Abstract
Pasteurella multocida (Pm) is a gram-negative bacterium able to infect different animal species, including human beings. This bacterium causes economic losses to the livestock industry because of its high morbidity and mortality in animals. In this work, we report the characterization of outer membrane vesicles (OMVs) released into the culture medium by different Pm serogroups. Purified OMVs in the range of 50–300 nm were observed by electron microscopy. Serum obtained from chickens infected with Pm recognized several proteins from Pm OMVs. Additionally, rabbit antiserum directed against a secreted protease from Actinobacillus pleuropneumoniae recognized a similar protein in the Pm OVMs, suggesting that OMVs from these bacterial species contain common immunogenic proteins. OmpA, a multifunctional protein, was identified in OMVs from different Pm serogroups, and its concentration was twofold higher in OMVs from Pm serogroups B and D than in OMVs from other serogroups. Three outer membrane proteins were also identified: OmpH, OmpW, and transferrin-binding protein. Three bands of 65, 110, and 250 kDa with proteolytic activity were detected in Pm OMVs of serogroups A and E. Additionally, β-lactamase activity was detected only in OMVs from Pm 12945 Ampr (serogroup A). Pm OMVs may be involved in different aspects of disease pathogenesis.
Keywords: Outer membrane vesicles, Pasteurella multocida, proteases, virulence factors, β-lactamase
Introduction
Pasteurella multocida is a gram-negative bacterium that infects different animal species and human beings. It causes many widespread infections, such as snuffles in rabbits; pneumonia in cattle, sheep, and goats; fowl cholera in chickens; and atrophic rhinitis in pigs (Carter 1967). In humans, it has been associated with diseases of the lower and the upper respiratory tracts, arthritis, osteomyelitis, meningitis, and infections due to scratches by dogs and cats, among others (Felix et al. 2003).
Based on soluble capsular polysaccharides, 5 serogroups of Pm (A, B, D, E, and F) and 16 somatic serotypes are currently differentiated, but none of them is completely restricted to a specific host (Mutters et al. 1989). The key virulence factors of Pm, which are required for a successful infection, are different types of fimbrial adhesins, such as type IV pili, filamentous hemagglutinin, and short pili, and nonfimbrial adhesins, such as OmpA, capsule, and LPS. Pm also carries other virulence determinants, such as iron acquisition proteins, siderophores, and a dermonecrotic toxin only expressed by serogroup D strains (Harper et al. 2006; Hatfaludi et al. 2010).
Gram-negative bacteria possess several secretion mechanisms including outer membrane vesicles (OMVs). OMVs can be released into the surrounding medium by commensal as well as pathogenic microorganisms (Beveridge 1999). The composition of the OMVs varies depending on the species and strain of the microorganism. The composition of the OMVs also varies when the microorganism is cultured under normal or stress conditions (Mashburn-Warren and Whiteley 2006; Schooling and Beveridge 2006; Amano et al. 2010). OMVs contain toxins, enzymes, adhesins, DNA, and other outer membrane (OM) and periplasmic virulence components (Amano et al. 2010). Based on their composition following distinct functions for OMVs have been suggested: (1) predatory functions because of their ability to merge with membranes from similar or different bacteria (Mashburn-Warren and Whiteley 2006; Mashburn-Warren et al. 2008); (2) generation of nutrients or free space at the colonization site; (3) as transport vehicles because they can contain virulence factors, DNA, antibiotics, and β-lactamase (Ciofu et al. 2000; Mashburn-Warren et al. 2008; Schaar et al. 2011); (4) as elements that mediate adherence by promoting the agglutination of the same bacterial species or related microorganisms; and (5) as main components of biofilms (Schooling and Beveridge 2006).
The aim of this work was to isolate OMVs from cultures of Pm to identify proteins that have been described as possible virulence factors and participate in the Pm pathogenesis.
Materials and Methods
Bacterial strains
The following Pm strains (ATCC) were used: 12945 (serogroup A, Ampr), 43017 (serogroup B); 12948 (serogroup D); 43020 (serogroup E); and C44 (a field isolate of serogroup A from rabbit) (Soriano-Vargas et al. 2012).
Isolation of outer membrane vesicles
All bacterial strains were grown in brain heart infusion broth (BHI; BD Bioxon, Cuautitlán Izcalli, México, México) at 37°C with agitation for 24 h. Bacterial cells were harvested by centrifugation (10,000g for 20 min at 4°C). Culture supernatants were filtered through a 0.22 μm pore size membrane (Millipore Billerica, MA, USA) to remove residual cells. OMVs were recovered by ultracentrifugation (150,000g for 3 h at 4°C) as described previously (Negrete-Abascal et al. 2000).
Electron microscopy
Whole bacterial cells or OMVs were placed on carbon-and Formvar-coated copper grids, negatively stained with 1% (w/v) phosphotungstic acid, and observed with a JEM 2000 EX transmission electron microscope (Peabody, MA USA) at 80 kV (Negrete-Abascal et al. 2000).
Electrophoresis and zymograms
Total protein (10–15 μg) that was obtained from Pm vesicles was loaded by well (Bradford, 1976) and separated by electrophoresis in the presence of 5% (v/v) β-mercaptoethanol on a 10% (w/v) SDS polyacrylamide gel, except when the analysis was carried out using gels copolymerized with 0.1% (w/v) gelatin or with 1% (w/v) casein, as described previously (Negrete-Abascal et al. 1998). Both types of gels were stained with Coomassie brilliant blue R-250 to visualize the protein patterns or proteolytic bands.
Immunoblotting
To identify immunogenic proteins present in Pm OMVs, samples were separated by electrophoresis and proteins were transferred to nitrocellulose membranes (Sigma, St Louis, MO) for 1 h at 400 mA according to the protocol of Towbin et al. (1979). Membranes were blocked with skim milk for 1 h at room temperature, incubated with either rabbit hyperimmune serum against an Actinobacillus pleuropneumoniae-purified protease (Negrete-Abascal et al. 1998) or a pool of serum samples from chickens infected with Pm. Membranes were processed as described previously by Ramón Rocha et al. (2006).
Presence of active β-lactamase in OMVs from Pm
Nitrocefin disks (Becton Dickinson of México, Cuautitlán Izcalli, México) were used to detect β-lactamase in Pm OMVs. As a positive control, a clinical isolate of β-lactam-resistant strain of Staphylococcus aureus was used, and for the negative control, Pm strains or OMVs of β-lactam-sensitive bacteria were used. According to the manufacturer's instructions, red color indicated resistance to the action of penicillin or cephalosporin.
Results and Discussion
Gram-negative bacteria secrete different components into their surrounding medium, including various virulence factors, such as toxins that damage host cells, proteases that degrade molecules involved in host defense, such as immunoglobulins and structural components of the host, or elemental molecules that capture iron, such as siderophores (Confer 2009). Bacteria are also able to release OMVs, which can carry different cargo molecules, such as proteases, toxins, hemagglutinins, and fimbriae proteins; these molecules can either be contained in the OMVs or displayed on the surface. In addition, OMVs are enriched in outer membrane proteins (Kuehn and Kesty 2005; Amano et al. 2010). In addition to being important components of biofilms, OMVs allow the microorganisms to interact with host tissues or other microorganisms in the same niche (Schooling and Beveridge 2006; Mashburn-Warren et al. 2008; Ellis and Kuehn 2010).
Negative staining of released OMVs or OMVs associated with Pm bacterial surface was performed to facilitate visualization by transmission electron microscopy. OMVs varied in size from 50 to 300 nm (Fig.1A and B), with an average size of 100 nm, which is similar to the size of OMVs that have been described for A. pleuropneumoniae, Avibacterium paragallinarum, and other Gram-negative bacteria (Amano et al. 2010). No bacterial growth in OMVs samples inoculated in liquid BHI confirmed the absence of viable cells in OMVs preparations. Upon electrophoresis, the molecular weight of the proteins from OMVs was found to range from 20 to 100 kDa (Fig.2). The molecular weight range of these proteins is similar to that described for OMV proteins from A. pleuropneumoniae and Av. paragallinarum, both of which are members of the Pasteurellaceae family (Negrete-Abascal et al. 2000; Ramón Rocha et al. 2006).
Figure 1.
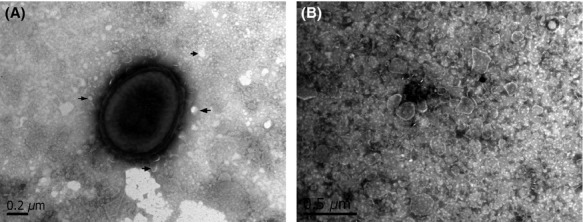
Transmission electron micrograph of negatively stained Pm 43020 strain. (A) The release of outer membrane vesicles (OMVs) from Pm can be seen in the micrograph. (B) Pm OMVs negatively stained. Arrows indicate some of the OMVs released
Figure 2.
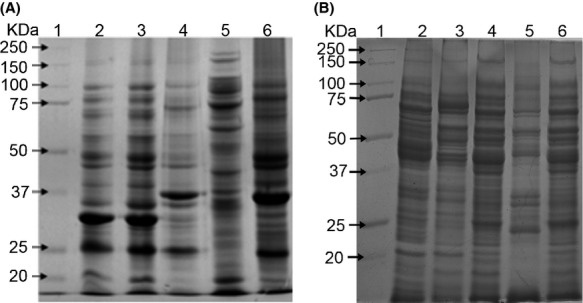
Protein pattern of outer membrane vesicles (OMVs) (A) and total cell extracts (B) from different strains of Pm as observed by electrophoresis in a 10% polyacrylamide gel. Lane 1: molecular weight markers (MWM); Lane 2: Pm 43017; Lane 3: Pm 43020; Lane 4: Pm C-44; Lane 5: Pm 12945; and Lane 6: Pm 12948. All samples were boiled in the presence of 5% β-mercaptoethanol.
Pasteurella multocida has the ability to infect a wide range of hosts. Its pathogenicity is complex, and different virulence factors, such as outer membrane proteins and porin proteins (Oma87, Psl, OmpH), type 4 fimbriae (PtfA), a filamentous hemagglutinin (PfhA), neuraminidases (NanB, NanH), iron acquisition-related factors (ExbBD-TonB, TbpA, HgbA, HgbB), a dermonecrotoxin (ToxA), and two superoxide dismutases (SodA, SodC) among others (Confer 2009; Hatfaludi et al. 2010), are involved in pathogenesis. Several of these virulence factors are located in the outer membrane and could be released in the OMVs.
An approximately 35–37 kDa protein was observed in OMVs from all the Pm serogroups that were studied. This protein was enriched in samples from Pm 12948 and Pm C44 strains (Fig.2). However, it migrated as a higher molecular weight protein if samples were not boiled or treated with 5% β-mercaptoethanol (Fig.3). Based on these properties, this protein was considered the OmpA of Pm (Hatfaludi et al. 2010). In some cases, specific proteins are enriched in OMVs while others are excluded, suggesting a specific sorting mechanism for these proteins (Kato et al. 2002).
Figure 3.
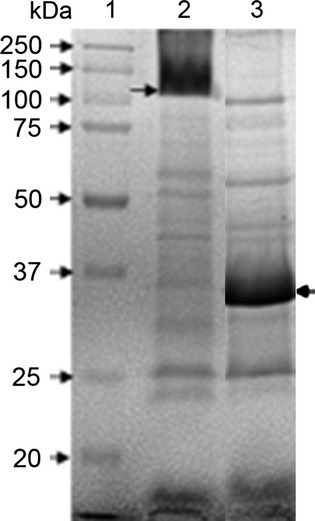
Protein patterns of outer membrane vesicles (OMVs) from Pm 12948 as observed by electrophoresis in a 10% polyacrylamide gel. Lane 1: molecular weight markers (MWM); Lane 2: sample without treatment; and Lane 3: sample was boiled in the presence of 5% β-mercaptoethanol. Arrows indicate the OmpA
Several OMV proteins obtained from Pm were immunorecognized by serum obtained from chickens infected with Pm (Fig.4). The 35–37 kDa heat-modified protein was also recognized by serum from animals infected with A. paragallinarum, Pm, or G. anatis, suggesting the presence of a similar protein and its in vivo expression in these microorganisms (data not shown).
Figure 4.
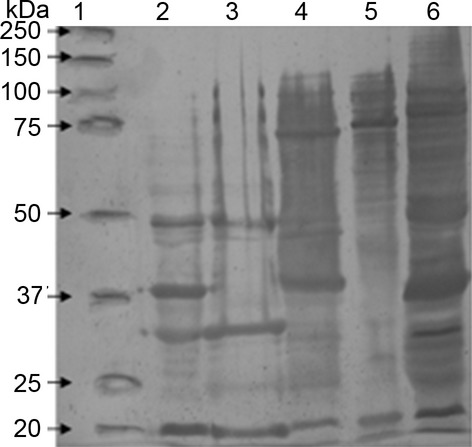
Immunorecognition of proteins from Pm OMVs using a pool of serum samples from chickens infected with Pm. The loading order and the amount of protein loaded are the same as in Figure2.
Due to its enrichment in OMVs, immunogenicity, and presence in different microorganisms, the 35–37 kDa protein from Pm 12948 was cut from the gel and submitted for mass spectrophotometric analysis. Six peptides were identified by in silico analysis as part of the outer membrane protein A (OmpA) from Pm and other microorganisms (Table1). This protein was characterized previously in Pm, and in addition to other functions, it is also involved in serum resistance and adhesion to host cells because of its interaction with heparin and fibronectin (Smith et al. 2007; Dabo et al. 2008; Hatfaludi et al. 2010). In Escherichia coli, OmpA is considered a multifunctional protein that exhibits both phage receptor activity and some porin activity; it maintains the integrity of the OM and is one of the major factors responsible for E. coli invasion of human brain microvascular endothelial cells (Smith et al. 2007). OmpA participates in biofilm formation and in the evasion of the immune system, and it is considered a target for development of vaccines. In addition, OmpA in Acinetobacter baumannii, an important nosocomial pathogen that causes a variety of human infections in critically ill patients, modulates the biogenesis and most likely the content of OMVs (Moon et al. 2012). OmpH, OmpW, and transferrin-binding protein were also identified into those outer membrane proteins immunorecognized by the sera used (Table1). OmpH is an outer membrane porin protein abundantly expressed in the P. multocida bacterial surface (Tan et al. 2010). OmpW is a scarcely studied protein in Pm; however, in E. coli this protein is important for avoiding the phagocytosis and its up-expression correlates with an increased bacterial survival during phagocytosis (Wu et al. 2013). This protein could be important for Pm virulence. Transferrin-binding proteins are considered virulence factors in several microorganisms, since they participate in iron uptake.
Table 1.
Peptide mass fingerprint of different immunogenic proteins from Pm OMVs.
| m/z | Residue | Sequence | |
|---|---|---|---|
| OmpA | 1871.7 | 42–59 | ASHDLGEGLSALAYAELR |
| 2142.7 | 143–163 | SAEFNGFTFGGATVFSAGADK | |
| 1094.5 | 171–180 | GFVVAGLYNR | |
| 2381.9 | 182–203 | NGDVGFALEAGYSQEYVTETAK | |
| 1108.5 | 254–262 | VYTDLIWAK | |
| 1284.5 | 286–296 | QVETFVEGGYR | |
| OmpH | 933.4 | 29–37 | GDLVDNGSR |
| 1873.9 | 42–59 | ASHDLGEGLSALAYAELR | |
| 2201.4 | 138–158 | SAEFNGFTFGGAYVFSADADK | |
| 1095.6 | 166–175 | GFVVAGLYNR | |
| 2250.0 | 177–197 | MGDVGFALEAGYSQKYVTVAK | |
| 1464.4 | 233–245 | ALEVGLNYDINDK | |
| OmpW | 1866.1 | 31–47 | GGPILVVPNASTNHDVFK |
| 2082.9 | 93–109 | TKHLPPSLYAQYYFLDK | |
| 2169.4 | 95–112 | HLPPSLYAQYYFLDKDAK | |
| 2007.2 | 113–130 | ARPYVGAGVNYTTFFSEK | |
| 1028.9 | 130–140 | AVLNGVTDLK | |
| 1483.3 | 191–203 | LDPTVFFVGLGYR | |
| Tbp | 2032.3 | 116–136 | VAVIVDGIPQAESTMSTSAR |
| 1945.5 | 142–158 | HNGNINNIEYENVSSLK | |
| 1884.2 | 251–266 | GKPNPLNYYTTSWLTK | |
| 1701.4 | 351–365 | NKLDSTMSFVYLQR | |
| 2758.6 | 526–549 | TSSQFLPNPDLQPETALNHEISYR | |
| 1650.7 | 718–731 | NVILNMGVFNLFNR |
Data about virulence genes have been reported mainly for two of the five strains studied here: Pm 12948 and Pm 12945 (Verma et al., 2013; Ewers et al. 2006).
Zymograms using porcine gelatin or casein as substrates revealed 65, 110, and 250 kDa bands of proteolytic activity in OMV samples prepared from Pm 43020 and Pm C44 (Fig.5A). The proteolytic activity was inactivated in the presence of 20 mmol/L EDTA, which is a characteristic of metalloproteases that have been previously described in field isolates of Pm from several animal species. These metalloproteases have been shown to degrade IgG molecules (Negrete-Abascal et al. 1999). Although only OMV samples from Pm 43020 and Pm C44 exhibited proteolytic activity, bands of approximately 60 kDa were immunorecognized by a rabbit polyclonal serum (raised against the purified protease from A. pleuropneumoniae) in OMV samples from all the Pm strains that were examined (Fig.5B), a similar recognition band was observed in a sample of A. pleuropneumoniae secreted proteins used as positive control of recognition. This immunorecognition corresponds to a band of proteolytic activity observed in the zymogram (Fig.5A). This result suggests that proteases are associated with OMVs in both active and inactive forms. The association of inactive proteases with OMVs has been described for A. pleuropneumoniae and A. paragallinarum (Negrete-Abascal et al. 2000; Ramón Rocha et al. 2006).
Figure 5.
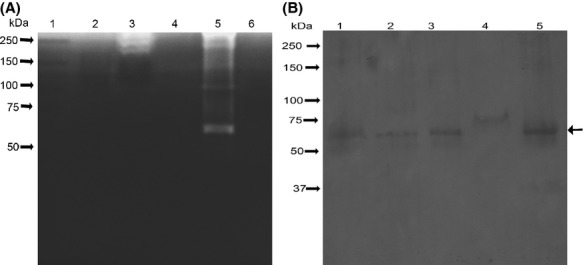
(A) Proteolytic activity of Pm outer membrane vesicles (OMVs) in 10% polyacrylamide gel copolymerized with 0.1% porcine gelatin. The loading order and the amount of sample loaded are the same as in Figure2. (B) Immunorecognition of putative proteases in Pm OMV samples using Actinobacillus pleuropneumoniae antiprotease serum. Lane 1: secreted proteins of A. pleuropneumoniae; Lane 2: Pm 43017; Lane 3: Pm 43020; Lane 4: Pm 12945; and Lane 5: Pm C-44. Arrow indicates the main band of recognition.
Of all the Pm strains tested for β-lactamase, only Pm 12945 was positive. This result is in good agreement with a previous study performed by Sellyei et al. (2009) to test the susceptibility of P. multocida isolated from chickens and pigs to antimicrobial agents. In that study, the authors found that the majority of P. multocida strains tested were susceptible to penicillin. OMVs derived from Pm 12945 strain tested positive on nitrocefin disks but a strong response was observed when the bacterial cells were included. This confirmed the presence of active β-lactamase enzymes in OMVs. A negative reaction was observed when culture medium or the OMVs from β-lactam antibiotic-sensitive Pm strains were used instead of Pm 12945 strain (Fig.6). Releasing β-lactamase activity in Pm OMVs may contribute to permanence into the host although not all the bacteria in situ contain a β-lactamase resistance gene in their genome. It has been shown that Moraxella catarrhalis, a human respiratory pathogen, releases OMVs containing active β-lactamase, which can rescue other amoxicillin-sensitive respiratory pathogens, including M. catarrhalis, Streptococcus pneumoniae, or Haemophilus influenzae (Schaar et al. 2011) from β-lactam killing. Presence of β-lactamase has also been described in Pseudomonas aeruginosa OMVs (Ciofu et al. 2000). In order to know if β-lactamase was a contamination from cell debris, OMVs were treated with proteinase K, to digest β-lactamase associated with the outer membrane of the OMVs, and permeabilized with 0.02% Triton X-100 and β-lactamase activity was measured on nitrocefin disks as before. Although activity diminished highly, it does not disappear, suggesting that β-lactamase activity was present externally and inside of OMVs (data not shown).
Figure 6.

Detection of β-lactamase associated with outer membrane vesicles (OMVs) using nitrocefin disk. (1) Penicillin-sensitive Pm 12948 strain; (2) OMVs; and (3) penicillin-resistant Pm 12945 cells.
These results demonstrate that the composition of OMVs is complex and distinct from the OM, and accentuates the potential for a diversity of biological functions for these vesicles. The different virulence factors in OMVs released by a pathogen could be responsible for damage caused during host infection. As with other microorganisms, they could also be good immunogens as components of vaccines.
Acknowledgments
This work was supported by DGAPA-UNAM. Projects: PAPIIT IN222313 and PAPCA-FES-Iztacala. We thank Jose Molina and Lourdes Rojas from the Electron Microscopy Laboratory of CINVESTAV-IPN for their technical assistance.
Conflict of Interest
None declared.
References
- Amano A, Takeuchi H. Furuta N. Outer membrane vesicles function as offensive weapons in host-parasite interactions. Microbes Infect. 2010;12:791–798. doi: 10.1016/j.micinf.2010.05.008. doi: 10.1016/j.micinf.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Beveridge TJ. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 1999;181:4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Carter GR. Pasteurellosis: Pasteurella multocida and Pasteurella haemolytica. Adv. Vet. Sci. Comp. Med. 1967;11:321–379. [PubMed] [Google Scholar]
- Ciofu O, Beveridge TJ, Kadurugamuwa J, Walther-Rasmussen J. Hoiby N. Chromosomal betalactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2000;45:9–13. doi: 10.1093/jac/45.1.9. [DOI] [PubMed] [Google Scholar]
- Confer AW. Update on bacterial pathogenesis in BRD. Anim. Health Res. Rev. 2009;10:145–148. doi: 10.1017/S1466252309990193. doi: 10.1017/S1466252309990193. [DOI] [PubMed] [Google Scholar]
- Dabo SM, Confer A, Montelongo M, York P. Wyckoff JH., III Vaccination with Pasteurella multocida recombinant OmpA induces strong but non-protective and deleterious Th2-type immune response in mice. Vaccine. 2008;26:4345–4351. doi: 10.1016/j.vaccine.2008.06.029. doi: 10.1016/j.vaccine.2008.06.029. [DOI] [PubMed] [Google Scholar]
- Ellis TN. Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. 2010;74:81–94. doi: 10.1128/MMBR.00031-09. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers C, Lübke-Decker A, Bethe A, Kiebling S, Filter M. Wieler LH. Virulence genotype pf Pasteurella multocida strains isolated from different hosts with various disease status. Vet. Microbiol. 2006;114:304–317. doi: 10.1016/j.vetmic.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Felix M, Tallon P, Salavert M, Navarro V, Breton JR, Pérez-Béllez C, et al. Bacteremia due to Pasteurella spp.: a rare process in our hospital over the last 8 years. Enferm. Infect. Microbiol. Clin. 2003;21:334–339. doi: 10.1016/s0213-005x(03)72959-0. [DOI] [PubMed] [Google Scholar]
- Harper M, Boyce JD. Adler B. Pasteurella multocida pathogenesis: 125 years after Pasteur. FEMS Microbiol. Lett. 2006;265:1–10. doi: 10.1111/j.1574-6968.2006.00442.x. [DOI] [PubMed] [Google Scholar]
- Hatfaludi T, Al-Hasani K, Boyce JD. Adler B. Outer membrane proteins of Pasteurella multocida. Vet. Microbiol. 2010;144:1–17. doi: 10.1016/j.vetmic.2010.01.027. doi: 10.1016/j.vetmic.2010.01.027. [DOI] [PubMed] [Google Scholar]
- Kato S, Kowashi Y. Demuth DR. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb. Pathog. 2002;32:1–13. doi: 10.1006/mpat.2001.0474. [DOI] [PubMed] [Google Scholar]
- Kuehn MJ. Kesty MC. Bacterial outer membrane vesicles and the host–pathogen interaction. Genes Dev. 2005;19:2645–2655. doi: 10.1101/gad.1299905. [DOI] [PubMed] [Google Scholar]
- Mashburn-Warren LM. Whiteley M. Special delivery: vesicle trafficking in prokaryotes. Mol. Microbiol. 2006;61:839–846. doi: 10.1111/j.1365-2958.2006.05272.x. [DOI] [PubMed] [Google Scholar]
- Mashburn-Warren LM, Mclean RJC. Whiteley M. Gram-negative outer membrane vesicles: beyond the cell surface. Geobiology. 2008;6:214–219. doi: 10.1111/j.1472-4669.2008.00157.x. doi: 10.1111/j.1472-4669.2008.00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon DC, Choi CH, Lee JH, Choi CW, Kim HY, Park JS, et al. Acinetobacter baumannii outer membrane protein A modulates the biogenesis of outer membrane vesicles. J. Microbiol. 2012;50:155–160. doi: 10.1007/s12275-012-1589-4. doi: 10.1007/s12275-012-1589-4. [DOI] [PubMed] [Google Scholar]
- Mutters R, Mannheim W. Bisgaard M. Taxonomy of the group. In: Adlam C, Rutter JM, editors; Pasteurella and Pasteurellosis. London: Academic Press; 1989. pp. 3–34. [Google Scholar]
- Negrete-Abascal E, Tenorio RV, Guerrero AL, García RM, Reyes ME. de la Garza M. Purification of a protease from Actinobacillus pleuropneumoniae serotype 1, an antigen common to all the serotypes. Can. J. Vet. Res. 1998;62:183–190. [PMC free article] [PubMed] [Google Scholar]
- Negrete-Abascal E, Tenorio RV. de la Garza M. Secretion of proteases from Pasteurella multocida isolates. Curr. Microbiol. 1999;38:64–67. doi: 10.1007/pl00006775. [DOI] [PubMed] [Google Scholar]
- Negrete-Abascal E, García RM, Reyes ME, Godinez D. de la Garza M. Membrane vesicles released by Actinobacillus pleuropneumoniae contain proteases and Apx toxins. FEMS Microbiol. Lett. 2000;191:109–113. doi: 10.1111/j.1574-6968.2000.tb09326.x. [DOI] [PubMed] [Google Scholar]
- Ramón Rocha M, García González O, Pérez Méndez A, Ibarra Caballero J, Pérez Márquez V, Vaca S, et al. Membrane vesicles released by Avibacterium paragallinarum contain putative virulence factors. FEMS Microbiol. Lett. 2006;257:63–68. doi: 10.1111/j.1574-6968.2006.00154.x. [DOI] [PubMed] [Google Scholar]
- Schaar V, Nordström T, Mörgelin M. Riesbeck K. Moraxella catarrhalis outer membrane vesicles carry ß-lactamase and promote survival of Streptococcus pneumoniae and Haemophilus influenzae by inactivating amoxicillin. Antimicrob. Agents Chemother. 2011;55:3845–3853. doi: 10.1128/AAC.01772-10. doi: 10.1128/AAC.01772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooling SR. Beveridge TJ. Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 2006;188:5945–5957. doi: 10.1128/JB.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellyei B, Varga Z, Szentesi-Samu K, Kaszanyitzky E. Magyar T. Antimicrobial susceptibility of Pasteurella multocida isolated from swine and poultry. Acta Vet. Hung. 2009;57:357–367. doi: 10.1556/AVet.57.2009.3.2. doi: 10.1556/AVet.57.2009.3.2. [DOI] [PubMed] [Google Scholar]
- Smith SG, Mahon V, Lambert MA. Fagan RP. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol. Lett. 2007;273:1–11. doi: 10.1111/j.1574-6968.2007.00778.x. [DOI] [PubMed] [Google Scholar]
- Soriano-Vargas E, Vega-Sánchez V, Zamora-Espinosa JL, Acosta-Dibarrat J, Aguilar-Romero F. Negrete-Abascal E. Identification of Pasteurella multocida capsular types isolated from respiratory diseased rabbits and other domestic animals in Mexico with respiratory diseases. Trop. Anim. Health Prod. 2012;44:935–937. doi: 10.1007/s11250-011-9995-x. doi: 10.1007/s11250-011-9995-x. [DOI] [PubMed] [Google Scholar]
- Tan HY, Nagoor NH. Sekaran SD. Cloning, expression and protective capacity of 37 kDa outer membrane protein gene (ompH) of Pasteurella multocida serotype B:2. Trop. Biomed. 2010;27:430–441. [PubMed] [Google Scholar]
- Towbin H, Staehelin T. Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Sharma M, Katoch S, Verma L, Kumar S, Dogra V, et al. Profiling of virulence associated genes of Pasteurella multocida isolated from cattle. Vet. Res. Commun. 2013;37:83–89. doi: 10.1007/s11259-012-9539-5. [DOI] [PubMed] [Google Scholar]
- Wu XB, Tian LH, Zou HJ, Wang CY, Yu ZQ, Tang CH, et al. Outer membrane protein OmpW of Escherichia coli is required for resistance to phagocytosis. Res. Microbiol. 2013;164:848–855. doi: 10.1016/j.resmic.2013.06.008. doi: 10.1016/j.resmic.2013.06.008. [DOI] [PubMed] [Google Scholar]


