Abstract
Aims/Introduction
Type 1 diabetes mellitus is a serious disorder characterized by destruction of pancreatic β-cells, culminating in absolute insulin deficiency. Genetic factors contribute to the susceptibility of type 1 diabetes mellitus. The aim of the present study was to identify more susceptibility genes of type 1 diabetes mellitus.
Materials and Methods
We carried out an initial gene-based genome-wide association study in a total of 4,075 type 1 diabetes mellitus cases and 2,604 controls by using the Gene-based Association Test using Extended Simes procedure. Furthermore, we carried out replication studies, differential expression analysis and functional annotation clustering analysis to support the significance of the identified susceptibility genes.
Results
We identified 452 genes associated with type 1 diabetes mellitus, even after adapting the genome-wide threshold for significance (P < 9.05E-04). Among these genes, 171 were newly identified for type 1 diabetes mellitus, which were ignored in single-nucleotide polymorphism-based association analysis and were not previously reported. We found that 53 genes have supportive evidence from replication studies and/or differential expression studies. In particular, seven genes including four non-human leukocyte antigen (HLA) genes (RASIP1, STRN4, BCAR1 and MYL2) are replicated in at least one independent population and also differentially expressed in peripheral blood mononuclear cells or monocytes. Furthermore, the associated genes tend to enrich in immune-related pathways or Gene Ontology project terms.
Conclusions
The present results suggest the high power of gene-based association analysis in detecting disease-susceptibility genes. Our findings provide more insights into the genetic basis of type 1 diabetes mellitus.
Keywords: Genome-wide Gene-Based Association Study, Knowledge-based mining system for Genome-wide Genetic studies, Type 1 diabetes mellitus
Introduction
Type 1 diabetes mellitus is a serious disorder characterized by destruction of pancreatic β-cells, leading to absolute insulin deficiency. Type 1 diabetes mellitus arises from uncontrolled inflammatory processes, and accounts for 5–10% of total cases of diabetes worldwide1. Diabetes mellitus is a major risk factor for micro- and macrovascular complications, and is associated with endothelial dysfunction, premature atherosclerosis and reduced capability of neovascularization in ischemic conditions. The increasing number of people developing diabetes might be associated with the changing environment in relation to diet and infection, but more with genetic factors2,3. Identification of genes predisposing to type 1 diabetes mellitus will increase our understanding of the genetic pathogenesis of type 1 diabetes mellitus, and contribute to the development of novel prevention and treatment of type 1 diabetes mellitus in the future.
Extensive evidence has shown that genetic factors play important roles in the development of type 1 diabetes mellitus3–5. However, identification of specific responsible genes and their variants has had limited success. The single-nucleotide polymorphism (SNP)-based genome-wide association studies (GWAS) have identified a long list of risk genes for type 1 diabetes mellitus6,7. However, the traditional GWAS ignored a large number of loci with moderate effects, because of the stringent significance thresholds used.
Gene-based analysis takes a gene as a basic unit for association analysis. As this method can combine genetic information given by all the SNPs in a gene to obtain more informative results8, it is being used as a novel method complementing SNP-based GWAS to identify disease susceptibility genes. Notably, this method can increase our chance of finding novel genes, which are usually ignored by SNP-based association analysis. In the present study, we presented a statistically robust gene-based GWAS, focusing on identifying ‘novel’ genes underlying susceptibility to type 1 diabetes mellitus.
Materials and Methods
Discovery Study Sample
The initial discovery sample included 4,075 cases and 2,604 controls. The case data came from ‘UK Genetic Resource Investigating Diabetes’ (available at www.childhood-diabetes.org.uk/grid.shtml). The control participants came from the 1958 British Birth Cohort. Genotyping, data-quality filter and SNP-based association analysis were detailed in the original publication in Nat Genet7, thus not elaborated here.
Replication Study Sample
Replication analyses were carried out in three independent study samples (Replication sample 1 [R1]: a total of 1,879 samples including 935 diabetic nephropathy cases and 944 normoalbuminuric controls; replication sample 2 [R2]: 486 trios including 223 affected trios and 263 unaffected trios; replication sample 3 [R3]: 685 white individuals with type 1 diabetes from the Diabetes Control and Complications Trial [DCCT]/Epidemiology of Diabetes Intervention and Complications [EDIC] study). Basic characteristics of the study participants, as well as the genotyping process, data quality control and SNP-based association analysis, have been detailed previously in the original publications9–11, thus not elaborated here.
Gene-Based Association Analysis
Raw data used in the present gene-based GWAS analysis and replication studies are P-values from genome-wide SNP-based GWAS. The data were downloaded from the publicly available dbGaP database (accession number: phs000180, phs000018, phs000086 and phs000088). Gene-based association analysis was carried out using Gene-based Association Test using Extended Simes procedure12, and the resultant gene-based P-value is a measure of statistical significance. The Gene-based Association Test using Extended Simes procedure was modeled in KGG, a systematic biological Knowledge-based mining system for Genome-wide Genetic studies (available at http://bioinfo1.hku.hk:13080/kggweb). The defined length of the extended gene region is from 5-kb upstream to 5-kb downstream of each gene.
Differential Expression Analyses of Type 1 Diabetes Mellitus-Associated Genes
Based on the normalized data available in the public databases (www.ncbi.nlm.nih.gov/geo), we tested differential expression of the aforementioned identified ‘novel’ candidate genes by comparing mean gene expression signals in peripheral blood mononuclear cells (PBMCs) or monocytes between type 1 diabetes mellitus cases and controls. Specifically, three gene expression datasets were downloaded from GEO Datasets (GSE number: GSE35725, GSE33440, GSE29142). Patients with type 1 diabetes mellitus show functional abnormalities of monocytes and monocyte-derived cells, which are assumed to promote the immunogenic potential of the cells13. Therefore, transcriptional signatures of patients' PBMCs might reflect inflammatory mechanisms14. The original studies were carried out to identify transcriptional signatures as a disease-specific and predictive inflammatory biomarker for type 1 diabetes mellitus14,15. Details on sample quality control and experimental procedures are previously described in the original publications14–16.
Functional Annotation Clustering Analysis
To gain insights into the functions of the identified genes, we tested the probability of the 452 identified genes clustering into a specific Gene Ontology (GO) project or a particular biological pathway that is defined by GO term or Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Specifically, a functional annotation clustering analysis was carried out by using the Database for Annotation, Visualization and Integrated Discovery (DAVID) integrated database query tools (http://david.abcc.ncifcrf.gov/)17. The enrichment can be quantitatively measured by using Fisher's exact test. The Bonferroni correction was used for multiple testing18.
Results
Discovery of Novel Genes Associated With Type 1 Diabetes Mellitus
A total of 24,984 genes were analyzed in the initial gene-based GWAS. Three quantile–quantile plots for gene-based P-values, SNP-based P-values inside genes and SNP-based P-values outside genes are shown in Figure1. We observed dramatic deviations at the tails of the distributions for the three plots. The deviation was much stronger for the plot of gene-based P-values than the other two plots, suggesting relatively higher power for gene-based association analysis.
Figure 1.
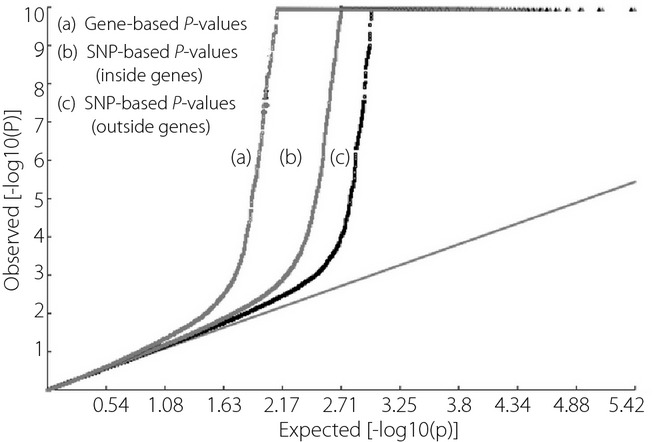
Q–Q plot of P-values. There are three quantile–quantile plots of the observed P-value distributions. As compared with the expected null P-value distributions, the tail of the distribution for gene-based P-values is the most significant deviation. SNPs, single-nucleotide polymorphisms.
The Manhattan plot of gene-level P-values across chromosomes is showed in Figure2. As expected, the majority of these associations were mapped to the HLA region, whose physical location lies in chromosome 6p21. To adjust for multiple testing, we used the Benjamini–Hochberg19 procedure to control false discovery rate (FDR) of the genome-wide association tests. To obtain a FDR of 0.05 across the whole genome, the significance level for a gene-based test is 9.05E-04. Accordingly, 452 genes were statistically significant (Table S1).
Figure 2.
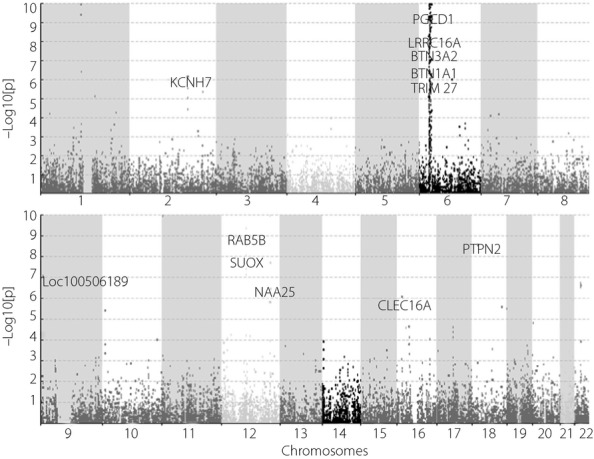
Manhattan plot of gene-based P-values on chromosomes.
In the original SNP-based GWAS analysis, according to the genome-wide P-value threshold of statistical significance (Bonferroni correction P < 9.94E-08), a total of 699 SNPs showed significant associations. These 699 SNPs corresponded to 269 genes. Comparatively, the current gene-based study detected 183 additional candidate genes for type 1 diabetes mellitus that were undetected by the SNP-based association analysis. To discover whether any of the 183 genes had been reported in other previous association studies, we searched the Phenotype-Genotype Integrator (PheGenI; www.ncbi.nlm.nih.gov/gap/PheGenI/), a database archiving previous association results. We compared the 183 genes and the list of genes with significant SNP-based P-values (P < 1.0E-07; Table S1). This comparison showed that just 12 among the total 183 genes were previously reported for an association with type 1 diabetes mellitus7,20–22. The rest of the 171 ‘novel’ genes were first detected for type 1 diabetes mellitus by the present study.
Confirmation of Type 1-Associated Genes by Replication Studies
The results of the replication analyses are summarized in Table S1 (P < 5.0E-02). For the 171 ‘novel’ genes, only one gene was replicated for associations with type 1 diabetes mellitus in study R1. In addition, eight genes were replicated in study R2 and 14 genes were replicated in study R3. In total, 23 of the 171 ‘novel’ genes (Table1) were confirmed for their association with type 1 diabetes mellitus. For the replication studies, the significance level of P < 5.0E-02 was used.
Table 1.
‘Novel’ type 1 diabetes mellitus-associated genes with supplementary evidence
| Gene symbol | Chr. | Start position | Length | SNP† | Gene P-value | Replication P-value | Differential expression P-value | ||
|---|---|---|---|---|---|---|---|---|---|
| RASIP1 | 19 | 49,218,842 | 30,128 | 6 | 4.83E –04 | 2.94E –02 | R2 | 3.18E –03 | ,S1 |
| STRN4 | 19 | 47,217,768 | 36,952 | 5 | 1.97E –04 | 2.67E –02 | R2 | 1.63E –02 | ,S1 |
| BCAR1 | 16 | 75,257,928 | 49,023 | 8 | 9.32E –05 | 2.27E –02 | R2 | 4.17E –02 | ,S1 |
| FYN | 6 | 111,977,485 | 222,142 | 48 | 3.08E –04 | 4.69E –03 | R2 | 2.01E –03 | ,S1 |
| MYL2 | 12 | 111,343,623 | 19,781 | 5 | 4.61E –04 | 8.53E –03 | R3 | 7.58E –03 | ,S1 |
| HLA-J | 6 | 29,968,748 | 13,985 | 1 | 3.49E –06 | 3.89E –02 | R3 | 1.63E –02 | ,S3 |
| PPP1R11 | 6 | 30,029,932 | 13,178 | 4 | 5.50E –06 | 2.99E –02 | R3 | 2.33E –03 1.69E –02 |
,S2 ,S3 |
| ITPR3 | 6 | 33,584,161 | 85,190 | 29 | 4.36E –05 | NS‡ | 4.29E –03 | ,S1 | |
| PLEKHA1 | 10 | 124,129,220 | 67,646 | 11 | 9.92E –05 | NS‡ | 1.04E –02 | ,S1 | |
| ATF7IP | 12 | 14,513,611 | 143,086 | 13 | 8.83E –05 | NS‡ | 2.10E –03 | ,S1 | |
| OR2B6 | 6 | 27,920,019 | 10,941 | 2 | 2.04E –06 | NS‡ | 2.65E –03 | ,S1 | |
| OR5V1 | 6 | 29,318,007 | 11,047 | 2 | 2.16E –06 | NA§ | 3.07E –03 | ,S1 | |
| HIST1H4E | 6 | 26,199,873 | 10,376 | 2 | 3.48E –06 | NA§ | 4.29E –03 | ,S1 | |
| HIST1H2BF | 6 | 26,194,787 | 10,429 | 1 | 6.65E –06 | NS‡ | 8.84E –03 | ,S1 | |
| GUSBL1 | 6 | 26,834,266 | 95,067 | 2 | 3.48E –07 | NA§ | 1.40E –02 | ,S1 | |
| HMGB1 | 13 | 31,027,877 | 17,204 | 2 | 2.07E –04 | NA§ | 1.99E –02 | ,S1 | |
| ZNF192 | 6 | 28,104,716 | 25,520 | 1 | 3.65E –06 | NS‡ | 3.92E –02 | ,S1 | |
| RING1 | 6 | 33,171,286 | 14,213 | 4 | 1.45E –04 | NS‡ | 4.40E –02 | ,S1 | |
| FAM46B | 1 | 27,326,511 | 17,822 | 1 | 6.36E –05 | NA§ | 4.97E –02 2.64E –02 |
,S1 ,S2 |
|
| OLFML3 | 1 | 114,517,030 | 12,845 | 2 | 2.13E –04 | NS‡ | 7.24E –03 1.54E –02 |
,S1 ,S3 |
|
| HIPK1 | 1 | 114,466,996 | 58,426 | 6 | 5.54E –04 | NS‡ | 3.02E –02 3.56E –02 |
,S1 ,S3 |
|
| NSL1 | 1 | 212,894,495 | 75,644 | 12 | 5.48E –05 | NS‡ | 2.27E –02 | ,S2 | |
| IL10 | 1 | 206,935,948 | 14,891 | 8 | 2.28E –04 | NS‡ | 1.05E –02 | ,S2 | |
| TRIM27 | 6 | 28,865,779 | 30,989 | 3 | 8.88E –07 | NS‡ | 8.79E –03 | ,S2 | |
| NCAPD2 | 12 | 6,598,298 | 47,834 | 10 | 1.36E –04 | NS‡ | 1.40E –04 | ,S2 | |
| PPP1R10 | 6 | 30,563,182 | 26,838 | 2 | 4.08E –04 | NS‡ | 3.95E –02 | ,S2 | |
| VPS52 | 6 | 33,213,049 | 31,613 | 2 | 6.73E –07 | NA§ | 1.32E –02 | ,S2 | |
| PHF1 | 6 | 33,373,773 | 15,457 | 2 | 3.14E –04 | NA§ | 6.98E –03 | ,S2 | |
| BAK1 | 6 | 33,535,323 | 17,747 | 7 | 5.18E –05 | NS‡ | 6.11E –03 1.60E –02 |
,S2 ,S3 |
|
| IKZF3 | 17 | 37,916,198 | 109,243 | 7 | 7.54E –05 | NS‡ | 1.87E –04 | ,S3 | |
| ZZEF1 | 17 | 3,902,739 | 148,514 | 23 | 3.55E –04 | NS‡ | 1.46E –03 | ,S3 | |
| GNS | 12 | 65,102,222 | 56,004 | 4 | 6.65E –05 | NS‡ | 1.83E –03 | ,S3 | |
| ORMDL3 | 17 | 38,072,296 | 16,558 | 3 | 7.22E –04 | NS‡ | 2.96E –02 | ,S3 | |
| BRAP | 12 | 112,074,950 | 53,840 | 3 | 1.16E –04 | NS‡ | 4.45E –02 | ,S3 | |
| CRYZL1 | 21 | 34,956,647 | 62,513 | 3 | 3.72E –04 | NA§ | 8.59E –03 | ,S3 | |
| SULT1A1 | 16 | 28,611,913 | 27,953 | 1 | 4.74E –04 | NA§ | 2.61E –02 | ,S3 | |
| TMEM129 | 4 | 1,712,679 | 15,405 | 3 | 7.99E –04 | NS‡ | 4.31E –02 | ,S3 | |
| IKZF1 | 7 | 50,439,231 | 38,568 | 15 | 6.66E –05 | 4.42E –02 | R1 | NA§ | |
| MICA | 6 | −1 | 0 | 3 | 7.52E –05 | 7.52E –05 | R2 | NA§ | |
| PLBD1 | 12 | 14,651,597 | 74,194 | 11 | 3.17E –04 | 3.26E –02 | R2 | NA§ | |
| DEXI | 16 | 11,017,748 | 23,509 | 3 | 8.72E –04 | 3.03E –03 | R2 | NA§ | |
| SBK1 | 16 | 28,298,840 | 41,330 | 7 | 2.28E –05 | 1.37E –02 | R2 | NA§ | |
| GCA | 2 | 163,195,583 | 28,566 | 4 | 9.67E –06 | 4.16E –02 | R3 | NA§ | |
| OR2B3 | 6 | 29,048,985 | 11,105 | 1 | 5.27E –07 | 3.31E –02 | R3 | NA§ | |
| HCP5P2 | 6 | 29,963,782 | 12,246 | 1 | 3.49E –06 | 3.89E –02 | R3 | NA§ | |
| HCG4P3 | 6 | 29,967,622 | 10,983 | 1 | 3.49E –06 | 3.89E –02 | R3 | NA§ | |
| OR2U1P | 6 | 29,225,436 | 11,420 | 5 | 1.05E –06 | 4.73E –02 | R3 | NA§ | |
| VN1R14P | 6 | 26,626,313 | 10,651 | 1 | 6.19E –06 | 2.94E –02 | R3 | NA§ | |
| HCG2P8 | 6 | 29,767,896 | 13,543 | 1 | 6.02E –07 | 3.80E –02 | R3 | NA§ | |
| HCGVIII-2 | 6 | 29,796,425 | 1,1279 | 2 | 4.51E –06 | 4.26E –02 | R3 | NA§ | |
| RPS10P1 | 6 | 26,197,351 | 10,592 | 2 | 3.48E –06 | 4.93E –02 | R3 | NA§ | |
| CCDC101 | 16 | 28,560,249 | 47,862 | 6 | 1.59E –04 | 2.98E –02 | R3 | NA§ | |
| FUT2 | 19 | 49,194,228 | 19,963 | 6 | 3.55E –04 | 3.28E –02 | R3 | NA§ | |
This table includes 53 significant type 1 diabetes mellitus-associated genes with supplemental evidence. Among these 53 genes, 37 were differentially expressed, and 23 genes have been replicated. The first seven genes in this table (RASIP1, STRN4, BCAR1, FYN, HLA-J, PPP1R1 and MYL2) have both differential expression and replication association. The numbers before and after ‘,’ are the differential expression P-value and sample number listed in Table2, respectively.
The number of single-nucleotide polymorphisms (SNP) included in a gene.
Not significant in R1, R2 and R3.
Not available in R1, R2 and R3.
Differential Expression Analyses of Type 1 Diabetes Mellitus Associated Genes
For the aforementioned 171 ‘novel’ genes, we used t-test to compare ribonucleic acid expression signals in PBMCs or monocytes between type 1 diabetes mellitus patients and healthy controls. We found that 37 genes, including 21 non-HLA genes (e.g. FAM46B, OLFML3 and HIPK1), were differentially expressed between type 1 diabetes mellitus patients and controls (Table2). For the differential expression study, the significance level of P < 5.0E-02 was used.
Table 2.
Differential expression analyses for ‘novel’ genes in type 1 diabetes mellitus-related cells
| Sample | S1 | S2 | S3 | |||||
|---|---|---|---|---|---|---|---|---|
| Target cells | PBMC | CD14+ Monocyte | PBMC | |||||
| Sample size | 46:44 | 16:6 | 9:10 | |||||
| Platform | [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | Illumina HumanHT-12 V3.0 expression beadchip | Phalanx Human One Array (version 4.3) | |||||
| References | 14 | 15 | 16 | |||||
| GSE NO. | GSE35725 | GSE33440 | GSE29142 | |||||
| Gene | Probe ID | t-test P-value | Gene | Probe ID | t-test P-value | Gene | Probe ID | t-test P-value |
| ATF7IP | 207728_at | 2.10E-03 | FYN | ILMN_1781207 | 2.64E-02 | HIPK1 | 14,914 | 3.56E-02 |
| BCAR1 | 223116_at | 4.17E-02 | BAK1 | ILMN_1805990 | 6.11E-03 | OLFML3 | 21,862 | 1.54E-02 |
| FAM46B | 229518_at | 4.97E-02 | IL10 | ILMN_2073307 | 1.05E-02 | BAK1 | 22,739 | 1.60E-02 |
| FYN | 1559101_at | 2.01E-03 | NCAPD2 | ILMN_1775008 | 1.40E-04 | PPP1R11 | 3205 | 1.69E-02 |
| GUSBL1 | 1555568_at | 1.40E-02 | NSL1 | ILMN_1739210 | 2.27E-02 | BRAP | 12,945 | 4.45E-02 |
| HIST1H2BF | 208490_x_at | 8.84E-03 | PHF1 | ILMN_1746968 | 6.98E-03 | CRYZL1 | 24,489 | 8.59E-03 |
| HIST1H4E | 206951_at | 4.29E-03 | PPP1R10 | ILMN_1659058 | 3.95E-02 | GNS | 8137 | 1.83E-03 |
| HMGB1 | 200679_x_at | 1.99E-02 | PPP1R11 | ILMN_1747598 | 2.33E-03 | HLA-J | 24,451 | 1.63E-02 |
| ITPR3 | 201187_s_at | 4.29E-03 | TRIM27 | ILMN_1655482 | 8.79E-03 | IKZF3 | 15,319 | 1.87E-04 |
| MYL2 | 209742_s_at | 7.58E-03 | VPS52 | ILMN_1666632 | 1.32E-02 | ORMDL3 | 10,233 | 2.96E-02 |
| OLFML3 | 218162_at | 7.24E-03 | SULT1A1 | 20,808 | 2.61E-02 | |||
| OR2B6 | 216522_at | 2.65E-03 | TMEM129 | 17,978 | 4.31E-02 | |||
| OR5V1 | 234840_s_at | 3.07E-03 | ZZEF1 | 16,832 | 1.46E-03 | |||
| PLEKHA1 | 219024_at | 1.04E-02 | ||||||
| RASIP1 | 220027_s_at | 3.18E-03 | ||||||
| RING1 | 208371_s_at | 4.40E-02 | ||||||
| STRN4 | 217903_at | 1.63E-02 | ||||||
| ZNF192 | 206579_at | 3.92E-02 | ||||||
| CRYZL1 | 1552347_at | 2.78E-02 | ||||||
Only the most significant probe was listed, even if more than one probe was tested or detected for a gene.
GSE NO, Gene Expression Omnibus Number (www.ncbi.nlm.nih.gov/geo/); PBMC, peripheral blood mononuclear cells.
In short, through a gene-based association study, we identified 183 type 1 diabetes mellitus-associated genes that were insignificant in the original SNP-based association tests. Among the 183 genes, 171 genes are ‘novel’ genes identified for type 1 diabetes mellitus. Replication studies and/or differential expression studies further supported the significance of 53 genes to type 1 diabetes mellitus. In particular, four non-HLA genes (RASIP1, STRN4, BCAR1 and MYL2) and three HLA genes (FYN, HLA-J and PPP1R11) were validated by both replication and differential expression studies.
Functional Annotation Clustering Analysis
Gene ontology analysis showed significant enrichment of 452 identified type 1 diabetes mellitus genes in particular biological terms and pathways. For example, we found a significant clustering (Bonferroni correction P = 1.50E-05) of 33 genes (e.g. HLA-DQB1, HLA-DMB, HLA-DMA) directly involved in immune response (Table S2). These genes tend to enrich in immune-related KEGG pathways (including hsa04940: type I diabetes mellitus, hsa04612: antigen processing and presentation, hsa04672: intestinal immune network for immunoglobulin A production; Table S2).
Discussion
Elucidation of the genetic basis of type 1 diabetes mellitus remains one of the huge challenges in the field of human genetics, which is largely because of the complex nature of the genetic determination for type 1 diabetes mellitus, including polygenic determinations, gene-by-gene and gene-by-environment interactions. Thus far, most of the published GWAS studies reported the results of single-marker-based analysis, where each SNP was analyzed individually6,21,23. Because of the large number of SNPs tested in a GWAS, stringent P-value thresholds for significance (typically P < 5.0E-08) are used to control false positive findings. Consequently, a large number of SNPs with moderate effects are missed. The gene-based association test is an important supplementary method for the SNP-based association test, which combines genetic information given by all the SNPs in a gene, thus obtaining a more informative result24. As we expected, the present gene-based association study identified more significant type 1 diabetes mellitus-susceptibility genes than the SNP-based test. Specifically, 171 ‘novel’ genes were identified for type 1 diabetes mellitus.
Compared with SNP/variant-based association tests, the gene-based association analyses have several distinct advantages. First, by combining the effects of all SNPs assigned to genes into a statistic analysis while correcting for linkage disequilibrium (LD), the gene-based analysis substantially alleviates the multiple-testing burden by reducing the number of tests in a GWAS. Second, the test unit is a gene, which is highly consistent across populations. Therefore, gene-based association analyses ignore confounding factors that are intrinsic in genetic variants-based association tests, such as allele frequencies, LD structure and heterogeneity across diverse human populations25. Finally, the gene-based tests are also ideally suitable for a network (or pathway) approach to interpret findings from GWAS26.
As previously reported2,3,27,28, more than half of the significant association signals for type 1 diabetes mellitus were identified within chromosome 6p21, which is the most important region in the vertebrate genome with respect to infection and autoimmunity, and is crucial in adaptive and innate immunity. The present study also found that more than 50% of the identified ‘novel’ genes (100 HLA genes/171 novel genes) are located at the HLA region. However, among 53 type 1 diabetes mellitus-associated genes validated by replication studies and/or differential expression studies, 28 genes are non-HLA genes. So far, the roles of these non-HLA genes directly in the pathogenesis of type 1 diabetes mellitus are largely unknown, but some of these genes play important roles in immune response. For example, BCAR1 might mediate its diabetogenic impact through impaired β-cell function29. IKZF1 and IKZF3 are members of the Ikaros family of zinc-finger proteins. The gene product is a transcription factor that is important in regulation of B lymphocyte proliferation and differentiation30,31. PLEKHA1 (also known as TAPP1) encodes a pleckstrin homology domain-containing adapter protein. The interaction of TAPP1 adapter proteins with phosphatidylinositol (3, 4)-bisphosphate could regulate B cell activation and autoantibody production. Several studies suggest that TAPP1 might play roles in B and T cell activation, which are necessary and sufficient conditions for immune response32. MYL2 can regulate the coordinated rearrangements of the actin–myosin cytoskeleton, and facilitate early and late events in T cell activation and signal transduction33. High mobility group box-1 (HMGB1) is an important component of the immune response, which can activate immune cells involved in immune process34. Interleukin-10 (IL-10) is a cytokine with anti-inflammatory and immunomodulatory function, which can regulate the biological functions of B and T cells35. GCA is a causal factor in autoimmune pancreatic β-cell destruction36. The aforementioned evidence supports that BCAR1, IKZF1, IKZF3, PLEKHA1, MYL2, HMGB1, IL-10 and GCA might have functional relevance to diabetes mellitus or immune response. In addition, some previous studies suggested that IL-10, ORMDL3 and FUT2 have been associated with type 1 diabetes mellitus7,37. Further studies are required to dissect the roles of these non-HLA genes in the pathogenesis of type 1 diabetes mellitus.
Our replication studies had a relatively low replication rate for the significant genes detected in the initial study. Small sample size (e.g., 486 trios subjects in R2 and 685 subjects in R3), and difference in demography (e.g., discover sample from the UK, whereas three replication study samples from New England, the USA and Canada) and genetic background could contribute to this. Another more important factor might be the difference in case identification. For R1, the case is s diabetic nephropathy patient. For R3, although the case is an individual with type 1 diabetes, the association analysis was carried out between the gene and type 1 diabetes-associated phenotype (E-selectin level in serum).
In conclusion, the findings presented in our study suggest high power for gene-based association analyses in detecting disease-susceptibility genes across the human genome. Our findings point to the involvement of new pathways in the pathogenesis of type 1 diabetes mellitus, and provide more insights into the genetic basis of type 1 diabetes mellitus.
Acknowledgments
The present study was supported by Natural Science Foundation of China (31271336, 31071097, 81373010), the Natural Science Foundation of Jiangsu Province (BK20130300), the Startup Fund from Soochow University (Q413900112, Q413900712), the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry, and a Project of the Priority Academic Program Development of Jiangsu Higher Education Institutions. There is no conflict of interest related to this study.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
| Information of 452 significant genes associated with type 1 diabetes mellitus.
| (a) Enrichment of Gene Ontology (GO) term of the 452 identified genes. (b) Enrichment of Kyoto Encyclopedia of Genes and Genomes pathways of the 452 identified genes.
References
- Maahs DM, West NA, Lawrence JM, et al. Epidemiology of type 1 diabetes. Endocrinol Metab Clin North Am. 2010;393:481–497. doi: 10.1016/j.ecl.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble JA, Valdes AM. Genetics of the HLA region in the prediction of type 1 diabetes. Curr Diab Rep. 2011;116:533–542. doi: 10.1007/s11892-011-0223-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Eisenbarth GS. Genetic susceptibility factors of Type 1 diabetes in Asians. Diabetes Metab Res Rev. 2001;171:2–11. doi: 10.1002/1520-7560(2000)9999:9999<::aid-dmrr164>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Sugihara S. Genetic susceptibility of childhood type 1 diabetes mellitus in Japan. Pediatr Endocrinol Rev. 2012;10(Suppl. 1):62–71. [PubMed] [Google Scholar]
- Mosaad YM, Auf FA, Metwally SS, et al. HLA-DQB1* alleles and genetic susceptibility to type 1 diabetes mellitus. World J Diabetes. 2012;38:149–155. doi: 10.4239/wjd.v3.i8.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AD, Waggott D, Boright AP, et al. A genome-wide association study identifies a novel major locus for glycemic control in type 1 diabetes, as measured by both A1C and glucose. Diabetes. 2010;592:539–549. doi: 10.2337/db09-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Clayton DG, Concannon P, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;416:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buil A, Martinez-Perez A, Perera-Lluna A, et al. A new gene-based association test for genome-wide association studies. BMC Proc. 2009;3(Suppl. 7):S130. doi: 10.1186/1753-6561-3-s7-s130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller PW, Rogus JJ, Cleary PA, et al. Genetics of Kidneys in Diabetes (GoKinD) study: a genetics collection available for identifying genetic susceptibility factors for diabetic nephropathy in type 1 diabetes. J Am Soc Nephrol. 2006;177:1782–1790. doi: 10.1681/ASN.2005080822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzolesi MG, Poznik GD, Mychaleckyj JC, et al. Genome-wide association scan for diabetic nephropathy susceptibility genes in type 1 diabetes. Diabetes. 2009;586:1403–1410. doi: 10.2337/db08-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AD, Lopes-Virella MF, Waggott D, et al. Genome-wide association identifies the ABO blood group as a major locus associated with serum levels of soluble E-selectin. Arterioscler Thromb Vasc Biol. 2009;2911:1958–1967. doi: 10.1161/ATVBAHA.109.192971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MX, Gui HS, Kwan JS, et al. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am J Hum Genet. 2011;883:283–293. doi: 10.1016/j.ajhg.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyan H, Drexhage RC, van der Heul Nieuwenhuijsen L, et al. Monocyte gene-expression profiles associated with childhood-onset type 1 diabetes and disease risk: a study of identical twins. Diabetes. 2010;597:1751–1755. doi: 10.2337/db09-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy H, Wang X, Kaldunski M, et al. Transcriptional signatures as a disease-specific and predictive inflammatory biomarker for type 1 diabetes. Genes Immun. 2012;138:593–604. doi: 10.1038/gene.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine KM, Gallego P, An X, et al. Peripheral blood monocyte gene expression profile clinically stratifies patients with recent-onset type 1 diabetes. Diabetes. 2012;615:1281–1290. doi: 10.2337/db11-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stechova K, Kolar M, Blatny R, et al. Healthy first degree relatives of patients with type 1 diabetes exhibit significant differences in basal gene expression pattern of immunocompetent cells compared to controls: expression pattern as predeterminant of autoimmune diabetes. Scand J Immunol. 2011;75:210–219. doi: 10.1111/j.1365-3083.2011.02637.x. [DOI] [PubMed] [Google Scholar]
- Sherman BT, da Huang W, Tan Q, et al. DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics. 2007;8:426. doi: 10.1186/1471-2105-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarone RE. A modified Bonferroni method for discrete data. Biometrics. 1990;462:515–522. [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;97:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- Todd JA, Walker NM, Cooper JD, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;397:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JD, Smyth DJ, Smiles AM, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. 2008;4012:1399–1401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagnol V, Howson JM, Smyth DJ, et al. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet. 2011;78:e1002216. doi: 10.1371/journal.pgen.1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakonarson H, Qu HQ, Bradfield JP, et al. A novel susceptibility locus for type 1 diabetes on Chr12q13 identified by a genome-wide association study. Diabetes. 2008;574:1143–1146. doi: 10.2337/db07-1305. [DOI] [PubMed] [Google Scholar]
- Howard TD, Koppelman GH, Xu J, et al. Gene-gene interaction in asthma: IL4RA and IL13 in a Dutch population with asthma. Am J Hum Genet. 2002;701:230–236. doi: 10.1086/338242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, McRae AF, Nyholt DR, et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;871:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranzini SE, Galwey NW, Wang J, et al. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum Mol Genet. 2009;1811:2078–2090. doi: 10.1093/hmg/ddp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugawan TL, Klitz W, Alejandrino M, et al. The association of specific HLA class I and II alleles with type 1 diabetes among Filipinos. Tissue Antigens. 2002;596:452–469. doi: 10.1034/j.1399-0039.2002.590602.x. [DOI] [PubMed] [Google Scholar]
- Sheehy MJ, Scharf SJ, Rowe JR, et al. A diabetes-susceptible HLA haplotype is best defined by a combination of HLA-DR and -DQ alleles. J Clin Invest. 1989;833:830–835. doi: 10.1172/JCI113965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder MN, Ribel-Madsen R, Justesen JM, et al. Type 2 diabetes risk alleles near BCAR1 and in ANK1 associate with decreased beta-cell function whereas risk alleles near ANKRD55 and GRB14 associate with decreased insulin sensitivity in the Danish Inter99 cohort. J Clin Endocrinol Metab. 2013;984:E801–E806. doi: 10.1210/jc.2012-4169. [DOI] [PubMed] [Google Scholar]
- Sun J, Matthias G, Mihatsch MJ, et al. Lack of the transcriptional coactivator OBF-1 prevents the development of systemic lupus erythematosus-like phenotypes in Aiolos mutant mice. J Immunol. 2003;1704:1699–1706. doi: 10.4049/jimmunol.170.4.1699. [DOI] [PubMed] [Google Scholar]
- Hu SJ, Wen LL, Hu X, et al. IKZF1: a critical role in the pathogenesis of systemic lupus erythematosus? Mod Rheumatol. 2013;232:205–209. doi: 10.1007/s10165-012-0706-x. [DOI] [PubMed] [Google Scholar]
- Landego I, Jayachandran N, Wullschleger S, et al. Interaction of TAPP adapter proteins with phosphatidylinositol (3,4)-bisphosphate regulates B-cell activation and autoantibody production. Eur J Immunol. 2012;4210:2760–2770. doi: 10.1002/eji.201242371. [DOI] [PubMed] [Google Scholar]
- Liu X, Lindberg R, Xiao BG, et al. CD24 and myosin light polypeptide 2 are involved in prevention of experimental autoimmune encephalomyelitis by myelin basic protein-pulsed dendritic cells. J Neuroimmunol. 2006;1721–2:137–144. doi: 10.1016/j.jneuroim.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Klune JR, Dhupar R, Cardinal J, et al. HMGB1: endogenous danger signaling. Mol Med. 2008;147–8:476–484. doi: 10.2119/2008-00034.Klune. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin YW, Hu AM, Sun QQ, et al. Association between interleukin 10 gene -1082 A/G polymorphism and the risk of type 2 diabetes mellitus: a meta-analysis of 4250 subjects. Cytokine. 2013;622:226–231. doi: 10.1016/j.cyto.2013.02.025. [DOI] [PubMed] [Google Scholar]
- Martinez A, Santiago JL, Cenit MC, et al. IFIH1-GCA-KCNH7 locus: influence on multiple sclerosis risk. Eur J Hum Genet. 2008;167:861–864. doi: 10.1038/ejhg.2008.16. [DOI] [PubMed] [Google Scholar]
- Smyth DJ, Cooper JD, Howson JM, et al. FUT2 nonsecretor status links type 1 diabetes susceptibility and resistance to infection. Diabetes. 2011;6011:3081–3084. doi: 10.2337/db11-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
| Information of 452 significant genes associated with type 1 diabetes mellitus.
| (a) Enrichment of Gene Ontology (GO) term of the 452 identified genes. (b) Enrichment of Kyoto Encyclopedia of Genes and Genomes pathways of the 452 identified genes.


