Abstract
ATP-binding cassette (ABC) transporters are a large superfamily of proteins that mediate diverse physiological functions by coupling ATP hydrolysis with substrate transport across lipid membranes. In insects, these proteins play roles in metabolism, development, eye pigmentation, and xenobiotic clearance. While ABC transporters have been extensively studied in vertebrates, less is known concerning this superfamily in insects, particularly hemipteran pests. We used RNA-Seq transcriptome sequencing to identify 65 putative ABC transporter sequences (including 36 full-length sequences) from the eight ABC subfamilies in the western tarnished plant bug (Lygus hesperus), a polyphagous agricultural pest. Phylogenetic analyses revealed clear orthologous relationships with ABC transporters linked to insecticide/xenobiotic clearance and indicated lineage specific expansion of the L. hesperus ABCG and ABCH subfamilies. The transcriptional profile of 13 LhABCs representative of the ABCA, ABCB, ABCC, ABCG, and ABCH subfamilies was examined across L. hesperus development and within sex-specific adult tissues. All of the transcripts were amplified from both reproductively immature and mature adults and all but LhABCA8 were expressed to some degree in eggs. Expression of LhABCA8 was spatially localized to the testis and temporally timed with male reproductive development, suggesting a potential role in sexual maturation and/or spermatozoa protection. Elevated expression of LhABCC5 in Malpighian tubules suggests a possible role in xenobiotic clearance. Our results provide the first transcriptome-wide analysis of ABC transporters in an agriculturally important hemipteran pest and, because ABC transporters are known to be important mediators of insecticidal resistance, will provide the basis for future biochemical and toxicological studies on the role of this protein family in insecticide resistance in Lygus species.
Introduction
ATP-binding cassette (ABC) proteins are an extensive family of transmembrane proteins that are ubiquitous to all organisms. The defining characteristic for most members of this superfamily is ATP hydrolysis driven unidirectional translocation of substrates (either import or export) across lipid membranes, typically in a thermodynamically unfavorable direction. However, ABC proteins also function as ion channels, regulators of ion channels, receptors, and in ribosome assembly and translation. They are structurally characterized by two highly conserved cytosolic nucleotide-binding domains (NBD) and two variable transmembrane domains (TMD) [1]–[4]. The NBDs, which are critical for ATP-binding and hydrolysis, provide the energy necessary for driving a substrate across the membrane. They are characterized by a catalytic core comprised of a Walker A motif (GXXGXGKS/T) and a Walker B motif (φφφφD; where φ represents a hydrophobic residue) separated by a conserved Q-loop and a Walker C motif. This latter component is a structurally diverse helical segment encompassing the ABC signature sequence (LSGGQ) that distinguishes ABC transporter family members from other ATP-binding proteins. Unlike the NBDs, TMDs vary in sequence, length, and helix number and are thought to provide initial substrate contact points. In eukaryotic organisms, the ABC transporter domains are organized as either full-transporters (FT) containing four domains (2 TMDs and 2 NBDs) or half-transporters (HT) comprised of only two domains (1 TMD and 1 NBD) that require homo- or heterodimerization for full functionality [2], [3].
Based on conserved domain homology and organization, the eukaryotic ABC family can be divided into eight distinct subfamilies (A-H) with most family members facilitating the movement of a diverse array of substrates (sugars, lipids, peptides, polysaccharides, metals, inorganic ions, amino acids, and xenobiotics) across membranes. The first characterized eukaryotic ABC transporter (P-glycoprotein, HsABCB1) was identified based on its multidrug efflux pump functionality in mammalian cancer cell lines [5]. Since then members of the ABCB, ABCC, and ABCG subfamilies have been reported to play roles in drug resistance and detoxification in a number of species across multiple phyla [6]–[8]. Unlike the other subfamilies, the ABCE and ABCF transporters lack TMDs and function in ribosome assembly and protein translation rather than substrate transport [9]–[13]. The ABCH subfamily, which so far has only been identified in arthropod genomes and zebrafish [8], [14], [15], has not been as extensively characterized as the other subfamilies.
Insect ABC transporters mediate diverse functions with critical roles in molting, cuticle differentiation, and egg development [16], eye pigmentation [17]–[19], uric acid uptake [20], germ cell migration [21], 20-hydroxyecdysone mediated circadian rhythmicity [22], phytochemical sequestration [23], and biogenic amine transport [24]. Insect ABC transporters also function in the clearance of xenobiotics including plant defensive compounds and numerous insecticides representing disparate chemical classes and modes of action [8], [25]. Resistance of some lepidopteran species to Bacillus thuringiensis (Bt) toxins has also been shown to involve novel interactions with ABC transporters independent of substrate transport [26]–[31].
Detailed genome/transcriptome wide analyses of ABC subfamilies have been conducted for a number of arthropods including water flea (Daphnia pulex) [32], spider mite (Tetranychus urticae) [33], fruitfly (Drosophila melanogaster) [34], olive fruit fly (Bactrocera oleae) [35], African malaria mosquito (Anopheles gambiae) [36], red flour beetle (Tribolium castaneum) [16], honeybee (Apis mellifera) [37], silkmoth (Bombyx mori) [37], [38], and poplar leaf beetle (Chrysomela populi) [39]. Among hemipterans, knowledge of ABC transporters is limited to unverified genome annotations for the pea aphid (Acyrthosiphon pisum) [40] and partial transcriptomic/EST analyses in whiteflies (Bemisia tabaci) [38], [41]–[43], bed bugs (Cimex lectularius) [44], and the brown planthopper (Nilaparvata lugens) [45].
The western tarnished plant bug (Lygus hesperus Knight) is a highly polyphagous hemipteran agricultural pest that primarily feeds on and injures the buds, flowers and seeds of many crops in the western United States and Canada [46]–[48]. Control strategies have traditionally relied on broad-spectrum insecticides. However, decreased efficacy has been reported for many of these compounds against field populations of Lygus [49]–[51]. Further compounding its pest status, our understanding of the molecular basis underlying Lygus resistance to insecticides is limited. To begin to address this issue, we performed a transcriptome-wide survey of ABC transporters by supplementing currently available transcriptomic data [52] with RNA-Seq analyses. We provide detailed sequence comparisons of the ABC subfamilies (ABCA-ABCH) in L. hesperus with those from four other arthropods and humans. In addition, the transcriptional profile of a subset of the assembled sequences was examined across developmental stages and in various sex-specific tissues. These findings will facilitate future exploration of the biological and physiological functions mediated by L. hesperus ABC transporters and their potential role in insecticide resistance.
Results and Discussion
RNA-Seq assembly and annotation
An earlier 454-based transcriptome of L. hesperus adults [52] contains 44 putative ABC transporter sequences. Here, we used Illumina RNA-Seq to obtain a comprehensive transcriptome that more extensively reflects ABC transporter transcription in Lygus adults. Furthermore, because some ABC transporters are associated with cellular stress [53]–[57], we combined the transcriptomes of L. hesperus exposed to cold and heat stress as well as non-stressed cohorts. Illumina HiSeq generated 144,898,116 raw 100 bp read pairs across nine libraries representing the three treatment groups. After quality filtering and in silico normalization, de novo assembly of 16,191,383 read pairs generated a raw assembly containing 132,802 isoforms across 77,246 unigenes with an N50 isoform length of 2,228 bp. Filtering this assembly by retaining only transcripts that have a predicted open reading frame reduced the assembly to 45,723 isoforms across 21,049 unigenes with an N50 isoform length of 2,989 bp.
Identification of L. hesperus ABC transporter transcripts
The L. hesperus RNA-Seq database was searched using protein sequences corresponding to the full complement of ABC transporters from seven metazoans as well as the 44 putative ABC transporter sequences from the previous transcriptome [52]. We identified 65 putative L. hesperus ABC transporter-like (LhABC) transcripts. Based on Transdecoder predictions, significant matches with the Pfam-A database, and manual inspection of sequences spanning the first in-frame Met and stop codons, 36 of the sequences are predicted to encompass complete open reading frames. The number of LhABCs identified is comparable to that reported for other arthropods (Table 1). However, this number may under-represent (exclusion of temporally or spatially restricted transcripts not expressed) or over-represent (multiple partial transcripts corresponding to different portions or isoforms of the same gene) the actual number of LhABC transporters.
Table 1. ABC subfamilies in L. hesperus and ten other species.
| phylum | Chordata | Nematoda | Arthropoda | ||||||||
| class | Insecta | ||||||||||
| order | Branchiopoda | Arachnida | Diptera | Coleoptera | Hymenoptera | Lepidoptera | Hemiptera | ||||
| H. sapiensa | C. elegansb | D. pulexc | T. urticaed | D. melanogastera | A. gambiaee | T. castaneumf | C. poulig | A. melliferah | B. morih,i | L. hesperus | |
| ABC subfamily | |||||||||||
| ABCA | 12 | 7 | 4 | 9 | 10 | 9 | 10 | 5 | 3 | 6, 9 | 11 |
| ABCB | 11 | 24 | 7 | 4 | 8 | 5 | 6 | 8 | 5 | 8, 9 | 6 |
| ABCC | 12 | 9 | 7 | 39 | 14 | 13 | 35 | 29 | 9 | 15, 15 | 12 |
| ABCD | 4 | 5 | 3 | 2 | 2 | 2 | 2 | 2 | 2 | 2, 2 | 2 |
| ABCE | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1, 1 | 1 |
| ABCF | 3 | 3 | 4 | 3 | 3 | 3 | 3 | 3 | 3 | 3, 3 | 3 |
| ABCG | 5 | 9 | 24 | 23 | 15 | 16 | 13 | 14 | 15 | 13, 12 | 19 |
| ABCH | 0 | 0 | 15 | 22 | 3 | 3 | 3 | 3 | 3 | 3, 2 | 11 |
| TOTAL | 48 | 58 | 65 | 103 | 56 | 52 | 73 | 65 | 41 | 51, 53 | 65 |
BLASTx (Table S1) and tBLASTn (Table S2) analyses revealed that the 65 putative LhABC transcripts represent all eight ABC transporter subfamilies (A-H). The most similar sequences from both BLAST analyses were putative transporters from A. pisum and T. castaneum (Figure S1). These results are consistent with previous transcriptome comparisons and not unexpected given the shared hemipteran lineage with A. pisum and the extensive genomic annotation in T. castaneum. The putative LhABC transcripts encode proteins or protein fragments ranging in size from 144 to 2237 amino acids. Although additional partial transcripts were identified in the dataset, only transcripts with coding sequences of >100 amino acids were further analyzed. Consistent with substrate translocation across the plasma membrane, 30 of the 36 full-length sequences identified are predicted to localize to the cell surface (Table 2). The six exceptions include sequences with highest similarity to ABCDs, which localize to the peroxisome, and ABCE/F subfamily members, which do not function in substrate transport. Multiple TMD prediction algorithms indicated the presence of numerous helical segments in 32 of the full-length sequences and 25 of the partial sequences (Table 2). No helices are predicted for the L. hesperus ABCE and ABCF transporters. ABC transporter motifs and/or NBDs are present in 62 of the 65 LhABC sequences (Table S3), and even for those lacking these domains there was significant sequence similarity (E-value<10−12) with genes annotated as ABC transporters (Table S1 and S2). LhABCC1A and LhABCC1B share the highest amino acid identity (96%), whereas sequence identity across the other LhABC transporters varies from 1% to 51% with highest levels of conservation observed within the respective subfamilies (Table S4).
Table 2. Bioinformatics analysis of putative LhABC transporters.
| Localization | Number of helical domains | ||||||
| L. hesperus id | Illumina assembly id | Size (aa) | Full/Partial CDS | WoLF PSORTa | TMHMMb | TopPredIIc | TopConsd |
| LhABCA1 | comp10339_c0_seq1 | 474 | partial | nde | 5 | 6 | 5 |
| LhABCA2 | comp9970_c0_seq1 | 663 | partial | nd | 5 | 5(7)f | 6 |
| LhABCA3 | comp35546_c0_seq1 | 637 | partial | nd | 5 | 6(8) | 6 |
| LhABCA4 | comp34679_c0_seq2 | 2237 | full | PM | 12 | 14(16) | 12 |
| LhABCA5 | comp28522_c0_seq4 | 1326 | full | PM | 14 | 16(19) | 12 |
| LhABCA6 | comp12312_c0_seq2 | 416 | partial | nd | 1 | 2 | 1 |
| LhABCA7 | comp33391_c0_seq1 | 585 | partial | nd | 5 | 6 | 5 |
| LhABCA8 | comp9781_c0_seq1 | 415 | partial | nd | 0 | 1 | 1 |
| LhABCA9 | comp38669_c0_seq1 | 275 | partial | nd | 0 | 0(1) | 0 |
| LhABCA10 | comp39633_c0_seq1 | 243 | partial | nd | 0 | 0 | 0 |
| LhABCA11 | comp28530_c0_seq1 | 795 | partial | nd | 5 | 5(6) | 6 |
| LhABCB1 | comp37357_c0_seq3 | 1191 | full | PM | 9 | 10(11) | 11 |
| LhABCB2 | comp30116_c0_seq1 | 830 | partial | nd | 6 | 6 | 6 |
| LhABCB3 | comp31442_c0_seq2 | 687 | partial | nd | 5 | 6(7) | 6 |
| LhABCB4 | comp35640_c0_seq1 | 834 | full | PM | 10 | 8(11) | 11 |
| LhABCB5 | comp37353_c1_seq7 | 735 | partial | nd | 5 | 7 | 6 |
| LhABCB6 | comp21601_c0_seq1 | 469 | partial | nd | 6 | 6(7) | 6 |
| LhABCC1A | comp37257_c0_seq2 | 944 | full | PM | 9 | 10(12) | 11 |
| LhABCC1B | comp19288_c0_seq1 | 944 | full | PM | 7 | 9(12) | 11 |
| LhABCC2 | comp36363_c3_seq3 | 589 | partial | nd | 5 | 5 | 6 |
| LhABCC3 | comp29144_c0_seq2 | 1407 | partial | nd | 9 | 11(16) | 12 |
| LhABCC4 | comp26101_c0_seq1 | 1226 | partial | nd | 9 | 7(10) | 12 |
| LhABCC5 | comp28277_c0_seq1 | 1316 | full | PM | 9 | 10(12) | 12 |
| LhABCC6 | comp28206_c0_seq2 | 633 | partial | nd | 4 | 4 | 6 |
| LhABCC7 | comp1105_c0_seq1/comp20506_c0_seq1 | 601 | partial | nd | 5 | 5(6) | 6 |
| LhABCC8 | comp36620_c0_seq1 | 1495 | full | PM | 16 | 14(17) | 17 |
| LhABCC9 | comp33891_c0_seq2 | 1415 | full | PM | 9 | 10(11) | 12 |
| LhABCC10 | comp14659_c0_seq1 | 256 | partial | nd | 0 | 0(1) | 0 |
| LhABCC11 | comp37326_c0_seq3 | 856 | full | PM | 10 | 9 | 10 |
| LhABCD1 | comp30627_c0_seq1 | 676 | full | cytosol | 3 | 4(5) | 5 |
| LhABCD2 | comp32676_c0_seq1 | 656 | full | mito | 5 | 4 | 5 |
| LhABCE1 | comp29836_c0_seq1 | 608 | full | cytosol | 0 | 1 | 0 |
| LhABCF1 | comp37052_c0_seq9 | 589 | full | cyto/nucleus | 0 | 0(1) | 0 |
| LhABCF2 | comp34354_c0_seq1 | 629 | full | nucleus | 0 | 1 | 0 |
| LhABCF3 | comp24795_c0_seq2 | 712 | full | cyto/nucleus | 0 | 0(1) | 0 |
| LhABCG1 | comp33145_c0_seq2 | 685 | full | PM | 6 | 6(7) | 6 |
| LhABCG2 | comp28267_c0_seq1 | 583 | partial | nd | 6 | 6(8) | 6 |
| LhABCG3 | comp36162_c0_seq6 | 622 | full | PM | 6 | 6(7) | 6 |
| LhABCG4 | comp3947_c0_seq1 | 226 | partial | nd | 0 | 0(1) | 0 |
| LhABCG5 | comp33057_c0_seq1 | 608 | full | PM | 5 | 5(8) | 6 |
| LhABCG6 | comp32204_c0_seq5 | 617 | full | PM | 6 | 6(7) | 6 |
| LhABCG7 | comp30007_c0_seq1 | 608 | full | PM | 5 | 4(6) | 6 |
| LhABCG8 | comp26890_c0_seq1 | 651 | partial | nd | 7 | 6(9) | 6 |
| LhABCG9 | comp31063_c0_seq2 | 479 | partial | nd | 4 | 4(7) | 6 |
| LhABCG10 | comp32700_c0_seq1 | 703 | full | PM | 5 | 6(7) | 6 |
| LhABCG11 | comp35069_c0_seq5 | 623 | full | PM | 6 | 4(8) | 6 |
| LhABCG12 | comp28984_c1_seq1 | 654 | full | PM | 5 | 9(11) | 6 |
| LhABCG13 | comp35811_c1_seq7 | 614 | full | PM | 7 | 7(11) | 6 |
| LhABCG14 | comp26007_c1_seq1 | 655 | partial | nd | 6 | 5(6) | 6 |
| LhABCG15 | comp34164_c0_seq8 | 601 | full | PM | 5 | 6(8) | 5 |
| LhABCG16 | comp32376_c3_seq1 | 606 | full | PM | 7 | 6(7) | 6 |
| LhABCG17 | comp32660_c1_seq2 | 611 | full | PM | 7 | 7(8) | 6 |
| LhABCG18 | comp9896_c0_seq1 | 319 | partial | nd | 0 | 1 | 1 |
| LhABCG19 | comp20817_c0_seq1 | 227 | partial | nd | 4 | 4(5) | 4 |
| LhABCH1 | comp36633_c0_seq2 | 795 | full | PM | 7 | 6(8) | 6 |
| LhABCH2 | comp37335_c0_seq4 | 748 | full | PM | 5 | 6(7) | 6 |
| LhABCH3 | comp34118_c0_seq2 | 683 | full | PM | 5 | 6(7) | 6 |
| LhABCH4 | comp32271_c0_seq13 | 768 | full | PM | 8 | 8(9) | 7 |
| LhABCH5 | comp31632_c0_seq1 | 680 | full | PM | 6 | 6(8) | 6 |
| LhABCH6 | comp6701_c0_seq1 | 673 | full | PM | 5 | 6(7) | 6 |
| LhABCH7 | comp35902_c0_seq6 | 685 | full | PM | 5 | 6(7) | 6 |
| LhABCH8 | comp37338_c0_seq18 | 144 | partial | nd | 3 | 3 | 4 |
| LhABCH9 | comp22796_c0_seq1 | 526 | partial | nd | 0 | 2(3) | 2 |
| LhABCH10 | comp31498_c1_seq1 | 693 | partial | nd | 7 | 7(9) | 6 |
| LhABCH11 | comp27033_c0_seq3 | 682 | full | PM | 6 | 6(7) | 6 |
ABCA subfamily
ABCA transporters are among the largest known ABCs and typically exhibit an extended extracellular domain between the TMDs, a dipolar diacidic motif downstream of the NBD region, and a conserved amino terminal sequence (XLXXKN) involved in post-Golgi trafficking [58], [59]. We found 11 putative ABCA transporter sequences (Table S1 and S2) exhibiting 3–50% amino acid identity (Table S5). LhABCA4 and LhABCA5 encompass full-length coding sequences with extended extracellular domains (653 aa in LhABCA4, 189 aa in LhABCA5) and the dipolar diacidic motif. The amino terminal post-Golgi trafficking motif is only found in LhABCA4. This motif is present in 11 of the 12 HsABCAs but only 6 of 38 ABCAs from T. urticae, D. melanogaster, B. mori, and T. castaneum, suggesting that arthropods may use an alternative post-Golgi targeting mechanism.
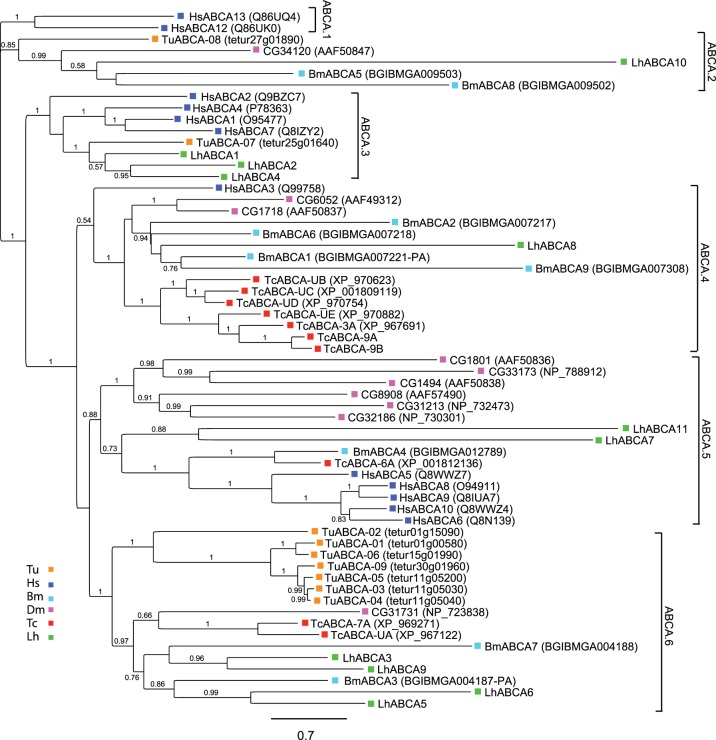
Alignment and phylogenetic analyses of putative LhABCAs with ABCAs from T. urticae, D. melanogaster, B. mori, T. castaneum and humans are consistent with those reported by other groups [8], [33], [37], [38] and indicate conserved subfamily clustering within six central clades that we have designated ABCA.1-ABCA.6 (Figure 1). LhABCA1, LhABCA2, and LhABCA4 aligned within clade ABCA.3, which is comprised of multiple HsABCAs that function in the transport of membrane lipids [60], [61]. LhABCA5, LhABCA6, LhABCA3, and LhABCA9 aligned with two BmABCAs to form one branch in clade ABCA.6. The first two LhABCAs are most similar to BmABCA3, while the other two share similarity with BmABCA7. Consistent with previous phylogenetic analyses [33], a second branch of clade ABCA.6 is comprised solely of T. urticae ABCAs. LhABCA7 and LhABCA11 form a separate branch in clade ABCA.5, which is dominated by D. melanogaster and human ABCAs. While no function has been assigned to the arthropod ABCAs in this clade, the HsABCAs function in lipid homeostasis [60], [61]. LhABCA10 aligned with sequences in clade ABCA.2, whereas LhABCA8 clustered with a group of BmABCAs to form a branch in clade ABCA.4. In addition to the LhABCA8 branch, the ABCA.4 clade is also characterized by gene expansion in T. castaneum. HsABCA3, a transporter involved in pulmonary surfactant secretion [60], [61], also sorted to ABCA.4. No arthropod sequences sorted to clade ABCA.1, which is comprised of HsABCA12, a keratinocyte lipid transporter [62], and HsABCA13, the biological function of which is not known.
Figure 1. Phylogenetic analysis of ABCA transporters from L. hesperus and five metazoan species.
Putative L. hesperus ABCA sequences and full-length ABCA proteins from five additional species were aligned using MUSCLE [159] and analyzed using the FastTree2 approximate likelihood method [160]. Numbers at the branch point of each node represent support values. Species abbreviations and color coding are: Bm, Bombyx mori (teal); Dm, Drosophila melanogaster (pink); Hs, Homo sapiens (blue); Lh, Lygus hesperus (green); Tc, Tribolium castaneum (red); and Tu, Tetranychus urticae (orange). Accession numbers are indicated in parentheses. The scale bar represents 0.7 amino acid substitutions per site. A full listing of the accession numbers for the five metazoan sequences is available in Table S11. LhABC transporter sequences are available in Table S12.
ABCB subfamily
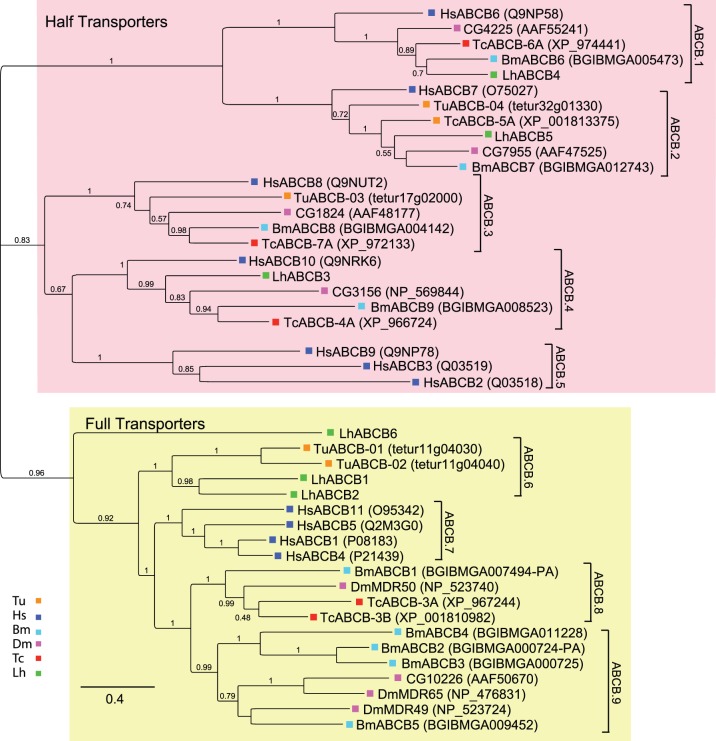
ABCBs are structurally organized as either HTs characterized by two domains (1 TMD, 1 NBD) or FTs that contain four domains (2 TMDs, 2 NBDs). Mammalian ABCBs transport diverse hydrophobic substrates including bile acids, peptides, steroids, drugs, and other xenobiotics. This broad substrate range of ABCBs likely contributes to their involvement in multidrug resistance phenotypes [63]–[65] and insecticide resistance [8], [25]. Based on BLAST analyses, we identified six LhABCB transcripts, which is similar to the number reported for other arthropods (Table 1). The LhABCB1 and LhABCB4 transcripts include full-length ORFs, whereas the other LhABCB transcripts encode protein fragments ranging in size from 469 to 830 amino acids (Table 2) that encompass predicted ABC transporter-like domains (Table S3). As a group, the LhABCBs, exhibit 12–51% sequence identity (Table S6). Consistent with the phylogenetic analyses of Dermauw et al.[8], the HTs aligned into five clades (ABCB.1-ABCB.5), whereas the FTs clustered into four clades (ABCB.6-ABCB.9) largely composed of lineage specific branches (Figure 2). The branching pattern in four of the HT clades (ABCB.1-ABCB.4) is suggestive of orthologous relationships. LhABCB4, LhABCB5, and LhABCB3 aligned with proteins in clades ABCB.1, ABCB.2, and ABCB.4 respectively. The expression of HsABCB6 (clade ABCB.1) has been correlated with increased drug resistance [65], [66] and arthropod transporters in clades ABCB.1 and ABCB.2 are reported to play roles in heavy metal detoxification [67], insecticide resistance [68], cold stress tolerance [69], and pupal-adult development in T. castaneum [16]. Despite orthologous sequences in the other arthropods examined, no LhABCB sequences aligned with proteins comprising clade ABCB.3. The three human transporters (HsABCB2, HsABCB3, HsABCB9) that function in antigen processing [70], [71] form a human specific clade (ABCB.5).
Figure 2. Phylogenetic analysis of ABCB transporters from L. hesperus and five metazoan species.
Clades corresponding to full-transporters and half-transporters are shaded light yellow and light red, respectively. The scale bar represents 0.4 amino acid substitutions per site. Analyses, abbreviations, and color-coding are as in Figure 1.
In contrast to the HTs, the FT sequences separated into clades with species-specific branches. Two LhABCBs (LhABCB1 and LhABCB2) and two T. urticae ABCBs form separate branches of the ABCB.6 clade, which is consistent with diversification arising from lineage specific gene duplication events [32]. Similar to the ABCB.5 clade, ABCB.7 is human specific with no arthropod sequences aligning with the four HsABCBs. The other two FT clades (ABCB.8 and ABCB.9) comprise sequences similar to D. melanogaster multiple drug resistance (MDR) proteins (i.e., DmMDR49, DmMDR50, and DmMDR65) that have been identified in abiotic stress and insecticide resistance [45], [53], [72]–[78]. Surprisingly, no L. hesperus sequences nor any T. urticae sequences clustered within either of these two clades. LhABCB6, which is a partial sequence, aligned with the FTs but did not sort with any of the clades.
ABCC subfamily
The ABCC subfamily consists of three functionally distinct classes of transporters: broad-specificity multidrug resistance-associated proteins (MRPs), sulfonylurea receptors (SUR), and the cystic fibrosis transmembrane conductance regulator (CFTR) [34], [79]–[81]. MRPs interact with a diverse array of substrates including a number of endogenous metabolites, xenobiotics, and various conjugated (glutathione, sulfate, and glucuronate) anions. Nine members of the mammalian ABCC subfamily are classified as MRPs: ABCC1–6 and ABCC10–12. In contrast to the xenobiotic transport activities of MRP-ABCCs, SURs (i.e. ABCC8 and ABCC9) function as regulators of specific potassium channels, and CFTR (i.e. ABCC7) functions as an ATP-gated chloride ion channel [34].
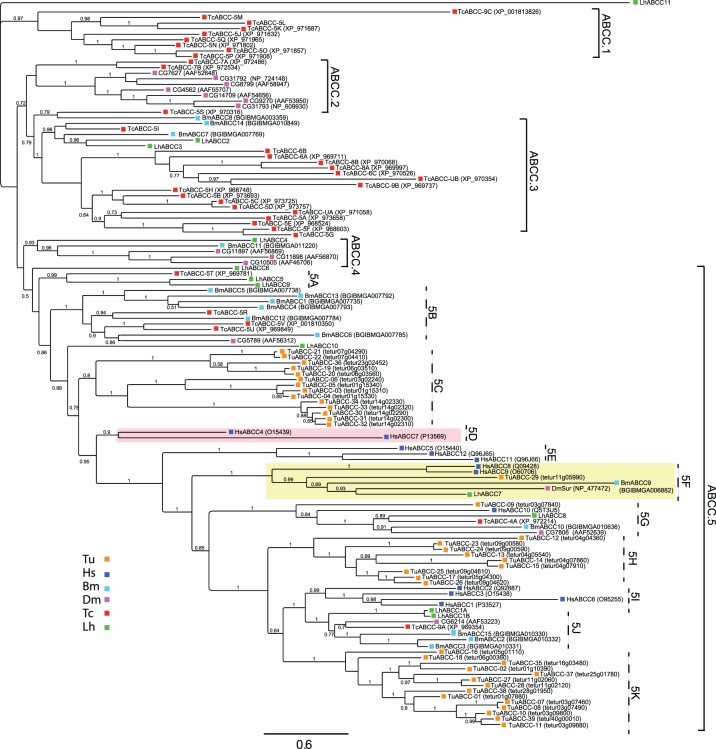
Based on sequence similarities, we identified 12 ABCC-like transcripts in L. hesperus, a number comparable to that reported for other arthropods with the exception of T. urticae, T. castaneum, and C. populi, all of which have undergone significant expansion of the ABCC subfamily (Table 1). Sequence identity among the LhABCC transcripts ranges from 2–96% (Table S7). Half of the transcripts comprise complete coding sequences, and the rest encode protein fragments (256 to 1407 amino acids) containing ABC transporter-like domains (Table S3). Our phylogenetic analysis generated five major clades (designated ABCC.1-ABCC.5) with 11 nested clades (designated ABCC.5A-5 K) branching from ABCC.5 (Figure 3). Consistent with other studies supporting gene duplication-based expansion of the ABCC subfamily [33], [82], our analysis clustered many of the T. castaneum and T. urticae sequences into lineage specific clades/branches (T. castaneum – clade ABCC.1 and two nested branches in clade ABCC.3; T. urticae – clades ABCC.5C, ABCC.5 H, and ABCC.5 K). As before, clear relationships in sequence alignment were seen for a number of the LhABCCs.
Figure 3. Phylogenetic analysis of ABCC transporters from L. hesperus and five metazoan species.
The scale bar represents 0.6 amino acid substitutions per site. Clades corresponding to SUR-like sequences and CFTR are shaded light yellow and light red, respectively. Analyses, abbreviations, and color-coding are as in Figure 1.
LhABCC1A and LhABCC1B sorted to nested clade ABCC.5J. The two transcripts encode full-length sequences that are 96% identical (Table S7) with sequence variation (86% pairwise sequence identity) primarily in the first 260 amino acids (Figure S2). Sanger sequencing of multiple clones confirmed the respective sequences. The sequence variation could reflect the heterogeneity of our L. hesperus colony. Alternatively, they could represent splice variants similar to that reported for D. melanogaster ABCC CG6214 [83] and CG6214 orthologs in A. gambiae [36] and Trichoplusia ni [84]. CG6214 is most similar with the two LhABCCs in ABCC.5J and is upregulated in response to xenobiotic feeding [75], [85]. The HsABCC specific clade (ABCC.5I) that branches from the same node as ABCC.5J is comprised of classic MRPs with broad substrate specificities and diverse resistance phenotypes [80], [86].
LhABCC8 aligned to clade ABCC.5G with HsABCC10 and four other arthropod ABCCs (Figure 3). HsABCC10 transports a wide range of substrates and has been linked with multiple multidrug resistance phenotypes [87]. While the biological function of the arthropod ABCCs in this grouping is unknown, ecdysone treatment has been shown to elevate expression of the D. melanogaster CG7806 transporter [88]. LhABCC4 sorts to clade ABCC.4 with BmABCC11 and three D. melanogaster ABCCs (CG11897, CG11898, and CG10505). The expression of CG11897 is strongly induced following immune challenge with an entomopathogenic bacterium [89], whereas that of CG10505 has been linked to heavy metal homeostasis [90] and alcohol exposure [91].
LhABCC2 and LhABCC3 aligned with two BmABCCs and numerous T. castaneum transporters in clade ABCC.3 (Figure 3), which is a sister clade to ABCC.2. The ABCC.2 clade is characterized by a cluster of six D. melanogaster ABCCs, two of which (CG14709 and CG8799) are expressed and/or upregulated in response to cellular stress [92]–[94]. A third ABCC (CG4562) in that clade was recently shown to be upregulated following knockdown of a detoxifying cytochrome P450 [95], suggesting potential compensatory cross talk occurs between the two detoxification mechanisms.
ABCC2 MRP-like transporters have been linked with resistance to Bacillus thuringiensis (Bt) Cry1 toxins in a number of lepidopterans [26]–[31]. This resistance does not appear to be linked to ATP-dependent transport but rather to the cell surface transporter functioning as a putative Bt Cry1A toxin receptor [96]. In our phylogenetic analysis, BmABCC13 (BGIBMGA007792) and BmABCC4 (BGIBMGA007793), which represent a single gene [28], correspond to the ABCC2 transporter linked to Bt resistance. Both B. mori sequences align within a lineage-specific branch of clade ABCC.5B, which also included several T. castaneum ABCCs and a D. melanogaster sequence. No LhABCC sequence aligned within that clade. Although a transgenic cotton plant expressing a hemipteran-active Bt toxin has been developed [97], it remains to be determined if LhABCCs will also function as Bt toxin receptors.
Well-supported alignment of LhABCC7 with the SUR ABCC transporters in clade ABCC.5F suggests possible conservation of function. These transporters assemble with other proteins to form ATP-sensitive potassium channels that function in a number of physiological processes [34], [98]. In insects, SUR is important for glucose homeostasis [99], protection against hypoxic stress [100], and has been proposed as the putative binding site for benzoylphenylureas, a class of insecticides that inhibit chitin synthesis [16], [101]. However, recent findings suggest that in some species SUR is dispensable for benzoylphenylurea action [8], [16], [102], [103].
The third functional class of ABCC transporters is CFTR, an ABC transporter that has undergone functional divergence to exhibit ATP-gated chloride channel activity [34]. In our phylogenetic analyses, HsABCC7 (i.e., CFTR) sorted to clade ABCC.5D with HsABCC4, the closest mammalian homolog of HsABCC7 [104]. While structurally similar, HsABCC4 is functionally differentiated by a broad substrate range that includes diverse xenobiotics [80]. Consistent with other studies, we did not find any L. hesperus sequences orthologous with the two HsABCCs. The top BLAST hits for LhABCC11, however, are with mammalian MRP4/ABCC4 sequences (Table S1 and S2). The LhABCC11 transcript encodes an 856 aa protein (Table1) containing characteristic ABC transporter domains (Table S3), but it did not align within any of the ABCC clades. A C. populi ABCC transporter, CpMRP, that functions in plant-derived phenolglucoside sequestration also shares similarity with MRP4 proteins [23]. Low sequence identity (∼15%) between LhABCC11 and CpMRP, however, suggests that the two transporters may interact with different substrates.
ABCD, ABCE, and ABCF subfamilies
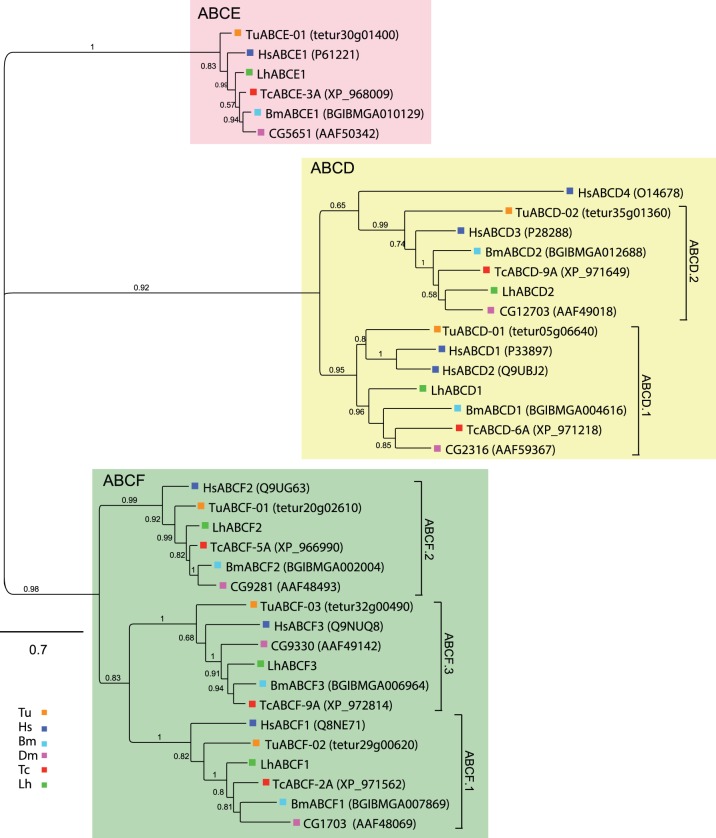
ABCD transporters function in peroxisomal import of long chain fatty acids and/or fatty acyl CoAs [105], [106]. We found that, like most other arthropods, L. hesperus has two ABCD transcripts (Table 1). LhABCD1 and LhABCD2 share 36% sequence identity (Table S8), which is comparable to the homology (27–35%) shared between the ABCDs in our phylogenetic analyses. Both LhABCDs have transporter domains (Table S3), as well as the two motifs, EEA-like (EEIAFYGG) and loop1 (LXXRT), that are considered essential for ABCD function [107]. LhABCD1 and LhABCD2 aligned with clades ABCD.1 and ABCD.2, respectively (Figure 4). Although little is known about arthropod ABCDs, the D. melanogaster ABCD transporter CG2316 (clade ABCD.1) is overexpressed in a cell line resistant to the insect growth regulator methoxyfenozide [108].
Figure 4. Phylogenetic analysis of ABCD, ABCE, and ABCF transporters from L. hesperus and five metazoan species.
Clades corresponding to the three subfamilies are shaded light red (ABCE), light yellow (ABCD), and green (ABCF). The scale bar represents 0.7 amino acid substitutions per site. Analyses, abbreviations, and color-coding are as in Figure 1.
ABCE and ABCF proteins contain the characteristic NBD but lack TMDs and do not transport substrates. Instead, ABCEs function in ribosome recycling and regulation of protein translation [13], [109], whereas ABCFs are involved in translation [10], [12]. Like other arthropods [8], L. hesperus has a single LhABCE and three LhABCF transcripts (Table 2). LhABCE shares ∼80% sequence identity with the other ABCEs examined in our analyses, supporting a possible evolutionarily conserved role. The three LhABCFs, which share 32–35% sequence identity (Table S8), sorted into distinct but well-supported clades (Figure 4). Sequence identity within each clade ranged from 45–84%, with highest identities in the LhABCF2 clade.
ABCG subfamily
ABCG transporters have a HT motif in which the lone NBD is localized on the amino terminal side of the TMD, in contrast to localization on the carboxyl terminal side as in other ABC transporters. In comparison with the ABCG subfamily in other metazoans, which frequently have 5–10 members [110], the arthropod lineage has undergone extensive gene expansion (Table 1). Consistent with this, we identified 19 LhABCG transcripts, including 12 full-length coding sequences (Table 2). The LhABCGs share low (10%) to moderate (58%) sequence identity (Table S9), and all but LhABCG19 have ABC transporter domains (Table S3). Despite the lack of known ABC domains, LhABCG19 shares sequence similarity (BLASTx E value<10−10) with other putative ABCG transporters (Table S1 and
S2).
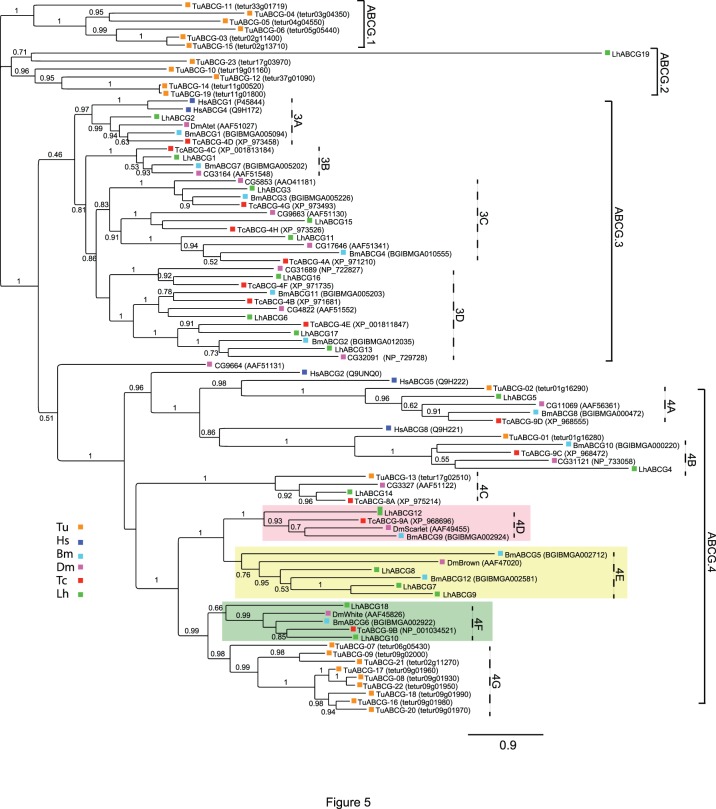
Our phylogenetic analysis resulted in four major clades (ABCG.1-ABCG.4) with a number of minor clades branching from ABCG.3 and ABCG.4 (Figure 5). As previously reported [33], T. urticae has multiple lineage-specific clades (ABCG.1 and ABCG.4G) indicating expansion by multiple gene duplication events [32]. Aside from these sequences, the clustering pattern of the other ABCGs within the respective clades suggests clear orthologous relationships (Figure 5). LhABCG2 aligned to clade ABCG.3A along with D. melanogaster ABCG CG2969 (i.e., DmAtet), reportedly a target of the transcriptional regulator gene, clock [111], and two HsABCGs involved in sterol homeostasis [112], [113]. LhABCG1 sorted to clade ABCG.3B with arthropod ABCGs that have been implicated in cuticular lipid transport [16]. LhABCG3, LhABCG11, and LhABCG15 sorted to separate branches of clade ABCG.3C with LhABCG15 aligning to the same branch as the D. melanogaster ABCG CG9663 and LhABCG11 aligning with D. melanogaster ABCG CG17646. CG9663 is a putative target of clock [111] that has also been linked to decreased susceptibility to oxidative stress [114]. CG17646 functions in triglyceride storage [115] and ethanol sensitivity [116]. LhABCG6, LhABCG13, LhABCG16, and LhABCG17 aligned to separate branches of clade ABCG.3D with arthropod ABCGs of unknown function. LhABCG4 and LhABCG5 aligned with potential orthologs of HsABCG8 and HsABCG5 in clades ABCG.4B and ABCG.4A respectively. The obligate heterodimerization of the two HsABCGs in sterol homeostasis [112], [113] suggests similar dimerization of LhABCG5 and LhABCG4 could be important for their functionality. LhABCG14 aligned to clade ABCG.4C with arthropod ABCGs potentially involved in ecdysteroid signaling [16], [22], [117]–[119].
Figure 5. Phylogenetic analysis of ABCG transporters from L. hesperus and five metazoan species.
Clades corresponding to transporters involved in eye pigmentation have been shaded light red (scarlet), light yellow (brown), and green (white). The scale bar represents 0.9 amino acid substitutions per site. Analyses, abbreviations, and color-coding are as in Figure 1.
Six LhABCGs sorted to three clades (ABCG.4D, ABCG.4E, and ABCG.4F) characterized by D. melanogaster ABC transporter genes (white, brown, and scarlet) that function in the import of eye pigment precursors [17], [18]. Heterodimers of the white and brown gene products transport red-pigmented pteridine precursors, whereas heterodimers of the white and scarlet gene products are crucial for the import of brown-pigmented monochrome precursors. Consequently, white mutants are characterized by white eyes (complete loss of pigmentation), brown mutants by dark brown eyes (loss of red pigments), and scarlet mutants by bright red eyes (loss of brown pigments). Similar roles in pigment transport have been described for homologs of the three genes in B. mori [19], [20], [120], [121]. These ABCGs also function in biogenic amine transport [24], uric acid uptake [20], [121], and courtship behavior in D. melanogaster [122]. In our analyses, LhABCG12 clustered within the scarlet clade (ABCG.4D), LhABCG10 and LhABCG18 within the white clade (ABCG.4F), and three LhABCGs (LhABCG7, LhABCG8, and LhABCG9) that share 40–58% sequence identity (Table S9) aligned to the brown clade (ABCG.4E). While red eye mutants of various plant bugs, including a species sympatric to L. hesperus (Lygus lineolaris), have been reported [123]–[126], the functional importance of scarlet in these phenotypes is unknown. The LhABCG19 partial sequence aligned with a group from T. urticae in clade ABCG.2.
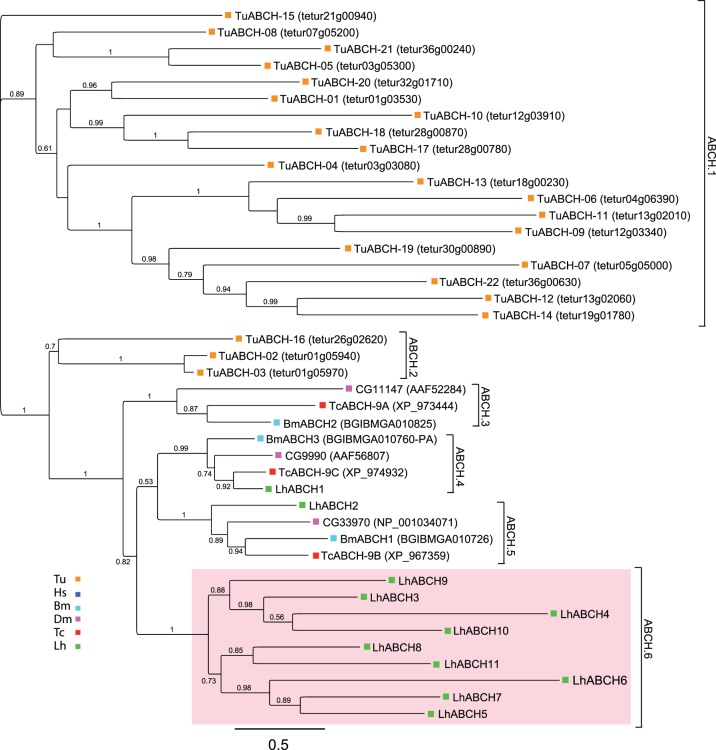
ABCH subfamily
Similar to ABCGs, ABCH transporters also have the inverted NBD-TMD configuration. ABCHs though, with the exception of zebrafish [15], [127], are specific to arthropods [8], [34]. While most arthropods have 3 ABCH transporters (Table 1), lineage-specific gene duplications have resulted in 22 and 14 ABCH genes in T. urticae and D. pulex respectively [32], [33]. We found similar expansion in L. hesperus with 11 LhABCH transcripts, 8 of which are full-length coding sequences. Although BLASTx analyses indicate that these transcripts share sequence similarity with ABCGs (Table S1 and S2), phylogenetic analyses clustered these transcripts in the ABCH clade (Figure S3). As reported previously [33], T. urticae sequences are unique and do not align well with other ABCHs (Figure 6). LhABCH1 aligned with transporters in clade ABCH.4 that likely function in cuticular lipid transport [16], [128], [129]. LhABCH2 sorted to the sister ABCH.5 clade along with the D. melanogaster ABCH CG33970, which is upregulated in response to cold hardening [130]. The remaining LhABCH sequences formed a separate clade, implying that, like T. urticae and D. pulex, independent lineage-specific gene duplication events have contributed to the expansion of the LhABCH subfamily. While the physiological functions of the ABCH subfamily remain largely uncharacterized, differential expression of ABCHs has been reported for T. urticae females in diapause [119] and some ABCH transcript levels are elevated in insecticide resistant strains of T. urticae [33] and P. xylostella [131].
Figure 6. Phylogenetic analysis of ABCH transporters from L. hesperus and four metazoan species.
The scale bar represents 0.5 amino acid substitutions per site. Analyses, abbreviations, and color-coding are as in Figure 1. Because the ABCH subfamily is largely restricted to the arthropod lineage, no representative sequences for H. sapiens are available and thus the analyses were performed using only arthropod sequences. L. hesperus ABCH sequences that have undergone gene expansion are shaded light red.
Expression profile of LhABC transcripts
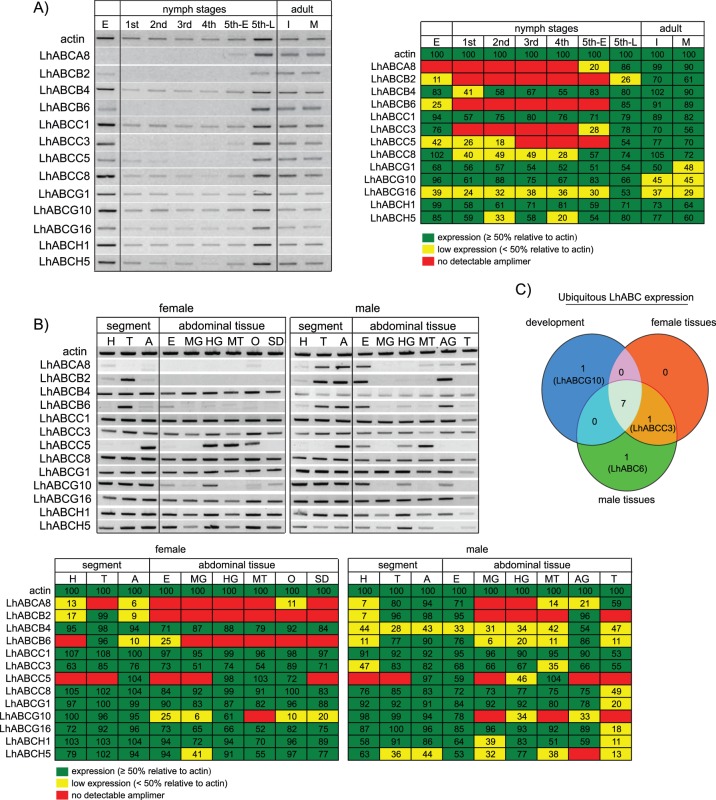
Many ABC transporters function in development [16], [21]. Consequently, we used end-point PCR to examine the developmental expression profile of a subset of 13 LhABCs representative of the ABCA, ABCB, ABCC, ABCG, and ABCH subfamilies. All of the transcripts were amplified from both reproductively immature and mature adults and all but LhABCA8 were expressed to some degree in eggs (Figure 7A). Five LhABCs had limited nymphal expression; LhABCA8 and LhABCC3 were detected in early and late stadium fifth instars, LhABCB2 and LhABCB6 in late stadium fifth instars, and LhABCC5 in first, second (albeit weakly) and late stadium fifth instars (Figure 7A). Orthologs of the eight LhABCs expressed throughout L. hesperus nymphal development have been reported to have similar expression profiles [16], suggesting potential roles in basic physiological functions.
Figure 7. Transcriptional expression profile of 13 LhABC transcripts.
A) Developmental profile. Expression profile of 13 LhABC transporters in eggs through 12-day-old adults was examined by end-point PCR using primers designed to amplify a ∼500 bp fragment of each transcript. Abbreviations: E, eggs; 1st, first instars; 2nd, second instars; 3rd, third instars; 4th, fourth instars; 5th-E, early stadium fifth instars; 5th-L, late stadium fifth instars; I, reproductively immature 1-day-old mixed sex adults; M, mature 12-day-old mixed sex adults. Products were analyzed on 1.5% agarose gels and stained with SYBR Safe. Actin was used as a positive control. Leftmost image - Representative gel image. For clarity, the negative image is shown. Rightmost image – Visual aid depicting semi-quantitative analysis of amplimers of interest compared to actin in the representative gel. Relative to the actin amplimer of each developmental stage, cells with amplimer intensity ≥50% are indicated in green, while cells <50% are indicated in yellow. Red cells indicate no detectable amplimer. Numbers inside individual cells denote the percentage of amplimer intensity compared to actin. The primer set used for LhABCC1 profiling amplifies a shared region of LhABCC1A and LhABCC1B. B) Adult tissue profile. The expression profile of the 13 LhABC transcripts was examined as above in adult body segments and various abdominal tissues prepared from 7-day-old adults. Abbreviations: H, head; T, thorax; A, abdomen; E, epidermis, MG, midgut; HG, hindgut; MT, Malpighian tubules; O, ovary; SD, seminal depository; AG, accessory glands (lateral and medial); T, testis. Top image - Representative gel image. For clarity, the negative image is shown. Lower image – Visual aid depicting semi-quantitative analysis of amplimers of interest compared to actin in the representative gel. Color shading is as above but relative to the intensity of actin amplimers within each respective tissue. C) Three-way Venn diagram comparing the transcriptional expression profile of LhABC transcripts ubiquitously expressed throughout L. hesperus development (blue) with those expressed in female (red) and male (green) tissues.
We next examined the transcription profiles of the 13 LhABC transporters in specific body segments (head, thorax, abdomen) and abdominal tissues (epidermis/cuticle, midgut, hindgut, Malpighian tubule, ovary, seminal depository, male medial and lateral accessory glands, and testis) from 7-day-old virgins of each sex (Figure 7B). Eight of the transcripts were amplified from all tissues/segments in both sexes with seven of them also constitutively transcribed throughout development (Figure 7C). In contrast, LhABCC3, which is present in all adult tissues/segments, was only amplified from eggs, fifth instar nymphs and adults (Figure 7A), suggesting a potential role in reproductive development. The inverse was observed with LhABCG10, a white-like gene that is transcribed throughout development but which exhibits tissue specific transcription in adults (Figure 7B). The low abundance of LhABCG10 transcripts in Malpighian tubules differs from that reported for white genes in D. melanogaster and B. mori [18], [120], [132], which play key roles in the uptake and concentration of excess tryptophan [133]. The undetectable transcript levels of LhABCG10 in this tissue suggests tryptophan transport in L. hesperus is mediated by some other mechanism or involves a different transporter such as LhABCG18, which also sorts with white-like genes in clade ABCG.4F (Figure 5).
Malpighian tubules are the main excretory and osmoregulatory organs in insects and are thus crucial in clearing toxic endogenous compounds and xenobiotics [134], [135]. In support of this role, ABC transporters and detoxification enzyme levels are frequently present at relatively high levels in Malpighian tubules [75], [85], [136]–[138]. LhABCC5 expression was specific to the abdomen of both sexes where it predominantly localized to the hindgut and Malpighian tubules (Figure 7B), suggesting a potential role in xenobiotic excretion.
The expression of LhABCA8, LhABCB2, and LhABCB6 was sex-biased with higher levels of the three transcripts in male abdomen compared to female abdomen (Figure 7B). Among the male abdominal tissues, the three transporters were enriched in reproductive tissues (LhABCA8– testis; LhABCB2 and LhABCB6– accessory gland). The developmental profile of the three transcripts is likewise similar with expression limited to eggs and fifth instars (Figure 7A). This latter period coincides with the development of male reproductive organs (Figure S4), suggesting an association with sexual maturation. A number of D. melanogaster ABC transporters are highly expressed in male reproductive tissues [84], [132], [139] and elevated testicular expression of ABC transporters has been reported for B. mori [37], [38], [77]. The elevated levels of ABC transporters in reproductive tissues may be critical for protection of spermatozoa [140]. Additionally, the ABC transporters might function in the loading of accessory glands and other male secretory reproductive tissues with seminal fluid components (e.g., prostaglandins, lipids, peptides, hormones, etc.) [141], [142].
The presence of LhABC transcripts in the abdominal epidermis of both males and females (Figure 7B) could indicate potential functions in integument coloration [19], [20], [143] or in the transport of cuticular lipids to prevent water loss [16]. Alternatively, the transporters may be expressed in oenocytes, polyploid insect cells found in close association with the epidermis that have been reported to function in xenobiotic detoxification, the synthesis of cuticle components, and innate immunity [144]. Elevated levels of ABC transporter transcripts in epidermis have been reported for some insecticide-resistant bed bug populations [145], suggesting a potential role in xenobiotic transport at the cuticular layer.
Conclusions
The genus Lygus encompasses more than 30 different species of polyphagous pests that attack crops worldwide. However, reports of insecticide resistance in field populations threaten the sustainability of insecticide-based management strategies. Consequently, there is growing interest in elucidating the molecular basis of resistance. While a number of studies have focused on identifying detoxification enzymes, the role of ABC transporters in insecticide clearance in hemipteran pests has been largely neglected. To address this, we used RNA-Seq to identify the ABC transporter superfamily in L. hesperus. Defining the functional relevance and substrate specificity of the 65 LhABC-like transcripts will be a future research priority. Initial efforts will focus on assessing the effects of insecticide exposure on expression levels of the LhABCs, in particular LhABCC5, the tissue localization of which is consistent with a role in insecticide/xenobiotic clearance. Furthermore, targeting ABC transporters by RNAi may facilitate the development of novel control strategies for L. hesperus and other hemipteran pests.
Materials and Methods
Insects
L. hesperus were obtained from an established laboratory colony. Stock insects were maintained at 27.5–29.0°C under 40% humidity with a L14:D10 photoperiod, and fed artificial diet packaged in Parafilm M [146]. Experimental nymphs and adults were generated from eggs deposited in oviposition packets (agarose gel packaged within Parafilm M) and maintained as described previously [147].
RNA isolation and Illumina sequencing
To induce expression of potential stress-related genes, 10-day-old L. hesperus adults from a single cohort were placed individually in covered glass Petri dishes (60×15 mm) along with a section of green bean. Dishes were transferred to environmental chambers and exposed to one of three temperatures (4°C, 25°C, or 39°C) for 4 hr. Insects were stored in RNALater (Ambion, Life Technologies, Carlsbad, CA) at −80°C. Total RNA from frozen samples was isolated by the University of Arizona Genomics Center (http://uagc.arl.arizona.edu; Tucson, AZ) using an RNeasy Mini Kit (Qiagen, Valencia, CA) followed by on-column DNAse digestion according to the manufacturer’s instructions. RNA samples were eluted in 30 µL RNAse-free H2O. RNA quality was assessed on a Fragment Analyzer Automated CE System (Advanced Analytical Technologies, Ames, IA) and RNA was quantified using RiboGreen (Molecular Probes, Eugene, OR). Triplicate RNA libraries for each of the three temperature regimens were constructed using a TruSeq RNA Sample Preparation Kit v2 (Illumina Inc., San Diego, USA) and sequenced on an Illumina HiSeq2500 in rapid run mode. CASAVA version 2.8 was used for base calling and de-multiplexing.
Transcriptome assembly
Raw de-multiplexed reads for each sample were assembled into single files and then trimmed and quality filtered with Trimmomatic version 0.30 [148] using the parameters LEADING:20, TRAILING:20, WINSIZE:5, WINCUTOFF:25, MINLEN:50, and ILLUMINACLIP:TruSeq3-PE.fa:2∶30∶10. Quality metrics were calculated for the unfiltered and filtered data using FASTQC version 0.10.1. After quality filtering, orphaned pairs were discarded while reads still having a read pair were used for assembly. Reads were normalized in silico using the “normalize_by_kmer_coverage.pl” script distributed with the Trinity transcriptome assembly pipeline (r2013_08_14) [149] and a kmer size of 25 and maximum read coverage of 40. Normalized reads were used to create a de novo transcriptome assembly with Trinity (Inchworm, Chrysalis, and Butterfly) using default parameters except with the jaccard_clip option used to compare paired-read consistency to reduce the creation of fused transcripts from non-strand specific data. The initial Trinity assembly was further filtered to maintain only transcripts exhibiting evidence of a coding region. Open reading frames (ORFs) were predicted using Transdecoder (r2012–08–15) with training against the 500 longest ORFs in the transcriptome. ORF transcripts were also identified based on significant matches to the Pfam-A database using a HMMER search [150]. Transcripts were only retained if they had an ORF predicted by Transdecoder with a length longer than 100 amino acids. The filtered transcriptome was annotated using InterProScan 5, and gene names assigned via BLASTp alignment to the UniProtKB/SwissProt database. The raw data was deposited in the NCBI sequence read archive under BioProject PRJNA238835, BioSamples SAMN02679940 - SAMN02679948, SRA Submission ID “PBARC: Lygus hesperus Heat Experiment”, SRA Study Accession SRP039607. To facilitate submission to the NCBI TSA database, transcript sequences were modified to put all coding sequence on the positive strand by reverse complementing when appropriate and the longest coding sequence for each transcript was submitted to TSA using an open source transcriptome preparation software package (http://genomeannotation.github.io/transvestigator/). The annotated assembly with putative gene name and functional annotations was submitted to NCBI under TSA submission GBHO00000000. The version described in this paper is the first version, GBHO01000000.
Annotation and bioinformatic analysis of the L. hesperus ABC transporter superfamily
ABC transporter annotations were performed on the longest isoform for each unigene. Putative ABC transporter sequences were identified by initially performing a BLASTn search (E value ≤10−10) of the assembly described above with queries consisting of sequences annotated previously as ABC transporters [52]. A BLASTx search (E value ≤10−10) of the RNA-Seq data was also performed using the full complement of ABC transporters from H. sapiens [34], C. elegans [151], D. pulex [32], T. urticae [33], D. melanogaster [34], T. castaneum [16], and B. mori [37], [38]. The longest isoform of the resulting L. hesperus sequence hits were then re-evaluated against the NCBI nr database using BLASTx and tBLASTn (E value ≤10−10). The sequence list of positive hits was curated to remove duplicates.
Identification of putative ABC transporter domains was performed using ScanProsite [152], [153] and the HMMscan module on the HMMER webserver [154]. Subcellular localization prediction of LhABC transporters was performed with WoLF PSORT [155]. Transmembrane domain predictions were performed using TMHMM v2.0 [156], TOPCONS [157], and TopPred II [158].
Putative LhABC transporters were initially assigned to the respective subfamilies (A-H) based on BLAST analyses, then assignments were refined based on phylogenetic inferences. Multiple sequence alignments consisting of the putative L. hesperus sequences and ABCs from five metazoans (H. sapiens, T. urticae, D. melanogaster, T. castaneum, and B. mori) were performed using default settings for MUSCLE [159]. Phylogenetic trees were constructed utilizing FastTree2 [160] implemented in Geneious 7.0.6 using default settings augmented by the Whelan-Goldman model and optimization for Gamma20 likelihood. Parallel analyses were also performed in MEGA5 [161] with bootstrap testing of 500 replicates using both the maximum parsimony method and the maximum likelihood method based on the JTT matrix-based model [162]. Phylogenetic analyses were performed using available sequences (both complete coding sequences and partial fragments) rather than specific domains as reported for other species [16], [33]. The clustering of subfamilies and orthologous genes was compared amongst the three phylogenetic methods and with previous analyses of ABC transporters [8], [16], [33], [37], [38]. Heat identity maps for the LhABC transporters were generated using Geneious 7.1.7 (Biomatters Ltd., Auckland, New Zealand) and MUSCLE-based sequence alignments. See Table S11 for the accession numbers of the proteins used in the phylogenetic analyses, and Table S12 for LhABC transporter amino acid sequences. T. castaneum sequences lacking accession numbers were downloaded as FASTA protein files directly from BeetleBase (http://beetlebase.org/) based on the reported GLEAN accessions [16].
End point PCR expression analyses
The expression profile of a subset of LhABC transporters was examined across three biological replicates throughout L. hesperus development and among sex-specific adult body segments/tissues. TRI Reagent Solution (Ambion) was used to isolate total RNA from pooled samples of eggs, first - fourth instars, early stadium fifth instars, late stadium fifth instars, reproductively immature adults (1-day-old) of each sex, and mature virgin adults (12-day-old) of each sex. Total RNA was also isolated from pooled 7-day-old adult virgin male and female body segments and abdominal tissues: 10×head, 3×thorax, 5×abdomen, 15×abdominal carcass, 5×midgut, 20×hindgut, 20×Malpighian tubules, 20×seminal depository, and 5 pairs each of ovaries, male medial and lateral accessory glands, and testes. First-strand cDNAs were generated using a Superscript III first-strand cDNA synthesis kit (Invitrogen) with custom-made random pentadecamers (IDT, San Diego, CA) and 500 ng of DNase I-treated total RNAs. PCR expression profiling was performed using 0.4 µL of the prepared cDNAs with Sapphire Amp Fast PCR Master Mix (Takara Bio Inc./Clontech, Madison, WI) and sequence-specific primers (Table 3) designed to amplify ∼500 bp fragments of the LhABC transcripts. Thermocycler conditions consisted of 95°C for 2 min followed by 35 cycles at 94°C for 20 s, 56°C for 20 s, and 72°C for 20 s, and finished with a 1 min incubation at 72°C. PCR products were analyzed by gel electrophoresis on 1.5% agarose gels stained with SYBR Safe (Life Tech.) and a Tris/acetate/EDTA buffer system. Representative amplimers of the expected sizes were gel excised using an EZNA Gel Extraction kit (Omega Bio-Tek Inc., Norcross, GA), sub-cloned into the pCR2.1TOPO TA cloning vector (Invitrogen), and sequenced at the Arizona State University DNA Core Lab (Tempe, AZ). In all cases, sequence variation of the cloned LhABC transporter fragments was minimal (>98% nucleotide identity) compared to the transcriptomic data, indicating that the assembled data accurately represent the sequences. The minor variations in sequence were likely attributable to the allelic heterogeneity of the L. hesperus laboratory colony, or to rare errors introduced during amplification.
Table 3. Oligonucleotide primers used in expression profiling and cloning.
| gene | sequence (5′-3′) | Amplimer size |
| Lygus actin F | ATGTGCGACGAAGAAGTTG | 555 |
| Lygus actin R1 | GTCACGGCCAGCCAAATC | |
| LhABCA8 8 F | AAGGCTGGTTTGCTGTGGCT | 533 |
| LhABCA8 541 R | AGGAGCTGGATCGATAGCTCG | |
| LhABCB2 444 F | CACCGCTCAGCAATGCAACC | 491 |
| LhABCB2 935 R | TGAGGACCTCGCCTGCGATA | |
| LhABCB4 1848 F | CTGGCTACGTCGTCCAGCTC | 511 |
| LhABCB4 2359 R | GTCAGCATTTCTCGCAGCGG | |
| LhABCB6 51 F | CCTCGCCTGGCTGAGAAGTC | 481 |
| LhABCB6 532 R | AATACAATCCGCCCGCCTCC | |
| LhABCC1 1240 F | GGCTGCACATACGGACTGCT | 468 |
| LhABCC1 1708 R | GCAGCACCGAAGTAGGCACT | |
| LhABCC3 1755 F | AGATGGCTGGGACCTTCCGT | 492 |
| LhABCC3 2247 R | TGGTTCAGAGCTGACACGGC | |
| LhABCC5 1143 F | GGTGGCCGAATGTCGAGTGT | 523 |
| LhABCC5 1666 R | GCGATGCTCCCCTTTCACCA | |
| LhABCC8 1210 F | CGTCGCAGGATTTTGCACCC | 496 |
| LhABCC8 1706 R | TGCGGTTGTCCTTGTGTTGC | |
| LhABCG1 220 F | TGACCATCACGCCGTGTCAG | 467 |
| LhABCG1 687 R | TCGTCTTCACGTGCTCCTGG | |
| LhABCG10 788 F | AAGCGTCTTGGTGGGCTCAG | 517 |
| LhABCG10 1305 R | AGGGCCCACTGACAAGGCTA | |
| LhABCG16 850 F | TCTGCACGATTCACCAGCCC | 483 |
| LhABCG16 1333 R | TTGCTACCGTCTTGTCCCGC | |
| LhABCH1 927 F | AGCTCCCCAAGTCCTGCTGA | 473 |
| LhABCH1 1400 R | CCTGTAGGGTCCCGACCGAT | |
| LhABCH5 934 F | ATAGGCACAGCAGTGCACCC | 495 |
| LhABCH5 1429 R | TATCCCCGGGACGTGATGCT | |
| LhABCC1A start F | ATGGCCGAGGATACGCTT | 767 |
| LhABCC1B start F | ATGGCAGAGGAAACACTTC | 767 |
| LhABCC1A/B 767 R | TCGCCAATGTGGCTTTCC |
Supporting Information
Distribution of the most highly represented species in BLASTx and tBLASTn analyses of L. hesperus ABC transporter sequences. BLAST analyses were performed using the NCBI non-redundant database with an E value ≤10−10.
(PS)
Amino acid sequence alignment of LhABCC1A and LhABCC1B. Alignment was performed using the default settings in MUSCLE [159]. Pairwise sequence identity for the full-length transporters is 96%. Pairwise sequence identity for the first 260 amino acids is 86% (223/260 aa), whereas identity over the remaining 684 amino acids is 99.6% (681/684 aa). Black shading is indicative of 100% amino acid sequence identity.
(EPS)
Phylogenetic analysis of ABCG and ABCH transporters from L. hesperus and four other species. The scale bar represents 1.0 amino acid substitutions per site. Analyses, abbreviations, and color-coding are as in Figure 1. Clades corresponding to the two subfamilies are indicated by tan shading (ABCH) or yellow shading (ABCG). As before, because the ABCH subfamily is restricted to the arthropod lineage, no representative sequences for H. sapiens were included in the analysis.
(EPS)
Length of male L. hesperus accessory glands and testes in fifth instar nymphs and adults. Testis length was measured from the base to the apical tip of the longest lobe. Accessory gland length was measured from the insertion at the common duct to its anterior end where the accessory gland folds over on itself. It should be noted that while primordial reproductive tissues are present in fourth instar nymphs they are smaller than that seen in early fifths and very poorly developed. Stage selection criteria were: early stadium fifth instars - small green abdomen and thin wing buds with light pigmentation; late stadium fifth instars - enlarged abdomen with yellow color and significant fatty deposits, thickened wing bugs with heavy pigmentation; adults – light body pigmentation, minimal body fat, wings not hardened, sampled within 12 h of eclosion. All specimens sampled were from the same cohort. Error bars represent standard deviation (n = 20 for each group).
(PDF)
Top five BLASTx hits from a search against the non-redundant protein database using the 65 putative LhABC transporter sequences as a query. Analysis performed with an E value ≤10−10.
(XLSX)
Top five tBLASTn hits from a search against the non-redundant database using the 65 putative LhABC transporter sequences as a query. Analysis performed with an E value ≤10−10.
(XLSX)
Identification of potential protein domains in the putative LhABC transporter sequences. Analyses were performed using default settings for ScanProsite [152] and HMMScan on the HMMER webserver [154] using default settings with protein databases set to Pfam, Gene3D, and Superfamily.
(XLSX)
MUSCLE based multiple sequence alignment heat map of the percent amino acid identities among the LhABC transporters. The matrix, which includes partial sequences, was generated from a MUSCLE alignment and indicates the percent identity across the predicted protein sequences. Cell shading is based on a sliding three color scale with lowest percent identities in red and highest percent identities in blue.
(XLSX)
MUSCLE based multiple sequence alignment heat map of the percent amino acid identities among the LhABCA transporters. The matrix and cell shading are as described in Table S4.
(XLSX)
MUSCLE based multiple sequence alignment heat map of the percent amino acid identities among the LhABCB transporters. The matrix and cell shading are as described in Table S4.
(XLSX)
MUSCLE based multiple sequence alignment heat map of the percent amino acid identities among the LhABCC transporters. The matrix and cell shading are as described in Table S4.
(XLSX)
MUSCLE based multiple sequence alignment heat map of the percent amino acid identities among the LhABCD, LhABCE, and LhABCF transporters. The matrix and cell shading are as described in Table S4.
(XLSX)
MUSCLE based multiple sequence alignment heat map of the percent amino acid identities among the LhABCG transporters. The matrix and cell shading are as described in Table S4.
(XLSX)
MUSCLE based multiple sequence alignment heat map of the percent amino acid identities among the LhABCH transporters. The matrix and cell shading are as described in Table S4.
(XLSX)
Gene accession/model numbers of ABC transporter protein sequences used in phylogenetics analyses.
(XLSX)
LhABC transporter protein sequences.
(XLS)
Acknowledgments
The authors thank Dr. David J. Hawthorne (University of Maryland) for initial discussions and critical reading of the manuscript. The authors also thank Daniel Langhorst and Lynn Forlow Jech (both from USDA-ARS ALARC) for maintaining the L. hesperus colony and assistance with tissue dissections, and Brian Hall (USDA-ARS Daniel K. Inouye Pacific Basin Agricultural Research Center) for assistance with NCBI data deposition. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U. S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. The raw sequence data was deposited in the NCBI sequence read archive under BioProject PRJNA238835, BioSamples SAMN02679940–SAMN02679948, SRA Submission ID “PBARC: Lygus hesperus Heat Experiment”, SRA Study Accession SRP039607. To facilitate submission to the NCBI TSA database, transcript sequences were modified to put all coding sequence on the positive strand by reverse complimenting when appropriate and the longest coding sequence for each transcript was submitted to TSA using an open source transcriptome preparation software package (http://genomeannotation.github.io/transvestigator/). The annotated assembly with putative gene name and functional annotations was submitted to NCBI under TSA submission GBHO00000000. The version described in this paper is the first version, GBHO01000000. Protein sequences of the Lygus hesperus ABC transporters are included as a Supporting Information file.
Funding Statement
This work was supported by CSB - Cotton Incorporated (project no. 12-373), by KC - Agriculture and Food Research Initiative Competitive Grant from the USDA National Institute of Food and Agriculture (no. 2011-38422-30955), and by SMG - National Science Foundation (no. OCI-1053575XSEDE under allocation TG-MCB140032). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Oldham ML, Davidson AL, Chen J (2008) Structural insights into ABC transporter mechanism. Curr Opin Struct Biol 18: 726–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rees DC, Johnson E, Lewinson O (2009) ABC transporters: the power to change. Nature 10: 218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jones PM, O’Mara ML, George AM (2009) ABC transporters: a riddle wrapped in a mystery inside an enigma. Trends Biochem Sci 34: 520–531. [DOI] [PubMed] [Google Scholar]
- 4. George AM, Jones PM (2012) Perspectives on the structure-function of ABC transporters: the Switch and Constant Contact models. Prog Biophys Mol Biol 109: 95–107. [DOI] [PubMed] [Google Scholar]
- 5. Kartner N, Riordan JR, Ling V (1983) Cell surface P-glycoprotein associated with multidrug resistance in mammalian cell lines. Science 221: 1285–1288. [DOI] [PubMed] [Google Scholar]
- 6. Lage H (2003) ABC-transporters: implications on drug resistance from microorganisms to human cancers. Int J Antimicrob Agents 22: 188–199. [DOI] [PubMed] [Google Scholar]
- 7. Leprohon P, Légaré D, Ouellette M (2011) ABC transporters involved in drug resistance in human parasites. Essays Biochem 50: 121–144. [DOI] [PubMed] [Google Scholar]
- 8. Dermauw W, Van Leeuwen T (2014) The ABC gene family in arthropods: Comparative genomics and role in insecticide transport and resistance. Insect Biochem Mol Biol 45: 89–110. [DOI] [PubMed] [Google Scholar]
- 9. Bisbal C, Martinand C, Silhol M, Lebleu B, Salehzada T (1995) Cloning and characterization of a RNAse L inhibitor. A new component of the interferon-regulated 2–5A pathway. J Biol Chem 270: 13308–13317. [DOI] [PubMed] [Google Scholar]
- 10. Tyzack JK, Wang X, Belsham GJ, Proud CG (2000) ABC50 interacts with eukaryotic initiation factor 2 and associates with the ribosome in an ATP-dependent manner. J Biol Chem 275: 34131–34139. [DOI] [PubMed] [Google Scholar]
- 11. Dong J, Lai R, Nielsen K, Fekete CA, Qiu H, et al. (2004) The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J Biol Chem 279: 42157–42168. [DOI] [PubMed] [Google Scholar]
- 12. Paytubi S, Wang X, Lam YW, Izquierdo L, Hunter MJ, et al. (2009) ABC50 promotes translation initiation in mammalian cells. J Biol Chem 284: 24061–24073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barthelme D, Dinkelaker S, Albers S-V, Londei P, Ermler U, et al. (2011) Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proc Natl Acad Sci USA 108: 3228–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dean M, Annilo T (2005) Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genomics Hum Genet 6: 123–142. [DOI] [PubMed] [Google Scholar]
- 15. Popovic M, Zaja R, Loncar J, Smital T (2010) A novel ABC transporter: the first insight into zebrafish (Danio rerio) ABCH1. Mar Environ Res 69 Suppl: S11–S13. [DOI] [PubMed] [Google Scholar]
- 16. Broehan G, Kroeger T, Lorenzen M, Merzendorfer H (2013) Functional analysis of the ATP-binding cassette (ABC) transporter gene family of Tribolium castaneum . BMC Genomics 14: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ewart GD, Cannell D, Cox GB, Howells AJ (1994) Mutational analysis of the traffic ATPase (ABC) transporters involved in uptake of eye pigment precursors in Drosophila melanogaster. Implications for structure-function relationships. J Biol Chem 269: 10370–10377. [PubMed] [Google Scholar]
- 18. Mackenzie SM, Brooker MR, Gill TR, Cox GB, Howells AJ, et al. (1999) Mutations in the white gene of Drosophila melanogaster affecting ABC transporters that determine eye colouration. Biochim Biophys Acta 1419: 173–185. [DOI] [PubMed] [Google Scholar]
- 19. Kômoto N, Quan G-X, Sezutsu H, Tamura T (2009) A single-base deletion in an ABC transporter gene causes white eyes, white eggs, and translucent larval skin in the silkworm w-3(oe) mutant. Insect Biochem Mol Biol 39: 152–156. [DOI] [PubMed] [Google Scholar]
- 20. Wang L, Kiuchi T, Fujii T, Daimon T, Li M, et al. (2013) Mutation of a novel ABC transporter gene is responsible for the failure to incorporate uric acid in the epidermis of ok mutants of the silkworm, Bombyx mori . Insect Biochem Mol Biol 43: 562–571. [DOI] [PubMed] [Google Scholar]
- 21. Ricardo S, Lehmann R (2009) An ABC transporter controls export of a Drosophila germ cell attractant. Science 323: 943–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Itoh TQ, Tanimura T, Matsumoto A (2011) Membrane-bound transporter controls the circadian transcription of clock genes in Drosophila. Genes Cells 16: 1159–1167. [DOI] [PubMed] [Google Scholar]
- 23. Strauss AS, Peters S, Boland W, Burse A (2013) ABC transporter functions as a pacemaker for the sequestration of plant glucosides in leaf beetles. eLife 2: e01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Borycz J, Borycz JA, Kubow A, Lloyd V, Meinertzhagen IA (2008) Drosophila ABC transporter mutants white, brown and scarlet have altered contents and distribution of biogenic amines in the brain. J Exp Biol 211: 3454–3466. [DOI] [PubMed] [Google Scholar]
- 25. Buss DS, Callaghan A (2008) Interaction of pesticides with p-glycoprotein and other ABC proteins: A survey of the possible importance to insecticide, herbicide and fungicide resistance. Pest Biochem Physiol 90: 141–153. [Google Scholar]
- 26. Gahan LJ, Pauchet Y, Vogel H, Heckel DG (2010) An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet 6: e1001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baxter SW, Badenes-Perez FR, Morrison A, Vogel H, Crickmore N, et al. (2011) Parallel evolution of Bacillus thuringiensis toxin resistance in Lepidoptera. Genetics 189: 675–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Atsumi S, Miyamoto K, Yamamoto K, Narukawa J, Kawai S, et al. (2012) Single amino acid mutation in an ATP-binding cassette transporter gene causes resistance to Bt toxin Cry1Ab in the silkworm, Bombyx mori . Proc Natl Acad Sci USA 109: E1591–E1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heckel DG (2012) Learning the ABCs of Bt: ABC transporters and insect resistance to Bacillus thuringiensis provide clues to a crucial step in toxin mode of action. Pest Biochem Physiol 104: 103–110. [Google Scholar]
- 30. Lei Y, Zhu X, Xie W, Wu Q, Wang S, et al. (2014) Midgut transcriptome response to a Cry toxin in the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Gene 533: 180–187. [DOI] [PubMed] [Google Scholar]
- 31. Park Y, González-Martínez RM, Navarro-Cerrillo G, Chakroun M, Kim Y, et al. (2014) ABCC transporters mediate insect resistance to multiple Bt toxins revealed by bulk segregant analysis. BMC Biology 12: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sturm A, Cunningham P, Dean M (2009) The ABC transporter gene family of Daphnia pulex . BMC Genomics 10: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dermauw W, Osborne EJ, Clark RM, Grbić M, Tirry L, et al. (2013) A burst of ABC genes in the genome of the polyphagous spider mite Tetranychus urticae . BMC Genomics 14: 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dean M, Rzhetsky A, Allikmets R (2001) The human ATP-binding cassette (ABC) transporter superfamily. Genome Res 11: 1156–1166. [DOI] [PubMed] [Google Scholar]
- 35. Pavlidi N, Dermauw W, Rombauts S, Chrisargiris A, Van Leeuwen T, et al. (2013) Analysis of the olive fruit fly Bactrocera oleae transcriptome and phylogenetic classification of the major detoxification gene families. PLoS ONE 8: e66533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roth CW, Holm I, Graille M, Dehoux P, Rzhetsky A, et al. (2003) Identification of the Anopheles gambiae ATP-binding cassette transporter superfamily genes. Mol Cells 15: 150–158. [PubMed] [Google Scholar]
- 37. Liu S, Zhou S, Tian L, Guo E, Luan Y, et al. (2011) Genome-wide identification and characterization of ATP-binding cassette transporters in the silkworm, Bombyx mori . BMC Genomics 12: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xie X, Cheng T, Wang G, Duan J, Niu W, et al. (2012) Genome-wide analysis of the ATP-binding cassette (ABC) transporter gene family in the silkworm, Bombyx mori . Mol Biol Rep 39: 7281–7291. [DOI] [PubMed] [Google Scholar]
- 39. Strauss AS, Wang D, Stock M, Gretscher RR, Groth M, et al. (2014) Tissue-specific transcript profiling for ABC transporters in the sequestering larvae of the phytophagous leaf beetle Chrysomela populi . PLoS ONE 9: e98637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. International Aphid Genomics Consortium (2010) Genome sequence of the pea aphid Acyrthosiphon pisum . PLoS Biol 8: e1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ye X-D, Su Y-L, Zhao Q-Y, Xia W-Q, Liu S-S, et al. (2014) Transcriptomic analyses reveal the adaptive features and biological differences of guts from two invasive whitefly species. BMC Genomics 15: 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang N, Xie W, Jones CM, Bass C, Jiao X, et al. (2013) Transcriptome profiling of the whitefly Bemisia tabaci reveals stage-specific gene expression signatures for thiamethoxam resistance. Insect Mol Biol 22: 485–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xia J, Zhang C-R, Zhang S, Li F-F, Feng M-G, et al. (2013) Analysis of whitefly transcriptional responses to Beauveria bassiana infection reveals new insights into insect-fungus interactions. PLoS ONE 8: e68185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mamidala P, Wijeratne AJ, Wijeratne S, Kornacker K, Sudhamalla B, et al. (2012) RNA-Seq and molecular docking reveal multi-level pesticide resistance in the bed bug. BMC Genomics 13: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bao Y-Y, Li B-L, Liu Z-B, Xue J, Zhu Z-R, et al. (2010) Triazophos up-regulated gene expression in the female brown planthopper, Nilaparvata lugens . J Insect Physiol 56: 1087–1094. [DOI] [PubMed] [Google Scholar]
- 46. Scott DR (1977) An annotated listing of host plants of Lygus hesperus Knight. Entomol Soc Am Bull 23: 19–22. [Google Scholar]
- 47. Young OP (1986) Host plants of the tarnished plant bug, Lygus lineolaris (Heteroptera: Miridae). Ann Entomol Soc Am 79: 747–762. [Google Scholar]
- 48.Wheeler AG (2001) Biology of the plant bugs (Hemiptera: Miridae): pests, predators, opportunists. Ithaca, NY: Comstock Publishing Associates. 528 p. [Google Scholar]
- 49. Snodgrass GL (1996) Insecticide resistance in field populations of the tarnished plant bug (Heteroptera: Miridae) in cotton in the Mississippi Delta. J Econ Entomol 89: 783–790. [DOI] [PubMed] [Google Scholar]
- 50. Snodgrass G, Scott W (2002) Tolerance to acephate in tarnished plant bug (Heteroptera: Miridae) populations in the Mississippi river delta. Southwestern Entomologist 27: 191–199. [Google Scholar]
- 51. Snodgrass G, Gore J, Abel C, Jackson R (2009) Acephate resistance in populations of the tarnished plant bug (Heteroptera: Miridae) from the Mississippi River Delta. J Econ Entomol 102: 699–707. [DOI] [PubMed] [Google Scholar]
- 52. Hull JJ, Geib SM, Fabrick JA, Brent CS (2013) Sequencing and de novo assembly of the western tarnished plant bug (Lygus hesperus) transcriptome. PLoS ONE 8: e55105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tapadia MG, Lakhotia SC (2005) Expression of mdr49 and mdr65 multidrug resistance genes in larval tissues of Drosophila melanogaster under normal and stress conditions. Cell Stress Chaperones 10: 7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. de Boussac H, Orbán TI, Várady G, Tihanyi B, Bacquet C, et al. (2012) Stimulus-induced expression of the ABCG2 multidrug transporter in HepG2 hepatocarcinoma model cells involves the ERK1/2 cascade and alternative promoters. Biochem Biophys Res Commun 426: 172–176. [DOI] [PubMed] [Google Scholar]
- 55. de Araujo Leite JC, de Vasconcelos RB, da Silva SG, de Siqueira-Junior JP, Marques-Santos LF (2013) ATP-binding cassette transporters protect sea urchin gametes and embryonic cells against the harmful effects of ultraviolet light. Mol Reprod Dev 81: 66–83. [DOI] [PubMed] [Google Scholar]
- 56. Kim Y, Park S-Y, Kim D, Choi J, Lee Y-H, et al. (2013) Genome-scale analysis of ABC transporter genes and characterization of the ABCC type transporter genes in Magnaporthe oryzae . Genomics 101: 354–361. [DOI] [PubMed] [Google Scholar]
- 57. Kulkarni SR, Donepudi AC, Xu J, Wei W, Cheng QC, et al. (2014) Fasting induces nuclear factor E2-related factor 2 and ATP-binding cassette transporters via protein kinase A and sirtuin-1 in mouse and human. Antioxid Redox Signal 20: 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Peelman F, Labeur C, Vanloo B, Roosbeek S, Devaud C, et al. (2003) Characterization of the ABCA transporter subfamily: identification of prokaryotic and eukaryotic members, phylogeny and topology. J Mol Biol 325: 259–274. [DOI] [PubMed] [Google Scholar]
- 59. Beers MF, Hawkins A, Shuman H, Zhao M, Newitt JL, et al. (2011) A novel conserved targeting motif found in ABCA transporters mediates trafficking to early post-Golgi compartments. J Lipid Res 52: 1471–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kaminski WE, Piehler A, Wenzel JJ (2006) ABC A-subfamily transporters: Structure, function and disease. Biochim Biophys Acta 1762: 510–524. [DOI] [PubMed] [Google Scholar]
- 61. Albrecht C, Viturro E (2007) The ABCA subfamily–gene and protein structures, functions and associated hereditary diseases. Pflugers Arch 453: 581–589. [DOI] [PubMed] [Google Scholar]
- 62. Akiyama M, Sugiyama-Nakagiri Y, Sakai K, McMillan JR, Goto M, et al. (2005) Mutations in lipid transporter ABCA12 in harlequin ichthyosis and functional recovery by corrective gene transfer. J Clin Invest 115: 1777–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bain LJ, LeBlanc GA (1996) Interaction of structurally diverse pesticides with the human MDR1 gene product P-glycoprotein. Toxicol Appl Pharmacol 141: 288–298. [DOI] [PubMed] [Google Scholar]
- 64. Gottesman MM, Fojo T, Bates SE (2002) Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer 2: 48–58. [DOI] [PubMed] [Google Scholar]
- 65. Szakács G, Annereau J-P, Lababidi S, Shankavaram U, Arciello A, et al. (2004) Predicting drug sensitivity and resistance: profiling ABC transporter genes in cancer cells. Cancer Cell 6: 129–137. [DOI] [PubMed] [Google Scholar]
- 66. Yasui K, Mihara S, Zhao C, Okamoto H, Saito-Ohara F, et al. (2004) Alteration in copy numbers of genes as a mechanism for acquired drug resistance. Cancer Res 64: 1403–1410. [DOI] [PubMed] [Google Scholar]
- 67. Sooksa-Nguan T, Yakubov B, Kozlovskyy VI, Barkume CM, Howe KJ, et al. (2009) Drosophila ABC transporter, DmHMT-1, confers tolerance to cadmium. DmHMT-1 and its yeast homolog, SpHMT-1, are not essential for vacuolar phytochelatin sequestration. J Biol Chem 284: 354–362. [DOI] [PubMed] [Google Scholar]
- 68. Bariami V, Jones CM, Poupardin R, Vontas J, Ranson H (2012) Gene amplification, ABC transporters and cytochrome P450s: unraveling the molecular basis of pyrethroid resistance in the dengue vector, Aedes aegypti . PLoS Negl Trop Dis 6: e1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Telonis-Scott M, Hallas R, McKechnie SW, Wee CW, Hoffmann AA (2009) Selection for cold resistance alters gene transcript levels in Drosophila melanogaster . J Insect Physiol 55: 549–555. [DOI] [PubMed] [Google Scholar]
- 70. Parcej D, Tampé R (2010) ABC proteins in antigen translocation and viral inhibition. Nature Chemical Biology 6: 572–580. [DOI] [PubMed] [Google Scholar]
- 71. Abele R, Tampé R (2011) The TAP translocation machinery in adaptive immunity and viral escape mechanisms. Essays Biochem 50: 249–264. [DOI] [PubMed] [Google Scholar]
- 72. Wu CT, Budding M, Griffin MS, Croop JM (1991) Isolation and characterization of Drosophila multidrug resistance gene homologs. Mol Cell Biol 11: 3940–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Vache C, Camares O, Cardoso-Ferreira M-C, Dastugue B, Creveaux I, et al. (2007) A potential genomic biomarker for the detection of polycyclic aromatic hydrocarbon pollutants: multidrug resistance gene 49 in Drosophila melanogaster . Environ Toxicol Chem 26: 1418–1424. [DOI] [PubMed] [Google Scholar]
- 74. Azad P, Zhou D, Russo E, Haddad GG (2009) Distinct mechanisms underlying tolerance to intermittent and constant hypoxia in Drosophila melanogaster . PLoS ONE 4: e5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chahine S, O’Donnell MJ (2009) Physiological and molecular characterization of methotrexate transport by Malpighian tubules of adult Drosophila melanogaster . J Insect Physiol 55: 927–935. [DOI] [PubMed] [Google Scholar]
- 76. Figueira-Mansur J, Ferreira-Pereira A, Mansur JF, Franco TA, Alvarenga ESL, et al. (2013) Silencing of P-glycoprotein increases mortality in temephos-treated Aedes aegypti larvae. Insect Mol Biol 22: 648–658. [DOI] [PubMed] [Google Scholar]
- 77. Tian L, Yang J, Hou W, Xu B, Xie W, et al. (2013) Molecular cloning and characterization of a P-glycoprotein from the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Intl J Mol Sci 14: 22891–22905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Luo L, Sun Y-J, Wu Y-J (2013) Abamectin resistance in Drosophila is related to increased expression of P-glycoprotein via the dEGFR and dAkt pathways. Insect Biochem Mol Biol 43: 627–634. [DOI] [PubMed] [Google Scholar]
- 79. Toyoda Y, Hagiya Y, Adachi T, Hoshijima K, Kuo MT, et al. (2008) MRP class of human ATP binding cassette (ABC) transporters: historical background and new research directions. Xenobiotica 38: 833–862. [DOI] [PubMed] [Google Scholar]
- 80. Keppler D (2010) Multidrug Resistance Proteins (MRPs, ABCCs): importance for pathophysiology and drug therapy. Drug Transporters. Handbook of Experimental Pharmacology. Berlin, Heidelberg: Springer Berlin Heidelberg, Vol. 201: 299–323. [DOI] [PubMed] [Google Scholar]
- 81. Slot AJ, Molinski SV, Cole SPC (2011) Mammalian multidrug-resistance proteins (MRPs). Essays Biochem 50: 179–207. [DOI] [PubMed] [Google Scholar]
- 82. Dassa E, Bouige P (2001) The ABC of ABCs: a phylogenetic and functional classification of ABC systems in living organisms. Res Microbiol 152: 211–229. [DOI] [PubMed] [Google Scholar]
- 83. Grailles M, Brey PT, Roth CW (2003) The Drosophila melanogaster multidrug-resistance protein 1 (MRP1) homolog has a novel gene structure containing two variable internal exons. Gene 307: 41–50. [DOI] [PubMed] [Google Scholar]
- 84. Labbé R, Caveney S, Donly C (2011) Genetic analysis of the xenobiotic resistance-associated ABC gene subfamilies of the Lepidoptera. Insect Mol Biol 20: 243–256. [DOI] [PubMed] [Google Scholar]
- 85. Chahine S, O’Donnell MJ (2011) Interactions between detoxification mechanisms and excretion in Malpighian tubules of Drosophila melanogaster. . J Exp Biol 214: 462–468. [DOI] [PubMed] [Google Scholar]
- 86. Luckenbach T, Epel D (2008) ABCB- and ABCC-type transporters confer multixenobiotic resistance and form an environment-tissue barrier in bivalve gills. Am J Physiol Regul Integr Comp Physiol 294: R1919–R1929. [DOI] [PubMed] [Google Scholar]
- 87. Kruh GD, Guo Y, Hopper-Borge E, Belinsky MG, Chen Z-S (2006) ABCC10, ABCC11, and ABCC12. Pflugers Arch 453: 675–684. [DOI] [PubMed] [Google Scholar]
- 88. Beckstead RB, Lam G, Thummel CS (2007) Specific transcriptional responses to juvenile hormone and ecdysone in Drosophila. Insect Biochem Mol Biol 37: 570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Vodovar N, Vinals M, Liehl P, Basset A, Degrouard J, et al. (2005) Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc Natl Acad Sci USA 102: 11414–11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yepiskoposyan H, Egli D, Fergestad T, Selvaraj A, Treiber C, et al. (2006) Transcriptome response to heavy metal stress in Drosophila reveals a new zinc transporter that confers resistance to zinc. Nucleic Acids Res 34: 4866–4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Morozova TV, Anholt RRH, Mackay TFC (2006) Transcriptional response to alcohol exposure in Drosophila melanogaster . Genome Biol 7: R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Monnier V, Girardot F, Cheret C, Andres O, Tricoire H (2002) Modulation of oxidative stress resistance in Drosophila melanogaster by gene overexpression. Genesis 34: 76–79. [DOI] [PubMed] [Google Scholar]
- 93. Huang H, Haddad GG (2007) Drosophila dMRP4 regulates responsiveness to O2 deprivation and development under hypoxia. Physiol Genomics 29: 260–266. [DOI] [PubMed] [Google Scholar]
- 94. Fernández-Ayala DJ, Chen S, Kemppainen E, O’Dell KMC, Jacobs HT (2010) Gene expression in a Drosophila model of mitochondrial disease. PLoS ONE 5: e8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Shah S, Yarrow C, Dunning R, Cheek B, Vass S, et al. (2011) Insecticide detoxification indicator strains as tools for enhancing chemical discovery screens. Pest Manag Sci 68: 38–48. [DOI] [PubMed] [Google Scholar]
- 96. Tanaka S, Miyamoto K, Noda H, Jurat-Fuentes JL, Yoshizawa Y, et al. (2013) The ATP-binding cassette transporter subfamily C member 2 in Bombyx mori larvae is a functional receptor for Cry toxins from Bacillus thuringiensis . FEBS J 280: 1782–1794. [DOI] [PubMed] [Google Scholar]
- 97. Baum JA, Sukuru UR, Penn SR, Meyer SE, Subbarao S, et al. (2012) Cotton plants expressing a hemipteran-active Bacillus thuringiensis crystal protein impact the development and survival of Lygus hesperus (Hemiptera: Miridae) nymphs. J Econ Entomol 105: 616–624. [DOI] [PubMed] [Google Scholar]
- 98. Aittoniemi J, Fotinou C, Craig TJ, de Wet H, Proks P, et al. (2009) SUR1: a unique ATP-binding cassette protein that functions as an ion channel regulator. Philos Trans R Soc Lond B Biol Sci 364: 257–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kim SK, Rulifson EJ (2004) Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. Nature 431: 316–320. [DOI] [PubMed] [Google Scholar]
- 100. Akasaka T, Klinedinst S, Ocorr K, Bustamante EL, Kim SK, et al. (2006) The ATP-sensitive potassium (KATP) channel-encoded dSUR gene is required for Drosophila heart function and is regulated by tinman. Proc Natl Acad Sci USA 103: 11999–12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Li Y, Qin Y, Yang N, Sun Y, Yang X, et al. (2013) Studies on insecticidal activities and action mechanism of novel benzoylphenylurea candidate NK-17. PLoS ONE 8: e66251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Merzendorfer H, Kim HS, Chaudhari SS, Kumari M, Specht CA, et al. (2012) Genomic and proteomic studies on the effects of the insect growth regulator diflubenzuron in the model beetle species Tribolium castaneum . Insect Biochem Mol Biol 42: 264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Meyer F, Flötenmeyer M, Moussian B (2013) The sulfonylurea receptor Sur is dispensable for chitin synthesis in Drosophila melanogaster embryos. Pest Manag Sci 69: 1136–1140. [DOI] [PubMed] [Google Scholar]
- 104. Jordan IK, Kota KC, Cui G, Thompson CH, McCarty NA (2008) Evolutionary and functional divergence between the cystic fibrosis transmembrane conductance regulator and related ATP-binding cassette transporters. Proc Natl Acad Sci USA 105: 18865–18870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wanders RJ, Visser WF, van Roermund CWT, Kemp S, Waterham HR (2007) The peroxisomal ABC transporter family. Pflugers Arch 453: 719–734. [DOI] [PubMed] [Google Scholar]
- 106. Morita M, Imanaka T (2012) Peroxisomal ABC transporters: structure, function and role in disease. Biochim Biophys Acta 1822: 1387–1396. [DOI] [PubMed] [Google Scholar]
- 107. Shani N, Sapag A, Valle D (1996) Characterization and analysis of conserved motifs in a peroxisomal ATP-binding cassette transporter. J Biol Chem 271: 8725–8730. [DOI] [PubMed] [Google Scholar]
- 108. Mosallanejad H, Badisco L, Swevers L, Soin T, Knapen D, et al. (2010) Ecdysone signaling and transcript signature in Drosophila cells resistant against methoxyfenozide. J Insect Physiol 56: 1973–1985. [DOI] [PubMed] [Google Scholar]
- 109. Khoshnevis S, Gross T, Rotte C, Baierlein C, Ficner R, et al. (2010) The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Reports 11: 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Liu S, Li Q, Liu Z (2013) Genome-wide identification, characterization and phylogenetic analysis of 50 catfish ATP-binding cassette (ABC) transporter genes. PLoS ONE 8: e63895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Abruzzi KC, Rodriguez J, Menet JS, Desrochers J, Zadina A, et al. (2011) Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev 25: 2374–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Tarr PT, Tarling EJ, Bojanic DD, Edwards PA, Baldán A (2009) Emerging new paradigms for ABCG transporters. Biochim Biophys Acta 1791: 584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kerr ID, Haider AJ, Gelissen IC (2011) The ABCG family of membrane-associated transporters: you don’t have to be big to be mighty. Br J Pharmacol 164: 1767–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Weber AL, Khan GF, Magwire MM, Tabor CL, Mackay TFC, et al. (2012) Genome-wide association analysis of oxidative stress resistance in Drosophila melanogaster . PLoS ONE 7: e34745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Buchmann J, Meyer C, Neschen S, Augustin R, Schmolz K, et al. (2007) Ablation of the cholesterol transporter adenosine triphosphate-binding cassette transporter G1 reduces adipose cell size and protects against diet-induced obesity. Endocrinology 148: 1561–1573. [DOI] [PubMed] [Google Scholar]
- 116. Morozova TV, Anholt RRH, Mackay TFC (2007) Phenotypic and transcriptional response to selection for alcohol sensitivity in Drosophila melanogaster . Genome Biol 8: R231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Hock T, Cottrill T, Keegan J, Garza D (2000) The E23 early gene of Drosophila encodes an ecdysone-inducible ATP-binding cassette transporter capable of repressing ecdysone-mediated gene activation. Proc Natl Acad Sci USA 97: 9519–9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Tan A, Palli SR (2008) Edysone receptor isoforms play distinct roles in controlling molting and metamorphosis in the red flour beetle, Tribolium castaneum . Mol Cell Endocrinol 291: 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Bryon A, Wybouw N, Dermauw W, Tirry L, Van Leeuwen T (2013) Genome wide gene-expression analysis of facultative reproductive diapause in the two-spotted spider mite Tetranychus urticae . BMC Genomics 14: 815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Abraham EG, Sezutsu H, Kanda T, Sugasaki T, Shimada T, et al. (2000) Identification and characterisation of a silkworm ABC transporter gene homologous to Drosophila white. Mol Gen Genet 264: 11–19. [DOI] [PubMed] [Google Scholar]
- 121. Tatematsu K-I, Yamamoto K, Uchino K, Narukawa J, Iizuka T, et al. (2011) Positional cloning of silkworm white egg 2 (w-2) locus shows functional conservation and diversification of ABC transporters for pigmentation in insects. Genes Cells 16: 331–342. [DOI] [PubMed] [Google Scholar]
- 122. Anaka M, MacDonald CD, Barkova E, Simon K, Rostom R, et al. (2008) The white gene of Drosophila melanogaster encodes a protein with a role in courtship behavior. J Neurogenet 22: 243–276. [DOI] [PubMed] [Google Scholar]
- 123. Snodgrass GL (2002) Characteristics of a red-eye mutant of the tarnished plant bug (Heteroptera: Miridae). Ann Entomol Soc Am 95: 366–369. [Google Scholar]
- 124. Allen ML (2013) Genetics of a sex-linked recessive red eye color mutant of the tarnished plant bug, Lygus lineolaris . Open J Anim Sci 3: 1–9. [Google Scholar]
- 125. Liu S-H, Yao J, Yao H-W, Jiang P-L, Yang B-J, et al. (2014) Biological and biochemical characterization of a red-eye mutant in Nilaparvata lugens (Hemiptera: Delphacidae). Insect Science 21: 469–476. [DOI] [PubMed] [Google Scholar]
- 126. Seo BY, Jung JK, Kim Y (2011) An orange-eye mutant of the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). J Asia-Pacific Entomol 14: 469–472. [Google Scholar]
- 127. Annilo T, Chen Z-Q, Shulenin S, Costantino J, Thomas L, et al. (2006) Evolution of the vertebrate ABC gene family: analysis of gene birth and death. Genomics 88: 1–11. [DOI] [PubMed] [Google Scholar]
- 128. Mummery-Widmer JL, Yamazaki M, Stoeger T, Novatchkova M, Bhalerao S, et al. (2009) Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature 458: 987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Zhang S, Feany MB, Saraswati S, Littleton JT, Perrimon N (2009) Inactivation of Drosophila Huntingtin affects long-term adult functioning and the pathogenesis of a Huntington’s disease model. Dis Model Mech 2: 247–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Qin W, Neal SJ, Robertson RM, Westwood JT, Walker VK (2005) Cold hardening and transcriptional change in Drosophila melanogaster . Insect Mol Biol 14: 607–613. [DOI] [PubMed] [Google Scholar]
- 131. You M, Yue Z, He W, Yang X, Yang G, et al. (2013) A heterozygous moth genome provides insights into herbivory and detoxification. Nat Genet 45: 220–225. [DOI] [PubMed] [Google Scholar]
- 132. Robinson SW, Herzyk P, Dow JAT, Leader DP (2012) FlyAtlas: database of gene expression in the tissues of Drosophila melanogaster . Nucleic Acids Res 41: D744–D750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Sullivan DT, Bell LA, Paton DR, Sullivan MC (1980) Genetic and functional analysis of tryptophan transport in Malpighian tubules of Drosophila. Biochem Genet 18: 1109–1130. [DOI] [PubMed] [Google Scholar]
- 134. Dow JAT, Davies S-A (2006) The Malpighian tubule: Rapid insights from post-genomic biology. J Insect Physiol 52: 365–378. [DOI] [PubMed] [Google Scholar]
- 135. Dow JAT (2009) Insights into the Malpighian tubule from functional genomics. J Exp Biol 212: 435–445. [DOI] [PubMed] [Google Scholar]
- 136. Wang J, Kean L, Yang J, Allan AK, Davies S-A, et al. (2004) Function-informed transcriptome analysis of Drosophila renal tubule. Genome Biol 5: R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Yang J, McCart C, Woods DJ, Terhzaz S, Greenwood KG, et al. (2007) A Drosophila systems approach to xenobiotic metabolism. Physiol Genomics 30: 223–231. [DOI] [PubMed] [Google Scholar]
- 138. Labbé R, Caveney S, Donly C (2011) Expression of multidrug resistance proteins is localized principally to the Malpighian tubules in larvae of the cabbage looper moth, Trichoplusia ni . J Exp Biol 214: 937–944. [DOI] [PubMed] [Google Scholar]
- 139. Wasbrough ER, Dorus S, Hester S, Howard-Murkin J, Lilley K, et al. (2010) The Drosophila melanogaster sperm proteome-II (DmSP-II). J Proteomics 73: 2171–2185. [DOI] [PubMed] [Google Scholar]
- 140. Jones SR, Cyr DG (2011) Regulation and characterization of the ATP-binding cassette transporter-B1 in the epididymis and epididymal spermatozoa of the rat. Toxicol Sci 119: 369–379. [DOI] [PubMed] [Google Scholar]
- 141. Gillott C (2003) Male accessory gland secretions: modulators of female reproductive physiology and behavior. Annu Rev Entomol 48: 163–184. [DOI] [PubMed] [Google Scholar]
- 142. Avila FW, Sirot LK, LaFlamme BA, Rubinstein CD, Wolfner MF (2011) Insect seminal fluid proteins: identification and function. Annu Rev Entomol 56: 21–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Quan GX, Kanda T, Tamura T (2002) Induction of the white egg 3 mutant phenotype by injection of the double-stranded RNA of the silkworm white gene. Insect Mol Biol 11: 217–222. [DOI] [PubMed] [Google Scholar]
- 144. Martins GF, Ramalho-Ortigao JM (2012) Oenocytes in insects. Inver Surv J 9: 139–152. [Google Scholar]
- 145. Zhu F, Gujar H, Gordon JR, Haynes KF, Potter MF, et al. (2013) Bed bugs evolved unique adaptive strategy to resist pyrethroid insecticides. Sci Rep 3: 1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Debolt JW (1982) Meridic diet for rearing successive generations of Lygus hesperus . Ann Entomol Soc Am 75: 119–122. [Google Scholar]
- 147. Brent CS, Hull JJ (2014) Characterization of male-derived factors inhibiting female sexual receptivity in Lygus hesperus . J Insect Physiol 60: 104–110. [DOI] [PubMed] [Google Scholar]
- 148. Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, et al. (2012) RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res 40: W622–W627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, et al. (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature Protocols 8: 1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Eddy SR (2011) Accelerated profile HMM searches. PLoS Comput Biol 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Sheps JA, Ralph S, Zhao Z, Baillie DL, Ling V (2004) The ABC transporter gene family of Caenorhabditis elegans has implications for the evolutionary dynamics of multidrug resistance in eukaryotes. Genome Biol 5: R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. de Castro E, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, et al. (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34: W362–W365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Sigrist CJA, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, et al. (2010) PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res 38: D161–D166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39: W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Horton P, Park K-J, Obayashi T, Fujita N, Harada H, et al. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35: W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580. [DOI] [PubMed] [Google Scholar]
- 157. Bernsel A, Viklund H, Hennerdal A, Elofsson A (2009) TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res 37: W465–W468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. von Heijne G (1992) Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol 225: 487–494. [DOI] [PubMed] [Google Scholar]
- 159. Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Price MN, Dehal PS, Arkin AP (2010) FastTree 2– approximately maximum-likelihood trees for large alignments. PLoS ONE 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of the most highly represented species in BLASTx and tBLASTn analyses of L. hesperus ABC transporter sequences. BLAST analyses were performed using the NCBI non-redundant database with an E value ≤10−10.
(PS)
Amino acid sequence alignment of LhABCC1A and LhABCC1B. Alignment was performed using the default settings in MUSCLE [159]. Pairwise sequence identity for the full-length transporters is 96%. Pairwise sequence identity for the first 260 amino acids is 86% (223/260 aa), whereas identity over the remaining 684 amino acids is 99.6% (681/684 aa). Black shading is indicative of 100% amino acid sequence identity.
(EPS)
Phylogenetic analysis of ABCG and ABCH transporters from L. hesperus and four other species. The scale bar represents 1.0 amino acid substitutions per site. Analyses, abbreviations, and color-coding are as in Figure 1. Clades corresponding to the two subfamilies are indicated by tan shading (ABCH) or yellow shading (ABCG). As before, because the ABCH subfamily is restricted to the arthropod lineage, no representative sequences for H. sapiens were included in the analysis.
(EPS)
Length of male L. hesperus accessory glands and testes in fifth instar nymphs and adults. Testis length was measured from the base to the apical tip of the longest lobe. Accessory gland length was measured from the insertion at the common duct to its anterior end where the accessory gland folds over on itself. It should be noted that while primordial reproductive tissues are present in fourth instar nymphs they are smaller than that seen in early fifths and very poorly developed. Stage selection criteria were: early stadium fifth instars - small green abdomen and thin wing buds with light pigmentation; late stadium fifth instars - enlarged abdomen with yellow color and significant fatty deposits, thickened wing bugs with heavy pigmentation; adults – light body pigmentation, minimal body fat, wings not hardened, sampled within 12 h of eclosion. All specimens sampled were from the same cohort. Error bars represent standard deviation (n = 20 for each group).
(PDF)
Top five BLASTx hits from a search against the non-redundant protein database using the 65 putative LhABC transporter sequences as a query. Analysis performed with an E value ≤10−10.
(XLSX)
Top five tBLASTn hits from a search against the non-redundant database using the 65 putative LhABC transporter sequences as a query. Analysis performed with an E value ≤10−10.
(XLSX)
Identification of potential protein domains in the putative LhABC transporter sequences. Analyses were performed using default settings for ScanProsite [152] and HMMScan on the HMMER webserver [154] using default settings with protein databases set to Pfam, Gene3D, and Superfamily.
(XLSX)
MUSCLE based multiple sequence alignment heat map of the percent amino acid identities among the LhABC transporters. The matrix, which includes partial sequences, was generated from a MUSCLE alignment and indicates the percent identity across the predicted protein sequences. Cell shading is based on a sliding three color scale with lowest percent identities in red and highest percent identities in blue.
(XLSX)
MUSCLE based multiple sequence alignment heat map of the percent amino acid identities among the LhABCA transporters. The matrix and cell shading are as described in Table S4.
(XLSX)
MUSCLE based multiple sequence alignment heat map of the percent amino acid identities among the LhABCB transporters. The matrix and cell shading are as described in Table S4.
(XLSX)
MUSCLE based multiple sequence alignment heat map of the percent amino acid identities among the LhABCC transporters. The matrix and cell shading are as described in Table S4.
(XLSX)
MUSCLE based multiple sequence alignment heat map of the percent amino acid identities among the LhABCD, LhABCE, and LhABCF transporters. The matrix and cell shading are as described in Table S4.
(XLSX)
MUSCLE based multiple sequence alignment heat map of the percent amino acid identities among the LhABCG transporters. The matrix and cell shading are as described in Table S4.
(XLSX)
MUSCLE based multiple sequence alignment heat map of the percent amino acid identities among the LhABCH transporters. The matrix and cell shading are as described in Table S4.
(XLSX)
Gene accession/model numbers of ABC transporter protein sequences used in phylogenetics analyses.
(XLSX)
LhABC transporter protein sequences.
(XLS)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. The raw sequence data was deposited in the NCBI sequence read archive under BioProject PRJNA238835, BioSamples SAMN02679940–SAMN02679948, SRA Submission ID “PBARC: Lygus hesperus Heat Experiment”, SRA Study Accession SRP039607. To facilitate submission to the NCBI TSA database, transcript sequences were modified to put all coding sequence on the positive strand by reverse complimenting when appropriate and the longest coding sequence for each transcript was submitted to TSA using an open source transcriptome preparation software package (http://genomeannotation.github.io/transvestigator/). The annotated assembly with putative gene name and functional annotations was submitted to NCBI under TSA submission GBHO00000000. The version described in this paper is the first version, GBHO01000000. Protein sequences of the Lygus hesperus ABC transporters are included as a Supporting Information file.