Significance
The rapid pace of evolution in bacteria is widely attributed to the promiscuous horizontal transfer and recombination of protein-coding genes. However, it has not been investigated if the same forces also drive the evolution of noncoding regulatory regions. Here, we establish that regulatory regions can “switch” between nonhomologous alternatives and that switching is ubiquitous, occurring across the bacterial domain. We show that regulatory switching has a strong impact on promoter architecture and expression divergence. Further, we demonstrate that regulatory transfer facilitates rapid phenotypic diversification of a human pathogen. This regulatory mobility enables bacterial genes to access a vast pool of potential regulatory elements, facilitating efficient exploration of the regulatory landscape.
Keywords: bacterial evolution, regulatory evolution, HRT, core genes
Abstract
Understanding the mechanisms that generate variation is a common pursuit unifying the life sciences. Bacteria represent an especially striking puzzle, because closely related strains possess radically different metabolic and ecological capabilities. Differences in protein repertoire arising from gene transfer are currently considered the primary mechanism underlying phenotypic plasticity in bacteria. Although bacterial coding plasticity has been extensively studied in previous decades, little is known about the role that regulatory plasticity plays in bacterial evolution. Here, we show that bacterial genes can rapidly shift between multiple regulatory modes by acquiring functionally divergent nonhomologous promoter regions. Through analysis of 270,000 regulatory regions across 247 genomes, we demonstrate that regulatory “switching” to nonhomologous alternatives is ubiquitous, occurring across the bacterial domain. Using comparative transcriptomics, we show that at least 16% of the expression divergence between Escherichia coli strains can be explained by this regulatory switching. Further, using an oligonucleotide regulatory library, we establish that switching affects bacterial promoter architecture. We provide evidence that regulatory switching can occur through horizontal regulatory transfer, which allows regulatory regions to move across strains, and even genera, independently from the genes they regulate. Finally, by experimentally characterizing the fitness effect of a regulatory transfer on a pathogenic E. coli strain, we demonstrate that regulatory switching elicits important phenotypic consequences. Taken together, our findings expose previously unappreciated regulatory plasticity in bacteria and provide a gateway for understanding bacterial phenotypic variation and adaptation.
The acquisition of genes from nonparental lineages through horizontal gene transfer (HGT) has been shown to transform bacterial capabilities radically, influencing key processes, including pathogenicity, antibiotic resistance, and utilization of novel energy substrates (1–4). These striking findings have led many to believe that changes in gene content underlie the rapid pace of bacterial evolution (5, 6). However, an overlooked corollary of this ubiquitous exchange of DNA (7, 8) is that noncoding regions can be similarly subject to transfer and recombination, enabling rapid rewiring of regulatory networks (9, 10). Consistent with this hypothesis, recent studies have uncovered several cases of regulatory rearrangements, whereby regulatory regions have “switched” to nonhomologous alternatives with remarkable phenotypic consequences (11–13). For example, the inversion of a single promoter was shown to convert a commensal to a pathogen (12). Similarly, in Escherichia coli, citrate utilization was shown to evolve through promoter capture, enabling expression of an otherwise silent transporter (13). These discoveries demonstrate that regulatory “switching” to divergent alternative sequences is possible and can produce functional transformations. Nonetheless, it remains unclear whether these intriguing observations reflect exceptional anecdotes restricted to highly mobile genes in unusual strains or early representatives of a broader paradigm.
Results
Regulatory Switching in E. coli Core Genes.
To assess the significance of regulatory switching on bacterial evolution, we first considered core genes, which are present in all members of a clade and typically encode basal cellular “housekeeping” functions. Core genes are subject to strong purifying selection and are viewed as islands of stability within the dynamic bacterial genome [although exceptions exist (14, 15)]. Accordingly, regulatory switching in core genes is particularly unexpected, and is also easily detectable against the background of sequence conservation in coding regions.
We compared multiple sequence alignments of the 1,479 core genes present in all 46 publicly available E. coli genomes and up to 300 base pairs of the upstream regulatory region for each gene. As expected, the regulatory regions of most core genes are highly conserved (median nucleotide identity of 94%); however, a significant minority (13%) appear to be nonhomologous, sharing less than 50% nucleotide identity (Fig. S1). Because such poor conservation is inconsistent with the traditional view that core genes are slow-evolving (5), we investigated this divergent subpopulation further.
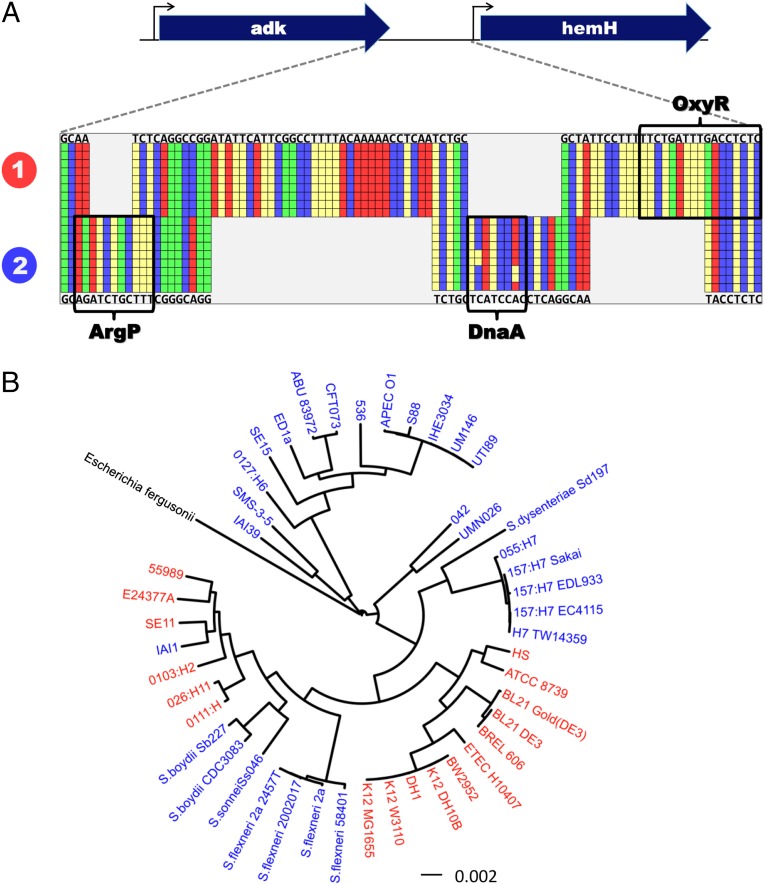
We first focused on hemH, as a representative of the nonhomologous upstream regulatory regions (Fig. 1A). hemH is a single gene operon that encodes ferrochelatase, the terminal enzyme in heme biosynthesis. hemH and its upstream gene, adk, display near-perfect conservation (>98% amino acid identity) across all 46 E. coli strains. However, the regulatory region between these genes comprises a 155-bp region that can be classified into two distinct, nonhomologous sequence types (less than 42% average pairwise nucleotide identity between clusters). In contrast, within clusters, there is almost perfect homology (>96% nucleotide identity). Thus, hemH represents a canonical example of regulatory switching between two alternative, nonhomologous regulatory sequences.
Fig. 1.
Regulatory switching and horizontal transfer of the hemH promoter. (A) Operonic structure and a representative multiple sequence alignment of the regulatory region of hemH (20 of 46 sequences are shown). The first line (E. coli K-12 MG1655) and the last line (E. coli O157:H7 Sakai) depict the nucleotide sequence of representative strains from each sequence type. Gray boxes represent gaps in the alignment. The nucleotides are colored red (Ade), yellow (Thy), green (Gua), and blue (Cys). Binding sites of ArgP, DnaA, and OxyR are boxed. The numbered labels in the left margin indicate the two alternative regulatory types that are found in hemH. (B) Two types mapped onto the E. coli species tree rooted with Escherichia fergusonii. Promoter type 1 is shown in red, and type 2 is shown in blue. (Scale bar: nucleotide substitutions per site.) The patchy distribution of these alternative sequence types is inconsistent with vertical transmission.
To determine the overall prevalence of such switching among E. coli core genes, we devised an algorithm that could systematically identify core genes with at least two distinct types of regulatory sequences (SI Text and Fig. S2). Remarkably, we found 166 unambiguous cases of regulatory switching (11% of all core genes in E. coli). The vast majority (83%) of these divergent regions contain bona fide promoters (16), as opposed to interoperonic regions, which is significantly more than expected by chance (Fisher’s exact test, P < 0.005), indicating that switching is enriched among promoters, where it can facilitate regulatory rewiring.
Moreover, we found that regulatory switching often creates new transcription factor binding sites. In 41% of the 44 diverged core genes for which high-quality transcription factor binding site annotations exist (17), alternative regulatory types were associated with divergent binding patterns (Table S1). For example, in hemH (Fig. 1A), all type 1 sequences contain an experimentally validated OxyR binding site (18) that is missing from all type 2 sequences. Type 2 sequences, instead, harbor canonical binding sites for both ArgP and DnaA (Fig. 1).
Horizontal Regulatory Transfer as a Switching Mechanism.
To elucidate the evolutionary mechanisms that lead to regulatory switching, we returned to our representative example of hemH and mapped its regulatory regions onto the E. coli species tree (Fig. S3; generated by concatenation of all core genes). We found that the distribution of the alternative promoter types is incongruent with the E. coli species phylogeny, consistent with evolution by horizontal regulatory transfer (HRT) (Fig. 1B). For the observed distribution to be explained by vertical transmission, multiple independent genomic rearrangement events with identical boundaries would have to be posited, with independent acquisition of the identical SNPs shared within each regulatory type; clearly, this alternative interpretation is implausible.
To determine if horizontal transfer has an impact on other regulatory regions in E. coli, we used the approximately unbiased (AU) test, a maximum-likelihood–based methodology (19). Specifically, we statistically tested for incongruence between the topology of the promoter sequences against the species tree. The null hypothesis of this test is vertical inheritance (as defined by the species tree); therefore, rejection of the null hypothesis is a strong indication of HRT. We found that 51% of all core gene promoters are incongruent with the species phylogeny, indicating that regulatory regions, similar to coding genes, are frequently transferred. However, in many of these cases, the promoter and its upstream gene might have been cotransferred. To tease out the cases in which the promoters were transferred independent of their genes, we compared the topology of each core gene with the topology of its associated upstream regulatory region (using the same methodology described above, with more details provided in SI Text). In the case of hemH, both the promoter history and the gene history significantly differed from the species tree, yet their topologies were not statistically different from each other. Therefore, we cannot exclude the possibility that these two regions were cotransferred. Nevertheless, for 32% of all promoters, we detected a clear signal that they were transferred independent of the gene they regulate.
Intergenera HRT Between E. coli and Enterobacter.
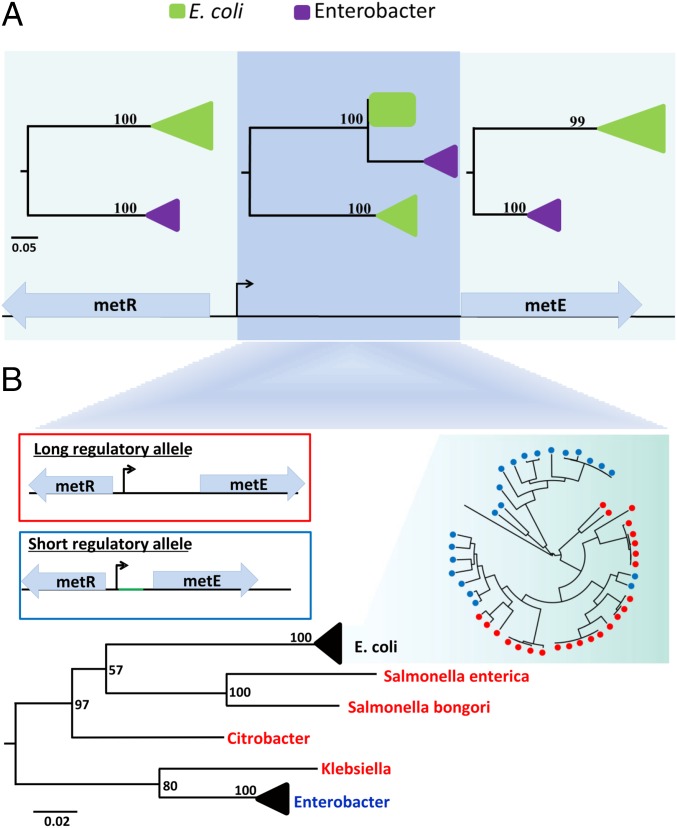
Given the frequency of HRT among E. coli strains, we expanded our analysis to investigate if HRT can cross species boundaries and discovered intergenera HRT between E. coli and Enterobacter. As shown in Fig. 2, we found that among 22 E. coli strains, the leader sequence of the biosynthesis gene metE exhibits a greater sequence similarity to the leader sequence found in Enterobacter than to its homologs in more closely related E. coli strains. Although most E. coli have a long leader sequence (169 bp), a subset of E. coli (most of which are uropathogenic E. coli) has, instead, a short (49 bp) AT-rich leader sequence that is shared with Enterobacter. In contrast to this incongruent regulatory region, phylogenies of the surrounding core genes match the species phylogeny, suggesting that the incongruence of the intervening regulatory sequence is best explained by horizontal transfer of the regulatory region alone (Fig. 2A). The direction of this regulatory transfer is most likely from Enterobacter to E. coli, because other Enterobacteriaceae species close to E. coli all harbor the long allele (Fig. 2B). Furthermore, all of the short E. coli regulatory alleles are nearly identical, suggesting a recent regulatory transfer.
Fig. 2.
HRT between E. coli and Enterobacter. (A) Phylogenetic tree for metR, metE, and the leader sequence of metE. Clades are collapsed into triangles or marked by a square (which represents that all sequences are 100% identical). Enterobacter is shown in purple, and E. coli is shown in green. For both protein-coding genes, E. coli and Enterobacter each form a monophyletic group. In contrast, the phylogeny of the intergenic region is incongruent with the phylogeny for the surrounding genes, suggesting horizontal transfer of the intergenic region independent of the surrounding genes. (B) Long and short regulatory alleles and their mapping onto the Enterobacteriaceae species tree. The long regulatory allele is shown in red, and the short regulatory allele is shown in blue. The phylogenetic pattern of the long and short alleles is consistent with HRT from Enterobacter to E. coli. The high AT content stretch of the short regulatory allele is marked in green. (Scale bar: nucleotide substitutions per site.) Statistical support for the internal branches was computed using 100 bootstrap repetitions.
Regulatory Switching Is Also Prevalent in the Accessory Genome.
Thus far, our analysis focused on core genes, for which regulatory switching was especially unexpected. Next, we examined the prevalence of regulatory switching among all gene classes. Among 2,286 noncore accessory genes in E. coli strain MG1655, we detected a similar level of switching (11.8%) to that observed across core genes in E. coli (11.2%). Moreover, we found that switching occurs across all functional categories, including global regulators (Fig. S4 and Table S2). The finding that global regulators exhibit regulatory switching is especially significant, because cis rewiring of a single regulatory protein could create large-scale downstream effects in trans. We also found that regulatory switching occurs more frequently in signal transduction pathways (Fisher’s exact test, P < 0.05). Regulatory switching in signal transduction pathways could help these vital environmental interfaces more rapidly align their response to environmental conditions upon shifts in ecological niches.
Regulatory Switching Affects E. coli Promoter Architecture.
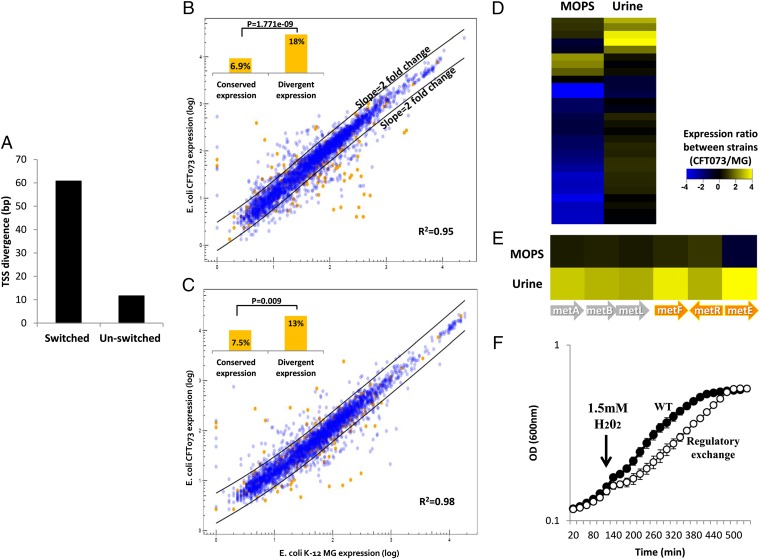
To assess the impact of regulatory switching, we first examined if promoter switching is associated with changes in the positioning of the gene transcription start site (TSS). To this end, we synthesized an E. coli promoter library, which allows detection of TSS from multiple bacterial strains in parallel. A similar approach was successfully applied to study TSS composition in E. coli (20). After filtering core genes for which the TSS could not be reliably determined due to annotation biases, we were left with 822 core gene clusters (SI Text). These core gene clusters were classified as either switched (166 core gene clusters) or unswitched (656 core gene clusters). From each core gene cluster, we selected at least two promoter regions, leading to a total of 1,693 promoters. The selected promoters were synthesized by Agilent Technologies using the oligo library synthesis method (21). This library was transformed into E. coli K-12 MG1655, and expression on LB was measured using RNA-sequencing (RNA-Seq). Expression data were used to accurately determine TSS positions of 485 promoter sequences from 40 different E. coli. Orthologous TSS positions were used to compute TSS divergence: average distance in base pairs between TSSs of orthologous genes. The mean divergence between switched orthologs was fivefold higher than that between unswitched orthologs (P < 0.01; Fig. 3A). Switched orthologs also exhibited significantly more TSS divergence than unswitched genes when multiple TSSs in a single gene were taken into account (P < 0.03; SI Text). Based on our results, we conclude that regulatory switching drives promoter architecture divergence.
Fig. 3.
Regulatory switching drives expression diversification and adaptation of E. coli. (A) Switched orthologs are more diverged with respect to their TSS position compared with unswitched genes. Expression diversification of E. coli strains grown on MOPS (B) and on pooled human urine (C). Each circle represents the average transcript level of an orthologous gene across three independent experiments. Genes that underwent regulatory switching are shown in orange, and those genes not affected by switching are shown in blue. The black lines, estimated by locally weighted scatterplot smoothing regression, indicate twofold change in expression between strains. (Insets) Level of regulatory switching in genes showing divergent expression vs. conserved expression. (D) Expression of switched genes exhibiting condition-specific expression divergence. (E) Condition-dependent divergence of the methionine biosynthesis pathways. Genes affected by regulatory switching are marked by orange arrows. Although both strains express the pathway in a similar manner when grown on MOPS, the pathogenic strain shows up to 16-fold higher expression of the pathway when strains are grown on urine. (F) Replacement of the short metE regulatory allele with the long ancestral allele renders the pathogenic bacteria more sensitive to oxidative stress. The strains were grown on MOPS media without methionine. After 2 h, oxidative stress was induced by adding H202 to a final concentration of 1.5 mM (marked by an arrow). Filled circles (●) denote CFT073 WT, and empty circles (○) denote CFT073 with a K-12 regulatory region. The data represent three independent experiments.
Regulatory Switching Drives Expression Diversification of E. coli Strains.
To test if regulatory switching alters the transcriptional response, we performed high-throughput RNA-Seq to compare the expression patterns of two E. coli strains that occupy distinct ecological niches: a gastrointestinal commensal (MG) and a urinary tract pathogen (CFT). We measured gene expression levels for all 3,293 orthologous genes present in both strains when grown on either defined minimal potassium morpholinopropane sulfonate (MOPS) media or pooled, sterile human urine (Fig. S5). Despite their ecological differences and more than 5 My of evolutionary divergence, most genes exhibited similar expression between strains exposed to the same conditions (Fig. 3; MOPS: R2 = 0.95, urine: R2 = 0.98). Nonetheless, as shown in Fig. 3, 266 genes in MOPS and 219 genes in urine exhibited statistically significant and substantial (over twofold change) expression divergence. The frequency of switched genes within this divergent expression group was found to be threefold higher than in the conserved expression group (Fig. 3B, Inset). The tendency of switched genes to exhibit higher expression divergence was also indicated by ∼1.4-fold higher median expression divergence compared with unswitched genes (MOPS: P = 9.65 × 10−9, urine: P = 6.75 × 10−5; Wilcoxon rank-sum test).
Notably, 45% of the genes exhibiting switching-associated expression divergence are condition-specific (i.e., their expression diverges in one condition only) (Fig. 3D). Thus, switching may alter the response of bacteria only in a subset of environmental conditions. For example, condition-dependent expression divergence was observed in genes belonging to the methionine biosynthesis pathway (Fig. 3E). These genes exhibited similar expression levels in both strains when grown on MOPS but displayed higher expression in the uropathogenic E. coli when grown on urine. Three of these genes underwent switching, including the regulator of this pathway (metR), metF, and the last enzyme in the pathway (metE), which exhibited the highest expression divergence (up to 16-fold) (Fig. 3E).
HRT Affects the Fitness of Pathogenic E. coli.
The gene which exhibits the greatest urine specific expression divergence, metE, is known for its high sensitivity to oxidation (22). Consequently, cells exposed to oxidative stress develop methionine auxotropy (23). This sensitivity poses a challenge to uropathogenic E. coli, which is often exposed to oxidative stress generated by host immune cells (24). We reasoned that the switching observed in the regulatory region of metE (common to all uropathogenic E. coli isolates) might confer a fitness advantage under oxidizing conditions. To test this hypothesis, we constructed an isogenic pathogenic strain that was identical to its parent strain except that the short metE regulatory allele was replaced with the longer ancestral allele found in commensal E. coli. The resulting strain exhibited a similar growth rate on MOPS media lacking methionine. In contrast, under oxidative stress, this replacement strain exhibited a marked growth defect relative to the WT strain harboring the shorter metE allele (Fig. 3F). These results demonstrate that a single regulatory switching event, in which the coding region remains unmodified, can confer a significant fitness advantage.
Regulatory Switching Is Ubiquitous Across the Bacterial Domain.
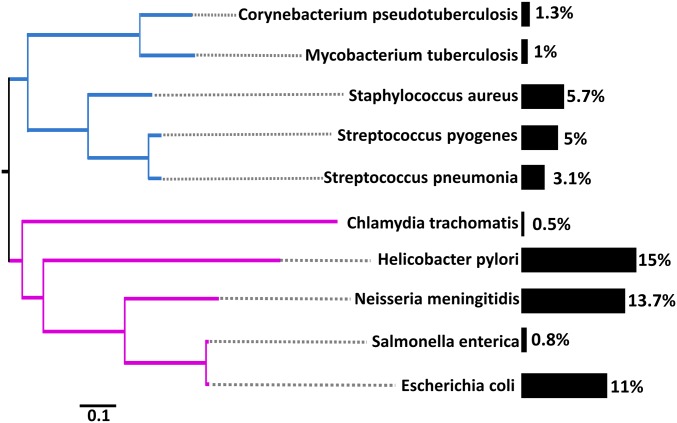
To determine if regulatory switching affects other clades beyond E. coli, we extended our analysis to nine additional taxa from across the bacterial domain with diverse physiological characteristics (Table S3). We found that all clades experienced switching, highlighting the phylogenetic breadth of this phenomenon (Fig. 4). Remarkably, the frequency of regulatory switching in core genes varies by more than an order of magnitude, from 0.5% in Chlamydia trachomatis, an obligate intracellular human pathogen, to more than 15% in Neisseria meningitidis, a highly recombinogenic pathogen that causes meningitis and septicemia. The variation in switching level among these bacterial clades could not be explained by sampling bias (Fig. S6) or phylogeny (Fig. 4).
Fig. 4.
Regulatory switching is ubiquitous across the bacterial domain. The bacterial species phylogeny based on 29 concatenated ribosomal proteins obtained from a study by Williams et al. (37) is shown. Bars indicate the level of regulatory switching observed across all genomes within each clade. Numbers at the end of each bar correspond to the percentage of core genes exhibiting regulatory switching. Gram-negative and Gram-positive taxa are shown in purple and blue, respectively. (Scale bar: substitutions per site.)
These findings raise the question as to what is driving variation in switching levels. Donor accessibility, ecology, and recombination efficiency were all found to affect gene transfer (25), and therefore are expected to affect regulatory transfer. Indeed, the level of switching is associated with the overall recombination-to-mutation (r/m) ratio (Table S4). Specifically, species with low r/m ratios are characterized by a low level of switching (e.g., C. trachomatis and Mycobacterium tuberculosis), whereas species with high r/m ratios are characterized by a high level of switching (e.g., Helicobacter pylori and N. meningitidis). However, this factor alone cannot explain the full extent of variation in the levels of regulatory switching. For instance, although Salmonella enterica and E. coli exhibit similar r/m ratios (0.14 and 0.38, respectively), E. coli exhibits more than a 10-fold higher level of regulatory switching. This difference might stem from the different lifestyle of the two species. Whereas S. enterica is an intracellular pathogen, E. coli is largely extracellular, and thus might be exposed to more foreign DNA during the course of its infection. Another factor that can affect the overall level of switching is the ability of bacteria to acquire DNA from the environment. Indeed, the highest levels of regulatory switching were found in the naturally competent bacteria H. pylori and N. meningitidis. Future work is needed to elucidate how mechanistic constraints and ecological barriers affect regulatory switching.
Discussion
Our observation that core genes exhibit ubiquitous regulatory switching contradicts the common assumption that core genes do not play a role in diversification (5). Previous studies have focused on protein-level conservation and overlooked regulatory switching as an orthogonal source of phenotypic variation in core genes. Switching enables a cell to bypass deleterious intermediates generated through the accumulation of point mutations, allowing even essential genes, such as hemH, to undergo regulatory modification. By enabling a “quantum leap” between the fitness peaks of functional regulatory elements, switching could facilitate efficient exploration of alternative promoter architectures.
The molecular mechanism most likely underlying the bacterial ability to switch from one regulatory sequence to another is homologous recombination. A short region of sequence identity is required to initiate this mechanism, and its efficiency decreases with increased sequence divergence between genomes (26, 27). Because core genes are highly conserved both between strains and often across distant species, they may enable regulatory switching between otherwise diverged bacteria. In line with this view, we find that 13.8% of the switched regulatory regions reside within a conserved region in which both the upstream and downstream genes are orthologous. Further support for the association between conservation and in situ replacement is the observation that xenologous recombination, the replacement of a gene by a distant homolog, was previously found to be prevalent within conserved operons (28). Of note, we expect regulatory switching to be even more frequent than in situ gene replacement, because regulatory regions are shorter than genes and can fit on a single E. coli recombination segment, which is, on average, 242 bp (SI Text).
We have shown that regulatory regions, similar to coding regions of bacteria, can be subjected to recombination and exchange. Several theories have been suggested to explain the differential frequencies with which genes undergo HGT. For example, the complexity hypothesis posits that HGT is rare in genes coding for proteins with many interactions compared with those genes coding for proteins with only a few interactions (29, 30). Other studies have detected functional and ecological barriers to horizontal transfer of protein-coding genes (25, 31). The barriers to HRT remain to be discovered, leaving many unanswered questions. Is it restricted by the number of regulatory interactions? Is it promoted by the availability of transcription factors that are shared between the donor and the acceptor? The sheer increase in the availability of fully sequenced bacterial genomes, together with the development of more specific tools for HRT analysis, should shed light on the evolutionary forces shaping the regulatory genome.
The ability of bacteria to tap a broad pool of regulatory sequences suggests that in addition to an environment-specific metagenome, there is an unexplored parallel pool of sequences, the metaregulome. In response to environmental changes, bacteria not only acquire new proteins; they may also acquire novel regulatory sequences to enable more appropriate control of their existing protein repertoire. Our results demonstrate the importance of mobile DNA in regulatory evolution, opening a new window for exploring the mechanisms that bacteria use to respond to environmental changes.
Materials and Methods
Additional details are available in SI Text.
Regulatory Switching Pipeline.
We detected orthologous genes using reciprocal Translated BLAST (tblastx) (32) best hits with at least 95% amino acid identity (for the core gene analysis; only genes that were shared among all strains of a given species were considered). Next, we detected orthologous gene clusters, requesting 90% identity among all members of a cluster. The regulatory region of each gene cluster, defined as 300 bp upstream of the TSS, was extracted. Last, the orthologous regulatory regions of each gene were clustered. Genes were considered switched if their regulatory regions formed more than one cluster.
HRT Detection.
HRT was detected by searching for statistical significant incongruence between the species tree and the regulatory region tree. Specifically, maximum-likelihood trees were reconstructed using PhyML (33) with the general time reversible model (34), and incongruence was tested using the AU (19) test as implemented in CONSEL software (35). To test whether a core gene and its regulatory region were independently transferred, we repeated this procedure comparing the core gene tree and the regulatory region tree.
Promoter Library TSS Determination.
We synthesized a library of 1,693 promoters from 40 E. coli strains and used the RNA-Seq–based approach described by Kosuri et al. (20) to determine the TSS of orthologous genes. For each of the 485 genes expressed under the experimental condition, we computed a distance score reflecting shifts in TSS positioning across strains. A bootstrap-based approach was used to test whether TSS shifts were significantly enriched among switched genes.
RNA-Seq.
E. coli CFT073 and E. coli K-12 MG1655 were grown with shaking at 37 °C in 12 mL of MOPS media supplemented with 0.2% tryptone and 0.2% glucose until the OD600 reached 0.2. Five milliliters of the bacterial media was then passed through a 0.2-mm pore-sized filter and resuspended in either urine (pooled from six healthy volunteers) or MOPS. The resuspended bacteria were grown for an additional 15 min with shaking at 37 °C and then harvested. Detailed information and sequences are available in the Gene Expression Omnibus (GEO) database (accession no. GSE59468).
Allelic Exchange and Exposure to Oxidative Stress.
MetE allelic exchange was achieved by using the λ-red recombination system (36). For the oxidative stress experiments, bacteria were grown for 2 h on minimal MOPS media with 0.2% glucose. After 2 h, H2O2 at a final concentration of 1.5 mM was added to the culture and growth was monitored.
Supplementary Material
Acknowledgments
We thank Sriram Kosuri for help in obtaining the Agilent oligo library. Y.O. is an Edmond J. Safra Fellow. T.P. is supported by Israeli Foundation Grant 1092/13. J.C. is supported by European Research Council Grant 239784 and by Academy of Finland Grant 251170. D.B. and E.Z.R. are supported by Deutsch-Israelische Projektkooperation Grant RO 2612/1-1. H.H.W. is supported by NIH Director's Early Independence Award 1DP5OD009172-01. N.I.J. is supported by a National Science Foundation (NSF) graduate research fellowship. E.J.A. is supported by the NSF Grant DEB-0936234.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE59468).
See Commentary on page 15865.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413272111/-/DCSupplemental.
References
- 1.Lester CH, Frimodt-Møller N, Sørensen TL, Monnet DL, Hammerum AM. In vivo transfer of the vanA resistance gene from an Enterococcus faecium isolate of animal origin to an E. faecium isolate of human origin in the intestines of human volunteers. Antimicrob Agents Chemother. 2006;50(2):596–599. doi: 10.1128/AAC.50.2.596-599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J, Novick RP. Phage-mediated intergeneric transfer of toxin genes. Science. 2009;323(5910):139–141. doi: 10.1126/science.1164783. [DOI] [PubMed] [Google Scholar]
- 3.Hehemann JH, et al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464(7290):908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 4.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405(6784):299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 5.Medini D, et al. Microbiology in the post-genomic era. Nat Rev Microbiol. 2008;6(6):419–430. doi: 10.1038/nrmicro1901. [DOI] [PubMed] [Google Scholar]
- 6.Treangen TJ, Rocha EP. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 2011;7(1):e1001284. doi: 10.1371/journal.pgen.1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dagan T, Artzy-Randrup Y, Martin W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci USA. 2008;105(29):10039–10044. doi: 10.1073/pnas.0800679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koonin EV, Makarova KS, Aravind L. Horizontal gene transfer in prokaryotes: Quantification and classification. Annu Rev Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ragan MA, Beiko RG. Lateral genetic transfer: Open issues. Philos Trans R Soc Lond B Biol Sci. 2009;364(1527):2241–2251. doi: 10.1098/rstb.2009.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matus-Garcia M, Nijveen H, van Passel MW. Promoter propagation in prokaryotes. Nucleic Acids Res. 2012;40(20):10032–10040. doi: 10.1093/nar/gks787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee DH, Palsson BO. Adaptive evolution of Escherichia coli K-12 MG1655 during growth on a Nonnative carbon source, L-1,2-propanediol. Appl Environ Microbiol. 2010;76(13):4158–4168. doi: 10.1128/AEM.00373-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Somvanshi VS, et al. A single promoter inversion switches Photorhabdus between pathogenic and mutualistic states. Science. 2012;337(6090):88–93. doi: 10.1126/science.1216641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blount ZD, Barrick JE, Davidson CJ, Lenski RE. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature. 2012;489(7417):513–518. doi: 10.1038/nature11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen SL, et al. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: A comparative genomics approach. Proc Natl Acad Sci USA. 2006;103(15):5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Retchless AC, Lawrence JG. Ecological adaptation in bacteria: Speciation driven by codon selection. Mol Biol Evol. 2012;29(12):3669–3683. doi: 10.1093/molbev/mss171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendoza-Vargas A, et al. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS ONE. 2009;4(10):e7526. doi: 10.1371/journal.pone.0007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gama-Castro S, et al. RegulonDB version 7.0: Transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor Units) Nucleic Acids Res. 2011;39(Database issue):D98–D105. doi: 10.1093/nar/gkq1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng M, et al. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol. 2001;183(15):4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51(3):492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 20.Kosuri S, et al. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc Natl Acad Sci USA. 2013;110(34):14024–14029. doi: 10.1073/pnas.1301301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeProust EM, et al. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process. Nucleic Acids Res. 2010;38(8):2522–2540. doi: 10.1093/nar/gkq163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leichert LI, Jakob U. Protein thiol modifications visualized in vivo. PLoS Biol. 2004;2(11):e333. doi: 10.1371/journal.pbio.0020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hondorp ER, Matthews RG. Oxidative stress inactivates cobalamin-independent methionine synthase (MetE) in Escherichia coli. PLoS Biol. 2004;2(11):e336. doi: 10.1371/journal.pbio.0020336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rama G, Chhina DK, Chhina RS, Sharma S. Urinary tract infections-microbial virulence determinants and reactive oxygen species. Comp Immunol Microbiol Infect Dis. 2005;28(5-6):339–349. doi: 10.1016/j.cimid.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Popa O, Dagan T. Trends and barriers to lateral gene transfer in prokaryotes. Curr Opin Microbiol. 2011;14(5):615–623. doi: 10.1016/j.mib.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 26.Shen P, Huang HV. Homologous recombination in Escherichia coli: Dependence on substrate length and homology. Genetics. 1986;112(3):441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vulić M, Dionisio F, Taddei F, Radman M. Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc Natl Acad Sci USA. 1997;94(18):9763–9767. doi: 10.1073/pnas.94.18.9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omelchenko MV, Makarova KS, Wolf YI, Rogozin IB, Koonin EV. Evolution of mosaic operons by horizontal gene transfer and gene displacement in situ. Genome Biol. 2003;4(9):R55. doi: 10.1186/gb-2003-4-9-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen O, Gophna U, Pupko T. The complexity hypothesis revisited: Connectivity rather than function constitutes a barrier to horizontal gene transfer. Mol Biol Evol. 2011;28(4):1481–1489. doi: 10.1093/molbev/msq333. [DOI] [PubMed] [Google Scholar]
- 30.Doolittle WF. Lateral genomics. Trends Cell Biol. 1999;9(12):M5–M8. [PubMed] [Google Scholar]
- 31.Sorek R, et al. Genome-wide experimental determination of barriers to horizontal gene transfer. Science. 2007;318(5855):1449–1452. doi: 10.1126/science.1147112. [DOI] [PubMed] [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 33.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 34. Simon T (1986) Some probabilistic and statistical problems in the analysis of DNA sequences. Some Mathematical Questions in Biology: DNA Sequence Analysis, Lectures on Mathematics in the Life Sciences, ed Minura RM (American Mathematical Society, Providence, RI), Vol 17, pp 57–86.
- 35.Shimodaira H, Hasegawa M. CONSEL: For assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17(12):1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- 36.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams D, et al. A rooted net of life. Biol Direct. 2011;6:45. doi: 10.1186/1745-6150-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.