Significance
Here we report a strategy for isolating and characterizing populations of proteins targeted to synapses. Using this approach, we isolated and characterized multiple protein transport complexes from mouse brain, providing novel insights into the synaptic targeting of proteins and composition of synaptic proteome.
Keywords: kinesin, protein transport, synapse, proteome, hippocampus
Abstract
Little is known regarding the identity of the population of proteins that are transported and localized to synapses. Here we describe a new approach that involves the isolation and systematic proteomic characterization of molecular motor kinesins to identify the populations of proteins transported to synapses. We used this approach to identify and compare proteins transported to synapses by kinesin (Kif) complexes Kif5C and Kif3A in the mouse hippocampus and prefrontal cortex. Approximately 40–50% of the protein cargos identified in our proteomics analysis of kinesin complexes are known synaptic proteins. We also found that the identity of kinesins and where they are expressed determine what proteins they transport. Our results reveal a previously unappreciated role of kinesins in regulating the composition of synaptic proteome.
A number of researchers have reported specific roles of proteins that are localized to synapses, termed the “synaptic proteome,” in determining synaptic function and plasticity (1–6). Identifying the composition and dynamics of the synaptic proteome is key to understanding the remodeling of existing synapses and growth of new synapses, as well as identifying novel therapeutic targets for synaptopathies (7, 8). The composition of the synaptic proteome is determined by proteins transported from the cell body and local synthesis of proteins at the synapses. Several studies have shown the identification and localization of RNAs at the synapse (9–12). Comprehensive knowledge of the proteins that are actively targeted to synapses will help elucidate the dynamics of signal transduction at the synapse, as well as the molecules that are critical for the remodeling of synapses and formation of new synapses that accompany long-term memory storage. Despite this perceived significance, however, there are no currently available methodologies for studying populations of proteins targeted to synapses.
To identify actively transported proteins, we focused on the protein complexes associated with kinesins, molecular motor proteins that move from the cell body to distal neuronal processes and mediate the transport of various gene products (9, 13). We considered that proteins associated with kinesins include both synaptic proteins and proteins localized to dendrites and axons. Our methodology involved isolating specific kinesin complexes by coimmunoprecipitation (co-IP) from different regions of the mouse brain and systematically identifying the protein cargos by mass spectrometry (Fig. 1A). Using this strategy, we have successfully isolated and characterized the Kif5C kinesin complex from hippocampus and prefrontal cortex (PFC) and the Kif3A kinesin complex from hippocampus. The cargos include organelles and proteins involved in translation, signaling, and ion channels. Several of these proteins are implicated in neuropsychiatric disorders (14).
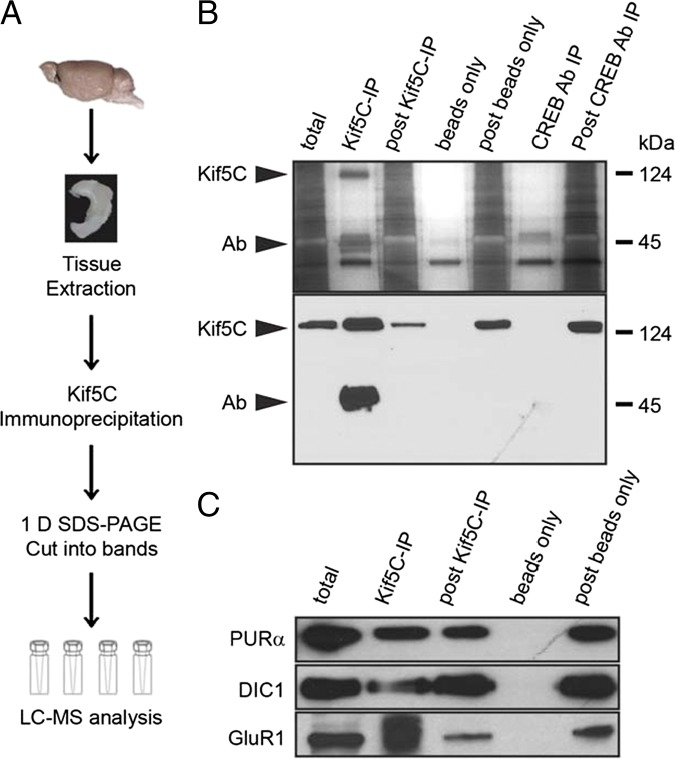
Fig. 1.
Identification of the Kif5C complex in mouse hippocampus. (A) Strategy used to analyze Kif5C-specific protein cargos in the mouse hippocampus. The schematic diagram shows the workflow for profiling Kif5C-transported proteins by proteomics. The processes include brain dissection, hippocampal tissue extraction and homogenization, Kif5C IP, complex separation by SDS/PAGE, isolation of specific bands followed by in-gel tryptic digestion, and LC-MS/MS analysis. (B) IP of Kif5C from mouse hippocampus. Kif5C complexes were immunoprecipitated from hippocampus using anti Kif5C antibody. (Upper) Protein complexes were then separated on SDS/PAGE and stained with silver. (Lower) Kif5C also was detected by Western blot analysis. Immunoprecipitation using CREB antibody and beads alone served as controls for IP. (C) Co-IP and Western blot analysis of Kif5C cargo proteins (PURα, DIC1, and GluR1) in mouse hippocampus. Total extract (input), IP, post-IP supernatant, beads-alone IP, and post-IP supernatant are shown. Arrows in B indicate the positions of Kif5C and antibody.
Results
Isolation and Characterization of Protein Complexes of Molecular Motor Kif5C in the Mouse Hippocampus.
To identify the population of synaptically targeted proteins, we first searched for kinesins that are abundant in mouse hippocampus. We studied the expression of 31 different kinesins in mouse hippocampus using specific primers (SI Appendix, Dataset S1). Quantitative real-time PCR (qRT-PCR) analysis of RNAs isolated from mouse hippocampus suggests that Kif5C is highly abundant in hippocampus (normalized expression level, 5.3 ± 0.14; SI Appendix, Fig. S1A). We then confirmed the expression of Kif5C by RNA in situ hybridization using digoxigenin-labeled sense and antisense riboprobes (SI Appendix, Fig. S1B) and by immunohistochemical and Western blot analyses using an anti-Kif5C antibody (SI Appendix, Fig. S1 C and D and Dataset S1).
Kif5C transport complexes were then immunoprecipitated from the hippocampus (Fig. 1A) following a previously published protocol (13) and analyzed by silver stain and Western blot analysis (Fig. 1B). Immunoprecipitated beads alone and immunoprecipitated anti-CREB antibody (SI Appendix, Dataset S1) served as specificity controls for the immunoprecipitation (IP). The specific protein band corresponding to Kif5C was present only in IP of Kif5C, and not in the beads alone or in antibody controls (Fig. 1B). We analyzed the protein bands corresponding to Kif5C by mass spectrometry, which identified ∼80% of the predicted tryptic peptides of Kif5C (SI Appendix, Fig. S1E).
To identify the population of proteins transported by Kif5C, we performed a systematic proteomic analysis of Kif5C complexes from hippocampus. We performed four biological replications of Kif5C IPs and four beads-alone control IPs. Scaffold software identified 72 proteins present only in the Kif5C IPs and not in the control IPs.
We considered two factors affecting the abundance of proteins associated with kinesin in individual IP biological replications: (i) the IP efficiency of samples and the stability of specific proteins interacting with kinesin and (ii) the physiological state of the neurons. Thus, to identify the maximum number of possible cargos associated with Kif5C in the hippocampus, we analyzed the proteomics dataset (SI Appendix, Dataset S1) to identify protein cargos that were 1.5-fold enriched in kinesin IPs compared with the control IPs. We believed that this analysis would offer a list of proteins for future experiments. We identified 202 proteins as possible cargos of Kif5C in hippocampus (SI Appendix, Dataset S1).
We then studied individual cargos for their possible synaptic localization by searching the National Center for Biotechnology Information (NCBI), Uni-Prot, Genecards, and OMIM databases. Among the 202 proteins, we identified 80 (highlighted in green in SI Appendix, Dataset S1) as having synaptic localization and function. Neuroligin-2 (Uni-Prot ID: Q69ZK9), cell adhesion molecule 2 (Q8BLQ9), ras GTPase-activating-like protein (Q3UQ44), ephrin type-A receptor 4 (Q03137), and myosin-Va (Q99104) are examples of the proteins selectively enriched in the Kif5C complex from hippocampus. Based on previously published work on kinesin complexes, we assumed that the Kif5C complex contains multiple proteins and is heterogeneous, with cargos expressed in different amounts in the complex. Consistent with this, we found that the Kif5C complex contains cargo proteins in varying amounts.
We selected three candidate protein cargos for detailed characterization: PURα, a nucleotide-binding protein; DIC, a dynein intermediate chain; and GluR1, glutamate receptor 1. We studied these cargos first by Western blot analyses of the Kif5C complex isolated from hippocampus and then by immunohistochemistry analyses after Kif5C knockdown in primary neurons. As shown in Fig. 1C, we found that these proteins are indeed specifically associated with the Kif5C complex and are not present in the beads-alone control IPs. Examination of the protein expression and distribution of Kif5C (SI Appendix, Fig. S1C) and the selected cargos in hippocampus by immunohistochemical staining (SI Appendix, Fig. S2) revealed that they are expressed in hippocampal neurons as well. We studied the expression levels of Kif5C in primary neuron cultures by qRT-PCR and found that, consistent with our data on Kif5C levels in dissected hippocampus, Kif5C is abundantly expressed in primary hippocampal neurons (SI Appendix, Fig. S1F).
Using specific siRNAs, we next knocked down the expression of Kif5C in primary hippocampal neurons, and evaluated the knockdown by Western blot and immunohistochemistry analyses. We found that Kif5C siRNA, but not the scrambled siRNA control, was effective in knocking down Kif5C protein levels in hippocampal neurons. Analysis of Western blot data by two-way ANOVA followed by the Tukey post hoc test indicated statistically significant Kif5C knockdown (Kif5C SiRNA, scrambled and no siRNA control, F(2, 63) = 16.866, P < 0.0001) at 72 h (0, 48, and 72 h, F(2, 63) = 12.622, P < 0.0001) after the siRNA transfection (SI Appendix, Fig. S3 A and B; 37.22 ± 4.69% knockdown, Kif5C siRNA vs. scrambled siRNA and control without siRNA transfection; n = 8), which was further supported by Kif5C immunohistochemistry data (SI Appendix, Fig. S3 C and D; F(2, 30) = 81.035, ***P < 0.001, one-way ANOVA).
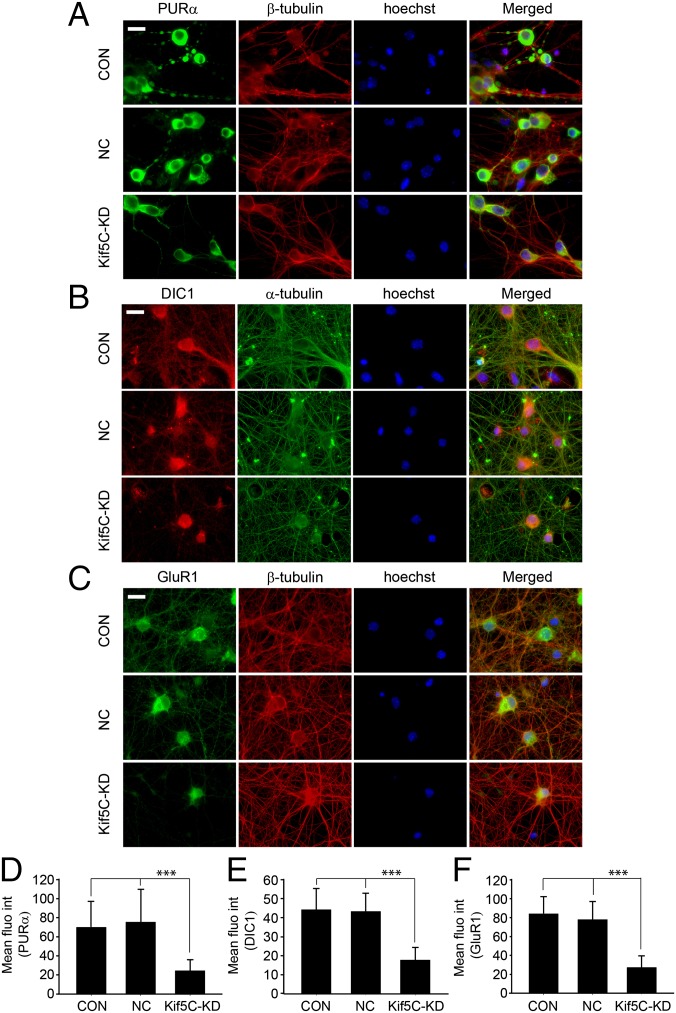
We then examined the distributions of PURα, DIC1, and GluR1 in primary neurons (Fig. 2). As shown in SI Appendix, Fig. S3 A–D, we observed an ∼40% decrease in Kif5C expression after siRNA transfection at 72 h. We measured the intensity level of these cargos on random spots in the neurites that were at least 10 μm away from the soma, and found that the mean fluorescence intensities of the three cargos of Kif5C in neurites were significantly decreased after Kif5C knockdown compared with those in the neurons without siRNA transfection and neurons transfected with scrambled siRNA duplex (Fig. 2; a 65.74 ± 3.2% decrease for PURα [F(2, 33) = 48.738, ***P < 0.001], a 58.51 ± 3.61% decrease for DIC1 [F(2, 27) = 99.292, ***P < 0.001], and a 61.87 ± 3.64% decrease for GluR1 [F(2, 30) = 94.041, ***P < 0.001], Kif5C siRNA vs. scrambled siRNA and control without siRNA transfection for each cargos; n = 10∼19 captured images for each group, 2∼4 neurons analyzed for each image, and 4∼5 random spots in the neurites, lying at least 10 μm away from the soma for each neuron; ***P < 0.001, one-way ANOVA followed by Tukey’s post hoc test). This finding indicates that Kif5C transports these proteins in primary hippocampal neurons, and that Kif5C IPs contain specific proteins as cargos.
Fig. 2.
Kif5C knockdown affects transport of protein cargos identified from proteomics. Immunohistochemical analyses of PURα, DIC1, and GluR1 (A–C) and bar graphs (D–F) showing quantitative analyses of immunohistochemical data shown in A–C. PURα/β-tubulin (A), DIC1/α-tubulin (B), and GluR1/β-tubulin (C) were coimmunostained with Hoechst (blue) in cultured hippocampal neurons with or without Kif5C siRNA transfection for 72 h. The fluorescence intensities of these protein cargos in neurites were quantified with Leica DM5500 software and statistically analyzed by one-way ANOVA, followed by Tukey’s post hoc test (D–F). KD, knockdown; NC, scrambled control siRNA; CON, vehicle-only control. Error bars represent SEM. ***P < 0.001. (Scale bar: 20 μm.)
Isolation and Characterization of Protein Complexes of Kif3A in Mouse Hippocampus.
We next asked whether the same neuron expresses two different kinesins, and whether they transport the same or a completely different set of proteins to the synapses. To address this question, we studied the Kif3A complex from the hippocampus and compared it with the Kif5C complex from the hippocampus. Kif3 motors constitute a functionally diverse subgroup of the microtubule-associated kinesin superfamily. Kif3A is also abundantly expressed in neurons (15), is a component of the presynaptic ribbon in vertebrate photoreceptors (16), and associates with membrane vesicles (17).
The data presented in SI Appendix, Fig. S1 A, D, and F show that Kif3A mRNA and protein are expressed in hippocampus. We used immunohistochemistry and confocal imaging analyses to study the distributions of Kif5C and Kif3A in hippocampal neurons. Our confocal imaging data suggest that Kif5C and Kif3A are expressed and localized in the same hippocampal neurons (SI Appendix, Fig. S4A). Similar to Kif5C, Kif3A protein is also expressed in the intact hippocampus (SI Appendix, Figs. S4B and S1D).
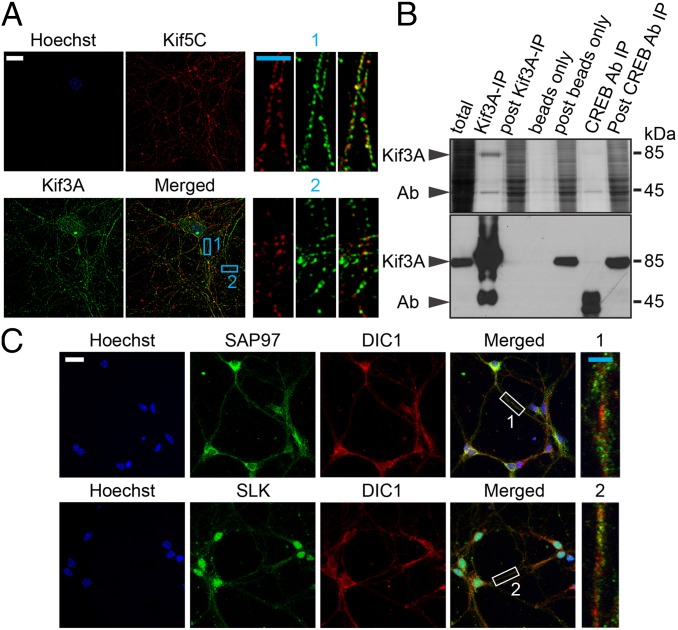
To obtain further evidence that Kif5C and Kif3A are indeed expressed in the same hippocampal neurons, we conducted a super-resolution immunocytochemical analysis of primary hippocampal neurons (Fig. 3A). Consistent with our confocal imaging data, we found that Kif5C and Kif3A were expressed simultaneously in the same neurons. The majority of Kif5C and Kif3A appeared as distinct spots (with <10% spot overlap).
Fig. 3.
Super-resolution imaging of Kif5C and Kif3A distribution in hippocampal neurons and characterization of Kif3A and Kif5C cargos. (A) Representative super-resolution images illustrate the distinct distribution of Kif5C (red) and Kif3A (green) proteins in the same neuron. The nucleus was stained with Hoechst. Digitally enlarged images (corresponding to Kif5C, Kif3A, and merged) show two representative sections from neurites marked 1 and 2 in blue. (B) IP of Kif3A complex from mouse hippocampus. Hippocampi from 7- to 11-wk-old mice were dissected out, and proteins were prepared as described in Fig. 1. (Upper) Kif3A complexes were immunoprecipitated using a specific antibody, and protein complexes were then separated on SDS/PAGE and stained with silver. (Lower) Kif3A also was detected by Western blot analysis. Immunoprecipitation using CREB antibody and beads alone served as controls for IP. Arrows in B indicate the positions of Kif3A and antibody. (C) Immunocytochemical analysis of cargos of Kif3A and Kif5C in cultured primary hippocampal neurons. Shown are confocal projection images of coimmunostaining of SAP97, a Kif3A cargo (green), and DIC1, a Kif5C cargo (red) and of SLK, a Kif3A cargo (green), and DIC1, a Kif5C cargo (red), as well as the merged images. Sections of representative neurites in the merged images are marked 1 and 2 (in white) and are digitally enlarged. (Insets) Expression of cargos in the same neurite. (White scale bar: 20 μm; blue scale bar: 5 μm.)
Following the similar sample preparation procedure used to isolate the Kif5C complex, we performed Kif3A IP using an anti Kif3A antibody (Fig. 3B and SI Appendix, Dataset S1), and evaluated the IP by silver stain and Western blot analyses. Beads-alone IP and anti-CREB antibody IP served as controls. We found that Kif3A was specifically present in the Kif3A IP, but not in the beads-alone or anti-CREB antibody IP controls. Proteomic analysis of the Kif3A IPs (n = 4) and beads-alone control IPs (n = 4) identified Kif3A and specific protein cargos (SI Appendix, Fig. S5A). Interestingly, kinesin light chains (KLC1 and KLC2) that were found in the Kif5C complex were absent in the Kif3A complex from hippocampus. Spectral counting analysis of the entire dataset suggested that 75 proteins were present only in the Kif3A complex.
We then analyzed the proteomics dataset (SI Appendix, Dataset S1) to identify protein cargos that were 1.5-fold enriched in kinesin IPs compared with control IPs (SI Appendix, Dataset S1). This analysis identified 74 proteins as specific cargos. We then studied individual cargos for their possible synaptic localization by searching the NCBI, Uni-Prot, Genecards, and OMIM databases and identified 37 proteins (highlighted in green in SI Appendix, Dataset S1) with synaptic localization and function among these 74 proteins. ARF GTPase-activating protein GIT1 (Q68FF6), piccolo (Q9QYX7), cytoplasmic FMR1-interacting protein 2 (Q5SQX6), and disks large-associated protein 1 (Q9D415) were among those identified as enriched in the Kif3A complex.
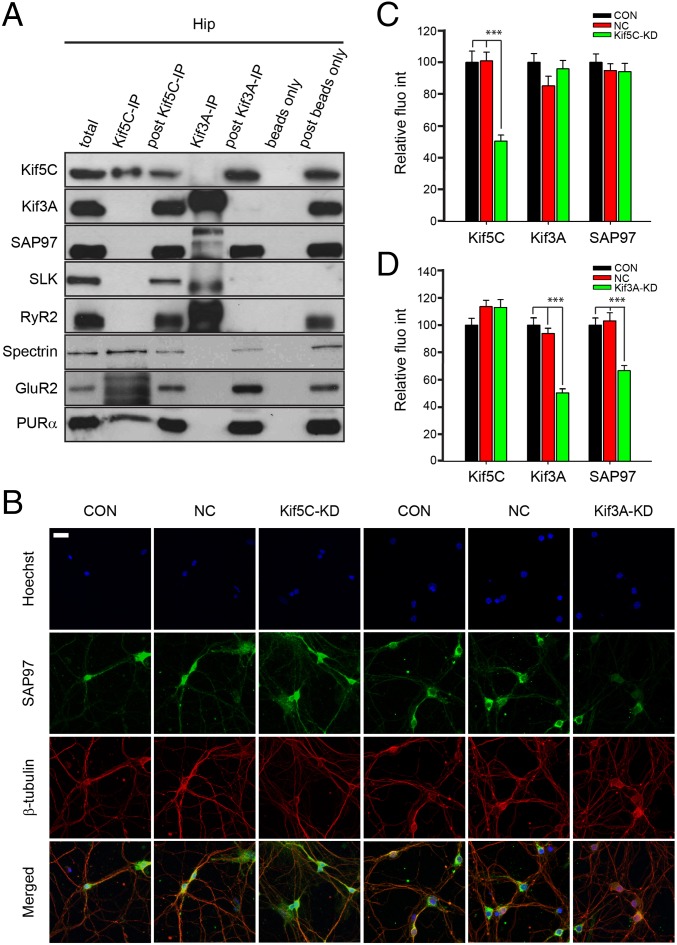
We next studied the selective association of cargos to Kif5C and Kif3A. Fig. 3C shows costaining of DIC, a cargo of Kif5C, and SAP97 and SLK, cargos of Kif3A in primary hippocampal neurons, suggesting that specific protein cargos of Kif5C and Kif3A are transported in the same hippocampal neuron. We then examined the distributions of SAP97, SLK, and RyR2 that were found in the Kif3A complex and of spectrin, GluR2, and PURα present in the Kif5C complex. SI Appendix, Fig. S5B shows the expression of Kif3A cargos (RyR2, SAP97, and SLK) in the CA1 hippocampal region. Western blot analyses of Kif5C and Kif3A complexes suggested that, consistent with proteomics data, these cargos are specifically associated with kinesin complexes and are not found in the beads-alone controls (Fig. 4A).
Fig. 4.
Selectivity of Kif5C and Kif3A cargos from the hippocampus. (A) Co-IP and Western blot analysis of Kif5C/Kif3A transported proteins in mouse hippocampus (Hip). The mouse hippocampal lysates were subjected to IP with anti-Kif5C or anti-Kif3A antibody. The complexes were analyzed by Western blot analysis and probed with anti-SAP97, SLK, RyR2, Spectrin, GluR2, and PURα antibodies. (B) Expression of Kif3A protein cargo SAP97 in neurites was decreased after Kif3A knockdown, but not after Kif5C knockdown, in cultured hippocampal neurons as determined by immunohistochemical characterization. After transfection of siRNA oligonucleotides or shRNA [Kif5C siRNA for 72 h, Kif3A shRNA for 48 h, NC (scrambled siRNA control), and CON (vehicle-only control)] in three to five DIV primary cultured mouse hippocampal neurons, immunohistochemical analyses of SAP97 (green) and β-tubulin (red) were carried out. Representative confocal projection images are shown. (C and D) Graphs showing the quantitative analyses of relative fluorescence intensities of Kif5C, Kif3A, and SAP97 in neurites after Kif5C (C) or Kif3A (D) knockdown. Error bars represent SEM. Data were analyzed by one-way ANOVA followed by Tukey’s post hoc test. ***P < 0.001. (Scale bar: 20 μm.)
We then tested whether knockdown of Kif3A or Kif5C affects the distribution of the other and their cargos. We evaluated the expression of Kif3A after Kif5C knockdown and the expression of Kif5C after Kif3A knockdown first by Western blot analysis [SI Appendix, Fig. S6 A and B; F(2, 36) = 5.432, P = 0.0087 for Kif3A shRNA transfection; F(2, 36) = 9.671, P = 0.0004 for 48 h of Kif3A shRNA transfection; n = 5) and then by immunocytochemistry (SI Appendix, Fig. S6C and Fig. 4 C and D). We found that knockdown of Kif3A [49.45 ± 3.18% decrease compared with scrambled shRNA; F(2, 132) = 44.736, ***P < 0.001] or Kif5C [48.26 ± 7.8% decrease compared with scrambled siRNA; F(2, 110) = 28.396, ***P < 0.001] does not affect the distribution of the other complex in primary hippocampal neurons.
We next analyzed the distribution of protein cargo SAP97, a Kif3A-specific cargo, in hippocampus in these experiments. We found that knockdown of Kif5C in cultured hippocampal neurons did not affect the transport of SAP97. As shown in Fig. 4 B–D, the SAP97 intensity in neurites decreased significantly [39.25 ± 5.1% decrease; F(2, 117) = 19.434, ***P < 0.001, Tukey’s post hoc test] with shRNA-mediated Kif3A knockdown (49.45 ± 3.18% decrease in Kif3A), whereas Kif5C knockdown (48.26 ± 7.8% decrease in Kif5C) produced no significant effect on SAP97 intensity in neurites [F(2, 96) = 0.411, P = 0.664; n = 4∼6 captured images for each group, at least 2 neurons analyzed for each image, and 8∼9 random spots in the neurites lying at least 10 μm away from the soma for each neuron; one-way ANOVA followed by Tukey’s post hoc test].
Isolation and Characterization of Kif5C Protein Complexes from Mouse PFC.
We asked whether the same kinesin motor could transport different cargos to different synapses. To address this question, we studied Kif5C complexes isolated from PFC and compared them with those from hippocampus. First, we examined the expression of Kif5C in PFC. Using qRT-PCR analysis of RNAs isolated from mouse PFC (sections taken through the PFC, relative to bregma (18): +3.08–0 mm), we found that Kif5C was highly abundant in PFC (normalized expression level, 6.8 ± 0.03; SI Appendix, Fig. S7A). Western blot analysis of proteins isolated from PFC suggested that Kif5C protein is expressed in PFC (SI Appendix, Fig. S1D).
We then examined the distribution of Kif5C mRNA in mouse PFC by in situ hybridization of Kif5C using digoxigenin-labeled riboprobes and immunohistochemistry analysis of PFC using specific antibodies for cortical layer markers (Ctip2, a marker of deep-layer subcortical projection, and CUX1, a marker of upper layer subcortical projection). Confocal imaging analysis suggested that Kif5C mRNA is abundantly expressed in mouse PFC, mainly colocalized with deep-layer marker Ctip2 (SI Appendix, Fig. S7B).
We next isolated Kif5C protein complexes from mouse PFC (SI Appendix, Fig. S7C). We analyzed the proteomics dataset (SI Appendix, Dataset S1) to identify the protein cargos that were 1.5-fold enriched in kinesin IPs compared with control IPs (SI Appendix, Dataset S1). This analysis identified 155 proteins as cargos. Neuromodulin (P06837), synaptojanin-1 (Q8CHC4), excitatory amino acid transporter 1 (P56564), and ADP ribosylation factor 3 (P61205) were among the proteins identified as enriched in the Kif5C complex from PFC. Interestingly, we also found that the Kif5C complex from PFC contains the same light chains (KLC1 and KLC2) besides the heavy chain, similar to the Kif5C complex from hippocampus, whereas KLC1 and KLC2 are absent in the Kif3A complex from hippocampus (SI Appendix, Dataset S1).
We studied individual cargos for their possible synaptic localization by searching the NCBI, Uni-Prot, Genecards, and OMIM databases. We identified 70 proteins (highlighted in green in SI Appendix, Dataset S1) with synaptic localization and function among 155 proteins evaluated.
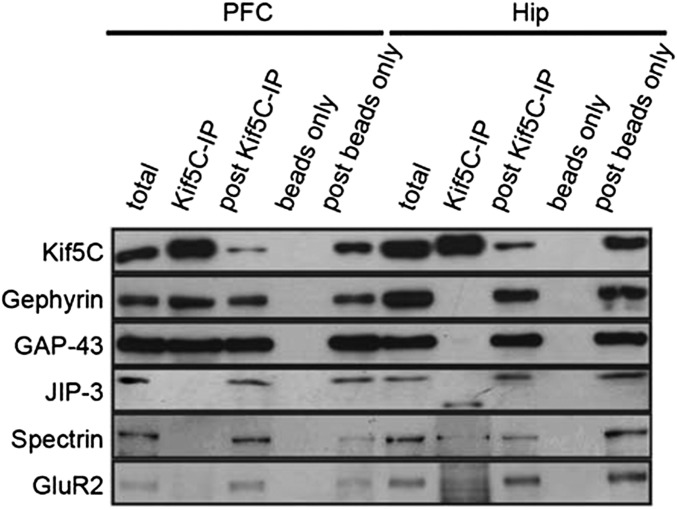
We next validated Kif5C cargos that were selective to PFC and hippocampus by Western blot analysis of Kif5C complexes (Fig. 5). We studied gephyrin and GAP43 cargos that our proteomics analysis identified as selective Kif5C cargos in PFC, and JIP3, spectrin, and GluR2 cargos identified as selective cargos in hippocampus (Fig. 5). Consistent with the proteomics data, our Western blot data suggest that these cargos are selective to brain regions.
Fig. 5.
Selectivity of Kif5C cargos from PFC and hippocampus (Hip). Co-IP and Western blot analysis of Kif5C-transported proteins in mouse PFC and hippocampus. Mouse PFC or hippocampus tissue lysates were subjected to co-IP with anti-Kif5C antibody, and the complexes were analyzed by Western blot analysis using anti-gephyrin, -GAP43, -JIP-3, -spectrin, and -GluR2 antibodies.
Discussion
Large-scale proteomic characterization of specific protein complexes localized to synapses has been used previously to obtain a broad view of synaptically localized proteins. For example, proteomic analysis of the composition of NMDAR multiprotein complexes isolated from the mouse brain (19, 20), as well as the postsynaptic density (21), have identified several proteins localized to the synapse. To date, however, no approach has proven effective in identifying actively transported proteins.
To address this issue, in the present work we focused on protein complexes associated with molecular motors that mediate active transport of gene products from the cell body to synapses. We specifically focused on Kif5C and Kif3A kinesin complexes and have identified several proteins that are potentially targeted to synapses. Previous studies identified some cargos of Kif5C, including GABA receptors (22), axonal synaptic vesicle precursors (23), APP-containing vesicles (24, 25), TrkB vesicles (26), mitochondria (23, 27–31), AMPAR vesicles (32), and mRNA/protein complexes (33, 34). Kif3 was shown to transport fodrin-associating vesicles in cultured superior cervical ganglion neurons (35), N-cadherin in the developing mouse brain (36) and axonal voltage-gated potassium channel in cultured hippocampal neurons (37). Approximately 40–50% of the protein cargos identified in our proteomics analysis of three kinesin complexes are known synaptic proteins (highlighted in SI Appendix, Dataset S1).
Cargos transported by either Kif5C or Kif3A identified in our present study have fundamental roles in neuronal function and are critically involved in neuronal disease pathogenesis. For example, among the cargos that we characterized, the Kif5C cargos PURα (38) and spectrin (39) have essential roles in synapse development; Kif3A cargos SAP97 (40) and RyR2 (41), and Kif5C cargos JIP-3 (42) from the hippocampus and gephyrin (43) from the PFC, are involved in Alzheimer’s disease; and abnormal expression of GAP-43 (Kif5C cargo in PFC) is implicated in schizophrenia (44), suggesting the importance of kinesin cargos in psychiatric disorders.
Kinesin Complexes from the Same Neurons.
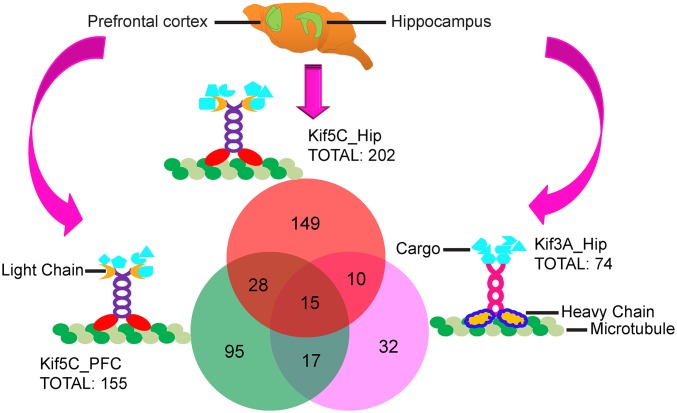
Variation in subdomains of kinesins has been identified as a strategy for achieving diversity in cargo transport (45). We studied the differential transport of cargos by comparing the proteins transported by two different kinesins expressed in the same neurons. The amino acid sequences of the C-terminal ends of Kif5C and Kif3A, which interact with cargos and light chains, are dissimilar. We found that in hippocampal neurons, ∼87% proteins among 202 Kif5C cargos identified are selective to Kif5C, and that ∼66% of the Kif3A-associated proteins among 74 Kif3A cargos identified are selective to Kif3A. This observation is consistent with the idea that the variable C-terminal end contributes to the diversity in cargo transport.
Kinesin Complexes from Two Different Brain Regions.
We found that ∼70–80% of cargos transported by Kif5C in hippocampus and PFC are selective to brain regions. Distinct groups of proteins, transported by the same motor in different brain regions, could be attributed to their characteristic function in a specific brain region. Kif5C complexes from hippocampus and PFC also share some cargos; for example, we identified components of mitochondria in PFC and hippocampus, indicating that mitochondria are transported by Kif5C in hippocampus and PFC. To gain insight into the significance of cargos transported, we carried out systematic bioinformatics analysis using ingenuity pathways analysis (IPA) (Fig. 6, and SI Appendix, Figs. S8–11 and Dataset S1). As revealed by associated network function analysis, the three kinesin cargo complexes in the present study are involved in neurologic diseases, shown in the top five networks of protein cargos identified (SI Appendix, Fig. S8 and Dataset S1). We identified disorders associated with specific kinesin complexes; for example, ∼60 cargos of Kif5C are implicated in psychiatric disorders, whereas ∼20 cargos of Kif3A are implicated in developmental disorders. This analysis also suggests that specific biological pathways might be regulated by kinesin complexes through the transport of specific cargos; for example, as shown in SI Appendix, Fig. S8, cell morphology, cellular assembly and organization, and neurologic disease excluding cancer are the top three biological functions assigned to Kif5C in hippocampus, whereas neurologic disease, cellular function and maintenance, and cellular assembly and organization are the top three functions assigned to the Kif5C complex from PFC. Consistent with our pathway analysis showing that Kif5C in hippocampus and PFC might regulate different functions, we find from signaling network analysis (top three networks for each complexes shown in SI Appendix, Figs. S9–S11) that cargos transported by Kif5C and Kif3A form networks with different topologies.
Fig. 6.
Comparison of the Kif5C and Kif3A complexes from hippocampus and PFC. The Venn diagram shows protein cargos that are 1.5-fold enriched in the three complexes. The Kif5C-Hip and Kif5C-PFC complexes contain the same light chains (KLC1 and KLC2) besides a heavy chain, whereas light chains are absent in the Kif3A complex from hippocampus (SI Appendix, Dataset S1).
Molecular Motors and Synaptic Targeting of Proteins.
More than 40 different kinesins are expressed in the mouse genome, and several hundred proteins are targeted to and localized to synapses. This begs the following questions: What confers specificity of transport? Is there a motor-cargo code that determines the transport of specific cargos? Goldstein et al. (30) suggested that the diversity of kinesins alone is not sufficient to account for the delivery of specific cargos, and that specific adaptor proteins might contribute to transport of specific cargos. As mentioned earlier, the C-terminal end is variable in different motors. Different kinesin light chains and adaptors also contribute to transport of specific cargos. Our analysis of three kinesin complexes suggests that there is much more than the variable C-terminal end and light chains contributing to specificity. Specifically, we found that the Kif5C complex from hippocampus and PFC contains the same light chains (KLC1 and KLC2), yet only 30% of cargos transported by Kif5C in PFC and hippocampus are similar. Thus, the environment in which they are expressed has a major role. Factors influencing cargo interactions with kinesins could include the expression of specific adaptor proteins or regulation of abundance of certain cargos. Our comparison of Kif5C and Kif3A complexes from hippocampus revealed that ∼80% of the cargos were distinct. Because Kif5C and Kif3A are expressed in the same neurons, neither the availability of adaptors nor the abundance of cargo proteins could determine the transport of selected proteins; thus, the difference in cargos associated with Kif5C and Kif3A in the hippocampus must come from selective motor–cargo interactions. These data support the existence of the motor–cargo code that determines the transport of selected proteins. Consistent with this idea, we found that components of retrograde transport machinery are differentially associated with Kif5C and Kif3A. We found Dynein heavy chain as a cargo of Kif3A in hippocampus, and dynein intermediate chain and dynein light chain as cargos of Kif5C in hippocampus (SI Appendix, Fig. S12).
In conclusion, our systematic analysis of multiple kinesin complexes provides insights into the mechanisms of protein targeting and composition of synaptic proteome. We have developed a new approach for elucidating the synaptic proteome. This approach, which has the potential to be extended to additional kinesins, various brain regions, and basal and activity-mediated states, will significantly advance the understanding of how synapses work, how their composition changes on neuronal activity and in different brain areas, and how pathological conditions of the brain affect these changes.
Materials and Methods
Animals, reagents, primers, antibodies, IP, proteomics, super-resolution imaging, Western blot analysis, immunohistochemistry, RNA in situ hybridization, qRT-PCR, cell culture and siRNA/shRNA knockdown experiments, bioinformatics, and statistical analyses are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by the Whitehall Foundation and the National Institute of Mental Health (Grant 1 R21MH096258-01A1).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401483111/-/DCSupplemental.
References
- 1.Choi YB, et al. Neurexin–neuroligin transsynaptic interaction mediates learning-related synaptic remodeling and long-term facilitation in aplysia. Neuron. 2011;70(3):468–481. doi: 10.1016/j.neuron.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang DO, et al. Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science. 2009;324(5934):1536–1540. doi: 10.1126/science.1173205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin KC, Ephrussi A. mRNA localization: Gene expression in the spatial dimension. Cell. 2009;136(4):719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey CH, Kandel ER. Synaptic remodeling, synaptic growth and the storage of long-term memory in Aplysia. Prog Brain Res. 2008;169:179–198. doi: 10.1016/S0079-6123(07)00010-6. [DOI] [PubMed] [Google Scholar]
- 5.Kandel ER. The molecular biology of memory storage: A dialogue between genes and synapses. Science. 2001;294(5544):1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 6.Antonova I, et al. Rapid increase in clusters of presynaptic proteins at onset of long-lasting potentiation. Science. 2001;294(5546):1547–1550. doi: 10.1126/science.1066273. [DOI] [PubMed] [Google Scholar]
- 7.Grant SG. Synaptopathies: Diseases of the synaptome. Curr Opin Neurobiol. 2012;22(3):522–529. doi: 10.1016/j.conb.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Kadakkuzha BM, Puthanveettil SV. Genomics and proteomics in solving brain complexity. Mol Biosyst. 2013;9(7):1807–1821. doi: 10.1039/c3mb25391k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puthanveettil SV, et al. A strategy to capture and characterize the synaptic transcriptome. Proc Natl Acad Sci USA. 2013;110(18):7464–7469. doi: 10.1073/pnas.1304422110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moccia R, et al. An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J Neurosci. 2003;23(28):9409–9417. doi: 10.1523/JNEUROSCI.23-28-09409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tubing F, et al. Dendritically localized transcripts are sorted into distinct ribonucleoprotein particles that display fast directional motility along dendrites of hippocampal neurons. J Neurosci. 2010;30(11):4160–4170. doi: 10.1523/JNEUROSCI.3537-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cajigas IJ, et al. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74(3):453–466. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puthanveettil SV, et al. A new component in synaptic plasticity: Up-regulation of kinesin in the neurons of the gill-withdrawal reflex. Cell. 2008;135(5):960–973. doi: 10.1016/j.cell.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu XA, Rizzo V, Puthanveettil SV. Pathologies of axonal transport in neurodegenerative diseases. Transl Neurosci. 2012;3(4):355–372. doi: 10.2478/s13380-012-0044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aizawa H, et al. Kinesin family in murine central nervous system. J Cell Biol. 1992;119(5):1287–1296. doi: 10.1083/jcb.119.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muresan V, Lyass A, Schnapp BJ. The kinesin motor KIF3A is a component of the presynaptic ribbon in vertebrate photoreceptors. J Neurosci. 1999;19(3):1027–1037. doi: 10.1523/JNEUROSCI.19-03-01027.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muresan V, et al. KIF3C and KIF3A form a novel neuronal heteromeric kinesin that associates with membrane vesicles. Mol Biol Cell. 1998;9(3):637–652. doi: 10.1091/mbc.9.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paxinos G, Watson C, Pennisi M, Topple A. Bregma, lambda and the interaural midpoint in stereotaxic surgery with rats of different sex, strain and weight. J Neurosci Methods. 1985;13(2):139–143. doi: 10.1016/0165-0270(85)90026-3. [DOI] [PubMed] [Google Scholar]
- 19.Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. 2000;3(7):661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- 20.Delint-Ramirez I, et al. In vivo composition of NMDA receptor signaling complexes differs between membrane subdomains and is modulated by PSD-95 and PSD-93. J Neurosci. 2010;30(24):8162–8170. doi: 10.1523/JNEUROSCI.1792-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walikonis RS, et al. Identification of proteins in the postsynaptic density fraction by mass spectrometry. J Neurosci. 2000;20(11):4069–4080. doi: 10.1523/JNEUROSCI.20-11-04069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Twelvetrees AE, et al. Delivery of GABAARs to synapses is mediated by HAP1-KIF5 and disrupted by mutant huntingtin. Neuron. 2010;65(1):53–65. doi: 10.1016/j.neuron.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pack-Chung E, Kurshan PT, Dickman DK, Schwarz TL. A Drosophila kinesin required for synaptic bouton formation and synaptic vesicle transport. Nat Neurosci. 2007;10(8):980–989. doi: 10.1038/nn1936. [DOI] [PubMed] [Google Scholar]
- 24.Kamal A, Stokin GB, Yang Z, Xia CH, Goldstein LS. Axonal transport of amyloid precursor protein is mediated by direct binding to the kinesin light chain subunit of kinesin-I. Neuron. 2000;28(2):449–459. doi: 10.1016/s0896-6273(00)00124-0. [DOI] [PubMed] [Google Scholar]
- 25.Kamal A, Almenar-Queralt A, LeBlanc JF, Roberts EA, Goldstein LS. Kinesin-mediated axonal transport of a membrane compartment containing beta-secretase and presenilin-1 requires APP. Nature. 2001;414(6864):643–648. doi: 10.1038/414643a. [DOI] [PubMed] [Google Scholar]
- 26.Arimura N, et al. Anterograde transport of TrkB in axons is mediated by direct interaction with Slp1 and Rab27. Dev Cell. 2009;16(5):675–686. doi: 10.1016/j.devcel.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 27.Cai Q, Gerwin C, Sheng ZH. Syntabulin-mediated anterograde transport of mitochondria along neuronal processes. J Cell Biol. 2005;170(6):959–969. doi: 10.1083/jcb.200506042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka Y, et al. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 1998;93(7):1147–1158. doi: 10.1016/s0092-8674(00)81459-2. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147(4):893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldstein AY, Wang X, Schwarz TL. Axonal transport and the delivery of pre-synaptic components. Curr Opin Neurobiol. 2008;18(5):495–503. doi: 10.1016/j.conb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006;173(4):545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Setou M, et al. Glutamate receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature. 2002;417(6884):83–87. doi: 10.1038/nature743. [DOI] [PubMed] [Google Scholar]
- 33.Ohashi S, et al. Identification of mRNA/protein (mRNP) complexes containing Puralpha, mStaufen, fragile X protein, and myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J Biol Chem. 2002;277(40):37804–37810. doi: 10.1074/jbc.M203608200. [DOI] [PubMed] [Google Scholar]
- 34.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: Isolation and characterization of an RNA-transporting granule. Neuron. 2004;43(4):513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 35.Takeda S, et al. Kinesin superfamily protein 3 (KIF3) motor transports fodrin-associating vesicles important for neurite building. J Cell Biol. 2000;148(6):1255–1265. doi: 10.1083/jcb.148.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teng J, et al. The KIF3 motor transports N-cadherin and organizes the developing neuroepithelium. Nat Cell Biol. 2005;7(5):474–482. doi: 10.1038/ncb1249. [DOI] [PubMed] [Google Scholar]
- 37.Gu C, et al. The microtubule plus-end tracking protein EB1 is required for Kv1 voltage-gated K+ channel axonal targeting. Neuron. 2006;52(5):803–816. doi: 10.1016/j.neuron.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 38.Khalili K, et al. Puralpha is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse. Mol Cell Biol. 2003;23(19):6857–6875. doi: 10.1128/MCB.23.19.6857-6875.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pielage J, Fetter RD, Davis GW. A postsynaptic spectrin scaffold defines active zone size, spacing, and efficacy at the Drosophila neuromuscular junction. J Cell Biol. 2006;175(3):491–503. doi: 10.1083/jcb.200607036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marcello E, et al. SAP97-mediated local trafficking is altered in Alzheimer disease patients' hippocampus. Neurobiol Aging. 2012;33(2):422 e421–410. doi: 10.1016/j.neurobiolaging.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 41.Adasme T, et al. Involvement of ryanodine receptors in neurotrophin-induced hippocampal synaptic plasticity and spatial memory formation. Proc Natl Acad Sci USA. 2011;108(7):3029–3034. doi: 10.1073/pnas.1013580108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muresan Z, Muresan V. c-Jun NH2-terminal kinase-interacting protein-3 facilitates phosphorylation and controls localization of amyloid-beta precursor protein. J Neurosci. 2005;25(15):3741–3751. doi: 10.1523/JNEUROSCI.0152-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hales CM, et al. Abnormal gephyrin immunoreactivity associated with Alzheimer disease pathologic changes. J Neuropathol Exp Neurol. 2013;72(11):1009–1015. doi: 10.1097/01.jnen.0000435847.59828.db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blennow K, Bogdanovic N, Gottfries CG, Davidsson P. The growth-associated protein GAP-43 is increased in the hippocampus and in the gyrus cinguli in schizophrenia. J Mol Neurosci. 1999;13(1-2):101–109. doi: 10.1385/JMN:13:1-2:101. [DOI] [PubMed] [Google Scholar]
- 45.Hesse WR, et al. Modular aspects of kinesin force generation machinery. Biophys J. 2013;104(9):1969–1978. doi: 10.1016/j.bpj.2013.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.