Significance
Nitric oxide (NO) plays key roles in coordinating plant growth and development with environmental cues. Potassium (K+) is an essential plant nutrient important for growth and stress tolerance. However, little is known about the molecular connection of NO to K+ homeostasis. Through a genetic approach, this study provides a detailed regulatory mechanism of NO to K+ channel AKT1-mediated K+ absorption, through its modulation on vitamin B6 biosynthesis in plants. This finding demonstrates a previously unidentified role of NO in the control of K+ content in plants, which may represent a normal plant adaptive response under unfavorable external conditions.
Keywords: genetic approach, electrophysiological studies, potassium nutrition
Abstract
Nitric oxide (NO), an active signaling molecule in plants, is involved in numerous physiological processes and adaptive responses to environmental stresses. Under high-salt conditions, plants accumulate NO quickly, and reorganize Na+ and K+ contents. However, the molecular connection between NO and ion homeostasis is largely unknown. Here, we report that NO lowers K+ channel AKT1-mediated plant K+ uptake by modulating vitamin B6 biosynthesis. In a screen for Arabidopsis NO-hypersensitive mutants, we isolated sno1 (sensitive to nitric oxide 1), which is allelic to the previously noted mutant sos4 (salt overly sensitive 4) that has impaired Na+ and K+ contents and overproduces pyridoxal 5′-phosphate (PLP), an active form of vitamin B6. We showed that NO increased PLP and decreased K+ levels in plant. NO induced SNO1 gene expression and enzyme activity, indicating that NO-triggered PLP accumulation mainly occurs through SNO1-mediated vitamin B6 salvage biosynthetic pathway. Furthermore, we demonstrated that PLP significantly repressed the activity of K+ channel AKT1 in the Xenopus oocyte system and Arabidopsis root protoplasts. Together, our results suggest that NO decreases K+ absorption by promoting the synthesis of vitamin B6 PLP, which further represses the activity of K+ channel AKT1 in Arabidopsis. These findings reveal a previously unidentified pivotal role of NO in modulating the homeostasis of vitamin B6 and potassium nutrition in plants, and shed light on the mechanism of NO in plant acclimation to environmental changes.
Nitric oxide (NO) acts as a crucial signaling molecule in various physiological processes in plants, such as seed germination and dormancy (1, 2), root development (3), leaf senescence (4, 5), floral transition (6), stomatal movement (7, 8), iron homeostasis (9, 10), and hormone responses (11, 12). NO production is altered when plants are subjected to abiotic or biotic stresses (13, 14). High salt, a major environmental factor that limits agriculture yield, induces a quick endogenous NO accumulation in plants (15, 16), and triggers enhanced Na+ influx and reduced K+ absorption in the root (17). Both endogenously produced NO and exogenously applied NO have been proposed to enhance plant salt tolerance (18–21) by attenuating high salt-induced increases in the Na+ to K+ ratio. Genetic analysis showed that K+ nutrition, but not Na+, plays critical role in plant salt tolerance (22). However, the molecular basis of NO effect on K+ or Na+ content is elusive.
K+ levels in plant tissues are determined by K+ uptake and translocation, which are mediated by a large number of transporters and channels. In Arabidopsis, the K+ channel AKT1 (23, 24) and K+ transporter AtHAK5 (25) are the two major molecular entities responsible for K+ absorption from the environment (26–28). AKT1 contributes to K+ acquisition over a wide range of external K+ concentrations (10 µM–10 mM), whereas AtHAK5 mediates limited uptake capacity at low external K+ concentrations (1–200 µM) (26–28). AKT1 is primarily and constitutively expressed in the root (29), and it functions as an inward-rectifying K+ channel (23) that displays a high selectivity for K+ over Na+ under physiological levels (30, 31). AKT1 activity is regulated mainly by protein modification through phosphorylation/dephosphorylation (30, 32). Plant K+ absorption is increased when Ca2+ sensors calcineurin B-like protein 1 (CBL1) and/or CBL9 recruit cytosolic protein kinase CIPK23 (CBL-interacting protein kinase 23) to the plasma membrane, where CIPK23 phosphorylates and activates the AKT1 channel (30, 32). Further, in this CBL–CIPK regulatory pathway, a set of CBLs were found to target multiple CIPKs to regulate the activity of AKT1 channel (33). The association of AKT1 with a 2C-type protein phosphatase (PP2C) AIP1, which dephosphorylates the channel (33), or AtKC1, which forms a ternary complex with the channel, inhibits AKT1 activity (34, 35). Different from AKT1, the AtHAK5 transporter is transcriptionally regulated, with remarkable induction seen in response to K+ deficiency (26). Environmental cues or cellular signals that modulate AKT1 channel activity or AtHAK5 expression may affect plant K+ uptake and therefore K+ content.
Recent studies suggested a potential link between K+ absorption and vitamin B6 homeostasis in plants. In a sos4 mutant containing low K+ and high Na+ relative to wild-type plants (36), vitamin B6 pyridoxal-5′-phosphate (PLP) levels were unusually high (37). Interestingly, in this NaCl-hypersensitive mutant, the less retention of K+ was more pronounced in roots than shoots, especially under salt stress (36), suggesting less K+ uptake in this high PLP-containing mutant. PLP was proposed to affect the activity of ion channels, particularly K+ channels, in plants (36) because PLP and its derivatives are known to antagonize the activation of some ATP-gated ion channels in animal cells (38). However, the effect of PLP on ion or K+ channels in plants has not been explored yet.
In this study, we screened and characterized a new NO-hypersensitive mutant, sno1, to dissect the regulatory role of NO in plant potassium absorption. SNO1 was identified as the same gene known as salt overly sensitive 4 (SOS4) (36) encoding a pyridoxal kinase that catabolizes the biosynthesis of an active form of vitamin B6, PLP. The sos4 mutant was reported to have unexpected elevated levels of PLP (37) and disturbed Na+ and K+ amounts (36). Here we show that NO induced accumulation of PLP. Furthermore, we found that PLP significantly inhibited the activity of inward-rectifying K+ channel AKT1, thereby resulting in decreased K+ content in plants. We propose that NO may lower plant K+ absorption to slow plant growth as an adaptive response to unfavorable environmental conditions.
Results
Isolation of sno1, an NO Hypersensitive Mutant.
Previously, Arabidopsis NO-hypersensitive mutants were screened using root growth on 5 µM sodium nitroprusside (SNP), an NO donor, as a selection phenotype (6). To further explore the roles of NO in plants, we carried out a similar screen using a higher dose of SNP (25 µM). A mutant designated sno1 (sensitive to nitric oxide 1) was identified from EMS-mutagenized M2 seeds and chosen for further characterization. Compared with wild-type seedlings, the root growth of sno1 was significantly inhibited by SNP treatment (Fig. 1A). Dose–response analysis confirmed the hypersensitivity of sno1 to SNP (Fig. 1B). To verify that the hypersensitive response of sno1 was specific to SNP-delivered NO gas but independent of cyanide, we tested the effect of potassium ferrocyanide [K4Fe(CN)6], a commonly used negative control for SNP as an NO donor (2), on sno1. We found that sno1 mutant displayed similar root growth as wild type under various concentrations of potassium ferrocyanide (Fig. S1A), confirming the independence of cyanide to the observed SNP response of sno1. In addition, we tested the response of sno1 to another NO donor, S-nitroso-N-acetylpenicillamine (SNAP), which has a distinct chemical structure from SNP, and found that sno1 exhibited hypersensitivity to SNAP treatment (Fig. S1B), and that the root growth inhibition of sno1 by SNAP was markedly reversed when NO scavenger 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3 oxide (c-PTIO) was added (Fig. S1C). Because NO rapidly reacts with molecular oxygen to produce nitrite and nitrate under aerobic conditions (39), we examined the influence of potassium nitrate (KNO3) on sno1. The result showed that potassium nitrate did not cause a clear root growth difference between sno1 and wild type (Fig. S1D), suggesting that the SNP or SNAP hypersensitivity of sno1 was the result of NO but not NO-generated by-products. Together, these results suggest that the sno1 mutant is hypersensitive to the gaseous signaling molecule NO.
Fig. 1.
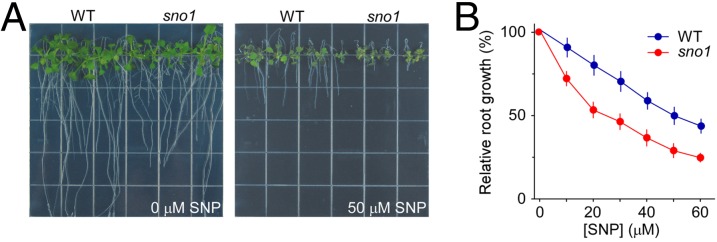
The root growth of sno1 is hyperresponsive to SNP. (A) Phenotypes of WT and sno1 seedlings grown on half-strength MS media supplemented with 0 and 50 μM SNP. Representative images are shown. (B) Sensitivity of sno1 seedlings to different doses of SNP as shown by relative root growth. Relative root growth shown as percentages is presented as a value relative to the primary root elongation on 0 μM SNP. Data shown are mean ± SD, n = 50 seedlings. In A and B, seeds were germinated on half-strength MS media for 3 d, and seedlings were transferred to the SNP containing media for 15 d before being analyzed.
We backcrossed sno1 with the Columbia (Col-0) wild type. When treated with SNP, all of the F1 seedlings showed a wild-type phenotype, and the F2 seedlings segregated at a ∼3:1 ratio (wild type vs. sno1; Fig. S2A), suggesting that sno1 harbors a monogenic recessive mutation.
SNO1 Encodes a Pyridoxal Kinase Involved in Vitamin B6 Synthesis.
The sno1 mutant (in Col-0 background) was crossed with wild-type Landsberg erecta (Ler), and the F2 seedlings that showed SNP hypersensitivity in root growth were selected for the mapping population to clone the SNO1 gene. A single nucleotide substitution from G to A at position 308 (G308A) from the translation start site (ATG) was found in At5G37850, a gene annotated as SOS4 (36), on chromosome 5 (Fig. S2B). This mutation would disrupt the normal splicing acceptor site of the second intron and lead to complete knockout expression of the gene.
To further confirm the identity of SNO1 to SOS4, we analyzed the SNP response of sos4-1 (36), a loss-of-function mutant. As expected, the sos4-1 mutant was hypersensitive to SNP treatment (Fig. S2 C and D). Moreover, the F1 seedlings generated from the sno1 × sos4-1 cross displayed similar sensitivity to SNP as the sos4-1 single mutant (Fig. S2E). Thus, we concluded that SNO1 is SOS4, a pyridoxal kinase that functions in the vitamin B6 synthesis pathway (36, 37) (Fig. S2F).
NO Induces PLP Accumulation in Plants.
To gain insight into the heightened responsiveness of sno1 to SNP, we checked the endogenous NO level of sno1 using an NO-sensitive dye diaminofluorescein-2-diacetate (DAF-2DA), as He et al. (6). No clear difference was found between sno1 and wild type (Fig. S3), so the possibility that higher NO caused greater SNP response in sno1 mutant was ruled out.
The sos4-1 mutant was reported to contain increased amount of several vitamers (37), especially PLP, which accumulated most significantly and unexpectedly. We confirmed this trend, and as shown in Fig. 2A, the sno1 mutant had approximately fourfold higher PLP than wild type (P < 0.01). We then hypothesized that the high level of PLP in sno1 mutant might be related to its NO hypersensitivity. To address this possibility, we checked the NO sensitivity of another mutant, pdx3 (37). PDX3 encodes pyridoxine/pyridoxamine 5′-phosphate oxidase, which functions in the other vitamin B6 salvage pathway parallel to SOS4 (SNO1 here; Fig. S2F), but losing function of this gene has no obvious effects on the levels of plant vitamers (37). Supporting our notion, pdx3 mutant showed similar SNP response as wild type (Fig. S4).
Fig. 2.
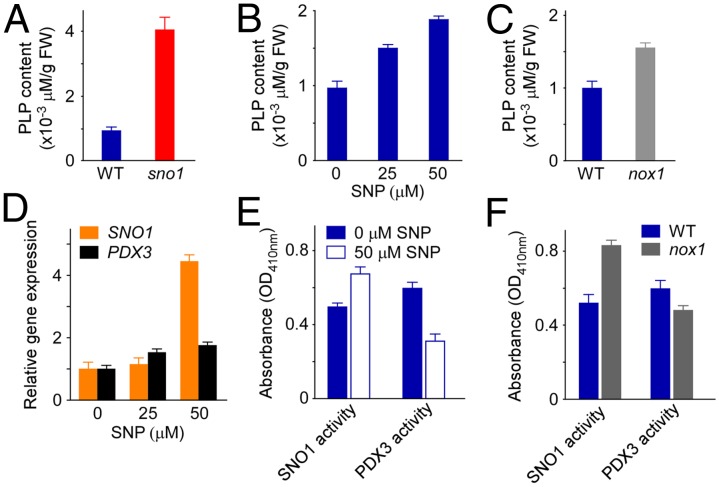
NO promotes PLP accumulation through up-regulation of SNO1, a vitamin B6 salvage pathway enzyme. (A) PLP levels in wild-type and sno1 mutant. (B) Effect of SNP on PLP content in WT. (C) PLP content in nox1 mutant. (D) Expression analyses of SOS4 and PDX3 in response to SNP treatment. The 1-wk-old WT seedlings were transferred to the media containing indicated concentrations of SNP for 2 d and collected for the analysis. Data shown are means ± SD, n = 3. (E) Enzyme activity of SOS4 and PDX3 in wild-type seedlings in response to SNP (50 μM) treatment. (F) Activity of SOS4 and PDX3 in wild-type and nox1 mutant. In A–C, E, and F, 3-d-old well-germinated seedlings were transferred to the media supplemented with or without indicated concentrations of SNP for 15 d and collected for the HPLC (A–C) and enzyme activity (E and F) analyses. Data shown are means ± SD, n = 3.
We then checked the effect of NO on PLP levels. Intriguingly, in wild-type seedlings, with the increase of SNP doses, PLP levels raised accordingly (Fig. 2B; P < 0.01 at all concentrations). To further determine the relevance of NO to the PLP content in vivo, we measured the PLP amount in nox1, a mutant that overproduces NO endogenously (6). The result showed that nox1 had more PLP than wild type (Fig. 2C; P < 0.01). Together, these observations suggest that NO significantly induces vitamin B6 PLP accumulation in plants, and that high levels of PLP in sno1 mutant might account for its hypersensitive response to NO. These findings also reveal a unique regulatory role of NO to the homeostasis of vitamin B6 PLP in Arabidopsis.
NO Up-Regulates the Expression and Activity of SNO1, a Key Enzyme that Functions in the Vitamin B6 Salvage Pathway.
We next questioned which components in the PLP synthesis pathways are affected by NO. In plants, vitamin B6 PLP can be synthesized by de novo or salvage pathways (40, 41). We analyzed the transcription of PDX1s and PDX2 in the de novo pathway, as well as SNO1 (or SOS4) and PDX3, the two major known genes in the salvage pathway (Fig. S2F), in responses to SNP treatment. The data showed that NO down-regulated PDX1s and up-regulated PDX2 transcript slightly (Fig. S5; P < 0.01). Consistently, the expression of one PDX1 was reported to be decreased slightly by high salt or abscisic acid stimulus (42), under which conditions NO production was supposed to be induced. Analyses of the two salvage pathway genes showed that PDX3 was briefly, but SNO1 dramatically, induced with the increase of SNP concentration (Fig. 2D; P < 0.01).
We further investigated the regulation of NO on the enzyme activity of SNO1 and PDX3. Compared with untreated wild-type controls, SNO1 activity was increased in both SNP-treated wild type and nox1 mutant (Fig. 2 E and F; P < 0.01); however, PDX3 activity was deceased in these samples (Fig. 2 E and F; P < 0.05). The opposite effect of NO on the activities of SNO1 and PDX3 we observed could be a competitive result between these two salvage pathway enzymes facing NO treatment, and the increase of SNO1 activity in response to SNP treatment may suggest a prominent role of SNO1 enzyme in stress responses because NO is a well-known stress-induced signaling molecule (43, 44). Consistent with this concept, previous study showed that the expression of SOS4 (or SNO1) is regulated by several environmental stimuli (36). However, both former study (37) and our result (Fig. S6; P < 0.01) showed that PDX3 activity was enhanced in sos4-1 mutant (or sno1 here), suggesting a possible compensatory mechanism between these two enzymes under normal physiological conditions. Together, the positive regulation of NO to SNO1 at both transcriptional and activity levels suggests that NO-stimulated PLP increases are primarily from SNO1-mediated vitamin B6 salvage biosynthetic pathways.
NO Decreases Plant K+ Content Through Its Repression on the K+ Channel AKT1.
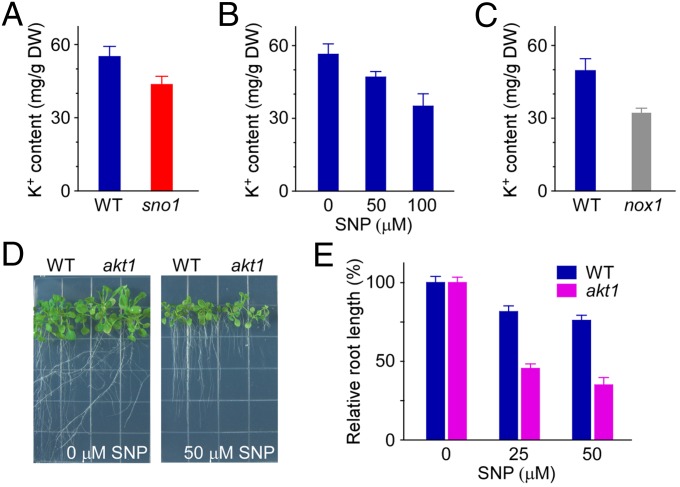
Previous studies have implicated NO in plant salt resistance regulation (18–21). Here, sno1, an NO hyperresponsive mutant, turned out to be identical to the salt overly sensitive mutant sos4-1 (36). The root growth of sno1 mutant is hypersensitive to NO (Fig. 1), whereas K+, a critical ion for salt tolerance (22) and an essential nutrient for plant growth, is reduced in the sos4-1 mutant (36). We therefore investigated the regulation of NO to plant K+ content. Similar to the K+ amount in sos4-1 (36), sno1 has less K+ content than wild type in roots and shoots (Fig. 3A; P < 0.01; Fig. S7A; P < 0.01). Most interestingly, NO decreased K+ content in both tissues of wild-type plants dose dependently (Fig. 3B; P < 0.01 at all concentrations; Fig. S7B; P < 0.05 at 50 µM SNP and P < 0.001 at 100 µM SNP). Furthermore, the K+ level was significantly lower in nox1 mutant than wild type (Fig. 3C; P < 0.001). Together, the negative correlation of NO to the K+ content in roots and shoots clearly suggest that NO decreases K+ absorption in Arabidopsis.
Fig. 3.
NO represses plant K+ uptake through its inhibition on AKT1 K+ channel. (A) Root K+ content in WT and sno1 mutant. (B) SNP effect on root K+ content in WT. (C) Root K+ content in nox1 mutant. (D) Phenotypes of akt1 mutant grown under 0 and 50 μM SNP for 15 d. (E) Relative root length of akt1 mutant in response to 0, 25, and 50 μM SNP treatment. Seeds were grown on SNP containing media for 15 d, and primary root length was measured. Relative root length is shown as a value relative to the root length on 0 μM SNP. Data shown are means ± SD, n = 35 seedlings. In A–C, 3-d-old seedlings were transferred to the new media for 15 d, and roots were collected for inductively coupled plasma–MS analysis. Data shown are means ± SD, n = 3.
We then surveyed which K+ transport proteins are regulated by NO under MS-level K+ conditions. Among the two main root K+ uptake systems (26, 27), AtHAK5 functions in relatively low concentrations of external K+ (26, 27) and is required for plant vitality in extreme low-K+ conditions (27), whereas AKT1 functions in both low and physiological levels of external K+ concentrations (29, 45). Disruption of AKT1 (akt1 mutant) reduces root K+ uptake (23) and tissue K+ content (30), and impairs seedling growth (23) under replete external K+ conditions. We determined the SNP responsiveness of both athak5 and akt1 mutants. The results showed that athak5 mutants had a similar root length as wild type under different concentrations of SNP treatment (Fig. S8), whereas the root growth of akt1 was hypersensitive to SNP (Fig. 3 D and E; P < 0.001 at all concentrations). These results suggest that the NO inhibition of plant K+ uptake may occur through AKT1 channels but not the AtHAK5 transporter.
Vitamin B6 PLP Represses the Activity of K+ Channel AKT1.
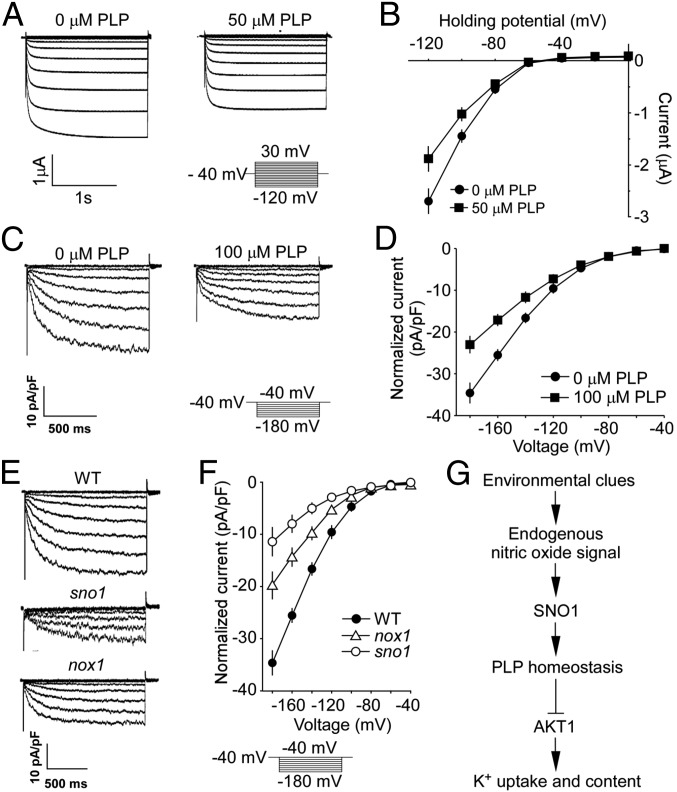
Our findings that NO has the opposite effect on plant PLP and K+ contents (Figs. 2B and 3B) and that the NO hypersensitive mutants sno1 and nox1 both have higher amounts of PLP but lower levels of K+ than wild type, together with the hypersensitive response of akt1 mutant to SNP, indicate a negative causality between PLP biosynthesis and plant K+ uptake that might be achieved through K+ channel AKT1. Because PLP has been proved to antagonize the activation of some ion channels in animal cells (38), we hypothesized that PLP may inhibit the activity of AKT1 channel in plants. To test this possibility, we carried out a series of electrophysiological studies using both Xenopus oocyte expression system and Arabidopsis root cell protoplasts. It is known that the activation of AKT1-mediated K+ current in oocytes requires coexpression of protein CBL1 (or CBL9) and CIPK23 (30), so we coinjected the mixed cRNAs of AKT1, CBL1, and CIPK23 in the oocytes, and obtained the canonical inward rectifying K+ currents (Fig. 4A). Intriguingly, the application of PLP (50 µM) notably suppressed these currents (Fig. 4 A and B). In contrast, currents conducted by a distinct K+ channel, KAT1 (46, 47), were unaffected by PLP, even at 100 µM (Fig. S9 A and B).
Fig. 4.
PLP represses AKT1-mediated inward-rectifying currents in Xenopus oocytes and Arabidopsis root cells. (A) PLP effect on AKT1 currents in Xenopus oocytes. Whole-cell currents were recorded in oocytes coinjected with mixed cRNAs of AKT1, CBL1, and CIPK23 in the absence (0 μM) or presence (50 μM) of PLP. (B) I–V relationship of AKT1 currents vs. applied voltages in the absence (0 μM) and presence (50 μM) of PLP in oocytes. Data shown are means ± SD, n = 6. (C) PLP effect on patch-clamp whole-cell recordings in protoplasts of Arabidopsis roots. (D) I–V relationship of the steady-state whole-cell currents vs. applied voltages with or without PLP treatment in root cell protoplasts. Data shown are means ± SD, n = 7. (E) Whole-cell AKT1 current recordings in root cells of wild type, sno1, and nox1. (F) I–V relationship of whole-cell currents in root cells. Data shown are means ± SD, n = 7 for WT, n = 5 for sno1, and n = 5 for nox1. (G) Schematic model for NO regulation of vitamin B6 and K+ homeostasis in Arabidopsis. Arrows denote positive regulation, and tee arrow means repression.
We further performed patch-clamp analysis using Arabidopsis root cell protoplasts to test the role of PLP to the AKT1 channel in vivo. The major inward-rectifying K+ channel detected in Arabidopsis root is AKT1, because the relative current disappears in the akt1 mutant (30). Consistent with the results we observed in Xenopus oocytes, whole-cell recordings from wild-type roots showed that the applied PLP significantly affected the AKT1-mediated inward K+ currents (Fig. 4 C and D). To better understand the specificity of PLP to the AKT1 channel, we tested the effects of other vitamers, pyridoxamine (PM) and pyridoxine (PN), and found that both PM and PN had no influences on the AKT1 currents (Fig. S9 C–E). Additionally, the inward-rectifying K+ currents in sno1 and nox1 mutants were significantly reduced in comparison with wild-type controls (Fig. 4 E and F), and the magnitudes of their currents negatively correlated with their PLP levels, with sno1 that contained more PLP than nox1 (Fig. 2 A and C) being more significantly reduced than nox1 (Fig. 4 E and F). Together, these data provide strong evidence showing that PLP repressed the function of AKT1 K+ channel in planta.
Discussion
In this study, using a combination of forward genetics with physiological, molecular, pharmacological, and electrophysiological approaches, we have unraveled a comprehensive signaling pathway for the molecular modulation of NO-to-potassium absorption in Arabidopsis. As shown in the schematic model of the pathway (Fig. 4G), NO up-regulates the activity of vitamin B6 salvage pathway gene SNO1, resulting in the accumulation of vitamin B6 PLP, which, in turn, represses the activity of K+ channel AKT1 and therefore reduces K+ uptake by roots. This report connects NO to the homeostasis of vitamin B6 and potassium, and provides evidence showing the direct regulation of vitamin B6 PLP to the activity of ion channel in plants. Our findings also support and provide useful information for the regulation of NO to K+ absorption and Na+ to K+ ratio under salt stresses.
In recent years, great attention has been paid to understand how NO fulfills its biological function at a molecular level in plants (48). Because of the bioactive feature, plant NO, rather than defining an NO-specific signaling pathway, tends to modify other signaling pathways to exert different roles (48). NO has been reported to inactivate the activity of outward-rectifying K+ channel in Vicia guard cell through direct S-nitrosylation of the not-yet identified channel or its closely attached protein (49). NO may also regulate K+ channel indirectly, in which case NO integrates with protein phosphorylation and intracellular Ca2+ release signals to inactivate the inward-rectifying K+ channels and close stomata (50, 51). Our finding here reveals a previously unidentified modulation of NO to the inward-rectifying K+ channel AKT1, the most important channel liable for K+ absorption in plant root under normal external K+ conditions, through effect of vitamin B6 PLP. In the NO hypersensitive mutant sno1, the losing function of SNO1 gene that catabolizes the biosynthesis of PLP surprisingly elevated PLP content rather than reducing the PLP amount (Fig. 2A), consistent with the PLP amount measured in sos4-1 mutant (identical to sno1 in this study) (37). The increase of PLP level in sno1 (or sos4-1) mutant might be caused by enhanced activities of PDX3 (37) (Fig. S6), the other known vitamin B6 salvage pathway enzyme parallel to SOS4, and PDX1 (37), a de novo pathway enzyme. In SNP-treated wild-type seedlings, the enhanced SNO1 gene expression and enzyme activity (Fig. 2 D and E) but not the decreased PDX3 activity (Fig. 2E) or PDX1s expression (Fig. S5) correlate with the increase of PLP content (Fig. 2B). Therefore, reasons for PLP accumulation in sno1 mutant and SNP-treated wild-type plant are different. The discovery that PLP directly represses AKT1 activity further illustrates the underlying connection of NO-enhanced PLP amount and reduced K+ content in plants. Overall, our study demonstrates that NO modifies vitamin B6 biosynthesis pathways to exert its role in plant K+ absorption from the environment.
Higher plants have evolved elaborate mechanisms to rapidly respond and adapt to the changing environment, by modifying their physiological and developmental programs. NO is a plant-endogenous messenger involved in growth and multiple stress responses (14, 44, 48). Here, we show that NO negatively regulates the absorption of K+, an ion that has marked impacts on cell metabolism, plant growth, and stress adaption (26, 52). Plant growth rate correlates with K+ uptake (26, 53), and less K+ uptake will retard plant growth. We therefore propose that one strategy that NO helps plant adapt to unfavorable conditions may be through its repression on K+ uptake from the environment to slow down growth. However, more external applied K+ has been proved to reduce the negative effects of several stresses (54), but the involvement and/or mechanism of NO under these high K+ situations is unclear. Recent reports suggest that NO enhances plant salt tolerance in different species through various mechanisms (18, 20, 55, 56). In a mangrove species resistant to high salinity, NO enhanced K+ content through decreasing the cytosolic K+ loss and increasing K+ uptake from the environment, and the transcript of AKT1 was increased under high salt condition (55). In this study we found that NO negatively regulates AKT1-mediated K+ uptake in Arabidopsis (Fig. 4G). Except for the species difference, it is also possible that NO functions differently in the absence and presence of salt stress. For example, NO may help plant maintain high levels of K+ to reduce the toxic effects of high salt, and it may lower plant K+ content to retard growth under nonsalinity stresses—i.e., NO may exert different strategies responding to changing conditions to precisely fulfill its roles in plant K+ homeostasis. Besides K+, NO, as a multifunctional signaling molecule, also implicates in the regulation of other ions, such as calcium (57), iron (9), cadmium (58), and aluminum (59), to adjust plant growth.
It has been known that AKT1 channel activity is tightly regulated. Our data that PLP represses the activity of AKT1 channel in Xenopus oocyte is obtained on the basis of coexpression of AKT1, CBL1, and CIPK23, and activation of the channel. The in vivo analysis of PLP repression to the AKT1 channel in wild-type Arabidopsis root cells also depends on the activation of the channel because the relative currents disappear in the akt1 mutant. Thus, our finding that NO-accumulated PLP represses AKT1 activity elaborates the regulatory mechanism of this channel, on the basis of the well-addressed CBL–CIPK pathway. Under adequate external K+ conditions, the activated AKT1 channel is antagonized by NO-elevated PLP. Further investigation is required to understand whether PLP can bind to the AKT1 channel and/or interfere with the conformation and activity of the channel. Alternatively, PLP may affect some ATP binding site containing proteins that regulate AKT1 channel activity indirectly, among which AKT1-associated regulatory proteins and H+-ATPases might be candidates, because PLP has been suggested to modulate the ATP binding sites of proteins, including ATP-gated ion channels (38), anion channels (60), and ion-transporting ATPases (61) in animal cells. Among the known plant AKT1-interacting proteins, CIPKs that control the phosphorylation and activity status of AKT1 channel might be potential PLP targets, with CIPK23 that contains an ATP-binding lysine critical for its kinase activity (32) being a good candidate. Plant K+ absorption against K+ concentration gradient by transport proteins in root cells is energized primarily by membrane potential across the PM produced by H+-ATPases (28, 62), and studies from Xenopus oocytes suggest a communication between AKT1 and H+-ATPases-mediated membrane potential (28, 34). It is possible that PLP disturbs the function of H+-ATPases by acting on the ATP binding sites, which then affects the membrane potential across the PM and lowers the AKT1 channel activity. Future investigation into the regulatory mechanism of PLP to AKT1 channel activity would help to better understand and detail the role of PLP in controlling plant physiology.
It is notable that PLP represses, but does not completely inactivate, AKT1 channel activity (Fig. 4 A–D). Conceivably, in SNP-treated wild-type seedlings where PLP accumulates, the function of AKT1 is partially inhibited, whereas in akt1 mutant, AKT1 activity is completely lost even in the absence of SNP; therefore, the akt1 mutant displays SNP hypersensitivity (Fig. 3 D and E). Expression analysis showed that the akt1 mutant transcribed a comparable level of SNO1 as wild type when SNP was absent, but expressed higher levels of SNO1 than wild type with increasing of SNP concentrations (Fig. S10; P < 0.01), indicating more PLP accumulation in akt1 mutant than wild type under SNP treatment. The akt1 mutant may lose the ability to sense PLP well, because PLP functions through affecting AKT1 activity (Fig. 4 A–D), which may then trigger the feedback biosynthesis of more PLP in akt1 than wild type under SNP treatment (Fig. S10). Thus, this expression result supports our main conclusion that NO-accumulated PLP represses AKT1 activity (Fig. 4G).
It would be interesting to know whether the PLP repression of AKT1 channels happens under K+-starved conditions with the absence or presence of salinity, where NO generation may vary. Studies showed that under K+-deprived saline conditions where plant K+ contents diminished dramatically, AKT1-mediated K+ efflux was promoted, and AtHAK5 transcription was also largely reduced (52), suggesting that the regulation of K+ contents in this situation happens at various aspects. The possible effect of NO on AtHAK5 transporter, especially in low K+ conditions, remains to be addressed.
It is significant that NO accumulates vitamin B6 PLP through the salvage pathway rather than the de novo synthetic pathway. The salvage pathway catalyzes interconversions among different forms of vitamins without generating a new pyridine ring (63, 64), so this pathway could trigger rapid responses and is cost-effective, thereby avoiding irreversible damage to plant cells. Thus, it is conceivable that, evolutionally, the vitamin B6 salvage pathway appeared earlier than the de novo pathway (63, 64), implicating the former pathway-derived vitamin in the harsh and volatile environments during long-term plant development. Considering the essential role of vitamin B6 PLP as an enzyme cofactor in basic cellular metabolism (37, 40), the NO-accumulated PLP here may represent more profound roles in plant growth besides its regulation of the AKT1 channel.
Studies have shown that NO regulates the Na+-to-K+ ratio under high-salt conditions (18–21). Na+ enters root cells mainly through nonselective cation channels (36), and maintaining a low cytosolic Na+ level is important for the normal physiology of plant cells. NO was reported to be involved in plant salt tolerance by increasing the activities of proton pump and Na+/H+ antiporter to exclude salt from cytoplasm (18, 20). In this study, we found that NO lowers plant K+ absorption through repressing the function AKT1 channel. Hence, NO may have multiple roles in modulating and balancing the cellular ion concentrations to help plants adapt to unfavorable environments.
Materials and Methods
The Arabidopsis thaliana Columbia genetic background was used as the wild type in this study. Plant materials and growth conditions; sno1 mutant isolation and map-based cloning of SNO1; PLP and K+ content measurements; analyses of SNO1 and PDX3 enzyme activity; electrophysiological studies in Xenopus oocytes and Arabidopsis root-cell protoplasts; and other procedures are fully described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Jian-Kang Zhu (Purdue University), Dr. Margaret E. Daub (North Carolina State University), and Dr. Wei-Hua Wu (China Agricultural University) for kindly providing the mutant seeds; Dr. Tobias I. Baskin (University of Massachusetts) and Dr. Heven Sze (University of Maryland) for critical reading of the manuscript; and Dr. Zhen-Ming Pei (Duke University) for helpful discussions. This work was supported by Ministry of Science and Technology of China Grants 2013CB967300 and 2007CB948201, and the Funding Project for Academic Human Resources Development in Institutions of Higher Learning under the Jurisdiction of Beijing Municipality [PHR (IHLB)] (to Y. He). D.K. was supported by National Science Foundation Grant MCB-1244303 to June M. Kwak (University of Maryland).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417473111/-/DCSupplemental.
References
- 1.Šírová J, Sedlářová M, Piterková J, Luhová L, Petřivalský M. The role of nitric oxide in the germination of plant seeds and pollen. Plant Sci. 2011;181(5):560–572. doi: 10.1016/j.plantsci.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 2.Bethke PC, Libourel IG, Reinöhl V, Jones RL. Sodium nitroprusside, cyanide, nitrite, and nitrate break Arabidopsis seed dormancy in a nitric oxide-dependent manner. Planta. 2006;223(4):805–812. doi: 10.1007/s00425-005-0116-9. [DOI] [PubMed] [Google Scholar]
- 3.Pagnussat GC, Simontacchi M, Puntarulo S, Lamattina L. Nitric oxide is required for root organogenesis. Plant Physiol. 2002;129(3):954–956. doi: 10.1104/pp.004036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mishina TE, Lamb C, Zeier J. Expression of a nitric oxide degrading enzyme induces a senescence programme in Arabidopsis. Plant Cell Environ. 2007;30(1):39–52. doi: 10.1111/j.1365-3040.2006.01604.x. [DOI] [PubMed] [Google Scholar]
- 5.Guo FQ, Crawford NM. Arabidopsis nitric oxide synthase1 is targeted to mitochondria and protects against oxidative damage and dark-induced senescence. Plant Cell. 2005;17(12):3436–3450. doi: 10.1105/tpc.105.037770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He Y, et al. Nitric oxide represses the Arabidopsis floral transition. Science. 2004;305(5692):1968–1971. doi: 10.1126/science.1098837. [DOI] [PubMed] [Google Scholar]
- 7.Neill S, et al. Nitric oxide, stomatal closure, and abiotic stress. J Exp Bot. 2008;59(2):165–176. doi: 10.1093/jxb/erm293. [DOI] [PubMed] [Google Scholar]
- 8.García-Mata C, Lamattina L. Nitric oxide induces stomatal closure and enhances the adaptive plant responses against drought stress. Plant Physiol. 2001;126(3):1196–1204. doi: 10.1104/pp.126.3.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramirez L, Simontacchi M, Murgia I, Zabaleta E, Lamattina L. Nitric oxide, nitrosyl iron complexes, ferritin and frataxin: A well equipped team to preserve plant iron homeostasis. Plant Sci. 2011;181(5):582–592. doi: 10.1016/j.plantsci.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Graziano M, Lamattina L. Nitric oxide and iron in plants: An emerging and converging story. Trends Plant Sci. 2005;10(1):4–8. doi: 10.1016/j.tplants.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Terrile MC, et al. Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J. 2012;70(3):492–500. doi: 10.1111/j.1365-313X.2011.04885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ribeiro DM, et al. Differential requirement for NO during ABA-induced stomatal closure in turgid and wilted leaves. Plant Cell Environ. 2009;32(1):46–57. doi: 10.1111/j.1365-3040.2008.01906.x. [DOI] [PubMed] [Google Scholar]
- 13.Leitner M, Vandelle E, Gaupels F, Bellin D, Delledonne M. NO signals in the haze: Nitric oxide signalling in plant defence. Curr Opin Plant Biol. 2009;12(4):451–458. doi: 10.1016/j.pbi.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Lamattina L, García-Mata C, Graziano M, Pagnussat G. Nitric oxide: The versatility of an extensive signal molecule. Annu Rev Plant Biol. 2003;54:109–136. doi: 10.1146/annurev.arplant.54.031902.134752. [DOI] [PubMed] [Google Scholar]
- 15.Bai X, et al. Deciphering the protective role of nitric oxide against salt stress at the physiological and proteomic levels in maize. J Proteome Res. 2011;10(10):4349–4364. doi: 10.1021/pr200333f. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Liang X, Wan Q, Wang X, Bi Y. Ethylene and nitric oxide are involved in maintaining ion homeostasis in Arabidopsis callus under salt stress. Planta. 2009;230(2):293–307. doi: 10.1007/s00425-009-0946-y. [DOI] [PubMed] [Google Scholar]
- 17.Zhu JK. Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol. 2003;6(5):441–445. doi: 10.1016/s1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
- 18.Chen J, et al. Nitric oxide enhances salt secretion and Na+ sequestration in a mangrove plant, Avicennia marina, through increasing the expression of H+-ATPase and Na+/H+ antiporter under high salinity. Tree Physiol. 2010;30(12):1570–1585. doi: 10.1093/treephys/tpq086. [DOI] [PubMed] [Google Scholar]
- 19.Zhao MG, Tian QY, Zhang WH. Nitric oxide synthase-dependent nitric oxide production is associated with salt tolerance in Arabidopsis. Plant Physiol. 2007;144(1):206–217. doi: 10.1104/pp.107.096842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, et al. Nitric oxide enhances salt tolerance in maize seedlings through increasing activities of proton-pump and Na+/H+ antiport in the tonoplast. Planta. 2006;224(3):545–555. doi: 10.1007/s00425-006-0242-z. [DOI] [PubMed] [Google Scholar]
- 21.Molassiotis A, Tanou G, Diamantidis G. NO says more than ‘YES’ to salt tolerance: Salt priming and systemic nitric oxide signaling in plants. Plant Signal Behav. 2010;5(3):209–212. doi: 10.4161/psb.5.3.10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu JK, Liu J, Xiong L. Genetic analysis of salt tolerance in Arabidopsis: Evidence for a critical role of potassium nutrition. Plant Cell. 1998;10(7):1181–1191. doi: 10.1105/tpc.10.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirsch RE, Lewis BD, Spalding EP, Sussman MR. A role for the AKT1 potassium channel in plant nutrition. Science. 1998;280(5365):918–921. doi: 10.1126/science.280.5365.918. [DOI] [PubMed] [Google Scholar]
- 24.Sentenac H, et al. Cloning and expression in yeast of a plant potassium ion transport system. Science. 1992;256(5057):663–665. doi: 10.1126/science.1585180. [DOI] [PubMed] [Google Scholar]
- 25.Rubio F, Santa-Maria GE, Rodriguez-Navarro A. Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol Plant. 2000;109(1):34–43. [Google Scholar]
- 26.Alemán F, Nieves-Cordones M, Martínez V, Rubio F. Root K+ acquisition in plants: The Arabidopsis thaliana model. Plant Cell Physiol. 2011;52(9):1603–1612. doi: 10.1093/pcp/pcr096. [DOI] [PubMed] [Google Scholar]
- 27.Pyo YJ, Gierth M, Schroeder JI, Cho MH. High-affinity K+ transport in Arabidopsis: AtHAK5 and AKT1 are vital for seedling establishment and postgermination growth under low-potassium conditions. Plant Physiol. 2010;153(2):863–875. doi: 10.1104/pp.110.154369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Wu WH. Potassium transport and signaling in higher plants. Annu Rev Plant Biol. 2013;64:451–476. doi: 10.1146/annurev-arplant-050312-120153. [DOI] [PubMed] [Google Scholar]
- 29.Lagarde D, et al. Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J. 1996;9(2):195–203. doi: 10.1046/j.1365-313x.1996.09020195.x. [DOI] [PubMed] [Google Scholar]
- 30.Xu J, et al. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell. 2006;125(7):1347–1360. doi: 10.1016/j.cell.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 31.Gaymard F, et al. The baculovirus/insect cell system as an alternative to Xenopus oocytes. First characterization of the AKT1 K+ channel from Arabidopsis thaliana. J Biol Chem. 1996;271(37):22863–22870. doi: 10.1074/jbc.271.37.22863. [DOI] [PubMed] [Google Scholar]
- 32.Li L, Kim BG, Cheong YH, Pandey GK, Luan S. A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc Natl Acad Sci USA. 2006;103(33):12625–12630. doi: 10.1073/pnas.0605129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee SC, et al. A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc Natl Acad Sci USA. 2007;104(40):15959–15964. doi: 10.1073/pnas.0707912104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, He L, Li HD, Xu J, Wu WH. Potassium channel alpha-subunit AtKC1 negatively regulates AKT1-mediated K+ uptake in Arabidopsis roots under low-K+ stress. Cell Res. 2010;20(7):826–837. doi: 10.1038/cr.2010.74. [DOI] [PubMed] [Google Scholar]
- 35.Geiger D, et al. Heteromeric AtKC1·AKT1 channels in Arabidopsis roots facilitate growth under K+-limiting conditions. J Biol Chem. 2009;284(32):21288–21295. doi: 10.1074/jbc.M109.017574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi H, Xiong L, Stevenson B, Lu T, Zhu JK. The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell. 2002;14(3):575–588. doi: 10.1105/tpc.010417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.González E, Danehower D, Daub ME. Vitamer levels, stress response, enzyme activity, and gene regulation of Arabidopsis lines mutant in the pyridoxine/pyridoxamine 5′-phosphate oxidase (PDX3) and the pyridoxal kinase (SOS4) genes involved in the vitamin B6 salvage pathway. Plant Physiol. 2007;145(3):985–996. doi: 10.1104/pp.107.105189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ralevic V, Burnstock G. Receptors for purines and pyrimidines. Pharmacol Rev. 1998;50(3):413–492. [PubMed] [Google Scholar]
- 39.Libourel IG, Bethke PC, De Michele R, Jones RL. Nitric oxide gas stimulates germination of dormant Arabidopsis seeds: Use of a flow-through apparatus for delivery of nitric oxide. Planta. 2006;223(4):813–820. doi: 10.1007/s00425-005-0117-8. [DOI] [PubMed] [Google Scholar]
- 40.Fitzpatrick TB, et al. Vitamin deficiencies in humans: Can plant science help? Plant Cell. 2012;24(2):395–414. doi: 10.1105/tpc.111.093120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tambasco-Studart M, et al. Vitamin B6 biosynthesis in higher plants. Proc Natl Acad Sci USA. 2005;102(38):13687–13692. doi: 10.1073/pnas.0506228102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen H, Xiong L. Pyridoxine is required for post-embryonic root development and tolerance to osmotic and oxidative stresses. Plant J. 2005;44(3):396–408. doi: 10.1111/j.1365-313X.2005.02538.x. [DOI] [PubMed] [Google Scholar]
- 43.Qiao W, Fan LM. Nitric oxide signaling in plant responses to abiotic stresses. J Integr Plant Biol. 2008;50(10):1238–1246. doi: 10.1111/j.1744-7909.2008.00759.x. [DOI] [PubMed] [Google Scholar]
- 44.Delledonne M. NO news is good news for plants. Curr Opin Plant Biol. 2005;8(4):390–396. doi: 10.1016/j.pbi.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 45.Cheong YH, et al. Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 2007;52(2):223–239. doi: 10.1111/j.1365-313X.2007.03236.x. [DOI] [PubMed] [Google Scholar]
- 46.Chérel I. Regulation of K+ channel activities in plants: From physiological to molecular aspects. J Exp Bot. 2004;55(396):337–351. doi: 10.1093/jxb/erh028. [DOI] [PubMed] [Google Scholar]
- 47.Schachtman DP, Schroeder JI, Lucas WJ, Anderson JA, Gaber RF. Expression of an inward-rectifying potassium channel by the Arabidopsis KAT1 cDNA. Science. 1992;258(5088):1654–1658. doi: 10.1126/science.8966547. [DOI] [PubMed] [Google Scholar]
- 48.Mur LA, et al. Nitric oxide in plants: An assessment of the current state of knowledge. AoB Plants. 2013;5:pls052. doi: 10.1093/aobpla/pls052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sokolovski S, Blatt MR. Nitric oxide block of outward-rectifying K+ channels indicates direct control by protein nitrosylation in guard cells. Plant Physiol. 2004;136(4):4275–4284. doi: 10.1104/pp.104.050344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sokolovski S, et al. Protein phosphorylation is a prerequisite for intracellular Ca2+ release and ion channel control by nitric oxide and abscisic acid in guard cells. Plant J. 2005;43(4):520–529. doi: 10.1111/j.1365-313X.2005.02471.x. [DOI] [PubMed] [Google Scholar]
- 51.Garcia-Mata C, et al. Nitric oxide regulates K+ and Cl- channels in guard cells through a subset of abscisic acid-evoked signaling pathways. Proc Natl Acad Sci USA. 2003;100(19):11116–11121. doi: 10.1073/pnas.1434381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nieves-Cordones M, Alemán F, Martínez V, Rubio F. The Arabidopsis thaliana HAK5 K+ transporter is required for plant growth and K+ acquisition from low K+ solutions under saline conditions. Mol Plant. 2010;3(2):326–333. doi: 10.1093/mp/ssp102. [DOI] [PubMed] [Google Scholar]
- 53.Rubio F, Alemán F, Nieves-Cordones M, Martínez V. Studies on Arabidopsis athak5, atakt1 double mutants disclose the range of concentrations at which AtHAK5, AtAKT1 and unknown systems mediate K uptake. Physiol Plant. 2010;139(2):220–228. doi: 10.1111/j.1399-3054.2010.01354.x. [DOI] [PubMed] [Google Scholar]
- 54.Cakmak I. The role of potassium in alleviating detrimental effects of abiotic stresses in plants. J Plant Nutr Soil Sci. 2005;168(4):521–530. [Google Scholar]
- 55.Chen J, et al. Nitric oxide mediates root K+/Na+ balance in a mangrove plant, Kandelia obovata, by enhancing the expression of AKT1-type K+ channel and Na+/H+ antiporter under high salinity. PLoS ONE. 2013;8(8):e71543. doi: 10.1371/journal.pone.0071543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao L, et al. Nitric oxide functions as a signal in salt resistance in the calluses from two ecotypes of reed. Plant Physiol. 2004;134(2):849–857. doi: 10.1104/pp.103.030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Courtois C, et al. Nitric oxide signalling in plants: Interplays with Ca2+ and protein kinases. J Exp Bot. 2008;59(2):155–163. doi: 10.1093/jxb/erm197. [DOI] [PubMed] [Google Scholar]
- 58.Arasimowicz-Jelonek M, Floryszak-Wieczorek J, Gwóźdź EA. The message of nitric oxide in cadmium challenged plants. Plant Sci. 2011;181(5):612–620. doi: 10.1016/j.plantsci.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 59.He H, He L, Gu M. Interactions between nitric oxide and plant hormones in aluminum tolerance. Plant Signal Behav. 2012;7(4):469–471. doi: 10.4161/psb.19312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ballatori N, Truong AT, Jackson PS, Strange K, Boyer JL. ATP depletion and inactivation of an ATP-sensitive taurine channel by classic ion channel blockers. Mol Pharmacol. 1995;48(3):472–476. [PubMed] [Google Scholar]
- 61.Hinz HR, Kirley TL. Lysine 480 is an essential residue in the putative ATP site of lamb kidney (Na,K)-ATPase. Identification of the pyridoxal 5′-diphospho-5′-adenosine and pyridoxal phosphate reactive residue. J Biol Chem. 1990;265(18):10260–10265. [PubMed] [Google Scholar]
- 62.Palmgren MG. Plant plasma membrane H+-ATPases: Powerhouses for nutrient uptake. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:817–845. doi: 10.1146/annurev.arplant.52.1.817. [DOI] [PubMed] [Google Scholar]
- 63.Fitzpatrick TB, et al. Two independent routes of de novo vitamin B6 biosynthesis: Not that different after all. Biochem J. 2007;407(1):1–13. doi: 10.1042/BJ20070765. [DOI] [PubMed] [Google Scholar]
- 64.Tanaka T, Tateno Y, Gojobori T. Evolution of vitamin B6 (pyridoxine) metabolism by gain and loss of genes. Mol Biol Evol. 2005;22(2):243–250. doi: 10.1093/molbev/msi011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.