Significance
Inhibitors of the FGF receptors (FGFRs) are currently under clinical investigation for the treatment of various cancers. All currently approved kinase inhibitors eventually are rendered useless by the emergence of drug-resistant tumors. We used structure-based drug design to develop the first, to our knowledge, selective, next-generation covalent FGFR inhibitors that can overcome the most common form of kinase inhibitor resistance, the mutation of the so-called “gatekeeper” residue located in the ATP-binding pocket. We also describe a novel kinase inhibitor design strategy that uses a single electrophile to target covalently cysteines that are located in different positions within the ATP-binding pocket. These results have important implications for the design of covalent FGFR inhibitors that can overcome clinical resistance.
Keywords: drug discovery, cancer drug resistance, kinase inhibitor, structure-based drug design
Abstract
The human FGF receptors (FGFRs) play critical roles in various human cancers, and several FGFR inhibitors are currently under clinical investigation. Resistance usually results from selection for mutant kinases that are impervious to the action of the drug or from up-regulation of compensatory signaling pathways. Preclinical studies have demonstrated that resistance to FGFR inhibitors can be acquired through mutations in the FGFR gatekeeper residue, as clinically observed for FGFR4 in embryonal rhabdomyosarcoma and neuroendocrine breast carcinomas. Here we report on the use of a structure-based drug design to develop two selective, next-generation covalent FGFR inhibitors, the FGFR irreversible inhibitors 2 (FIIN-2) and 3 (FIIN-3). To our knowledge, FIIN-2 and FIIN-3 are the first inhibitors that can potently inhibit the proliferation of cells dependent upon the gatekeeper mutants of FGFR1 or FGFR2, which confer resistance to first-generation clinical FGFR inhibitors such as NVP-BGJ398 and AZD4547. Because of the conformational flexibility of the reactive acrylamide substituent, FIIN-3 has the unprecedented ability to inhibit both the EGF receptor (EGFR) and FGFR covalently by targeting two distinct cysteine residues. We report the cocrystal structure of FGFR4 with FIIN-2, which unexpectedly exhibits a “DFG-out” covalent binding mode. The structural basis for dual FGFR and EGFR targeting by FIIN3 also is illustrated by crystal structures of FIIN-3 bound with FGFR4 V550L and EGFR L858R. These results have important implications for the design of covalent FGFR inhibitors that can overcome clinical resistance and provide the first example, to our knowledge, of a kinase inhibitor that covalently targets cysteines located in different positions within the ATP-binding pocket.
Receptor tyrosine kinases (RTKs) serve as critical sensors of extracellular cues that activate a myriad of intracellular signaling pathways to regulate cell state. There are 58 receptor tyrosine kinases in the human genome, and many have been demonstrated to be constitutively activated through amplification or mutation in particular cancers. The signals emanating from these RTKs, such as epidermal growth factor receptor (EGFR), FGF receptor (FGFR), platelet-derived growth factor receptor (PDGFR), protein kinase Kit (KIT), and protein kinase c-Met (MET), have been pharmacologically proven to be essential to the survival of cancers expressing mutant forms of these proteins. However, rapid resistance to monotherapy with first-generation RTK inhibitors has been universally observed. Resistance typically arises from the emergence of cancer cells expressing mutant forms of RTKs that are impervious to the action of first-generation drugs or from the activation of by-pass signaling mechanisms. Resistance can be overcome by developing new inhibitors that target the mutant RTK directly or target bypass signaling mechanisms. Indeed this approach has been deployed successfully in the case of resistance to first-generation inhibitors of EGFR in nonsmall cell lung cancer (NSCLC) and of Abelson tyrosine-protein kinase (ABL) in chronic myelogenous leukemia (CML) (1–4).
Human FGFRs are a family of four RTKs (FGFR1–4) which are sensors of a diverse family of 18 FGF ligands. FGFRs are key regulators of fibrogenesis, embryogenesis, angiogenesis, metabolism, and many other processes of proliferation and differentiation (5, 6). The fundamental importance of FGFR to development is well proven by gain-of-function mutations that result in dwarfism in model organisms and in humans (7–10). Deregulation of FGFR signaling through mutation, chromosomal translocation, and gene amplification or overexpression has been documented abundantly in numerous cancers (11). Activation of FGFR-dependent signaling pathways can stimulate tumor initiation, progression, and resistance to therapy. Translocation events implicating the FGFR1 gene and various fusions of FGFR1 are found in myeloproliferative syndromes (12); chromosomal translocations of FGFR1 or FGFR3 and the transforming acidic coiled-coil genes (TACC1 or TACC3) are oncogenic in glioblastoma multiforme, bladder cancer, head and neck cancer, and lung cancer (13–16); oncogenic mutations of FGFR2 and FGFR3 are observed in lung squamous cell carcinoma; FGFR2 N549K is observed in 25% of endometrial cancers; FGFR3 t(4;14) alterations are reported in 15–20% of multiple myeloma (17–19); FGFR4 Y367C mutation in the transmembrane domain drives constitutive activation and enhanced tumorigenic phenotypes in a breast carcinoma cell line (20–22); and K535 and E550 mutants are reported to activate FGFR4 in rhabdomyosarcoma (23). FGFR amplification is reported in various cancers (24, 25): FGFR1 is amplified in colorectal, lung, and renal cell cancers (26, 27); FGFR2 is amplified in gastric cancer and colorectal cancer (28, 29); FGFR3 is commonly amplified in bladder cancer and also is reported for cervical, oral, and hematological cancers (30–32); and FGFR4 is amplified in hepatocellular carcinoma, gastric cancer, pancreatic cancer, and ovarian cancer (33–37). FGFR also is involved in autocrine activation of STAT3 as a positive feedback in many drug-treated cancer cells which are driven by diverse oncogenes such as EGFR, ALK, MET, and KRAS (38).
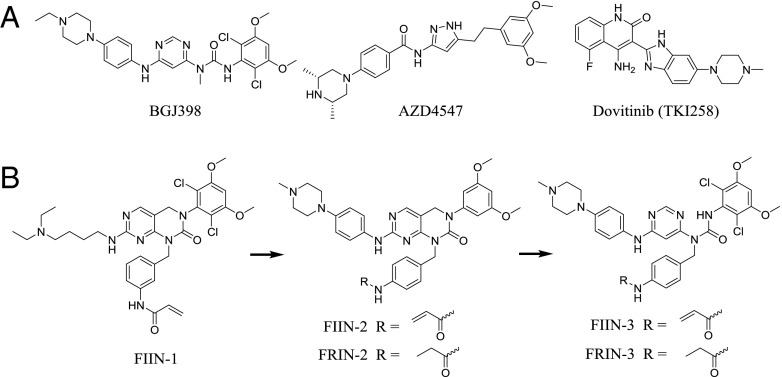
Currently known inhibitors of kinases can target a variety of conformational states and binding pockets and can be either reversible or covalent. Several potent and selective ATP-competitive, small-molecule FGFR inhibitors have been reported, with BGJ398 and AZD4547 being the clinically most advanced compounds (Fig. 1A) (39–42). We previously reported the first (to our knowledge) covalent FGFR irreversible inhibitor (FIIN-1), which targets a cysteine residue conserved in all four FGFR kinases and which inhibits the proliferation of Ba/F3 cells engineered to be dependent on FGFR1, FGFR2, or FGFR3 with EC50s in the 10-nM range, a potency comparable to that exhibited by BGJ398 and AZD4547 (43). All FGFR kinases have a valine at the gatekeeper position, in contrast to ABL, EGFR, KIT, and PDGFR, which all possess a threonine gatekeeper in which resistance can be conferred by mutation of the threonine to a larger hydrophobic valine, isoleucine, or methionine residue in response to first-generation inhibitors of these kinases (44–46). The FGFR V561M mutation was reported to induce strong resistance to PD173074 and FIIN-1 (43, 47); later the gatekeeper mutant FGFR3 V555M emerged as a mechanism of resistance to AZ8010 in KMS-11 myeloma cells and also was demonstrated to confer resistance to other FGFR inhibitors, including PD173074 and AZD4547 (48). The FGFR2 V564I gatekeeper mutant was isolated as a resistant clone in a FGFR2 Ba/F3 screen of dovitinib and also was reported to confer resistance to the multitargeted drug ponatinib (19). In humans the FGFR4 V550L gatekeeper mutation was detected in 9% (4/43) of embryonal rhabdomyosarcoma tumors (49), and the FGFR4 V550M mutation was detected in 13% (2/15) of neuroendocrine breast carcinomas (50). To overcome gatekeeper mutations found in primary FGFR-driven cancers and those that likely will arise in FGFR inhibitor-treated tumors in the future, we developed next-generation covalent FGFR inhibitors. Here we describe the identification and characterization of the covalent FGFR inhibitors FIIN-2 and FIIN-3, which to our knowledge are the first FGFR inhibitors that are capable of potently inhibiting the gatekeeper mutants of FGFRs. We also demonstrate that FIIN-3 is capable of covalently inhibiting both FGFR and EGFR by using distinct binding modes to target different cysteine residues.
Fig. 1.
(A) Chemical structures of clinical-stage FGFR inhibitors. (B) Evolution of FIIN-2 and FIIN-3 from FIIN-1. Structures of the reversible counterparts FRIN-2 and FRIN-3 are shown also.
Results
FIIN-1 was designed based on the structure of the noncovalent FGFR inhibitor PD173074 (51). It possesses a benzamide arm that can reach Cys488 located in the P-loop but possesses weak activity against the V561M gatekeeper mutant of FGFR1, possibly as a result of steric crowding between the dichlorodimethoxyphenyl substituent and the methionine at the gatekeeper position (43). To explore whether analogs of FIIN-1 could potently inhibit the gatekeeper mutants of FGFR kinases, we synthesized derivatives in which both the side chains and core scaffolds were diversified. These new derivatives then were tested for their ability to inhibit the proliferation of Ba/F3 cells that were engineered to be dependent on WT FGFRs and the V564M gatekeeper mutant of FGFR2. Two compounds which emerged from this effort were FIIN-2 and FIIN-3. FIIN-2 maintains the pyrimido[4,5-d]pyrimidinone core of FIIN-1 but removes the two chlorine atoms, which are crucial for FIIN-1’s potency against FGFR. Replacing the aliphatic amine of FIIN-1 with a 4-(4-methylpiperazin-1-yl)aniline group, which is a stronger hinge binder, compensates for the loss of potency, and FIIN-2 potently inhibits WT FGFRs (EC50s in the 1- to 93-nM range) and the gatekeeper mutant of FGFR2 (EC50 of 58 nM). FIIN-3 incorporates a pyrimidyl urea core that forms an intramolecular H-bond, thereby forming a pseudo six-membered ring, a design feature of BGJ398 (39, 52). The H-bond of this pseudo ring was envisioned to provide greater rotatory flexibility to the dichlorodimethoxylphenyl group of FIIN-3, which could better tolerate the methionine gatekeeper. FIIN-3 potently inhibits both WT FGFRs (EC50 in the 1- to 41-nM range) and the gatekeeper mutant of FGFR2 (EC50 of 64 nM). Both FIIN-2 and FIIN-3 possess a 4-acrylamidebenzyl group in contrast to the 3-acrylamidebenzyl group in FIIN-1 and maintain the ability to form a covalent bond with the P-loop cysteine but alter the selectivity profile versus other kinases. Noncovalent analogs of both compounds were prepared with the approximately isosteric propionamide replacing the acrylamide motif to yield FGFR reversible inhibitor 2 and 3 (FRIN-2 and FRIN-3) (Fig. 1B).
To assess the kinase selectivity of FIIN-2 and FIIN-3 broadly, they were profiled against a diverse panel of 456 kinases [DiscoveRX, KinomeScan (53, 54)] using an in vitro ATP-site competition binding assay at a concentration of 1.0 μM. Compared with FIIN-1, both compounds displayed strong binding to FGFRs and exhibited good overall kinase selectivity (SI Appendix, Table S1). FIIN-2 and FIIN-3 have selectivity scores of 10 and 15, respectively (SI Appendix, Fig. S1), which are defined as the number of kinases with scores less than 1 [S(1)] (54). To our surprise, FIIN-3 also potently bound to WT EGFR and to a subset of EGFR mutants, but FIIN-2 exhibited much less affinity for EGFR. To corroborate these findings using an independent biochemical assay, the IC50 values of both compounds also were determined using Z’-lyte enzyme assays [SelectScreen; Life Technology (55)]. FIIN-2 inhibited FGFR1–4 with IC50s of 3.1, 4.3, 27, and 45 nM, respectively, and FIIN-3 displayed IC50s of 13, 21, 31, and 35 nM, respectively. Consistent with the kinase-binding assays, FIIN-3 strongly inhibited EGFR, with an EC50 of 43 nM, and FIIN-2 moderately inhibited EGFR, with an IC50 of 204 nM. FRIN-2 and FRIN-3 also were profiled with KinomeScan assays and, except for EGFR and Bruton’s tyrosine kinase (BTK), exhibited selectivity similar to that of their respective counterparts (SI Appendix, Fig. S1 and Table S1).
We evaluated the potency with which FIIN-2 and FIIN-3 could inhibit the proliferation of Ba/F3 cells engineered to be dependent on FGFR and EGFR kinase activity. Ba/F3 cells are a murine pre-B cell that can be transformed readily with activated kinases to allow growth in the absence of IL-3 and frequently are used to evaluate the activity of compounds against kinases of interest in a cellular context (56). FIIN-2 and FIIN-3 inhibited proliferation of FGFR1-4 Ba/F3 cells with EC50s in the single- to double-digit nanomolar range and were especially potent against FGFR2, with EC50s in the 1-nM range (Table 1). This activity was not caused by general cytotoxicity, because the parental Ba/F3 cells were not inhibited at the highest concentration tested (3.3 µM). FIIN-2 and FIIN-3 also inhibited the FGFR2 V564M gatekeeper mutant Ba/F3 cells, with EC50s of 58 and 64 nM, respectively, whereas FIIN-1 and BGJ398 had EC50s of over 1.0 µM against this mutant. We also applied these inhibitors on four other FGFR2 mutant Ba/F3 cell lines (V564F, E565K, M538I, and K659N) expressing previously isolated FIIN-1 resistance alleles. [K659N already had been reported in patients (17, 57).] Both FIIN-2 and FIIN-3 showed good potency against gatekeeper mutant V564F; FIIN-3 also was potent against the gatekeeper-plus-1 mutant E565K; but BGJ398 was efficient only against the M538I and K659N mutants. FIIN-3 also displayed antiproliferative activity (with an EC50 of 135 nM) against Ba/F3 cells transformed by the EGFR vIII fusion protein, which has a WT EGFR kinase domain. In contrast, FIIN-2 was fourfold less potent (EC50 of 506 nM). FIIN-3 showed even better activity against EGFR L858R (EC50 of 17 nM) and moderate activity, displaying an EC50 of 231 nM, against the EGFR L858R/T790M mutant, which is resistant to first-generation EGFR inhibitors, whereas FIIN-2 was inactive up to a concentration of 1.8 µM. Two covalent EGFR inhibitors, BIBW2992 (58) and WZ4002 (4), were tested on the FGFR2-dependent Ba/F3 cell lines and showed either no or weak potency (SI Appendix, Table S2). The corresponding noncovalent analogs FRIN-2 and FRIN-3 also were profiled against a subset of the FGFR- and EGFR-transformed Ba/F3 cell lines. Interestingly, they maintained similar potency relative to the covalent inhibitors against WT FGFR1-3. This finding is consistent with the results reported for FIIN-1 and also with the notion that these scaffolds are very potent noncovalent binders. However, FRIN-2 and FRIN-3 lost potency against FGFR4, as did FIIN-1 and BGJ398 (no inhibition was detected at 1.0 µM) and were at least 20-fold less potent than their covalent counterparts against the V564M and V564F FGFR2 mutants. FRIN-3 also lost potency against EGFR, suggesting that covalence is required to achieve potency against EGFR, as is consistent with reports for other covalent inhibitors such as WZ4002 (4, 46). Taken together, our assays in Ba/F3 cells show that the new-generation covalent inhibitors FIIN-2 and FIIN3 show strong inhibitory activity against WT (including FGFR4) and gatekeeper mutant FGFR kinases. FIIN-2 and FIIN-3 also were profiled on several other transformed Ba/F3 cell lines to validate their possible off-targets. Some potential off-targets identified using KinomeScan, such as BTK and KIT, were not confirmed, and FIIN-2 showed rather poor potency against protein kinase FLT1 (FLT1); FIIN-3 was not potent against either FLT1 or FLT4 (SI Appendix, Table S2).
Table 1.
Antiproliferative activity of FGFR inhibitors on transformed Ba/F3 cells
| Ba/F3 cell lines | EC50, nM | |||||
| FIIN-1 | BGJ398 | FIIN-2 | FIIN-3 | FRIN-2 | FRIN-3 | |
| Parental | >3,300 | >3,300 | >3,300 | 2,970 | >3,300 | >3,300 |
| FGFR1 | 14 | 3 | 4 | 1 | ||
| FGFR2 | 7 | 4 | 1 | <1 | 10 | 3 |
| FGFR2 (V564M) | 1,000 | 1,500 | 58 | 64 | 2,810 | 1,380 |
| FGFR2 (V564F) | >3,300 | 3,145 | 100 | 71 | ||
| FGFR2 (E565K) | >3,300 | 490 | 273 | 69 | ||
| FGFR2 (K659N) | 3,710 | 25 | 9 | 6 | ||
| FGFR2 (M538I) | 839 | 43 | 27 | 30 | ||
| FGFR2 (C491A) | 168 | 1 | 8 | 3 | 195 | 26 |
| FGFR2(C491A/V564M) | >3,300 | 2,100 | 3,140 | 1,000 | ||
| FGFR3 (S249C) | 10 | 67 | 93 | 41 | ||
| FGFR4 | >1,000 | >1,000 | 32 | 22 | >1,000 | >1,000 |
| EGFR (VIII) | >3,300 | >3,300 | 506 | 135 | >3,300 | 1,840 |
| EGFR (L858R) | >3,300 | >3,300 | 231 | 16.8 | 2,590 | 2,538 |
| EGFR DEL(E746-A750) | 240 | |||||
| EGFR (T790M/L858R) | >3,300 | >3,300 | 1,773 | 231 | >3,300 | >3,300 |
| EGFR (DEL/T790M) | 687 | |||||
| EGFR (C797S/L858R) | >3,300 | >3,300 | >3,300 | |||
To investigate the requirement for covalence in a complementary fashion and to prove the site of covalent modification, we also investigated the activity of the compounds against mutant forms of EGFR and FGFR2 in which the putatively reactive cysteine was mutated to a serine or alanine, respectively. Both FIIN-2 and FIIN-3 maintained their ability to inhibit FGFR2 C491A potently, but FIIN-3 lost its ability to inhibit EGFR C797S. However, when we constructed Ba/F3 cells transformed with the FGFR2 C491A/V564M dual mutant, both compounds lost potency on this dual mutant, thereby demonstrating the requirement for the formation of a covalent bond to Cys491 in the presence of V564M mutant (Table 1).
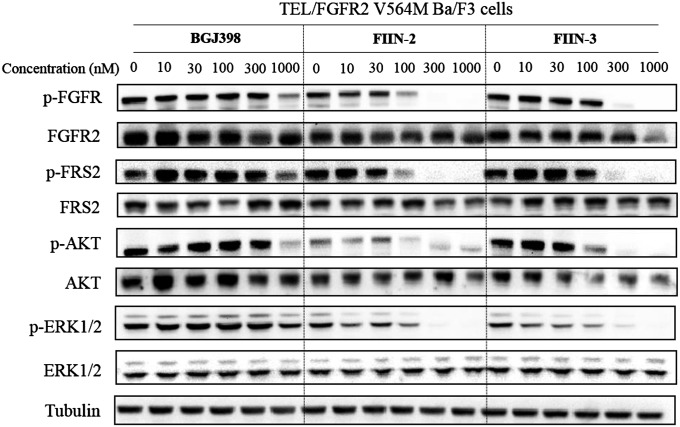
We investigated the effect of FIIN-2 and FIIN-3 relative to established inhibitors, such as BGJ398, on FGFR phosphorylation and on FGFR-dependent signaling. In WT FGFR2 Ba/F3 cells, FIIN-2, FIIN-3, and BGJ398 all completely inhibited the FGFR2 autophosphorylation on Tyr656/657 at concentrations as low as 3 nM (SI Appendix, Fig. S2). In FGFR2 V564M Ba/F3 cells, FIIN-2 and FIIN-3 were capable of inhibiting the FGFR2 V564M autophosphorylation with partial inhibition at 100 nM and complete inhibition observed at 300 nM; this effect also was associated with inhibition of phosphorylation of the FRS2, AKT, and ERK1/2 effector proteins (Fig. 2). The reference compounds BGJ398 and FIIN-1 showed only partial inhibition of FGFR2 V564M at concentrations of 1.0 µM (Fig. 2 and SI Appendix, Fig. S2).
Fig. 2.
Inhibition of FGFR-dependent signaling by BGJ398, FIIN-2, and FIIN-3 in Tel/FGFR2 V564M Ba/F3 cells. Cells were treated with a dose escalation of inhibitors for 6 h and then were lysed and subjected to Western blot for the indicated proteins or phosphoproteins.
As we showed above, Ba/F3 cells expressing the FGFR2 C491A/V564M dual mutant were resistant to inhibition by FIIN-2 and FIIN-3, strongly suggesting that both inhibitors require covalent binding to FGFR to achieve potency. We independently corroborated that FIIN-2 and FIIN-3 are indeed covalent inhibitors by performing cellular wash-out experiments. WT FGFR2 Ba/F3 cells were treated with the reversible inhibitor BGJ398 or with FIIN-2 or FIIN-3 at 20 nM for 3 h and then were washed extensively with PBS and were allowed to recover for 4 h. Western blot of the cellular lysates revealed that, as expected, FIIN-2 and FIIN-3 were capable of sustained inhibition of FGFR2 autophosphorylation after the washout, but the reversible inhibitor BGJ398 was not (SI Appendix, Fig. S3A). An analogous experiment was performed using FGFR2 C491A Ba/F3 cells in which reversible inhibition was demonstrated for all three inhibitors, as expected. To monitor the degree of FGFR target engagement, the lysates also were treated with a biotinylated version of FIIN-1, FIIN-1-biotin (43), which covalently labels FGFR and allows streptavidin-mediated affinity chromatography. Consistent with the signaling studies, FIIN-1-biotin strongly labeled FGFR2 in BGJ398- but not in FIIN-2– or FIIN-3–treated and washed cells (SI Appendix, Fig. S3B). Cumulatively these results provide strong evidence that FIIN-2 and FIIN-3 are irreversible, covalent inhibitors and that Cys491 of FGFR2 is the primary labeled site.
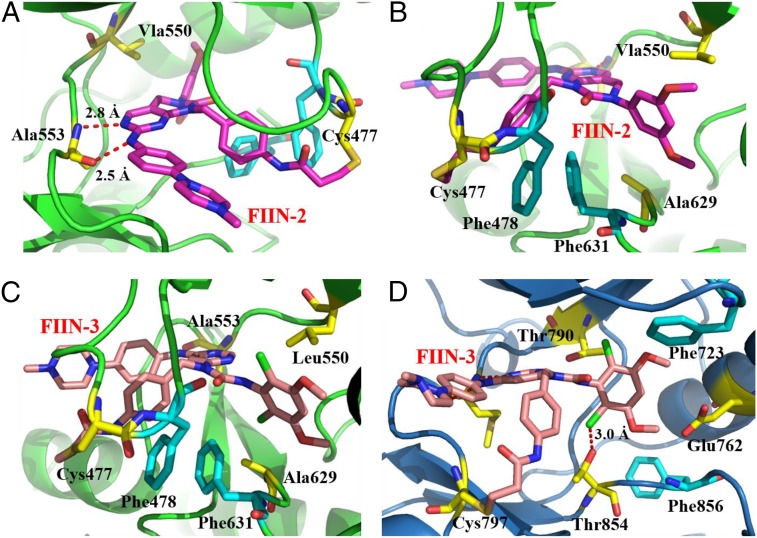
To study the binding modes and structure-affinity relationship of our inhibitors, we solved the cocrystal structure of FGFR4 kinase domain bound to FIIN-2 [Protein Data Bank (PDB) ID code 4QQC] at 2.35-Å resolution (Fig. 3 A and B). In the structure, the two nitrogen atoms from the pyrimidine moiety of FIIN-2 form two hydrogen bonds with Ala553 in the hinge-binding region. Consistent with our biochemical data, a covalent bond is formed between the reactive acrylamide group of FIIN-2 and Cys477 in the kinase P-loop. This covalent bonding pulls down the adjoining Phe478 from the P-loop, allowing it to engage in aromatic contacts with the acrylamidobenzyl group of the compound. Importantly, this conformational change creates favorable intramolecular π–π stacking contacts between Phe478 from the P-loop and Phe631 from the DFG motif, permitting Phe631 to interact with the 3,5-dimethoxylphenyl group and the 4-acrylamidobenzyl group in a “π-stacking sandwich” fashion. As a result of Phe631 being stabilized in a DFG-out conformation, FGFR4 adopts an inactive conformation upon binding with FIIN-2; this conformation has not been observed in any previously reported FGFR crystal structures (Fig. 3B) (59). This observation was surprising, because FIIN-2 was designed to be a type I inhibitor and does not possess the typical benzamide moiety that allows prototypical type II inhibitors, such as imatinib and ponatinib, to occupy the hydrophobic pocket created by the flip of the DFG motif that characterizes the inactive conformation (60). We also solved the FGFR4V550L/FIIN-3 cocrystal structure (PDB ID code 4R6V), which exhibited a conformation and binding mode very similar to that of FGFR4WT/FIIN-2. The pseudo six-membered ring next to the 4,6-pyrimidine core in FIIN-3 adopts a conformation almost identical to the bicyclic core of FIIN-2 (Fig. 3C). To elucidate the binding modes that enable FIIN-3 to be a dual inhibitor of both FGFR and EGFR, we solved the cocrystal structure of EGFR L858R kinase domain with FIIN-3 (PDB ID code 4R5S) (Fig. 3D). EGFR L858R is an oncogenic mutant that commonly is found in NSCLC and that is very similar structurally to WT EGFR (61). As expected, FIIN-3 forms a covalent bond to Cys797 of EGFR (Cys797 is the site of covalent modification for all the reported covalent EGFR inhibitors). As in FGFR family kinases, EGFR has an equivalently positioned Phe723 in the P-loop. However, this Phe723 does not partake in inhibitor binding, nor does the Phe856 of the DFG motif, and EGFR adopts a DFG-in conformation upon binding with FIIN-3. These observations indicate that the formation of a covalent bond with a cysteine residue in the P-loop is necessary for the formation of hydrophobic contacts between these phenylalanines and the drug. The chlorine of FIIN-3 is within hydrogen-bonding distance of Thr854 in EGFR (and of Ala629, at the same position, in FGFR); this interaction may explain why FIIN-3 showed stronger potency than FIIN-2 against EGFR. The 4-acrylamidobenzyl group of FIIN-3, which is longer than the 3-acrylamidophenyl substituents present in other reported EGFR covalent inhibitors (4, 62), provides the flexibility and proper distance for the covalent binding to distinctive cysteines of EGFR and FGFR.
Fig. 3.
(A and B) FIIN-2 (purple stick) covalently binds to Cys477 in the P-loop of FGFR4 (green ribbons) and results in the DFG-out conformation of FGFR4. (C) FIIN-3 (pink stick) binds to Cys477 of FGFR4 V550L (green ribbons) with a similar binding mode. (D) FIIN-3 binds to Cys797 of EGFR L858R (blue ribbons) covalently and in a DFG-in conformation.
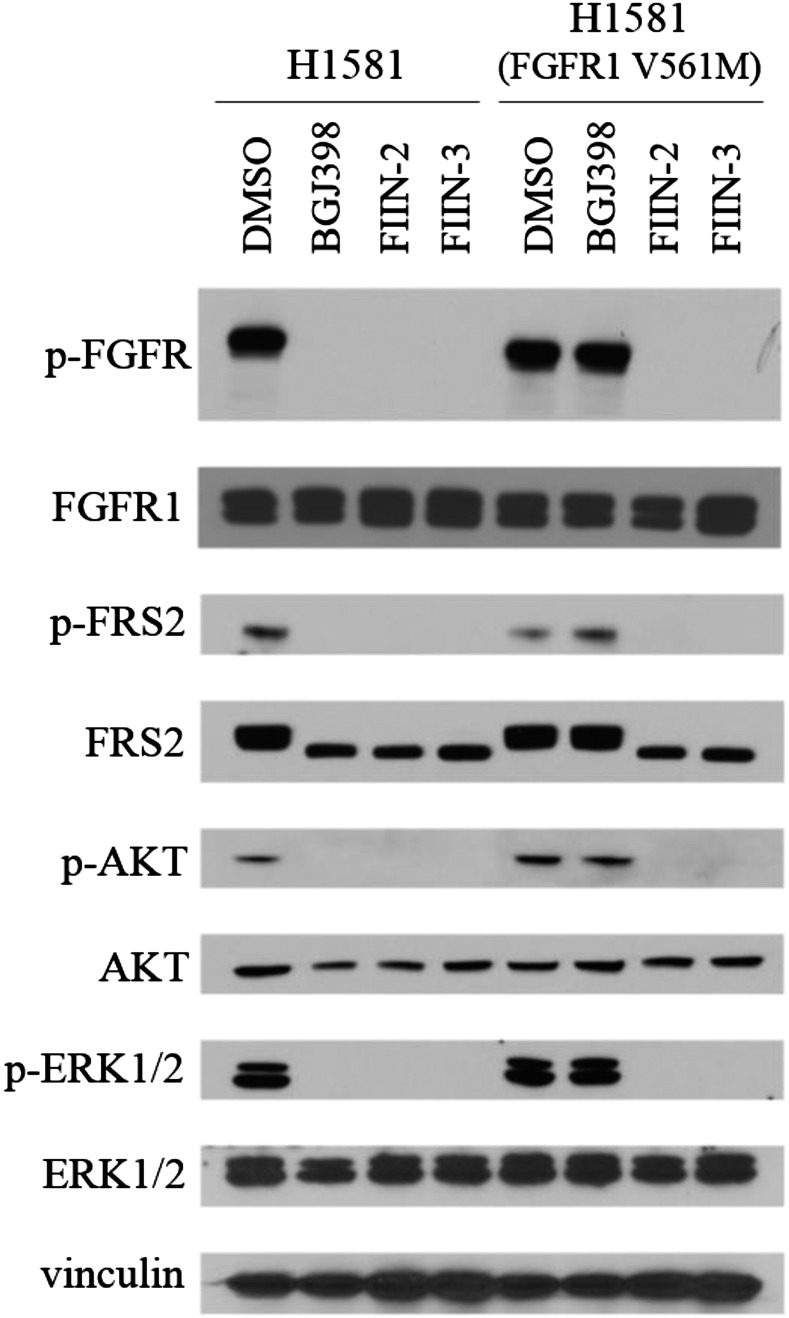
To evaluate the antiproliferative activity more broadly, FIIN-2, FIIN-3, and BGJ398 were profiled on several established cancer cell lines known to be dependent on FGFR signaling for survival (Table 2). As expected, all three inhibitors displayed similarly potent inhibition of cells, such as the RT112 bladder cancer cell line, that harbor the FGFR3/TACC3 fusion (39). To confirm that the resistance conferred by the gatekeeper mutation in FGFR2 also would be observed for the gatekeeper mutation in FGFR1 in the context of a cancer cell line, we generated FGFR1 V561M gatekeeper mutants in both H2077 and H1581 cells, two cell lines derived from a patient with a lung cancer with high-level FGFR1 amplification. These mutations caused a >50-fold shift in the EC50 of BGJ398, whereas FIIN-2 and FIIN-3 maintained good potency, with EC50s reduced less than 10-fold relative to WT. The downstream prosurvival signaling pathways of FGFR also were examined in these cell lines, showing that all three inhibitors effectively suppressed p-FRS2, p-FGFR, p-AKT, and p-ERK in these FGFR-activated cells at 1.0 µM, except that BGJ398 failed in the FGFR1 V561M H1581 cells (Fig. 4 and SI Appendix). In biochemical assays [SelectScreen; Life Technology (55)], FIIN-2 and FIIN-3 inhibited FGFR1 V561M with IC50s of 89 and 109 nM, respectively. In H1581 cells they also inhibited FGFR V561M in a dose-responsive manner, with both of them inhibiting most autophosphorylation of FGFR1 V561M at 333 nM, whereas BGJ398 still was inactive at 1.0 µM. (SI Appendix, Fig. S4). FIIN-2 and FIIN-3 also inhibited the proliferation of A2780 ovarian carcinoma cells, which were reported to be FGFR4-dependent (36). Their potency was at least 10-fold stronger than that of BGJ398 and at least 60-fold stronger than that of their respective noncovalent counterparts. Both FIIN-2 and FIIN-3 also were very potent against the 4T1 breast cancer cell line, which is reported to be pan-FGFR–dependent (63), being at least 15-fold more potent than their respective noncovalent counterparts (Table 2). We conclude that FIIN-2 and FIIN-3 show excellent antiproliferative activity in a variety of backgrounds, including cell lines that have gatekeeper mutations in FGFR1 and that are dependent on FGFR4.
Table 2.
Antiproliferative activity of FGFR inhibitors against various cancer cell lines
| Cell line | Genotype | Cancer type | EC50, nM | ||||
| BGJ398 | FIIN-2 | FIIN-3 | FRIN-2 | FRIN-3 | |||
| H2077 | NSCLC | 7 | 3.9 | 5.3 | |||
| H2077 (V561M) | FGFR1 amplification | Transgenic | >1,000 | 16.1 | 1.4 | ||
| H2077 (vector) | 3.5 | 3.9 | 2.4 | ||||
| H1581 | NSCLC | 10.5 | 4.8 | 2.5 | |||
| H1581 (V561M) | FGFR1 amplification | Transgenic | 507.5 | 23.6 | 11.8 | ||
| H1581 (vector) | 9 | 3.3 | 2.15 | ||||
| H520 | FGFR1 amplification | NSCLC | 121.2 | 109.3 | 98.9 | ||
| Kato III | FGFR2 amplification | Gastric carcinoma | 3.4 | 1.9 | 2.5 | ||
| AN3 CA | FGFR2 N549K activating mutation | Endometrial adenocarcinoma | 26.6 | 23.9 | 26.2 | ||
| RT112 | FGFR3/TACC3 fusion and FGFR3 amplification | Bladder carcinoma | 2.7 | 6.8 | 15.9 | ||
| A2780 | FGFR4 amplification | Ovarian carcinoma | 0.5 | 0.04 | 0.01 | 3.2 | 0.6 |
| 4T1 | FGFRs amplification | Breast carcinoma | 125 | 23 | 46 | 455 | 669 |
| SKOV-3 | FGFRs and EGFR amplification | Bladder carcinoma | 1,633 | 925 | 499 | 1,654 | 1,649 |
Fig. 4.
Inhibition of FGFR-dependent signaling by BGJ398, FIIN-2, and FIIN-3 in H1581 (FGFR1 WT or V561M) cells. Cells were treated with indicated inhibitors at 1.0 µM for 12 h and then were lysed and subjected to Western blot for the indicated proteins or phosphoproteins.
The ability to inhibit simultaneously both FGFR and EGFR kinase activity covalently while still maintaining good overall kinase selectivity is a unique property of FIIN-3. To validate the dual inhibitory activity of FIIN-3, we picked the SKOV-3 ovarian carcinoma cell line, which is reported to overexpress both EGFR and FGFR and whose proliferation could be inhibited only partially by selective FGFR or EGFR inhibitors (36, 64, 65). SKOV-3 cells were treated with FIIN-2, FIIN-3, and BGJ398 in the presence or absence of FGF or EGF ligands, and the growth response was assessed. Without any stimulation FIIN-3 inhibited proliferation of the SKOV-3 cells with an EC50 of 499 nM, whereas the EC50 of FIIN-2 was 925 nM. FRIN-2 and FRIN-3 showed potency comparable to that of BGJ398, with EC50s around 1.6 µM (Table 2). The addition of 10 ng/mL FGF1 increased the total cell number by 20–30%, but this increase was abolished by all three inhibitors at concentrations above 100 nM. The addition of 10 ng/mL EGF stimulated similar increases in cell number which were observable for all three inhibitors at all concentrations tested; FIIN-3 again was the most potent inhibitor (SI Appendix, Fig. S5). Next we evaluated the inhibitory effects of the three compounds at a concentration of 1.0 µM on the FGFR-dependent signaling pathway with or without exogenous FGF1 stimulation (SI Appendix, Fig. S6). Consistent with the biochemical and cellular evaluation of EGFR and FGFR activities, FIIN-3 was uniquely capable of inhibiting phosphorylation of both EGFR and FGFR regardless of FGF1 stimulation. In the absence of FGF1, all three compounds were capable of inhibiting the phosphorylation of the downstream effectors FRS2, AKT, and ERK1/2; however, after FGF1 stimulation, p-AKT and p-ERK1/2 were significantly restored for BGJ398, partially restored for FIIN-2, and were not restored for FIIN-3. Cumulatively these results show that only FIIN-3 is an effective cellular inhibitor of both EGFR and FGFR signaling in SKOV-3 cells, whereas FIIN-2 might partially inhibit EGFR at 1.0 µM.
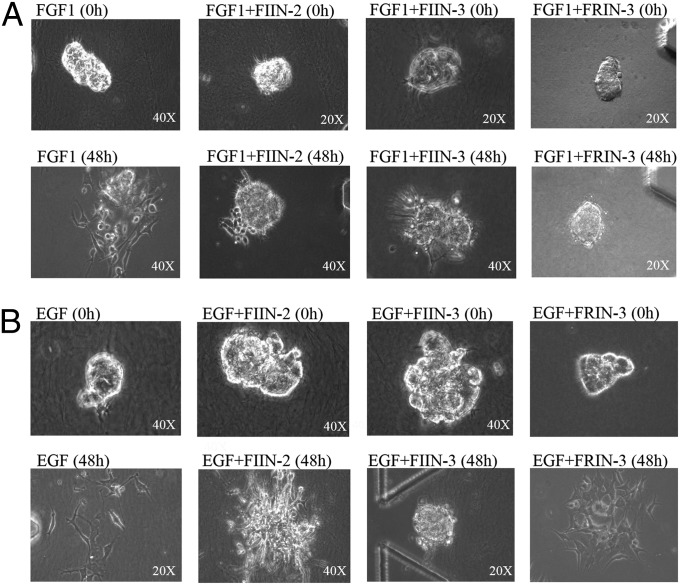
FIIN-2 and FIIN-3 also were evaluated using 3D dispersion assays in a microfluidic device. Compared with conventional 2D assays The 3D assay creates a 3D microenvironment that allows better evaluation of drugs that inhibit cell migration or epithelial–mesenchymal transition (66). SKOV-3 spheroids were suspended in the gel region of the microfluidic system and cultured in the presence or absence of FGF or EGF (20 ng/mL). SKOV-3 spheroids did not dissociate within a 48-h period in the absence of a high concentration of growth factors (SI Appendix, Fig. S7A); however, many cells had dissociated and spread by 48 h in the presence of FGF1 (20 ng/mL). FIIN-2, FIIN-3, and FRIN-3 at 1.0 µM almost fully suppressed this dissociation up to 48 h after treatment (Fig. 5A). FIIN-3, but not FIIN-2 or FRIN-3 at 1.0 µM, could also block most dissociation of SKOV-3 spheroids in the presence of EGF (Fig. 5B). The numbers of dispersed cells under each condition were used to compute percentages of inhibition (SI Appendix, Fig. S7). The results of the 3D assay support the conclusion from 2D cultures that, although both FIIN-2 and FIIN-3 can inhibit FGFR signaling, only FIIN-3 is an effective inhibitor of EGF-induced signaling.
Fig. 5.
Effects of FIIN-2, FIIN-3, and FRIN-3 on FGF- and EGF-induced dispersion of SKOV-3 cells in a 3D microfluidic device. Images show representative spheroids of SKOV-3 cells after indicated treatment with FGF (A) or EGF (B), in the presence or absence of the FIIN-2 and FIIN-3. Objective magnification power is indicated in the lower right corner. (A) Phase-contrast images of SKOV-3 spheroids at the indicated times after the addition of FGF. In contrast to the cell dispersal observed in control-treated devices, treatment with 1.0 μM of FIIN-2, FIIN-3, or FRIN-2 inhibited FGF1-induced dispersion of SKOV-3 cells. (B) Phase-contrast images of the SKOV-3 spheroids induced to disperse with EGF and subjected to control, FIIN-2, or FIIN-3 treatment. At 1.0 µM FIIN-3, but not FIIN-2, fully inhibited the EGF-induced dispersion of SKOV-3 cells.
Finally, the in vivo efficacy of FIIN-2 and FIIN-3 was examined using a zebrafish developmental model. Treatment of fish in the embryonic state with either FIIN-2 or FIIN-3 caused defects to the posterior mesoderm similar to the phenotypes reported following genetic knockdown of FGFR or treatment with other reported FGFR inhibitors (9, 67). FIIN-2 and FIIN-3 caused mild or severe phenotypes to the tail morphogenesis in all treated embryonic zebrafish. The efficiencies were lower than that of BGJ398 but higher than that of AZD4547 and PD173074 (SI Appendix, Fig. S8).
Discussion
Based on the structure of the first (to our knowledge) covalent FGFR inhibitor, FIIN-1, we developed a second generation of FGFR inhibitors exemplified by FIIN-2 and FIIN-3. These two inhibitors maintained the ability to form a covalent bond with a conserved cysteine residue in the P-loop of all FGFR kinases while improving the affinity for WT and mutant forms of FGFR1–4, including gatekeeper mutants. Mutation of the reactive cysteine residue, washout, and biotin probe-labeling experiments demonstrated that the ability of FIIN-2 and FIIN-3 to inhibit FGFR gatekeeper mutants requires the formation of a covalent bond with the P-loop cysteine residue, and this finding was validated further by the WT and gatekeeper-mutated FGFR4 cocrystal structures. Surprisingly, these cocrystal structures also demonstrated a unique mode of inducing the DFG-out flip without conforming to the pharmacophore typically used to achieve this binding mode (60). Comparison with the EGFRL858R/FIIN-3 cocrystal structure demonstrated how the 4-acrylamidobenzyl moiety of FIIN-3 provides the flexibility and spacing required to target spatially distinct cysteines in FGFR and EGFR.
Both inhibitors exhibited excellent potency against typical cancer cell lines harboring FGFR amplifications or mutations, including the FGFR4-dependent cell line A2780, which is resistant to many current FGFR inhibitors. FGFR4 has been reported to play a very important role in metastasis, drug resistance, and poor prognosis (23, 68–70); therefore FIIN-2 and FIIN-3, with good FGFR4 potency, show promising potential for application in many FGFR-dependent cancer types such as breast cancer (63, 71) and hepatocellular carcinoma (72, 73). In addition, they are capable of overcoming the valine-to-methionine gatekeeper mutation in H2077 and H1581 cell lines, although similar mutations found in patients’ specimens have been demonstrated experimentally to confer resistance to the leading clinical FGFR inhibitors (19, 48, 50).
FGFR and EGFR both signal primarily through the PI3K/AKT/mTOR and RAS/MAPK networks; therefore compensation from either receptor is possible (74–76). Both EGFR and FGFR have been reported to be overexpressed and likely contributors in ovarian cancer (36, 64, 65), and FGF3/EGFR co-overexpression also was reported in NSCLC (77). In addition, the activation of FGFR autocrine pathways were found to be individually responsible for acquired resistance to gefitinib (an EGFR inhibitor) in NSCLC, and combination treatments such as PD173074 and gefitinib were required to restore effective growth inhibition (78, 79). Likewise, EGFR/ErbBs recently were reported to limit the sensitivity to FGFR inhibition in FGFR3-mutated or -translocated cell lines such as RT112 (FGFR3/TACC3 fusion) and MGHU3 (Y375C) (80, 81), and the PD173074 and gefitinib combination displayed an obvious synergistic effect (80). It was reported that p-EGFR level was much higher in 4T1 tumors than in cell cultures, suggesting potential crosstalk with exogenous growth factors and cytokines in the in vivo tumor environment and that combinatorial targeting of FGFR and EGFR might be advantageous (38, 82). Although combination therapy with selective inhibitors is possible when available, rationally designed dual and multitargeted inhibitors have the potential advantage of possessing the desired polypharmacology engineered into a single compound, thereby avoiding potential drug–drug interactions that can arise with combination treatment (83). Dual covalent inhibitors of EGFR and VEGFR have been designed previously, but these compounds possessed two electrophilic groups. In contrast, FIIN-3 exploits a single acrylamide group that is capable of accessing two spatially distinct cysteine residues in EGFR and FGFR (84). FIIN-3 presents the potential advantage of circumventing resistance that could arise from either FGFR- or EGFR-induced bypass signaling. As a dual FGFR and EGFR inhibitor, FIIN-3 was more potent than FIIN-2 and BGJ398 in inhibiting the AKT and ERK1/2 phosphorylation and proliferation of SKOV-3 cells, especially when stimulated with exogenous growth factors such as FGF and EGF, which are present in vivo. We used a newly developed 3D assay to observe the effect of FIIN-2 and FIIN-3 on migration of SKOV-3 cells when stimulated with growth factors and further confirmed FIIN-3 as an effective dual inhibitor. There are great opportunities for developing dual inhibitors with even better EGFR potency based on the cocrystal of EGFR with FIIN-3; for example, the side chain of Glu762 in EGFR is located very close to the 2,6-dichloro-3,5-dimethoxyphenyl group of FIIN-3, providing the possibility of H-bond interaction after proper modification. We anticipate that FIIN-2 and FIIN-3 will serve as prototype covalent FGFR inhibitors that will enable further preclinical validation and will inspire the development of next-generation FGFR-directed therapy.
Materials and Methods
Cloning Full-Length TEL-FGFR2.
Full-length TEL oncogene (TEL)-FGFR2 cDNA was constructed by RT-PCR amplifying the N-terminal fragment of TEL containing a unique ApaLI restriction site and the C-terminal fragment of FGFR2, using a forward primer containing the complementary TEL ApaLI region. PCR sequences used to generate the overlapping fragments are
TEL-F: 5′-ATACGAAGTTATCAGTCGACATGTCTGAGACTCCTGCTCAGT-3′
TEL-R: 5′-ATTTGTCGTGATAGGTGACCTGGA-3′
FGFR2-F: 5′-GGATAATGTGCACCATAACCCTGTTTCGGCTGAGTCCAGCTC-3′
FGFR2-R: 5′- ACGAATGGTCTAGAAAGCTTTCATGTTTTAACACTGCCGTTTATG-3′
The fusion DNA was inserted in a pDNR-Dual vector (BD Biosciences) using SalI/HindIII sites and was recombined into the JP1520 retroviral vector as previously described (4). Full-length cDNA was confirmed by sequencing.
Ba/F3 Cell-Viability Assays.
TEL-FGFR2–transformed Ba/F3 cells were seeded in a 96-well plate and were treated with each concentration of the compounds. After 72 h the cells were assessed by MTS tetrazolium assay. The IC50 values were calculated using GraphPad Prism version 5.0 (GraphPad Software Inc.). To generate FGFR2 mutants, V564M, C491A, or C491A/V564M were introduced into Tel-FGFR2 WT cells using site-directed mutagenesis (Agilent) followed by introduction into Ba/F3 cells using retroviral infection. For other mutants, Ba/F3 cells expressing TEL-FGFR2 WT were exposed to 50 μg/mL N-ethyl-N-nitrosourea (Sigma Aldrich) for 24 h, followed by culture in 96-well plates in the presence of FIIN-1 (0.1 μM). Resistant clones were sequenced using RT-PCR for secondary FGFR2 mutations. M538I, E565K, V564F, or K659N mutations were detected in the drug-resistant cells. Each mutation was introduced into TEL-FGFR2 WT cells using site-directed mutagenesis and then was introduced into Ba/F3 cells using retroviral infection. The EGFR- and RET-transformed Ba/F3 cells were generated as previously described (4, 85). BTK, KIT, FLT1/4 transformed Ba/F3 cells were from Carna Biosciences.
Immunoblotting Analysis and Washout Experiment.
Cells grown under the previously specified conditions were lysed in lysis buffer (Cell Signaling Technology). For washout experiments, cells were incubated with FGFR inhibitors for 3 h, washed with PBS three times, maintained in Roswell Park Memorial Institute (RPMI) medium with 10% (vol/vol) FBS for 4 h, and then harvested. The resulting lysates were analyzed for immunoblotting or immunoprecipitation. Western blotting analyses were conducted after separation by SDS/PAGE electrophoresis and transfer to nitrocellulose membranes. Immunoblotting was conducted according to the antibody manufacturer’s recommendations (Fig. 2 and SI Appendix). For immunoprecipitation, the lysates were treated with FIIN-1-biotin (5.0 µM) for 1 h and were immunoprecipitated with anti-FGFR2 antibody (SI Appendix, Fig. S3). Anti–p-FGFR (Tyr653/654), anti–p-FRS2 (Tyr436), anti–p-Akt (Ser473), anti-AKT, and anti–α-tubulin were obtained from Cell Signaling Technology; anti–p-ERK1/2 (pT185/pY187) and anti-ERK1/2 were purchased from Invitrogen; anti-FGFR2 and anti-FRS2 were from Santa Cruz.
FGFR4-FIIN-2 and FGFR4 V550L-FIIN-3 Crystallization and Structure Determination.
FGFR constructs spanning residues L445–E753 and bearing the V550L mutation were prepared, and the protein was expressed and purified as previously reported (86). All crystals were grown by the hanging-drop vapor diffusion method at 18 °C. Purified FGFR4KWT and FGFR4KV550L protein were concentrated to ∼20 mg/mL using Centricon-10 (Millipore). To generate cocrystals, kinases and inhibitors were mixed at a molar ratio of 1:1.2 and were incubated at 4 °C overnight to allow the formation of covalent bonds between the compound and cysteine 477 in the kinase. FGFR4KWT/FIIN-2, FGFR4KV550L/FIIN-2, and FGFR4KV550L/FIIN-3 complexes were crystallized using crystallization buffer composed of 0.1 M Hepes (pH 7.5), ∼1.0–1.2 M (NH4)2SO4, and 10 mM Yttrium (III) chloride hexahydrate. Crystals grew in about 7–15 d at 18 °C, were stabilized in mother liquor by increasing the glycerol concentration stepwise to 25% (vol/vol), and then were flash-frozen in liquid nitrogen. Diffraction data were collected at Beamline X-4C at the National Synchrotron Light Source, Brookhaven National Laboratory, Upton, NY. All diffraction data were processed using the HKL2000 suite (87). All crystal structures were solved using the maximum likelihood molecular replacement program Phaser in the PHENIX software suite (88). The crystal structure of WT FGFR2 kinase (PDB ID code 2PSQ) (89) was used as the search model. The A-loop, the b2–b3 loop, and the kinase insert region were removed from the search model. Model building was carried out using Coot (90), and refinements were done using phenix.refine in the PHENIX suite (88). Data collection and structure refinement statistics are listed in SI Appendix. Atomic superimpositions were performed using the lsqkab program (91) in the CCP4 suite (92), and structural representations were prepared using PyMOL (93).
EGFR L858R-FIIN-3 Crystallization and Structure Determination.
The EGFR construct spanning residues 696–1022 and bearing the L858R mutation was prepared, and the protein was expressed and purified as previously reported (61). The apo-EGFR 696–1022 L858R crystals were prepared in 40% (vol/vol) PEG400, 150 mM NaCl, 0.1 M Hepes (pH 8.0), 5 mM Tris(2-carboxyethyl)phosphine, and 0.1 M nondetergent sulfobetaine (NDSB)-211. The compound was incorporated by soaking the crystals in the crystallization reservoir solution supplemented with 0.5 mM FIIN-3 for 4–6 h, and then the complex crystals were flash-frozen in the same solution, which served as a cryo-protectant. The diffraction data were collected at The Advanced Photon Source (APS) ID19 at 100 K and were processed using the HKL-3000 program (87). The structure was solved by the difference Fourier method using the previously reported EGFRL858R/AMP-PnP structure (PDB ID code 2ITV) (61) with PHENIX software (88) and then was refined using Coot (90) and PHENIX. The inhibitor was modeled into the closely fitting positive Fo-Fc electron density and then was included in following refinement and fitting cycles. Topology and parameter files for the inhibitors were generated using PRODRG (47). Data collection and structure refinement statistics are listed in SI Appendix.
Cancer Cell Proliferation Assays and Immunoblotting Analysis.
NCI-H2077, NCI-H1581, H520, Kato III, AN3CA, RT112, A2780, 4T1, and SKOV-3 cells were treated with inhibitors 1 d after being plated at a density of 1,500 cells per well in 96-well plates. The gatekeeper mutation cell lines were generated by ectopically overexpressing FGFR1 V561M in either NCI-H2077 or NCI-H1581 cells via lentiviral transduction. Cell survival was assessed at 96 h following the addition of inhibitor using the Cell-Titer-Glo reagent (Promega) according to the manufacturer’s instructions. EC50 values were calculated using GraphPad Prism 5 (GraphPad Software) (Table 2). SKOV-3 cells also were treated in the presence of FGF or EGF ligand. Proliferation measurements were made after 96 h using a luminometer. Data are shown as relative values: The luminescence of cells with indicated inhibitor dose is compared with that of untreated cells (SI Appendix, Fig. S5). For immunoblotting analysis, H1581, H1581 (FGFR1 V561M), KATO III, RT112, and SKOV3 cells (1 million cells per well) were seeded and serum-starved for 12 h with either DMSO or the indicated doses of inhibitors. After 12 h of pretreatment, SKOV3 cells were treated with FGF ligands at 10 ng/mL for another 15 min or were left untreated; then all cells were lysed in RIPA. Equal amounts of protein were analyzed by SDS/PAGE (Fig. 4 and SI Appendix). Primary antibodies used were as follows: p-FRS2-α (Tyr436, #3861S, and Tyr196, #3864), p-EGFR (Y1068, 1H12, #2236S), AKT (#9272S), p-AKT (Ser473, #4060S), p-ERK1/2 (T202/Y204, #4370S), and ERK1/2 (#4695S) were from Cell Signaling Technologies. FRS2 (H-91, sc-8318) was from Santa Cruz. EGFR (#A300-388A) was from Bethyl Antibodies.
3D Dispersion Assays.
For 3D dispersion assays, SKOV-3 cells were allowed to grow in spheroids by resuspending cells at low density (2,000–4,000 cells∕mL) and were cultured for 10–14 d in ultra-low-attachment dishes (Corning Inc.). Spheroids of 40- to 70-μm diameter were selected with sieves as reported (66). Spheroids were seeded in the central region of a microfluidic device using standard soft lithography techniques (94, 95). The central region of the device is flanked by two channels. For our assays, the channel surfaces were coated with poly-d-lysine and dried and then were loaded at low pressure with SKOV-3 spheroids suspended in 2.5 mg∕mL type I collagen (30–50 spheroids∕200 μL). After gel polymerization, medium with or without FGF or EGF and with or without FIIN-2 or FIIN-3 was added to the channels flanking the gel region, and the devices were incubated in a humid environment at 37 °C with 5% CO2. Images of the spheroids were captured on an Olympus CKX41 microscope equipped with a QIClick camera (QImaging).
Zebrafish Embryo Study.
WT Danio rerio (zebrafish) Tübingen/AB strain embryos were collected from male–female crosses and were incubated at 28 °C. At 2 h postfertilization (hpf), 15 embryos were placed in each well of a 24-well plate in 1 mL of E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4). Vehicle (DMSO) control and stock solutions of FIIN compounds or known FGFR inhibitors were added to the wells at a final concentration of 25 μM, except for NVP-BGJ398, which was tested at 5.0 μM. The treated embryos were incubated at 28 °C until 50 hpf, when the phenotype of abnormal posterior mesoderm was scored. Results shown were combined from two independent experiments. Images were captured using Leica Wild M10 dissecting microscope and a SPOT Insight camera.
Supplementary Material
Acknowledgments
We thank Liping Wang for technical assistance and Dr. Sara Buhrlage for proofreading. This work was supported by Lung Specialized Programs of Research Excellence Grant 5 P50 CA090578-10 (to P.A.J. and N.S.G.) and National Institute of Dental and Craniofacial Research Grant R01 DE13686 (to M.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.P. is a guest editor invited by the Editorial Board.
Data deposition: Crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4QQC, 4R6V, and 4R5S).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403438111/-/DCSupplemental.
References
- 1.Breccia M, Alimena G. Nilotinib: A second-generation tyrosine kinase inhibitor for chronic myeloid leukemia. Leuk Res. 2010;34(2):129–134. doi: 10.1016/j.leukres.2009.08.031. [DOI] [PubMed] [Google Scholar]
- 2.O’Hare T, et al. Inhibition of wild-type and mutant Bcr-Abl by AP23464, a potent ATP-based oncogenic protein kinase inhibitor: Implications for CML. Blood. 2004;104(8):2532–2539. doi: 10.1182/blood-2004-05-1851. [DOI] [PubMed] [Google Scholar]
- 3.Solca F, et al. Target binding properties and cellular activity of afatinib (BIBW 2992), an irreversible ErbB family blocker. J Pharmacol Exp Ther. 2012;343(2):342–350. doi: 10.1124/jpet.112.197756. [DOI] [PubMed] [Google Scholar]
- 4.Zhou W, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462(7276):1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tiong KH, Mah LY, Leong CO. Functional roles of fibroblast growth factor receptors (FGFRs) signaling in human cancers. Apoptosis. 2013;18(12):1447–1468. doi: 10.1007/s10495-013-0886-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks AN, Kilgour E, Smith PD. Molecular pathways: Fibroblast growth factor signaling: A new therapeutic opportunity in cancer. Clin Cancer Res. 2012;18(7):1855–1862. doi: 10.1158/1078-0432.CCR-11-0699. [DOI] [PubMed] [Google Scholar]
- 7.Richette P, Bardin T, Stheneur C. Achondroplasia: From genotype to phenotype. Joint Bone Spine. 2008;75(2):125–130. doi: 10.1016/j.jbspin.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Harding MJ, Nechiporuk AV. Fgfr-Ras-MAPK signaling is required for apical constriction via apical positioning of Rho-associated kinase during mechanosensory organ formation. Development. 2012;139(17):3130–3135. doi: 10.1242/dev.082271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ota S, Tonou-Fujimori N, Yamasu K. The roles of the FGF signal in zebrafish embryos analyzed using constitutive activation and dominant-negative suppression of different FGF receptors. Mech Dev. 2009;126(1-2):1–17. doi: 10.1016/j.mod.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 10.Böttcher RT, Niehrs C. Fibroblast growth factor signaling during early vertebrate development. Endocr Rev. 2005;26(1):63–77. doi: 10.1210/er.2003-0040. [DOI] [PubMed] [Google Scholar]
- 11.Dieci MV, Arnedos M, Andre F, Soria JC. Fibroblast growth factor receptor inhibitors as a cancer treatment: From a biologic rationale to medical perspectives. Cancer Discov. 2013;3(3):264–279. doi: 10.1158/2159-8290.CD-12-0362. [DOI] [PubMed] [Google Scholar]
- 12.Etienne A, et al. Combined translocation with ZNF198-FGFR1 gene fusion and deletion of potential tumor suppressors in a myeloproliferative disorder. Cancer Genet Cytogenet. 2007;173(2):154–158. doi: 10.1016/j.cancergencyto.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Singh D, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337(6099):1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y, et al. Integrative and comparative genomic analysis of lung squamous cell carcinomas in East Asian patients. J Clin Oncol. 2014;32(2):121–128. doi: 10.1200/JCO.2013.50.8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu YM, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3(6):636–647. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majewski IJ, et al. Identification of recurrent FGFR3 fusion genes in lung cancer through kinome-centred RNA sequencing. J Pathol. 2013;230(3):270–276. doi: 10.1002/path.4209. [DOI] [PubMed] [Google Scholar]
- 17.Liao RG, et al. Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung squamous cell carcinoma. Cancer Res. 2013;73(16):5195–5205. doi: 10.1158/0008-5472.CAN-12-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gatius S, et al. FGFR2 alterations in endometrial carcinoma. Mod Pathol. 2011;24(11):1500–1510. doi: 10.1038/modpathol.2011.110. [DOI] [PubMed] [Google Scholar]
- 19.Byron SA, et al. The N550K/H mutations in FGFR2 confer differential resistance to PD173074, dovitinib, and ponatinib ATP-competitive inhibitors. Neoplasia. 2013;15(8):975–988. doi: 10.1593/neo.121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roidl A, et al. The FGFR4 Y367C mutant is a dominant oncogene in MDA-MB453 breast cancer cells. Oncogene. 2010;29(10):1543–1552. doi: 10.1038/onc.2009.432. [DOI] [PubMed] [Google Scholar]
- 21.Shen YY, et al. Fibroblast growth factor receptor 4 Gly388Arg polymorphism in Chinese gastric cancer patients. World J Gastroenterol. 2013;19(28):4568–4575. doi: 10.3748/wjg.v19.i28.4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ezzat S, et al. The cancer-associated FGFR4-G388R polymorphism enhances pancreatic insulin secretion and modifies the risk of diabetes. Cell Metab. 2013;17(6):929–940. doi: 10.1016/j.cmet.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Taylor JG, 6th, et al. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest. 2009;119(11):3395–3407. doi: 10.1172/JCI39703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh M, Nakagama H. FGF receptors: Cancer biology and therapeutics. Med Res Rev. 2014;34(2):280–300. doi: 10.1002/med.21288. [DOI] [PubMed] [Google Scholar]
- 25.Weiss J, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2(62):62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsimafeyeu I, Demidov L, Stepanova E, Wynn N, Ta H. Overexpression of fibroblast growth factor receptors FGFR1 and FGFR2 in renal cell carcinoma. Scand J Urol Nephrol. 2011;45(3):190–195. doi: 10.3109/00365599.2011.552436. [DOI] [PubMed] [Google Scholar]
- 27.Sato T, et al. Overexpression of the fibroblast growth factor receptor-1 gene correlates with liver metastasis in colorectal cancer. Oncol Rep. 2009;21(1):211–216. [PubMed] [Google Scholar]
- 28.Cottoni F, et al. Overexpression of the fibroblast growth factor receptor 2-IIIc in Kaposi’s sarcoma. J Dermatol Sci. 2009;53(1):65–68. doi: 10.1016/j.jdermsci.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 29.Matsuda Y, Hagio M, Seya T, Ishiwata T. Fibroblast growth factor receptor 2 IIIc as a therapeutic target for colorectal cancer cells. Mol Cancer Ther. 2012;11(9):2010–2020. doi: 10.1158/1535-7163.MCT-12-0243. [DOI] [PubMed] [Google Scholar]
- 30.Gómez-Román JJ, et al. Fibroblast growth factor receptor 3 is overexpressed in urinary tract carcinomas and modulates the neoplastic cell growth. Clin Cancer Res. 2005;11(2 Pt 1):459–465. [PubMed] [Google Scholar]
- 31.Henson BJ, Gollin SM. Overexpression of KLF13 and FGFR3 in oral cancer cells. Cytogenet Genome Res. 2010;128(4):192–198. doi: 10.1159/000308303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalff A, Spencer A. The t(4;14) translocation and FGFR3 overexpression in multiple myeloma: Prognostic implications and current clinical strategies. Blood Cancer J. 2012;2:e89. doi: 10.1038/bcj.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho HK, et al. Fibroblast growth factor receptor 4 regulates proliferation, anti-apoptosis and alpha-fetoprotein secretion during hepatocellular carcinoma progression and represents a potential target for therapeutic intervention. J Hepatol. 2009;50(1):118–127. doi: 10.1016/j.jhep.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 34.Motoda N, et al. Overexpression of fibroblast growth factor receptor 4 in high-grade pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Int J Oncol. 2011;38(1):133–143. [PubMed] [Google Scholar]
- 35.Poh W, et al. Klotho-beta overexpression as a novel target for suppressing proliferation and fibroblast growth factor receptor-4 signaling in hepatocellular carcinoma. Mol Cancer. 2012;11(14):1–10. doi: 10.1186/1476-4598-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaid TM, et al. Identification of FGFR4 as a potential therapeutic target for advanced-stage, high-grade serous ovarian cancer. Clin Cancer Res. 2013;19(4):809–820. doi: 10.1158/1078-0432.CCR-12-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye YW, et al. Fibroblast growth factor receptor 4 regulates proliferation and antiapoptosis during gastric cancer progression. Cancer. 2011;117(23):5304–5313. doi: 10.1002/cncr.26207. [DOI] [PubMed] [Google Scholar]
- 38.Lee HJ, et al. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell. 2014;26(2):207–221. doi: 10.1016/j.ccr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 39.Guagnano V, et al. Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. J Med Chem. 2011;54(20):7066–7083. doi: 10.1021/jm2006222. [DOI] [PubMed] [Google Scholar]
- 40.Wasag B, Lierman E, Meeus P, Cools J, Vandenberghe P. The kinase inhibitor TKI258 is active against the novel CUX1-FGFR1 fusion detected in a patient with T-lymphoblastic leukemia/lymphoma and t(7;8)(q22;p11) Haematologica. 2011;96(6):922–926. doi: 10.3324/haematol.2010.036558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao G, et al. A novel, selective inhibitor of fibroblast growth factor receptors that shows a potent broad spectrum of antitumor activity in several tumor xenograft models. Mol Cancer Ther. 2011;10(11):2200–2210. doi: 10.1158/1535-7163.MCT-11-0306. [DOI] [PubMed] [Google Scholar]
- 42.Gavine PR, et al. AZD4547: An orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 2012;72(8):2045–2056. doi: 10.1158/0008-5472.CAN-11-3034. [DOI] [PubMed] [Google Scholar]
- 43.Zhou W, et al. A structure-guided approach to creating covalent FGFR inhibitors. Chem Biol. 2010;17(3):285–295. doi: 10.1016/j.chembiol.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bixby D, Talpaz M. Mechanisms of resistance to tyrosine kinase inhibitors in chronic myeloid leukemia and recent therapeutic strategies to overcome resistance. Hematology (Am Soc Hematol Educ Program) 2009;2009:461–476. doi: 10.1182/asheducation-2009.1.461. [DOI] [PubMed] [Google Scholar]
- 45.Weisberg E, et al. Discovery of a small-molecule type II inhibitor of wild-type and gatekeeper mutants of BCR-ABL, PDGFRalpha, Kit, and Src kinases: Novel type II inhibitor of gatekeeper mutants. Blood. 2010;115(21):4206–4216. doi: 10.1182/blood-2009-11-251751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yun CH, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci USA. 2008;105(6):2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schüttelkopf AW, van Aalten DM. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 8):1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 48.Chell V, et al. Tumour cell responses to new fibroblast growth factor receptor tyrosine kinase inhibitors and identification of a gatekeeper mutation in FGFR3 as a mechanism of acquired resistance. Oncogene. 2013;32(25):3059–3070. doi: 10.1038/onc.2012.319. [DOI] [PubMed] [Google Scholar]
- 49.Shukla N, et al. Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clin Cancer Res. 2012;18(3):748–757. doi: 10.1158/1078-0432.CCR-11-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ang D, et al. Novel mutations in neuroendocrine carcinoma of the breast: Possible therapeutic targets. Diagn Mol Pathol. 2014 doi: 10.1097/PDM.0b013e3182a40fd1. [DOI] [PubMed] [Google Scholar]
- 51.Kammasud N, et al. Novel inhibitor for fibroblast growth factor receptor tyrosine kinase. Bioorg Med Chem Lett. 2007;17(17):4812–4818. doi: 10.1016/j.bmcl.2007.06.058. [DOI] [PubMed] [Google Scholar]
- 52.Jansma A, et al. Verification of a designed intramolecular hydrogen bond in a drug scaffold by nuclear magnetic resonance spectroscopy. J Med Chem. 2007;50(24):5875–5877. doi: 10.1021/jm700983a. [DOI] [PubMed] [Google Scholar]
- 53.Goldstein DM, Gray NS, Zarrinkar PP. High-throughput kinase profiling as a platform for drug discovery. Nat Rev Drug Discov. 2008;7(5):391–397. doi: 10.1038/nrd2541. [DOI] [PubMed] [Google Scholar]
- 54.Miduturu CV, et al. High-throughput kinase profiling: A more efficient approach toward the discovery of new kinase inhibitors. Chem Biol. 2011;18(7):868–879. doi: 10.1016/j.chembiol.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lebakken CS, et al. Development and applications of a broad-coverage, TR-FRET-based kinase binding assay platform. J Biomol Screen. 2009;14(8):924–935. doi: 10.1177/1087057109339207. [DOI] [PubMed] [Google Scholar]
- 56.Warmuth M, Kim S, Gu XJ, Xia G, Adrián F. Ba/F3 cells and their use in kinase drug discovery. Curr Opin Oncol. 2007;19(1):55–60. doi: 10.1097/CCO.0b013e328011a25f. [DOI] [PubMed] [Google Scholar]
- 57.Reintjes N, et al. Activating somatic FGFR2 mutations in breast cancer. PLoS ONE. 2013;8(3):e60264. doi: 10.1371/journal.pone.0060264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li D, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008;27(34):4702–4711. doi: 10.1038/onc.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leproult E, Barluenga S, Moras D, Wurtz JM, Winssinger N. Cysteine mapping in conformationally distinct kinase nucleotide binding sites: Application to the design of selective covalent inhibitors. J Med Chem. 2011;54(5):1347–1355. doi: 10.1021/jm101396q. [DOI] [PubMed] [Google Scholar]
- 60.Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat Chem Biol. 2006;2(7):358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- 61.Yun CH, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11(3):217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang S, et al. Design, synthesis, and biological evaluation of novel conformationally constrained inhibitors targeting epidermal growth factor receptor threonine790 → methionine790 mutant. J Med Chem. 2012;55(6):2711–2723. doi: 10.1021/jm201591k. [DOI] [PubMed] [Google Scholar]
- 63.Dey JH, et al. Targeting fibroblast growth factor receptors blocks PI3K/AKT signaling, induces apoptosis, and impairs mammary tumor outgrowth and metastasis. Cancer Res. 2010;70(10):4151–4162. doi: 10.1158/0008-5472.CAN-09-4479. [DOI] [PubMed] [Google Scholar]
- 64.Cole C, et al. Inhibition of FGFR2 and FGFR1 increases cisplatin sensitivity in ovarian cancer. Cancer Biol Ther. 2010;10(5):495–504. doi: 10.4161/cbt.10.5.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sewell JM, Macleod KG, Ritchie A, Smyth JF, Langdon SP. Targeting the EGF receptor in ovarian cancer with the tyrosine kinase inhibitor ZD 1839 (“Iressa”) Br J Cancer. 2002;86(3):456–462. doi: 10.1038/sj.bjc.6600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aref AR, et al. Screening therapeutic EMT blocking agents in a three-dimensional microenvironment. Integr Biol (Camb) 2013;5(2):381–389. doi: 10.1039/c2ib20209c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Itoh N. The Fgf families in humans, mice, and zebrafish: Their evolutional processes and roles in development, metabolism, and disease. Biol Pharm Bull. 2007;30(10):1819–1825. doi: 10.1248/bpb.30.1819. [DOI] [PubMed] [Google Scholar]
- 68.Turkington RC, et al. Fibroblast growth factor receptor 4 (FGFR4): A targetable regulator of drug resistance in colorectal cancer. Cell Death Dis. 2014;5:e1046. doi: 10.1038/cddis.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miura S, et al. Fibroblast growth factor 19 expression correlates with tumor progression and poorer prognosis of hepatocellular carcinoma. BMC Cancer. 2012;12(56):1–15. doi: 10.1186/1471-2407-12-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peláez-García A, et al. FGFR4 role in epithelial-mesenchymal transition and its therapeutic value in colorectal cancer. PLoS ONE. 2013;8(5):e63695. doi: 10.1371/journal.pone.0063695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Agarwal D, et al. kConFab Investigators Australian Ovarian Cancer Study Group GENICA Network TNBCC FGF receptor genes and breast cancer susceptibility: Results from the Breast Cancer Association Consortium. Br J Cancer. 2014;110(4):1088–1100. doi: 10.1038/bjc.2013.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheng AL, Shen YC, Zhu AX. Targeting fibroblast growth factor receptor signaling in hepatocellular carcinoma. Oncology. 2011;81(5-6):372–380. doi: 10.1159/000335472. [DOI] [PubMed] [Google Scholar]
- 73.French DM, et al. Targeting FGFR4 inhibits hepatocellular carcinoma in preclinical mouse models. PLoS ONE. 2012;7(5):e36713. doi: 10.1371/journal.pone.0036713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ito F. Foreword. Target therapy for cancer: Anti-cancer drugs targeting growth-factor signaling molecules. Biol Pharm Bull. 2011;34(12):1773. doi: 10.1248/bpb.34.1773. [DOI] [PubMed] [Google Scholar]
- 75.Slattery ML, et al. Associations with growth factor genes (FGF1, FGF2, PDGFB, FGFR2, NRG2, EGF, ERBB2) with breast cancer risk and survival: The Breast Cancer Health Disparities Study. Breast Cancer Res Treat. 2013;140(3):587–601. doi: 10.1007/s10549-013-2644-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kono SA, Marshall ME, Ware KE, Heasley LE. The fibroblast growth factor receptor signaling pathway as a mediator of intrinsic resistance to EGFR-specific tyrosine kinase inhibitors in non-small cell lung cancer. Drug Resist Updat. 2009;12(4-5):95–102. doi: 10.1016/j.drup.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tai AL, et al. Co-overexpression of fibroblast growth factor 3 and epidermal growth factor receptor is correlated with the development of nonsmall cell lung carcinoma. Cancer. 2006;106(1):146–155. doi: 10.1002/cncr.21581. [DOI] [PubMed] [Google Scholar]
- 78.Terai H, et al. Activation of the FGF2-FGFR1 autocrine pathway: A novel mechanism of acquired resistance to gefitinib in NSCLC. Mol Cancer Res. 2013;11(7):759–767. doi: 10.1158/1541-7786.MCR-12-0652. [DOI] [PubMed] [Google Scholar]
- 79.Ware KE, et al. Rapidly acquired resistance to EGFR tyrosine kinase inhibitors in NSCLC cell lines through de-repression of FGFR2 and FGFR3 expression. PLoS ONE. 2010;5(11):e14117. doi: 10.1371/journal.pone.0014117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Herrera-Abreu MT, et al. Parallel RNA interference screens identify EGFR activation as an escape mechanism in FGFR3-mutant cancer. Cancer Discov. 2013;3(9):1058–1071. doi: 10.1158/2159-8290.CD-12-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang J, et al. Ligand-associated ERBB2/3 activation confers acquired resistance to FGFR inhibition in FGFR3-dependent cancer cells. Oncogene. 2014 doi: 10.1038/onc.2014.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Issa A, et al. Combinatorial targeting of FGF and ErbB receptors blocks growth and metastatic spread of breast cancer models. Breast Cancer Res. 2013;15(R8):1–16. doi: 10.1186/bcr3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shimizu T, et al. The clinical effect of the dual-targeting strategy involving PI3K/AKT/mTOR and RAS/MEK/ERK pathways in patients with advanced cancer. Clin Cancer Res. 2012;18(8):2316–2325. doi: 10.1158/1078-0432.CCR-11-2381. [DOI] [PubMed] [Google Scholar]
- 84.Wissner A, et al. Dual irreversible kinase inhibitors: Quinazoline-based inhibitors incorporating two independent reactive centers with each targeting different cysteine residues in the kinase domains of EGFR and VEGFR-2. Bioorg Med Chem. 2007;15(11):3635–3648. doi: 10.1016/j.bmc.2007.03.055. [DOI] [PubMed] [Google Scholar]
- 85.Lipson D, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18(3):382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang Z, et al. Structural mimicry of a-loop tyrosine phosphorylation by a pathogenic FGF receptor 3 mutation. Structure. 2013;21(10):1889–1896. doi: 10.1016/j.str.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: The integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 8):859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 88.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen H, et al. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol Cell. 2007;27(5):717–730. doi: 10.1016/j.molcel.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kabsch W. Solution for Best Rotation to Relate 2 Sets of Vectors. Acta Crystallogr A. 1976;32(Part 5):922–923. [Google Scholar]
- 92.Anonymous Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 93.DeLano WL. The PyMOL User's Manual. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 94.Farahat WA, et al. Ensemble analysis of angiogenic growth in three-dimensional microfluidic cell cultures. PLoS ONE. 2012;7(5):e37333. doi: 10.1371/journal.pone.0037333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Duffy DC, McDonald JC, Schueller OJ, Whitesides GM. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane) Anal Chem. 1998;70(23):4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.