Significance
All organisms have systems in place to ensure that aberrant nascent polypeptide chains are promptly dealt with before being released from ribosomes and posing harm to the cell. The ribosome-associated quality control complex (RQC), composed of the Ltn1 E3 ubiquitin ligase catalytic subunit and cofactors, has become a paradigm for understanding quality control in eukaryotes. However, exactly how RQC functions has remained unknown. Here, we determine the structure of the 60S subunit-bound RQC complex. The data provide critical insights into how RQC is able to selectively target aberrant nascent chains, while ignoring nascent chains in normally translating ribosomes. Furthermore, the structure shows the architecture of a ribosome-bound E3 ligase poised to mark nascent chains for degradation.
Keywords: Tae2/Nemf, translational surveillance, protein quality control, cryo-EM, listerin/Ltn1 E3 ubiquitin ligase
Abstract
All organisms have evolved mechanisms to manage the stalling of ribosomes upon translation of aberrant mRNA. In eukaryotes, the large ribosomal subunit-associated quality control complex (RQC), composed of the listerin/Ltn1 E3 ubiquitin ligase and cofactors, mediates the ubiquitylation and extraction of ribosome-stalled nascent polypeptide chains for proteasomal degradation. How RQC recognizes stalled ribosomes and performs its functions has not been understood. Using single-particle cryoelectron microscopy, we have determined the structure of the RQC complex bound to stalled 60S ribosomal subunits. The structure establishes how Ltn1 associates with the large ribosomal subunit and properly positions its E3-catalytic RING domain to mediate nascent chain ubiquitylation. The structure also reveals that a distinguishing feature of stalled 60S particles is an exposed, nascent chain-conjugated tRNA, and that the Tae2 subunit of RQC, which facilitates Ltn1 binding, is responsible for selective recognition of stalled 60S subunits. RQC components are engaged in interactions across a large span of the 60S subunit surface, connecting the tRNA in the peptidyl transferase center to the distally located nascent chain tunnel exit. This work provides insights into a mechanism linking translation and protein degradation that targets defective proteins immediately after synthesis, while ignoring nascent chains in normally translating ribosomes.
During the canonical termination and recycling steps of translation, stop codon recognition triggers factor-mediated hydrolysis of the nascent peptidyl-tRNA conjugate, nascent chain release, and ribosome splitting (1–3). Conversely, translation of aberrant mRNA, such as mRNA lacking stop codons (“nonstop mRNA”), renders 80S ribosomes stalled with nascent polypeptides (1–3). Furthermore, “nonstop proteins” cannot be corrected by quality control chaperones and have the potential to interfere with cellular function (3, 4). Not surprisingly, defective termination and recycling are under surveillance by a variety of mechanisms (1–3). In eukaryotes, “rescue factors” homologous to termination factors promote dissociation of translationally halted ribosomes in a stop codon-independent manner (5). However, because rescue factors lack peptidyl-tRNA hydrolase activity, their action results in nascent chains remaining stalled on the released 60S subunit.
Ltn1 is the critical E3 ligase mediating ubiquitylation of aberrant proteins that become stalled on ribosomes during translation (4). Mutation of the Ltn1 mouse ortholog, listerin, causes neurodegeneration (6), suggesting an important function for this process. Ltn1 works together with several cofactors as part of the ribosome-associated quality control complex (RQC) (7–9) and appears to first associate with nascent chain-stalled 60S subunits together with two proteins of unknown function, Tae2 and Rqc1 (7, 9). Ltn1-mediated ubiquitylation of the stalled polypeptide then results in the recruitment of the AAA ATPase Cdc48/p97/VCP. This recruitment also requires Rqc1 and Tae2, and is followed by Cdc48-mediated nascent chain extraction and delivery to the proteasome for degradation (7–9).
Exactly how RQC recognizes stalled ribosomes and performs its functions has not been understood, and the only structure available in this system is that of free Ltn1 at 40-Å resolution (10). To elucidate mechanisms underlying RQC function, we set out to solve the structure of the endogenous complex purified from yeast. To reduce sample heterogeneity so as to facilitate structural characterization, RQC assembly was blocked in a preubiquitylation step by using cells expressing endogenous Ltn1 with a deletion of the E3-catalytic RING domain (Ltn1-∆R). This Ltn1 mutant is competent for binding to 60S subunits but fails to ubiquitylate substrates (4), thus preventing downstream events from being triggered.
Results
Purification and EM Processing of the RQC Complex.
A Flag tag added to the C terminus of the Ltn1-∆R subunit enabled affinity purification of the complex, hereby referred to as RQC∆R. Anti-Flag immunoprecipitation (IP) of prefractionated extracts followed by elution with Flag peptide recovered, as expected, Ltn1, 60S ribosomal subunit proteins, Tae2, and Rqc1, according to both silver staining of SDS/PAGE gels (Fig. S1) and mass spectrometry (MS) analyses (Dataset S1). This sample, containing both ribosomal and RQC components, was vitrified for imaging by single-particle cryoelectron microscopy (cryo-EM). Individual raw particles of the sample (Fig. S2A) were summed into homogeneous groups of class averages (Fig. S2B). From these class averages and using a recently described method for generating ab initio 3D reconstructions from single-particle data (11), we computed an initial model that clearly resembled the 60S ribosomal subunit (Fig. S2C), with strong agreement between the reference-free class averages and the reprojections from the computed model (Fig. S2D). Taken together, these results indicated that complexes with the 60S subunit were the primary constituent of the sample.
The initial model of the 60S subunit was further refined using a conventional single-model refinement protocol to a nominal resolution of 8.5 Å (Fig. S3 A and B); the resolution is in part limited by the preferred particle orientation (Fig. S3C) and background from the thin carbon support. The local resolution (12) was also calculated for this map, which agreed well with the global resolution estimation and showed that the majority of the 60S ribosomal subunit core is resolved to ∼8–9 Å. However, this analysis additionally indicated that a lower resolution was observed for the peripheral regions (Fig. S3D). To obtain more insight regarding regions of heterogeneity, we performed a codimensional principal components analysis (13) of the entire dataset. The resulting variance map indeed showed that the peripheral regions were highly heterogeneous (Fig. S3E). Using likelihood-based 3D classification (14), we were able to subclassify the data and account for some of the heterogeneity (Fig. S3F). One of the resulting classes gathered poor particles (presumably due to unaccounted heterogeneity in the data and/or improperly assigned Euler angles); two more recovered classes contained a tRNA, apparently attached to the 60S subunit; the fourth recovered class revealed a large mass extending from around the ribosomal peptidyl site (P site) to the nascent polypeptide tunnel exit (TE) at a nominal resolution of 9.6 Å (Fig. S3F and Fig. 1).
Fig. 1.

(A) RQC∆R complex structure generated after enriching for nonribosomal density using a likelihood-based classification algorithm (14) (see also Fig. S3), displayed in solid gray, and (B) colored by local resolution. (C) Global Fourier shell correlation curve and nominal resolution value of 9.6 Å for the same map.
The Ltn1 E3 Ubiquitin Ligase Bridges the Ribosomal Factor-Binding Site and the TE.
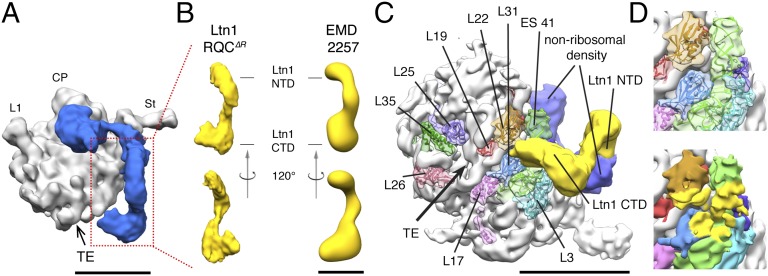
Ltn1 is readily identified and oriented within the nonribosomal density by docking the structure of isolated purified Ltn1 protein (10) into a similarly shaped region of the RQC∆R complex (Fig. 2 A and B). The E3 corresponds to a large, elongated mass spanning the periphery of the ribosome. Ltn1’s evolutionarily conserved N terminus (4, 10) contacts the vicinity of the canonical translational GTPase interaction site (Fig. 2 A and C). Meanwhile, Ltn1’s evolutionarily conserved C terminus binds to the ribosome in the vicinity of the TE, placed so that the RING domain would be in an appropriate position for direct ubiquitylation of stalled nascent chains (Fig. 2C). Accordingly, ribosomal protein L25 (Rpl25), which is also exposed in proximity of the TE, is likewise ubiquitylated in an Ltn1-dependent manner (15). Rpl22, Rpl31, Rpl3, and Rpl24, as well as the rRNA expansion segment ES-41, reside in the immediate vicinity of Ltn1’s C-terminal attachment site, with the ribosomal TE alongside (Fig. 2 C and D). This attachment region has been previously implicated in the binding of other factors involved in nascent chain metabolism (16).
Fig. 2.
The Ltn1 E3 ubiquitin ligase bridges the ribosomal factor-binding site and the TE. (A) Crown view of the RQC∆R complex structure from the intersubunit surface, filtered to 15 Å to emphasize nonribosomal density that is resolved at lower resolution. Gray, 60S subunit; blue, RQC∆R density. The opening of the nascent chain tunnel (tunnel exit, TE), ribosomal protein Rpl1 (L1), central protuberance (CP), and stalk (St) are indicated. (B) Comparison between the Ltn1 density (yellow) segmented from the RQC∆R complex (Left) and a negative stain reconstruction of purified Ltn1 from the Electron Microscopy Data Bank (EMD 2257) (10) (Right). (C) View of the surface surrounding the TE, with ribosomal proteins and rRNA expansion segment-41 (ES-41) indicated. PDB IDs 3U5D and 3U5E were used for ribosomal RNA and proteins, respectively. Ltn1 is shown in yellow. Additional nonribosomal density is shown in blue. (D) Close-up view of the Ltn1 interaction site on the 60S ribosomal subunit. Ribosomal proteins and RNA in closest proximity are docked into the experimental density (Top); their surfaces within a distance of 15 Å from Ltn1 are shaded yellow (Bottom). (Scale bars, 100 Å.)
Ltn1’s middle region, unlike the ends, exhibits no obvious direct contacts. Rather, this region, predicted to be composed of a long stretch of HEAT- or ARM-type α-helical repeats (10), appears to function as a linker mediating communication between Ltn1’s ends bound to distal sites on the ribosome. This observation may explain the apparent lack of sequence conservation of the middle region among Ltn1 orthologs (4) and why the ENU-induced 13-aa internal deletion in the corresponding region of mouse listerin results in a hypomorphic allele and neurodegeneration (6).
Structural Basis for Ltn1’s Selective Binding to the 60S Ribosomal Subunit.
Ltn1 was found only in association with 60S ribosomal subunits in the samples analyzed, and there was no indication that it can bind to 80S ribosomes. This observation supports previous data showing that Ltn1 is predominantly found in the 60S fraction of sucrose gradients (4), copurifies with 60S subunits (4, 7, 9), and relies on ribosome splitting for function (17). To understand the structural elements underlying this selectivity, the segmented Ltn1 density from RQC∆R was docked onto an 80S ribosome. The results show that Ltn1 would not be able to bind to 80S ribosomes in the configuration observed here for the RQC∆R complex, as this would result in extensive clashes between the 40S subunit and the E3 (Fig. 3 A and B and Fig. S4). Notably, with regard to overall shape and size, Ltn1 shows striking similarities with the signal recognition particle (SRP), another factor that binds to and connects distal sites on the 60S subunit, but in the context of 80S ribosomes (18). Comparison of 60S subunit complexes with Ltn1 and SRP shows that SRP follows a different trajectory on the ribosomal surface that allows it to also connect the TE and GTPase sites, but without clashing with the 40S subunit (Fig. 3 C and D and Fig. S5).
Fig. 3.

Structural basis for Ltn1’s selective binding to the 60S ribosomal subunit. (A) Overlay of the segmented Ltn1 density from the RQC∆R complex with the 80S ribosome structure as observed in the RQC∆R complex. Ltn1, bright yellow; 60S subunit, gray; 40S subunit, light yellow. (B) The overlap of the Ltn1 and the 40S subunit densities is indicated in red (Ltn1 removed for clarity). (C) SRP-80S complex from Halic et al. (35). SRP is shown in red. (D) Overlay of the segmented Ltn1 structure (yellow) from the RQC∆R complex with the SRP-80S complex from Halic et al. (35).
Given that Ltn1’s ∼50-residue–long RING domain suffices for E3 activity (10), it has remained to be understood why Ltn1 orthologs are all large proteins, in the 150- to 190-kDa range. The above data suggest that Ltn1’s size, elongated shape, and ribosome interaction mode can potentially be explained by the need to mediate communication between distal sites on the 60S subunit, as well as by the need for its RING domain to bind in proximity to the TE in a stalled 60S subunit while avoiding binding to 80S ribosomes and ubiquitylating normally elongating nascent chains.
Tae2 Recognizes the Exposed tRNA Moiety in a Stalled 60S⋅Peptidyl-tRNA Structure.
We next investigated the nature of the RQC∆R density connected to Ltn1’s N terminus and surrounding the ribosomal P site. To determine whether the density corresponds to Rqc1 or Tae2, we analyzed Ltn1-ΔR IPs from strains that had been deleted for one or the other factor. Comparison between 3D reconstructions of RQC∆R (Fig. 1) and RQC∆R/tae2∆ (Fig. 4A) revealed that ∼85% of the density at the 60S intersubunit surface disappeared upon TAE2 deletion (Fig. 4B and Fig. S6). In contrast, no clear density differences could be identified between structures of complexes purified from wild-type and rqc1∆ strains (Fig. S6). We conclude that the P-site density region of RQC∆R corresponds largely to Tae2, although Tae2’s precise boundaries cannot be determined, and the contribution of additional factors cannot be excluded without an atomic structure of the complex. The RQC∆R/tae2∆ structure in Fig. 4 A and B and Fig. S6 also confirms that Ltn1 is able to bind to ribosomes in cells deficient for Tae2 (7, 9), although Ltn1 binding is less stable (9) and the ubiquitylation and degradation of nonstop substrates is reduced in Tae2’s absence (7, 9).
Fig. 4.
Tae2 recognizes the exposed tRNA moiety in a stalled 60S-peptidyl-tRNA structure. (A) Refinement of RQC∆R/tae2∆ after 3D classification and enrichment for nonribosomal density, displayed in solid gray (Top) and colored by local resolution (Middle). The global resolution curve is shown (Bottom) and indicates a nominal value of 11.2 Å. (B) Refinement of RQC∆R (Left) and RQC∆R/tae2∆ (Middle) after enriching for all nonribosomal density, both filtered to 15Å. (Right) comparison between the two structures, with nonribosomal densities present in RQC∆R but not in RQC∆R/tae2∆ shown in red, filtered to 15 Å, and displayed at 5σ above the mean pixel value. (C) Segmented density of the RQC∆R structure in Fig. 1A showing the P-site tRNA (green) and surrounding Tae2-dependent density (orange). Both the tRNA and Tae2 components were segmented using a watershed algorithm implemented in Segger (36). Without atomic resolution, precise localization of Tae2 domain boundaries is not possible. (D) Tae2-dependent density contacts in the vicinity of the ribosomal P site. PDB IDs 3U5D and 3U5E were used for ribosomal RNA and proteins, respectively. P-tRNA (green), ribosomal proteins (teal and blue), and rRNA helices (“H”; violet, red, and purple) in the Tae2-dependent density binding site are indicated (Left); regions within 15 Å of the density are indicated by checkered orange shading (Right). (E) Ltn1-∆R co-IPs with peptidyl-tRNA. Cells expressed Ltn1-∆R-Flag in the presence or absence of a protein A reporter encoded by nonstop mRNA (PtANS). Lysates were used for anti-Flag IP (lanes 4–6), and co-IP’ed PtANS and its tRNA-conjugated form was detected by anti-protein A immunoblot. (Lanes 1–3) Whole cell extract (input). Samples for lanes 3 and 6 were treated with RNase A before loading. (F–I) Steric hindrance prevents simultaneous binding of Tae2 and the 40S ribosomal subunit to the 60S subunit. (F) 80S ribosome structure, with the 60S subunit in gray and the 40S subunit in yellow. (G and H) Only the contour of the 60S subunit is presented. (G) 40S subunit as in F. (H) The Tae2-tRNA density as observed in the RQC∆R complex (Tae2, orange; tRNA, green). (I) The overlap of Tae2 on the 40S subunit density is indicated in red. Tae2 and tRNA were removed for clarity.
Notably, the RQC∆R structure reveals that a tRNA is present in the P site and that this tRNA is contacted by Tae2 (Fig. 4 C and D). Interestingly, bioinformatics analyses predict a tRNA-binding function for Tae2 (19). In support of this observation, our results indicate a strong preference for Tae2 to be associated with tRNA within the cryo-EM data (Fig. S7). Whereas tRNA would not be expected to be found associated with the P site in free 60S subunits, unhydrolyzed peptidyl-tRNA is a product of defective translation termination (5) and has been shown to accumulate in the absence of Ltn1 function (8), suggesting that it can be targeted by Ltn1-mediated ubiquitylation and degradation. We thus presume the Tae2-bound P-tRNA shown in Fig. 4 C and D is conjugated to 60S-stalled nascent chains, although those chains cannot be visualized in the structure at this resolution. To further test the model that peptidyl-tRNA is present in RQC∆R complexes, Flag IP products from cells expressing Ltn1-∆R-Flag along with a nonstop reporter protein (protein A nonstop) were analyzed by immunoblot against protein A. The results revealed that peptidyl-tRNA indeed co-IP’ed with Ltn1-ΔR (Fig. 4E).
In addition to binding to the P-site tRNA, Tae2 binds to the surrounding 28S rRNA helix H69 (Fig. 4D). This contact site is also shared by the 40S ribosomal subunit. Given that the Tae2-dependent density is large, and that Tae2 and 40S subunits share a common binding region on the 60S intersubunit surface (Fig. 4 F–I), Tae2 and the 40S subunit cannot bind simultaneously to the 60S subunit. This observation explains Tae2’s reported 60S subunit binding selectivity (7, 9), as is also the case with Ltn1 (Figs. 3 and 4).
Discussion
Principles Underlying Ribosome-Associated Protein Quality Control.
E3 ligases that function in protein quality control must target heterogeneous substrates in a broad yet specific manner. In this regard, the structure of the RQC∆R complex presented here supports the evidence that stalled nascent chains are presented as substrates for ubiquitylation on ribosomes (4, 7–9, 17) and shows that Ltn1 is able to target a broad diversity of such quality control substrates by using the 60S subunit as an adapter. Furthermore, our results suggest that, due to steric hindrance preventing simultaneous binding of Ltn1 and 40S subunits to 60S subunits, Ltn1 can specifically target nascent chains that are stalled on 60S subunits for ubiquitylation without interfering with nascent chains on translating ribosomes.
The RQC∆R structure provides evidence that an exposed tRNA moiety of peptidyl-tRNA serves as a signal that is unique to stalled 60S subunits, as hypothesized previously (20), and that Tae2 is the RQC subunit recognizing the tRNA. We propose a model (Fig. 5 and Fig. S8) in which RQC assembly would optimally begin with Tae2-mediated tRNA recognition. Accordingly, a yeast cell is estimated to have ∼3,000 free 60S particles and a comparable number of free 40S particles, ∼8,000 Tae2 molecules (21), and only ∼200 Ltn1 molecules (21). In recognizing the stalled tRNA, Tae2 would additionally prevent the reassociation of free 40S subunits to the stalled 60S-peptidyl-tRNA complex due to steric hindrance. An expected consequence of Tae2’s antiassociation activity is to facilitate Ltn1 binding, because Ltn1 appears to be unable to bind to 80S ribosomes. In support of this model, in the absence of Tae2, RQC substrates are inefficiently degraded and found predominantly in 80S ribosomal complexes (9). The RQC∆R structure also shows a continuous density connecting Tae2 and Ltn1, so in addition to recognition and antiassociation functions, Tae2 may act in stabilizing Ltn1 binding to the complex, as also supported by biochemical evidence (9). Once recruited, the elongated Ltn1 molecule can then extend to the vicinity of the TE and position the RING domain to ubiquitylate stalled polypeptides (Fig. 5). Subsequently, downstream cofactors would extract and deliver stalled chains to the proteasome for degradation (7–9). Such a mechanism ensures that stalled nascent chains encoded by defective messages are promptly recognized and dealt with before being released and posing harm to the cell.
Fig. 5.
Model for RQC function. (A) Ribosomes can halt translation for a variety of reasons. For example, ribosomes translating aberrant mRNA lacking stop codons (“nonstop mRNA”) eventually reach the poly(A) tail, which encodes poly(Lys); nascent polybasic tracts can cause translation to slow down or pause altogether (4). (B) 80S ribosomes that pause during translation and fail to either resume elongation or terminate translation are rescued by factors that promote splitting of the subunits (e.g., ref. 5). Ribosomal rescue generates free 60S subunits that remain stalled with peptidyl-tRNA (e.g., ref. 5). (C) The Tae2 subunit of the RQC complex recognizes peptidyl-tRNA-stalled 60S subunits via direct interaction with the exposed tRNA. Tae2 also stabilizes the binding of Ltn1, the E3-catalytic subunit of RQC, to the complex. Ltn1 is positioned such that the RING domain sits next to the ribosomal tunnel exit (TE). The RING domain binds to ubiquitin (Ub)-carrying E2 conjugases and stimulates Ub transfer to substrates. The RQC∆R structure implies that the C-terminal poly-Lys tracts characteristic of a subset of Ltn1 substrates would remain buried within the nascent chain exit tunnel, supporting the model that such sequences act as destabilizing motifs (“degrons”) by mediating translational stalling, rather than by acting directly as ubiquitin acceptor sites (4). Ubiquitylation happens while nascent chains are still 60S subunit associated and provides two functions in this pathway. First, it mediates extraction of the stalled protein by recruiting the AAA ATPase Cdc48/VCP/p97. Second, it signals proteasomal degradation of the aberrant polypeptide. (D) In a competing reaction with Tae2 binding, stalled 60S subunits can reassociate with free 40S subunits (17). Because Ltn1 is unable to bind to 80S ribosomes, Tae2-mediated P-tRNA recognition also facilitates Ltn1 binding by preventing ribosomal subunit reassociation.
Note that while this manuscript was under review, a report including the cryo-EM structure of a mammalian listerin⋅60S subunit complex was published (22). The structure was generated by a different approach from the one used in this work—a minimal complex was reconstituted in vitro using components isolated from rabbit reticulocyte lysate. Listerin’s overall shape and mode of 60S subunit binding in that complex are remarkably similar to the results described here for the yeast ortholog. Likewise, the authors’ interpretations of how listerin binds to 60S subunits and how it distinguishes stalled 60S subunits from normally translating 80S ribosomes are in line with the interpretations presented here. However, because stalled tRNA is not visible in that structure, and the Tae2 ortholog was absent in the reconstitution reaction, the structure of the RQC∆R complex purified from cells presented here also reveals how stalled ribosomes can be selectively recognized via the tRNA-Tae2 interaction and provides insight into Tae2’s roles in the context of ribosome-associated protein quality control.
Accession Codes.
The EM reconstructions of the RQC∆R and RQC∆R/tae2∆ complex were deposited in the Electron Microscopy Data Bank under accession codes EMD-2797 and EMD-2798, respectively.
Materials and Methods
Reagents.
The primary antibodies used were: mouse monoclonal anti-Flag tag (M2; Sigma) and the agarose conjugated anti-Flag M2 (Sigma, A2220) and rabbit anti-protein A (Sigma; P3775-1VL). The secondary antibodies used were HRP-conjugated anti-mouse and anti-rabbit (Molecular Probes). The 3× Flag peptide was obtained from Sigma (F4799). The protein A-nonstop (PtANS) construct was a gift from A. van Hoof (University of Texas, Houston) (23).
Yeast Strains.
All strains used in the work are isogenic to BY4741. Yeast media preparation, growth, transformation, and manipulations were performed according to standard protocols (24). The wild-type and single deletion strains were obtained at Open Biosystems. The Ltn1-Flag and Ltn1-ΔR-Flag strains were generated by the insertion of the kanMX6 selectable marker as previously described (4). Deletion of TAE2 or RQC1 in the Ltn1-ΔR-Flag background was generated by insertion of the His3MX6 cassette.
Coimmunoprecipitation Assay.
Cells starting at OD600 = 0.3 were grown until OD600 = 1.6. Pelleted cells were resuspended in the cold IP buffer and lysed using a mechanical bead beater (Savant Fast prep; FP120). Total lysates were centrifuged once at 21,000 × g for 5 min and the collected supernatant was submitted to another centrifugation of 1 h. After quantification of the protein concentration, 20 mL of bead-conjugated anti-Flag M2 antibody slurry was incubated with 1.5 mL of each sample (∼4 mg total protein) for 2 h at 4 °C, washed four times with IP buffer, and eluted with 50 mL of 3× Flag peptide (2 mg/mL) in IP buffer.
Immunoaffinity Purification of RQC∆R Complexes.
Reseeded cells grown until OD600 = 1.6 were centrifuged at 2,800 × g for 10 min, washed with cold water, and flash frozen. Pelleted cells were resuspended in cold IP buffer [50 mM Hepes-KOH pH 6.8, 150 mM KOAc, 5 mM MgOAc, 15% (vol/vol) glycerol, 1 mM EDTA, 2 mM DTT, 2× protease inhibitors, 0.1% Igepal CA-630] (7) and disrupted using a French press at 18,000 psi (Thermo Fisher Scientific; FA-078A). Total lysates were first centrifuged at 2,800 × g for 10 min, then collected supernatants were centrifuged at 210,000 × g for 70 min. The protein concentration of the final supernatants was determined by the bicinchoninic acid assay (BCA) method (Pierce). A total of 80 mL of bead-conjugated anti-Flag M2 antibody slurry was incubated with 12 mL of each sample (∼24 mg total protein) for 2 h at 4 °C, washed four times with IP buffer, and eluted with 100 mL of 3× Flag peptide (2 mg/mL) in IP buffer containing 100 mM KOAc. Samples prepared for cryo-EM analyses were concentrated and further purified using 0.5-mL centrifugal filters with 100-kDa cutoff (Amicon/Millipore).
Peptidyl-tRNA Analysis.
Cells were seeded at OD600 = 0.2 and grown until OD600 = 0.8. Whole cell extract preparation and immunoprecipitation were carried out as above, except that 1× ribonuclease inhibitor (RNAseOUT; Invitrogen) was added unless samples were to be treated with RNase A. RNase A (Roche) was used at 2 mg per 30 mg of total protein (determined by the BCA method) for 10 min at 37 °C. To better preserve peptidyl-tRNA conjugates, protein electrophoresis was conducted in NuPAGE 4–12% Bis-Tris pH 6.4 gels (Invitrogen) at 4 °C.
Immunoblotting.
Samples were separated in 4–20% Tris-glycine gradient gel (Novex; Life Technologies) and transferred to PVDF membranes (Immonobilon-P; Millipore) using a semidry system (WEP Company). Membranes were blocked with 5% (vol/vol) milk and used for probing with anti-Flag antibody diluted 1:20,000.
Silver Staining.
Silver staining of proteins separated in NuPAGE 4–12% gradient gels (Novex; Life Technologies) was performed with the Silver-Quest kit (Life Technologies; LC6070).
Mass Spectrometry.
Mass Spectrometry analysis was analyzed by LC-MS/MS using a Q-Exactive mass spectrometer (Thermo Scientific) and is described in detail in SI Materials and Methods.
Specimen Vitrification for Cryoelectron Microscopy.
RQC∆R, RQC∆R/tae2∆, and RQC∆R/rqc1∆ specimens were prepared for cryo-EM by applying 3 μL of sample to a freshly plasma-cleaned holey carbon C-flat grid (Protochips) that had been overlaid with 2 nm thin carbon film, allowing the sample to adsorb to the grid for 30 s, followed by blotting with filter paper and plunge freezing into liquid ethane using a manual cryoplunger in an ambient environment at 4 °C.
Cryoelectron Microscopy Data Collection.
For all samples, data were acquired using the Leginon software (25) installed on a Tecnai F20 Twin transmission electron microscope operating at 200 kV, with a dose of 32e−/Å2. For the RQC∆R and RQC∆R/tae2∆ samples, a TVIPS Tietz F416 CMOS CCD 4K × 4K camera was used to record the images at a nominal magnification of 50,000×, corresponding to a pixel size of 1.69 Å at the specimen level. A total of 3,037 images were collected for RQC∆R and 2,048 images were collected for RQC∆R/tae2∆. For the RQC∆R/rqc1∆ sample, a Gatan K2 Summit direct detection device was used to record 20 raw frames collected over a 5-s exposure time (250 ms per frame), with each frame receiving a dose of ∼9.4 e−/pixel/s, as previously described (26). A total of 839 such “movies” were automatically collected and recorded at a nominal magnification of 29,000×, corresponding to a pixel size of 1.21 Å at the specimen level. The individual frames were aligned as previously described (26). For all datasets, the estimated underfocus ranged from 0.5 to 4.0 µm and was distributed in an approximately Gaussian manner while centered at 2.0–2.25 µm.
Image Processing and Single-Model Refinement of RQC∆R Data.
Unless otherwise noted, all of the following image processing steps were performed within the Appion pipeline (27). The contrast transfer function (CTF) for all micrographs was estimated using the CTFFind3 package (28). All micrographs were also manually masked with the “manual masking” tool in Appion to remove regions that were over thick carbon and/or containing large amounts of disordered or aggregated particles. Particles were initially selected using Dog Picker (29) from 100 micrographs. A phase-flipped, contrast-inverted, and 4× binned stack of 5k particles was created from these picks using a box size of 64 pixels and pixel size of 6.76 Å. The stack was subjected to reference-free 2D alignment and clustering using CL2D (30) to obtain 64 2D classes. The classes were inspected visually, and 20 of these classes were used as templates to select particles over the entire set of micrographs using FindEM (31), providing 276,411 particles located over thin carbon. Micrographs were phase flipped, contrast inverted, and binned by 4, and individual particles were extracted using a box size of 64 pixels, corresponding to a pixel size of 6.76 Å. We performed 2D alignment and clustering using CL2D (30) to eliminate any particles that did not display identifiable features. We then performed another round of 2D alignment and clustering using ISAC (32), outside of Appion to eliminate particles that did not consistently produce stable class averages, as previously described (26). After manual micrograph masking, CL2D, and ISAC, 77,962 particles were retained for all subsequent analyses. The omitted particles likely suffered from drift, radiation damage, and/or poorly formed quaternary structure. An ab initio model was created from reference-free 2D classes obtained from the 4× binned particles using OptiMod (11). All subsequent steps were performed outside of Appion. Initial Euler angles were determined using the projection-matching protocol implemented within the Xmipp package using an unbinned, phase-flipped, and contrast-inverted stack (33). After determining the initial angles and shifts, the particle parameters were converted for use in Frealign (34). An unbinned stack that had not been phase flipped or contrast inverted was used in the parameter refinement in Frealign. The resolution to which the data were refined in Frealign was always limited to 15 Å. We additionally performed a gold-standard refinement of the unclassified data, as described previously (26), and show that the global resolution in both cases is similar (Fig. S3B). Following this step, wherein a single set of Euler angles was obtained with a single-model refinement protocol, we proceeded to iterative 3D classification procedures, as described below.
Unsupervised 3D Classification of the RQC∆R Dataset.
After establishing a set of Euler angles using a single-model refinement protocol, we next proceeded with unsupervised 3D classification using the likelihood-based approach implemented in Frealign9 (14) to characterize regions of heterogeneity. The general strategy follows the theoretical and practical guidelines that have been previously proposed (14). Details are described in SI Materials and Methods. The results of k = 4 are shown in Fig. S3F.
Three-Dimensional Sorting of the RQC∆R Dataset.
To obtain a better understanding and quantification of particle occupancies corresponding to heterogeneous regions identified using Frealign9, we performed a 3D sorting strategy, similar to that which has been previously described (26), and is further explained in SI Materials and Methods. The described sorting procedure enabled us to calculate and quantify coassociations between the different components, a procedure that is describe in detail in SI Materials and Methods.
Image Processing and Refinement of RQC∆R/tae2∆ and RQC∆R/rqc1∆ Datasets.
Data were processed in a conceptually identical manner as for RQC∆R described above, except that the number of particles in the RQC∆R/tae2∆ dataset was 37,819 and in the RQC∆R/rqc1∆ dataset, 16,784.
Supplementary Material
Acknowledgments
We thank the members of the C.A.P.J. laboratory and the C.S.P.–B.C. laboratory for discussions, O. Brandmann for advice, A. van Hoof for reagents, and Jean-Cristophe Ducom (The Scripps Research Institute, TSRI) for support with computational resources. The EM work was conducted at the National Resource for Automated Molecular Microscopy at TSRI, which is supported by the National Institutes of Health’s National Institute of General Medical Sciences (NIGMS) Biomedical Technology Research Center program (GM103310) (B.C. and C.S.P.). Work in the C.A.P.J. laboratory is supported by R01 Grant NS075719 from the National Institute of Neurological Disorders and Stroke and Grant CA152103 from the National Cancer Institute. Some of this work was also supported by grants from the Ellison Medical Foundation and the Sidney Kimmel Cancer Research foundation (to E.J.B.) and by the Leona M. and Harry B. Helmsley Charitable Trust Grant 2012-PG-MED002 (to D.L.). Molecular graphics and analyses were performed with the University of California San Francisco chimera package (supported by NIGMS P41-GM103311). Raw EM data will be provided upon request. This is manuscript 27018 from The Scripps Research Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: EM maps have been deposited in the Electron Microscopy Data Bank (EMD-2797 and EMD-2798 for the RQC-delta-RING and RQC-delta-RING/Tae2D reconstructions, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413882111/-/DCSupplemental.
References
- 1.Graille M, Séraphin B. Surveillance pathways rescuing eukaryotic ribosomes lost in translation. Nat Rev Mol Cell Biol. 2012;13(11):727–735. doi: 10.1038/nrm3457. [DOI] [PubMed] [Google Scholar]
- 2.Petry S, Weixlbaumer A, Ramakrishnan V. The termination of translation. Curr Opin Struct Biol. 2008;18(1):70–77. doi: 10.1016/j.sbi.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 4.Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467(7314):470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330(6002):369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu J, et al. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc Natl Acad Sci USA. 2009;106(7):2097–2103. doi: 10.1073/pnas.0812819106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandman O, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151(5):1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verma R, Oania RS, Kolawa NJ, Deshaies RJ. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. elife. 2013;2:e00308. doi: 10.7554/eLife.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Defenouillère Q, et al. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc Natl Acad Sci USA. 2013;110(13):5046–5051. doi: 10.1073/pnas.1221724110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyumkis D, et al. Single-particle EM reveals extensive conformational variability of the Ltn1 E3 ligase. Proc Natl Acad Sci USA. 2013;110(5):1702–1707. doi: 10.1073/pnas.1210041110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyumkis D, Vinterbo S, Potter CS, Carragher B. Optimod—an automated approach for constructing and optimizing initial models for single-particle electron microscopy. J Struct Biol. 2013;184(3):417–426. doi: 10.1016/j.jsb.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11(1):63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Penczek PA, Kimmel M, Spahn CM. Identifying conformational states of macromolecules by eigen-analysis of resampled cryo-EM images. Structure. 2011;19(11):1582–1590. doi: 10.1016/j.str.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyumkis D, Brilot AF, Theobald DL, Grigorieff N. Likelihood-based classification of cryo-EM images using FREALIGN. J Struct Biol. 2013;183(3):377–388. doi: 10.1016/j.jsb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ossareh-Nazari B, et al. Ubiquitylation by the Ltn1 E3 ligase protects 60S ribosomes from starvation-induced selective autophagy. J Cell Biol. 2014;204(6):909–917. doi: 10.1083/jcb.201308139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leidig C, et al. Structural characterization of a eukaryotic chaperone—the ribosome-associated complex. Nat Struct Mol Biol. 2013;20(1):23–28. doi: 10.1038/nsmb.2447. [DOI] [PubMed] [Google Scholar]
- 17.Shao S, von der Malsburg K, Hegde RS. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol Cell. 2013;50(5):637–648. doi: 10.1016/j.molcel.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halic M, et al. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427(6977):808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- 19.Burroughs AM, Aravind L. A highly conserved family of domains related to the DNA-glycosylase fold helps predict multiple novel pathways for RNA modifications. RNA Biol. 2014;11(4):360–372. doi: 10.4161/rna.28302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodrigo-Brenni MC, Hegde RS. Design principles of protein biosynthesis-coupled quality control. Dev Cell. 2012;23(5):896–907. doi: 10.1016/j.devcel.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 22.Shao S, Hegde RS. Reconstitution of a minimal ribosome-associated ubiquitination pathway with purified factors. Mol Cell. 2014;55(6):880–890. doi: 10.1016/j.molcel.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson MA, Meaux S, van Hoof A. A genomic screen in yeast reveals novel aspects of nonstop mRNA metabolism. Genetics. 2007;177(2):773–784. doi: 10.1534/genetics.107.073205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams A, Gottschling D, Kaiser C. Methods in Yeast Genetics. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1997. [Google Scholar]
- 25.Suloway C, et al. Automated molecular microscopy: The new Leginon system. J Struct Biol. 2005;151(1):41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Lyumkis D, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342(6165):1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lander GC, et al. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166(1):95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142(3):334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 29.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166(2):205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorzano CO, et al. A clustering approach to multireference alignment of single-particle projections in electron microscopy. J Struct Biol. 2010;171(2):197–206. doi: 10.1016/j.jsb.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roseman AM. FindEM—a fast, efficient program for automatic selection of particles from electron micrographs. J Struct Biol. 2004;145(1–2):91–99. doi: 10.1016/j.jsb.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Yang Z, Fang J, Chittuluru J, Asturias FJ, Penczek PA. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 2012;20(2):237–247. doi: 10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sorzano CO, et al. XMIPP: A new generation of an open-source image processing package for electron microscopy. J Struct Biol. 2004;148(2):194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Grigorieff N. FREALIGN: High-resolution refinement of single particle structures. J Struct Biol. 2007;157(1):117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 35.Halic M, et al. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature. 2006;444(7118):507–511. doi: 10.1038/nature05326. [DOI] [PubMed] [Google Scholar]
- 36.Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.