Significance
Stem cells make lineage-choice decisions based on a combination of internal and external signals, including mechanical cues from the surrounding environment. Here we show that Piezo1, an ion channel opened by membrane tension, plays an important role in transducing matrix mechanical information to intracellular pathways affecting differentiation in neural stem cells. Piezo1 activity influences whether neural stem cells differentiate along a neuronal or astrocytic lineage. One of the barriers to successful neural stem cell transplantation therapy for neurological disorders lies in directing the fate of transplanted cells. Pharmacological agents aimed at modulating Piezo1 activity may be useful in directing the fate of transplanted neural stem cells toward the desired lineage.
Keywords: matrix mechanics, calcium signaling, myosin II, Yap/Taz, blebbistatin
Abstract
Neural stem cells are multipotent cells with the ability to differentiate into neurons, astrocytes, and oligodendrocytes. Lineage specification is strongly sensitive to the mechanical properties of the cellular environment. However, molecular pathways transducing matrix mechanical cues to intracellular signaling pathways linked to lineage specification remain unclear. We found that the mechanically gated ion channel Piezo1 is expressed by brain-derived human neural stem/progenitor cells and is responsible for a mechanically induced ionic current. Piezo1 activity triggered by traction forces elicited influx of Ca2+, a known modulator of differentiation, in a substrate-stiffness–dependent manner. Inhibition of channel activity by the pharmacological inhibitor GsMTx-4 or by siRNA-mediated Piezo1 knockdown suppressed neurogenesis and enhanced astrogenesis. Piezo1 knockdown also reduced the nuclear localization of the mechanoreactive transcriptional coactivator Yes-associated protein. We propose that the mechanically gated ion channel Piezo1 is an important determinant of mechanosensitive lineage choice in neural stem cells and may play similar roles in other multipotent stem cells.
Mechanical properties of the cellular environment are powerful modulators of stem cell behavior. For instance, local mechanical cues such as extracellular matrix elasticity and nanotopology affect stem cell lineage choice (1, 2). Mechanical effects on fate are particularly relevant for stem cell transplant therapy, because stem cells encounter diverse mechanical signals upon engraftment (3, 4). Moreover, before transplantation, stem cells are grown in vitro, where the mechanical properties of culture conditions can affect their behavior. Recent studies show that stem cells may possess a memory of past mechanical environments (5) and that the mechanical properties of the culturing environment before transplantation can influence the outcome of in vivo stem cell transplants (6). Hence, a molecular and mechanistic understanding of how stem cells process mechanical cues and how this processing results in downstream signaling events and ultimately in fate decisions is needed for greater control over the fate of transplanted cells.
Studies in mesenchymal and neural stem cells have revealed the involvement of focal adhesion zones and cytoskeletal proteins, such as integrins, nonmuscle myosin II (7), Rho GTPases (8–10), and vinculin (11), that participate in the generation of cellular traction forces. Recent work also has identified the nucleoskeletal protein lamin-A (12) and the transcriptional coactivators Yap (Yes-associated protein) and Taz (transcriptional coactivator with PDZ-binding motif) (13) in mechanotransduction in mesenchymal stem cells. However, the mechanisms by which mechanical cues detected by cellular traction forces are transduced to downstream intracellular pathways of differentiation remain unclear.
Ion channels are involved, directly or indirectly, in the transduction of all forms of physical stimuli—including sound, light, temperature, mechanical force, and even gravity—into intracellular signaling pathways. Hence, we wondered whether ion channels could be involved in transducing matrix mechanical cues to intracellular signaling pathways linked to lineage specification. In particular, we focused here on cationic stretch-activated channels (SACs) because they are known to detect mechanical forces with high sensitivity and broad dynamic range and because they are permeable to Ca2+, an important second messenger implicated in cell fate (14, 15). We examined the role of SACs in neural stem cells, for which mechanical cues influence specification along the neuronal–glial lineage (8, 16, 17). We find that human neural stem/progenitor cells (hNSPCs) express the SAC Piezo1 and that the activity of the channel is mediated by cell-generated traction forces. Piezo1 activation elicits transient Ca2+ influx in a substrate-stiffness–dependent manner, favors nuclear localization of the mechanoreactive transcription coactivator Yap, and influences neuronal vs. glial specification.
Results
We used cultures of the hNSPCs SC23 and SC27 derived from the cerebral cortices of two separate postmortem fetal brains (18, 19) to test the role of SACs in mechanical modulation of stem cell behavior. Both cultures express standard neural stem cell markers, including Sox2, nestin, and the cell surface marker CD133, and have the potential to differentiate into the three major neural cell types: astrocytes, neurons, and oligodendrocytes (18–22). When transplanted into a mouse model of Sandhoff disease, these hNSPCs stably integrated into the host brain and delayed disease onset, reflecting their functional and therapeutic potential (23). Similar fetal brain-derived hNSPCs also have been shown to differentiate into neurons, astrocytes, and oligodendrocytes upon transplantation into the central nervous system (24) and currently are in clinical trials for multiple neurological conditions (25, 26).
hNPSCs Exhibit Stretch-Activated Ionic Currents.
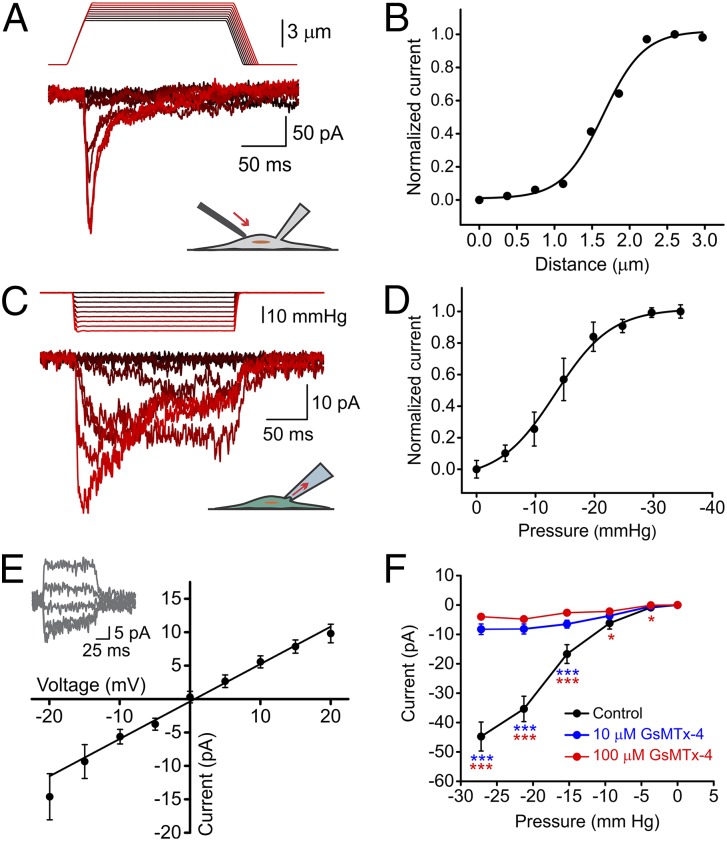
To test whether SC23 and SC27 cells possess SAC activity, we mechanically stimulated their plasma membranes using a piezoelectric actuator while simultaneously measuring ionic currents with whole-cell patch clamp. We consistently observed ionic currents in response to mechanical stimuli (Fig. 1A). Peak amplitude increased with stimulus intensity along a Boltzmann curve as expected for a SAC (Fig. 1B). We confirmed this result using a different patch-clamp assay, which measures currents from a small patch of membrane rather than from the whole cell. In this cell-attached configuration, the membrane is stretched by negative suction pulses applied with a high-speed pressure clamp (Fig. 1 C and D and Fig. S1). Using this technique, we measured ionic currents in response to mechanical stimulation in 91 of 107 cells (85%).
Fig. 1.
Mechanically induced currents in hNSPCs. (A) Representative traces of mechanically induced inward currents from SC23 hNSPC cells. Cells were subjected to a series of mechanical stimuli by indenting the plasma membrane with a stimulation probe. The probe was moved in 0.25-μm steps while recording in the whole-cell patch configuration with a holding potential of −80 mV. Data shown are representative of n = 6 cells. (B) Current amplitude in response to indentation by probe displacement from the traces shown in A. (C) Representative currents in an SC27 hNSPC cell induced by negative pipette pressure (0 to −30 mmHg in 3-mmHg steps) administered by a pressure clamp. Holding potential: −80 mV. (D) Normalized current–pressure relationship of SACs at a holding potential of −80 mV fitted with a Boltzmann equation with P50 = −13.4 mmHg and s = 4.75 mmHg. Results are shown as mean ± SEM (n = 8 cells). (E) Average current–voltage relationships of SACs in SC27 hNSPC cells. Results are shown as mean ± SEM (n = 7 cells). (Inset) Current traces in response to voltage steps from −40 mV to 40 mV in 20-mV steps, applied 250 ms before a negative pressure pulse of −30 mmHg. (F) Mean current amplitudes elicited by negative suction pulses in cell-attached patch-clamp mode in the absence (black, n = 29 cells) and presence of 10 μM (blue, n = 69 cells) and 100 μM (red, n = 42 cells) extracellular GsMTx-4. Error bars indicate SEM and are smaller than data points in some cases. *P < 0.05, **P < 0.01, ***P < 0.001 by two-sample t test. See also Fig. S1.
Ionic currents measured by both techniques displayed hallmarks of SACs: in standard extracellular and intracellular solutions (SI Methods) and at a holding potential of −80 mV, the currents were inward in direction and reduced in magnitude exponentially when presented with a persisting mechanical stimulus (Fig. 1 A and C). Ionic currents reversed direction near 0 mV (reversal potential = −0.75 ± 0.67 mV) (Fig. 1E), as is consistent with the activity of a nonspecific cation channel. We next tested the susceptibility of hNSPC SAC currents to GsMTx-4, a peptide isolated from the venom of the Chilean rose tarantula spider, Grammostola rosea. GsMTx-4 is the only known drug to inhibit cationic SACs specifically without inhibiting other ion-channel families such as voltage-gated sodium, potassium, and calcium channels (27) or potassium-selective SACs (28). It acts as a gating modifier by partitioning into the cell membrane and affecting force transfer to the channel protein (29). hNSPC SAC currents were inhibited by extracellular GsMTx-4 (Fig. 1F), as is consistent with their generation by a stretch-activated cationic channel.
Molecular Identification of hNSPC SAC.
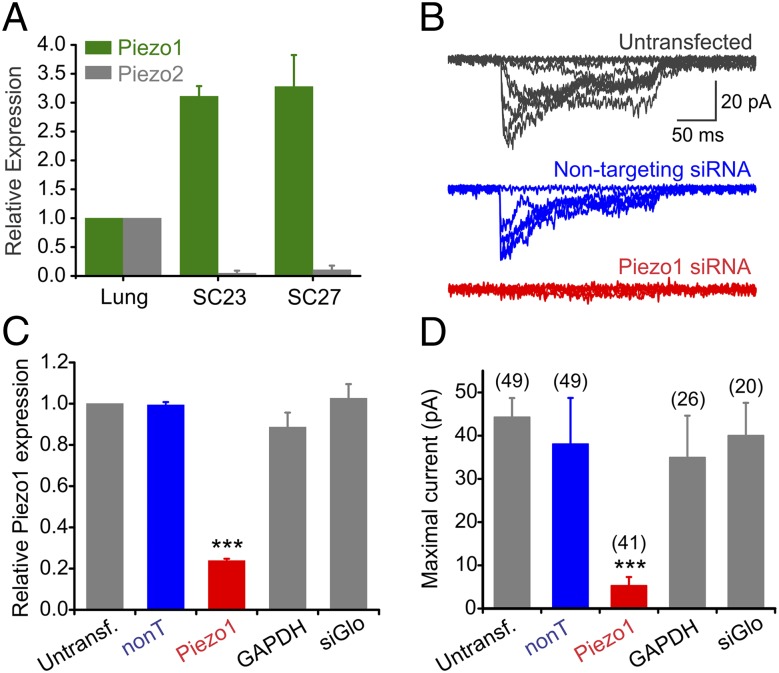
We next asked which ion channel(s) could be responsible for the measured mechanically induced currents. Based on the electrophysiological properties of the ionic currents, we examined nine known cationic mechanosensitive channels for expression in SC27 hNSPCs using quantitative real-time PCR (qRT-PCR) (Fig. S2A). Of these, Piezo1 was expressed at much greater levels than the other channels. The Piezo channels recently were shown to mediate physiologically relevant mechanically activated and nonspecific cationic currents in mammals (30–34). Like the endogenous hNSPC SAC, Piezo1 is inactivated by persisting mechanical stimuli (31) and is sensitive to GsMTx-4 (35). Although Piezo1 was highly expressed in SC23 and SC27 cells, its paralogue, Piezo2, was not (Fig. 2A).
Fig. 2.
Piezo1 is essential for mechanically induced currents in hNSPCs. (A) mRNA expression of Piezo1 (green) and Piezo2 (gray) in SC23 and SC27 hNSPCs determined by qRT-PCR with 18S as the reference gene. Human lung was used as the tissue calibrator by the 2–∆∆CT method, because previously it was shown to express both channels (31). n = 6 independent experiments for Piezo1 and n = 4 independent experiments for Piezo2. Data are shown as mean ± SEM. (B) Representative mechanotransduction currents from an untransfected (Top, gray; same cell as in Fig. 1C), a cell transfected with a control nontargeting pool of siRNAs (Middle, blue), and a cell transfected with four siRNAs against Piezo1 (Bottom, red). The holding potential was −80 mV. (C) Quantification of Piezo1 transcripts by qRT-PCR from three independent transfection experiments. Parallel samples were used for patch-clamp measurements in D. ***P < 0.001 by ANOVA. (D) Mean maximal mechanotransduction currents recorded from untransfected cells, from cells transfected with a nontargeting pool of four siRNAs (nonT, 20 nM, blue), siGlo transfection marker alone (20 nM) or with a pool of four siRNAs against Piezo1 (20 nM, red) or GAPDH (25 nM). Numbers in parentheses refer to the numbers of cells patched for each condition. Data in C and D are from three independent transfection experiments. ***P < 0.001, Kruskal–Wallis test. See also Figs. S2 and S8.
To determine whether Piezo1 underlies the hNSPC mechanically induced currents, we tested the effect of siRNA-mediated gene knockdown on the ionic current. We transfected SC27 hNSPCs with a pool of four siRNAs against Piezo1. We performed qRT-PCR to assess transcript levels and cell-attached patch-clamp recordings to examine ionic current amplitude 36–72 h after transfection. Treatment with Piezo1 siRNAs reduced Piezo1 transcripts and caused an attenuation of ionic currents (Fig. 2). Pools of nontargeting control siRNAs or GAPDH siRNAs and the fluorescent reporter of transfection (siGlo) alone did not show a significant reduction of the Piezo1 RNA or of mechanotransduction currents (Fig. 2 and Fig. S2B). Among the cells transfected with Piezo1 siRNA, Piezo1 transcripts were reduced by 76.2 ± 0.9%, and mean maximal current amplitude was reduced by 89% relative to untransfected cells, with 27 of 42 cells (64%) showing no measurable current. The nearly complete loss of mechanically evoked currents in response to Piezo1 siRNA indicates that Piezo1 underlies mechanotransduction currents in hNSPCs.
Piezo1 Activity Elicits Spontaneous Ca2+ Transients.
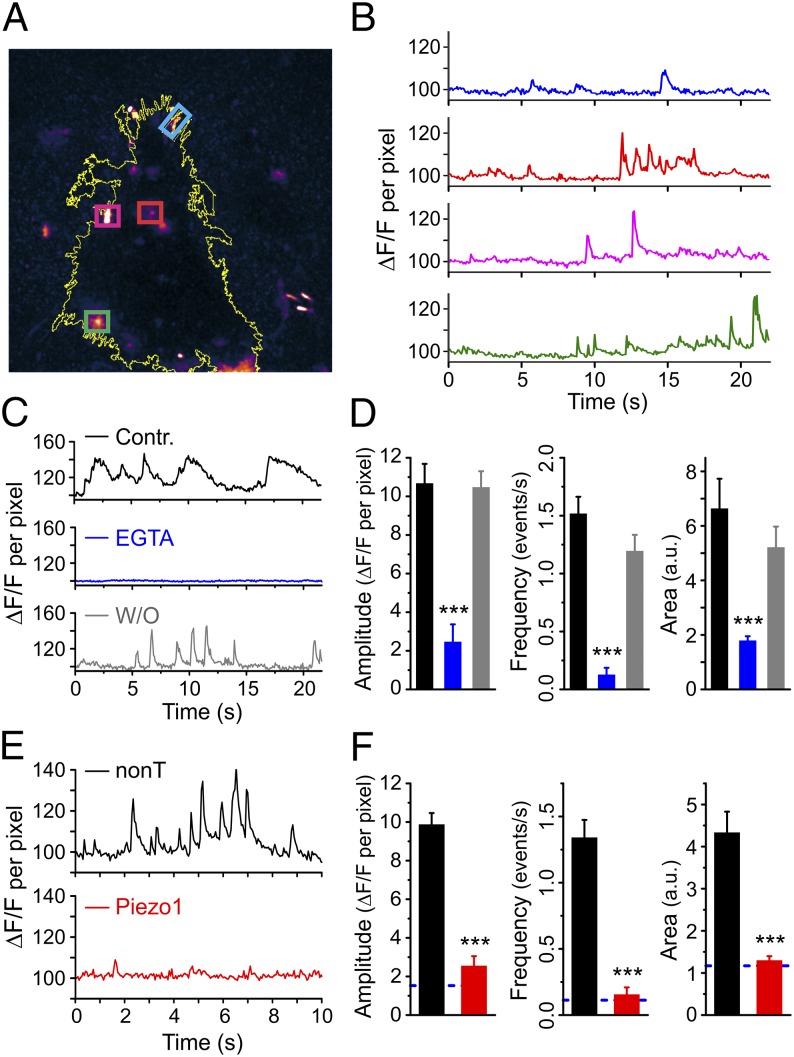
Piezo1 has been implicated in cellular processes in which cells respond to an external mechanical force, e.g., a stretch stimulus, osmotic pressure, or shear stress (31, 33, 34, 36–38). We asked whether cell-generated forces, such as traction forces, also can activate Piezo1. Our patch-clamp assays described above preferentially stimulate cells on the dorsal surface of the cell, whereas traction forces are generated more prominently at the cell’s ventral surface, where the cell contacts the substrate through focal adhesion zones. To assay the activity of Piezo1 at the ventral surface, we used the Piezo1 channel’s preference for conducting Ca2+ (31). We imaged spontaneous Ca2+ dynamics of hNSPCs grown on glass coverslips and loaded with the fluorescent Ca2+ indicator Fluo-4 AM. We selectively imaged plasma membrane events in the absence of intracellular background fluorescence by using total internal reflection fluorescence microscopy (TIRFM). We observed many spontaneous Ca2+ transients over a time scale of seconds (Fig. 3 A and B and Movie S1). Spontaneous Ca2+ signals were reversibly abolished by chelating external Ca2+ with EGTA, indicating that Ca2+ influx across the plasma membrane is required for the generation of the signals (Fig. 3 C and D). We quantified the spontaneous Ca2+ transients by computing the amplitude of the transients, their frequency, and the area under the fluorescence curve. All three measures showed reversible and statistically significant reduction in the presence of EGTA.
Fig. 3.
Piezo1 activity elicits spontaneous Ca2+ signals. (A) A maximum projection TIRFM image of an hNSPC cell loaded with Fluo-4 AM from a 600-frame video showing hotspots of Ca2+ activity. Four regions of interest are marked by colored boxes. The yellow line depicts the outline of the cell. See also Movie S1. (B) Plots of fluorescence intensity over time in the hotspots highlighted in A. The color of the trace matches the corresponding box color in A. Data are shown as signal per pixel with the fluorescence intensity at the start of the recording normalized to 100 (SI Methods). (C and D) Spontaneous Ca2+ transients are reversibly inhibited by addition of 6 mM EGTA to the bath solution. n = 22 for control (Contr.), 28 for EGTA, and 27 for washout (W/O). (E and F) hNSPCs transfected with Piezo1 siRNA show fewer and smaller spontaneous Ca2+ transients than cells transfected with nontargeting siRNA. The blue dashed line in F represents the levels expected in the absence of external calcium (based on D). n = 44 for nontargeting siRNA, and n = 53 for Piezo1 siRNA. Error bars represent SEM. ***P < 0.001 by two-sample t test. a.u., arbitrary units. See also Fig. S3.
To test whether spontaneous Ca2+ transients arise from Piezo1 activity, we examined the effect of Piezo1 knockdown on spontaneous Ca2+ transients. Cells transfected with Piezo1 siRNA showed a strong reduction in Ca2+ transients compared with control-transfected cells, as evidenced by analysis of amplitude, frequency of events, and the area under the curve (Fig. 3 E and F). The magnitude of the residual calcium signals measured in Piezo1 knockdown cells was similar in magnitude to that measured in EGTA-treated cells. A significant reduction in spontaneous Ca2+ transients also was obtained with extracellular application of the SAC inhibitor GsMTx-4 (Fig. S3). Taken together, these observations carried out in the absence of an externally applied mechanical force indicate that Piezo1 activity generates spontaneous Ca2+ transients in hNSPCs.
Piezo1 Is Activated by Traction Forces.
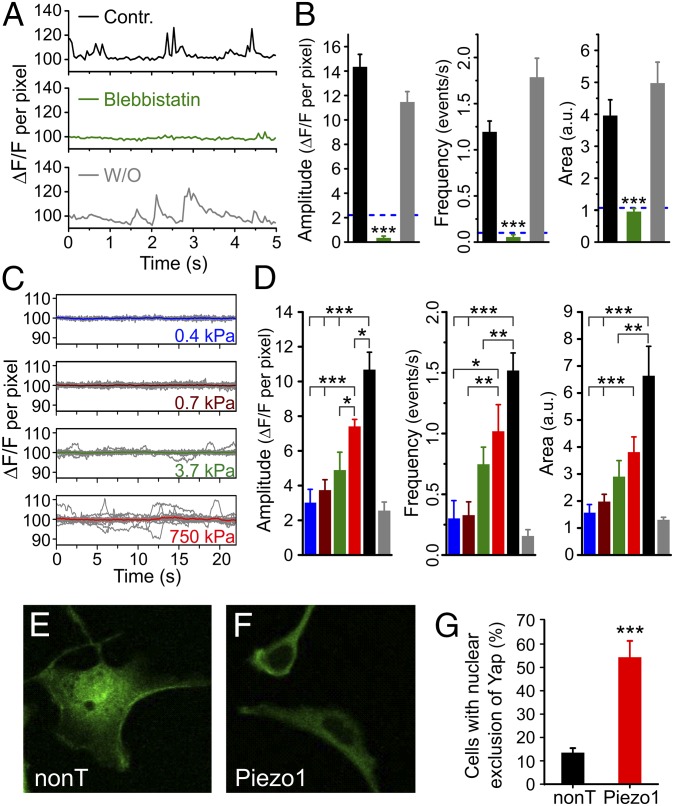
Traction forces allow cells to sense the stiffness, geometry, and topography of their environment and play an important role in cell adhesion, migration, extracellular matrix reorganization, and differentiation (39). These forces are generated in an ATP-dependent process by nonmuscle myosin II along actin fibers and are transmitted to the substrate through focal adhesion zones. Blebbistatin is known to inhibit myosin II specifically (40) and, as a result, to inhibit traction forces (7). To determine whether Piezo1 is activated by traction forces, we examined whether blebbistatin inhibits the spontaneous Ca2+ transients generated by Piezo1. We found that blebbistatin treatment abolishes spontaneous Ca2+ transients to a similar extent as EGTA (Fig. 4 A and B). These results indicate that Piezo1 can be activated by cell-generated forces such as traction forces without application of an external mechanical force.
Fig. 4.
Piezo1 activity is linked to traction forces, substrate stiffness, and Yap localization. (A and B) Spontaneous Ca2+ transients are reversibly inhibited by the traction force inhibitor blebbistatin (50 μM), which acts by blocking the ATPase activity of myosin II. n = 57 for control (Contr.); n = 49 for blebbistatin; and n = 28 for washout (W/O). The dashed blue line in B represents levels expected in the absence of external calcium (based on Fig. 3D). (C and D) Spontaneous Ca2+ transients measured from hNSPCs grown on high-refractive-index silicone elastomers scale with substrate stiffness. n = 14 for 0.4 kPa; n = 26 for 0.7 kPa; n = 15 for 3.7 kPa; n = 14 for 750 kPa; and n = 22 for glass. The colors of the columns in D are matched to the traces in C: blue, 0.4 kPa; brown, 0.7 kPa; green, 3.7 kPa; red, 750 kPa; black, glass; gray, data from Piezo1 siRNA-transfected cells from Fig. 3F (reproduced for comparison). (E–G) hNSPCs grown on glass coverslips show nuclear exclusion of Yap at a higher frequency when transfected with Piezo1 siRNA than when transfected with nontargeting siRNA. Error bars represent SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by two-sample t test. a.u., arbitrary units.
Because traction forces are known to vary with substrate stiffness, an important modulator of differentiation in several different stem cell types (7, 8), we asked whether Piezo1 activity varies with substrate stiffness. We performed TIRFM imaging of spontaneous Ca2+ transients of hNSPCs grown on high-refractive-index Qgel silicone elastomers of varying stiffness (41), fabricated as described in SI Methods. We found that spontaneous Ca2+ activity scaled with substrate stiffness, with minimal or no transients on soft substrates (0.4 and 0.7 kPa) and increasing activity on stiffer substrates (3.7 kPa and 750 kPa) and on glass (Fig. 4 C and D). Thus, Piezo1 activity varies with substrate stiffness.
Piezo1 Knockdown Evokes Nuclear Exclusion of the Mechanoreactive Transcriptional Coactivator Yap.
Because Piezo1 activity varies with substrate stiffness, we asked whether it affects a molecule known to relay information about substrate stiffness to the nucleus. The transcriptional coactivators Yap and Taz (also known as “WWTR1”) were first identified as part of the Hippo pathway that controls organ size (42). Recent findings in epithelial cells and in mesenchymal stem cells show that Yap and Taz localize to the nucleus on stiff substrates, but on soft substrates they are shuttled to the cytoplasm (13). The same study also showed that Yap and Taz influence mechanosensitive lineage specification in mesenchymal stem cells. The molecular mechanism by which matrix stiffness regulates Yap/Taz localization remains unknown.
We first examined Yap localization in hNSPCs on soft (0.7 kPa) and stiff (750 kPa) substrates using immunostaining. As seen for other cell types, in hNSPCs grown on soft substrates the Yap protein is excluded from the nucleus (Fig. S4). Next, we examined Yap localization in hNSPCs grown on glass coverslips and transfected with either nontargeting or Piezo1 siRNA. Glass has a stiffness in the gigapascal range, at which Yap is expected to localize to the nucleus. We observed that cells transfected with Piezo1 siRNA displayed nuclear exclusion more frequently than cells transfected with nontargeting siRNA (Fig. 4 E–G). This result suggests that Piezo1 knockdown can override the mechanical cue for localizing Yap to the nucleus and that Yap could be a downstream effector of Piezo1 activity.
Effect of Substrate Stiffness on hNSPC Differentiation.
Earlier studies on rodent neural stem cells have shown the importance of matrix elasticity in lineage specification (8, 16, 17). However, the effect of substrate stiffness on hNSPC differentiation has not been investigated previously. When we differentiated hNSPCs on substrates of different stiffness (0.7 kPa vs. 750 kPa), we observed a greater number of microtubule-associated protein 2 (MAP2)-positive neuronal cells on the stiffer substrate (Fig. S5). In rodent neural stem cells, substrate stiffness was found to have the opposite effect, with soft substrates (<1 kPa) supporting more neurogenesis than stiffer substrates (>5 kPa) (8, 16, 17). Because we performed our differentiation assays on Qgel-based substrates, whereas earlier studies used polyacrylamide or methacrylamide chitosan as supporting material (8, 16, 17), we wondered whether this difference could be responsible for the discrepant results. However, when we differentiated rat adult hippocampal neural stem cells [rahNSCs, which generated more neurons on soft polyacrylamide substrates (17)] on Qgels (0.7 kPa vs. 750 kPa), we similarly observed more neurogenesis on softer substrates (Fig. S6). Hence, we conclude that the difference in mechanosensitive differentiation between hNSPCs and rahNSCs is likely caused by the different origin of the two types of cells (human vs. rat, fetal vs. adult, cortical vs. hippocampal).
Piezo1 Directs Neuronal–Glial Lineage Choice of hNSPCs.
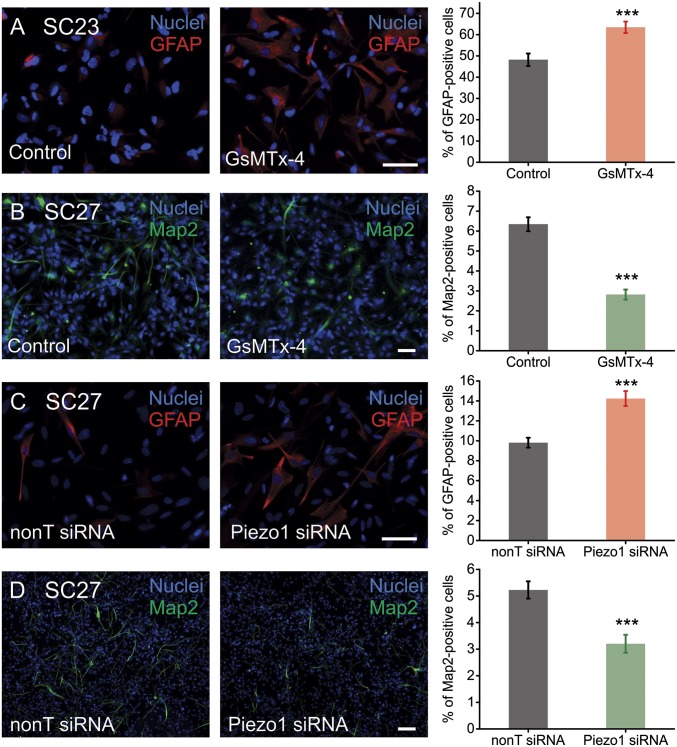
The results presented in Fig. 4 show that Piezo1-mediated Ca2+ transients increase on stiffer substrates. We also found that stiffer substrates favor the neuronal differentiation of hNSPCs (Fig. S5). Hence, we asked whether Piezo1 activity influences hNSPC lineage choice. We used two different approaches to investigate this question. We first determined whether pharmacological inhibition of Piezo1 by extracellular application of GsMTx-4 alters hNSPC lineage choice. hNSPCs were differentiated in the presence and absence of extracellular GsMTx-4, and differentiation was assayed using Map2 and doublecortin (Dcx) as neuronal markers and glial fibrillary acidic protein (GFAP) as an astrocytic marker (SI Methods). In the presence of GsMTx-4, SC23 hNSPCs showed an increase in astrocyte formation, as evidenced by the percentage of GFAP+ cells, from 48.2 ± 2.9% to 63.5 ± 2.7% (a 35% increase relative to control) (Fig. 5A). We showed earlier that SC27 hNSPCs generate neurons at higher percentages than SC23s (21); hence tests on neurogenesis were performed with the SC27 hNSPCs. These hNSPCs showed a reduction in neuron formation from 6.4 ± 0.3% to 2.8 ± 0.3% (a 54% decrease relative to control) when differentiated in the presence of GsMTx-4, as evidenced by the percentage of Map2+ cells (Fig. 5B). Similar results were obtained for Dcx+ cells (Fig. S7A). Thus, pharmacological inhibition of SACs appears to promote astrocyte formation and to reduce neuron formation.
Fig. 5.
Piezo1 directs neuronal–glial lineage choice in human neural stem cells. (A) SC23 hNSPCs display an increase in astrogenesis (GFAP+ cells) in the presence of 5 μM free GsMTx-4 (SI Methods). n = 3 independent experiments. (B) SC27 hNSPCs display a reduction in the percentage of Map2+ cells when differentiated in the presence of 5 μM free GsMTx-4. n = 4 biological repeats from three independent experiments. (C) SC27 hNSPCs transfected with 20 nM Piezo1 siRNA differentiate into astrocytes more efficiently than SC27 hNSPCs transfected with 20 nM control nontargeting (nonT) siRNA. n = 3 biological repeats from two independent transfection experiments. (D) SC27 hNSPCs transfected with 20 nM Piezo1 siRNA show reduced neurogenesis as compared with cells transfected with 20 nM control nontargeting siRNA. n = 3 biological repeats from two independent transfection experiments. (Scale bars: 20 μm.) ***P < 0.001 with two-sample t test. See also Fig. S7.
To determine whether this effect on lineage choice specifically involved Piezo1, we examined differentiation in the context of siRNA-mediated knockdown of Piezo1. SC27 hNSPCs were transfected with either Piezo1 siRNA or nontargeting siRNA and differentiated into neurons or astrocytes after allowing 24 h for siRNA action. qRT-PCR analysis indicated that Piezo1 knockdown is 78–86% at 1–4 d and 71% at 7 d posttransfection. Immunostaining analysis of the differentiated progeny showed that Piezo1 knockdown increased astrocyte formation from 9.8 ± 0.5% to 14.2 ± 0.7% (a 48% increase relative to control) (Fig. 5C) while reducing neuron formation from 5.2 ± 0.3% to 3.2 ± 0.3% (a 42% decrease relative to control) (Fig. 5D and Fig. S7B). SC23 hNSPCs also showed an increase in astrogenesis with Piezo1 knockdown (Fig. S7C). These results recapitulate the effect of pharmacological SAC inhibition on lineage choice and demonstrate that Piezo1 activity is involved in neuronal–glial specification of hNSPCs.
Discussion
Matrix mechanics is a strong determinant of stem cell behavior with important implications for regenerative medicine and stem cell therapy. Using a multidisciplinary approach, we have identified a new molecular player, the SAC Piezo1, which detects matrix mechanics and directs lineage specification of hNSPCs. The Piezo1 protein was discovered recently and was shown to be the pore-forming subunit of a nonspecific cation channel that is intrinsically gated by membrane tension (31, 32). Recent studies demonstrate a role for Piezo1 in regulating erythrocyte volume (43), epithelial crowding and homeostasis (36), cell migration (44), and detection of blood flow (33, 34). Our results suggest a role for Piezo1 in detecting matrix elasticity and determining lineage choice. In addition, we find that Piezo1 activation can be triggered by cell-generated forces in the absence of an external mechanical force, thus bringing to light a previously unidentified way to regulate Piezo1 gating.
Piezo1 also is identified in public gene-expression databases from human embryonic stem (hES) cells (45), mesenchymal stem cells (46), and neural stem cells (47, 48). Of particular interest, van de Leemput et al. (48) differentiated hES cells into brain organoids to study changes in gene expression accompanying human corticogenesis. Their RNA-seq database (cortecon.neuralsci.org) shows Piezo1 expression throughout the differentiation process. Temporal analysis of Piezo1 expression especially correlated with the periods of pluripotency (day 0 of the differentiation protocol) and of upper-layer neuron generation during corticogenesis (day 63 onwards). These findings are consistent with our observations in cortical fetal hNSPCs that Piezo1 activity supports neuron formation.
The identification of mechanosensors and transducers involved in stem cell fate has been an area of intense investigation since the discovery that mechanical cues can direct differentiation outcomes. Most of the molecules discovered to date are cytoskeletal proteins such as myosin II (7), the Rho/ROCK system (8, 10), and vinculin (11). Although cytoskeletal proteins and cellular contractility regulators are essential for generating traction forces and sensing environmental mechanics, the molecular mechanisms connecting these signaling molecules to intracellular pathways that influence differentiation have remained elusive. Spontaneous and evoked Ca2+ transients are known to regulate gene expression and differentiation in several types of stem cells (for recent reviews of Ca2+ signaling in stem cells see refs. 14 and 15). In this context, a stretch-activated ion channel that is activated by traction forces and elicits Ca2+ influx is an attractive candidate for transducing mechanosensitive lineage specification. In support of this role for Piezo1, we find that stiff substrates increase Piezo1 activity and favor neuronal specification of hNSPCs, whereas inhibition of Piezo1, either by genetic knockdown or by pharmacological inhibition, has the opposite effect and reduces neuronal specification (Fig. 5 and Fig. S7).
We also find that Piezo1 activity influences nucleo–cytoplasmic localization of the mechano-reactive transcriptional coactivator Yap (Fig. 4 E–G). Yap and its partner Taz have been studied in mesenchymal stem cells, where their nucleo–cytoplasmic localization governs lineage choice (13). However, similar roles for Yap and Taz in neural stem cells have not yet been investigated. Our observations lay the foundation for further mechanistic elucidation of the Piezo1–Yap/Taz connection in the context of stem cell differentiation.
The identification of an ion channel involved in mechanosensitive lineage specification of neural stem cells raises the question of whether Piezo1 may play a similar role in other multipotent stem cells. Interestingly, mesenchymal stem cells, which also strongly display mechanosensitive differentiation, express Piezo1 as well (Fig. S8; ref. 46), suggesting that Piezo1 could have a similar role in multiple types of stem cells.
The role of Piezo1 in lineage specification has potential therapeutic implications for neural stem cell transplant therapy. One of the bottle-necks of this therapeutic approach, which currently is under investigation for the treatment of neurological disorders such as Alzheimer's disease, Parkinson’s disease, stroke, and spinal cord injury, is directing cell fate after injection. Pharmacological agents aimed at modulating Piezo1 activity may be useful in directing lineage choice during neural stem cell transplant therapy.
In conclusion, we propose that myosin II-dependent traction forces involved in the detection of matrix elasticity generate local membrane tension that triggers the activation of Piezo1. Channel activation results in transient Ca2+ influx through the plasma membrane, nuclear localization of Yap, and altered neuronal–glial specification of hNSPCs (Fig. S9). Our findings point to Piezo1 as an important determinant of mechanosensitive lineage choice in neural stem cells and possibly other multipotent stem cells, providing a link between extracellular matrix mechanics and intracellular signaling pathways. Future work will determine the mechanism by which Piezo1 activity regulates mechanosensitive lineage commitment in the stem cell niche and how these results translate to the in vivo environment.
Methods
Informed written consent was obtained for all human subjects. All human cell research involved cells with no patient identifiers and was approved by the University of California, Irvine Institutional Review Board and the Human Stem Cell Research Oversight Committee.
Patch clamp measurements, TIRFM Ca2+ imaging, and differentiation assays were performed on two brain-derived human fetal hNSPC cultures (SC23 and SC27) isolated from the cerebral cortices of two separate fetuses of 23-wk gestational age (18–21). Further details on cell-culture conditions, siRNA knockdown experiments, electrophysiological measurements, calcium imaging, Qgel substrate fabrication, and data analysis are presented in SI Methods.
Supplementary Material
Acknowledgments
We thank Dr. Phillip Schwartz for the gift of hNSPCs; Dr. David Schaffer for the gift of rat adult hippocampal neural stem cells; Dr. Stephen White for the use of his spin coater; Drs. Ian Parker and Joseph Dynes for advice with TIRFM experiments; Ms. Lisa McDonnell for technical assistance; the Sue and Bill Gross Stem Cell Research Center at the University of California, Irvine (UCI) for support; and Drs. Peter Donovan and Mathew Blurton-Jones for comments on the manuscript. This work was supported by a Council on Research, Computing, and Libraries Award from UCI and National Institutes of Health Grant GM098973 (to F.T.); Undergraduate Research Opportunities Program funding (to T.T.); a seed grant from the UCI Center for Autism Research and Translation (to L.A.F. and F.T.); and by National Science Foundation CAREER Award IOS-1254060, Grant UL1 TR000153 from the National Center for Research Resources and the National Center for Advancing Translational Sciences, and a gift by Pearl Tze Hosfiel and Keith Hosfiel (to L.A.F.). This work was made possible, in part, through access to the Optical Biology Shared Resource funded by the UCI Cancer Center Grant CA-62203. M.M.P. was supported in part by a postdoctoral research fellowship from the Helen Hay Whitney Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409802111/-/DCSupplemental.
References
- 1.Sun Y, Chen C, Fu J. Forcing stem cells to behave: A biophysical perspective of the cellular microenvironment. Ann Review Biophys. 2012;41:519–542. doi: 10.1146/annurev-biophys-042910-155306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watt FM, Huck WT. Role of the extracellular matrix in regulating stem cell fate. Nat Rev Mol Cell Biol. 2013;14(8):467–473. doi: 10.1038/nrm3620. [DOI] [PubMed] [Google Scholar]
- 3.Jones DL, Wagers AJ. No place like home: Anatomy and function of the stem cell niche. Nat Rev Mol Cell Biol. 2008;9(1):11–21. doi: 10.1038/nrm2319. [DOI] [PubMed] [Google Scholar]
- 4.Tyler WJ. The mechanobiology of brain function. Nat Rev Neurosci. 2012;13(12):867–878. doi: 10.1038/nrn3383. [DOI] [PubMed] [Google Scholar]
- 5.Yang C, Tibbitt MW, Basta L, Anseth KS. Mechanical memory and dosing influence stem cell fate. Nat Mater. 2014;13(6):645–652. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert PM, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329(5995):1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 8.Keung AJ, de Juan-Pardo EM, Schaffer DV, Kumar S. Rho GTPases mediate the mechanosensitive lineage commitment of neural stem cells. Stem Cells. 2011;29(11):1886–1897. doi: 10.1002/stem.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim T-J, et al. Substrate rigidity regulates Ca2+ oscillation via RhoA pathway in stem cells. J Cell Physiol. 2009;218(2):285–293. doi: 10.1002/jcp.21598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6(4):483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 11.Holle AW, et al. In situ mechanotransduction via vinculin regulates stem cell differentiation. Stem Cells. 2013;31(11):2467–2477. doi: 10.1002/stem.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swift J, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341(6149):1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474(7350):179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 14.Leclerc C, Neant I, Moreau M. The calcium: An early signal that initiates the formation of the nervous system during embryogenesis. Front Mol Neurosci. 2012;5:3. doi: 10.3389/fnmol.2012.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tonelli FM, et al. Stem cells and calcium signaling. Adv Exp Med Biol. 2012;740:891–916. doi: 10.1007/978-94-007-2888-2_40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leipzig ND, Shoichet MS. The effect of substrate stiffness on adult neural stem cell behavior. Biomaterials. 2009;30(36):6867–6878. doi: 10.1016/j.biomaterials.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Saha K, et al. Substrate modulus directs neural stem cell behavior. Biophys J. 2008;95(9):4426–4438. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer TD, et al. Cell culture. Progenitor cells from human brain after death. Nature. 2001;411(6833):42–43. doi: 10.1038/35075141. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz PH, et al. Isolation and characterization of neural progenitor cells from post-mortem human cortex. J Neurosci Res. 2003;74(6):838–851. doi: 10.1002/jnr.10854. [DOI] [PubMed] [Google Scholar]
- 20.Flanagan LA, Rebaza LM, Derzic S, Schwartz PH, Monuki ES. Regulation of human neural precursor cells by laminin and integrins. J Neurosci Res. 2006;83(5):845–856. doi: 10.1002/jnr.20778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labeed FH, et al. Biophysical characteristics reveal neural stem cell differentiation potential. PLoS ONE. 2011;6(9):e25458. doi: 10.1371/journal.pone.0025458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pistollato F, Chen H-L, Schwartz PH, Basso G, Panchision DM. Oxygen tension controls the expansion of human CNS precursors and the generation of astrocytes and oligodendrocytes. Mol Cell Neurosci. 2007;35(3):424–435. doi: 10.1016/j.mcn.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Lee J-P, et al. Stem cells act through multiple mechanisms to benefit mice with neurodegenerative metabolic disease. Nat Med. 2007;13(4):439–447. doi: 10.1038/nm1548. [DOI] [PubMed] [Google Scholar]
- 24.Salazar DL, Uchida N, Hamers FP, Cummings BJ, Anderson AJ. Human neural stem cells differentiate and promote locomotor recovery in an early chronic spinal cord injury NOD-scid mouse model. PLoS ONE. 2010;5(8):e12272. doi: 10.1371/journal.pone.0012272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta N, et al. Neural stem cell engraftment and myelination in the human brain. Sci Transl Med. 2012;4(155):155ra137. doi: 10.1126/scitranslmed.3004373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsukamoto A, Uchida N, Capela A, Gorba T, Huhn S. Clinical translation of human neural stem cells. Stem Cell Res Ther. 2013;4(4):102. doi: 10.1186/scrt313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suchyna TM, et al. Identification of a peptide toxin from Grammostola spatulata spider venom that blocks cation-selective stretch-activated channels. J Gen Physiol. 2000;115(5):583–598. doi: 10.1085/jgp.115.5.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowman C, Gottlieb P, Suchyna T, Murphy Y, Sachs F. Mechanosensitive ion channels and the peptide inhibitor GsMTx-4: History, properties, mechanisms and pharmacology. Toxicon. 2007;49(2):249–270. doi: 10.1016/j.toxicon.2006.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suchyna TM, et al. Bilayer-dependent inhibition of mechanosensitive channels by neuroactive peptide enantiomers. Nature. 2004;430(6996):235–240. doi: 10.1038/nature02743. [DOI] [PubMed] [Google Scholar]
- 30.Kim SE, Coste B, Chadha A, Cook B, Patapoutian A. The role of Drosophila Piezo in mechanical nociception. Nature. 2012;483(7388):209–212. doi: 10.1038/nature10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coste B, et al. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010;330(6000):55–60. doi: 10.1126/science.1193270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coste B, et al. Piezo proteins are pore-forming subunits of mechanically activated channels. Nature. 2012;483(7388):176–181. doi: 10.1038/nature10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, et al. Piezo1 integration of vascular architecture with physiological force. Nature. 2014 doi: 10.1038/nature13701. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ranade SS, et al. Piezo1, a mechanically activated ion channel, is required for vascular development in mice. Proc Natl Acad Sci USA. 2014;111(28):10347–10352. doi: 10.1073/pnas.1409233111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bae C, Sachs F, Gottlieb PA. The mechanosensitive ion channel Piezo1 is inhibited by the peptide GsMTx4. Biochemistry. 2011;50(29):6295–6300. doi: 10.1021/bi200770q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eisenhoffer GT, et al. Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature. 2012;484(7395):546–549. doi: 10.1038/nature10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faucherre A, Kissa K, Nargeot J, Mangoni ME, Jopling C. Piezo1 plays a role in erythrocyte volume homeostasis. Haematologica. 2014;99(1):70–75. doi: 10.3324/haematol.2013.086090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyamoto T, et al. Functional role for Piezo1 in stretch-evoked Ca2+ influx and ATP release in urothelial cell cultures. J Biol Chem. 2014;289(23):16565–16575. doi: 10.1074/jbc.M113.528638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310(5751):1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 40.Straight AF, et al. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299(5613):1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 41.Gutierrez E, et al. High refractive index silicone gels for simultaneous total internal reflection fluorescence and traction force microscopy of adherent cells. PLoS ONE. 2011;6(9):e23807. doi: 10.1371/journal.pone.0023807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: An updated version. Genes Dev. 2010;24(9):862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zarychanski R, et al. Mutations in the mechanotransduction protein PIEZO1 are associated with hereditary xerocytosis. Blood. 2012;120(9):1908–1915. doi: 10.1182/blood-2012-04-422253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McHugh BJ, Murdoch A, Haslett C, Sethi T. Loss of the integrin-activating transmembrane protein Fam38A (Piezo1) promotes a switch to a reduced integrin-dependent mode of cell migration. PLoS ONE. 2012;7(7):e40346. doi: 10.1371/journal.pone.0040346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Theunissen TW, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15(4):471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barberi T, Willis LM, Socci ND, Studer L. Derivation of multipotent mesenchymal precursors from human embryonic stem cells. PLoS Med. 2005;2(6):e161. doi: 10.1371/journal.pmed.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fathi A, et al. Comprehensive gene expression analysis of human embryonic stem cells during differentiation into neural cells. PLoS ONE. 2011;6(7):e22856. doi: 10.1371/journal.pone.0022856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van de Leemput J, et al. CORTECON: A temporal transcriptome analysis of in vitro human cerebral cortex development from human embryonic stem cells. Neuron. 2014;83(1):51–68. doi: 10.1016/j.neuron.2014.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.