Significance
Long noncoding RNAs (lncRNAs) play important roles in chromatin regulation in higher eukaryotes. Studies analysing how lncRNAs influence Polycomb silencing suggested they specifically bind and recruit Polycomb Repressive Complex 2 (PRC2) to defined targets. However, the promiscuous binding of PRC2 to RNA has raised questions as to the mechanism by which lncRNA contributes to Polycomb silencing. We investigate the function of a set of cold-induced antisense transcripts in the Polycomb-dependent epigenetic silencing of a floral repressor gene in Arabidopsis. Through analysis of a transgene, in which antisense transcripts are no longer cold-induced, we show that these antisense transcripts are required for coordinated switching of opposing chromatin states. This mechanism links transcriptional shutdown by cold to long-term epigenetic silencing.
Keywords: flowering, long noncoding RNA, histone modifications, Polycomb, Arabidopsis
Abstract
Long noncoding RNAs (lncRNAs) have been proposed to play important roles in gene regulation. However, their importance in epigenetic silencing and how specificity is determined remain controversial. We have investigated the cold-induced epigenetic switching mechanism involved in the silencing of Arabidopsis thaliana FLOWERING LOCUS C (FLC), which occurs during vernalization. Antisense transcripts, collectively named COOLAIR, are induced by prolonged cold before the major accumulation of histone 3 lysine 27 trimethylation (H3K27me3), characteristic of Polycomb silencing. We have found that COOLAIR is physically associated with the FLC locus and accelerates transcriptional shutdown of FLC during cold exposure. Removal of COOLAIR disrupted the synchronized replacement of H3K36 methylation with H3K27me3 at the intragenic FLC nucleation site during the cold. Consistently, genetic analysis showed COOLAIR and Polycomb complexes work independently in the cold-dependent silencing of FLC. Our data reveal a role for lncRNA in the coordinated switching of chromatin states that occurs during epigenetic regulation.
An important developmental transition in plants is the switch from vegetative growth to flowering. Several pathways that control flowering converge to regulate expression of the gene encoding the floral repressor FLOWERING LOCUS C (FLC) (1, 2). Vernalization, the quantitative acceleration of flowering by prolonged cold, leads to the epigenetic silencing of FLC (1, 3). Polycomb Repressive Complex 2 (PRC2) is required for vernalization (4) with the core PRC2 recruited to the FLC locus before silencing. Cold exposure induces formation of a modified PRC2, PHD-PRC2 involving the Plant Homeodomain (PHD) proteins (VRN5 and VIN3), at an intragenic nucleation site (5). Upon return to warm conditions, PHD-PRC2 spreads across the whole FLC locus, resulting in high levels of histone 3 lysine 27 trimethylation (H3K27me3) and stable epigenetic repression. The quantitative nature of vernalization was found to be the result of a cell autonomous switch between bistable epigenetic states (6). The silent state, which is reflected by H3K27me3 accumulation and increasing cold, progressively increases the proportion of cells in which this switch has occurred. A detailed temporal and spatial analysis exploring the opposing expression state showed H3K36me3 is the most likely modification with an antagonistic role to H3K27me3 (7). However, the lack of an absolute mirror profile between H3K27me3 and H3K36me3 at FLC suggests additional factors are necessary for epigenetic switching at FLC (7).
COOLAIR, a group of long antisense RNAs expressed from the FLC locus, has an important role in mediating FLC expression in nonvernalized plants (8–10). COOLAIR is alternatively spliced and alternatively polyadenylated with two major classes in the warm, the proximally polyadenylated class I and the distally polyadenylated class II (Fig. 1 A and B and refs. 11 and 12). Increased proximal polyadenylation enhances H3K4me2 demethylase activity and reduces FLC transcription, resulting in a positive feedback mechanism that reinforces proximal polyadenylation and low expression of FLC (9, 10). Distal polyadenylation is associated with a high expression state of the locus, but it is still unclear whether use of the distal site causes, or is a consequence of, increased FLC transcription (9, 12). In addition to this processing of the COOLAIR transcript, the COOLAIR promoter is regulated by the presence of an R loop (13). Stabilization of the R loop by the homeodomain protein NDX1 reduces COOLAIR transcription (13). COOLAIR transcription is also regulated by cold, with a transient expression peak after 2–3 wk of cold exposure (11). Addition of COOLAIR promoter sequences as the 3′ terminator of a GFP sensor construct was sufficient for cold-induced reduction of GFP expression. However, the role of COOLAIR in the cold-mediated epigenetic silencing during vernalization remained unclear because plants carrying insertions into the COOLAIR promoter that attenuate COOLAIR expression were found to vernalize (14).
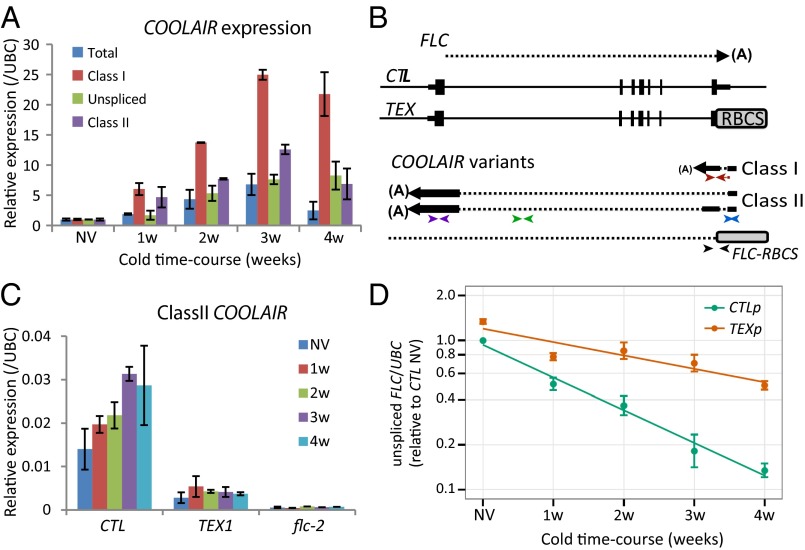
Fig. 1.
COOLAIR accelerates FLC transcriptional down-regulation during vernalization. (A) Quantitative RT-PCR (qRT-PCR) analysis showing COOLAIR forms are differentially induced during a time-course of cold exposure, nonvernalized material (NV), 1 wk cold exposure (1w), 2 wk cold (2w), 3 wk cold (3w), and 4 wk cold (4w). Values are normalized to NV (set as 1), means ± SEM of three biological replicates. (B) Schematic representation of the FLC genomic locus and sense and antisense RNA transcripts in the control (CTL) and Terminator EXchange lines (TEX). CTL lines were generated by transformation of FLC genomic construct (15 kb of the FLC locus, FLC-15) into flc-2 FRI background. TEX lines were generated as described (9, 10). For antisense transcripts in TEX lines, a combination of FLC and RBCS primers were used. (A), pA sites; RBCS, Rubisco gene terminator. The position of the primers used in qRT-PCR analysis to assess class I (red arrows), class II (purple arrows), unspliced antisense (green arrows), total COOLAIR (blue arrows), or antisense RNA transcript derived from TEX construct (black arrows) are shown; sequences are listed in SI Appendix, Table S2. (C) qRT-PCR analysis of class II COOLAIR relative to UBC in CTL and TEX. Values are means ± SEM of three biological replicates. (D) Down-regulation of FLC unspliced RNA is significantly abrogated (P < 0.005) in TEX compared with control during cold exposure. CTLp and TEXp correspond to a mix of 50 T3 transgenic lines (Materials and Methods). Values are means ± SEM of five biological replicates and are plotted on a log scale. NV levels are also significantly different (P < 0.001).
Here, we have sought to clarify the role of COOLAIR in vernalization by analyzing an FLC gene carrying a replacement 3′ region, so lacking the COOLAIR promoter. We show that COOLAIR significantly accelerates FLC transcriptional repression in the cold. COOLAIR RNA was found to physically associate with FLC chromatin in two important regulatory regions. Disruption of COOLAIR expression prevented the cold-induced reduction of H3K36 methylation, without changing the H3K27me3 accumulation dynamics, at the intragenic FLC nucleation site. This work reveals a role for COOLAIR in the coordinated switching of chromatin states that occurs during cold, linking transcriptional shutdown with epigenetic silencing.
Results
COOLAIR Isoforms Accumulate Differentially in the Cold.
To further understand roles of COOLAIR, we first undertook a thorough analysis of the accumulation of COOLAIR isoforms over a time course of cold exposure (Fig. 1A). The unspliced, class I and class II isoforms of COOLAIR all accumulated during cold with, in this experimental setup, a maximum induction at 3 wk cold. However, induction of the class I isoform was several fold higher than the others, consistent with proximal polyadenylation correlating with reduced levels of FLC transcription. To distinguish whether the accumulation in the cold was due to transcriptional induction or higher transcript stability in the cold, we measured COOLAIR steady-state levels in the presence of cordycepin, a chain termination adenosine analog (3′-deoxyadenosine) (15). The half-life of the COOLAIR transcript increased slightly in the cold (t1/2 NV = 100 min, t1/2 2w = 240 min; SI Appendix, Fig. S1A), but this change only partly explained the robust increase (seven- to eightfold) in COOLAIR RNA levels (Fig. 1A). Accumulation of COOLAIR during cold is therefore a consequence of both transcript stabilization and transcriptional induction.
COOLAIR Accelerates Transcriptional Shutdown of FLC During Cold Exposure.
To investigate the biological functions of COOLAIR in FLC regulation, we had generated plant lines impaired in FLC antisense transcription by exchanging the FLC terminator/COOLAIR promoter with the strong RBCS terminator (RBCS3B At5G38410) in the flc-2 background to form Terminator EXchange lines, TEX (9, 10) (Fig. 1B). flc-2 is a loss-of-function FLC genotype, which has a deletion/rearrangement within the endogenous FLC gene (1). The TEX lines were compared with transgenic plant lines carrying an intact FLC gene (control line, CTL; ref. 16) (Fig. 1B). Because expression of transgenes vary depending on the genomic context of their integration, the analysis was carried out both on selected representative lines (CTL and TEX1) or a pool of 50 individual T3 lines (referred as CTLp and TEXp). First, we confirmed that the level of COOLAIR transcription in TEX lines is lower than wild-type and is not cold inducible (Fig. 1C and SI Appendix, Fig. S2 A–C and S3 A and C). Next, we assessed the effect of COOLAIR disruption on FLC transcription by measuring the level of FLC unspliced transcript in TEX samples. Transcription of FLC is significantly higher in nonvernalized plants (pooled samples, Fig. 1D) and is reduced by cold exposure much more slowly compared with lines carrying an intact FLC expressing COOLAIR (Fig. 1D, and in single line SI Appendix, Fig. S3B). To analyze how COOLAIR exerts this effect, we compared FLC unspliced transcript stability in TEX1 versus CTL line. No significant differences in FLC half-life were detected (t1/2CTL = 72 min, t1/2TEX1 = 88 min; SI Appendix, Fig. S1D). The difference in the amount of unspliced FLC transcript levels between the TEX and CTL lines therefore is likely to be the consequence of increased COOLAIR transcriptional activity.
The FLC mRNA (fully spliced) levels did not closely follow the dynamics of the unspliced FLC RNA during cold exposure, with almost no decrease in FLC mRNA for the first 2 wk of cold exposure (SI Appendix, Fig. S2D). These results suggest that FLC mRNA has a long half-life of many days that is not significantly affected by cold exposure (SI Appendix, Fig. S1C). The multiple stability motifs (17) found in the 3′ UTR of FLC may contribute to this long half-life (SI Appendix, Fig. S1F). The FLC-TEX transcript (missing the FLC 3′ UTR) was found to be slightly less stable (SI Appendix, Fig. S1E). The higher levels of FLC-TEX in both before and during vernalization are thus likely due to increased transcription, reinforcing the view that COOLAIR accelerates transcriptional shutdown of FLC during cold exposure.
COOLAIR Associates with FLC Genomic DNA.
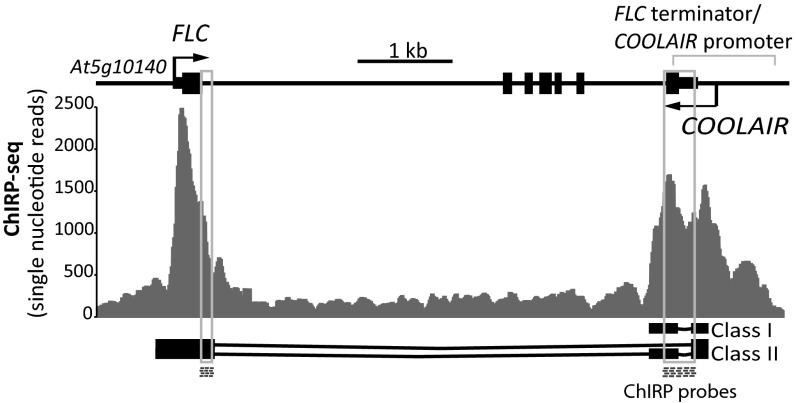
We have shown that COOLAIR up-regulation does not depend on FLC sense transcriptional down-regulation (11). However, if sense and antisense transcription occur simultaneously at the same locus, increased antisense transcription could function to transcriptionally interfere with sense transcription. With this question in mind, we compared data from custom tiling arrays that contained both strands of the entire FLC genomic region at single nucleotide resolution (SI Appendix, Fig. S4, see also ref. 11). The arrays had been hybridized with RNA from both warm and 2-wk cold-treated plants. There was no indication that COOLAIR transcription caused transcriptional interference with FLC transcription, i.e., that FLC unspliced transcripts were more abundant at the 5′ end of the gene as COOLAIR transcription increased (SI Appendix, Fig. S4). In the absence of data supporting a transcriptional interference mechanism, we searched for other mechanisms. Many lncRNAs have been shown to localize to nuclei and induce chromatin changes on their target genes (18). We therefore checked the subcellular localization of COOLAIR and found that whereas spliced forms are efficiently exported, both spliced and unspliced COOLAIR were found in nuclear fractions (SI Appendix, Fig. S5). To address whether COOLAIR has ON-chromatin roles, Chromatin Isolation by RNA Purification (ChIRP) was performed (Fig. 2 and SI Appendix, Figs. S6 and S7; ref. 19). COOLAIR RNA but not UBC RNA was efficiently precipitated by using biotinylated antisense DNA oligo probes (the location of the DNA probes are shown on Fig. 2; sequences listed in SI Appendix, Table S1) and consistent with its induction by cold, higher amounts of COOLAIR RNA could be precipitated from cold-treated samples (SI Appendix, Fig. S7 A–C). Sequencing of the coprecipitated DNA showed COOLAIR enriched over the whole FLC locus (SI Appendix, Fig. S6) but concentrated in two discrete regions of the FLC genomic DNA, the nucleation region and the 3′ region of the gene/COOLAIR promoter (Fig. 2 and PCR validation in SI Appendix, Fig. S7D). These regions are colinear with COOLAIR exon locations but not to the biotin oligo locations used in the ChIRP (Fig. 2). The DNA signal was reduced when samples were pretreated with RNaseA/H, differentially affected by proteinase K (SI Appendix, Fig. S7D) and was completely abolished when glutaraldehyde cross-linking was omitted. This result suggests the different COOLAIR/DNA interactions have different requirements for protein factors. When we analyzed the amount of associated DNA that copurified with COOLAIR in nonvernalized and 2-wk cold-treated samples, only a minor difference was observed. We conclude therefore that most of COOLAIR induced in cold is in the OFF-chromatin fraction. The genomic locations of the ON-chromatin COOLAIR have both been implicated as important for FLC regulation. The nucleation region is where the localized increase of H3K27me3 and parallel decrease of H3K36me3 occurs (6, 7). The 3′ region of the gene/COOLAIR promoter has been shown to contain an R-loop that suppresses COOLAIR transcription (13). This region also shows dynamic changes in histone modifications during vernalization (7), suggesting that COOLAIR might facilitate chromatin modification of FLC chromatin.
Fig. 2.
COOLAIR associates in cis with FLC chromatin. ChIRP deep sequencing analysis data shows COOLAIR associates with FLC chromatin in two distinct regions. Locations of biotinylated DNA probes used for COOLAIR ChIRP are shown (dashed boxes). A schematic of the FLC locus is shown with the structure of the class I and class II COOLAIR transcripts.
COOLAIR Effect on FLC Transcriptional Shutdown During Vernalization Is Independent of Polycomb Machinery and H3K27me3 Accumulation.
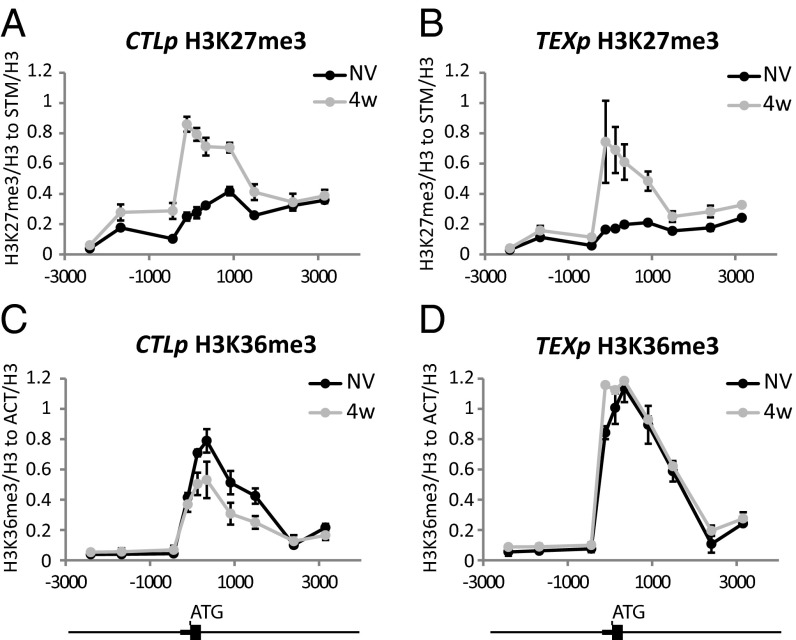
Cold exposure gradually increases H3K27me3 accumulation at the nucleation site within the FLC gene through the activity of the PHD–PRC2 complex (5, 20). Upon return to warm, a modified PHD-PRC2 and H3K27me3 spread to cover the length of the FLC locus (6). To test the role of COOLAIR in these changes, we compared H3K27me3 levels at FLC in TEX compared with control lines (Fig. 3 A and B). No significant differences could be observed in the nucleation of H3K27me3 at FLC during vernalization (Fig. 3 A and B). This data suggests that COOLAIR is not essential for the deposition of the H3K27me3 modification.
Fig. 3.
COOLAIR differentially affects the dynamics of H3K36me3 and H3K27me3 during cold-induced silencing at FLC. ChIP analysis of H3K27me3 in control lines (A) and TEX lines (B). ChIP analysis of H3K36me3 in control lines (C) and TEX lines (D). Nonvernalized (NV) and 4 wk vernalized (4w) samples were compared. Small and big boxes on the schematic below show the FLC 5′ UTR and exon1, respectively. Numbers define positions from translation start site (ATG). Values are means ± SEM of three biological replicates.
We also asked whether nascent RNAs derived from FLC could interact with the PHD–PRC2 complex. RNA-immunoprecipitation (RIP) experiments using GFP-tagged VERNALIZATION 5 (VRN5) and SWINGER (SWN) (21) were undertaken. No enrichment with sense or antisense RNA originating from FLC was found (SI Appendix, Fig. S8 A–F). Based on the ChIP and RIP data, we conclude that COOLAIR does not accelerate the reduction in FLC transcription during vernalization by targeting PRC2-PHD or modifying its H3K27me3 activity.
COOLAIR Mediates Reduction in H3K36me3 at FLC.
We have recently shown that for many phases of the vernalization process, H3K36me3 and H3K27me3 show opposing profiles in the FLC nucleation region (7). H3K36me3 and H3K27me3 were found to rarely coexist on the same histone tail, an antagonism that was shown to be functionally important (7). We therefore compared H3K36me3 levels at FLC between CTL and TEX lines. H3K36me3 levels were higher in the nucleation region before cold and did not decrease during the 4 wk of cold exposure in the TEX lines (Fig. 3D), in contrast to CTL where levels decreased over the 4-wk cold exposure (Fig. 3C). Whereas H3K27me3 accumulation occurred normally in the TEX lines, the removal of COOLAIR disrupted the coordinated changes in H3K27me3/H3K36me3 at the nucleation region. H3K4me3 levels often parallel H3K36me3 so we also analyzed H3K4me3 dynamics in CTL and TEX lines. Removal of COOLAIR also disrupted cold-induced reduction of H3K4me3 levels in the nucleation region (SI Appendix, Fig. S9).
H3K36me3 and H3K27me3 Can Be Modulated Independently in FLC Silencing.
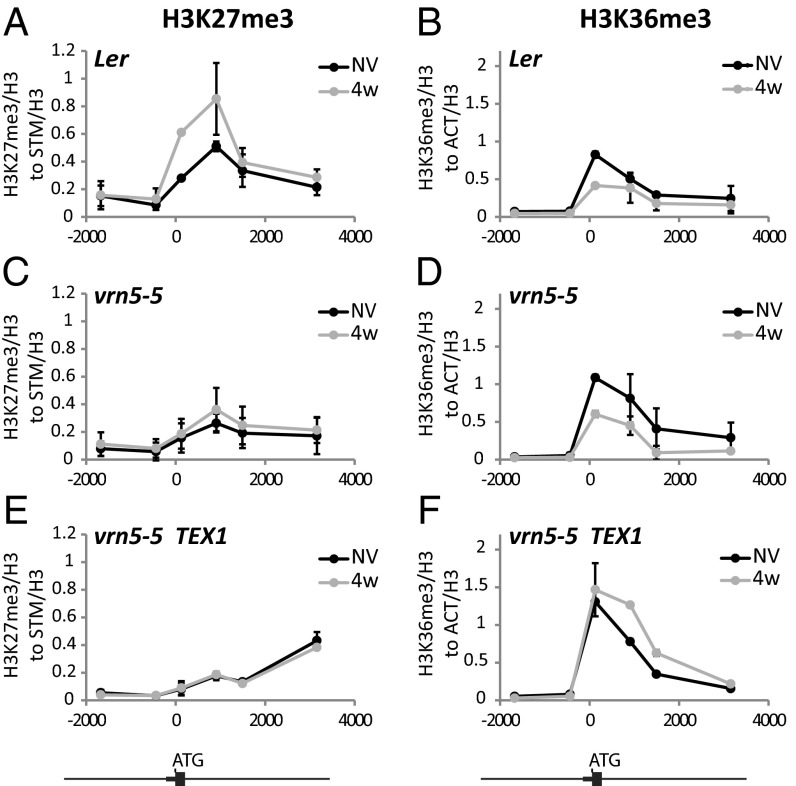
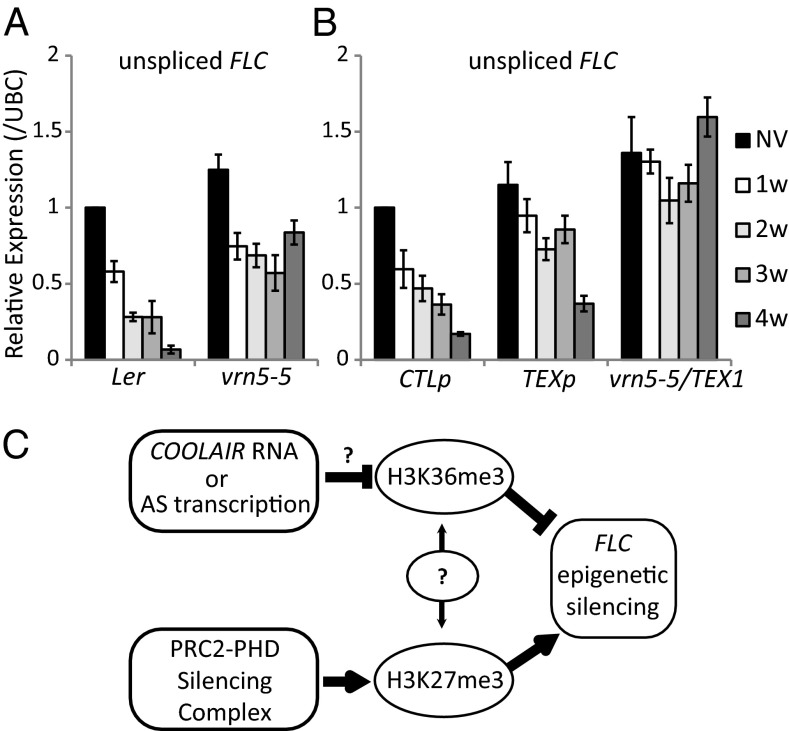
Interestingly, the high levels of H3K36me3 at the nucleation region in the TEX lines did not limit accumulation of H3K27me3 modifications (Fig. 3 B and D). Mutually exclusive histone modification states are predicted to result from linked positive feedback mechanisms connecting production of one modification with removal of the opposing one (22). The ability to disconnect the changes in H3K36me3 and H3K27me3 in the TEX lines suggests that their opposing profiles result from coordinated, but not obligatory, linked methylation/demethylation processes. To further understand these connections we exploited vrn5, a mutation in a component of the PRC2–PHD complex (23). In vrn5, H3K27 methylation is impaired, so H3K27me3 levels at FLC remain low during vernalization (Fig. 4 A and C). Consequently, levels of unspliced FLC (Fig. 5A) and spliced variants of COOLAIR are elevated because of a higher transcriptional activity in the absence of Polycomb-mediated repression (SI Appendix, Fig. S10). vrn5, however, did not affect the dynamics of cold induction of COOLAIR (SI Appendix, Fig. S10). Although H3K27 methylation is impaired, H3K36me3 reduction during cold was not affected by vrn5 (Fig. 4 B and D). These data indicate that removal of H3K36me3 during vernalization is not a direct consequence of H3K27me3. Because the TEX transgene is affected in H3K36me3 removal but not H3K27me3 deposition (Fig. 3 A–D), the H3K36me3 and H3K27me3 changes are likely to be coordinated processes linking transcriptional shutdown with epigenetic silencing. We predicted that the vrn5 mutation would have a dominant effect on H3K27me3 modification in a vrn5/TEX genotype. Similarly, TEX modification would be dominant in defining H3K36me3 in vrn5/TEX during cold. The pattern of chromatin modifications in vrn5/TEX behaved as predicted (Fig. 4 E and F), reinforcing the view that reduction in H3K36me3 and increase in H3K27me3 act in parallel to regulate FLC silencing. We tested the consequence of disrupting both processes on the transcriptional down-regulation of FLC during vernalization. Reduction of FLC transcription, as assayed by unspliced transcript levels, occurred more slowly in vrn5 and in TEX compared with wild-type and control lines, but was completely abolished in the vrn5/TEX double mutant (Fig. 5 A and B). FLC down-regulation and silencing in the cold is therefore a consequence of parallel activities that result in H3K27 trimethylation and H3K36me3 demethylation (Fig. 5C).
Fig. 4.
H3K36me3 and H3K27me3 change independently of each other. ChIP showing H3K27me3 dynamics in Landsberg erecta (Ler) (A), vrn5-5 (C), and vrn5-5/TEX1 plants (E), nonvernalized (NV), after 4 wk cold (4w). ChIP showing H3K36me3 dynamics in Ler (B), vrn5-5 (D), and vrn5-5/TEX1 plants (F) nonvernalized (NV), after 4 wk cold (4w). Numbers define positions from translation start site (ATG). Values are means ± SEM of three biological replicates.
Fig. 5.
Loss of cold-induced transcriptional repression of FLC is additive when vrn5 is combined with a TEX transgene. qRT-PCR analysis of unspliced FLC RNA through a time-course of cold exposure in Ler and vrn5-5 plants (A) and CTL, TEX lines or vrn5-5/TEX (B). Values are means ± SEM of three biological replicates. (C) Proposed model: COOLAIR negatively affects H3K36me3-methylation status and functions independently of PHD-PRC2–mediated H3K27 trimethylation to promote FLC silencing during vernalization.
Discussion
We have investigated the role of the antisense COOLAIR in the epigenetic silencing mechanism underlying vernalization by analyzing an FLC gene lacking the COOLAIR promoter. We found that COOLAIR plays a major role during the cold accelerating FLC transcriptional repression in a mechanism involving reduction of H3K36 methylation. This mechanism functions in parallel to the Polycomb processes that result in accumulation of H3K27me3 at the intragenic FLC nucleation site. COOLAIR is thus important in the coordinated switching of chromatin states that occurs during cold, linking transcriptional shutdown with epigenetic silencing. How could COOLAIR lead to reduction in H3K36me3? COOLAIR RNA itself, as an ON-chromatin or OFF-chromatin fraction, may be responsible for H3K36me3 demethylation. This function could be achieved through a positive effect on a demethylase (by directing remodeling complexes) or due to a negative effect on a methyltransferase (inhibition of activity, obstruction of its binding by the ON-chromatin fraction, or by a decoy effect of the OFF-chromatin fraction). Candidate COOLAIR interacting H3K36 methyltransferase enzymes could be EARLY FLOWERING IN SHORT DAYS (EFS) (24, 25), AtPAF1c-mutant VIP4 methyltransferase (26), or Ash1 homologs. Reports have shown that the conserved Trithorax group protein Ash1 is an H3K36-specific dimethylase (27, 28). In Arabidopsis, the Ash1 homolog SDG4 was shown to methylate H3K4 and H3K36 (29). Another Ash1 homolog, SDG8, was shown to contribute to flowering time regulation through changed H3K36 methylation at FLC (30). Although H3K36 demethylase are known in animal systems (31), their identification in A. thaliana will be necessary to test potential positive effects of COOLAIR on their activity.
Association of COOLAIR at the FLC chromatin, as evidenced by the ChIRP experiments, might influence association of chromatin complexes with the FLC nucleation site. The RNA-IP experiments showed no specific association of COOLAIR with the PHD–PRC2 complex (SI Appendix, Fig. S8). However, these type of analyses need to be undertaken with the appropriate H3K36 methyltransferases and demethylases. How lncRNAs generally lead to chromatin complex activity in specific chromosomal domains is unclear (32). An influence on local association/dissociation rates of chromatin complexes to their respective modifications may be an important contributing factor (33).
Our finding that the opposing profiles of H3K27me3 and H3K36me3 during vernalization can be disconnected has important implications in understanding epigenetic switching mechanisms. Bistable epigenetic states are thought to arise from positive feedback, reinforcing mutually exclusive histone modifications (22). The ability to disconnect the changes in H3K36me3 and H3K27me3 in the TEX lines suggests that their opposing profiles result from coordinated, but not obligatory linked, COOLAIR–PHD-PRC2 activities. Separately, modifying the generation of active and repressive histone modifications may provide a “fail-safe” mechanism where each one reinforces the other. In vrn5 or TEX, where only one of the pathways is faulty, FLC transcriptional silencing still occurs, just at a slower rate, whereas in the vrn5/TEX, it is completely abolished. How exactly these two modification systems are integrated with each other remains an open question (Fig. 5C).
An apparently contradictory answer with respect to the importance of COOLAIR in vernalization has been found through the analysis of Arabidopsis plants containing T-DNA insertions in the COOLAIR promoter, or FLC gene modules missing the 3′ end region (14, 34). Vernalization was found to still accelerate flowering in these plants, although in one of the T-DNA lines (SALK_140021), FLC transcriptional repression was slower (14). In fact, we also find that after >4 wk cold regimes, FLC silencing is reasonably advanced in the TEX lines and sufficient for early flowering to occur when the plants are returned to the warm (SI Appendix, Fig. S11). One complication in this comparison, however, is that flowering time may be affected by the slightly shorter half-life of the FLC-TEX transcript compared with the FLC transcript (SI Appendix, Fig. S1E) or an altered polyadenylation of the FLC-TEX mRNA. However, we would still conclude that COOLAIR RNA or transcription is not absolutely essential for vernalization in laboratory vernalization conditions. In natural conditions where temperature fluctuates widely over both daily and seasonal cycles or in different FLC alleles (16) that have different silencing dynamics, the quantitative contributions of processes reducing FLC transcription, including COOLAIR, may differ.
A surprising finding from this study that needs to be pursued is the high stability of the FLC mRNA. It may be physiologically important in natural conditions because any reduction in temperature could immediately influence FLC transcription. High stability of FLC mRNA could buffer fluctuating temperatures, avoiding large changes in FLC activity that would precipitate precocious flowering. The RNA stability may also play an important role in the time averaging mechanisms the plant must use to monitor the weeks and months required for full vernalization. Our knowledge is limited about how environmental inputs are integrated with epigenetic-modification pathways to regulate stress or adaptation responses. Deciphering chromatin modifications events, their interactions, and dynamics on FLC is providing a paradigm for environmental epigenetic regulation generally.
Materials and Methods
Plant Material, Growth Conditions, and Cold Treatment.
Seeds were sown on GM-glucose media plates and stratified for 2 d. For nonvernalized samples, seedlings were grown in long-day conditions for 10 d (16-h light at 20 °C, 8-h darkness at 16 °C). For vernalization, seeds were pregrown for 10 d at standard warm growing conditions (16-h light at 20 °C, 8-h darkness at 16 °C) before being transferred to cold (8-h light and 16-h darkness at 5 °C) for 2 wk, and then returned to warm conditions for 7 d.
Terminator Exchange Lines.
Generation of TEX lines has been fully described in ref. 10. We used the mixed samples from approximately 50 individual transgenic lines, Basta-resistant T3 generation heterozygote/homozygote mix, to analyze RNA expression in both CTL (named as CTLp) and TEX constructs (named as TEXp), (method has been described in ref. 16). One homozygous T3 representative line (TEX577, here renamed as TEX1) was chosen to generate the vrn5-5/TEX1 genotype (9). Similarly, we randomly selected one homozygous T3 transgenic line from the CTL pool (CTL).
Chromatin Immunoprecipitation.
ChIP experiments were done as described (6), using anti-H3 (Abcam; ab1791), anti-H3K27me3 (Millipore; 07-449), and anti-H3K36me3 (Abcam; ab9050). The ChIP data were quantified by quantitative PCR, normalizing to internal reference genes. STM (At1g62360) was used as the reference gene for H3K27me3 and ACTIN (At5g09810) for H3K36me3.
Chromatin Isolation by RNA Purification.
ChIRP was performed as described (19). A detailed protocol can be found in SI Appendix, SI Materials and Methods.
GFP Fusion Proteins and Western Blotting.
The GFP-SWN line is described in ref. 21. VRN5-EYFP is described in ref. 23. Magnetic GFP-Trap beads (ChromoTek) were used for pull-down YFP tagged VRN5 and SWN, and anti-GFP antibody (Roche) was used for the Western blot shown in SI Appendix, Fig. S8A.
Supplementary Material
Acknowledgments
We thank The Sainsbury Lab (Norwich) for making the ChIRP-seq DNA library; Martin Trick for help with ChIRP-seq data analysis; Scott Berry for helping to plot Fig. 1D; Andrew Angel for analysis of tiling array data and plotting SI Appendix, Fig. S4; and Timothy Wells for the assistant of plant growth. Discussion with all C.D. laboratory members is much appreciated. This work is supported by a UK Biotechnology and Biological Sciences Research Council Institute Strategic Programme Grant BB/J004588/1 to John Innes Centre, and a European Research Council Advanced Investigator grant (ENVGENE) (to C.D.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419030111/-/DCSupplemental.
References
- 1.Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11(5):949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC) Proc Natl Acad Sci USA. 2000;97(7):3753–3758. doi: 10.1073/pnas.060023597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheldon CC, et al. The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11(3):445–458. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gendall AR, Levy YY, Wilson A, Dean C. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell. 2001;107(4):525–535. doi: 10.1016/s0092-8674(01)00573-6. [DOI] [PubMed] [Google Scholar]
- 5.De Lucia F, Crevillen P, Jones AM, Greb T, Dean C. A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci USA. 2008;105(44):16831–16836. doi: 10.1073/pnas.0808687105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angel A, Song J, Dean C, Howard M. A Polycomb-based switch underlying quantitative epigenetic memory. Nature. 2011;476(7358):105–108. doi: 10.1038/nature10241. [DOI] [PubMed] [Google Scholar]
- 7.Yang H, Howard M, Dean C. Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr Biol. 2014;24(15):1793–1797. doi: 10.1016/j.cub.2014.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu F, Marquardt S, Lister C, Swiezewski S, Dean C. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010;327(5961):94–97. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- 9.Marquardt S, et al. Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol Cell. 2014;54(1):156–165. doi: 10.1016/j.molcel.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang ZW, Wu Z, Raitskin O, Sun Q, Dean C. Antisense-mediated FLC transcriptional repression requires the P-TEFb transcription elongation factor. Proc Natl Acad Sci USA. 2014;111(20):7468–7473. doi: 10.1073/pnas.1406635111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462(7274):799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- 12.Hornyik C, Duc C, Rataj K, Terzi LC, Simpson GG. Alternative polyadenylation of antisense RNAs and flowering time control. Biochem Soc Trans. 2010;38(4):1077–1081. doi: 10.1042/BST0381077. [DOI] [PubMed] [Google Scholar]
- 13.Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science. 2013;340(6132):619–621. doi: 10.1126/science.1234848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helliwell CA, Robertson M, Finnegan EJ, Buzas DM, Dennis ES. Vernalization-repression of Arabidopsis FLC requires promoter sequences but not antisense transcripts. PLoS ONE. 2011;6(6):e21513. doi: 10.1371/journal.pone.0021513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutierrez RA, Ewing RM, Cherry JM, Green PJ. Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: Rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci USA. 2002;99(17):11513–11518. doi: 10.1073/pnas.152204099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coustham V, et al. Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science. 2012;337(6094):584–587. doi: 10.1126/science.1221881. [DOI] [PubMed] [Google Scholar]
- 17.Narsai R, et al. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell. 2007;19(11):3418–3436. doi: 10.1105/tpc.107.055046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batista PJ, Chang HY. Long noncoding RNAs: Cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finnegan EJ, Dennis ES. Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr Biol. 2007;17(22):1978–1983. doi: 10.1016/j.cub.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 21.Wang D, Tyson MD, Jackson SS, Yadegari R. Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc Natl Acad Sci USA. 2006;103(35):13244–13249. doi: 10.1073/pnas.0605551103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dodd IB, Micheelsen MA, Sneppen K, Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129(4):813–822. doi: 10.1016/j.cell.2007.02.053. [DOI] [PubMed] [Google Scholar]
- 23.Greb T, et al. The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr Biol. 2007;17(1):73–78. doi: 10.1016/j.cub.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 24.Dong G, Ma DP, Li J. The histone methyltransferase SDG8 regulates shoot branching in Arabidopsis. Biochem Biophys Res Commun. 2008;373(4):659–664. doi: 10.1016/j.bbrc.2008.06.096. [DOI] [PubMed] [Google Scholar]
- 25.Kim SY, et al. Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell. 2005;17(12):3301–3310. doi: 10.1105/tpc.105.034645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu L, et al. Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol Cell Biol. 2008;28(4):1348–1360. doi: 10.1128/MCB.01607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan W, et al. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem. 2011;286(10):7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka Y, Katagiri Z, Kawahashi K, Kioussis D, Kitajima S. Trithorax-group protein ASH1 methylates histone H3 lysine 36. Gene. 2007;397(1-2):161–168. doi: 10.1016/j.gene.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 29.Cartagena JA, et al. The Arabidopsis SDG4 contributes to the regulation of pollen tube growth by methylation of histone H3 lysines 4 and 36 in mature pollen. Dev Biol. 2008;315(2):355–368. doi: 10.1016/j.ydbio.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Z, Yu Y, Meyer D, Wu C, Shen WH. Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat Cell Biol. 2005;7(12):1256–1260. doi: 10.1038/ncb1329. [DOI] [PubMed] [Google Scholar]
- 31.Pedersen MT, Helin K. Histone demethylases in development and disease. Trends Cell Biol. 2010;20(11):662–671. doi: 10.1016/j.tcb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 32.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keller C, Kulasegaran-Shylini R, Shimada Y, Hotz HR, Bühler M. Noncoding RNAs prevent spreading of a repressive histone mark. Nat Struct Mol Biol. 2013;20(8):994–1000. doi: 10.1038/nsmb.2619. [DOI] [PubMed] [Google Scholar]
- 34.Sheldon CC, Conn AB, Dennis ES, Peacock WJ. Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell. 2002;14(10):2527–2537. doi: 10.1105/tpc.004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seeley KA, Byrne DH, Colbert JT. Red light-independent instability of oat phytochrome mRNA in vivo. Plant Cell. 1992;4(1):29–38. doi: 10.1105/tpc.4.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.