Abstract
Background
The endosteum of the bone marrow provides a specialized hypoxic niche that may serve to preserve the integrity, pluripotency, longevity and stemness of resident mesenchymal stem cells (MSCs). To explore the molecular genetic consequences of such a niche we subjected human (h) MSCs to a pO2 of 4 mmHg and analyzed global gene expression and alternative splicing (AS) by genome-exon microarray and RT-qPCR, and phenotype by western blot and immunostaining.
Results
Out of 446 genes differentially regulated by >2.5-fold, down-regulated genes outnumbered up-regulated genes by 243:203. Exon analyses revealed 60 hypoxia-regulated AS events with splice indices (SI) >1.0 from 53 genes and a correlation between high SI and degree of transcript regulation. Parallel analyses of a publicly available AS study on human umbilical vein endothelial cells (HUVECs) showed that there was a strong cell-specific component with only 11 genes commonly regulated in hMSCs and HUVECs and 17 common differentially spliced genes. Only 3 genes were differentially responsive to hypoxia at the gene (>2.0) and AS levels in both cell types. Functional assignments revealed unique profiles of gene expression with complex regulation of differentiation, extracellular matrix, intermediate filament and metabolic marker genes. Antioxidant genes, striated muscle genes and insulin/IGF-1 signaling intermediates were down-regulated. There was a coordinate induction of 9 out of 12 acidic keratins that along with other epithelial and cell adhesion markers implies a partial mesenchymal to epithelial transition.
Conclusions
We conclude that severe hypoxia confers a quiescent phenotype in hMSCs that is reflected by both the transcriptome profile and gene-specific changes of splicosome actions. The results reveal that severe hypoxia imposes markedly different patterns of gene regulation of MSCs compared with more moderate hypoxia. This is the first study to report hypoxia-regulation of AS in stem/progenitor cells and the first molecular genetic characterization of MSC in a hypoxia-induced quiescent immobile state.
Keywords: Hypoxia, Microarray, Alternative splicing, Stem cell Niche
Background
The stem cell niche refers to a well-defined physiological compartment that includes cellular and acellular components and serves to integrate systemic and local signals to regulate the biology of stem cells (reviewed in [1,2]). Like other such niches, the bone marrow provides highly specialized and heterogeneous microenvironments that determine the self-renewal, multipotency, survival and migration of residing hematopoietic and progenitor cells including mesenchymal stem cells (MSCs). Recently oxygen tension (hypoxia) has been recognized as an important component of stem cell niches that exerts control over the proliferation, differentiation and pluripotency of resident cells [3-5]. The oxygen tension of the endosteum, a narrow compartment of the bone marrow directly adjacent to the bone, is less than 10 mmHg, while that of the sinusoidal cavity ranges between 30–60 mmHg; therefore bone marrow cells are subject to a gradient of hypoxia the severity of which depends on their location within the niche [3,6]. Studies of embryonic stem cells (ESCs) as well as induced pluripotential stem cells (iPSC) indicate that oxygen gradients control stem cell functions. Culture of ESCs under an aerobic pO2 of 160 mmHg causes spontaneous differentiation that is suppressed by more physiological pO2 within the range of 14–36 mmHg. Further reduction of pO2 to <10 mmHg, equivalent to the bone marrow endosteum suppresses both differentiation and proliferation of ESCs while retaining pluripotency [7,8]. Studies on bone marrow or adipose derived MSCs have shown similarly that moderate hypoxic culture equivalent to the central BM sinusoidal niche enhances proliferation and protects against senescence while more severe hypoxia may block proliferation and induce cell death [9-16].
Culture of MSCs under moderate hypoxia has been shown to modulate gene expression by HIF-1/2-dependent and independent mechanisms [8,17-21]. In addition to the predicted HIF-1α target genes such as those required for anaerobic metabolism (glycolytic enzymes, glucose transporters), cell cycle (p21, p53), and angiogenesis (VEGF), moderate hypoxia was shown to mediate increased expression of Oct4 and telomerase activity of human bone marrow MSCs [16,22,23]. When cultured under moderate hypoxia (20–40 mmHg O2), MSCs display enhanced proliferation and migratory activity that has been attributed to increased Akt phosphorylation, expression of c-MET, VEGF, chemokine receptors CXCR4 and CXCR1, and increased phosphorylation of focal adhesion kinase [10,18,19,24-26]. Suppression of stem cell differentiation by hypoxia has been linked to Notch pathway signaling wherein hypoxia promotes recruitment of HIF-1α to the Notch intracellular domain and subsequently to Notch-dependent promoters thereby enhancing their expression [27]. Recently, HIF-1α was shown to regulate MSC proliferation through the enhancement of TWIST expression, which down-regulates the E2A-p21 pathway, inhibits senescence and increases proliferation [28]. Oxygen tensions between 20 and 40 mmHg enhance proliferation and pluripotency of stem cells whereas tension below 10 mmHg (<1%) inhibit proliferation and may promote apoptosis [8-12,17-19].
Up to 95% of all human genes are alternatively spliced [29,30]. AS results in changes in composition of an mRNA produced from a given gene, brought about by changes in splice site choice and thence the production of proteins with different properties. AS is regulated by cell specific, developmental, and extracellular signal-regulated cues and pathways (Reviewed in [31]). Aberrant alternative splicing can cause disease and may contribute to cancer and neurodegenerative disease [32,33]. Exon arrays allow for detection and quantification of AS on a genome-wide scale. There are currently only 2 such reports of genome-wide analyses of hypoxia-related changes in pre-mRNA splicing. One identified Lama3 as a hypoxia-related splice variant in head and neck cancers [34]. Another analyzed the effects of hypoxia on AS in human umbilical vein endothelial cells (HUVECs) and identified multiple alternative splice events [35].
Here we investigated the effects of severe hypoxia on gene expression, exon splicing, and phenotype of human (h) MSCs. The results reveal for the first time unique sets of (severe) hypoxia-activated and repressed genes, many of which differ from those reported previously for more moderate hypoxia. We report for the first time a coordinate increase in expression of acidic keratins perhaps indicating a partial mesenchymal to epithelial (epidermal) transition (MET), a decrease in insulin/IGF-1 signaling with lower phosphor-Akt, and decreased expression of anti-oxidant-related genes that suggests lower metabolism and growth compared with aerobic culture. The expression of differentiation-related markers is consistent with enhancement of osteogenic and angiogenic pathways perhaps at the expense of myogenesis and adipogenesis. We also identify a novel set of hypoxia-regulated alternatively spliced transcripts in hMSCs. To our knowledge this is the first study to report on patterns of hypoxia-mediated alternative splicing in stem cells. The results provide a molecular framework for understanding the role of severe hypoxia in preserving bone marrow progenitor cell integrity and perhaps insights into the role of hypoxia in regulating cell biology in hypoxic niche environments such as the endosteum.
Results
Isolation and characterization of human MSCs
Human bone marrow MSCs were isolated as described in Methods and used at passage 8. At this time cells were visually homogeneous, fibroblast-like and positive for the expression of mesenchymal-specific markers CD29 (99.0%) and CD166 (41.8%) and negative for the expression of hematopoietic lineage marker CD34 (0.4%). These characteristics were unaltered after exposure to hypoxia for 24 h (Figure 1A and 1B).
Figure 1.
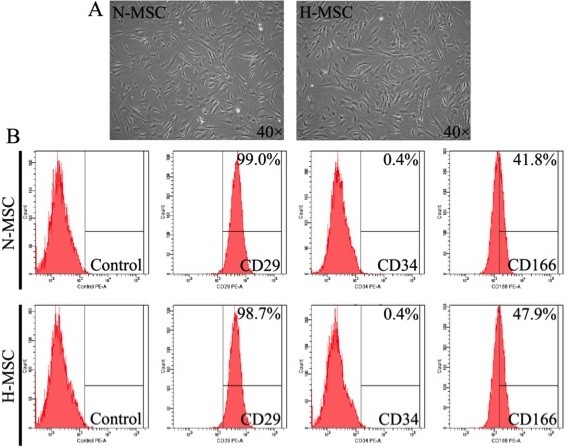
Characterization of hMSCs cultures under air or hypoxia. (A) Morphology and (B) surface antigen profiling of normoxic (N-MSC) and hypoxic hMSCs (H-MSC). Conditions and procedures are described in Methods. Representative of n = 3.
Gene expression array
Gene expression profiles comparing normoxia and hypoxia were obtained using Agilent Human 4 × 180 K Exon and 8 × 60 K-GE microarrays as described in Methods. Only the Exon arrays are described in the present analysis and the GE arrays were used for confirmation of some gene transcripts. Hierarchical clustering of the Exon arrays confirmed high reproducibility between samples (Additional file 1: Figure S1). A robust response to hypoxia was confirmed by quantifying HIF-1α-regulated transcripts. As shown in Table 1, multiple well-characterized HIF-1α-regulated genes were represented including carbonic anhydrase (>5-fold), metallothionein (>4-fold) and VEGF-A (>4-fold). Most of these genes have been reported previously in similar high throughput analyses of MSCs exposed to hypoxia in the range 1-5% [14,17-19]. Noteworthy in our analyses are the strong inductions of leptin and insulin-like growth factor binding protein 1 transcripts, confirmed in both Exon and K-GE arrays (latter data not shown) and relatively low induction of Bnip3 and glucose transporters. Glycolytic enzyme genes are widely recognized as markers of hypoxia with at least 8 of 11 glycolytic enzymes genes responsive through the HIF-1α pathway (reviewed in [36]). Surprisingly, glycolytic gene transcripts were not represented in the 2.5-fold cut-off groupings used for our array analyses, therefore we used RT-PCR to quantify phosphofructokinase (PFK) a major rate-limiting enzyme [37], phosphoglycerate kinase (PGK) a strong hypoxia responder [38], phosphoglycerate mutase (PGAM), a moderate responder [39] and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), that is regulated by hypoxia in a tissue-specific manner [40-42]. As indicated in the Table, transcript levels of PFK and PGK increased, there was no significant change of GAPDH (p = 0.20, n = 7) and PGAM transcripts decreased (p = 0.02, n = 7). The results suggest mixed responses of individual genes. Glycolytic flux increased significantly under hypoxia; aerobic cultures used glucose at a rate of 2.83 ± 0.2 mg/dL/h compared with 3.33 ± 0.1 mg/dL/h for hypoxia (p > 0.05; n = 3), and there was a parallel increased rate of lactic acid generation over 24 h (data not show). Glycolytic flux is largely regulated by small molecule binding and allosteric control of PFK, the main rate-limiting enzyme and most of the pathway enzymes are present in large excess. Induction of gene expression by hypoxia is likely to be a chronic adaptation that is not required for the acute response to substrates and energy level.
Table 1.
HIF-1 regulated transcripts
| Gene description | Gene symbol | Fold change HX/NX |
|---|---|---|
| Leptin |
LEP |
74.3 |
| Insulin-like growth factor binding protein 1 |
IGFBP1 |
8.93 |
| Phopshoglycerate kinase |
PGK |
7.10* |
| Lysyl oxidase-like 4 |
LOXL4 |
6.5 |
| Carbonic anhydrase IX |
CA9 |
5.55 |
| Metallothionein 3 |
MT3 |
4.78 |
| Vascular Endothelial Growth Factor A |
VEGFA |
4.61 |
| Carbonic anhydrase XII |
CA12 |
4.62 |
| Lysyl Oxidase |
LOX |
3.99 |
| Basic helix-loop-helix family member 40 |
BHLHE40 |
3.97 |
| Adrenomedullin |
ADM |
3.39 |
| Placental Growth Factor |
PGF |
3.28 |
| Phosphofructokinase |
PFK |
3.20* |
| Insulin-like growth factor binding protein 3 |
IGFBP3 |
3.10 |
| Angiopoietin-like 4 |
ANGPTL4 |
2.88 |
| BCL2/adenovirus E1B 19 kDa interacting protein 3 |
Bnip3 |
2.17 |
| Lactate Dehydrogenase-A |
LDH-A |
2.10 |
| Glyceraldehyde-3-Phosphate Dehydrogenase |
GAPDH |
1.48* |
| PhopshoGlycerate Mutase |
PGAM |
0.70* |
| Solute carrier family 2 (facilitated glucose transporter), member 1 |
SLC2A1 (GLUT 1) |
2.01 |
| Solute carrier family 2 (facilitated glucose transporter), member 3 | SLC2A1 (GLUT 3) | 2.11 |
Genes were identified from the gene expression array and all values are significant (p < 0.05, n = 3). For genes with (*), transcripts were quantified by QPCR using 3 different preparations of hMSCs run in duplicate; (PGK p < 0.001; PFK, p < 0.01; GAPDH, p = 0.21; PGAM, p = 0.02; all n = 7).
A total of 446 genes were found to have >2.5-fold change (p < 0.05). In agreement with previous reports on the effects of hypoxia on human MSC and endothelial cells [17-19,35] we found that more genes were down- than up-regulated (243 vs. 203). In order to confirm the microarray results RT-PCR was implemented on 3 known HIF-1α target genes in addition to the glycolytic transcripts described above, and four hypoxia-induced genes not previously identified as HIF targets. As shown in Figure 2, leptin (LEP), metallothionein-3 (MET3) and lysyl oxidase (LOX) were all confirmed to increase in a manner that reflected the array data. Similarly transcripts of non-HIF-regulated genes, keratin-16 (KRT16), Serpin peptidase inhibitor-1 (PAI-1), RAS p21 protein activator (RASA1) and Immediate Early Response 3 (IER3) were also confirmed to increase in the hypoxic samples in a manner that reflected the array data. Western blot and ELISA further confirmed that the levels of secreted hypoxia-marker genes VEGF and leptin were significantly increased by hypoxia in the spent media (Figure 3A and B). Figure 4 and Tables 2 and 3 show some of the most strongly hypoxia-regulated genes separated into functional categories. GO analyses identified 3 categories including [1] antioxidant pathways, NAD(P)H quinone-1, aldo-keto reductase family members and thioredoxin reductase were markedly down-regulated [2] extracellular matrix structural constituents in particular collagens were subject to up and down regulation suggesting dynamic changes of the ECM and [6] glucose transporters that were also subject to both up and down-regulation (see Figure 4 and Tables 2 and 3).
Figure 2.
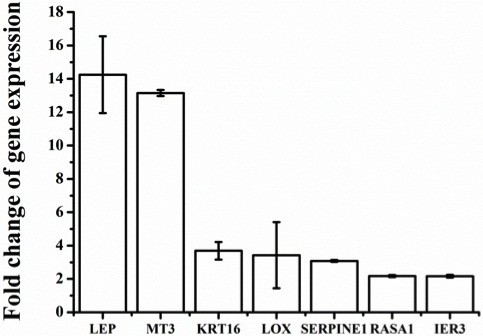
Real-time PCR (RTqPCR) validation of differentially expressed genes. RTqPCR was implemented on the same mRNA samples used for microarray as described in Methods. Results are mean ± SEM of n = 3.
Figure 3.
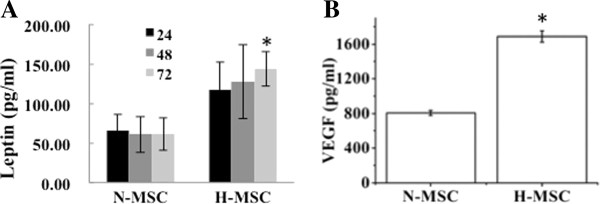
Quantification of leptin and VEGF protein in MSC spent media during exposure to hypoxia. (A) Numbers in left panel refer to time (h) under hypoxia. (B) VEGF incubation time was 24 h. All results are representative of 3 separate experiments; *p < 0.05, n = 3.
Figure 4.
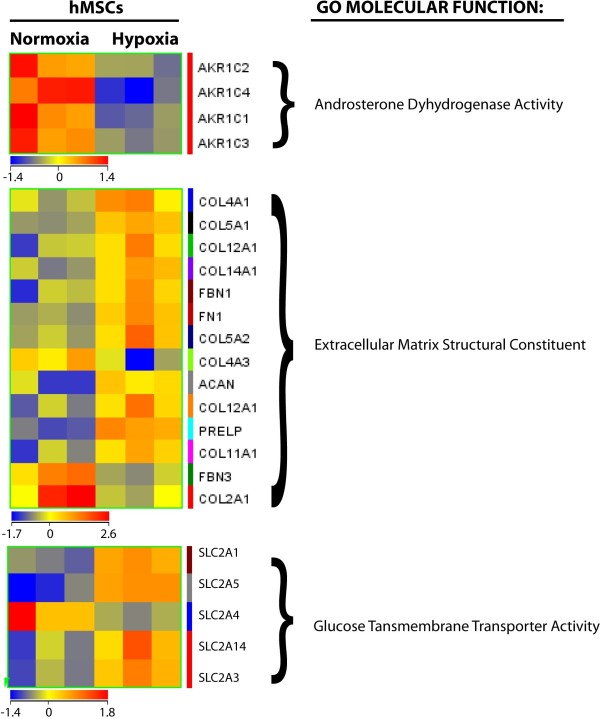
Heatmaps of differentially expressed genes in 2 main Gene Ontology (GO) molecular functions. Differentially expressed genes in human MSCs under hypoxia were subjected to GO analysis. A significance cut-off of p < 0.05 was used.
Table 2.
Up-regulated genes and functional group allocations
| Class | Gene description | Gene symbol | HX/NX |
|---|---|---|---|
|
Differentiation | |||
| |
Leptin |
LEP |
74.3 |
| Early Growth Response 2 |
EGR2 |
6.78 |
|
| Inhibin, beta B |
INHBB |
6.25 |
|
| Interleukin 11 |
IL11 |
5.5 |
|
| Growth differentiation factor 6 |
GDF6 |
5.29 |
|
| Hemopoietic cell kinase |
HCK |
4.82 |
|
| Podoplanin |
PDPN |
3.65 |
|
| Ephrin-A3 |
EFNA3 |
3.65 |
|
| Semaphorin A7 |
SEMA7 |
2.70 |
|
| Activin A receptor type IIA |
ACVR2A |
2.70 |
|
| Matrix metalloproteinase 11 |
MMP11 |
2.52 |
|
| Aggrecan |
ACAN |
2.50 |
|
| Sparc/osteonectin (testican) |
SPOCK1 |
2.2 |
|
| Frizzled homologue-8 |
FZD8 |
3.2 |
|
| Secreted frizzled related protein 4 |
SFRP4 |
2.3 |
|
| Cadherin 11, type 2 (osteoblast) |
CDH11 |
2.0 |
|
|
Survival/Apoptosis | |||
| |
Transient receptor potential cation channel |
TRPM7 |
6.63 |
| Interleukin 11 |
IL11 |
5.5 |
|
| Metallothionein 3 |
MT3 |
4.80 |
|
| Stanniocalcin 1 |
STC1 |
4.8 |
|
| Regucalcin |
RGN |
4.04 |
|
| Vascular endothelial growth factor A |
VEGFA |
3.56 |
|
| RAS p21 protein activator 1 |
RASA1 |
2.92 |
|
| Immediate Early Response 3 |
IER3 |
2.52 |
|
|
Proliferation/Survival | |||
| |
Insulin-like growth factor binding protein 1 |
IGFBP1 |
8.93 |
| Insulin-like growth factor binding protein 3 |
IGFBP3 |
3.1 |
|
| Insulin-like growth factor binding protein 5 |
IGFBP5 |
3.1 |
|
| Interleukin 11 |
IL11 |
5.5 |
|
| Placental Growth Factor |
PGF |
3.28 |
|
| Transcription elongation factor A3 |
TCEA3 |
2.82 |
|
| Hepatocyte growth factor receptor |
MET |
2.70 |
|
| Heparin-binding EGF-like GF |
HBEGF |
2.64 |
|
| Jun B oncogene |
JUNB |
3.07 |
|
| c-Jun oncogene |
JUN |
2.66 |
|
| Inhibin, beta B |
INHBB |
6.25 |
|
| Growth differentiation factor 6 |
GDF6 |
5.29 |
|
| Inhibitor of growth family member 3 |
ING3 |
3.53 |
|
|
ECM migration/adhesion/cytoskeleton | |||
| |
Early growth response 2 |
EGR2 |
6.78 |
| Interleukin 11 |
IL11 |
5.5 |
|
| Activated leukocyte cell adhesion molecule |
ALCAM |
2.89 |
|
| Junctional adhesion molecule 2 |
JAM2 |
3.10 |
|
| Semaphorin 5A |
SEMA5A |
4.02 |
|
| Connective tissue growth factor |
CTGF |
2.40 |
|
| Protein tyrosine phosphatase, receptor type, F |
PTPRF |
2.66 |
|
| Signal-induced proliferation-associated 1 like 1 |
SIPA1L1 |
4.0 |
|
| Intergrins alpha- 1, 3, 5, 6, 7 |
ITGA |
>2.2 |
|
| Integrin beta-1 |
ITGB |
2.1 |
|
|
Intermediate filament | |||
| |
Periplakin |
PPL |
7.88 |
| Desmoplakin |
DSP |
3.86 |
|
| Keratin 16 |
KRT16 |
6.39 |
|
| Keratin 14 |
KRT14 |
4.04 |
|
| Keratin 20 |
KRT20 |
4.40 |
|
| Keratin 19 |
KRT19 |
3.5 |
|
| Keratin 15 |
KRT15 |
3.35 |
|
| Keratin 13 |
KRT13 |
3.37 |
|
| Keratin 24 |
KRT24 |
3.09 |
|
| Keratin 17 |
KRT17 |
2.70 |
|
| Keratin 12 |
KRT12 |
2.60 |
|
| Mucin 1 |
MUC1 |
2.23 |
|
| KIAA1199 |
KIAA1199 |
5.2 |
|
| RAS p21 protein activator |
RASA1 |
2.92 |
|
| Microtubule-associated protein 1B |
MAO1B |
2.70 |
|
| Microtubule-actin cross-linking factor 1 |
MACF1 |
2.4 |
|
| Lysyl oxidase-like 4 |
LOX4 |
6.5 |
|
| Lysyl oxidase-like 2 |
LOX2 |
2.5 |
|
|
Angiogenesis | |||
| |
Leptin |
LEP |
74.3 |
| Stanniocalcin 1 |
STC1 |
4.70 |
|
| Vascular endothelial growth factor A |
VEGFA |
4.61 |
|
| Placental growth factor |
PGF |
3.28 |
|
| Hepatocyte growth factor receptor |
MET |
2.70 |
|
| PDGF receptor B |
PDGFRB |
2.1 |
|
| PDGF receptor A |
PDGFRA |
2.0 |
|
| Angiopoietin-like 4 |
ANGPTL4 |
2.80 |
|
| Serpin peptidase inhibitor member 1 |
SERPINE1 |
4.92 |
|
| Serpin peptidase inhibitor member 2 |
SERPINE2 |
2.4 |
|
| Tissue plasminogen activator |
PLAT |
2.3 |
|
| Endothelial tyrosine kinase, Ang 1 receptor |
TEK |
5.16 |
|
| Lysyl oxidase-like 4 |
LOX4 |
6.5 |
|
| Desmoplakin |
DSP |
3.86 |
|
| Noggin |
NOG |
3.91 |
|
|
Glucose Transport | |||
| Glucose transporter member 1 |
SLC2A1 |
2.01 |
|
| Glucose transporter member 3 |
SLC2A3 |
2.11 |
|
| Glucose/fructose transporter member 5 |
SLC2A5 |
2.0 |
|
| Glucose transporter, member 14 | SLC2A14 | 2.88 | |
Genes were identified from the gene expression array using Genespring software and GO analysis to assign functional categories as described in Methods and manually from inspection of the arrays. All values are significant (p < 0.05, n = 3).
Table 3.
Down-regulated genes and functional group allocations
| Class | Gene description | Gene symbol | HX/NX |
|---|---|---|---|
|
Antioxidant | |||
| |
NAD(P)H dehydrogenase, Quinone 1 |
NQO1 |
−5.63 |
| Aldo-Keto Reductase 1C1 |
AKR1C1 |
−2.71 |
|
| Aldo-Keto Reductase 1C2 |
AKR1C2 |
−2.61 |
|
| Aldo-Keto Reductase 1C3 |
AKR1C3 |
−2.60 |
|
| Aldo-Keto Reductase 1C4 |
AKR1C4 |
−4.28 |
|
| Aldo-Keto Reductase1D1 |
AKR1D1 |
−3.04 |
|
| Aldo-Keto Reductase 1B10 |
AKR1B10 |
−3.07 |
|
| Aldo-Keto Reductase 1B15 |
AKR1B15 |
−2.10 |
|
| Glucose 6 phosphate DH |
G6PDH |
−2.00 |
|
| Thioredoxin reductase |
TXNRD1 |
−3.77 |
|
|
Proliferation/growth associated | |||
| |
Insulin-like growth factor 1 |
IGF-1 |
−3.2 |
| PI3-kinase regulatory subunit 2 |
PIK3R2 |
−2.9 |
|
| Platelet derived growth factor receptor-like |
PDGFRL |
−2.85 |
|
| Fibroblast growth factor 7 |
FGF7 |
−2.1 |
|
| Cell cycle arrest checkpoint |
RAD9B |
−4.4 |
|
| Mediator of DNA damage checkpoint 1 |
MDC1 |
−3.39 |
|
| Helicase (DNA) B |
HELB |
−4.4 |
|
| Inhibitor of DNA binding 1 |
ID1 |
−3.26 |
|
| Inhibitor of DNA binding 2 |
ID2 |
−3.0 |
|
| Inhibitor of DNA binding 4 |
ID4 |
−3.0 |
|
| TNF superfamily, member 14 |
TNFSF14 |
−2.22 |
|
|
Migration-associated | |||
| |
Phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
PREX1 |
−2.23 |
| Coronin 7 |
CORO7 |
−5.10 |
|
| Formin homology 2 domain containing 1 |
FHOD1 |
−2.15 |
|
| Actin filament associated protein 1 like 2 |
AFPAP1L2 |
−3.3 |
|
| Palmdelphin |
PALMD |
−2.8 |
|
| Tubulin, alpha 3D |
TUBA3D |
−3.9 |
|
|
Glucose metabolism | |||
| |
Glucose 6 phosphate DH |
G6PDH |
−2.00 |
| Pyruvate dehydrogenase (liver RBC) |
PKLR |
−2.61 |
|
| Phosphoglycerate mutase 2 |
PGAM2 |
−3.00 |
|
| Phosphoglycerate mutase 5 |
PGAM5 |
−2.27 |
|
| Phosphogluconate dehydrogenase |
PGD |
−2.55 |
|
| Glucose transporter, member 4 |
SLC2A4 |
−2.7 |
|
| Glucose transporter, member 8 |
SLC2A8 |
−2.5 |
|
| Glucose/fructose transporter, member 11 |
SLC2A11 |
−2.24 |
|
|
Muscle/myogenesis/structural | |||
| |
Actin alpha-1 (Sk) |
ACTA1 |
−4.33 |
| Actin gamma-2 (SM) |
ACTG2 |
−5.83 |
|
| Fer-1-like 5 (myotube formation) |
FER1L5 |
−5.30 |
|
| Tripartite motif containing 16-like |
TRIM16L |
−4.95 |
|
| Myosin heavy chain 2 |
MYH2 |
−4.0 |
|
| Myosin heavy chain 7 |
MYH7 |
−3.0 |
|
| Troponin T type 2 (cardiac) |
TNNT2 |
−2.8 |
|
| ATPase calcium channel (cardiac) |
ATP2A1 |
−2.94 |
|
| Myosin binding protein B (fast) |
MYBPC2 |
−2.79 |
|
|
Miscellaneous | |||
| Lysine acetyl transferase 2A |
KAT2A |
−3.45 |
|
| Galactosidase beta-1-like (senescence) |
GLB1L3 |
−3.0 |
|
| Eukaryotic translation initiating factor 2B subunit gamma 3 |
EIF2B3 |
−2.6 |
|
| Eukaryotic translation elongating factor 1 epsilon 1 |
EEF1E1 |
−2.67 |
|
| Osteocrin |
OSTN |
−2.3 |
|
| Elongation factor RNA Pol II, 2 |
ELL2 |
−2.0 |
|
| Telomerase associated protein-1 |
TEP1 |
−2.67 |
|
| Death associated protein kinase 2 |
DK2 |
−2.4 |
|
| Protein kinase C delta |
PRKCD |
−3.37 |
|
| Interleukin-8 |
IL8 |
−3.05 |
|
| Interleukin-19 |
IL19 |
−2.61 |
|
| Interleukin-27 |
IL27 |
−3.30 |
|
| Interleukin-28A |
IL28A |
−2.24 |
|
| Chemokine (C-C motif) ligand 2 | CCL2 | −3.19 | |
Genes were identified from the gene expression array using Genespring software and GO analysis to assign functional categories as described in Methods; additional genes were identified manually from inspection of the arrays. All values are significant (p < 0.05, n = 3).
Intermediate and cytoskeleton filaments
Intermediate filaments and microfilaments provide scaffolding that allows cells to rapidly remodel in response to environmental or intracellular signals, for example to increase or decrease movement and/or cell adhesion [43,44]. Intermediate filaments and their keratin structural components are markers of epithelium where in combination with cell junction components, desmosomes, hemidesmosomes and integrins, they contribute to the adhesion of epithelial cells to the basement membrane [43]. The keratins that compromise intermediate filaments are typically selected from 12 acidic (type 1) and 8 basic (type 2) cytokeratins that usually dimerize with specific partners from the opposite group to form organized filaments. Acidic keratins are coded on chromosome 17q whereas the basic keratins are clustered on chromosome 12q. The gene expression analysis revealed a remarkable increase in the expression of 9 out of 12 acidic keratins and decreased transcript levels of 3 (K2, K4, K5) out of 8 basic keratins with no change of the other members (Tables 2 and 3). To determine whether these changes in gene expression were reflected at the protein and cell structural levels, we measured K16 protein by western blot and the organization of intermediate and microfilaments respectively by keratin and F-actin immunostaining. As shown in Figure 5, K16 levels were increased by 2 ± 0.1-fold (p < 0.05) in MSC after exposure to hypoxia and this was associated with a change in the organization of intermediate filaments and altered cellular morphology (Figure 5B). The changes in cell shape and organization of cytoplasmic filaments indicated by keratin staining were also reflected by similar cell images imaged by F-actin immunostaining (Figure 5C). Other proteins related to intermediate filaments structure and function that were also increased by hypoxia include periplakin and desmoplakin, junctional adhesion molecule 2 (JAM2), adipocyte-specific adhesion molecule (ASAM) adhesion molecule with Ig-like domain 2 (AMIGO2) and podoplanin (PDPN) (see Table 2). In addition, six integrins (α-1,-3,-5,-6,-7, β-1) were increased by >2-fold. These changes may reflect a partial mesenchymal to epithelial transition (MET) that is a reversal of epithelial to mesenchymal transition (EMT) a well-established pathway associated with development and carcinogenesis. MET is predicted to involve reduced cell mobility and proliferative quiescence [45]. Interestingly hypoxia also increased the transcript levels of the HGF receptor c-MET that is normally expressed by cells of epithelial origin (Table 2).
Figure 5.
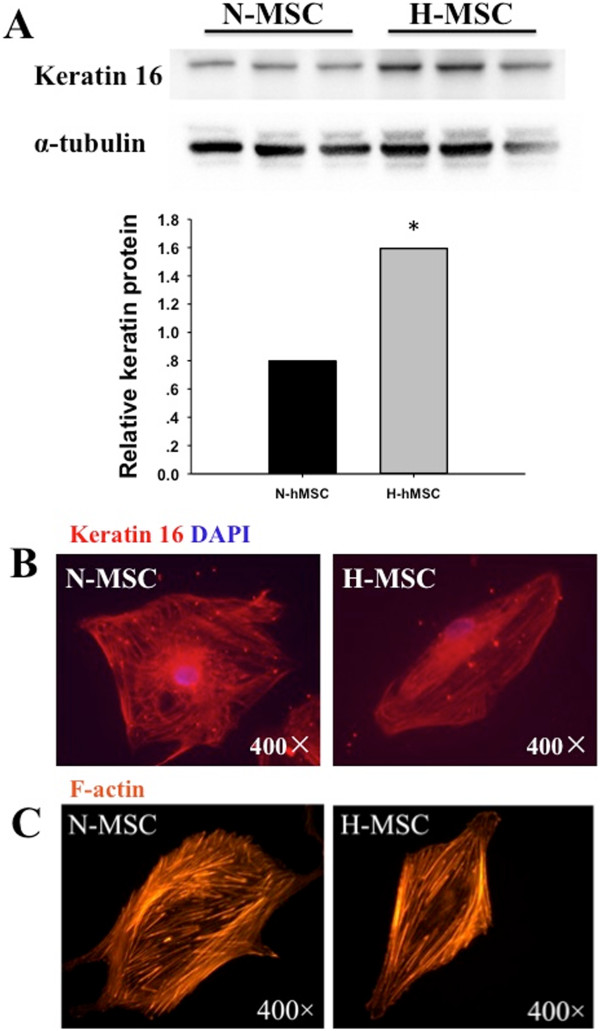
Keratin induction and reorganization of intermediate- and micro-filaments under hypoxia. (A) Western blot of Keratin-16 expression in normoxic (N-MSC) and hypoxic (H-MSC) human MSCs. (B) Keratin immunostaining and (C) F-actin immunostaining of MSCs cultured for 24 h under normoxia (N-MSC) or hypoxia (H-MSC). Western blots and immunostaining are described in Methods. All results are representative of 3 separate experiments; *p < 0.05, n = 3.
Regulation of differentiation
MSCs are pluripotent with the capacity to differentiate into adipocytes, osteocytes, chondrocytes, endothelium, neurons and muscle as well as hematopoietic cells. It has been reported that moderate hypoxia >10 mmHg increased [19,46] or decreased [23,47,48] adipogenic differentiation and increased [10,19,47,48] or decreased [20,23] osteogenic differentiation of MSCs. In our studies, combined GO and manual analyses revealed trends of hypoxia-regulated differentiation markers that suggest repression of adipogenesis and myogenesis in favor of osteogenesis, angiogenesis and hematopoiesis. Leptin, interleukin-11 (Il-11), growth differentiation factor-6 (GDF6), and MMP11, all strongly induced by hypoxia, are secreted factors that favor osteogenesis over adipogenesis [49-51]. Collagens are major constituents of the bone matrix and are highly expressed in MSCs. Type 1 collagen, the main osteogenic collagen was not amongst the regulated genes, however, multiple other osteogenic markers genes including aggrecan (ACAN), sparc/osteonectin (SPOCK1), osteoblast cadherin (CDH11) and Wnt pathway members Frizzled homologue-8 (FZD8) and Secreted frizzled related protein 4 (SFRP4) were induced in parallel with decreased expression of the negative osteogenic regulator osteocrin (Tables 2 and 3) [52,53]. Four of the most strongly induced genes including early growth response 2 (EGR2 > 6-fold) [54], interleukin 11 (Il-11 > 5-fold), growth differentiation factor 6 (GDF6 > 5-fold) and hemopoietic cell kinase (HCK ~5-fold) as well as activin-A receptor (2-fold [55]) are associated with hematopoiesis. Transcript levels of multiple genes associated with angiogenesis were increased by hypoxia [56] while there were marked decreases of multiple striated muscle markers (Tables 2 and 3). The coordinate decrease of 3 Inhibitor of DNA binding factors ID1, ID2, and ID4 (Table 3) is also consistent with a switch in differentiation patterns. Taken together the results support inductions of multiple marker genes for osteogenesis, angiogenesis, and hematopoiesis by severe hypoxia that may predispose cells to these lineages.
Proliferation and metabolism
As discussed above, oxygen tensions above 15 mmHg enhance proliferative and migratory potential of MSCs while tensions below10 mmHg decrease proliferation [8-12,17-19]. In agreement with this we found that proliferation of hMSCs was decreased under a pO2 of 4 mmHg and the cultures became stationary after 3 days (data not shown). Enhanced proliferation under moderate hypoxia has been attributed to down-regulation of the p21-Ras pathway and increased activity of PI3-kinase-Akt while increased migration was attributed to increased expression of c-MET, VEGF, CXCR4 and CXCR1 (reviewed in [16]). Whereas we also observed increased expression of c-MET and VEGF-A we found no change in expression of CXCR receptors, increased expression of p21-Ras and decreased activity of the PI3-kinase-Akt pathway (see Tables 2 and 3). PI3-kinase-Akt is a central regulator of cell growth and survival. We found that the expression of both IGF-1 and PI3-kinase was significantly decreased under hypoxia while multiple IGF-1BPs were increased (Tables 2 and 3). To determine whether these changes were reflected by parallel changes in pathway activity we measured the phosphorylation of Akt-Thr308 after culture under normoxia or hypoxia. As shown in Figure 6, phosphor-Akt-Thr308 levels were significantly lower after hypoxia. This contrasts with the effects of moderate hypoxia where Akt phosphorylation is increased [26]. Other down-regulated transcripts related to growth and survival included senescence marker galactosidase beta-1-like (−3.0) and death associated kinase-2 (−2.4). There were also decreases of several interleukins and the cell migration cytokine CCL2, also known as MCP-1.
Figure 6.
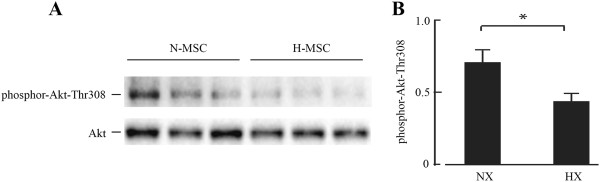
Western blot and quantification of phosphor-Akt expression in normoxic (N-MSC) and hypoxic (H-MSC) human MSCs (A and B). Western blot procedures are described in Methods. Akt-P-Thr308 quantification was by NIH image using total Akt as loading control; *p < 0.05, n = 3.
Glucose metabolism
During hypoxic exposure, cells switch from oxidative metabolism to anaerobic glycolysis for energy production. Glycolysis is less efficient than oxidative phosphorylation and more glucose is required to sustain the same level of cell function. Glycolytic enzyme and glucose transporter genes are regulated by HIF-1α and previous studies have reported their induction by moderate hypoxia in the range of 20–40 mmHg [13,14,17-19]. We found increased transcript levels of at least 2 key glycolytic pathway genes, PFK and PGK, decrease levels of PGAM, no change of GAPDH and decreased transcripts of glucose-6-phosphate dehydrogenase (G6PDH) and pyruvate dehydrogenase PDH; the latter two enzymes are involved in the pentose phosphate pathway (PPP) and acetyl-CoA production respectively. In addition, out of 7 regulated glucose transporters 4 were induced and 3 repressed by hypoxia. The results indicate mixed responses of glucose metabolizing genes. Decreased flux through the PPP is predicted to decrease antioxidant capacity through lower NADPH production, an effect consistent with the down-regulation of other anti-oxidant pathways discussed above.
Hypoxia-related AS events
Exon Microarrays identified 53 genes that were subjected to hypoxia-dependent AS; the arrays also revealed a correlation between high splice index and genes that responded the most strongly at the level of gene transcripts. As shown in Figure 7, eight of the most strongly induced genes and 5 of those that were most strongly repressed also had the highest splicing indices. This suggests a possible mechanistic link between gene regulation and control of AS by hypoxia. LEP, IL-11, IGFBP1, TEK, CA9, LOX4, HCK and EGR2 were each induced by more than 5-fold by hypoxia whereas EFNA3, CORO7, FER1L5, MYH2 and ACAT1 were each repressed (preferentially expressed in air) by greater than 4-fold under hypoxia. Four of the induced genes are HIF-1 targets and EFNA3 a down-regulated transcript, is an inhibitor of angiogenesis and target of the HIF-1-regulated microRNA-210 [57]. GO analyses indicated that the predicted functions of genes with hypoxia-regulated AS included cell adhesion, migration, apoptosis, angiogenesis and oxidation-reduction (Table 4). In terms of exon use, 8 of the identified AS events are described in the human genome annotation (5 C-terminus, 2 cassette-exon, 1 alt-3′ split site), and 16 involve constitutive exons. The remainder are novel AS events.
Figure 7.
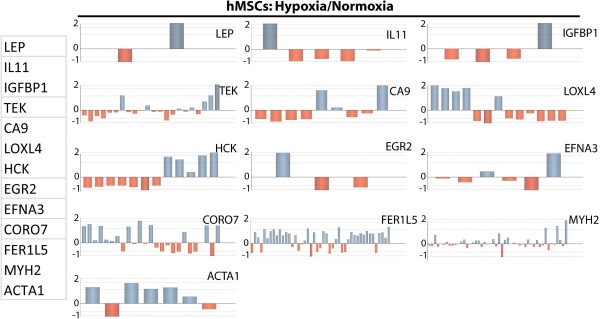
Splicing maps of highly regulated genes with high splice indices. Genes were selected based on highest positive (LEP, IL11, IGFBP1, TEK, CA9, LOX4, HCK, ERG2) and negative (EFNA3, CORO7, FER1L5, MYH2, ACTA1) fold change of gene expression hypoxia vs. normoxia. Bar graphs indicate hypoxic/normoxic differential exonic expression levels (n = 3).
Table 4.
GO enrichment classification of alternatively spliced genes after hypoxia
| Pathways | Gene symbol | Accession number | Splicing Index | Probe location | Gene description |
|---|---|---|---|---|---|
|
Apoptosis-associated |
|
|
|
|
|
| Induction of apoptosis by intracellular signals |
SART1 |
NM_005146 |
0.46 |
Exon11 |
Squamous cell carcinoma antigen recognized by T cells |
| Positive regulation of neuron apoptosis |
PTPRF |
NM_002840 |
2.56 |
Exon34 |
Protein tyrosine phosphatase, receptor type, F |
| Apoptosis |
PPP1R13L |
NM_006663 |
2.01 |
Exon13 |
Protein phosphatase 1, regulatory (inhibitor) subunit 13 like |
| |
STEAP3 |
NM_182915 |
2.22 |
Exon6 |
STEAP family member 3 |
| TNFRSF14 |
NM_003820 |
2.39 |
Exon8 |
Tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
|
| Negative regulation of apoptosis |
ERCC2 |
NM_001130867 |
0.42 |
Exon8 |
Excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| Anti-apoptosis |
SERPINB2 |
NM_002575 |
0.48 |
Exon8 |
Serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
|
Proliferation-associated |
|
|
|
|
|
| Cell growth |
ACTA1 |
NM_001100 |
0.22 |
Exon6 |
Actin, alpha 1, skeletal muscle |
| NDRG4 |
NM_001130487 |
2.62 |
Exon5 |
NDRG family member 4 |
|
| Cell cycle checkpoint |
ERCC2 |
NM_001130867 |
0.42 |
Exon8 |
Excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| Multicellular organism growth |
ERCC2 |
NM_001130867 |
0.42 |
Exon8 |
Excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| Cell proliferation |
ERCC2 |
NM_001130867 |
0.42 |
Exon8 |
Excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| MT3 |
NM_005954 |
2.72 |
Exon3 |
Metallothionein 3 |
|
| Cell cycle arrest |
SART1 |
NM_005146 |
0.46 |
Exon11 |
Squamous cell carcinoma antigen recognized by T cells |
| Positive regulation of cell proliferation |
IL11 |
NM_000641 |
7.99 |
Exon5 |
Interleukin 11 |
| Negative regulation of cell proliferation |
PTPRF |
NM_002840 |
2.56 |
Exon34 |
Protein tyrosine phosphatase, receptor type, F |
|
Migration-associated |
|
|
|
|
|
| Axon guidance |
SLIT3 |
NM_003062 |
0.44 |
Exon1 |
Slit homolog 3 (Drosophila) |
| SEMA5A |
NM_003966 |
2.90 |
Exon23 |
Sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
|
| Motor axon guidance |
EGR2 |
NM_000399 |
5.08 |
Exon2 |
Early growth response 2 |
| Cell migration |
PVR |
NM_001135770 |
0.49 |
Exon3 |
Poliovirus receptor |
| Positive regulation of cell migration |
LAMB1 |
NM_002291 |
0.46 |
Exon9 |
Laminin, beta 1 |
| Regulation of cell shape |
ARAP1 |
NM_001135190 |
0.45 |
Exon5 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
|
Angiogenesis-associated |
|
|
|
|
|
| Positive regulation of angiogenesis |
RUNX1 |
NM_001122607 |
0.40 |
Exon1 |
Runt-related transcription factor 1 |
| Wound healing | SERPINB2 | NM_002575 | 0.48 | Exon8 | Serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
Differentially spliced genes in human MSCs under hypoxia, were subjected to GO analysis as described in Methods.
We chose two alternatively spliced genes, ALDH3A2 and NDRG4 for further analysis and qPCR confirmation; ALDH3A2 transcripts were decreased by hypoxia whereas NDRG4 were not regulated. ALDH3A2 is a member of the ALDH superfamily of NAD(P) + −dependent enzymes that catalyze the oxidation of a wide variety of aliphatic and aromatic aldehydes. ALDH3A2 is also known as fatty-aldehyde dehydrogenase (FALDH) because of its role in protecting against lipid peroxidation [58]; loss of ALDH3A2 activity is the cause of Sjogren-Larsson syndrome [59]. As shown in Figure 8A, hypoxia/normoxia dictate the use of AS to generate alternative isoforms of the ALDH3A2 gene. Exon array analysis revealed that the AS event occurred in the last exon, while PCR validation confirmed that the unique exon 10 was not expressed in hypoxic MSCs. This indicates preferentially expression of the M2 isoform under hypoxia (Figure 8B). Previous work has shown that hypoxia can repress the expression of both constitutive and induced ALDH3 isoforms [60] and it is also known that ALDH3A2 gene expression is positively regulated by PPARα [61]. Our finding that PPARα expression is also decreased by hypoxia in parallel with ALDH3A2 (Table 3) suggests a possible mechanism for the transcriptional repression of ALDH3A2 by hypoxia. Other work has shown that AS of the ALDH3A2 gene may determine its subcellular localization with physiological implications for function [62]. Our results are the first to show that hypoxia regulates AS of the FALDH gene.
Figure 8.
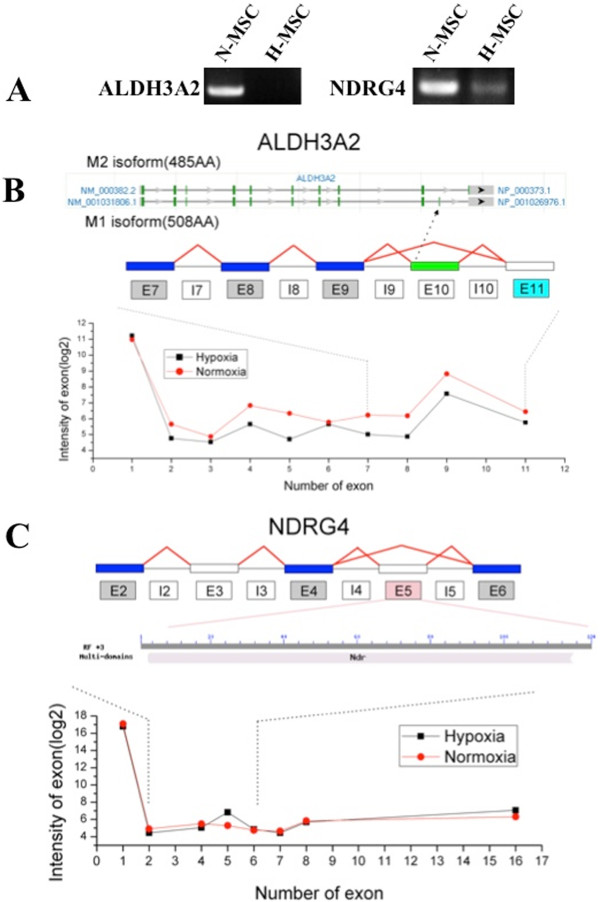
Oxygen-dependent differential splicing of the ALDH3A2 and NDRG4 genes. (A) Expression of splice isoforms confirmed by RT-PCR of genes with prior evidence of AS. (B) Annotation of each alternative isoform (M2 and M1, top graphic), exon structure (middle graphic) and expression profiles (bottom graphic) for ALDH3A2. Light blue boxes indicate down-regulation for hypoxic (H) versus normoxic (N) MSCs; gray boxes indicate no significant change; white boxes indicate no probe detection above expression threshold. The green exon (exon10) indicates the unique exon of isoform M1. Exon expression values (log2) are displayed for both H-MSC (black data points) and N-MSC (red data points), ranked in order of genomic position on the x-axis. The various isoforms can be validated in the PCR result although there is no probe annotated to the isoform-specific exon. (C) Exon structure (top graphic) modified domain of AS (middle graphic) and expression profiles (bottom graphic) for NDRG4. Light red boxes indicate up-regulation for hypoxic (H) versus normoxic MSCs (N) MSCs; gray boxes indicate no significant change; white boxes indicate no probe detection above expression threshold. A Modified or disrupted conserved domain of differentially expressed exon is from the CDD database (NCBI). Exon expression values (log2) are displayed for both H-MSC (black data points) and N-MSC (red data points), ranked in order of genomic position on the x-axis.
The N-myc downstream-regulated gene (NDRG) family belongs to the alpha/beta hydrolase super family of genes that regulate survival, growth and differentiation of host cells. The NDRG4 isoform is preferentially expressed in brain and heart and its overexpression has been linked with an aggressive behavior of meningioma tumors [63]. AS variants of NDRG4 include heart (H) and two brain (B- and Bvar) isoforms [64]. As shown in Figure 8C, exon 5 of the NDRG4 gene was subject to hypoxia-dependent AS according to the exon microarray, and this was validated by qPCR. Alternatively spliced transcripts of the NDRG genes have been described although the functional significance is unknown [64,65].
Tissue-specific regulation of gene expression and AS by hypoxia
Weigand et al. [35] recently reported results of their global analyses of hypoxia-regulated AS using human umbilical vein endothelial cells (HUVECs). Because our results as well as those of others suggest that many of the effects of hypoxia are tissue-specific we compared our gene expression and AS array data with that of Weigand et al. These results are shown in Supplement Figures 2, 3 and 4. Out of 446 and 113 genes regulated >2.5-fold by hypoxia in hMSCs and HUVECs respectively only 9 were commonly regulated including ANGPTL4, VEGFA, STC, SLC2A3, PGF, EGLN3, ADM, BHLHE40 and NQO1 (Additional file 2: Figure S2). Out of 541 and 239 gene isoforms created by hypoxia-regulated AS in hMSCs and HUVECs respectively, only 17 isoforms were common (Additional file 3: Figure S3). As shown in Additional file 4: Figure S4 comparing all of the genes in hMSCs and HUVECs regulated by hypoxia at the levels of gene expression and AS only 3 genes were common. These included Egln3, a prolyl hydroxylase involved in HIF-1 proteolytic targeting, stanniocalcin-1 (STC) a pro-angiogenic anti-apoptosis gene product [66,67] (both up-regulated) and NAD(P) Quinone oxidoreductase 1 (NQO1) a Phase II redox detoxification enzyme [68], (down-regulated). It should be noted that Weigand et al. exposed HUVECs to 1% oxygen for 48 h compared with our 0.5% for 24 h therefore the differences may be due in part to different oxygen tension as well as cell type.
Discussion
Our studies suggest that severity of hypoxia within a range of 4–10 mmHg is a critical determinant of global gene expression that has important implications for the biology and function of human MSCs. Transcriptional profiling revealed altered patterns of gene expression particularly involving glucose metabolism, insulin/IGF-1 signaling, intermediate filaments, extracellular matrix, anti-oxidant enzymes, and differentiation-markers. The results are consistent with the quiescent, immobile state conferred by severe hypoxia along with reduced oxidative stress and a switch in favor of osteogenic, angiogenic and perhaps hematopoietic programs over those of adipogenesis and myogenesis. The gene expression profiles are distinct in many respects from those described previously for MSCs subjected to more moderate hypoxia that mediates enhanced proliferation [9,10,17-19].
Salient features of the present study that define the role of severe hypoxia and distinguish it from moderate hypoxia include the following: [1] Mixed responses of genes involved in glucose metabolism including the pentose phosphate and glycolysis pathways, and moderately induced or decreased levels of glucose transporter gene transcripts. Although glycolytic flux increased acutely under hypoxia, a lower than a maximal induction of all genes may be physiologically beneficial in the long term for a closed environment such as the endosteum where high glycolytic activity would generate localized acidosis and cell death. An analogous condition may be represented in anoxic turtles where the PGAM gene is also repressed and may contribute to the hypometabolic state that is necessary for the turtle to survive extended anoxia [69]. Notable also is the weak induction of Bnip3, a programmed cell death protein normally strongly induced by hypoxia through HIF-1α and co-regulated by hypoxia and acidosis [70]. Also noteworthy is the strong induction of carbonic anhydrase (CA9) suggesting a pre-emptive adaptation to acidosis [2]. IGF-1 and PI3-kinase transcripts were decreased while IGFBP-1, −3 and −5 transcripts were increased, and this was associated with depressed phosphorylation of Akt-Thr308 (Figure 6). The IGF-PI3-kinase pathway regulates cell growth and survival through key targets including mTOR and p70-S6-kinase, and its depressed activity by severe hypoxia is consistent with reduced growth and metabolism. PI3-kinase signaling and phosphor-Akt were reported to be activated in MSCs by moderate hypoxia [26] therefore the reduced activity by severe hypoxia again distinguishes the two conditions [6]. Acidic keratins were markedly increased by hypoxia and this correlated with a more polar organization of intermediate filaments and cell elongation. The acidic keratin genes are clustered on chromosome 17q and the coordinately increase levels of multiple transcripts suggests a common regulatory mechanism. Regulation of keratin expression by the HIF pathway has not been reported. It is also noteworthy that whereas acidic keratin transcripts were all induced, 3 basic keratin gene transcripts decreased. Cytosolic keratins are markers of epithelium and the strong inductions are reminiscent of mesenchymal to epithelial transition (MET) that is associated with reduced rates of growth and migration [43-45]. Such a partial transition is again consistent with the quiescent immobile state predicted by the severely hypoxic environment of the endosteum. Changes in expression of multiple collagen and actin genes provides additional evidence for dynamic changes in cytoskeleton and extracellular matrix that contribute to cell growth and mobility [3]. Reduced expression of antioxidant pathway genes in particular NAD(P)H quinone-1 and aldo-keto reductase family members is consistent with lower oxidative stress created by severe hypoxia and may be associated with lower electron leakage and free radical production by mitochondria. Lower ROS and oxidative stress may also contribute to reduced proliferation [4]. Enhanced expression of osteogenic and angiogenic marker genes and depressed expression of adipogenic and myogenic markers is consistent with some but not all previous studies. Mayer et al. [48] and Hung et al. [10] found that osteogenesis was induced in MSCs by an oxygen tension of 10 mmHg. Hung et al. also reported that adipogenesis was reduced by 10 mmHg whereas Fink et al. [46] and Kato et al. [16] reported the opposite effect with increased adipogenesis under the same pO2. The differences may be due to incubation conditions and whether the cell are exposed to reoxygenation during treatments; Hung et al. [12] reported that the effects of hypoxia on proliferation and differentiation were fully reversed by reoxygenation.
The induction of numerous markers of angiogenesis is consistent with many previous reports describing angiogenic activation by hypoxia. The selective decrease of multiple myogenic gene markers by severe hypoxia is a novel observation of this study.
This is the second report to describe global changes in the alternative splicing of genes mediated by hypoxia and the first to describe such an effect in stem cells. There was only a small amount of overlap of hypoxia-regulated alternatively spliced genes in HUVECs versus MSCs with only 17 commonly regulated exons. It is noteworthy that multiple genes that were highly regulated at the transcript level in hMSCs were also subject to differential exon inclusion suggesting possible common regulatory factors in both pathways. Regulators of AS include specific RNA binding proteins and transcription factors (reviewed in [32]). It is known that transcription factor binding can influence AS perhaps by influencing the concentration of direct AS regulators within the transcription complex, or by altering the rate of RNA polymerase II elongation, leading indirectly to AS. Extracellular signals can also modify AS events by changing the activity of regulators for example, hnRNP-A1 (heterogeneous nuclear ribonucleoprotein A1) an RNA binding protein involved in nuclear pre-RNA processing that inhibits the inclusion of multiple alternative exons becomes phosphorylated upon osmotic shock resulting in cytoplasmic accumulation with consequent effects on AS [71]. HIF-1 was shown to regulate differential splicing of the LDH gene in shrimps [72], as well as AS of the hTERT gene that regulates telomerase activity in mammalian cells [73]. Of the most strongly and dually (transcripts and AS) regulated genes LEP, IGFBP1, CA9 and LOXL4 are all HIF-1α regulated. Therefore HIF-1α may contribute to the AS of dependent genes providing dual and coordinated regulation of transcription and AS by hypoxia. It is difficult to assess the physiological significance of most of the AS responding genes without information on the functional consequences, however it was possible to group the genes into functional categories of cell survival, proliferation, mobility and differentiation (Table 4).
Conclusion
We report on molecular genetic and phenotypic changes conferred on hMSC by severe hypoxia. The gene expression changes are largely distinct from those reported previously for more moderate hypoxia that support enhanced proliferation, and the results are consistent with a quiescent, immobile phenotype with reduced metabolic activity and lower oxidative stress. Hypoxia-mediated AS may contribute importantly to gene regulation and protein function during adaptation to a severely hypoxic environment such as that imposed by the bone marrow endosteum.
Methods
Isolation, culture and characterization of human MSC
Normal human bone marrow aspirates were obtained with written consent from healthy donors in accordance with the Declaration of Helsinki and with the approval of the Human subjects Ethics Committee of Second Affiliated Hospital of Zhejiang University. MSCs from 3 such donors were cultured as described previously [10,12,19,74]. Cell surface markers were profiled using a BD FACS CantoTM II Flow Cytometry System after 3–5 passages as described previously [75] with the following human specific monoclonal antibodies: CD29-phycoerythrin (PE) (eBioscience, San Diego, CA, USA), CD34-PE (MACS, Miltenyi Biotec, Auburn, CA, USA) and CD166-PE (BD Biosciences Pharmingen, San Diego, CA, USA), respectively.
Hypoxia
MSCs were plated at 1 × 105 cells/cm2 in complete culture medium and incubated under hypoxia (0.5% O2, 5% CO2) or normoxia (21% O2, 5% CO2) for 24 hours using a ProOX Model C21 system (BioSpherix, Redfield, NY, USA).
RNA extraction
Total RNA from normoxic and hypoxic MSCs (3 independent donor samples each) was extracted using a Kit from Biochain, (Hayward, CA, USA), according to the manufacturer’s instructions. The RNA quality was assessed by formaldehyde agarose gel electrophoresis and quantified using a spectrophotometer (Nanodrop, Wilmington, DE, USA).
RNA amplification and labeling
Gene expression microarray: RNA was amplified, reverse transcribed and labeled as described previously [36]. Briefly, 1 μg of total RNA was amplified using a Message AmpTM II RNA Amplification kit (Life Technologies, Austin, TX, USA). The RNA was reverse transcribed in the presence of cy3-dCTP or cy5-dCTP using Klenow enzyme. For exon microarray, RNA from 3 separate replicate samples of normoxic or hypoxia MSCs was amplified using a Low Input Quick Amp WT Labeling Kit from Agilent.
Array hybridization and data acquisition
Hybridization, scanning and washing were performed on Agilent’s Microarray Platform according to standard protocols. Raw data were acquired using an Agilent DNA microarray scanner and Agilent feature extraction software. All data is MIAME compliant and raw files from the 6 arrays (n = 3 per group) are deposited in the GEO database at NCBI (GEO accession # GSE55875). In addition, we downloaded from the NCBI GEO database the 6 raw data files (n = 3 per group) from the AS study by Weigand et al. [35].
Gene and AS analyses
All raw text files (from hMSC Agilent arrays) and CEL files (from HUVEC Affymetrix arrays) were imported into GeneSpring GX 11 software (Silicon Genetics, Redwood City, CA) for either global gene analysis or alternative splicing. A total of four analytical experiments were performed with 3 replicate arrays for each condition. Normalized expression values were calculated by the Robust Multi-array Average (RMA) method. The resultant signal information was analyzed using one-way analysis of variance (ANOVA) (p < 0.05), assuming normality and equal variances. Multiple Testing Correction (MTC) of p-values by Benjamini Hochberg screened out >90% of regulated genes (33/446) including many of the known HIF-1α-regulated genes such as metallothionein (MT3), carbonic anhydrase XII (CA12; 4.6-fold), (VEGFA; 4.6-fold) and placental growth factor, (PGF; 3.28-fold) (Table 1), as well as many of the gene transcripts that were subsequently confirmed by QPCR including phosphoglycerate kinase (PGK; 7.1-fold), phosphofructokinase (PFK; 3.2-fold), RAS p21 protein activator (RASA1; 2.9-fold) and Immediate Early Response 3 (IER3; 2.5-fold). MTC is known to screen out false positives as well as many true positives, therefore as in our previous micro-array studies [74,76] it was not employed here. The GeneSpring Cross Gene Error model was applied; this model determines the likelihood of observing a specific fold change to the likelihood of observing a fold measurement by the 50.0th percentile of all measurements in the sample. The average value of expression level for each gene across the samples is set to 1.0 and the resulting normalized signal value plotted for each sample. Lists of differentially expressed genes from different experiments were compared within GeneSpring and displayed as Venn diagrams to show overlapping and non-overlapping genes. Heatmaps and graphs were also generated within GeneSpring.
Conserved domain function annotation of AS exons
To identify protein domains of genes modified by AS, a conserved domain database (CCD) from NCBI web services was used. By default, domain predictions are derived by comparing two protein isoform sequences: one that aligns to the alternative exon and another in which the exon is absent from the corresponding mRNA sequences (competitive isoform).
Validation of the differentially expressed genes
Differentially expressed genes-of-interest were selected based on function and confirmed using quantitative real-time (RT-q) PCR. 2 μg of total RNA respectively from normoxic and hypoxic MSCs was reverse transcribed with oligo (dT)18 using M-MLV reverse transcriptase (TAKARA, Japan) in a volume of 40 μL. Following reverse transcription, 1 μl of this cDNA mixture was employed for a qPCR program of 40 cycles (10 s at 95°C/32 s at 60°C) with SYBR®Premix Ex TaqTM. Data were generated using Perfect Real Time (TAKARA, Japan) and Real-Time PCR (Applied Biosystems, Foster City, CA, USA). Data were analyzed by the 2-∆∆Ct method and results are shown as fold change relative to control.
Confirmation of alternative exon expression
24 alternative exon sequences were selected for confirmation by RT-qPCR. The sequences of the AS exon and its neighboring exons were acquired from the Human Genome UCSC Genome Database. If AS of the last exon of a gene was indicated, the last two-exon sequences of this gene were accessed. Constitutive exon-specific primers were designed and after reverse transcription, qPCR products were separated in 1% agarose gels supplemented with ethidium bromide, and visualized by UV.
Enzyme-linked immunosorbent assay (ELISA)
Leptin and VEGF concentrations in culture medium of normoxic and hypoxic MSCs were measured by ELISA (R&D System, Minneapolis, MN, USA), according to the manufacturer’s instructions. Assays were conducted on a SoftMax Pro® multiplate reader (Molecular Devices, Inc., Downingtown, PA, USA).
Western blot
Our western blot procedures are described in detail elsewhere [77,78]. Briefly, equal amounts of protein were separated by 10-15% SDS polyacrylamide gel electrophoresis and electro-transferred onto an Immobilon-P Transfer Membrane (Millipore, Billerica, MA, USA). Membranes were blocked with 5% BSA in TBS-T and incubated with VEGF (Santa Cruz Biotechnology, Santa Cruz, CA, USA), leptin, α-tubulin, or Akt (R&D System), antibodies overnight at 4°C. Horseradish peroxide–conjugated secondary antibodies were hybridized by standard procedures. β-actin was used as loading control.
F-actin and keratin intermediate filament staining
Normoxic and hypoxic MSCs were examined for actin filaments organization using Alexa Fluor 555 phalloidin (Invitrogen). Briefly, MSCs were fixed in 4% formaldehyde for 10 min, permeabilized with 0.1% Triton X-100 for 5 min, and blocked with PBS containing 1% BSA for 25 min. Cells were stained with diluted phalloidin in PBS for 20 min at room temperature and viewed under a fluorescence microscope.
Statistical analysis
One-way ANOVA was used to compare experimental groups. Data are expressed as mean ± standard deviation (SD), and a p value < 0.05 was considered as statistically significant.
Abbreviations
AS: Alternative splicing; MSC: Mesenchymal stem cell; MET: Mesenchymal to epidermal transition; VEGF: Vascular endothelial growth factor.
Competing interest
The authors declare that they have no competing interests financial or non-financial related to the content of this article.
Authors’ contributions
XH: conception, design of experiments and manuscript writing; RW: implement experiments, data analysis, manuscript writing; LS: data analysis, bioinformatics, statistics, manuscript writing; LZ, XH, FG, XL: implement experiments; HY: design of experiments and manuscript writing; KAW: conception, design of experiments, data analysis, manuscript writing, corresponding author; JW, conception, design of experiments, data analysis, manuscript writing, corresponding author. All authors read and approved the final manuscript.
Supplementary Material
Differential MSC gene expression. Heatmap of the top differentially expressed genes by at least 2.0-fold (p < .01) in hMSCs under hypoxia relative to normoxia.
Venn diagram and heatmap of differentially expressed genes that overlap in hMSCs and HUVECs under hypoxia. (A). Differentially expressed genes determined by our analyses of HUVECs and human MSCs under hypoxia were compared to find common differentially expressed transcripts. A 2.0 fold change and p < 0.01 significance cut-offs were used. (B). Selected genes, including VEGF-A, from the 9 transcripts overlapping in HUVEC and hMSCs, are shown by heatmap displaying expression levels in normoxic and hypoxic MSCs. Color bar shown is Log2.
Venn diagram and graphs of differentially expressed isoforms overlapping in hMSCs and HUVECs under hypoxia. (A). Differentially expressed isoforms determined by our analysis of HUVECs and human MSCs under hypoxia were compared to find common differentially expressed exons. A 1.0 splicing index and p < 0.01 significance cutoffs were used. (B). Exonic expression of 17 isoforms overlapping in HUVECs and hMSCs are shown in the graphs displaying the hypoxic vs. normoxic exonic expression levels.
Venn Diagrams and heatmap of differentially expressed genes overlapping genes in hMSCs versus HUVECs. Differentially expressed genes and isoforms determined by our analysis of HUVECs and human MSCs under hypoxia were compared to find common or unique differentially expressed genes and isoforms. A 1.0 splicing index, 2.0 fold change, and p < 0.01 significance cut-offs were used.
Contributor Information
Xinyang Hu, Email: hxy0507@hotmail.com.
Rongrong Wu, Email: worealagatha55@aliyun.com.
Lina A Shehadeh, Email: LShehadeh@med.miami.edu.
Qing Zhou, Email: zhouqing8888@126.com.
Cizhong Jiang, Email: czjiang@tongji.edu.cn.
Xin Huang, Email: xh@zju.edu.cn.
Ling Zhang, Email: zhanglingwws@163.com.
Feng Gao, Email: gaofeng0408@hotmail.com.
Xianbao Liu, Email: liuxb2009@hotmail.com.
Hong Yu, Email: yuvascular@163.com.
Keith A Webster, Email: kwebster@med.miami.edu.
Jian’an Wang, Email: jian_an_wang@yahoo.com.
Acknowledgements
This work was supported by grants from the National High Technology Research and Development Program (“863” Program) of China (2011AA020102), National Natural Science Foundation of China (31000650; 31101052; 31171418), Zhejiang Provincial Natural Science Foundation of China (Y2090308; Y2110254), Florida State (3KN05 to LAS), and the National Institute of Health (K01AG040468 to LAS and RO1 HL072924-01 to KAW).
References
- Jones DL, Wagers AJ. No place like home: anatomy and function of the stem cell niche. Nat Rev Mol Cell Biol. 2008;9:11–21. doi: 10.1038/nrm2319. [DOI] [PubMed] [Google Scholar]
- Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- Eliasson P, Rehn M, Hammar P, Larsson P, Sirenko O, Flippin LA, Cammenga J, Jönsson JI. Hypoxia mediates low cell-cycle activity and increases the proportion of long-term reconstituting hematopoietic stem cells during in vitro culture. Exp Hematol. 2010;38:301–310. doi: 10.1016/j.exphem.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Panchision DM. The role of oxygen in regulating neural stem cells in development and disease. J Cell Physiol. 2009;220:562–568. doi: 10.1002/jcp.21812. [DOI] [PubMed] [Google Scholar]
- Silván U, Díez-Torre A, Arluzea J, Andrade R, Silió M, Aréchaga J. Hypoxia and pluripotency in embryonic and embryonal carcinoma stem cell biology. Differentiation. 2009;78:159–168. doi: 10.1016/j.diff.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Eliasson P, Jonsson JI. The hematopoietic stem cell niche: low in oxygen but a nice place to be. J Cell Physiol. 2010;222:17–22. doi: 10.1002/jcp.21908. [DOI] [PubMed] [Google Scholar]
- Ezashi T, Das P, Roberts RM. Low O2 tensions and the prevention of differentiation of hES cells. Proc Natl Acad Sci. 2005;102:4783–4788. doi: 10.1073/pnas.0501283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong CH, Lee HJ, Cha JH, Kim JH, Kim KR, Kim JH, Yoon DK, Kim KW. Hypoxia-inducible factor-1 alpha inhibits self-renewal of mouse embryonic stem cells in Vitro via negative regulation of the leukemia inhibitory factor-STAT3 pathway. J Biol Chem. 2007;282:13672–13679. doi: 10.1074/jbc.M700534200. [DOI] [PubMed] [Google Scholar]
- Basciano L, Nemos C, Foliguet B, de Isla N, de Carvalho M, Tran N, Dalloul A. Long term culture of mesenchymal stem cells in hypoxia promotes a genetic program maintaining their undifferentiated and multipotent status. BMC Cell Biol. 2011;12:12. doi: 10.1186/1471-2121-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung SP, Ho JH, Shih YR, Lo T, Lee OK. Hypoxia promotes proliferation and osteogenic differentiation potentials of human mesenchymal stem cells. J Orthop Res. 2012;30:260–266. doi: 10.1002/jor.21517. [DOI] [PubMed] [Google Scholar]
- Follmar KE, Decroos FC, Prichard HL, Wang HT, Erdmann D, Olbrich KC. Effects of glutamine, glucose, and oxygen concentration on the metabolism and proliferation of rabbit adipose-derived stem cells. Tissue Eng. 2006;12:3525–3533. doi: 10.1089/ten.2006.12.3525. [DOI] [PubMed] [Google Scholar]
- Hung SC, Pochampally RR, Hsu SC, Sanchez C, Chen SC, Spees J, Prockop DJ. Short-term exposure of multipotent stromal cells to low oxygen increases their expression of CX3CR1 and CXCR4 and their engraftment in vivo. PLoS One. 2007;2(5):e416. doi: 10.1371/journal.pone.0000416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CC, Yew TL, Yang DC, Huang WH, Hung SC. Benefits of hypoxic culture on bone marrow multipotent stromal cells. Am J Blood Res. 2012;2:148–159. [PMC free article] [PubMed] [Google Scholar]
- Martin-Rendon E, Hale SJ, Ryan D, Baban D, Forde SP, Roubelakis M, Sweeney D, Moukayed M, Harris AL, Davies K, Watt SM. Transcriptional profiling of human cord blood CD133+ and cultured bone marrow mesenchymal stem cells in response to hypoxia (1.5%) Stem Cells. 2007;25:1003–1012. doi: 10.1634/stemcells.2006-0398. [DOI] [PubMed] [Google Scholar]
- Nazmul H, Mohammad Tariqur R, Noor Hayaty Abu K, Aied Mohammed A. Hypoxic culture conditions as a solution for mesenchymal stem cell based regenerative therapy. Sci World J. 2013;2013:632972. doi: 10.1155/2013/632972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Kato T, Furu M, Nasu A, Kajita Y, Mitsui H, Ueda M, Aoyama T, Nakayama T, Nakamura T, Toguchida J. Mesenchymal stem cells cultured under hypoxia escape from senescence via down-regulation of p16 and extracellular signal regulated kinase. Biochem Biophys Res Commun. 2010;39:1471–1476. doi: 10.1016/j.bbrc.2009.12.096. [DOI] [PubMed] [Google Scholar]
- Ohnishi S, Yasuda T, Kitamura S, Nagaya N. Effect of hypoxia on gene expression of bone marrow-derived mesenchymal stem cells and mononuclear cells. Stem Cells. 2007;25:1166–1177. doi: 10.1634/stemcells.2006-0347. [DOI] [PubMed] [Google Scholar]
- Nekanti U, Dastidar S, Venugopal P, Totey S, Ta M. Increased proliferation and analysis of differential gene expression in human Wharton’s jelly-derived mesenchymal stromal cells under hypoxia. Int J Biol Sci. 2010;6:499–512. doi: 10.7150/ijbs.6.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson WL, Zhao F, Bunnell B, Ma T. Hypoxia enhances proliferation and tissue formation of human mesenchymal stem cells. Biochem Biophys Res Commun. 2007;358:948–953. doi: 10.1016/j.bbrc.2007.05.054. [DOI] [PubMed] [Google Scholar]
- Salim A, Nacamuli RP, Morgan EF, Giaccia AJ, Longaker MT. Transient changes in oxygen tension inhibit osteogenic differentiation and Runx2 expression in osteoblasts. J Biol Chem. 2004;279:40007–40016. doi: 10.1074/jbc.M403715200. [DOI] [PubMed] [Google Scholar]
- Robins JC, Akeno N, Mukherjee A, Dalal RR, Aronow BJ, Koopman P, Clemens TL. Hypoxia induces chondrocyte-specific gene expression in mesenchymal cells in association with transcriptional activation of Sox9. Bone. 2005;37:313–322. doi: 10.1016/j.bone.2005.04.040. [DOI] [PubMed] [Google Scholar]
- Tsai CC, Su PF, Huang YF, Yew TL, Hung SC. Oct4 and Nanog directly regulate Dnmt1 to maintain self-renewal and undifferentiated state in mesenchymal stem cells. Mol Cell. 2012;47:169–182. doi: 10.1016/j.molcel.2012.06.020. [DOI] [PubMed] [Google Scholar]
- Fehrer C, Brunauer R, Laschober G, Unterluggauer H, Reitinger S, Kloss F, Gülly C, Gassner R, Lepperdinger G. Reduced oxygen tension attenuates differentiation capacity of human mesenchymal stem cells and prolongs their lifespan. Aging Cell. 2007;6:745–757. doi: 10.1111/j.1474-9726.2007.00336.x. [DOI] [PubMed] [Google Scholar]
- Hu X, Wei L, Taylor TM, Wei J, Zhou X, Wang JA, Yu SP. Hypoxic preconditioning enhances bone marrow mesenchymal stem cell migration via Kv2.1 channel and FAK activation. Am J Physiol Cell Physiol. 2011;301:C362–C372. doi: 10.1152/ajpcell.00013.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Lee YJ, Song CH, Ahn YK, Han HJ. Role of FAK phosphorylation in hypoxia-induced hMSCS migration: involvement of VEGF as well as MAPKS and eNOS pathways. Am J Physiol Cell Physiol. 2010;298:C847–C856. doi: 10.1152/ajpcell.00418.2009. [DOI] [PubMed] [Google Scholar]
- Rosová I, Dao M, Capoccia B, Link D, Nolta JA. Hypoxic preconditioning results in increased motility and improved therapeutic potential of human mesenchymal stem cells. Stem Cells. 2008;26:2173–2182. doi: 10.1634/stemcells.2007-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MV, Zheng X, Pereira T, Gradin K, Jin S, Lundkvist J, Ruas JL, Poellinger L, Lendahl U, Bondesson M. Hypoxia requires Notch signaling to maintain the undifferentiated cell state. Dev Cell. 2005;9:617–628. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Tsai CC, Chen YJ, Yew TL, Chen LL, Wang JY, Chiu CH, Hung SC. Hypoxia inhibits senescence and maintains mesenchymal stem cell properties through down-regulation of E2A-p21 by HIF-TWIST. Blood. 2011;117:459–469. doi: 10.1182/blood-2010-05-287508. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Manley JL. The search for alternative splicing regulators: new approaches offer a path to a splicing code. Genes Dev. 2008;22:279–285. doi: 10.1101/gad.1643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JD, Janitz M. Alternative splicing of mRNA in the molecular pathology of neurodegenerative diseases. Neurobiol Aging. 2011;33:1012 e1011–1024. doi: 10.1016/j.neurobiolaging.2011.10.030. [DOI] [PubMed] [Google Scholar]
- Moller-Levet CS, Betts GN, Harris AL, Homer JJ, West CM, Miller CJ. Exon array analysis of head and neck cancers identifies a hypoxia related splice variant of LAMA3 associated with a poor prognosis. PLoS Comput Biol. 2009;5:e1000571. doi: 10.1371/journal.pcbi.1000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigand JE, Boeckel JN, Gellert P, Dimmeler S. Hypoxia-induced alternative splicing in endothelial cells. PLoS One. 2012;7:e42697. doi: 10.1371/journal.pone.0042697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest. 2013;123:3664–3671. doi: 10.1172/JCI67230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchenko O, Opentanova I, Minchenko D, Ogura T, Esumi H. Hypoxia induces transcription of 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase-4 gene via hypoxia-inducible factor-1alpha activation. FEBS Lett. 2004;576:14–20. doi: 10.1016/j.febslet.2004.08.053. [DOI] [PubMed] [Google Scholar]
- Li H, Ko HP, Whitlock JP. Induction of phosphoglycerate kinase 1 gene expression by hypoxia. Roles of Arnt and HIF1alpha. J Biol Chem. 1996;271:21262–21267. doi: 10.1074/jbc.271.35.21262. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Takahashi S, Yoshimi T, Miura T. Hypoxia-induced expression of phosphoglycerate mutase B in fibroblasts. Eur J Biochem. 1998;254:497–504. doi: 10.1046/j.1432-1327.1998.2540497.x. [DOI] [PubMed] [Google Scholar]
- Lu S, Gu X, Hoestje S, Epner DE. Identification of an additional hypoxia responsive element in the glyceraldehyde-3-phosphate dehydrogenase gene promoter. Biochim Biophys Acta. 2002;1574:152–156. doi: 10.1016/S0167-4781(01)00359-1. [DOI] [PubMed] [Google Scholar]
- Said HM, Hagemann C, Stojic J, Schoemig B, Vince GH, Flentje M, Roosen K, Vordermark D. GAPDH is not regulated in human glioblastoma under hypoxic conditions. BMC Mol Biol. 2007;8:55. doi: 10.1186/1471-2199-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashimura Y, Nakajima Y, Yamaji R, Harada N, Shibasaki F, Nakano Y, Inui H. Up-regulation of glyceraldehyde-3-phosphate dehydrogenase gene expression by HIF-1 activity depending on Sp1 in hypoxic breast cancer cells. Arch Biochem Biophys. 2011;509:1–8. doi: 10.1016/j.abb.2011.02.011. [DOI] [PubMed] [Google Scholar]
- Chung BM, Rotty JD, Coulombe PA. Networking galore: intermediate filaments and cell migration. Curr Opin Cell Biol. 2013;25:600–612. doi: 10.1016/j.ceb.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windoffer R, Beil M, Magin TM, Leube RE. Cytoskeleton in motion: the dynamics of keratin intermediate filaments in epithelia. J Cell Biol. 2011;194:669–678. doi: 10.1083/jcb.201008095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S, Subramanyam D, Blelloch R, Derynck R. Regulation of epithelial- mesenchymal and mesenchymal-epithelial transitions by microRNAs. Curr Opin Cell Biol. 2013;25:200–207. doi: 10.1016/j.ceb.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink T, Abildtrup L, Fogd K, Abdallah BM, Kassem M, Ebbesen P, Zachar V. Induction of adipocyte-like phenotype in human mesenchymal stem cells by hypoxia. Stem Cells. 2004;22:1346–1355. doi: 10.1634/stemcells.2004-0038. [DOI] [PubMed] [Google Scholar]
- Wagegg M, Gaber T, Lohanatha FL, Hahne M, Strehl C, Fangradt M, Tran CL, Schonbeck K, Hoff P, Ode A, Perka C, Duda GN, Buttgereit F. Hypoxia promotes osteogenesis but suppresses adipogenesis of human mesenchymal stromal cells in a hypoxia-inducible factor-1 dependent manner. PLoS One. 2012;7(9):e46483. doi: 10.1371/journal.pone.0046483. doi:10.1371/journal.pone.0046483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer H, Bertram H, Lindenmaier W, Korff T, Weber H, Weich H. Vascular endothelial growth factor (VEGF-A) expression in human mesenchymal stem cells: autocrine and paracrine role on osteoblastic and endothelial differentiation. J Cell Biochem. 2005;95:827–839. doi: 10.1002/jcb.20462. [DOI] [PubMed] [Google Scholar]
- Avecilla ST, Hattori K, Heissig B, Tejada R, Liao F, Shido K, Jin DK, Dias S, Zhang F, Hartman TE, Hackett NR, Crystal RG, Witte L, Hicklin DJ, Bohlen P, Eaton D, Lyden D, De Sauvage F, Rafii S. Chemokine-mediated interaction of hematopoietic progenitors with the bone marrow vascular niche is required for thrombopoiesis. Nat Med. 2004;10:64–71. doi: 10.1038/nm973. [DOI] [PubMed] [Google Scholar]
- Turner KJ, Neben S, Weich N, Schaub RG, Goldman SJ. The role of recombinant interleukin 11 in megakaryocytopoiesis. Stem Cells. 1996;14(Suppl 1):53–61. doi: 10.1002/stem.5530140707. [DOI] [PubMed] [Google Scholar]
- Motrescu ER, Rio MC. Cancer cells, adipocytes and matrix metalloproteinase 11: a vicious tumor progression cycle. Biol Chem. 2008;389:1037–1041. doi: 10.1515/BC.2008.110. [DOI] [PubMed] [Google Scholar]
- Eijken M, Meijer IM, Westbroek I, Koedam M, Chiba H, Uitterlinden AG, Pols HA, van Leeuwen JP. Wnt signaling acts and is regulated in a human osteoblast differentiation dependent manner. J Cell Biochem. 2008;104(2):568–579. doi: 10.1002/jcb.21651. [DOI] [PubMed] [Google Scholar]
- Yerges LM, Zhang Y, Cauley JA, Kammerer CM, Nestlerode CS, Wheeler VW, Patrick AL, Bunker CH, Moffett SP, Ferrell RE, Zmuda JM. Functional characterization of genetic variation in the Frizzled 1 (FZD1) promoter and association with bone phenotypes: more to the LRP5 story? J Bone Miner Res. 2009;24:87–96. doi: 10.1359/jbmr.080816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Miao T, Sebastian M, Bhullar P, Ghaffari E, Liu M, Symonds AL, Wang P. The transcription factors Egr2 and Egr3 are essential for the control of inflammation and antigen-induced proliferation of B and T cells. Immunity. 2012;37:685–696. doi: 10.1016/j.immuni.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y, Zipori D. The role of activin a in regulation of hemopoiesis. Activin A expression is induced during hemopoietic differentiation. Stem Cells. 2002;20:493–500. doi: 10.1634/stemcells.20-6-493. [DOI] [PubMed] [Google Scholar]
- Sadanandam A, Rosenbaugh EG, Singh S, Varney M, Singh RK. Semaphorin 5A promotes angiogenesis by increasing endothelial cell proliferation, migration, and decreasing apoptosis. Microvasc Res. 2010;79:1–9. doi: 10.1016/j.mvr.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Huang M, Li Z, Jia F, Ghosh Z, Lijkwan MA, Fasanaro P, Sun N, Wang X, Martelli F, Robbins RC, Wu JC. MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation. 2010;122(11 Suppl):S124–S131. doi: 10.1161/CIRCULATIONAHA.109.928424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demozay D, Mas JC, Rocchi S, Van Obberghen E. FALDH reverses the deleterious action of oxidative stress induced by lipid peroxidation product 4-hydroxynonenal on insulin signaling in 3T3-L1 adipocytes. Diabetes. 2008;57:1216–1226. doi: 10.2337/db07-0389. [DOI] [PubMed] [Google Scholar]
- De Laurenzi V, Rogers GR, Hamrock DJ, Marekov LN, Steinert PM, Compton JG, Markova N, Rizzo WB. Sjögren-Larsson syndrome is caused by mutations in the fatty aldehyde dehydrogenase gene. Nat Genet. 1996;12:52–57. doi: 10.1038/ng0196-52. [DOI] [PubMed] [Google Scholar]
- Reisdorph R, Lindahl R. Hypoxia exerts cell-type-specific effects on expression of the class 3 aldehyde dehydrogenase gene. Biochem Biophys Res Commun. 1998;249:709–712. doi: 10.1006/bbrc.1998.9216. [DOI] [PubMed] [Google Scholar]
- Motojima K, Hirai T. Peroxisome proliferator-activated receptor alpha plays a vital role in inducing a detoxification system against plant compounds with crosstalk with other xenobiotic nuclear receptors. FEBS J. 2006;273:292–300. doi: 10.1111/j.1742-4658.2005.05060.x. [DOI] [PubMed] [Google Scholar]
- Ashibe B, Hirai T, Higashi K, Sekimizu K, Motojima K. Dual subcellular localization in the endoplasmic reticulum and peroxisomes and a vital role in protecting against oxidative stress of fatty aldehyde dehydrogenase are achieved by alternative splicing. J Biol Chem. 2007;282:20763–20773. doi: 10.1074/jbc.M611853200. [DOI] [PubMed] [Google Scholar]
- Kotipatruni RP, Ferraro DJ, Ren X, Vanderwaal RP, Thotala DK, Hallahan DE, Jaboin JJ. NDRG4, the N-Myc downstream regulated gene, is important for cell survival, tumor invasion and angiogenesis in meningiomas. Integr Biol (Camb) 2012;4:1185–1197. doi: 10.1039/c2ib20168b. [DOI] [PubMed] [Google Scholar]
- Zhou RH, Kokame K, Tsukamoto Y, Yutani C, Kato H, Miyata T. Characterization of the human NDRG gene family: a newly identified member, NDRG4, is specifically expressed in brain and heart. Genomics. 2001;73:86–97. doi: 10.1006/geno.2000.6496. [DOI] [PubMed] [Google Scholar]
- Svensson E, Vidovic K, Olofsson T, Vallon-Christersson J, Borg A, Gullberg U. The Wilms’ tumor gene 1 (WT1) induces expression of the N-myc downstream regulated gene 2 (NDRG2) DNA Cell Biol. 2007;26:589–597. doi: 10.1089/dna.2007.0586. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Ko JH, Yun JH, Kim JA, Kim TE, Lee HJ, Kim SH, Park KH, Oh JY. Stanniocalcin-1 protects retinal ganglion cells by inhibiting apoptosis and oxidative damage. PLoS One. 2013;8(5):e63749. doi: 10.1371/journal.pone.0063749. doi:10.1371/journal.pone.0063749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law AY, Wong CK. Stanniocalcin-1 and −2 promote angiogenic sprouting in HUVECs via VEGF/VEGFR2 and angiopoietin signaling pathways. Mol Cell Endocrinol. 2013;374:73–81. doi: 10.1016/j.mce.2013.04.024. [DOI] [PubMed] [Google Scholar]
- Harada S, Fujii C, Hayashi A, Ohkoshi N. An association between idiopathic Parkinson’s disease and polymorphisms of phase II detoxification enzymes: glutathione S-transferase M1 and quinone oxidoreductase 1 and 2. Biochem Biophys Res Commun. 2001;288:887–892. doi: 10.1006/bbrc.2001.5868. [DOI] [PubMed] [Google Scholar]
- Zhang J, Biggar KK, Storey KB. Regulation of p53 by reversible post-transcriptional and post-translational mechanisms in liver and skeletal muscle of an anoxia tolerant turtle. Trachemys Scr Elegans Gene. 2013;513:147–155. doi: 10.1016/j.gene.2012.10.049. [DOI] [PubMed] [Google Scholar]
- Kubasiak LA, Hernandez OM, Bishopric NH, Webster KA. Hypoxia and acidosis activate cardiac myocyte death through the Bcl-2 family protein BNIP3. Proc Natl Acad Sci. 2002;99:12825–12830. doi: 10.1073/pnas.202474099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allemand E, Guil S, Myers M, Moscat J, Caceres JF, Krainer AR. Regulation of heterogenous nuclear ribonucleoprotein A1 transport by phosphorylation in cells stressed by osmotic shock. Proc Natl Acad Sci. 2005;102:3605–3610. doi: 10.1073/pnas.0409889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soñanez-Organis JG, Rodriguez-Armenta M, Leal-Rubio B, Peregrino-Uriarte AB, Gómez-Jiménez S, Yepiz-Plascencia G. Alternative splicing generates two lactate dehydrogenase subunits differentially expressed during hypoxia via HIF-1 in the shrimp Litopenaeus vannamei. Biochimie. 2012;94:1250–1260. doi: 10.1016/j.biochi.2012.02.015. [DOI] [PubMed] [Google Scholar]
- Anderson CJ, Hoare SF, Ashcroft M, Bilsland AE, Keith WN. Hypoxic regulation of telomerase gene expression by transcriptional and post-transcriptional mechanisms. Oncogene. 2006;25:61–69. doi: 10.1038/sj.onc.1209011. [DOI] [PubMed] [Google Scholar]
- Wilson A, Shehadeh L, Yu H, Webster KA. Age-related changes in the molecular genetic profiles of murine bone marrow mesenchymal stem cells. BMC Genomics. 2010;11:229. doi: 10.1186/1471-2164-11-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Gu B, Zhao X, Tan Z, Chen L, Zhu J, Zhang M. Derivation of multipotent nestin(+)/CD271 (−)/STRO-1 (−) mesenchymal-like precursors from human embryonic stem cells in chemically defined conditions. Hum Cell. 2013;26:19–27. doi: 10.1007/s13577-011-0022-3. [DOI] [PubMed] [Google Scholar]
- Shehadeh LA, Webster KA, Hare JM, Vazquez-Padron RI. Dynamic regulation of vascular myosin light chain (MYL9) with injury and aging. PLoS One. 2011;6(10):e25855. doi: 10.1371/journal.pone.0025855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Wang W, Chopra I, Li HF, Dougherty CJ, Adi J, Adi N, Wang H, Webster KA. c-Jun N-terminal kinase (JNK-1) confers protection against brief but not extended ischemia during acute myocardial infarction. J Biol Chem. 2011;286:13995–14006. doi: 10.1074/jbc.M110.211334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Xu Q, Wu Q, Ma Q, Salgueiro L, Wang J, Eton D, Webster KA, Yu H. Defective CXCR4 expression in aged bone marrow cells impairs vascular regeneration. J Cell Mol Med. 2011;15:2046–2056. doi: 10.1111/j.1582-4934.2010.01231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differential MSC gene expression. Heatmap of the top differentially expressed genes by at least 2.0-fold (p < .01) in hMSCs under hypoxia relative to normoxia.
Venn diagram and heatmap of differentially expressed genes that overlap in hMSCs and HUVECs under hypoxia. (A). Differentially expressed genes determined by our analyses of HUVECs and human MSCs under hypoxia were compared to find common differentially expressed transcripts. A 2.0 fold change and p < 0.01 significance cut-offs were used. (B). Selected genes, including VEGF-A, from the 9 transcripts overlapping in HUVEC and hMSCs, are shown by heatmap displaying expression levels in normoxic and hypoxic MSCs. Color bar shown is Log2.
Venn diagram and graphs of differentially expressed isoforms overlapping in hMSCs and HUVECs under hypoxia. (A). Differentially expressed isoforms determined by our analysis of HUVECs and human MSCs under hypoxia were compared to find common differentially expressed exons. A 1.0 splicing index and p < 0.01 significance cutoffs were used. (B). Exonic expression of 17 isoforms overlapping in HUVECs and hMSCs are shown in the graphs displaying the hypoxic vs. normoxic exonic expression levels.
Venn Diagrams and heatmap of differentially expressed genes overlapping genes in hMSCs versus HUVECs. Differentially expressed genes and isoforms determined by our analysis of HUVECs and human MSCs under hypoxia were compared to find common or unique differentially expressed genes and isoforms. A 1.0 splicing index, 2.0 fold change, and p < 0.01 significance cut-offs were used.


