantial alteration in the cardiac transcriptional response to cigarette smoke of the DIO heart as compared with that of the RW heart (Table 1); at the same time, a portion of the adaptive response to high dietary fat and obesity was altered by repeated smoke exposure (Table 2). In particular, the regulation of sensitive genes involved in protective responses to smoke (i.e., hypoxia, inflammation, oxidative stress and DNA damage responses) was impaired in the DIO heart. Similarly, a number of DIO gene adaptations that were lost after smoke exposure appear to reflect responses to the oxidative stress inherent in a heart utilizing high levels of dietary fats, as previously demonstrated by increases in lipid and protein oxidation, NADPH oxidase activity, and in mitochondrial hydrogen peroxide in DIO rodent hearts.56–58 Another smokeinduced loss of the DIO phenotype with important implications for the vasculature is that of Serpine1, encoding the plasminogen activator inhibitor-1 (PAI-1), which is normally considered a profibrotic protein based on its normal role inhibiting vascular clot degradation. However, recent evidence from PAI-1 knock-out mice has suggested that this protein plays a unique and protective role in the heart where it functions to maintain microvascular integrity.79 Thus, although transcriptional responses of the heart to DIO and smoke are extensive and diverse, a common theme observed from the smoke-exposed DIO heart is the impaired regulation of stress response genes.
In particular, impairment of HIF-1α gene responses in the presence of a strong induction of PPARα genes in the DIO heart is consistent with studies identifying inhibitory interactions between these two transcription factors, and their essential and complementary roles in regulating fuel and oxygen utilization in the contracting heart. The underlying mechanism is suggested by a recent study showing that activation of PPAR-α in cancer cells enhances binding of HIF-1α to von Hippel-Lindau tumor suppressor, a protein that mediates HIF-1α degradation through the ubiquitin-proteasome pathway; thus HIF-1α levels and hypoxia responses were reduced. 59 Impaired hypoxia responses have been observed in diabetic rat hearts, which share common characteristics with DIO hearts, e.g., activation of PPAR-α, enhanced fatty acid oxidation, and decreased glucose utilization.60–63 The human heart, too, exhibits reduced hypoxia responses as evidenced by significantly lower mRNA and protein levels of HIF-1α found in biopsies from ischemic (hypoxic) heart regions of diabetic patients as compared with those of non-diabetic patients.60 Such deficits in hypoxia responses may have serious consequences for the maintenance of efficient cardiac energy production under normal fluctuations in oxygen levels as well as during recovery from the acute hypoxia of ischemia.63, 64
Summary and Future Work
The present work has provided a comprehensive characterization of the early transcriptional response to smoke and diet within the collection of tissues that make up the murine heart. In view of multiple levels of documented post-transcriptional regulation (e.g., microRNA, protein modification) of smoking responses, future work should extend this knowledge towards defining responses at the level of the functional proteome.65–68 Further, parsing the sensitivity and kinetics of individual responses in specific cardiac cell types will go a long way towards the ability to predict the implications of smoking, obesity and dietary fat content on the development of CVD and other cardiac pathologies. Nevertheless, the present study of the whole transcriptome in multiple tissues suggests hypotheses regarding possible roles for HIF-1α and PPAR-α interactions in the impaired stress response of the DIO heart.
Supplementary Material
Acknowledgements
The work was performed at Pacific Northwest National Laboratory (PNNL), a multi-program laboratory operated by Battelle for the U.S. Department of Energy under Contract DE-AC05-76RK01830 and at Battelle Toxicology Northwest. We thank Matthew Gaffrey who performed all RNA isolations and Rick Zangar for insightful discussions.
Funding This work is supported by the National Institutes of Health (NIEHS) under grant U54 ES016015.
ABBREVIATIONS
- BAL
bronchoalveolar lavage
- CO
carbon monoxide
- CO-Hb
carboxyhemoglobin
- CVD
cardiovascular disease
- DIO
diet-induced obese/obesity
- GO
gene ontology
- HDL
high density lipoprotein
- HIF-1α
hypoxia inducible factor-1alpha
- LDL
low density lipoprotein
- MS
mainstream
- PCA
principle component analysis
- PPARα
peroxisome proliferator activated-receptor-alpha
- RW
regular weight
- SC
sham control
- SS
sidestream
- WTPM
wet total particulate material
Footnotes
Supporting Information Characteristics of study mice (body weight, blood COHb, BAL fluid cytology); exposure conditions; hierarchical clustering of genes; analysis of gene processes; spreadsheets of all significant genes and genes involved in significant enriched GO processes in the heart. This material is available free of charge via the Internet at http://pubs.acs.org
Raw data from Affymetrix microarrays is available at the Gene Expression Omnibus (GEO) site (accession number GSE47022).
REFERENCES
- 1.Raupach T, Schafer K, Konstantinides S, Andreas S. Secondhand smoke as an acute threat for the cardiovascular system: a change in paradigm. Eur. Heart J. 2006;27:386–392. doi: 10.1093/eurheartj/ehi601. [DOI] [PubMed] [Google Scholar]
- 2.Schick S, Glantz S. Philip Morris toxicological experiments with fresh sidestream smoke: more toxic than mainstream smoke. Tob. Control. 2005;14:396–404. doi: 10.1136/tc.2005.011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.U.S. Department of Health and Human Services. How tobacco causes disease: the biology and behavioral basis for smoking-attributable disease: a report of the Surgeon General. Atlanta GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health; 2010. [Google Scholar]
- 4.U.S. Department of Health and Human Services. The health consequences of involuntary exposure to tobacco smoke: a report of the Surgeon General. Atlanta GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health; 2006. [Google Scholar]
- 5.Schick S, Glantz S. Scientific analysis of second-hand smoke by the tobacco industry, 1929–1972. Nicotine Tob. Res. 2005;7:591–612. doi: 10.1080/14622200500185082. [DOI] [PubMed] [Google Scholar]
- 7.Moir D, Rickert WS, Levasseur G, Larose Y, Maertens R, White P, Desjardins S. A comparison of mainstream and sidestream marijuana and tobacco cigarette smoke produced under two machine smoking conditions. Chem. Res. Toxicol. 2008;21:494–502. doi: 10.1021/tx700275p. [DOI] [PubMed] [Google Scholar]
- 8.Barnoya J, Glantz SA. Cardiovascular effects of secondhand smoke: nearly as large as smoking. Circulation. 2005;111:2684–2698. doi: 10.1161/CIRCULATIONAHA.104.492215. [DOI] [PubMed] [Google Scholar]
- 9.Howard G, Thun MJ. Why is environmental tobacco smoke more strongly associated with coronary heart disease than expected? A review of potential biases and experimental data. Environ. Health Perspect. 1999;107(Suppl. 6):853–858. doi: 10.1289/ehp.99107s6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vazzana N, Santilli F, Sestili S, Cuccurullo C, Davi G. Determinants of increased cardiovascular disease in obesity and metabolic syndrome. Curr. Med. Chem. 2011;18:5267–5280. doi: 10.2174/092986711798184299. [DOI] [PubMed] [Google Scholar]
- 11.Cascio G, Schiera G, Di Liegro I. Dietary fatty acids in metabolic syndrome, diabetes and cardiovascular diseases. Curr. Diabetes Rev. 2012;8:2–17. doi: 10.2174/157339912798829241. [DOI] [PubMed] [Google Scholar]
- 12.Taube A, Schlich R, Sell H, Eckardt K, Eckel J. Inflammation and metabolic dysfunction: links to cardiovascular diseases. Am. J. Physiol. Heart Circ. Physiol. 2012;302:H2148–H2165. doi: 10.1152/ajpheart.00907.2011. [DOI] [PubMed] [Google Scholar]
- 13.Harwood HJ., Jr The adipocyte as an endocrine organ in the regulation of metabolic homeostasis. Neuropharmacology. 2012;63:57–75. doi: 10.1016/j.neuropharm.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 14.Park M, Sweeney G. Direct effects of adipokines on the heart: focus on adiponectin. Heart Fail. Rev. 2012 Aug 15; doi: 10.1007/s10741-012-9337-8. [DOI] [PubMed] [Google Scholar]
- 15.Turer AT, Malloy CR, Newgard CB, Podgoreanu MV. Energetics and metabolism in the failing heart: important but poorly understood. Curr. Opin. Clin. Nutr. Metab. Care. 2010;13:458–465. doi: 10.1097/MCO.0b013e32833a55a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leone TC, Kelly DP. Transcriptional control of cardiac fuel metabolism and mitochondrial function. Cold Spring Harb. Symp. Quant. Biol. 2011;76:175–182. doi: 10.1101/sqb.2011.76.011965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tekin D, Dursun AD, Xi L. Hypoxia inducible factor 1 (HIF-1) and cardioprotection. Acta Pharmacol. Sin. 2010;31:1085–1094. doi: 10.1038/aps.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Semenza GL. O2-regulated gene expression: transcriptional control of cardiorespiratory physiology by HIF-1. J. Appl. Physiol. 2004;96:1173–1177. doi: 10.1152/japplphysiol.00770.2003. discussion 1170–1172. [DOI] [PubMed] [Google Scholar]
- 19.Obot C, Lee K, Fuciarelli A, Renne R, McKinney W. Characterization of mainstream cigarette smoke-induced biomarker responses in ICR and C57Bl/6 mice. Inhal. Toxicol. 2004;16:701–719. doi: 10.1080/08958370490476604. [DOI] [PubMed] [Google Scholar]
- 20.Lee KM, Renne RA, Harbo SJ, Clark ML, Johnson RE, Gideon KM. 3-week inhalation exposure to cigarette smoke and/or lipopolysaccharide in AKR/J mice. Inhal. Toxicol. 2007;19:23–35. doi: 10.1080/08958370600985784. [DOI] [PubMed] [Google Scholar]
- 21.Pounds JG, Flora JW, Adkins JN, Lee KM, Rana GS, Sengupta T, Smith RD, McKinney WJ. Characterization of the mouse bronchoalveolar lavage proteome by micro-capillary LC-FTICR mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2008;864:95–101. doi: 10.1016/j.jchromb.2008.01.044. [DOI] [PubMed] [Google Scholar]
- 22.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 23.Kerr MK, Martin M, Churchill GA. Analysis of variance for gene expression microarray data. J. Comput. Biol. 2000;7:819–837. doi: 10.1089/10665270050514954. [DOI] [PubMed] [Google Scholar]
- 24.Benjamini Y a. H. Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 25.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 26.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Rossmeisl M, Rim JS, Koza RA, Kozak LP. Variation in type 2 diabetes--related traits in mouse strains susceptible to diet-induced obesity. Diabetes. 2003;52:1958–1966. doi: 10.2337/diabetes.52.8.1958. [DOI] [PubMed] [Google Scholar]
- 29.Collins S, Martin TL, Surwit RS, Robidoux J. Genetic vulnerability to dietinduced obesity in the C57BL/6J mouse: physiological and molecular characteristics. Physiol. Behav. 2004;81:243–248. doi: 10.1016/j.physbeh.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Surwit RS, Feinglos MN, Rodin J, Sutherland A, Petro AE, Opara EC, Kuhn CM, Rebuffe-Scrive M. Differential effects of fat and sucrose on the development of obesity and diabetes in C57BL/6J and A/J mice. Metabolism. 1995;44:645–651. doi: 10.1016/0026-0495(95)90123-x. [DOI] [PubMed] [Google Scholar]
- 31.Meng QR, Gideon KM, Harbo SJ, Renne RA, Lee MK, Brys AM, Jones R. Gene expression profiling in lung tissues from mice exposed to cigarette smoke, lipopolysaccharide, or smoke plus lipopolysaccharide by inhalation. Inhal. Toxicol. 2006;18:555–568. doi: 10.1080/08958370600686226. [DOI] [PubMed] [Google Scholar]
- 32.Obot CJ, Lee KM, Fuciarelli AF, Renne RA, McKinney WJ. Characterization of mainstream cigarette smoke-induced biomarker responses in ICR and C57Bl/6 mice. Inhal. Toxicol. 2004;16:701–719. doi: 10.1080/08958370490476604. [DOI] [PubMed] [Google Scholar]
- 33.D'Hulst A I, Maes T, Bracke KR, Demedts IK, Tournoy KG, Joos GF, Brusselle GG. Cigarette smoke-induced pulmonary emphysema in scid-mice. Is the acquired immune system required? Respir. Res. 2005;6:147. doi: 10.1186/1465-9921-6-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang da W, Sherman BT, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID gene ID conversion tool. Bioinformation. 2008;2:428–430. doi: 10.6026/97320630002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wouters BG, van den Beucken T, Magagnin MG, Koritzinsky M, Fels D, Koumenis C. Control of the hypoxic response through regulation of mRNA translation. Semin. Cell. Dev. Biol. 2005;16:487–501. doi: 10.1016/j.semcdb.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 36.Brostrom CO, Brostrom MA. Regulation of translational initiation during cellular responses to stress. Prog. Nucleic Acid Res. Mol. Biol. 1998;58:79–125. doi: 10.1016/s0079-6603(08)60034-3. [DOI] [PubMed] [Google Scholar]
- 37.Sheikh MS, Fornace AJ., Jr Regulation of translation initiation following stress. Oncogene. 1999;18:6121–6128. doi: 10.1038/sj.onc.1203131. [DOI] [PubMed] [Google Scholar]
- 38.Patel J, McLeod LE, Vries RG, Flynn A, Wang X, Proud CG. Cellular stresses profoundly inhibit protein synthesis and modulate the states of phosphorylation of multiple translation factors. Eur. J. Biochem. 2002;269:3076–3085. doi: 10.1046/j.1432-1033.2002.02992.x. [DOI] [PubMed] [Google Scholar]
- 39.Shimazu T, Ikeuchi H, Sugimoto H, Goodwin CW, Mason AD, Jr, Pruitt BA., Jr Half-life of blood carboxyhemoglobin after short-term and long-term exposure to carbon monoxide. J. Trauma. 2000;49:126–131. doi: 10.1097/00005373-200007000-00019. [DOI] [PubMed] [Google Scholar]
- 40.Vallejo S, Romacho T, Angulo J, Villalobos LA, Cercas E, Leivas A, Bermejo E, Carraro R, Sanchez-Ferrer CF, Peiro C. Visfatin impairs endothelium-dependent relaxation in rat and human mesenteric microvessels through nicotinamide phosphoribosyl-transferase activity. PloS one. 2011;6:e27299. doi: 10.1371/journal.pone.0027299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding H, Howarth AG, Pannirselvam M, Anderson TJ, Severson DL, Wiehler WB, Triggle CR, Tuana BS. Endothelial dysfunction in Type 2 diabetes correlates with deregulated expression of the tail-anchored membrane protein SLMAP. Am. J. Physiol. Heart Circ. Physiol. 2005;289:H206–H211. doi: 10.1152/ajpheart.00037.2005. [DOI] [PubMed] [Google Scholar]
- 42.Jeyaraj D, Haldar SM, Wan X, McCauley MD, Ripperger JA, Hu K, Lu Y, Eapen BL, Sharma N, Ficker E, Cutler MJ, Gulick J, Sanbe A, Robbins J, Demolombe S, Kondratov RV, Shea SA, Albrecht U, Wehrens XH, Rosenbaum DS, Jain MK. Circadian rhythms govern cardiac repolarization and arrhythmogenesis. Nature. 2012;483:96–99. doi: 10.1038/nature10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chatham JC, Young ME. Regulation of myocardial metabolism by the cardiomyocyte circadian clock. J. Mol. Cell. Cardiol. 2012;55:139–146. doi: 10.1016/j.yjmcc.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Satoh T, Hosokawa M. Structure, function and regulation of carboxylesterases. Chem. Biol. Interact. 2006;162:195–211. doi: 10.1016/j.cbi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 45.Vasu VT, Cross CE, Gohil K. Nr1d1, an important circadian pathway regulatory gene, is suppressed by cigarette smoke in murine lungs. Integr. Cancer Ther. 2009;8:321–328. doi: 10.1177/1534735409352027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujita M, Momose A, Ohtomo T, Nishinosono A, Tanonaka K, Toyoda H, Morikawa M, Yamada J. Upregulation of fatty acyl-CoA thioesterases in the heart and skeletal muscle of rats fed a high-fat diet. Biol. Pharm. Bull. 2011;34:87–91. doi: 10.1248/bpb.34.87. [DOI] [PubMed] [Google Scholar]
- 47.Ren H, Vallanat B, Brown-Borg HM, Currie R, Corton JC. Regulation of Proteome Maintenance Gene Expression by Activators of Peroxisome Proliferator-Activated Receptor alpha. PPAR Res. 2010;2010:1–14. doi: 10.1155/2010/727194. 727194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vallanat B, Anderson SP, Brown-Borg HM, Ren H, Kersten S, Jonnalagadda S, Srinivasan R, Corton JC. Analysis of the heat shock response in mouse liver reveals transcriptional dependence on the nuclear receptor peroxisome proliferator-activated receptor alpha (PPARalpha) BMC Genomics. 2010;11:16. doi: 10.1186/1471-2164-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huss JM, Kelly DP. Nuclear receptor signaling and cardiac energetics. Circ. Res. 2004;95:568–578. doi: 10.1161/01.RES.0000141774.29937.e3. [DOI] [PubMed] [Google Scholar]
- 50.Lefebvre P, Chinetti G, Fruchart JC, Staels B. Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J. Clin. Invest. 2006;116:571–580. doi: 10.1172/JCI27989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hwang B, Jeoung NH, Harris RA. Pyruvate dehydrogenase kinase isoenzyme 4 (PDHK4) deficiency attenuates the long-term negative effects of a high-saturated fat diet. Biochem. J. 2009;423:243–252. doi: 10.1042/BJ20090390. [DOI] [PubMed] [Google Scholar]
- 52.Lichtenstein L, Mattijssen F, de Wit NJ, Georgiadi A, Hooiveld GJ, van der Meer R, He Y, Qi L, Koster A, Tamsma JT, Tan NS, Muller M, Kersten S. Angptl4 protects against severe proinflammatory effects of saturated fat by inhibiting fatty acid uptake into mesenteric lymph node macrophages. Cell. Metab. 2010;12:580–592. doi: 10.1016/j.cmet.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Georgiadi A, Lichtenstein L, Degenhardt T, Boekschoten MV, van Bilsen M, Desvergne B, Muller M, Kersten S. Induction of cardiac Angptl4 by dietary fatty acids is mediated by peroxisome proliferator-activated receptor beta/delta and protects against fatty acidinduced oxidative stress. Circ. Res. 2010;106:1712–1721. doi: 10.1161/CIRCRESAHA.110.217380. [DOI] [PubMed] [Google Scholar]
- 54.Buettner R, Scholmerich J, Bollheimer LC. High-fat diets: modeling the metabolic disorders of human obesity in rodents. Obesity (Silver Spring) 2007;15:798–808. doi: 10.1038/oby.2007.608. [DOI] [PubMed] [Google Scholar]
- 55.McNamara P, FitzGerald GA. Smoking-induced vascular disease: a new twist on an old theme. Circ. Res. 2001;89:563–565. [PubMed] [Google Scholar]
- 56.Noeman SA, Hamooda HE, Baalash AA. Biochemical study of oxidative stress markers in the liver, kidney and heart of high fat diet induced obesity in rats. Diabetol. Metab. Syndr. 2011;3:17. doi: 10.1186/1758-5996-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sonta T, Inoguchi T, Tsubouchi H, Sekiguchi N, Kobayashi K, Matsumoto S, Utsumi H, Nawata H. Evidence for contribution of vascular NAD(P)H oxidase to increased oxidative stress in animal models of diabetes and obesity. Free Radic.Biol. Med. 2004;37:115–123. doi: 10.1016/j.freeradbiomed.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 58.Rindler PM, Plafker SM, Szweda LI, Kinter M. High Dietary Fat Selectively Increases Catalase Expression within Cardiac Mitochondria. J. Biol. Chem. 2013;288:1979–1990. doi: 10.1074/jbc.M112.412890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou J, Zhang S, Xue J, Avery J, Wu J, Lind SE, Ding WQ. Activation of Peroxisome Proliferator-activated Receptor alpha (PPARalpha) Suppresses Hypoxia-inducible Factor-1alpha (HIF-1alpha) Signaling in Cancer Cells. J. Biol. Chem. 2012;287:35161–35169. doi: 10.1074/jbc.M112.367367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marfella R, Esposito K, Nappo F, Siniscalchi M, Sasso FC, Portoghese M, Di Marino MP, Baldi A, Cuzzocrea S, Di Filippo C, Barboso G, Baldi F, Rossi F, D'Amico M, Giugliano D. Expression of angiogenic factors during acute coronary syndromes in human type 2 diabetes. Diabetes. 2004;53:2383–2391. doi: 10.2337/diabetes.53.9.2383. [DOI] [PubMed] [Google Scholar]
- 61.Marfella R, D'Amico M, Di Filippo C, Piegari E, Nappo F, Esposito K, Berrino L, Rossi F, Giugliano D. Myocardial infarction in diabetic rats: role of hyperglycaemia on infarct size and early expression of hypoxia-inducible factor 1. Diabetologia. 2002;45:1172–1181. doi: 10.1007/s00125-002-0882-x. [DOI] [PubMed] [Google Scholar]
- 62.Chou E, Suzuma I, Way KJ, Opland D, Clermont AC, Naruse K, Suzuma K, Bowling NL, Vlahos CJ, Aiello LP, King GL. Decreased cardiac expression of vascular endothelial growth factor & its receptors in insulin-resistant and diabetic States: a possible explanation for impaired collateral formation in cardiac tissue. Circulation. 2002;105:373–379. doi: 10.1161/hc0302.102143. [DOI] [PubMed] [Google Scholar]
- 63.Holloway C, Cochlin L, Codreanu I, Bloch E, Fatemian M, Szmigielski C, Atherton H, Heather L, Francis J, Neubauer S, Robbins P, Montgomery H, Clarke K. Normobaric hypoxia impairs human cardiac energetics. FASEB J. 2011;25:3130–3135. doi: 10.1096/fj.11-183426. [DOI] [PubMed] [Google Scholar]
- 64.Heather LC, Clarke K. Metabolism, hypoxia and the diabetic heart. J. Mol. Cell. Cardiol. 2011;50:598–605. doi: 10.1016/j.yjmcc.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 65.Izzotti A, Calin GA, Steele VE, Croce CM, De Flora S. Relationships of microRNA expression in mouse lung with age and exposure to cigarette smoke and light. FASEB J. 2009;23:3243–3250. doi: 10.1096/fj.09-135251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Izzotti A, Larghero P, Longobardi M, Cartiglia C, Camoirano A, Steele VE, De Flora S. Dose-responsiveness and persistence of microRNA expression alterations induced by cigarette smoke in mouse lung. Mutat. Res. 2011;717:9–16. doi: 10.1016/j.mrfmmm.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 67.Yang SR, Chida AS, Bauter MR, Shafiq N, Seweryniak K, Maggirwar SB, Kilty I, Rahman I. Cigarette smoke induces proinflammatory cytokine release by activation of NF-kappaB and posttranslational modifications of histone deacetylase in macrophages. Am. J. Physiol. Lung Cell. Mol. Physiol. 2006;291:L46–L57. doi: 10.1152/ajplung.00241.2005. [DOI] [PubMed] [Google Scholar]
- 68.Kuipers I, Bracke KR, Brusselle GG, Wouters EF, Reynaert NL. Smoke decreases reversible oxidations S-glutathionylation and S-nitrosylation in mice. Free Radical Res. 2012;46:164–173. doi: 10.3109/10715762.2011.647011. [DOI] [PubMed] [Google Scholar]
- 69.Chepelev NL, Willmore WG. Regulation of iron pathways in response to hypoxia. Free Radic. Biol. Med. 2011;50:645–666. doi: 10.1016/j.freeradbiomed.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 70.Natsuizaka M, Naganuma S, Kagawa S, Ohashi S, Ahmadi A, Subramanian H, Chang S, Nakagawa KJ, Ji X, Liebhaber SA, Klein-Szanto AJ, Nakagawa H. Hypoxia induces IGFBP3 in esophageal squamous cancer cells through HIF-1alpha-mediated mRNA transcription and continuous protein synthesis. FASEB J. 2012;26:2620–2630. doi: 10.1096/fj.11-198598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Corradetti MN, Inoki K, Guan KL. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J. Biol. Chem. 2005;280:9769–9772. doi: 10.1074/jbc.C400557200. [DOI] [PubMed] [Google Scholar]
- 72.Chai TF, Leck YC, He H, Yu FX, Luo Y, Hagen T. Hypoxia-inducible factor independent down-regulation of thioredoxin-interacting protein in hypoxia. FEBS Lett. 2011;585:492–498. doi: 10.1016/j.febslet.2010.12.033. [DOI] [PubMed] [Google Scholar]
- 73.Khan AR, Birbach M, Cohen MS, Ittenbach RF, Spray TL, Levy RJ, Gaynor JW. Chronic hypoxemia increases ventricular brain natriuretic peptide precursors in neonatal swine. Ann. Thorac. Surg. 2008;85:618–623. doi: 10.1016/j.athoracsur.2007.08.041. [DOI] [PubMed] [Google Scholar]
- 74.Shi GX, Andres DA, Cai W. Ras family small GTPase-mediated neuroprotective signaling in stroke. Cent. Nerv. Syst. Agents Med. Chem. 2011;11:114–137. doi: 10.2174/187152411796011349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.